Abstract
Hepatic disease is common in severe COVID-19. This study compared the histologic/molecular findings in the liver in fatal COVID-19 (n = 9) and age-matched normal controls (n = 9); three of the fatal COVID-19 livers had pre-existing alcohol use disorder (AUD). Controls showed a high resident population of sinusoidal macrophages that had variable ACE2 expression. Histologic findings in the cases included periportal/lobular inflammation. SARS-CoV2 RNA and nucleocapsid protein were detected in situ in 2/9 COVID-19 livers in low amounts. In 9/9 cases, there was ample in situ SARS-CoV-2 spike protein that co-localized with viral matrix and envelope proteins. The number of cells positive for spike/100× field was significantly greater in the AUD/COVID-19 cases (mean 5.9) versus the non-AUD/COVID-19 cases (mean 0.4, p < 0.001) which was corroborated by Western blots. ACE2+ cells were 10× greater in AUD/COVID-19 livers versus the other COVID-19/control liver samples (p < 0.001). Co-expression experiments showed that the spike protein localized to the ACE2 positive macrophages and, in the AUD cases, hepatic stellate cells that were activated as evidenced by IL6 and TNFα expression. Injection of the S1, but not S2, subunit of spike in mice induced hepatic lobular inflammation in activated macrophages. It is concluded that endocytosed viral spike protein can induce hepatitis in fatal COVID-19. This spike induced hepatitis is more robust in the livers with pre-existing AUD which may relate to why patients with alcohol abuse are at higher risk of severe liver disease with SARS-CoV2 infection.
Keywords: COVID-19, Liver, Spike protein, Hepatic stellate cells, Alcohol liver disease
1. Introduction
The COVID-19 pandemic has of this writing infected over 250 million people with more than 5 million deaths. Although the nasopharynx and lung are the epicenters of infectious SARS-CoV-2, viral RNA, presumably infectious, has also been reported in many sites including the blood, placenta, brain, liver, and heart [1], [2], [3], [4]. This led to the hypothesis that severe/fatal COVID-19 represents systemic infection that often involves the endothelial cells of microvessels [1], [2], [3], [4]. Alternatively, the systemic cytokine and coagulopathy of severe/fatal COVID-19 have been postulated to be responsible for the major systemic clinical/pathologic effects in the disease, including in the CNS, heart, and liver [1], [2], [3], [4].
Liver disease is a well-documented and common problem in severe COVID-19 [5], [6], [7], [8]. Pathologic changes in the liver ascribed to SARS-CoV2 include portal tract/lobular inflammation, microvascular thrombi, and hepatocyte necrosis [6], [7], [8]. However, several studies have indicated that SARS-CoV2 RNA is either not detectable or present in a minority of COVID-19 related hepatitis cases [6], [8], [9] though abundant in the lung [9], [10], [11], [12]. Thus, some have speculated that the liver disease in severe COVID-19 may represent an indirect effect of the cytokine storm or therapy [5], [6], [7], [8], [9]. It has also been established that pre-existing liver disease is associated with a worse outcome in COVID-19 [5]. One such pre-existing condition is excessive alcohol intake (AUD – alcohol use disorder). It has been reported that patients with a history of alcohol abuse that were infected by SARS-CoV2 were 5.5 times more at risk of needing an ICU admission and 10 times more likely to have liver failure [13], [14], [15]. What is not well understood is the molecular mechanisms whereby ethanol abuse potentiates liver damage in people infected with SARS-CoV2.
The purpose of this study was to examine the liver tissues from 9 people who died of COVID-19 including 3 with AUD and compare the molecular/histologic/viral findings in a blinded fashion to 9 pre-COVID controls.
2. Materials and methods
2.1. COVID-19 autopsies and controls
Autopsy material was available from nine people who died of COVID-19. They ranged in age from 50 to 72 (mean 64) with five men and four women. Salient clinical information included that 5/9 cases had pre-existing liver disease (three AUD and two non-alcoholic steatohepatitis) (NASH) (each mild). The liver tissues from nine aged matched people who died prior to 2016 served as negative controls where, for six people, the liver pathology was described as unremarkable and the other three had mild NASH. For qRT-PCR and Western blot analyzes, five ten-micron paraffin ribbons were placed in sterile Eppendorf tubes, the paraffin removed, and the RNA or protein extracted with the appropriate Qiagen kit as per the manufacturer's recommendations.
2.2. Immunohistochemistry
Immunohistochemistry was done as previously reported [10], [11], [12]. In brief, optimal conditions for each antibody were determined by testing various dilutions and pretreatment conditions (Table 1 ). The immunohistochemistry protocol used the Leica Bond Max automated platform with both the Fast red (DS 9820) and the DAB (DS 9800) detection kits from Leica Biosystems (Buffalo Grove, IL) used with equivalent results. As evident in Table 1, the host response was interrogated with antibodies against activated Caspase 3, IL6, TNFα, Complement terminal complex C5b-9, and Complement component 6 (C6). The HRP conjugate from Enzo Life Sciences (Farmingdale, New York, USA) was used in cases in place of the equivalent reagent from Leica in the DAB kit as this has been shown to reduce background for some primary antibodies [10], [11], [12].
Table 1.
