Abstract
Purpose of review
Transcription of erythroid-specific genes is regulated by the three-dimensional (3D) structure and composition of chromatin, which dynamically changes during erythroid differentiation. Chromatin organization and dynamics are regulated by several epigenetic mechanisms involving DNA (de-)methylation, post-translational modifications (PTMs) of histones, chromatin-associated structural proteins, and higher-order structural changes and interactions. This review addresses examples of recent developments in several areas delineating the interface of chromatin regulation and erythroid-specific lineage transcription.
Recent findings
We survey and discuss recent studies that focus on the erythroid chromatin landscape, erythroid enhancer-promotor interactions, super-enhancer functionality, the role of chromatin modifiers and epigenetic crosstalk, as well as the progress in mapping red blood cell (RBC) trait-associated genetic variants within cis-regulatory elements (CREs) identified in genome-wide association study (GWAS) efforts as a step towards determining their impact on erythroid-specific gene expression.
Summary
As one of the best characterized and accessible cell differentiation systems, erythropoiesis has been at the forefront of studies aiming to conceptualize how chromatin dynamics regulates transcription. New emerging technologies that bring a significantly enhanced spatial and temporal resolution of chromatin structure, and allow investigation of small cell numbers, have advanced our understanding of chromatin dynamics during erythroid differentiation in vivo.
Keywords: erythropoiesis, epigenetics, enhancer, chromosome conformation capture, genome-wide association study, chromatin modifiers
Introduction
Tissue differentiation is driven by the expression of lineage-specific genes expression of which is critically dependent on the structure and spatial organization of the genome [1,2]. For example, for transcription to be initiated, enhancers as CREs must be recognized within chromatin and engaged, and the chromatin opened into a nucleosome-free region, which is facilitated by the formation of enhancer-promoter loops. Indeed, a recent comprehensive study of 220 transcription factor (TF) DNA binding domains (eDBDs) and 13 full-length TFs revealed that most preferentially bound to naked DNA, and their binding was inhibited by the presence of a nucleosome [3].
During terminal differentiation of the erythroid lineage from the proerythroblast stage onwards, each cell division produces daughter cells of a distinct differentiation stage, with discrete morphology and gene expression profile [4–7], which demands tightly controlled gene regulation. While several essential TFs driving erythropoiesis have been known since long [8], the elucidation of chromatin determinants has lagged behind. This review gives an update of recently emerging principles of erythroid-specific epigenetic mechanisms. Due to space limitations, the examples discussed are not meant to be all-encompassing, and we apologize to all colleagues whose work we could not mention.
The erythroid chromatin landscape
Integrating genome-wide expression with epigenetic dynamics across erythropoiesis promotes the understanding of gene regulatory principles and serves as a framework that genetic variation and disease can be related to. A number of combined epigenomic-transcriptomic time courses of erythroid differentiation have been undertaken in both mice and humans [9–18]. Two comprehensive studies of human erythropoiesis examined transcriptome dynamics in conjunction with chromatin accessibility (employing the ‘assay for transposase-accessible chromatin using sequencing’ or ATAC-seq, [10,16]) and DNA methylome [16] analysis by differentiating and sorting primary umbilical cord-derived [16] or adult CD34+ [10] hematopoietic stem and progenitor cells (HSPCs) at eight erythropoiesis stages. Despite some differences in erythroid differentiation trajectories and transcriptional profiles between neonatal and adult peripheral-blood derived HSPCs [19], a theme emerging from these studies, as well as another study charting chromatin conformation dynamics during murine erythropoiesis at high temporal resolution, is that DNA demethylation and chromatin opening precedes gene transcription [10,15,16]. Future studies could further improve this resolution by leveraging a newly developed technology that simultaneously profile chromatin accessibility and gene expression in the same cell (SHARE-seq) as shown for another lineage [20]. In addition, the erythroid time course studies show that both chromatin accessibility and transcriptional activity continue to gradually increase over several differentiation stages before reaching a plateau. Of note, while some of these studies showed that the GATA2-to-GATA1 switch in human primary CD34+ cells undergoing in-vitro differentiation towards the erythroid lineage occurs at the proerythroblast stage [5,11,12], a study of the dynamic chromatin landscape during murine erythropoiesis suggests that, in vivo, the GATA2-to-GATA1 switch occurs already in a subpopulation of earlier CMPs [9]. It will be interesting to compare these findings with epigenomic-transcriptomic time courses of human erythroid differentiation of primary human cells directly sorted from healthy bone marrow.
Importantly, combined erythroid epigenomic-transcriptomic analysis enables the identification of new essential and stage-specific regulators of erythroid differentiation. For example, ATAC-seq analysis allowed the identification of an erythroid-specific enhancer that becomes accessible specifically in the PolyE and OrthoE stages, and drives expression of a shorter, erythroid-specific isoform of TMCC2 [10], an ApoE-binding protein [21]. Many datasets exploring erythroid and other hematopoietic cells have been uniformly processed and made available to the general public in the framework of the VISION project [13], which comprises over 200,000 candidate CREs, including almost all experimentally verified erythroid CREs. This and other databases (for example, the ENCODE project) are an extremely valuable resource for the discovery of new CREs, as exemplified by a newly found functional enhancer upstream of the Gγ-globin gene [22].
