Cryo-EM reveals the structure of the mitochondrial ABC transporter Atm1 from yeast in the occluded state.
Abstract
The mitochondrial inner membrane ABC transporter Atm1 exports an unknown substrate to the cytosol for iron-sulfur protein biogenesis, cellular iron regulation, and tRNA thio-modification. Mutations in the human relative ABCB7 cause the iron storage disease XLSA/A. We determined 3D structures of two complementary states of Atm1 in lipid nanodiscs by electron cryo-microscopy at 2.9- to 3.4-Å resolution. The inward-open structure resembled the known crystal structure of nucleotide-free apo-Atm1 closely. The occluded conformation with bound AMP-PNP-Mg2+ showed a tight association of the two nucleotide-binding domains, a rearrangement of the C-terminal helices, and closure of the putative substrate-binding cavity in the homodimeric transporter. We identified a hydrophobic patch on the C-terminal helices of yeast Atm1, which is unique among type IV ABC transporters of known structure. Truncation mutants of yeast Atm1 suggest that the C-terminal helices stabilize the dimer, yet are not necessary for closure of the nucleotide-binding domains.
INTRODUCTION
The ABC [adenosine 5-′triphosphate (ATP)–binding cassette] transporter of mitochondria 1 (Atm1) from Saccharomyces cerevisiae is essential for the biogenesis of cytosolic iron-sulfur proteins, cellular iron regulation, and cytosolic tRNA thiolation (1–3). Similar functions have been reported for Atm1 homologs in mitochondria of, e.g., man, mice, plants, and other fungi (4–7). Depletion of the mitochondrial sulfur-donating cysteine desulfurase Nfs1 or of glutathione (GSH) elicits a similar cytosolic phenotype as Atm1 deficiency (1, 8–10). These cell biological and recent biochemical studies have therefore suggested that Atm1 exports a molecule containing sulfur, iron, and GSH from mitochondria for utilization in the cytosol (3, 11). We have previously observed that reduced GSH binds to Atm1 both in solution and in a crystal structure (12). Various thiol compounds including GSH stimulate the ATP hydrolysis activity of purified Atm1 in detergent solution or in proteoliposomes, while their oxidized disulfide forms do not, with the exception of oxidized glutathione (GSSG) (13). Atm1 expressed in Lactococcus lactis was reported to transport GSSG, but not GSH, into membrane vesicles (14). A homolog of yeast Atm1 from the Gram-negative bacterium Novosphingobium aromaticivorans (NaAtm1) is implicated in heavy metal detoxification. Crystal structures of NaAtm1 have shown the binding of GSH, GSSG, and mercury-complexing GSH (Hg-GSH2) in a region close to the GSH-binding site in yeast Atm1 (15). Point mutations in the human Atm1 ortholog ABCB7 cause the recessive disorder X-linked sideroblastic anemia and cerebellar ataxia (XLSA/A). Affected cells contain iron-loaded mitochondria, the so-called sideroblasts (16–20). Similarly, yeast mitochondria lacking intact Atm1 accumulate free iron in response to a deregulated iron uptake system (2, 21).
The common architecture of ABC transporters consists of a set of two nucleotide-binding domains (NBDs) and two transmembrane domains (TMDs) (22). As an exporter belonging to the group of type IV ABC transporters (23), the putative substrate cavity of Atm1 is expected to be largely defined by the transmembrane (TM) helices of the TMDs (24). While substrate transport is fueled by ATP hydrolysis in the NBDs, basal levels of futile ATP hydrolysis in the absence of cargo are a common feature of ABC transporters (25–27). According to the alternate access model, the transport cycle of an ABC exporter consists of at least two conformations (24, 28). In the inward-facing conformation of a mitochondrial transporter, cargo binds from the matrix, where the NBDs are located (29). Cargo release to the intermembrane space requires a different, outward-facing conformation (30, 31). Various additional structures of ABC exporters in detergent and detergent-free ambiences have been solved that represent intermediate states of the transport cycle between the two complementary open conformations (32–34). So far, for yeast Atm1, only the x-ray structure of the open, inward-facing conformation in the detergent n-dodecyl-β-d-maltoside (DDM) is known (12).
In the absence of nucleotides, the crystal structures of both yeast and bacterial Atm1 indicate that, in detergent, they adopt an inward-open conformation (12, 15). Although the structures are similar and the proteins share 45% sequence identity in the overlapping range, the distance between the NBDs differs by roughly 6 Å. In the yeast Atm1 homodimer, the C-terminal α helices of both protomers interact with and presumably stabilize one another, as C-terminal truncations diminish Atm1 levels in vivo, which in turn impairs cytosolic iron-sulfur protein biogenesis (12). Interactions between the two C-terminal α helices in the yeast Atm1 dimer are extensive and need to break to allow the NBDs to closely associate and form the two ATP-binding sites. The two helices are positioned on opposite sides relative to the corresponding helices in NaAtm1 (15, 34). Although the mode of helix dissociation and their location after NBD closure are unknown, this observation suggests that the yeast Atm1 helices need to move around each other in the process of NBD association, in contrast to those of NaAtm1. A similar helix rearrangement has been reported for TmrAB from Thermus thermophilus, where the comparably short C-terminal “zipper helices” end up on opposite sides of each other when the NBDs close (33, 35).
Here, we present structures of yeast Atm1 in lipid nanodiscs in the nucleotide-free inward-open and nucleotide-bound occluded conformations determined by electron cryo–microscopy (cryo-EM). Cryo-EM, negative-stain EM, and biochemical data for Atm1 from yeast and Chaetomium thermophilum confirm that lipid nanodiscs in a membrane scaffold protein (MSP) are an ideal environment for structural and functional studies of ABC transporters. Our results provide new insights into the stabilization of the yeast Atm1 dimer through its C-terminal helices and into their relative movement during the transport cycle.
RESULTS
Cryo-EM structure of nucleotide-free apo-Atm1 in MSP1D1 lipid nanodiscs
S. cerevisiae Atm1 expressed in Escherichia coli was purified as described (12, 13) and reconstituted into MSP1D1 and MSP1E3D1 lipid nanodiscs without added nucleotides. Negative-stain EM indicates that the wild-type transporter inserts into MSP1D1 nanodiscs independently of the presence or absence of cardiolipin (CL) (fig. S1A). Additional density around the TMDs of Atm1 is most likely due to the nanodisc scaffold, as this density is absent in two-dimensional (2D) class averages of ABC transporters in DDM micelles (36). Atm1 reconstituted into nanodiscs supported continuous ATP hydrolysis (fig. S1B). The specific ATP hydrolysis activity of Atm1 of 1.1 μmol min−1 mg−1 in the smaller, MSP1D1 lipid nanodisc is roughly half of that measured for Atm1 reconstituted into proteoliposomes (13). Atm1 in the wider MSP1E3D1 lipid nanodiscs has intermediate activity. The mitochondria-specific lipid CL enhanced the specific ATPase activity, confirming our previous ATP hydrolysis assays of Atm1 in DDM detergent micelles with added phospholipids (13). The detected ATPase activity was specific for Atm1, because hardly any activity was observed for the mutant Atm1 E598Q in lipid nanodiscs that carries a mutation in the Walker B motif. Atm1 ATPase activity depended on Mg2+ and was strongly inhibited by either vanadate or AMP-PNP, as preincubation with these compounds before the initiation of the ATPase reaction resulted in little release of free phosphate.
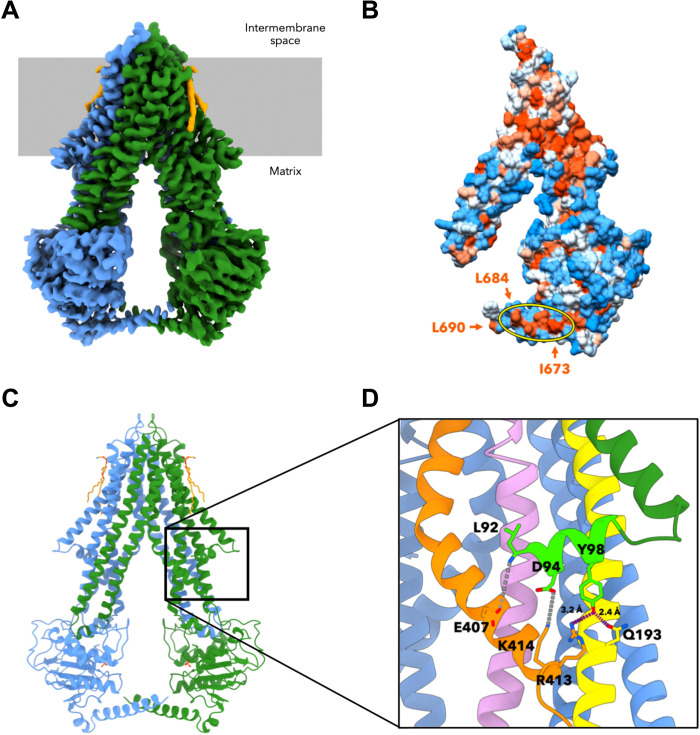
The single-particle cryo-EM structure of apo-Atm1 in MSP1D1 nanodiscs attained a resolution of 3.3 Å [Protein Data Bank (PDB) ID: 7psl; Fig. 1, A and C; figs. S2 and S3A; and table S1]. The map definition was best (up to 2.8-Å resolution) in the TMDs (fig. S4, A and B), as is typical for cryo-EM structures of ABC transporters (33, 36, 37). Atm1 adopts an inward-facing conformation that is overall very similar [root mean square deviation (RMSD), 1.3 Å] to the previously determined Atm1 crystal structure (fig. S4C) (12). Neither crystal contacts nor the difference in chemical environments of lipid nanodiscs or detergent micelles affects the overall structure of Atm1, in particular the distance between the NBDs (fig. S5). The quality of the cryo-EM map allowed all side chains to be built, including the previously unresolved amphipathic elbow helix at the N terminus of mature Atm1 (residues 92 to 100) (Fig. 1, C and D). This short helix is located on the matrix side of the inner membrane and runs perpendicular to the TM helices, comparable to N-terminal elbow helices of other ABC transporters (29, 30). Each elbow helix interacts with TM helices 2, 3, and 6 of the same protomer through hydrogen bonds (Y98, Q193, and R413), hydrophobic contacts, and up to two possible salt bridges involving the side chains of D94, E407, and K414 (Fig. 1D). The hydrogen bond between Y98 and Q193 is N/Q conserved in ScAtm1 and NaAtm1, where the corresponding residues are Y26 and N119 (15). Compared to the arrangement in ScAtm1, the NaAtm1 elbow helices connect to TM helix 6 through a similar, yet charge-reversed ionic bond between R22 and D340. In a broader context, interactions between TM and elbow helices are a common feature of type IV ABC transporters, although they differ in detail (29, 30, 38). One bound phospholipid molecule per protomer was resolved near Q127 of TM helix 1 and near Y252 and Q253 of TM helix 3 (fig. S4D).