Probes and antibodies used for the detection of SARS-CoV2 RNA and proteins and the concomitant host response.
| Reagent | Source/catalogue# | Dilution/pretreatment |
|---|---|---|
| SARS-CoV2 RNA probe | ACD/848561-C3 | RTU/AR and proteasea |
| ACE2 Ab | ProSci/3215 | 1:25,000/ARb |
| SARS-COV2 Spike S1 Ab | ProSci/9083 | 1:6000/AR |
| SARS-COV2 Spike S2 Ab | ProSci/9123 | 1:12,000/AR |
| SARS-COV2 envelope Ab | ProSci/3521 | 1:800/AR |
| SARS-COV2 matrix Ab | ProSci/3527 | 1:4000/AR |
| SARS-COV2 nucleocapsid Ab | ProSci/9099 | 1:13,000/AR |
| Caspase 3 Ab | Abcam/ab 4051 | 1:1200/AR |
| IL6 Ab | Abcam/ab6672 | 1: 7000/AR |
| TNFα Ab | Abcam/ab270264 | 1:5000/AR |
| Smooth muscle actin Ab | Ventana/790-2334 | RTU/AR |
| Complement component 6 | Proteintech/17239-1-AP | 1:1000/AR |
| CD11b | Abcam/ab 133657 | 1:7000/AR |
| CD163 | Abcam/182422 | 1:7000/AR |
| CD206 | Abcam/64693 | 1:6000/AR |
| CD68 | Ventana/790-2931 | RTU/AR |
| Type I collagen | Abcam/34710 | 1:600/AR |
| S-100 | Ventana/790-2914 | RTU/AR |
Each nonviral antibody was optimized in tonsil except for Complement component 6 which was optimized in COVID-19 lung with severe microangiopathy and pre-COVID normal lung.
Each viral antibody/probe was optimized in COVID-19 lung with high copy viral RNA in parallel with pre-COVID unremarkable lung.
As per ACD recommended protocol.
AR = antigen retrieval 30 min with Leica EDTA antigen retrieval solution; RTU = ready to use.
2.3. In situ hybridization
Detection of SARS-CoV-2 RNA was done using the ACD RNAscope (Newark, California, USA) probe (Cat No. 848561-C3) following the manufacturers recommended protocol as previously published [11], [12]. MicroRNA in situ hybridization for miR-128 and miR-155 was done as previously published using the Exiqon digoxigenin tagged locked nucleic acid probes [16].
2.4. Co-expression and statistical analyses
Co-expression analyses were done using the Nuance/InForm system whereby each chromogenic signal is separated, converted to a fluorescence-based signal, then mixed to determine the percentage of co-localization as previously described [10], [11], [12]. The number of positive cells/100× field was counted with the InForm software or manually in 10 fields/tissue. Statistical analysis was done using the InStat Statistical Analysis Software (version 3.36) and a paired t-test (also referred to as a “repeated measure t-test”). The null hypothesis was rejected if the significance level was below 5%.
2.5. qRT-PCR and Western blot analyses
For qRT-PCR, equal amounts of RNA extracted from each sample was reverse transcribed and pre-amplified in a Specific Target Amplification solution (COVID19 N1, N2, and Human RNase P targets) for CoVid19 detection. The reverse transcribed and pre-amplified cDNA was diluted and then detected by qPCR on the Fluidigm microfluidics platform using the CDC developed assays for CoVid19 N1, N2, and Human RNase P.
Western blot analyses were done using equal amounts of protein extracted from the formalin fixed paraffin embedded tissues using the Qiagen Qproteome FFPE tissue kit (cat # 37623, Germantown, MD). Proteins were separated on Mini-PROTEAN TGX premade 4:20% acrylamide gradients gels according to the manufacturer's recommendations (BioRad, Hercules CA).
2.6. Mouse IV injection studies
C57BL/6 female mice (9 months old) purchased from Jackson laboratory, Maine, were tail vein injected with different spike peptides, as described [11]. Specifically, 10 μg/mouse of Spike S1 (10–300; ProSci) and 3 μg/mouse of Spike S2 (10−303), which served as the negative control were tail vein injected. All procedures were approved by and performed in accordance with The Ohio State University's Institutional Lab Animal Care and Use Committee (Columbus, Ohio).
3. Results
3.1. Clinical/pathologic correlation
Nine autopsies from people who died of COVID-19 were studied: age range 50–72 (mean 64 vs mean 61 for controls) with 5 men/4 women. Pre-existing liver disease was evident in five cases: two NASH and three AUD; another person had several metastatic nodules in the liver from a neuroendocrine tumor of the pancreas. The cause of death in each case was viral induced bronchopneumonia and in six cases disseminated intravascular coagulation (DIC)/multiorgan failure was documented. In one AUD case, hepatic failure was listed as a cause of death.
The hematoxylin and eosin stains were reviewed blinded to the clinical information. Three cases showed centrolobular inflammation with admixed neutrophils/mononuclear cells and Mallory bodies consistent with AUD and one of these also showed cirrhosis. The AUD cases also showed variable macrosteatosis in zones 2–3 that was associated with many large, stellate cells which at times contained fat droplets; i.e., hepatic stellate cells (Fig. 1A, B).