Enhancer-promoter contacts
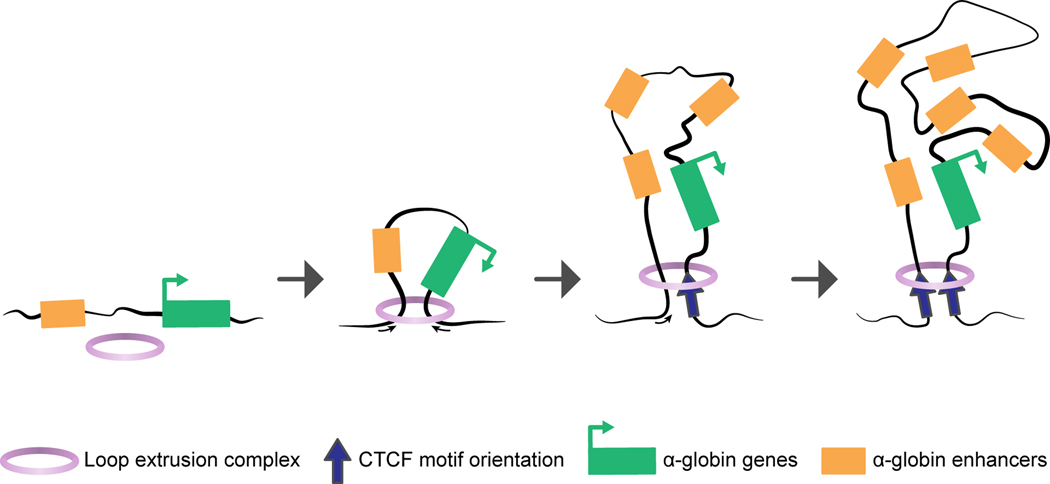
Over the last decade, chromosome conformation capture (3C)-based methods have revealed that chromosomes are organized into topologically associating domains (TADs) that self-associate and whose boundaries are demarcated by the CCCTC-binding zinc finger protein CTCF and the cohesin complex, which insulate them from chromatin of neighboring TADs. Within TADs or smaller sub-TADs, chromatin loops form – possibly by extrusion (Fig. 1) [23,24] – that bring enhancers in proximity of gene promoters [25]. Important outstanding questions are how the 3D genome architecture influences transcription during differentiation and how specific enhancer-promoter interactions are achieved in gene-dense regions. The investigation of the α- and β-globin clusters continues to be instrumental in elucidating this regulation. Recently, the dynamics of chromatin organization at the α-globin cluster has been examined during early erythropoiesis in vivo with the help of low-input 3C technology (Tiled-C, [15]). This study found that while the ~165-kb large TAD encompassing the α-globin cluster is already present in HSPCs, a smaller ~70-kb erythroid sub-TAD contained within it, harboring the enhancer-promoter interactions, only gradually appeared after the α-globin enhancers became accessible. Further chromatin accessibility and sub-TAD formation was progressive and took place in parallel with gradual α-globin gene activation. Similar observations were made at other erythroid gene loci and also in an in-vitro study using the murine G1ER erythroid cell differentiation model [14]. Methodological innovations now allow the assessment of multi-way (so far, primarily three-way) rather than just two-way genome contacts [26–28]. Their use led to the emergence of a hub model (Fig. 1) for both the α- and β-globin loci in which several promoters interact with the enhancers in a common compartment, as predicted also in models of transcription factories [29].
Figure 1: Chromatin loop extrusion model for the α-globin locus-containing chromatin domain.
In this model of α-globin locus-containing chromatin domain formation a cohesin-containing extrusion complex is loaded onto chromatin, through which the chromatin fiber is extruded until convergently orientated CTCF motifs are encountered. Model based on [23,24,27,31].
In mouse embryonic stem cells (mESCs), conditional degradation of CTCF causes insulation defects at >80% of TAD boundaries, as well as loss of chromatin loops [30]. Similarly, polymer physics models based on high-resolution Capture-C data of the α-globin locus in mESCs showed the inactive α-globin locus to reside within a highly intermingled chromatin conformation [31]. While in differentiating primary mouse erythroid cells, the α-globin locus forms a folded hairpin-like structure (more complex than a simple TAD), the flanking CTCF-sites interestingly do not participate in this hairpin, but rather remain part of the surrounding higher-order structure. Since the boundary elements within the α-globin locus are bound by CTCF and cohesin in both mESCs and differentiating erythroblasts [32], it is likely that other yet to be elucidated mechanisms must mediate hairpin domain formation around the α-globin locus in erythroid cells. Emerging technologies such as CAPTURE (CRISPR affinity purification in situ of regulatory elements)-proteomics that allow the unbiased identification of locus-specific protein complexes, as shown for the locus control region (LCR), the β-globin enhancer, might be a promising approach to identify other CTCF-interacting factors at boundaries [33].
Enhancer-promoter interactions seem not to be required for establishment of the active α-globin erythroid sub-TAD, and active transcription of the α-globin locus not required for maintenance of these TADs [34]. In contrast, forced looping between the LCR and the β-globin promoter by zinc finger-mediated tethering of the transcription co-factor LBD1 to the β-globin locus can substantially induce β-globin expression in Gata1-null mouse (G1E) erythroblasts [35], and a similar mechanism can also re-activate the fetal βh1-globin gene in adult erythroid cells [36]. Recently, these forced LCR-promoter contacts have been shown by single-molecule quantitative RNA FISH to affect transcriptional burst fraction (the number of alleles transcribing per cell) but not burst size (the number of RNA molecules produced per burst), even though the LCR itself modestly affect burst size as well, suggesting that the mechanism of how the LCR contributes to the production of transcripts is independent of enhancer-promoter contact [37].
Super-enhancers
Super-enhancers (SEs) are defined as clusters of single enhancers, are larger in size than regular enhancers, and contain a much higher density of bound TFs and transcriptional coactivators, in particular the Mediator complex. They are thought to drive expression of key cell-type specific genes [38]. However, whether SE activity is the result of synergistic or additive action of single enhancers has been debated [39,40]. Mouse in-vivo studies of the α-globin regulatory region that deleted single enhancer elements or combinations thereof and assayed globin expression and hemoglobin levels showed that individual enhancer elements functioned additively, with some of them (R1 and R2) having more pronounced effects on the α:β-globin ratio than the other enhancer elements [41]. Similarly, by computationally merging publicly available Hi-C (3C method that detects all possible pairwise chromatin interactions) and ChIP-seq data, Huang and colleagues found that ~25% of all 843 SEs in K562 erythroleukemia cells had a hierarchical structure consisting of ‘hub’ and ‘non-hub’ enhancer elements. ‘Hub’ elements are characterized by a higher frequency of chromatin interactions with more cohesin and CTCF bound, and were in closer linear physical proximity to genes associated with blood cell identity [42]. In the same study, CRISPR/Cas9-mediated knockout of ‘hub’ enhancer elements led to more pronounced downregulation of the affected genes than that of ‘non-hub’ elements within the SE-containing TAD. The fact that only a subset of SEs is hierarchically structured might in part explain why different groups have arrived at different conclusions about SE behavior. On the other hand, the hierarchical organization of SEs does not per se imply additive or synergistic functions of individual SE elements. Rather, to establish a correlation between enhancer structure and function systematic knockout or CRISPR interference analysis of many enhancers and SEs would be needed. Intriguingly, in a recent comparative study, so-called hyperacetylated chromatin domains – continuous regions marked by H3K27Ac as well as H3K4Me2 selected by peak breadth rather than signal intensity – in mouse fetal liver erythroid cells proved to be better suited for annotating genes involved in erythroid biological processes by GO analysis compared to SEs [43]. This study suggests that extending the criteria for identifying cell-type specific enhancers might be beneficial.