Fig. 1. Cryo-EM structure of apo-Atm1 in MSP1D1 nanodiscs.
(A) At 3.3-Å resolution, the cryo-EM map of yeast apo-Atm1 reveals an inward-facing conformation, and the densities for both monomers are represented in blue and green. The lipids are orange, and the membrane is gray. (B) Surface representation of one monomer of the cryo-EM structure colored by hydrophobicity indicating a hydrophobic patch on the C-terminal helix between I673 and L684 [arrows and yellow ellipse; structure turned by 20° with respect to (A)]. Note L690 at the end of the C-terminal helix. The hydrophobicity was calculated according to the Kyte-Doolittle scale (73) and is represented by an orange (hydrophobic) to white (neutral) to blue (hydrophilic) color gradient. (C) The atomic model built on the basis of the cryo-EM data comprises all residues from 92 to 690 [color coding as in (A)]. (D) The N-terminal elbow helix was resolved in the cryo-EM map (lime green). L92 as well as the side chains of D94 and Y98 form contacts to TM helices 2 (yellow), 3 (pink), and 6 (orange; hydrogen bonds in purple, and potential ionic bonds in gray). The structure is rotated by 35° with respect to (C).
In both the cryo-EM and the crystal structures, the distance between the two NBDs is defined by side-to-side interactions of the two C-terminal helices [this paper and (12)]. In particular, the hydrophobic side chains of I673, L677, L680, and L684 form a hydrophobic patch spanning four helix turns on either side of their twofold symmetrical interface (Fig. 1B). The patches point toward their counterpart residues on the C-terminal helices of the opposite protomer in an antiparallel manner. The patch starts at I673 within 4.5 Å of another hydrophobic residue, L690, which is the C terminus of yeast Atm1. This pattern minimizes the solvent exposure of the aliphatic side chains and is likely to confine the distance between both NBDs to a narrow range. Hydrophilic interactions appear to play a minor role, as the only likely hydrogen bond involving the C-terminal helices could possibly be formed by the Q687 side-chain amide and either I673 (main chain) or the D676 side chain.
Comparing the cryo-EM and x-ray structures of apo-Atm1, the side-chain staggering is shifted by 1.2 Å. By contrast, in NaAtm1, the gap between the NBDs in nanodiscs and in crystals differed by ~11 Å (fig. S6) (15, 34). The NBD distances in the NaAtm1 structures differed not only from each other but also from yeast Atm1. NaAtm1 lacks a hydrophobic patch on its C-terminal helices, as does the Atm1 homolog from the thermophilic fungus C. thermophilum (CtAtm1) (fig. S7) (12, 15). We therefore asked whether the NBD arrangement was similar in ScAtm1 and CtAtm1. Purified CtAtm1 was reconstituted into MSP1E3D1 nanodiscs, and a comparable, yet smaller cryo-EM dataset was collected. Image processing as for yeast Atm1 resulted in three separately refined maps of nucleotide-free CtAtm1 (figs. S3B, S8, A to C, and S9; and table S1). The three maps revealed inward-facing states that vary in their degrees of NBD separation. The mutual stabilization of the C-terminal helices does not apply to CtAtm1, as judged by the 3D maps and 2D class averages (figs. S8, A to C, and S9). In contrast to yeast Atm1, the C-terminal helices of CtAtm1 do not interact with one another and may be more flexible. The varying degrees of NBD separation found for the three apo states are not associated with noticeable differences in the TM helix 6 conformations near the putative cargo-binding site (fig. S8, D and E). Within the TMDs, the largest positional shifts are observed in TM helices 4 and 5, which reach across to the opposite protomer (fig. S8F).
In general, NBD separation in nucleotide-free structures of other ABC transporters varies. Different degrees of NBD separation were observed in crystal structures of the pseudoheterodimeric mouse multidrug transporter ABCB1 (P-gp) (39, 40) and of the homodimeric lipid-linked oligosaccharide flippase PglK from Campylobacter jenuni (41), where the distance appeared to depend on the choice of detergent and lattice contacts. The homodimeric lipid flippase MsbA from E. coli adopts multiple different apo conformations in crystal structures in detergent and in cryo-EM structures in MSP1D1 lipid nanodisc and peptidisc environments (31, 36, 42). The nucleotide-free cryo-EM structures of the heterodimeric siderophore importer YbtPQ from E. coli in the detergent lauryl maltose neopentyl glycol and MSP1D1 nanodiscs are very similar with regard to NBD separation, yet the YbtPQ C-terminal helices do not interact with each other in a comparable manner (43). The only similar case of largely invariable NBD separation determined by C-terminal helices is the heterodimeric TmrAB without bound nucleotides (33, 35). In TmrAB, both C-terminal zipper helices are connected through a combination of hydrophilic and hydrophobic interactions. In summary, the hydrophobic patch found along the C-terminal helices of yeast Atm1 stands out among the currently available apo structures of type IV ABC transporters (23) and also other fungal Atm1 homologs (fig. S7).
Cryo-EM structures of Atm1 with bound AMP-PNP in MSP lipid nanodiscs
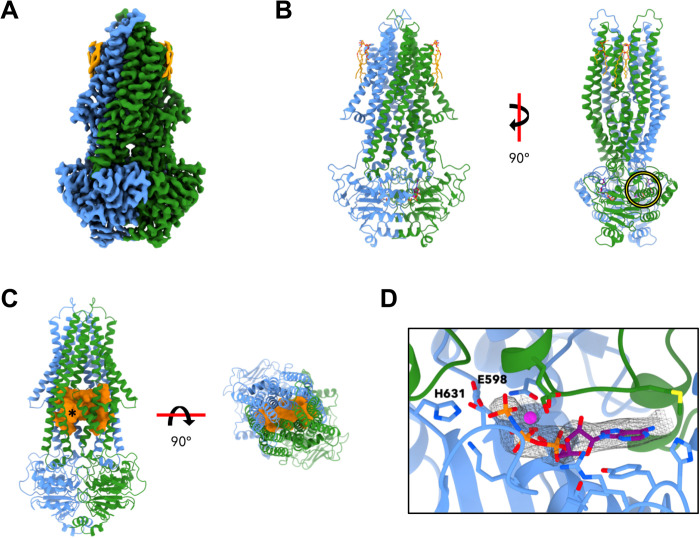
To obtain alternative conformations, purified yeast Atm1 was incubated with the nonhydrolyzable ATP analog AMP-PNP after reconstitution into either MSP1D1 or MSP1E3D1 nanodiscs. Cryo-EM structures attained resolutions of 2.9 Å (Atm1-MSP1E3D1; PDB ID: 7psn) and 3.4 Å (Atm1-MSP1D1; PDB ID: 7psm) (Fig. 2, A and B; figs. S3A and S10 to S12; and table S1). Structures in both nanodiscs comprise residues 92 to 676 and were very similar (RMSD, 0.58 Å), including bound lipids, which were found in the same locations as in the inward-open structure (Fig. 2B and figs. S4D and S10). Addition of AMP-PNP-Mg2+ induced a major conformational change relative to nucleotide-free Atm1. The NBDs interacted closely, resulting in an occluded conformation of the dimeric transporter (Fig. 2, A and B, and fig. S10). The TMDs of the two monomers approached one another, defining a large, 700-Å3 cavity at the dimer interface (Fig. 2C and fig. S13). The cavity is almost fully enclosed, apart from a narrow channel at the membrane surface on the matrix side. The gap is only wide enough for the passage of water molecules or small ions and vanishes when the probe radius for calculating the cavity volume is increased. The cavity is bordered by TM helices 2 to 6 of both Atm1 protomers and is lined by a number of charged and polar residues (fig. S13). The channels connecting the cavity to the mitochondrial matrix run past charged and polar side chains including R284 of TM helix 4 and Q401 of TM helix 6. Residues Y396, R397, and D398 are found at the hinge region of TM helix 6, and D398 corresponds to one of the human ABCB7 mutations that cause XLSA/A, E433 (12, 17).
Fig. 2. Cryo-EM structure of Atm1 in MSP1E3D1 nanodiscs with bound AMP-PNP-Mg2+.
(A) The 2.9-Å cryo-EM map reveals an occluded conformation of Atm1 trapped in a prehydrolysis state by addition of AMP-PNP-Mg2+ (color coding as in Fig. 1). (B) The model comprises two bound AMP-PNP molecules (purple) and Mg2+ ions (magenta) at the dimer interface of the NBDs, and two lipids per monomer (orange). A yellow circle identifies the ATP-binding site presented in greater detail in (D). (C) The cavity (orange) is lined by the TM helices and extends across the dimer [left: turned by 40° with respect to (B); right: viewed from the intermembrane space]. At the membrane surface on the matrix side, the asterisk indicates a narrow passage to the matrix. (D) Enlargement of encircled region in (B), with bound Mg2+ and AMP-PNP (gray mesh). Protein color coding as in Fig. 1. The side chains lining the ATP-binding site are shown as sticks. The Walker B E598 mutated in Atm1 E598Q and the switch histidine H631 are pointed out.