Fig. 1.
Histologic findings and detection of viral RNA/proteins in the liver in fatal COVID-19. Panels A/B show at low and high magnification, respectively, features typical of AUD in the liver/COVID-19 with a hematoxylin and eosin stain. Note the macrosteatosis, abundant Mallory bodies (rectangle) and hepatic stellate cells (oval). In comparison, panel C shows the histologic findings in a NASH/COVID-19 liver where steatosis without Mallory bodies is evident. Panel D shows the histology of another COVID-19 liver without NASH/AUD where lobular inflammation is evident (rectangle). Panel E shows the high copy number SARS-CoV2 RNA in the lung (brown signal); note that the liver from the same person showed only 1 viral RNA positive cell was seen in the entire section (arrow, panel F) (panel G, a viral negative COVID-19/non AUD liver). Panel H shows a Western blot of the lung protein extracts in which the COVID-19 cases show one to several bands when analyzed for the spike S1 and S2 subunit. Panel I: Only the COVID-19 lung samples, and not the liver or heart, showed a band of the expected size (50 kDa) when analyzed for the nucleocapsid protein. Panel J: Most of the COVID-19 liver samples showed a band when analyzed for the viral spike protein; samples 1 and 2 showed the strongest bands and were each AUD/COVID-19 cases. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
The most common histologic change in the non-AUD cases was lobular inflammation; portal inflammation was also evident. Lobular inflammation, as defined by variable sized groups of lymphocytes and macrophages in the sinusoidal areas, ranged from 0 to 4.5/10 hepatic lobules (mean 2,1) in the non AUD COVID-19 cases (Fig. 1C, D) and was not evident in the controls.
3.2. qRT-PCR/in situ hybridization and Western blot analyses
The distribution of SARS-CoV2 RNA and protein were next assessed in the liver samples. The lung tissues from five of the cases were used as the positive controls as each were assessed to have high viral copy load by qRT-PCR. Each of the lung tissues showed high copy RNA as well by in situ hybridization (Fig. 1E) with a mean of >25 positive cells/200× field; no signal was seen in the pre-COVID19 lung tissues. Based on these controls, all 18 liver samples were tested by in situ hybridization and scored in a blinded fashion. None of the controls were positive (data not shown, but see Fig. 1G for equivalent result) and viral RNA was only seen in 2/9 COVID-19 cases. Importantly, the two liver cases positive for viral RNA had only rare (1–3/cm of tissue) positive cells that would not explain the widespread histologic changes. The rare presence of infectious virus in the liver was corroborated by qRT-PCR as cases were negative or showed high Ct values (data not shown).
Next, the total proteins were extracted from the formalin fixed, paraffin embedded tissues and subjected to Western blot analyses. Two normal skin biopsies were divided into half and either fixed in 10% buffered formalin overnight or frozen after which the extracted proteins were compared by Western blot for type I and III collagen. Bands of the expected size were seen after testing equal amounts of protein but the signal intensity was reduced about 50% in the formalin fixed, paraffin embedded tissues samples (data not shown).
Proteins extracted from COVID-19 formalin fixed, paraffin embedded tissues were then tested for viral spike (subunits 1 and 2), matrix, nucleocapsid, and envelope proteins. The five COVID-19 lung samples positive by in situ hybridization for SARS-CoV2 were the positive controls and each showed discrete single bands for the matrix, envelope, and nucleocapsid proteins on Western blot, with no band in the controls (Fig. 1I). However, the lung tissues that were positive for the viral spike protein typically showed multiple bands that ranged from the expected sizes of 180 (full length) and 90 KDa (cleaved upon viral entry), to other bands as small as 16 kDa (Fig. 1H & J). Also note that only the lung samples, and not the liver COVID-19 protein extracts, had detectable nucleocapsid protein (Fig. 1I). Fig. 1J demonstrates some of the 6/9 liver extracts that contained detectable viral spike and matrix proteins (data not shown) and that the most intense bands were evident for the AUD/COVID-19 liver samples (Fig. 1J, first two lanes).
3.3. Immunohistochemistry for viral proteins
The data above suggested that the infectious virus (viral RNA and associated nucleocapsid protein), though routinely detected in the lung samples, was rarely present in the matched liver tissues. However, viral spike and matrix proteins were detected in the majority of liver samples by Western blot, with data suggesting that they may be localizing in the liver, especially the AUD livers. To address this and determine if the viral capsid proteins were in the blood or the liver parenchyma per se, each liver sample was tested for the viral spike, nucleocapsid, matrix, and envelope proteins by immunohistochemistry and scored blinded to the clinical information.