Making sense of GWAS hits
Great efforts are being undertaken to precisely map and assign mechanistic relevance to disease-associated genetic variants identified in GWAS, including in erythropoiesis [10,11,16,42,44,45]. ~80–90% of genetic variants locate to the non-coding genome [46], predominantly to enhancers [47]. One of the above mentioned time course studies addressing the dynamics of chromatin accessibility during erythroid differentiation has uncovered that variants associated with hematocrit and hemoglobin levels are localized in CREs that are most accessible at the earlier CFU-E and ProE stages, while variants affecting mean corpuscular volume and red blood cell (RBC) count localize to CREs most accessible at the BasoE and PolyE stages [10]. Thus, chromatin accessibility dynamics of regions encompassing pathogenic variants can inform about when during erythroid development the regulatory region is functionally required. Interestingly, another time course study found that 55% of erythroid CREs that are already accessible and thus ‘bookmarked’ in HSPCs contain GATA1 motifs, and almost half of a collection of 497 single-nucleotide polymorphisms (SNPs) associated with RBC traits localized to bookmarked CREs [16].
Genetic variants are localized non-randomly within CREs: Within SEs, the above described ‘hub’ elements are enriched for GWAS SNPs and blood-cell-associated expression quantitative trait loci (eQTLs) compared to ‘non-hub’ elements and regular enhancers. A large-scale study, in which TF occupancy was mapped at nucleotide-resolution by DNase I footprinting in 243 different primary and transformed cell and tissue samples including primary CD34+ cells subjected to erythroid differentiation, revealed functional genetic variants to be preferentially localized with nucleotide-precision within the core of TF footprints [48]. An interesting recent study found that SNPs associated with RBC traits are specifically enriched within enhancers co-bound by signal-induced effector TFs such as SMAD1/2 and TCF7L2 with roles in the BMP/TGFβ- and WNT signaling pathways, respectively [11], suggesting that the phenotypes induced by pathogenic genetic variants could be due to a compromised integration of extracellular signals.
It is intriguing that the use of CREs driving erythroid-cell specific gene expression changes during ontogeny with genes in primitive circulating murine erythroblasts (E10.5) being regulated by promoters, while genes in adult fetal liver erythroblasts at (E13.5) being regulated by distal enhancers [49]. In the same study, a broader survey of 48 human cell types found enhancers active at adult stages that are specific for the interrogated cell type to be enriched for disease-associated GWAS SNPs. Embryonic stage-specific enhancers lacked this enrichment of pathogenic SNPs, which could be possibly due to them causing prenatal lethality and thus underrepresentation in GWAS cohorts.
Chromatin modifiers
Several writers, readers, and erasers of the “histone code” [50] have been implicated in the regulation of erythropoiesis. Yet, outstanding important questions are (1) how sometimes ubiquitously expressed chromatin modifiers enact erythroid-specific gene expression programs and, importantly, only at specific loci; (2) whether the resulting chromatin modifications are causal or correlative; and (3) whether chromatin-modifying enzymes indeed act on histones or rather on non-histone targets to impact erythropoiesis.
Erythroid lineage specificity likely is conferred by a combination of erythroid-specific expression of genes or specific isoforms [10] and alternative TF-chromatin-modifying complexes that form at specific CREs at specific differentiation stages. For example, a model posits that in megakaryocyte-erythroid progenitor cells (MEPs), that give rise to both cell types, the arginine methyltransferase PRMT6 is recruited by RUNX1 as part of a SIN3A-HDAC1-corepressor complex to promote erythroid fate by repressing megakaryopoiesis genes [51]. In MEPs committed to megakaryocytic differentiation, a differently composed complex containing RUNX1-PRMT6 represses erythroid genes, such as GYPA and KLF1. As a consequence, the promoters of those genes are associated with high H3R2me2a and low H3K4me3 levels. Interestingly, pharmacological inhibition of PRMT6 reverses the methylation status at both of these histones and stimulates erythroid differentiation [52,53].
The demethylase LSD1, which demethylates H3K4me1/2 and H3K9me1/2, is also part of several multi-subunit protein complexes with both co-activator and repressor functions. Recently, binding of LSD1 to GFI1B, an essential TF for erythroid differentiation in one of these complexes, was found to be necessary for GFI1B’s function. Using a strategy based on LSD1-binding competent and -defective GFI1B variants and biotin proximity labeling, recent work demonstrated that GFI1B recruits the BRAF-histone deacetylase (BHC) complex via LSD1 to promote erythroid differentiation in K562 cells at the expense of megakaryocyte differentiation [54].