In one of the x-ray structures of the Atm1 inward-facing conformation, a map density near this location was assigned to bound GSH (12). The GSH density was only found in one of three protomers of the asymmetric unit, and GSH had not been added but carried along during purification from E. coli. In all cryo-EM structures, the putative substrate-binding cavity is empty, although the calculated cavity volume is large enough to accommodate GSH or a GSH conjugate. We also looked for evidence of bound GSH in reconstructions without applied twofold symmetry but did not detect any density for GSH in the potential binding site. Most likely, the additional purification steps following lipid nanodisc reconstitution removed any residual GSH. Supplying the potential cargo molecules GSH or GSSG in the absence of nucleotides did not result in NBD closure (fig. S14).
Two molecules of AMP-PNP-Mg2+ bound in the canonical nucleotide binding sites between the two NBDs were well resolved in the map (Fig. 2D). Other characteristic structural features of ABC transporters were similarly well resolved, in particular the contacts to AMP-PNP and the Mg2+ ion formed by the Walker A and B motifs, the A-, H-, and Q-loop residues, and the signature motif of the opposite protomer (ISGGE). Since the putative substrate-binding cavity was empty, the transition to the occluded state did not require bound substrate or ligands. The change in Atm1 conformation upon AMP-PNP-Mg2+ binding was almost quantitative, because only a small proportion of particles were found in the inward-facing conformation (<10%). These particles were removed during data processing (figs. S11 and S12).
Nucleotide binding triggered an extensive rearrangement of the TM helices. The movement of TM helices 4 and 5 toward the core of the respective monomer followed the common pattern observed in other type IV ABC transporters, and the close proximity of the two pairs of TM helices 3 and 6 at the twofold dimer axis helps define the putative substrate-binding cavity (fig. S13). This rearrangement leaves the TM helices poised for the transition into the outward-facing conformation (31, 33, 36). We did not observe particles in the outward-facing conformation, not even with Atm1 in the larger nanodisc that could presumably accommodate the characteristic splaying of the TM helices away from each other (36).
The yeast Atm1 cryo-EM structures with bound AMP-PNP-Mg2+ closely resembles the recently published posthydrolysis structures of bacterial NaAtm1 with bound adenosine 5′-diphosphate (ADP)–Mg2+–vanadate (34). Both AMP-PNP–bound yeast Atm1 structures differ from the corresponding crystal structure of bacterial NaAtm1, in particular with regard to TM helix 6, which is kinked by roughly 50° in yeast Atm1, a feature reminiscent of the NaAtm1 post- rather than prehydrolysis state (fig. S15) (34). The differences between the occluded states of ScAtm1 and NaAtm1 further extend to the cargo-binding cavity in yeast Atm1. As the posthydrolysis NaAtm1 structure is in good agreement with our structures, a cavity of similar size and shape should be present in the bacterial Atm1 structure with bound ADP-Mg2+-vanadate. On the basis of the available structural data of yeast Atm1, the assembly of a model transport cycle for ScAtm1 remains difficult. Ideally, such a model would be based on structures obtained in chemically similar environments (33) and in the presence of the cargo molecule, which has yet to be identified.
NBD interaction does not require the C-terminal helices
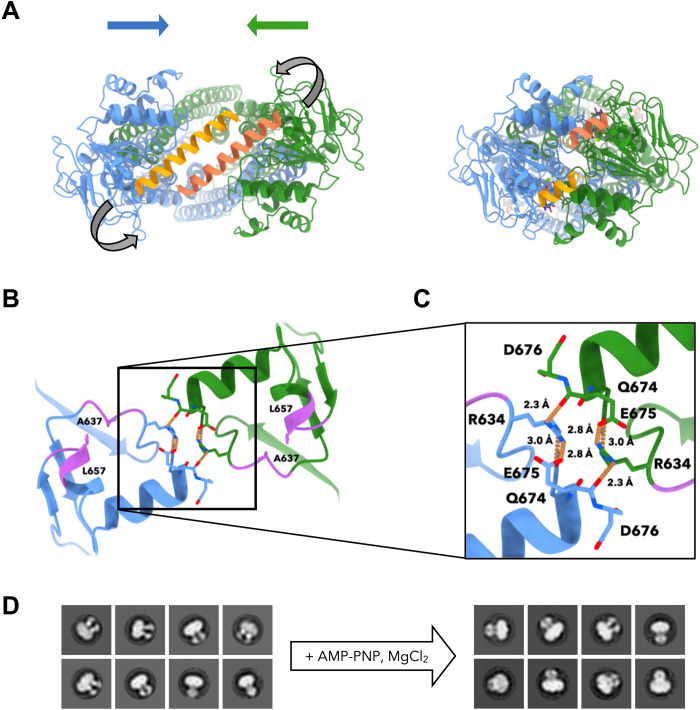
During transition from the nucleotide-free to the AMP-PNP-Mg2+–bound state, the overall architecture of the NBDs was maintained (RMSD of 1.3 Å for residues 419 to 676 of both conformations). The close similarity did not extend to the C-terminal α helices (residues 666 to 690) and to the x-loop upstream of the ABC signature motif (30), which appears to undergo a rotation upon NBD interaction. The inward-facing structures of apo-Atm1 have raised questions about the movement of the C-terminal helices during nucleotide-induced NBD association and about their location within the structure after NBD closure. The C-terminal helices interact with each other in the apo-Atm1 homodimer (Fig. 3A, left) (12). In the occluded conformation, the α helices were resolved up to residue D676. The resolved helix segments had moved to the far side of the corresponding helix of the opposite protomer (Fig. 3A, right). Their C-terminal residues form a network of hydrogen bonds and salt bridges around the twofold symmetry axis of the homodimer (Fig. 3, B and C). On the immediate dimer interface, the R634 guanidinium group forms one hydrogen bond each to the backbone oxygen of E675 of the opposite protomer and to the side chain of Q674 of the same protomer. In addition, an intraprotomer ionic bond to the side chain of E675 is formed (Fig. 3C). Notably, R634 is three residues downstream of the switch histidine H631 of the H-loop, establishing a possible link to the ATP-binding site (Fig. 2D). R634 and E675 are conserved only in fungal Atm1 (fig. S7). Their interactions may contribute to the tight association of the two NBDs and help to keep the ordered segments of the C-terminal helices in position, thereby leaving the remaining 14 residues (and the Strep-tag) solvent exposed. This involves a change in orientation and a partial unfolding of the two C-terminal α helices of apo-Atm1, because otherwise the helices would clash with polypeptide segments around A637 and L657 in the occluded state (Fig. 3B).
Fig. 3. The C-terminal segments of Atm1 move around each other upon nucleotide-dependent NBD association.
(A) Cryo-EM structures of Atm1 in the absence (left) and presence of AMP-PNP-Mg2+ (right, MSP1E3D1 structure) in the same colors as in Fig. 1 except for the C-terminal helices (orange and coral; viewed from the matrix). Blue and green arrows denote the movement of the NBDs toward each other, and gray arrows indicate a possible path for the helix rearrangement. (B) At the C terminus of AMP-PNP-Mg2+–bound Atm1, Q674, E675, and D676 and the side chains of R634 (shown as sticks) form intra- and interchain hydrogen and ionic bonds (dotted brown lines). Side chains of A637 and L657 and the segments around them (purple) would clash if the C-terminal helices were to move as rigid bodies. (C) Detail of (B). (D) Negative-stain EM 2D class averages show the Atm1-L657 truncation mutant reconstituted into MSP1D1 nanodiscs with or without added AMP-PNP and MgCl2.
A partial truncation of the C-terminal helices (residues 676 to 690; Atm1-D676) to disrupt the helical contacts does not impair the in vivo function of Atm1 in cytosolic iron-sulfur protein biogenesis (12). In contrast, truncation of the entire C-terminal helix at L666 (residues 666 to 690; Atm1-L666) significantly impaired iron-sulfur protein maturation, mainly due to reduced Atm1 protein stability. Yeast cells expressing Atm1-L666 instead of wild-type Atm1 show a slight growth defect at 37°C and diminished cytosolic iron-sulfur protein formation, while cells expressing Atm1-D676 do not (12). The interactions involving R634, Q674, and E675 stabilize the occluded conformation (Fig. 3, B and C) and may act as a counterforce to the hydrophobic interactions of the C-terminal helices in the inward-open conformation, in which the NBDs have separated. Thus, as the cargo transport cycle requires stably associated NBDs in both the occluded and the outward-facing conformation, the interactions of these residues may explain the observed differences between Atm1-D676 and Atm1-L666 with regard to iron-sulfur protein biogenesis and yeast cell growth.