The viral spike, matrix, and envelope proteins were detected in 9/9 of the COVID-19 livers, 5/5 of the corresponding lung tissues, and none of the controls. Representative data for the viral spike, nucleocapsid, matrix and envelope proteins is provided are Fig. 2 . Note the absence of the viral nucleocapsid protein (Fig. 2E) which was detected in only 2/9 of the liver samples, and, in the positive cases, only 1 and 4 positive cells were evident/cm. Also note that the viral spike, envelope, and matrix proteins showed equivalent distributions in serial sections and, thus, showed strong co-localization to each other (data not shown). Quantification of the number of cells positive for viral spike protein/100× field yielded interesting results. The 3 AUD/COVID-19 liver cases had an average of 5.9 cells positive for spike protein/100× field (SEM = 1.1) as compared to 0.4 positive cells (SEM 0.1) for the two NASH/COVID-19 cases, and 1.9 positive cells (SEM = 0.4) for the COVID-19 liver cases without AUD/NASH. This represents a significant increase in the density of viral spike protein in the AUD/COVID-19 livers versus both the NASH and non-NASH livers (each p < 0.001). The immunohistochemistry data for the viral spike protein in the COVID-19 liver samples correlated well with the Western blot data as the three cases negative by Western blot (Fig. 1J, lanes 3, 4 and 6) showed the lowest density of viral spike protein by immunohistochemistry whereas the three AUD cases (Fig. 1J shows 2 cases, lanes 1, 2) showed the strongest Western blot bands.
Fig. 2.
In situ detection of viral proteins in the liver in fatal COVID-19.
Note the strong signal for the viral spike protein (red color) in a COVID-19 lung (panel A) and the negative result in a pre-COVID-19 liver (panel B). Panels C-E came from the same patient; note the strong signal for nucleocapsid protein (red color) in the lung (panel C), the strong signal for spike (red color) in the AUD liver (panel D) and the lack of signal in the same liver for the nucleocapsid protein (panel E). Panels F/G show at higher magnification that the same liver is also positive for the viral matrix and envelope proteins (brown color). The NASH COVID-19 livers (panel H) showed much less of the spike protein (red color) than the AUD livers (panel D). Panels I and J are serial sections of the same COVID-19 liver without AUD or NASH; note the equivalent distribution of the viral spike (panel I, brown) and, at higher magnification, the envelope (panel J) protein. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
With regards to the cellular targets of the viral capsid proteins in the liver, the positive cells were not hepatocytes but, rather, had the cytology of macrophages (ample cytoplasm with branching forms) and, in the AUD livers, hepatic stellate cells (Fig. 2D, F-J). Given that ACE2 is the main receptor for the SARS-CoV2 spike protein, ACE2 immunohistochemistry and co-localization experiments were done next.
3.4. Immunohistochemistry for ACE2 and hepatic stellate cells
The ACE2 protein localized to three cell types based on co-expression data (Fig. 3B, C): Kupffer cells, endothelial cells in the sinusoids and, in the AUD cases, the hepatic stellate cells. The co-expression data for CD68 (Fig. 3C), other macrophage markers (CD11b, CD163, CD206) and CD31 (endothelial cells, data not shown) in the non-AUD/COVID-19 cases showed that about 90% of the cells with ACE2 in the liver were the former. However, by far the greatest density of ACE2-positive cells in the COVID-19 livers were seen in the three AUD/COVID-19 cases (Fig. 3B vs Fig. 3E). The 3 AUD/COVID-19 liver cases had an average of 27.9 cells positive for ACE2 protein/100× field (SEM = 3.1) as compared to 2.8 positive cells (SEM = 0.8) for the COVID-19 liver cases without AUD (p < 0.001) and 2.2 positive cells (SEM = 0.9) for the controls; there was no significant difference between the two latter groups. Fig. 3F further documents the expression of smooth muscle actin, typical of hepatic stellate cells, as further evidenced by their star-like cytology. Also note the strong co-localization of SARS-CoV2 spike protein in these ACE2 positive cells (Fig. 3D). The majority of the hepatic stellate cells also expressed type I collagen and S100 which are other indicators of an activated state (data not shown). In the non-AUD livers, including the NASH cases (both COVID-19 and controls), the hepatic stellate cells showed none to rare type I collagen, smooth muscle actin, or S100 expression (data not shown), consistent with a non-activated state.
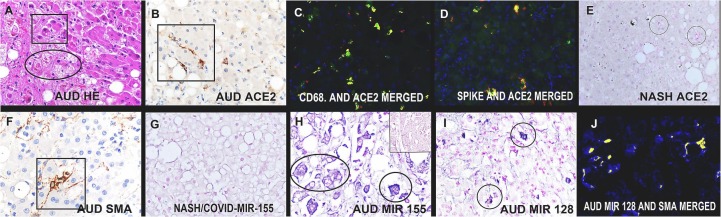
Fig. 3.
Histologic and molecular findings in the hepatic stellate cells in fatal AUD/COVID-19.