Erythroid specificity through preferential expression of a chromatin-modifying enzyme is exemplified by the only known H4K20 mono-methyltransferase SETD8 [55]. SETD8 expression is highly specific for CD71+ erythroblasts that, in addition, exhibit low levels of the H4K20me1 demethylase PHF8, and EpoR-driven SETD8 deletion in mice causes embryonic lethality at E12.5 due to severe anemia [56]. In cultures of primary erythroid progenitors isolated from E14.5 fetal livers, Setd8 knockdown resulted in a de-repression of Gata2 specifically in R2-proerythroblasts [57–59]. Mechanistically, SETD8 represses Gata2 expression by binding to the essential +9.5 intronic enhancer, which is associated with an increase of H4K20me1 at nearby nucleosomes and shields SCL/TAL1, but not GATA1 and LDB1 from this enhancer [57]. Interestingly, knockdown of Gata2 could only modestly rescue the erythroid block upon Setd8 knockdown [58], which could be due to other essential SETD8 targets. Maturing Setd8−/− erythroblasts also showed a severe defect in chromatin condensation and heterochromatin formation [56], which could hint towards a direct role for SETD8 in higher order chromatin organization. Indeed, two condensin II subunits have been shown to read H4K20me1 [60], but which proteins are binding to H4K20me1 in erythroid cells is unknown. Of note, also non-histone SETD8 methylation targets, including TP53, PCNA and NUMB [61] could mediate its erythroid loss-of-function phenotypes. Thus, a direct comparison of phenotypes incurred by mutation of Setd8 with those of H4K20me1 would address the question of the relevant SETD8 target(s) in erythropoiesis.
Such a comparison has recently been provided by two independent studies focusing on another histone residue, H3K36. NSD1 is a H3K36 mono-/dimethylase, although in its absence not only levels of H3K36me1, me2, but also H3K36me3 are decreased [62]. Ablation of Nsd1 in the hematopoietic system, leads to completely penetrant erythroleukemia-like disease with benzidine-positive yet serially replatable BFU-E-like colonies in the bone marrow, anemia, thrombocytopenia and splenomegaly [63]. Based on the observation that introduction of exogenous histone K-to-M mutants that can no longer be methylated outcompete endogenous histones [64], mice were recently generated that inducibly and ubiquitously express H3K36M [65]. Young adult mice that were treated with doxycycline for 4–7 weeks, developed a severe anemia with an accumulation of predominantly CFU-Es and pro-erythroblasts which, however, were not replatable. Despite not exactly matching phenotypes, their overall similarities are striking evidence for proper regulation of H3K36 methylation being essential for erythropoiesis.
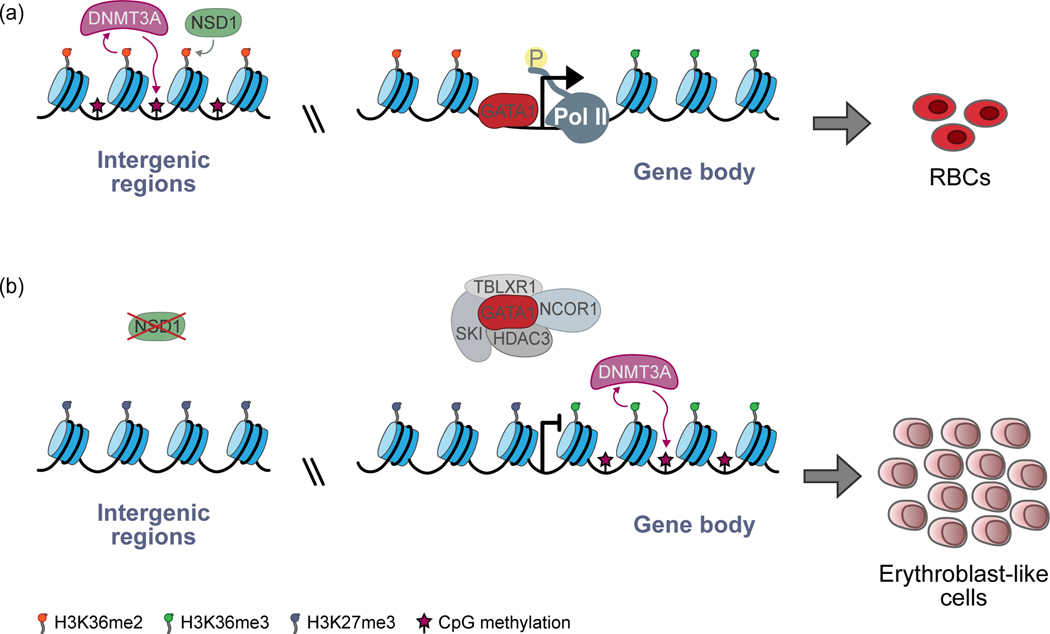
A subset of mice that only express 5% of normal Gata1 levels also develop an erythroleukemia-like disease [66]. Accordingly, in primary Nsd1−/− erythroblasts only wild-type Nsd1, but not the Nsd1N1918Q mutation rendering the SET methyltransferase domain catalytically inactive allowed for efficient GATA1 recruitment to chromatin [63]. Interestingly, the GATA1-interactome in erythroblasts expressing Nsd1N1918Q was enriched for several transcriptional co-repressors, most significantly, the v-Ski avian sarcoma viral oncogene homolog (SKI), knockdown of which intriguingly rescued the differentiation defect of Nsd1−/− erythroblasts (Fig. 2).
Figure 2: Model of epigenetic crosstalk with impact on erythroid differentiation.
A. In this simplified model, H3K36me2 catalyzed by NSD1 leads to the recruitment of DNMT3A to and methylation of intergenic CpGs. GATA1 activates transcription of erythroid-specific genes. B. In the absence of NSD1 and thus loss of H3K36me2, DNMT3A re-localizes to gene bodies marked by H3K36me3, leading to CpG methylation, decreased GATA1 chromatin occupancy and association of GATA1 with members of the nuclear co-repressor (NCoR) complex (NCOR1, HDAC3, TBLXR1) and the transcriptional repressor protein SKI, which together impairs erythroid differentiation with an accumulation of erythroblast-like cells. Model modified from [71,79] with data from [63,71].