To further investigate the role of the C-terminal region of Atm1 in NBD association, we generated the Atm1-L657 truncation mutant, in which both the C-terminal α helix and the preceding perpendicular helix (residues 656 to 661) had been deleted (Fig. 3B). Yields of Atm1-L657 purified from E. coli were fivefold lower than for wild-type Atm1, suggesting that the truncated protein is less stable. The thermostability of Atm1-L657 was reduced by 4.6°C compared to wild-type Atm1 in nano–differential scanning fluorimetry (nanoDSF) assays (Table 1), consistent with the lower stability of Atm1-L666 in yeast (12). Examination of Atm1-L657 reconstituted into MSP1D1 nanodiscs by negative-stain EM did not indicate a substantial effect of the missing C-terminal region on NBD association (Fig. 3D and fig. S16). Similarly, addition of AMP-PNP and MgCl2 to Atm1-L657 in detergent solution induces NBD closure, so that most transporter dimers adopt the occluded state (fig. S17). Consistent with our finding that Q674 and E675 stabilize the Atm1 C terminus in the occluded state, the unfolding transition temperature difference between Atm1-L657 and wild-type Atm1 was similar (5.4°C) to that of apo-Atm1, as the truncation mutant lacks these residues (Table 1 and Fig. 3, B and C). Measurement of the specific ATPase activity of Atm1-L657 confirmed that the truncation mutant was able to hydrolyze ATP. The ATPase activity even increased by a factor of two, compared to wild-type Atm1, possibly because the network of interactions formed by R634, Q674, and E675 (missing in Atm1-L657) reduced the level of futile ATP hydrolysis (Table 1). The apo forms of both wild-type Atm1 and Atm1-L657 showed a similar inter-NBD distance in detergent solution and MSP1D1 nanodiscs (Fig. 3D and figs. S14A and S17). In particular, no wide-open conformations were detected (figs. S14A and S17), whereas such conformations were found in other, more distantly related ABC transporters (31, 36, 41, 44). Our negative-stain EM experiments suggest that the NBDs can associate independently of the C-terminal helices and the chemical environment of the TM region (Fig. 3D and fig. S17). In conclusion, the combination of negative-stain EM, cryo-EM, and nanoDSF assays confirm that the occluded state is stabilized by residues in the C-terminal α helices.
Table 1. Thermal stability of Atm1 decreases upon truncation of the C-terminal helix.
Thermal stability of wild-type Atm1 and the C-terminal L657 truncation mutant protein was measured by nano–differential scanning fluorimetry (nanoDSF) in DDM detergent solution. Measurements were performed in the absence (apo) or presence of AMP-PNP and Mg2+. The unfolding transition temperature was determined from the inflection point of the fluorescence signal (mean ± SD; n = 3). As a control, the specific ATPase activity of the proteins from above and the Walker B E598Q Atm1 mutant protein was determined by an NADH-coupled assay (mean ± SD; n ≥ 3).
| Thermal stability (Tm) (°C) | ||||
| apo | AMP-PNP-Mg2+ | |||
| Atm1 wild type | 42.5 | ±0.4 | 61.5 | ±2.7 |
| Atm1-L657 | 37.9 | ±1.4 | 56.1 | ±0.8 |
| Specific ATPase activity (nmol ATP min−1 mg−1 Atm1) | ||||
| Atm1 wild type | 175.9 | ±39.9 | ||
| Atm1 E598Q | 22.2 | ±10.0 | ||
| Atm1-L657 | 389.5 | ±39.9 | ||
Different functional states of Atm1 in MSP1D1 nanodiscs
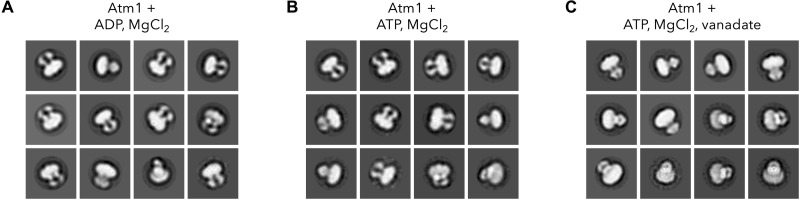
As cargo transport driven by nucleotide hydrolysis depends on NBD closure, we asked what conformational spectrum of Atm1 would result from the addition of ATP or ADP. The conformation distribution of Atm1 reconstituted into MSP1D1 nanodiscs upon addition of ADP-Mg2+ suggests the quantitative presence of inward-open dimers (Fig. 4A). Although in negative-stain EM, front views of Atm1 with interacting NBDs are difficult to distinguish from side views of any expected Atm1 conformation, our data clearly contain dimers viewed from varying orientations. Hence, the ADP-Mg2+–bound state of yeast Atm1 presumably resembles the inward-facing conformation of apo-Atm1. In the presence of ATP and Mg2+, Atm1 also adopts mostly this conformation (Fig. 4B). The 2D class averages in negative-stain EM capture Atm1 under turnover conditions. They likely show both front and side views of Atm1, hence suggesting separated NBDs in most molecules. In contrast, in the presence of both ATP and vanadate, Atm1 adopts a conformation with the two NBDs in close contact (Fig. 4C). Under these conditions, Atm1 appears to be almost quantitatively trapped in the transition state with bound ADP-Mg2+, where vanadate occupies the γ-phosphate position of the ATP-binding site immediately after nucleotide hydrolysis and phosphate release (45). However, we cannot rule out that some of the dimers are in an outward-facing state. Together, our ATP hydrolysis, negative-stain EM, and cryo-EM findings suggest that ATP-Mg2+ and not ADP-Mg2+ causes the NBDs of Atm1 in lipid nanodiscs to interact. Since a large proportion of dimers with dissociated NBDs can only be identified when the transporter is not trapped in a pre- or posthydrolysis state, ATP hydrolysis in the absence of cargo appears to be a fast process, and the conformation of associated NBDs has a short lifetime.
Fig. 4. Negative-stain EM of Atm1 in MSP1D1 nanodiscs.
Representative negative stain EM 2D class averages of Atm1 in MSP1D1 nanodiscs in the presence of ADP and MgCl2 (A), ATP and MgCl2 (B), and ATP, MgCl2, and vanadate (C). The most populated class averages are shown, representing 77% (ADP-Mg2+), 79% (ATP-Mg2+), and 75% (ATP-Mg2+ and vanadate) of all particles. Concentrations applied were 4 mM ADP or ATP, 10 mM MgCl2, 1 mM vanadate, and Atm1 in MSP1D1 nanodiscs (0.02 mg/ml).
DISCUSSION
The cryo-EM structures of yeast Atm1 capture this ABC transporter at near-atomic resolution in a lipid bilayer environment. Our 3.3-Å map of the inward-facing conformation allowed us to build all protein residues ranging from the previously unresolved N-terminal elbow helices to the characteristic C terminus (Fig. 1, A, C, and D). The AMP-PNP-Mg2+–bound structures in two different-sized nanodiscs at 2.9- and 3.4-Å resolution cover the full homodimer to residue D676, where the straight continuation of the two C-terminal helices is sterically hindered near a salt bridge conserved in fungal Atm1 (Figs. 2, A and B, and 3, A and C, and figs. S7 and S10).
The nucleotide-free cryo-EM structure of Atm1 is in excellent agreement with our previously published x-ray structure of Atm1 in DDM detergent (fig. S4C) (12). This consistency includes the interactions of the C-terminal helices that form contacts to each other in an antiparallel, crossover fashion (Figs. 1C and 3A, left). A hydrophobic patch forming interdimer interactions (Fig. 1B), thus far found only on the C-terminal helices of yeast Atm1, appears to stabilize the transporter and to confine the degree of NBD separation to a narrow range (fig. S2). To our knowledge, such a binding mode of mostly hydrophobic contacts has not been found in any other type IV ABC transporter before and is different in fungal CtAtm1 and bacterial NaAtm1 (figs. S6 and S8) (15, 34).
AMP-PNP-Mg2+ binding (Fig. 2D) converts yeast Atm1 into an occluded conformation. During the transition to the nucleotide-bound state, the C-terminal helices move around each other (Fig. 3A), reminiscent of the C-terminal zipper helices of TmrAB (33, 35). The rearrangement requires the opening of hydrophobic interactions between the helices and break part of the α-helical structure of the helices themselves. The resulting state of associated NBDs, in turn, is stabilized by a network of hydrophilic inter- and intrachain contacts, including conformation-specific interactions between residues R634 and E675 that are conserved only in the fungal homologs (Fig. 3, A to C, and fig. S7). As neither R634 nor E675 is conserved in NaAtm1, and as the NaAtm1 C-terminal helices become disordered beyond residue A598 (corresponding to yeast Atm1 D676) (34), the contacts involving R634 may not be required to break up the α-helical structures. Our previously published in vivo data revealed that C-terminal truncation affected protein levels, but not the function of Atm1, resulting in decreased cell growth and diminished cytosolic iron-sulfur protein biogenesis (12). This phenotype was evident only for the shorter mutant protein Atm1-L666, but not the longer Atm1-D676 that maintains the R634-E675 salt bridge. Our nanoDSF findings of the apo and occluded states of wild-type and C-terminally truncated Atm1 indicate that this salt bridge enhances the stability of Atm1 (Table 1).
Our ATPase assays further confirm a stimulatory effect of CL on the ATP hydrolysis activity of Atm1 [fig. S1B and (13)]. A bound lipid was identified in the same site of all three ScAtm1 cryo-EM maps (Figs. 1, A and C, and 2, A and B, and figs. S4D and S10, A and B). In the case of the AMP-PNP-Mg2+–bound state in MSP1E3D1 lipid nanodiscs, a second lipid was found in a neighboring site (Fig. 2, A and B). While none of the lipid molecules were clearly identified as CL, Atm1 in lipid nanodiscs is surrounded by several additional, unresolved lipids. It is therefore quite possible that CL binds directly to Atm1, stimulating its ATPase activity. Cells deficient in CL through CRD1 gene knockout showed a mild iron-sulfur protein biogenesis defect in mitochondria and the cytosol, which, however, was not alleviated by Atm1 overexpression (46). The stimulatory effect of CL on the in vitro ATPase activity of Atm1 is therefore unlikely to be physiologically relevant.