Panel A shows a high magnification of the AUD/COVID-19 liver (H&E staining); note the many Mallory bodies (rectangle) and hepatic stellate cells (oval). The same liver showed many ACE2 positive cells with a stellate/branching morphology (panel B, brown color). Co-expression analyses showed co-localization (seen as fluorescent yellow) of CD68 and ACE2 (panel C) as well as ACE2 and the viral spike protein (panel D). Far fewer cells expressed ACE2 (red color) in the NASH liver (panel E, circles). The hepatic stellate cells expressed SMA (F, brown color), as well as miR-155 (blue color, panel H with the insert showing negative hepatocytes from the same case) and miR-128 (blue color, panel I). None of the control livers of COVID-19 non-AUD cases showed either miR-128 or miR-155 expression (panel G, blue color). Panel J shows the co-expression (seen as fluorescent yellow) of miR-128 with SMA. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.5. In situ detection of miR-128 and miR-155 in COVID-19 livers
Our preliminary data showed that global knockout of miR-128 in a mouse model of COVID-19 [11] results in less systemic disease after IV injection of the spike protein whereas overexpression of miR-155 results in worse disease (Tili E and Nuovo GJ, unpublished data). In addition, miR-155 plays a critical role in the inflammatory response and cytokine production [17], [18], regulates the activation of hepatic stellate cells [19], [20], and has been implicated in AUD [21], [22]. Also, miR-128 has been associated with liver fibrosis via activation of hepatic stellate cells [23]. Thus, the liver samples were tested for miR-128 and miR-155 by in situ hybridization in a blinded fashion. Neither miRNA was detected in situ in any of the liver controls, including NASH, and only rare positive cells (0.5–0.7/100× field) for each miRNA were seen in the liver tissues from the COVID-19 cases without AUD (Fig. 3G). However, miR-155 and miR-128 each localized to the majority of the hepatic stellate cells in AUD/COVID-19 livers as documented by co-expression with SMA (Fig. 3H-J); this translated to 21.2 positive cells/100× field (miRNA -128) and 23,8 positive cells/100× field for miR-155, each significantly greater than controls or the non-AUD COVID-19 livers (p < 0.001).
3.6. Immunohistochemistry for the host response to the viral spike protein
Serial sections of the liver tissues tested for the viral capsid proteins were examined for complement activation (C5b-9 and complement component = C6), IL6, TNFα, and activated caspase 3. Caspase 3, C 6, C5b-9, IL6 and TNFα each showed a distribution pattern equivalent to that for the viral membrane/spike proteins (Fig. 4A-F). Also note that co-expression experiments corroborated that the same cells that had the viral spike protein also contained the pro-inflammatory cytokines (Fig. 4F-H). Thus, the expression of these cytokines and activated Caspase 3 was significantly higher in the AUD/COVID-19 livers compared to the COVID-19 livers that were not AUD associated. The 3 AUD/COVID-19 liver cases had an average of 4.1–6.9 cells positive for C6, Caspase 3, IL6, and TNFα/100× field (SEM = 0.9–1.3) as compared to 0.9–1.7 positive cells (SEM = 0.4–0.5) for the COVID-19 liver cases without AUD (Fig. 4A vs B for Caspase 3).
Fig. 4.
Host response in the liver in fatal COVID-19. Note the strong expression of activated Caspase 3 in the AUD/COVID-19 liver (panel A, brown color) compared to the NASH/COVID-19 liver (panel B, red color). Panels C and D show strong complement activation in the AUD/COVID-19 liver as evidenced as Complement component 6 and C5b-9 (brown colors), respectively. Also note that IL6 (panel E, red) and TNFα (panels F-H) are also strongly expressed in these tissues in the large, stellate cells; the TNFα (green) co-localizes (yellow) with the viral spike protein (red). Panels I and J show that the liver in mice injected with the S1 subunit of the spike protein demonstrate lobular inflammation which contains both the spike subunit (brown color) and TNFα (brown color), respectively, in the same distribution. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.7. Mouse liver data after IV injection of spike S1 subunit
We have previously shown that injection of spike S1 subunit in the mouse tail vein induces neurologic pathology as seen in fatal COVID-19 patients [11]. Thus, the livers in these mice were examined for evidence of spike induced pathologic changes. In agreement with the human data, scattered foci of lobular inflammation were seen in the livers of the mice injected with the S1 subunit of the viral spike protein and both S1 (Fig. 4I) as well as TNFα (Fig. 4J) and IL6 (data not shown) were documented in these foci (Fig. 4I, J). In contrast, injecting spike S2 subunit had no detectable effect on the mouse liver (data not shown).
4. Discussion
Abnormal liver function tests are common in moderate/severe COVID-19 and often are evidenced as liver failure in the setting of pre-existing AUD. The main issue addressed by this study was the mechanism(s) whereby there is liver damage in severe COVID-19. Two tenable hypotheses are: 1) hepatic dysfunction is an indirect effect originating outside of the liver reflecting the high levels of circulating cytokines (cytokine storm) and coagulopathy; and 2) the virus per se is able to directly damage the liver. The data in this manuscript documents that the latter mechanism is indeed operative. Interestingly, the infectious virus was not responsible for the liver damage since SARS-CoV2 RNA and the associated nucleocapsid protein were either rarely evident or not detectable. This is consistent with other studies that have indicated that infectious virus is rarely/not evident in the liver in fatal COVID-19 [6], [8], [9]. Rather, it was the SARS-CoV2 spike protein that was commonly detected in the liver. The spike protein was presumably in the circulation with the two associated viral proteins envelope and matrix (which comprise the viral membrane) since each strongly co-localized with spike. It is reasonable to assume that the source of these circulating viral proteins was the very high copy infectious virus in the lung where the microangiopathy led to viral degeneration and entry of the spike protein in the circulation [10]. Preliminary data suggests that these proteins are not free floating in the circulation, but rather carried by macrophages (Nuovo GJ and Tili, E, unpublished data). The finding of viral membrane proteins including spike without the infectious virus in the liver was unexpected and, thus, three different methods (in situ hybridization, immunohistochemistry with co-localization and Western blot analyses) were done which gave equivalent data. Although the specific mechanism of spike presentation in the liver needs to be elucidated, once endocytosed by ACE2 positive sinusoidal macrophages, including hepatic stellate cells in the AUD cases, the spike protein induced a pro-inflammatory and hypercoagulable state that would cause hepatocyte dysfunction. Importantly, spike protein alone has been shown to be cytotoxic by multiple groups including in the liver [11], [24], [25]. This is not to say that circulating cytokines do not also play a role in hepatic dysfunction, but rather that in situ production of these cytokines clearly is a feature of hepatic dysfunction in severe COVID-19.