Two recent studies have uncovered erythroid-specific functions of polycomb repressive complex 2 (PRC2), the reader/writer of the repressive histone PTM H3K27me3. PRC2 is comprised of 3 proteins, Suz12, Eed, and either Ezh1 or Ezh2 which play different roles during hematopoiesis [67]. As none of these core proteins have DNA binding activity, accessory proteins such as Mtf2 target PRC2 to DNA. One study found that Mtf2-null mice die at E15.5 with severe anemia [68]. Intersecting genes depleted for H3K27me3 in Mtf2-null fetal liver erythroblasts with Mtf2-target genes in their wild-type counterparts revealed canonical Wnt signaling among the top upregulated pathways. Supporting a causal role for excess Wnt activity in the phenotype of Mtf2-deficient erythroblasts, inhibition of Wnt signaling rescued their erythroid differentiation block in vitro. A second study found through a CRISPR/Cas9 screen that the ubiquitin ligase FBXO11 is required for erythropoiesis [69]. FBXO11 targets the PRC2-complex-interacting protein BAHD1 for degradation, thus relieving PRC2-mediated repression and activating erythroid gene expression. These opposite erythroid phenotypes caused by PRC2 loss could be explained by the use of different model systems (murine primary versus human immortalized erythroblasts [70]) and suggest that PRC2 complex composition might dynamically vary across erythroid differentiation stages.
DNA methylation and epigenetic crosstalk
A challenge ahead is to not just consider single epigenetic marks, but also the crosstalk between different epigenetic modifications and the responsible modifiers, including crosstalk between chromatin-regulatory pathways such as histone PTMs and DNA methylation. A case in point is the finding that H3K36me2 induced by nuclear receptor-binding SET domain protein 1 (NSD1) is required for DNMT3A recruitment specifically to intergenic CpG dinucleotides and their methylation [71] (Fig. 2). Importantly, DNMT3A’s PWWP ‘reader’ domain recognizes both H3K36me3 and, with higher affinity, H3K36me2 in gene bodies and intergenic regions, respectively. In the absence of NSD1, H3K36me2 marks are lost, leading to the re-localization of DNMT3A to H3K36me3-enriched gene bodies and their CpG methylation. A study investigating DNA methylation changes upon loss of Dnmt3a in mice found CpG-rich TF binding motifs to be preferentially affected [72]. Interestingly, the binding motifs of lineage-determining erythroid TFs such as Tal1 and Klf1 are CpG-rich, explaining why genome-wide hypomethylation upon Dnmt3a loss leads to the specific phenotype of erythroid over myelomonocytic lineage skewing. Of note, Gata1, in addition to its canonical T/AGATA motif, has been recently shown to recognize CGATA sites, however, also only in the context of an unmethylated CpG dinucleotide [73]. Despite the development of new technologies [74], there are experimental limits of how many PTMs and/or DNA methylation states can be combinatorically examined; thus, in the future, advanced machine learning approaches might be able to systematically tackle the crosstalk between different types of epigenetic modifications [75].
Conclusion and outlook
In the last few years, a series of technological advances has enabled significant insights into the interplay between chromatin dynamics and erythroid-specific gene expression, and erythropoiesis remains a key model system for studying developmental genome organization. But there is still a lot to learn about how the 3D genome is dynamically shaping erythroid-specific transcriptional output. For example, higher temporal resolution shows that chromatin interactions (3C) seem to be much more dynamic than can be captured by chromatin accessibility (ATAC) studies alone [14], emphasizing the importance of studying the 3D chromatin structure, in particular enhancer-promoter interactions to understand gene regulation. And even though it is now possible to monitor spatial chromatin organization during in-vivo erythroid differentiation with as few as 2000 cells using newer 3C technologies [15], they are still only providing a snapshot of steady-state population-average chromatin structure. Additional assays are needed to assess higher chromatin organization in single cells. Imaging-based approaches are an alternative but have their own limitations when it comes to resolution and throughput. In this regard, it will be interesting to address how dynamically changing chromatin organization is coupled with the concept of liquid-liquid phase separation during erythroid differentiation [76]. Last but not least, a key future endeavor that seems within reach [14,33,74,77] will be the systematic and unbiased identification of proteins bound to CREs in vivo during erythroid differentiation. This could shed light on the question of how protein complex composition and organization translates to specificity of transcriptional programs. Such studies should also be extended to yet to be revealed enhancer RNAs that might dynamically stabilize enhancer-promoter loops in cis [78]. We are still a ways from completely understanding how to enhance red cell fate.
KEY POINTS.
Erythropoiesis is an invaluable model to understand the functional impact of nuclear chromatin dynamics on a lineage differentiation process
New lower-input chromosome conformation capture (3C)-based methods allow to interrogate 3D chromatin structure during erythroid differentiation in vivo, and to investigate ≥3-way chromatin contacts
High-resolution chromatin accessibility maps combined with databases comprised of fine-mapped SNPs and functional assays are a great step forward in discerning causal disease-associated genetic variants
Elucidating the extensive crosstalk between different histone post-translational modifications (PTMs) and other epigenetic modifications such as different DNA methylation states will require systematic and unbiased approaches
Acknowledgements
We thank J. Xu for discussion and B. Boettner for critical reading of the manuscript.
Funding
Funding for this study was provided by the National Heart, Lung, and Blood Institute (NHLBI, 4R01HL048801, 5P01HL032262, 5U01HL134812, 1P01HL131477), the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK, 1U54DK110805, 3R24DK092760), and Harvard Catalyst to L.I.Z.
Footnotes
Competing interests
L.I.Z. is a founder of and holds stock in Fate Therapeutics, Camp4 Therapeutics, and Scholar Rock; and is a consultant to Celularity. M.P.R. declares no competing interests.
References
*Oudelaar Nat Commun. 2019
*Allahyar
The studies by Oudelaar et al. and Allahyar et al. describe new methods to detect multi-way chromatin interactions, rather than the two-way interactions detected by previous 3C methods.
*Fox
This study provides an interesting new approach to stratify enhancers to better predict key enhancers that drive erythroid-specific gene transcription.
**Leonards
**Brumbaugh
Brumbaugh et al. and Leonards et al. demonstrate largely overlapping phenotypes induced by expressing a histone H3K36M mutant or by conditionally knocking out NSD1, respectively.
*Izzo
This study describes a mechanism for why global DNA methylation changes in Dnmt3a and Tet2 knockout mice result in specific erythroid or myelomonocytic lineage skewing phenotypes.