Under all tested conditions, most of yeast Atm1 in lipid nanodiscs appears to adopt one predominant state (Fig. 4 and figs. S1A, S2, S11, and S12). In the absence of nucleotides, in the presence of ADP-Mg2+, and under turnover conditions (ATP-Mg2+), the NBDs are separated. By contrast, addition of AMP-PNP-Mg2+ or ATP-Mg2+ plus vanadate resulted in almost quantitative NBD closure. The latter conditions involve the binding of non-native substrates and hence may point toward a short-lived closed state within the ATP hydrolysis and transport cycle, in which the NBDs are normally dissociated and the C-terminal helices intertwined. Conversely, in the event of cargo binding, an additional stabilization of the transporter with closed NBDs might drive the conformational equilibrium toward breaking the hydrophobic interactions of the C-terminal helices. Together, both the inward-open and occluded conformations of yeast Atm1 appear to be stabilized by conformation-specific features, in particular the hydrophobic patch on the C-terminal helices and the interactions involving the conserved R634. It remains unknown, however, why the ionic bond between R634 and E675 is conserved in fungal Atm1 homologs, while the hydrophobic patch on the C-terminal helices is only found in S. cerevisiae Atm1 (fig. S7). Other open questions include whether the substantial rearrangement of the C-terminal helices in yeast Atm1 serves a particular purpose (Fig. 3A) and whether it is yeast-specific or also occurs in other Atm1 homologs.
MATERIALS AND METHODS
Expression and purification of S. cerevisiae Atm1 and its variants
S. cerevisiae Atm1-Strep encoded in vector pASK-IBA2 was expressed in the E. coli BL21–derived strain C43 (47) and purified as described before (12, 13). The quantity of pure Atm1 was ca. 900 μg protein per liter of cell culture. The ATM1 Walker B (amino acid residue exchange E598Q) mutant gene was generated by site-directed mutagenesis of wild-type ATM1 in pASK-IBA2 by polymerase chain reaction–mediated mutagenesis using the primer pair ACCAGGCCACTAGTGCCTTGGACACACACACAG and AGGCACTAGTGGCCTGGTCAAAAAACATGATCC. The mutant gene encoding C-terminally Strep-tagged Atm1-L657 (residues 92 to 656) was generated using fragment synthesis. The desired sequences of the mutants were validated by DNA sequencing.
Cloning, expression, and purification of C. thermophilum Atm1
The gene encoding Atm1 from C. thermophilum (UniProt, G0SBE6) starting from codon 26 was cloned into the pASK-IBA2 vector that contains a C-terminal Strep-tag. At the N terminus, an E. coli OmpA signal sequence for membrane targeting was introduced (13). The correctness of the fusion construct was verified by DNA sequencing. C. thermophilum Atm1 was produced in E. coli BL21 cells and purified as previously described for Atm1 from S. cerevisiae (13).
MSP1D1 expression and purification
A preculture of 90 ml of E. coli BL21 harboring the pMSP1D1 plasmid (Addgene #20061) in LB medium containing 2 mM MgSO4 and kanamycin sulfate (50 μg/ml) was grown at 37°C and stored overnight at 4°C after reaching an OD600nm (optical density at 600 nm) of 0.7. The preculture was used to inoculate 6 liters of TB buffer with 2 mM MgSO4 and kanamycin sulfate (50 μg/ml). Isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 1 mM at an OD600nm of 2. Cells were harvested after 3 hours of protein overexpression.
The cell pellet (13 g) was resuspended in 100 ml of 40 mM KH2PO4/K2HPO4 (pH 7.4) with one tablet of cOmplete EDTA-free Protease Inhibitor Cocktail (Roche #05056489001), lysozyme (1 mg/ml), and 1 mg of deoxyribonuclease (DNase) I and passed through a Constant Systems T5 cell disruptor. Centrifugation was followed by addition of 1% (w/v) Triton X-100 and imidazole to a final concentration of 20 mM. MSP1D1 was purified using a 5-ml HisTrap FF column essentially according to Bayburt et al. (48). The relevant fractions were pooled, concentrated, and dialyzed against 20 mM tris-HCl, 0.1 M NaCl, and 0.5 mM EDTA (pH 7.4; volume ratio, 1:300) using 6 to 8 MWCO (molecular weight cutoff) Spectra/Por membranes (Thermo Fisher Scientific #132650), yielding 5.2 ml of MSP1D1 (7.6 mg/ml).
MSP1E3D1 expression and purification
A preculture of 50 ml of E. coli BL21 harboring the pMSP1E3D1 plasmid (Addgene #20066) in LB medium containing kanamycin sulfate (50 μg/ml) was grown at 37°C and stored overnight at 4°C after reaching an OD600nm of 0.7. The preculture was used to inoculate 6 liters of TB buffer with 2 mM MgSO4 and kanamycin sulfate (50 μg/ml). IPTG was added to a final concentration of 1 mM at an OD600nm of 2. Cells were harvested after 3 hours of protein overexpression.
The cell pellet (9.4 g) was resuspended in 100 ml of 40 mM KH2PO4/K2HPO4 (pH 7.4) with one tablet of cOmplete EDTA-free Protease Inhibitor Cocktail (Roche #05056489001), lysozyme (1 mg/ml), and 1 mg of DNAse I and passed through a Constant Systems T5 cell disruptor. Centrifugation was followed by addition of 1% (w/v) Triton X-100 and imidazole to a final concentration of 20 mM. MSP1E3D1 was purified using a 5-ml HisTrap FF column essentially according to Bayburt et al. (48). The relevant fractions were pooled, concentrated, and dialyzed against 20 mM tris-HCl, 0.1 M NaCl, and 0.5 mM EDTA (pH 7.4; volume ratio, 1:300) using 6 to 8 MWCO Spectra/Por membranes (Thermo Fisher Scientific #132650), yielding 7 ml of MSP1E3D1 (10.9 mg/ml).
Lipid nanodisc reconstitution and purification
Lipids were prepared as a 2:2:1 molar mixture of POPC:POPE:CL (1-palmitoyl-2-oleoyl-glycero-3-phosphocholine, 1-palmitoyl-2-oleoyl-glycero-3-ethanolamine, and CL, respectively; Avanti Polar Lipids #850457, #850757, and #710335, respectively) in the presence of 50 mM NaCl, 20 mM tris-HCl (pH 7.5), and 0.57% (w/v) DDM, otherwise following the procedures reported by Dijkman and Watts (49). Atm1 for cryo-EM was reconstituted into lipid nanodiscs at a molar ratio of Atm1monomer:MSP:lipids of 1:2:40 (nucleotide-free state in MSP1D1 nanodiscs), 1:2.5:21 (AMP-PNP–bound state; MSP1D1), or 1:2:100 (AMP-PNP–bound state; MSP1E3D1) in the presence of 1.4-fold molar excess of DDM (50). CtAtm1 was reconstituted at an CtAtm1monomer:MSP1E3D1:lipids ratio of 1:2:140, to account for additional N-terminal residues. Incubation with roughly 1.5 mg of Bio-Beads SM-2 Adsorbent Media (Bio-Rad #152-3920) per 1 mg of DDM was followed by a two-step purification consisting of a column filled with 0.5 ml of Strep-Tactin Superflow resin (IBA #2-1206-025; procedure according to the instructions of the manufacturer), and size exclusion chromatography on a Superdex 200 Increase 5/150 GL column using 50 mM NaCl and 20 mM tris-HCl (pH 7.5), and in the case of the MSP1D1 samples, 10 mM MgCl2.
Nanodisc reconstitutions of Atm1 wild-type, Atm1 E598Q, and Atm1-L657 for negative-stain EM and ATP hydrolysis assays were prepared at Atm1monomer:MSP:lipid molar ratios of 1:2:40 (MSP1D1) and 1:2:100 (MSP1E3D1) and purified in the absence of MgCl2, as explained above. In the case of nanodiscs without CL, lipid mixtures of a 1:1 molar ratio of POPC and POPE were used. Empty lipid nanodiscs were prepared using a 70-fold molar excess of the lipid mixture over MSP1D1 and purified in a single step using the same size exclusion chromatography setup as for Atm1-containing nanodiscs.
Negative-stain EM
All negative-stain EM data were recorded from 3.05-mm G400 copper grids (SPI Supplies) carbon coated in a Leica EM ACE600 and glow discharged in a PELCO easiGlow (Ted Pella, Inc.). Three microliters of protein sample were added onto each grid and blotted away after 30 s, followed by three 2.5-μl applications of 2% uranyl formate following established protocols (51, 52). All images were recorded on an FEI Tecnai G2 Spirit BioTwin operated at 120 kV equipped with a Gatan US4000 4k × 4k charge-coupled device camera at a nominal magnification of 49,000 (2.2-Å pixel size) and processed using EMAN2 and RELION 3.1 (53, 54).
Molybdate-based ATP hydrolysis assay
All protein concentrations were measured as absorbances at 280 nm on a NanoDrop One (Thermo Fisher Scientific), and the contributions of the MSPs were taken into account assuming two MSP copies per lipid nanodisc. The ATPase activities were measured as triplicates following a molybdate-based assay (55). Atm1 (0.25 mg/ml) or empty MSP1D1 lipid nanodiscs (0.09 mg/ml) were incubated for 20 min at 30°C. The reaction conditions included 80 mM tris-HCl, 50 mM NaCl, 1 mM ATP, and 2 mM MgCl2, pH 7.5. In the case of the inhibition samples, Atm1 was preincubated with 1 mM Na3VO4 or 0.7 mM AMP-PNP. All samples were analyzed on a SpectraMax M2 (Molecular Devices) at 850 nm, and the activities were standardized to the Atm1 wild-type in MSP1D1 condition representing 100%.