The other primary finding of this study related to the clinical observation that pre-existing AUD can potentiate the liver damage in severe COVID-19. It was observed that pre-existing AUD was associated with a 10-fold increase in the numbers of ACE2-positive cells in the liver, mainly in the form of activated hepatic stellate cells. This was, in turn, associated with a similar increase in the density of cells in the liver that had endocytosed spike protein and showed a concomitant in situ expression of IL6 and TNFα. Hepatic stellate cells are well documented to have innate immune functions including macrophage-type endocytosis of viruses. Further, they also function as antigen presenting cells and their activation, marked by SMA, type I collagen, and S100 expression, plays a key role in the inflammation and fibrosis typical of progressive liver disease [26], [27], [28]. It can thus be postulated that other pre-existing liver conditions in which there are many activated hepatic stellate cells should also be predisposed to more severe disease in moderate/severe COVID-19, which will await further study.
miRNAs, in particular miR-128 and miR-155, are well documented to play roles in inflammation in general and hepatic stellate cell activation in particular [17], [18], [19], [20], [21], [22], [23]. It was noted in this study that neither of these microRNAs were evident using in situ hybridization in control, unremarkable pre-COVID-19 livers, or in pre-COVID NASH livers, and were rare in COVID-19 livers which did not show AUD but rather, at most, had low NASH scores with minimal inflammation. Pre-COVID-19 AUD cases were not available for study. Still, it was evident that in the three AUD/COVID-19 cases there was dramatic expression of miR-128 and miR-155 where each localized mostly to the hepatic stellate cells. miR-155 is a well-known proinflammatory microRNA needed for the mounting of the immune responses [17], [18]. Thus, miR-155 inactivation attenuates liver injury after ischemia/reperfusion by decreasing the expression of pro-inflammatory makers such as CD80, CD86, and major histocompatibility complex class II expression in Kupffer cells, resulting in suppression of proinflammatory cytokine secretion and increased secretion of anti-inflammatory IL-10 [29]. The role of miR-155 in liver disease, however, is complex as it has been reported to play a protective role in the development of non-alcoholic hepatosteatosis in mice [30], and to behave as an anti-fibrotic agent by attenuating the activation of hepatic stellate cell by simultaneously preventing epithelial-mesenchymal transition and ERK1 signaling pathway [19]. In addition, miR-155 activity is protective by decreasing T-lymphocyte-mediated liver injury and promoting an alteration of inflammatory cell recruitment [31]. Thus, miR-155 activity might well prove protective in the context of AUD however deleterious in COVID-19, depending on the context and its level of expression. Indeed, it has been shown that miR-155 target different sets of transcripts depending on its level of expression [32]. Thus, miR-155 enhanced expression in hepatic stellate cells in AUD/COVID-19 livers might well result from both pro-inflammatory effects due to COVID-19 infection and protective activity in the context of AUD.
In sum, the data suggests that in situ production of pro-inflammatory cytokines with complement activation in the liver induced by endocytosis of circulating spike protein primarily in ACE2-positive/CD68-positive cells (macrophages and hepatic stellate cells) is an important mechanism of hepatic disease complication in severe COVID-19. Thus, it is possible that anti-spike antibodies such as REGEN-COV may be able to ameliorate the liver disease, especially in people at high risk for such, including those with AUD. Since the levels of circulating spike protein in fatal COVID-19 are many folds higher than post-vaccination, it is not surprising that there is no liver dysfunction post-vaccination. The work also underscores the key role that the diagnostic anatomic pathologist can play in dissecting the mechanism of COVID-19 disease by combining histologic analyses with basic immunohistochemistry testing.
Acknowledgments
The authors greatly appreciate the help of Dr. Margaret Nuovo with the photomicroscopy, Dr. Saul Suster who provided many of the samples, Dr. Cynthia Magro who provided samples and much advice/comments and Ms. Eva Matys who performed most of the Western blots.
Footnotes
The authors have no conflict of interests to report. Financial support: OSU Dept of Anesthesiology bridge fund (ET).