- 1.Chen T, Dent SY: Chromatin modifiers and remodellers: regulators of cellular differentiation. Nat Rev Genet 2014, 15:93–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Spielmann M, Lupianez DG, Mundlos S: Structural variation in the 3D genome. Nat Rev Genet 2018, 19:453–467. [DOI] [PubMed] [Google Scholar]
- 3.Zhu F, Farnung L, Kaasinen E, Sahu B, Yin Y, Wei B, Dodonova SO, Nitta KR, Morgunova E, Taipale M, et al. : The interaction landscape between transcription factors and the nucleosome. Nature 2018, 562:76–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.An X, Schulz VP, Li J, Wu K, Liu J, Xue F, Hu J, Mohandas N, Gallagher PG: Global transcriptome analyses of human and murine terminal erythroid differentiation. Blood 2014, 123:3466–3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen K, Liu J, Heck S, Chasis JA, An X, Mohandas N: Resolving the distinct stages in erythroid differentiation based on dynamic changes in membrane protein expression during erythropoiesis. Proc Natl Acad Sci U S A 2009, 106:17413–17418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu J, Liu J, Xue F, Halverson G, Reid M, Guo A, Chen L, Raza A, Galili N, Jaffray J, et al. : Isolation and functional characterization of human erythroblasts at distinct stages: implications for understanding of normal and disordered erythropoiesis in vivo. Blood 2013, 121:3246–3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu J, Zhang J, Ginzburg Y, Li H, Xue F, De Franceschi L, Chasis JA, Mohandas N, An X: Quantitative analysis of murine terminal erythroid differentiation in vivo: novel method to study normal and disordered erythropoiesis. Blood 2013, 121:e43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cantor AB, Orkin SH: Transcriptional regulation of erythropoiesis: an affair involving multiple partners. Oncogene 2002, 21:3368–3376. [DOI] [PubMed] [Google Scholar]
- 9.Heuston EF, Keller CA, Lichtenberg J, Giardine B, Anderson SM, Center NIHIS, Hardison RC, Bodine DM: Establishment of regulatory elements during erythro-megakaryopoiesis identifies hematopoietic lineage-commitment points. Epigenetics Chromatin 2018, 11:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ludwig LS, Lareau CA, Bao EL, Nandakumar SK, Muus C, Ulirsch JC, Chowdhary K, Buenrostro JD, Mohandas N, An X, et al. : Transcriptional States and Chromatin Accessibility Underlying Human Erythropoiesis. Cell Rep 2019, 27:3228–3240 e3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choudhuri A, Trompouki E, Abraham BJ, Colli LM, Kock KH, Mallard W, Yang ML, Vinjamur DS, Ghamari A, Sporrij A, et al. : Common variants in signaling transcription-factor-binding sites drive phenotypic variability in red blood cell traits. Nat Genet 2020, 52:1333–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang J, Liu X, Li D, Shao Z, Cao H, Zhang Y, Trompouki E, Bowman TV, Zon LI, Yuan GC, et al. : Dynamic Control of Enhancer Repertoires Drives Lineage and Stage-Specific Transcription during Hematopoiesis. Dev Cell 2016, 36:9–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiang G, Keller CA, Heuston E, Giardine BM, An L, Wixom AQ, Miller A, Cockburn A, Sauria MEG, Weaver K, et al. : An integrative view of the regulatory and transcriptional landscapes in mouse hematopoiesis. Genome Res 2020, 30:472–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu X, Chen Y, Zhang Y, Liu Y, Liu N, Botten GA, Cao H, Orkin SH, Zhang MQ, Xu J: Multiplexed capture of spatial configuration and temporal dynamics of locus-specific 3D chromatin by biotinylated dCas9. Genome Biol 2020, 21:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oudelaar AM, Beagrie RA, Gosden M, de Ornellas S, Georgiades E, Kerry J, Hidalgo D, Carrelha J, Shivalingam A, El-Sagheer AH, et al. : Dynamics of the 4D genome during in vivo lineage specification and differentiation. Nat Commun 2020, 11:2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schulz VP, Yan H, Lezon-Geyda K, An X, Hale J, Hillyer CD, Mohandas N, Gallagher PG: A Unique Epigenomic Landscape Defines Human Erythropoiesis. Cell Rep 2019, 28:2996–3009 e2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Romano O, Petiti L, Felix T, Meneghini V, Portafax M, Antoniani C, Amendola M, Bicciato S, Peano C, Miccio A: GATA Factor-Mediated Gene Regulation in Human Erythropoiesis. iScience 2020, 23:101018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shearstone JR, Pop R, Bock C, Boyle P, Meissner A, Socolovsky M: Global DNA demethylation during mouse erythropoiesis in vivo. Science 2011, 334:799–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yan H, Hale J, Jaffray J, Li J, Wang Y, Huang Y, An X, Hillyer C, Wang N, Kinet S, et al. : Developmental differences between neonatal and adult human erythropoiesis. Am J Hematol 2018, 93:494–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma S, Zhang B, LaFave LM, Earl AS, Chiang Z, Hu Y, Ding J, Brack A, Kartha VK, Tay T, et al. : Chromatin Potential Identified by Shared Single-Cell Profiling of RNA and Chromatin. Cell 2020, 183:1103–1116 e1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hopkins PC, Sainz-Fuertes R, Lovestone S: The impact of a novel apolipoprotein E and amyloid-beta protein precursor-interacting protein on the production of amyloid-beta. J Alzheimers Dis 2011, 26:239–253. [DOI] [PubMed] [Google Scholar]