Cryo-EM grid freezing and data collection
C-flat 2/2-3C holey carbon-coated copper grids (Science Services ECF-223C-50) were glow discharged in a PELCO easiGlow (Ted Pella Inc.). AMP-PNP (TOCRIS #6086) was added 40 min (MSP1D1 data; 3.7 mM final concentration) or 70 min (MSP1E3D1 data; 3.6 mM final concentration) before freezing and incubated on ice. In an attempt to avoid preferred orientation, DDM was added to the sample in the case of the AMP-PNP–bound state (MSP1D1) immediately before freezing to a final concentration of 0.02 mM. The final sample concentrations including the contributions of the MSP molecules were 0.98 mg/ml (nucleotide-free state; MSP1D1, with 10 mM MgCl2), 0.72 mg/ml (AMP-PNP–bound state; MSP1D1, with 9.3 mM MgCl2), 0.7 mg/ml (AMP-PNP–bound state; MSP1E3D1, with 3.6 mM MgCl2), and 0.8 mg/ml (CtAtm1). Three microliters of the sample were applied to each grid and blotted using filter papers of the 595 type (Schleicher & Schuell #10311607) and a FEI Vitrobot Mark IV.
Cryo-EM datasets were recorded on an FEI Krios G3i microscope equipped with a Gatan K3 direct electron detector (yeast Atm1 nucleotide-free state: 0.837 Å per pixel, 25-eV energy filter slit width, 64 electrons/Å2 total dose, 3-s exposure time, 50 frames per movie, 2430 + 1690 micrographs; CtAtm1: 0.837 Å per pixel, 30-eV energy filter slit width, 63 electrons/Å2 total dose, 3-s exposure time, 50 frames per movie, 2066 images) or an FEI Falcon 3EC camera (AMP-PNP–bound state, MSP1D1; 0.833 Å per pixel, 50 electrons/Å2 total dose, 41-s exposure time, 53 frames per movie, 4610 + 904 micrographs), or on an FEI Krios G2 microscope with a Gatan K2 direct electron detector (AMP-PNP–bound state, MSP1E3D1; 0.828 Å per pixel, 30-eV energy filter slit width, 68 electrons/Å2 total dose, 8-s exposure time, 40 frames per movie, 1885 micrographs), all using the automated data acquisition software EPU (Thermo Fisher Scientific).
Data processing
All micrographs were drift corrected using MotionCor2 (56), for the AMP-PNP–bound state in MSP1E3D1 nanodiscs together with gain reference correction. The contrast transfer function (CTF) was estimated using Gctf (57). Following visual examination, 7.2%, 5.4% (yeast Atm1 nucleotide-free state, first and second data collections, respectively), 7.7%, 7.5% (AMP-PNP–bound state in MSP1D1 nanodiscs, first and second data collections, respectively), 11% (AMP-PNP–bound state in MSP1E3D1 nanodiscs), and 13% (CtAtm1) of all micrographs were considered unsuitable for data processing and not used further. Micrographs ranging between −1.2- and −3.5-μm defocus with CTF fits extending beyond 4.5 Å were included for downstream processing of the yeast Atm1 nucleotide-free datasets (excluding another 7.4 and 12%). Similarly, an additional 5.4 and 7.9% of the AMP-PNP–bound state in MSP1D1 nanodiscs datasets were excluded (−1.0- to −3.5-μm defocus and 4.5-Å CTF fit). All yeast Atm1 particles were picked in RELION 3.0 (58), while Topaz was used for the CtAtm1 data (59). All other RELION steps were carried out with version 3.1 (53). The original 3D references for ScAtm1 data processing were lower-resolution maps of the inward-open and occluded conformations obtained from test datasets recorded on an FEI Polara equipped with a Gatan K2 direct electron detector. They were fourfold binned and lowpass filtered to 20-Å resolution. Similarly, the original 3D reference map for the CtAtm1 data was an ScAtm1 map of the nucleotide-free state that had been fourfold binned and lowpass filtered to 15-Å resolution. Initial particle extractions used a fourfold-binned 256 pixel box, followed by one round of 3D classification before working with unbinned data. As for all 3D classifications, class inclusion was based not only on their visual appearance and statistics but also on cryoSPARC 2D classifications of every class to assess classification success [all ScAtm1 cryoSPARC processing with v2.15.0 (60)].
As for the yeast Atm1 nucleotide-free state, picked particles were cleaned up and classified by two more rounds of 3D classification. Bayesian polishing, CTF refinement, and 2D classification in cryoSPARC were carried out before data merging. The TMDs of the resulting 724,000 particles were masked (see fig. S2) for focused 3D classification. A class of 439,015 particles was extracted at a box size of 320 pixels. 3D refinement, Bayesian polishing, and CTF refinements were carried out before data merging. The final map was obtained after another round of CTF refinement with cryoSPARC Non-uniform Refinement.
In the case of the AMP-PNP–bound state in MSP1D1 nanodiscs, particles of both datasets underwent Bayesian polishing and CTF refinement before merging for a second round of 3D classification and subsequent 2D classification in cryoSPARC. A bias due to overrepresented views along the twofold symmetry axis of the dimer was successfully avoided using cryoSPARC Ab-Initio Reconstructions for further 3D classification. The resulting class of 186,526 particles was extracted at a box size of 320 pixels in RELION and processed like the nucleotide-free dataset. Particles with a defocus above −2.9 μm were excluded, yielding a final dataset of 178,134 particles used for cryoSPARC Non-uniform Refinement.
In the case of the AMP-PNP–bound state in MSP1E3D1 nanodiscs, one round of 3D classification of the unbinned particles preceded two short 3D classification rounds to remove particles in the inward-facing state and hot pixels using a previously obtained hot pixel 3D map and the previously obtained maps of the nucleotide-free and AMP-PNP–bound states as references. The dataset was cleaned up further by a fifth round of 3D classification and cryoSPARC 2D classification. The resulting 209,295 particles were extracted at a box size of 320 pixels in RELION, followed by Bayesian polishing and CTF refinement. Particles with a defocus above −2.5 μm were excluded, yielding a final dataset of 129,849 particles used for cryoSPARC Non-uniform Refinement.
For CtAtm1 data, the initial, binned 3D classification reduced the total number of particles to 54% of the initially picked 374,012 particles. The particles from the two selected classes were merged and polished. Cleaning with cryoSPARC 2D classification resulted in a dataset of 181,901 particles. Subsequent particle subtraction and 3D classification were carried out using the same mask as for the yeast Atm1 nucleotide-free data. All three classes were refined using cryoSPARC Non-uniform Refinement (all CtAtm1 cryoSPARC processing with v3.0.1).
All final maps were calculated applying C2 symmetry. Transfer of particles from cryoSPARC back to RELION 3.1 and creation of the angular distribution plots were greatly aided by pyem (61).
Model building
The model of the nucleotide-free state of Atm1 was built using the previously published crystal structure [PDB ID: 4myc; (12)] following PHENIX Dock in map (62) and alternating steps of manual model building in Coot (63) and phenix.real_space_refine (64) using restraints files created by PHENIX eLBOW (65). The model of the AMP-PNP–bound structures started with manual and automated model docking of pieces of the nucleotide-free state model using UCSF Chimera (66) and PHENIX Dock in map before proceeding as described above. RMSD values between different sets of coordinates were calculated using CCP4 GESAMT (67). Figures were drawn using UCSF Chimera and UCSF ChimeraX (66, 68). Cavities were calculated and drawn using CASTp 3.0 and HOLLOW (69, 70) with a 1.4-Å probe radius, using the coordinates of the MSP1E3D1 variant of the AMP-PNP-Mg2+–bound Atm1 structures with plugs added to block the narrow passages to the matrix (asterisk, Fig. 2D). The sequence alignment was prepared by Clustal Omega and ESPript 3.0 (71, 72).
Nanodifferential scanning fluorimetry
The thermostability of purified Atm1 and its variants was measured by nanoDSF in gel filtration buffer (100 mM Tris-HCl, 150 mM NaCl, 10% sorbitol, and 0.025% DDM) using a Prometheus NT.48 (Nanotemper, Munich). Atm1 (1 mg/ml final concentration) was supplemented without or with 5 mM AMP-PNP and 8 mM MgCl2 in a final volume of 40 μl. Triplicate samples were introduced into 10-μl capillaries and subjected to a temperature increase of 1°C per minute from 20°C to 95°C. A wavelength scan (ratio of 330 nm versus 300 nm at an overall laser power of 10% to 30%) was recorded to obtain an unfolding curve of the protein in presence or absence of AMP-PNP-Mg2+. Measurements were performed for at least three independent protein purifications. The unfolding transition temperature was calculated according to the manufacturer’s instructions.
NADH-coupled ATP hydrolysis assay
The specific ATPase activity of Atm1 and variants was analyzed essentially as described (12) using an NADH (reduced form of nicotinamide adenine dinucleotide)–coupled assay in which the hydrolysis product ADP was detected via the sequential action of pyruvate kinase (PK) and lactate dehydrogenase (LDH) in the presence of phosphoenolpyruvate (PEP). The reaction mixtures contained 450 μM NADH, LDH (55 U/ml), PK (184 U/ml), 1.38 mM PEP, 0.5 mM DDM, 5.3 mM ATP, and 8 mM MgCl2. ATPase activity was measured in a Tecan Infinite pro plate reader at 30°C. Each sample was produced in quadruplicate for at least three independent protein purifications.
Acknowledgments
We thank D. Mills and S. Prinz for valuable assistance with cryo-EM specimen preparation and data collection, A. Möller for advice, and A. R. Mehdipour and J. Vonck for discussions.
Funding: This work was funded by the Max Planck Society (W.K. and T.L.E.) and the LOEWE program of the State of Hessen. R.L. and V.S. acknowledge generous financial support from the German Research Foundation DFG (LI 415/5, LI 415/6-1, and SR 113/1) and the LOEWE program of the State of Hessen.