References
- 1.Zhang Y., Geng X., Tan Y., Li Q., Xu C., Xu J., et al. New understanding of the damage of SARS-CoV-2 infection outside the respiratory system. Biomed Pharmacother. 2020;127 doi: 10.1016/j.biopha.2020.110195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li H., Liu L., Zhang D., Xu J., Dai H., Tang N., et al. SARS-CoV-2 and viral sepsis: observations and hypotheses. Lancet. 2020;395:1517–1520. doi: 10.1016/S0140-6736(20)30920-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou L., Zhang M., Wang J., Gao J. Sars-Cov-2: underestimated damage to nervous system. Travel Med Infect Dis. 2020;36 doi: 10.1016/j.tmaid.2020.101642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Varga Z., Flammer A.J., Steiger P., Haberecker M., Andermatt R., Zinkernagel A.S., et al. Endothelial cell infection and endotheliitis in COVID-19. Lancet. 2020;395:1417–1418. doi: 10.1016/S0140-6736(20)30937-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martinez M.A., Franco S. Impact of COVID-19 in liver disease progression. Hepatol Commun. 2021;5:1138–1150. doi: 10.1002/hep4.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chornenkyy Y., Mejia-Bautista M., Brucal M., Blanke T., Dittmann D., Yeldandi A., et al. Liver pathology and SARS-CoV-2 detection in formalin-fixed tissue of patients with COVID-19. Am J Clin Pathol. 2021;155:802–814. doi: 10.1093/ajcp/aqab009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lagana S.M., Kudose S., Iuga A.C., et al. Hepatic pathology in patients dying of COVID-19: a series of 40 cases including clinical, histologic, and virologic data. Mod Pathol. 2020;33(11):2147–2155. doi: 10.1038/s41379-020-00649-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fassan M., Mescoli C., Sbaraglia M., Guzzardo V., Russo F.P., Fabris R., Trevenzoli M., Pelizzaro F., Cattelan A.M., Basso C., Navalesi P., Farinati F., Vettor R., Dei Tos A.P. Liver histopathology in COVID-19 patients: a mono-institutional series of liver biopsies and autopsy specimens. Pathol Res Pract. 2021 May;221 doi: 10.1016/j.prp.2021.153451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delorey T.M., Ziegler C.G.K., Heimberg G., Normand R., Yang Y., Segerstolpe Å., et al. COVID-19 tissue atlases reveal SARS-CoV-2 pathology and cellular targets. Nature. 2021;595:107–113. doi: 10.1038/s41586-021-03570-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Magro C., Mulvey J.J., Berlin D., Nuovo G., Salvatore S., Harp J., Baxter-Stoltzfus A., Laurence J. Complement associated microvascular injury and thrombosis in the pathogenesis of severe COVID-19 infection: a report of five cases. Transl Res. 2020–06;220:1–13. doi: 10.1016/j.trsl.2020.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nuovo G.J., Magro C., Shaffer T., Awad H., Suster D., Mikhail S., et al. Endothelial cell damage is the central part of COVID-19 and a mouse model induced by injection of the S1 subunit of the spike protein. Ann Diagn Pathol. 2021;51:12–18. doi: 10.1016/j.anndiagpath.2020.151682. 151682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Magro C.M., Mulvey J.J., Laurence J., Seshan S., Crowson A.N., Dannenberg A.J., Salvatore S., Harp J., Nuovo G.J. Docked severe acute respiratory syndrome coronavirus 2 proteins within the cutaneous and subcutaneous microvasculature and their role in the pathogenesis of severe coronavirus disease 2019. Hum Pathol. 2020;12(106):106–116. doi: 10.1016/j.humpath.2020.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cargill Z., Kattiparambil S., Hansi N., Barnsabas A., Shawcross D.L., Williams R., et al. Severe alcohol-related liver disease admissions post-COVID-19 lockdown: canary in the coal mine? Frontline Gastroenterol. 2020;12:354–355. doi: 10.1136/flgastro-2020-101693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moon A.M., Curtis B., Mandrekar P., Singal A.K., Verna E.C., Fix O.K. Alcohol-associated liver disease before and after COVID-19-an overview and call for ongoing investigation. Hepatol Commun. 2021;5:1616–1621. doi: 10.1002/hep4.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cholankeril G., Goli K., Rana A., Hernaez R., Podboy A., Jalal P., et al. Impact of COVID-19 pandemic on liver transplantation and alcohol-associated liver disease in the USA. Hepatology. 2021;10:1002. doi: 10.1002/hep.32067. Apr 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nuovo G.J., Nana-Sinkam P., Elton T., Croce C., Volinia S., Schmittgen T. A methodology for the combined in situ analyses of the precursor and mature forms of microRNAs and correlation with their putative targets. Nat Protoc. 2009:107–115. doi: 10.1038/nprot.2008.215. –4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tili E., Michaille J.J., Cimino A., Costinean S., Dumitru C.D., Adair B., Fabbri M., Alder H., Liu C.G., Calin G.A., Croce C.M. Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNF-alpha stimulation and their possible roles in regulating the response to endotoxin shock. J Immunol. 2007 Oct 15;179(8):5082–5089. doi: 10.4049/jimmunol.179.8.5082. [DOI] [PubMed] [Google Scholar]