- 22.Shen Y, Bassett MA, Gurumurthy A, Nar R, Knudson IJ, Guy CR, Perez A, Mellen RW, Ikeda M, Hossain MA, et al. : Identification of a Novel Enhancer/Chromatin Opening Element Associated with High-Level gamma-Globin Gene Expression. Mol Cell Biol 2018, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fudenberg G, Imakaev M, Lu C, Goloborodko A, Abdennur N, Mirny LA: Formation of Chromosomal Domains by Loop Extrusion. Cell Rep 2016, 15:2038–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanborn AL, Rao SS, Huang SC, Durand NC, Huntley MH, Jewett AI, Bochkov ID, Chinnappan D, Cutkosky A, Li J, et al. : Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc Natl Acad Sci U S A 2015, 112:E6456–6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eagen KP: Principles of Chromosome Architecture Revealed by Hi-C. Trends Biochem Sci 2018, 43:469–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Allahyar A, Vermeulen C, Bouwman BAM, Krijger PHL, Verstegen M, Geeven G, van Kranenburg M, Pieterse M, Straver R, Haarhuis JHI, et al. : Enhancer hubs and loop collisions identified from single-allele topologies. Nat Genet 2018, 50:1151–1160. [DOI] [PubMed] [Google Scholar]
- 27.Oudelaar AM, Harrold CL, Hanssen LLP, Telenius JM, Higgs DR, Hughes JR: A revised model for promoter competition based on multi-way chromatin interactions at the alpha-globin locus. Nat Commun 2019, 10:5412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oudelaar AM, Davies JOJ, Hanssen LLP, Telenius JM, Schwessinger R, Liu Y, Brown JM, Downes DJ, Chiariello AM, Bianco S, et al. : Single-allele chromatin interactions identify regulatory hubs in dynamic compartmentalized domains. Nat Genet 2018, 50:1744–1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sutherland H, Bickmore WA: Transcription factories: gene expression in unions? Nat Rev Genet 2009, 10:457–466. [DOI] [PubMed] [Google Scholar]
- 30.Nora EP, Goloborodko A, Valton AL, Gibcus JH, Uebersohn A, Abdennur N, Dekker J, Mirny LA, Bruneau BG: Targeted Degradation of CTCF Decouples Local Insulation of Chromosome Domains from Genomic Compartmentalization. Cell 2017, 169:930–944 e922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chiariello AM, Bianco S, Oudelaar AM, Esposito A, Annunziatella C, Fiorillo L, Conte M, Corrado A, Prisco A, Larke MSC, et al. : A Dynamic Folded Hairpin Conformation Is Associated with alpha-Globin Activation in Erythroid Cells. Cell Rep 2020, 30:2125–2135 e2125. [DOI] [PubMed] [Google Scholar]
- 32.Hanssen LLP, Kassouf MT, Oudelaar AM, Biggs D, Preece C, Downes DJ, Gosden M, Sharpe JA, Sloane-Stanley JA, Hughes JR, et al. : Tissue-specific CTCF-cohesin-mediated chromatin architecture delimits enhancer interactions and function in vivo. Nat Cell Biol 2017, 19:952–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu X, Zhang Y, Chen Y, Li M, Zhou F, Li K, Cao H, Ni M, Liu Y, Gu Z, et al. : In Situ Capture of Chromatin Interactions by Biotinylated dCas9. Cell 2017, 170:1028–1043 e1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brown JM, Roberts NA, Graham B, Waithe D, Lagerholm C, Telenius JM, De Ornellas S, Oudelaar AM, Scott C, Szczerbal I, et al. : A tissue-specific self-interacting chromatin domain forms independently of enhancer-promoter interactions. Nat Commun 2018, 9:3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deng W, Lee J, Wang H, Miller J, Reik A, Gregory PD, Dean A, Blobel GA: Controlling long-range genomic interactions at a native locus by targeted tethering of a looping factor. Cell 2012, 149:1233–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deng W, Rupon JW, Krivega I, Breda L, Motta I, Jahn KS, Reik A, Gregory PD, Rivella S, Dean A, et al. : Reactivation of developmentally silenced globin genes by forced chromatin looping. Cell 2014, 158:849–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bartman CR, Hsu SC, Hsiung CC, Raj A, Blobel GA: Enhancer Regulation of Transcriptional Bursting Parameters Revealed by Forced Chromatin Looping. Mol Cell 2016, 62:237–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA: Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 2013, 153:307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dukler N, Gulko B, Huang YF, Siepel A: Is a super-enhancer greater than the sum of its parts? Nat Genet 2016, 49:2–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pott S, Lieb JD: What are super-enhancers? Nat Genet 2015, 47:8-12. [DOI] [PubMed] [Google Scholar]
- 41.Hay D, Hughes JR, Babbs C, Davies JOJ, Graham BJ, Hanssen L, Kassouf MT, Marieke Oudelaar AM, Sharpe JA, Suciu MC, et al. : Genetic dissection of the alpha-globin super-enhancer in vivo. Nat Genet 2016, 48:895–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang J, Li K, Cai W, Liu X, Zhang Y, Orkin SH, Xu J, Yuan GC: Dissecting super-enhancer hierarchy based on chromatin interactions. Nat Commun 2018, 9:943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fox S, Myers JA, Davidson C, Getman M, Kingsley PD, Frankiewicz N, Bulger M: Hyperacetylated chromatin domains mark cell type-specific genes and suggest distinct modes of enhancer function. Nat Commun 2020, 11:4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ulirsch JC, Lareau CA, Bao EL, Ludwig LS, Guo MH, Benner C, Satpathy AT, Kartha VK, Salem RM, Hirschhorn JN, et al. : Interrogation of human hematopoiesis at single-cell and single-variant resolution. Nat Genet 2019, 51:683–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ulirsch JC, Nandakumar SK, Wang L, Giani FC, Zhang X, Rogov P, Melnikov A, McDonel P, Do R, Mikkelsen TS, et al. : Systematic Functional Dissection of Common Genetic Variation Affecting Red Blood Cell Traits. Cell 2016, 165:1530–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, Manolio TA: Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A 2009, 106:9362–9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maurano MT, Humbert R, Rynes E, Thurman RE, Haugen E, Wang H, Reynolds AP, Sandstrom R, Qu H, Brody J, et al. : Systematic localization of common disease-associated variation in regulatory DNA. Science 2012, 337:1190–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vierstra J, Lazar J, Sandstrom R, Halow J, Lee K, Bates D, Diegel M, Dunn D, Neri F, Haugen E, et al. : Global reference mapping of human transcription factor footprints. Nature 2020, 583:729–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cai W, Huang J, Zhu Q, Li BE, Seruggia D, Zhou P, Nguyen M, Fujiwara Y, Xie H, Yang Z, et al. : Enhancer dependence of cell-type-specific gene expression increases with developmental age. Proc Natl Acad Sci U S A 2020, 117:21450–21458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jenuwein T, Allis CD: Translating the histone code. Science 2001, 293:1074–1080. [DOI] [PubMed] [Google Scholar]