Author contributions: V.S., R.L., and W.K. initiated the study; T.M. expressed and purified wild-type and mutant Atm1 proteins and performed DSF and NADH-coupled ATP hydrolysis assays; T.L.E. performed EM sample preparation, data collection and image processing, model building and interpretation, molybdate-based ATP hydrolysis assays, nanodisc protein expression, purification and reconstitution, and drew the figures; T.L.E., R.L., and W.K. analyzed the data and wrote the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: The cryo-EM maps have been deposited in the Electron Microscopy Data Bank with accession codes EMD-13613 (nucleotide-free yeast Atm1), EMD-13614 (AMP-PNP-Mg2+–bound yeast Atm1 in MSP1D1 nanodiscs), EMD-13615 (AMP-PNP-Mg2+–bound yeast Atm1 in MSP1E3D1 nanodiscs), EMD-13616 (nucleotide-free CtAtm1 class 1), EMD-13617 (nucleotide-free CtAtm1 class 2), and EMD-13618 (nucleotide-free CtAtm1 class 3). The atomic models of yeast Atm1 have been deposited in the Protein Data Bank under accession codes 7psl (nucleotide-free yeast Atm1), 7psm (AMP-PNP-Mg2+–bound yeast Atm1 in MSP1D1 nanodiscs), and 7psn (AMP-PNP-Mg2+–bound yeast Atm1 in MSP1E3D1 nanodiscs). All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S17
Table S1
REFERENCES AND NOTES
- 1.Kispal G., Csere P., Prohl C., Lill R., The mitochondrial proteins Atm1p and Nfs1p are essential for biogenesis of cytosolic Fe/S proteins. EMBO J. 18, 3981–3989 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hausmann A., Samans B., Lill R., Mühlenhoff U., Cellular and mitochondrial remodeling upon defects in iron-sulfur protein biogenesis. J. Biol. Chem. 283, 8318–8330 (2008). [DOI] [PubMed] [Google Scholar]
- 3.Pandey A., Pain J., Dziuba N., Pandey A. K., Dancis A., Lindahl P. A., Pain D., Mitochondria export sulfur species required for cytosolic tRNA thiolation. Cell Chem. Biol. 25, 738–748.e3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cavadini P., Biasiotto G., Poli M., Levi S., Verardi R., Zanella I., Derosas M., Ingrassia R., Corrado M., Arosio P., RNA silencing of the mitochondrial ABCB7 transporter in HeLa cells causes an iron-deficient phenotype with mitochondrial iron overload. Blood 109, 3552–3559 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Pondarré C., Antiochos B. B., Campagna D. R., Clarke S. L., Greer E. L., Deck K. M., McDonald A., Han A. P., Medlock A., Kutok J. L., Anderson S. A., Eisenstein R. S., Fleming M. D., The mitochondrial ATP-binding cassette transporter Abcb7 is essential in mice and participates in cytosolic iron-sulfur cluster biogenesis. Hum. Mol. Genet. 15, 953–964 (2006). [DOI] [PubMed] [Google Scholar]
- 6.Kushnir S., Babiychuk E., Storozhenko S., Davey M. W., Papenbrock J., de Rycke R., Engler G., Stephan U. W., Lange H., Kispal G., Lill R., van Montagu M., A mutation of the mitochondrial ABC transporter Sta1 leads to dwarfism and chlorosis in the Arabidopsis mutant starik. Plant Cell 13, 89–100 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garcia-Santamarina S., Uzarska M. A., Festa R. A., Lill R., Thiele D. J., Cryptococcus neoformans iron-sulfur protein biogenesis machinery is a novel layer of protection against Cu stress. MBio 8, e01742-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li J., Kogan M., Knight S. A., Pain D., Dancis A., Yeast mitochondrial protein, Nfs1p, coordinately regulates iron-sulfur cluster proteins, cellular iron uptake, and iron distribution. J. Biol. Chem. 274, 33025–33034 (1999). [DOI] [PubMed] [Google Scholar]
- 9.Biederbick A., Stehling O., Rösser R., Niggemeyer B., Nakai Y., Elsässer H. P., Lill R., Role of human mitochondrial Nfs1 in cytosolic iron-sulfur protein biogenesis and iron regulation. Mol. Cell. Biol. 26, 5675–5687 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sipos K., Lange H., Fekete Z., Ullmann P., Lill R., Kispal G., Maturation of cytosolic iron-sulfur proteins requires glutathione. J. Biol. Chem. 277, 26944–26949 (2002). [DOI] [PubMed] [Google Scholar]
- 11.Pearson S. A., Wachnowsky C., Cowan J. A., Defining the mechanism of the mitochondrial Atm1p [2Fe-2S] cluster exporter. Metallomics 12, 902–915 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Srinivasan V., Pierik A. J., Lill R., Crystal structures of nucleotide-free and glutathione-bound mitochondrial ABC transporter Atm1. Science 343, 1137–1140 (2014). [DOI] [PubMed] [Google Scholar]
- 13.Kuhnke G., Neumann K., Mühlenhoff U., Lill R., Stimulation of the ATPase activity of the yeast mitochondrial ABC transporter Atm1p by thiol compounds. Mol. Membr. Biol. 23, 173–184 (2006). [DOI] [PubMed] [Google Scholar]
- 14.Schaedler T. A., Thornton J. D., Kruse I., Schwarzländer M., Meyer A. J., van Veen H. W., Balk J., A conserved mitochondrial ATP-binding cassette transporter exports glutathione polysulfide for cytosolic metal cofactor assembly. J. Biol. Chem. 289, 23264–23274 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee J. Y., Yang J. G., Zhitnitsky D., Lewinson O., Rees D. C., Structural basis for heavy metal detoxification by an Atm1-type ABC exporter. Science 343, 1133–1136 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Allikmets R., Raskind W. H., Hutchinson A., Schueck N. D., Dean M., Koeller D. M., Mutation of a putative mitochondrial iron transporter gene (ABC7) in X-linked sideroblastic anemia and ataxia (XLSA/A). Hum. Mol. Genet. 8, 743–749 (1999). [DOI] [PubMed] [Google Scholar]
- 17.Bekri S., Kispal G., Lange H., Fitzsimons E., Tolmie J., Lill R., Bishop D. F., Human ABC7 transporter: Gene structure and mutation causing X-linked sideroblastic anemia with ataxia with disruption of cytosolic iron-sulfur protein maturation. Blood 96, 3256–3264 (2000). [PubMed] [Google Scholar]
- 18.Maguire A., Hellier K., Hammans S., May A., X-linked cerebellar ataxia and sideroblastic anaemia associated with a missense mutation in the ABC7 gene predicting V411L. Br. J. Haematol. 115, 910–917 (2001). [DOI] [PubMed] [Google Scholar]
- 19.D’Hooghe M., Selleslag D., Mortier G., Van Coster R., Vermeersch P., Billiet J., Bekri S., X-linked sideroblastic anemia and ataxia: A new family with identification of a fourth ABCB7 gene mutation. Eur. J. Paediatr. Neurol. 16, 730–735 (2012). [DOI] [PubMed] [Google Scholar]
- 20.Protasova M. S., Grigorenko A. P., Tyazhelova T. V., Andreeva T. V., Reshetov D. A., Gusev F. E., Laptenko A. E., Kuznetsova I. L., Goltsov A. Y., Klyushnikov S. A., Illarioshkin S. N., Rogaev E. I., Whole-genome sequencing identifies a novel ABCB7 gene mutation for X-linked congenital cerebellar ataxia in a large family of Mongolian ancestry. Eur. J. Hum. Genet. 24, 550–555 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kispal G., Csere P., Guiard B., Lill R., The ABC transporter Atm1p is required for mitochondrial iron homeostasis. FEBS Lett. 418, 346–350 (1997). [DOI] [PubMed] [Google Scholar]
- 22.Dawson R. J., Hollenstein K., Locher K. P., Uptake or extrusion: Crystal structures of full ABC transporters suggest a common mechanism. Mol. Microbiol. 65, 250–257 (2007). [DOI] [PubMed] [Google Scholar]
- 23.Thomas C., Aller S. G., Beis K., Carpenter E. P., Chang G., Chen L., Dassa E., Dean M., Duong van Hoa F., Ekiert D., Ford R., Gaudet R., Gong X., Holland I. B., Huang Y., Kahne D. K., Kato H., Koronakis V., Koth C. M., Lee Y., Lewinson O., Lill R., Martinoia E., Murakami S., Pinkett H. W., Poolman B., Rosenbaum D., Sarkadi B., Schmitt L., Schneider E., Shi Y., Shyng S. L., Slotboom D. J., Tajkhorshid E., Tieleman D. P., Ueda K., Váradi A., Wen P. C., Yan N., Zhang P., Zheng H., Zimmer J., Tampé R., Structural and functional diversity calls for a new classification of ABC transporters. FEBS Lett. 594, 3767–3775 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rees D. C., Johnson E., Lewinson O., ABC transporters: The power to change. Nat. Rev. Mol. Cell Biol. 10, 218–227 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eytan G. D., Regev R., Assaraf Y. G., Functional reconstitution of P-glycoprotein reveals an apparent near stoichiometric drug transport to ATP hydrolysis. J. Biol. Chem. 271, 3172–3178 (1996). [DOI] [PubMed] [Google Scholar]
- 26.Shukla S., Rai V., Banerjee D., Prasad R., Characterization of Cdr1p, a major multidrug efflux protein of Candida albicans: Purified protein is amenable to intrinsic fluorescence analysis. Biochemistry 45, 2425–2435 (2006). [DOI] [PubMed] [Google Scholar]
- 27.Thomas C., Tampé R., Structural and mechanistic principles of ABC transporters. Annu. Rev. Biochem. 89, 605–636 (2020). [DOI] [PubMed] [Google Scholar]
- 28.Jardetzky O., Simple allosteric model for membrane pumps. Nature 211, 969–970 (1966). [DOI] [PubMed] [Google Scholar]
- 29.Shintre C. A., Pike A. C. W., Li Q., Kim J. I., Barr A. J., Goubin S., Shrestha L., Yang J., Berridge G., Ross J., Stansfeld P. J., Sansom M. S. P., Edwards A. M., Bountra C., Marsden B. D., von Delft F., Bullock A. N., Gileadi O., Burgess-Brown N. A., Carpenter E. P., Structures of ABCB10, a human ATP-binding cassette transporter in apo- and nucleotide-bound states. Proc. Natl. Acad. Sci. U.S.A. 110, 9710–9715 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dawson R. J. P., Locher K. P., Structure of a bacterial multidrug ABC transporter. Nature 443, 180–185 (2006). [DOI] [PubMed] [Google Scholar]