- 18.Tili E., Michaille J.J., Croce C.M. MicroRNAs play a central role in molecular dysfunctions linking inflammation with cancer. Immunol Rev. 2013 May;253(1):167–184. doi: 10.1111/imr.12050. [DOI] [PubMed] [Google Scholar]
- 19.Dai W., Zhao J., Tang N., Zeng X., Wu K., Ye C., Shi J., Lu C., Ning B., Zhang J., Lin Y. MicroRNA-155 attenuates activation of hepatic stellate cell by simultaneously preventing EMT process and ERK1 signaling pathway. Liver Int. 2015 Apr;35(4):1234–1243. doi: 10.1111/liv.12660. [DOI] [PubMed] [Google Scholar]
- 20.Ezhilarasan D. MicroRNA interplay between hepatic stellate cell quiescence and activation. Eur J Pharmacol. 2020 Oct;15(885) doi: 10.1016/j.ejphar.2020.173507. [DOI] [PubMed] [Google Scholar]
- 21.Torres J.L., Novo-Veleiro I., Manzanedo L., Alvela-Suárez L., Macías R., Laso F.J., Marcos M. Role of microRNAs in alcohol-induced liver disorders and non-alcoholic fatty liver disease. World J Gastroenterol. 2018 Sep 28;24(36):4104–4118. doi: 10.3748/wjg.v24.i36.4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu L., Ren T., Zhu Z., Cheng M., Mou Q., Mu M., Liu Y., Yao Y., Cheng Y., Zhang B., Cheng Z. Thymosin-β4 mediates hepatic stellate cell activation by interfering with CircRNA-0067835/miR-155/FoxO3 signaling pathway. Cell Physiol Biochem. 2018;51(3):1389–1398. doi: 10.1159/000495556. [DOI] [PubMed] [Google Scholar]
- 23.Povero D., Panera N., Eguchi A., Johnson C.D., Papouchado B.G., de Araujo Horcel L., Pinatel E.M., Alisi A., Nobili V., Feldstein A.E. Lipid-induced hepatocyte-derived extracellular vesicles regulate hepatic stellate cell via microRNAs targeting PPAR-γ. Cell Mol Gastroenterol Hepatol. 2015 Nov 1;1(6):646–663. doi: 10.1016/j.jcmgh.2015.07.007. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rhea E.M., Logsdon A.F., Hansen K.M., et al. The S1 protein of SARS-CoV-2 crosses the blood–brain barrier in mice. Nat Neurosci. 2021;24:368–378. doi: 10.1038/s41593-020-00771-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li C., Chen Y., Zhao Y., Lung D.C., Ye Z., Song W., Liu F.F., Cai J.P., Wong W.M., Yip C.C., Chan J.F., To KK. Sridhar S., Hung I.F., Chu H., Kok K.H., Jin D.Y., Zhang A.J., Yuen K.Y. Intravenous injection of COVID-19 mRNA vaccine can induce acute myopericarditis in mouse model. Clin Infect Dis. 2021 doi: 10.1093/cid/ciab707. Aug 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Winau F., Hegasy G., Weiskirchen R., Weber S., Cassan C., Sieling P.A., et al. Ito cells are liver-resident antigen-presenting cells for activating T cell responses. Immunity. 2007;26(1):117–129. doi: 10.1016/j.immuni.2006.11.011. [DOI] [PubMed] [Google Scholar]
- 27.Yang A., Yan X., Fan X., Shi Y., Huang T., Li W., et al. Hepatic stellate cells-specific LOXL1 deficiency abrogates hepatic inflammation, fibrosis, and corrects lipid metabolic abnormalities in non-obese NASH mice. Hepatol Int. 2021 May;15(5):1122–1135. doi: 10.1007/s12072-021-10210-w. [DOI] [PubMed] [Google Scholar]
- 28.Viñas O., Bataller R., Sancho-Bru P., Ginès P., Berenguer C., Enrich C., et al. Human hepatic stellate cells show features of antigen-presenting cells and stimulate lymphocyte proliferation. Hepatology. 2003;38:919–929. doi: 10.1053/jhep.2003.50392. [DOI] [PubMed] [Google Scholar]
- 29.Li Y., Ma D., Wang Z., Yang J. MicroRNA-155 deficiency in kupffer cells ameliorates liver ischemia-reperfusion injury in mice. Transplantation. 2017 Jul;101(7):1600–1608. doi: 10.1097/TP.0000000000001765. [DOI] [PubMed] [Google Scholar]
- 30.Miller A.M., Gilchrist D.S., Nijjar J., Araldi E., Ramirez C.M., Lavery C.A., Fernández-Hernando C., McInnes I.B., Kurowska-Stolarska M. MiR-155 has a protective role in the development of non-alcoholic hepatosteatosis in mice. PLoS One. 2013 Aug 21;8(8) doi: 10.1371/journal.pone.0072324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blaya D., Aguilar-Bravo B., Hao F., Casacuberta-Serra S., Coll M., Perea L., Vallverdú J., Graupera I., Pose E., Llovet L., Barquinero J., Cubero F.J., Caballería J., Ginès P., Sancho-Bru P. Expression of microRNA-155 in inflammatory cells modulates liver injury. Hepatology. 2018 Aug;68(2):691–706. doi: 10.1002/hep.29833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Michaille J.J., Awad H., Fortman E.C., Efanov A.A., Tili E. miR-155 expression in antitumor immunity: the higher the better? Genes Chromosomes Cancer. 2019 Apr;58(4):208–218. doi: 10.1002/gcc.22698. [DOI] [PubMed] [Google Scholar]