- 31.Ward A., Reyes C. L., Yu J., Roth C. B., Chang G., Flexibility in the ABC transporter MsbA: Alternating access with a twist. Proc. Natl. Acad. Sci. U.S.A. 104, 19005–19010 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Choudhury H. G., Tong Z., Mathavan I., Li Y., Iwata S., Zirah S., Rebuffat S., van Veen H. W., Beis K., Structure of an antibacterial peptide ATP-binding cassette transporter in a novel outward occluded state. Proc. Natl. Acad. Sci. U.S.A. 111, 9145–9150 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hofmann S., Januliene D., Mehdipour A. R., Thomas C., Stefan E., Brüchert S., Kuhn B. T., Geertsma E. R., Hummer G., Tampé R., Moeller A., Conformation space of a heterodimeric ABC exporter under turnover conditions. Nature 571, 580–583 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fan C., Kaiser J. T., Rees D. C., A structural framework for unidirectional transport by a bacterial ABC exporter. Proc. Natl. Acad. Sci. U.S.A. 117, 19228–19236 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nöll A., Thomas C., Herbring V., Zollmann T., Barth K., Mehdipour A. R., Tomasiak T. M., Brüchert S., Joseph B., Abele R., Oliéric V., Wang M., Diederichs K., Hummer G., Stroud R. M., Pos K. M., Tampé R., Crystal structure and mechanistic basis of a functional homolog of the antigen transporter TAP. Proc. Natl. Acad. Sci. U.S.A. 114, E438–E447 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mi W., Li Y., Yoon S. H., Ernst R. K., Walz T., Liao M., Structural basis of MsbA-mediated lipopolysaccharide transport. Nature 549, 233–237 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alam A., Küng R., Kowal J., McLeod R. A., Tremp N., Broude E. V., Roninson I. B., Stahlberg H., Locher K. P., Structure of a zosuquidar and UIC2-bound human-mouse chimeric ABCB1. Proc. Natl. Acad. Sci. U.S.A. 115, E1973–E1982 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jin M. S., Oldham M. L., Zhang Q., Chen J., Crystal structure of the multidrug transporter P-glycoprotein from Caenorhabditis elegans. Nature 490, 566–569 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ward A. B., Szewczyk P., Grimard V., Lee C. W., Martinez L., Doshi R., Caya A., Villaluz M., Pardon E., Cregger C., Swartz D. J., Falson P. G., Urbatsch I. L., Govaerts C., Steyaert J., Chang G., Structures of P-glycoprotein reveal its conformational flexibility and an epitope on the nucleotide-binding domain. Proc. Natl. Acad. Sci. U.S.A. 110, 13386–13391 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li J., Jaimes K. F., Aller S. G., Refined structures of mouse P-glycoprotein. Protein Sci. 23, 34–46 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perez C., Gerber S., Boilevin J., Bucher M., Darbre T., Aebi M., Reymond J. L., Locher K. P., Structure and mechanism of an active lipid-linked oligosaccharide flippase. Nature 524, 433–438 (2015). [DOI] [PubMed] [Google Scholar]
- 42.Angiulli G., Dhupar H. S., Suzuki H., Wason I. S., Duong van Hoa F., Walz T., New approach for membrane protein reconstitution into peptidiscs and basis for their adaptability to different proteins. eLife 9, e53530 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Z., Hu W., Zheng H., Pathogenic siderophore ABC importer YbtPQ adopts a surprising fold of exporter. Sci. Adv. 6, eaay7997 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moeller A., Lee S. C., Tao H., Speir J. A., Chang G., Urbatsch I. L., Potter C. S., Carragher B., Zhang Q., Distinct conformational spectrum of homologous multidrug ABC transporters. Structure 23, 450–460 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sharma S., Davidson A. L., Vanadate-induced trapping of nucleotides by purified maltose transport complex requires ATP hydrolysis. J. Bacteriol. 182, 6570–6576 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Patil V. A., Fox J. L., Gohil V. M., Winge D. R., Greenberg M. L., Loss of cardiolipin leads to perturbation of mitochondrial and cellular iron homeostasis. J. Biol. Chem. 288, 1696–1705 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miroux B., Walker J. E., Over-production of proteins in Escherichia coli: Mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol. 260, 289–298 (1996). [DOI] [PubMed] [Google Scholar]
- 48.Bayburt T. H., Grinkova Y. V., Sligar S. G., Self-assembly of discoidal phospholipid bilayer nanoparticles with membrane scaffold proteins. Nano Lett. 2, 853–856 (2002). [Google Scholar]
- 49.Dijkman P. M., Watts A., Lipid modulation of early G protein-coupled receptor signalling events. Biochim. Biophys. Acta 1848, 2889–2897 (2015). [DOI] [PubMed] [Google Scholar]
- 50.Ritchie T. K., Grinkova Y. V., Bayburt T. H., Denisov I. G., Zolnerciks J. K., Atkins W. M., Sligar S. G., Reconstitution of membrane proteins in phospholipid bilayer nanodiscs. Methods Enzymol. 464, 211–231 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tao H., Lee S. C., Moeller A., Roy R. S., Siu F. Y., Zimmermann J., Stevens R. C., Potter C. S., Carragher B., Zhang Q., Engineered nanostructured β-sheet peptides protect membrane proteins. Nat. Methods 10, 759–761 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gewering T., Januliene D., Ries A. B., Moeller A., Know your detergents: A case study on detergent background in negative stain electron microscopy. J. Struct. Biol. 203, 242–246 (2018). [DOI] [PubMed] [Google Scholar]
- 53.Zivanov J., Nakane T., Scheres S. H. W., Estimation of high-order aberrations and anisotropic magnification from cryo-EM data sets in RELION-3.1. IUCrJ 7, 253–267 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tang G., Peng L., Baldwin P. R., Mann D. S., Jiang W., Rees I., Ludtke S. J., EMAN2: An extensible image processing suite for electron microscopy. J. Struct. Biol. 157, 38–46 (2007). [DOI] [PubMed] [Google Scholar]
- 55.Chifflet S., Torriglia A., Chiesa R., Tolosa S., A method for the determination of inorganic phosphate in the presence of labile organic phosphate and high concentrations of protein: Application to lens ATPases. Anal. Biochem. 168, 1–4 (1988). [DOI] [PubMed] [Google Scholar]
- 56.Zheng S. Q., Palovcak E., Armache J. P., Verba K. A., Cheng Y., Agard D. A., MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang K., Gctf: Real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zivanov J., Nakane T., Forsberg B. O., Kimanius D., Hagen W. J. H., Lindahl E., Scheres S. H. W., New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bepler T., Morin A., Rapp M., Brasch J., Shapiro L., Noble A. J., Berger B., Positive-unlabeled convolutional neural networks for particle picking in cryo-electron micrographs. Nat. Methods 16, 1153–1160 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Punjani A., Rubinstein J. L., Fleet D. J., Brubaker M. A., cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 61.D. Asarnow, E. Palovcak, Y. Cheng, in UCSF pyem v0.5. Zenodo. (2019); 10.5281/zenodo.3576630. [DOI]
- 62.Liebschner D., Afonine P. V., Baker M. L., Bunkóczi G., Chen V. B., Croll T. I., Hintze B., Hung L. W., Jain S., McCoy A. J., Moriarty N. W., Oeffner R. D., Poon B. K., Prisant M. G., Read R. J., Richardson J. S., Richardson D. C., Sammito M. D., Sobolev O. V., Stockwell D. H., Terwilliger T. C., Urzhumtsev A. G., Videau L. L., Williams C. J., Adams P. D., Macromolecular structure determination using X-rays, neutrons and electrons: Recent developments in Phenix. Acta Crystallogr. D Struct. Biol. 75, 861–877 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Afonine P. V., Poon B. K., Read R. J., Sobolev O. V., Terwilliger T. C., Urzhumtsev A., Adams P. D., Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. D Struct. Biol. 74, 531–544 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Moriarty N. W., Grosse-Kunstleve R. W., Adams P. D., electronic Ligand Builder and Optimization Workbench (eLBOW): A tool for ligand coordinate and restraint generation. Acta Crystallogr. D Biol. Crystallogr. 65, 1074–1080 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E., UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 67.Krissinel E., Enhanced fold recognition using efficient short fragment clustering. J. Mol. Biochem. 1, 76–85 (2012). [PMC free article] [PubMed] [Google Scholar]
- 68.Pettersen E. F., Goddard T. D., Huang C. C., Meng E. C., Couch G. S., Croll T. I., Morris J. H., Ferrin T. E., UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tian W., Chen C., Lei X., Zhao J., Liang J., CASTp 3.0: Computed atlas of surface topography of proteins. Nucleic Acids Res. 46, W363–W367 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ho B. K., Gruswitz F., HOLLOW: Generating accurate representations of channel and interior surfaces in molecular structures. BMC Struct. Biol. 8, 49 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sievers F., Wilm A., Dineen D., Gibson T. J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., Thompson J. D., Higgins D. G., Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Robert X., Gouet P., Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kyte J., Doolittle R. F., A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157, 105–132 (1982). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S17
Table S1