- 51.Herglotz J, Kuvardina ON, Kolodziej S, Kumar A, Hussong H, Grez M, Lausen J: Histone arginine methylation keeps RUNX1 target genes in an intermediate state. Oncogene 2013, 32:2565–2575. [DOI] [PubMed] [Google Scholar]
- 52.Herkt SC, Kuvardina ON, Herglotz J, Schneider L, Meyer A, Pommerenke C, Salinas-Riester G, Seifried E, Bonig H, Lausen J: Protein arginine methyltransferase 6 controls erythroid gene expression and differentiation of human CD34(+) progenitor cells. Haematologica 2018, 103:18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kuvardina ON, Herglotz J, Kolodziej S, Kohrs N, Herkt S, Wojcik B, Oellerich T, Corso J, Behrens K, Kumar A, et al. : RUNX1 represses the erythroid gene expression program during megakaryocytic differentiation. Blood 2015, 125:3570–3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McClellan D, Casey MJ, Bareyan D, Lucente H, Ours C, Velinder M, Singer J, Lone MD, Sun W, Coria Y, et al. : Growth Factor Independence 1B-Mediated Transcriptional Repression and Lineage Allocation Require Lysine-Specific Demethylase 1-Dependent Recruitment of the BHC Complex. Mol Cell Biol 2019, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fang J, Feng Q, Ketel CS, Wang H, Cao R, Xia L, Erdjument-Bromage H, Tempst P, Simon JA, Zhang Y: Purification and functional characterization of SET8, a nucleosomal histone H4-lysine 20-specific methyltransferase. Curr Biol 2002, 12:1086–1099. [DOI] [PubMed] [Google Scholar]
- 56.Malik J, Lillis JA, Couch T, Getman M, Steiner LA: The Methyltransferase Setd8 Is Essential for Erythroblast Survival and Maturation. Cell Rep 2017, 21:2376–2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.DeVilbiss AW, Sanalkumar R, Hall BD, Katsumura KR, de Andrade IF, Bresnick EH: Epigenetic Determinants of Erythropoiesis: Role of the Histone Methyltransferase SetD8 in Promoting Erythroid Cell Maturation and Survival. Mol Cell Biol 2015, 35:2073–2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Malik J, Getman M, Steiner LA: Histone methyltransferase Setd8 represses Gata2 expression and regulates erythroid maturation. Mol Cell Biol 2015, 35:2059–2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Myers JA, Couch T, Murphy Z, Malik J, Getman M, Steiner LA: The histone methyltransferase Setd8 alters the chromatin landscape and regulates the expression of key transcription factors during erythroid differentiation. Epigenetics Chromatin 2020, 13:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu W, Tanasa B, Tyurina OV, Zhou TY, Gassmann R, Liu WT, Ohgi KA, Benner C, Garcia-Bassets I, Aggarwal AK, et al. : PHF8 mediates histone H4 lysine 20 demethylation events involved in cell cycle progression. Nature 2010, 466:508–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Milite C, Feoli A, Viviano M, Rescigno D, Cianciulli A, Balzano AL, Mai A, Castellano S, Sbardella G: The emerging role of lysine methyltransferase SETD8 in human diseases. Clin Epigenetics 2016, 8:102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bennett RL, Swaroop A, Troche C, Licht JD: The Role of Nuclear Receptor-Binding SET Domain Family Histone Lysine Methyltransferases in Cancer. Cold Spring Harb Perspect Med 2017, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Leonards K, Almosailleakh M, Tauchmann S, Bagger FO, Thirant C, Juge S, Bock T, Mereau H, Bezerra MF, Tzankov A, et al. : Nuclear interacting SET domain protein 1 inactivation impairs GATA1-regulated erythroid differentiation and causes erythroleukemia. Nat Commun 2020, 11:2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lewis PW, Muller MM, Koletsky MS, Cordero F, Lin S, Banaszynski LA, Garcia BA, Muir TW, Becher OJ, Allis CD: Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science 2013, 340:857–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brumbaugh J, Kim IS, Ji F, Huebner AJ, Di Stefano B, Schwarz BA, Charlton J, Coffey A, Choi J, Walsh RM, et al. : Inducible histone K-to-M mutations are dynamic tools to probe the physiological role of site-specific histone methylation in vitro and in vivo. Nat Cell Biol 2019, 21:1449–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shimizu R, Kuroha T, Ohneda O, Pan X, Ohneda K, Takahashi S, Philipsen S, Yamamoto M: Leukemogenesis caused by incapacitated GATA-1 function. Mol Cell Biol 2004, 24:10814–10825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Di Carlo V, Mocavini I, Di Croce L: Polycomb complexes in normal and malignant hematopoiesis. J Cell Biol 2019, 218:55–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rothberg JLM, Maganti HB, Jrade H, Porter CJ, Palidwor GA, Cafariello C, Battaion HL, Khan ST, Perkins TJ, Paulson RF, et al. : Mtf2-PRC2 control of canonical Wnt signaling is required for definitive erythropoiesis. Cell Discov 2018, 4:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xu P, Scott DC, Xu B, Yao Y, Feng R, Cheng L, Mayberry K, Wang YD, Bi W, Palmer LE, et al. : FBXO11-mediated proteolysis of BAHD1 relieves PRC2-dependent transcriptional repression in erythropoiesis. Blood 2021, 137:155–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Daniels DE, Downes DJ, Ferrer-Vicens I, Ferguson DCJ, Singleton BK, Wilson MC, Trakarnsanga K, Kurita R, Nakamura Y, Anstee DJ, et al. : Comparing the two leading erythroid lines BEL-A and HUDEP-2. Haematologica 2020, 105:e389–e394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Weinberg DN, Papillon-Cavanagh S, Chen H, Yue Y, Chen X, Rajagopalan KN, Horth C, McGuire JT, Xu X, Nikbakht H, et al. : The histone mark H3K36me2 recruits DNMT3A and shapes the intergenic DNA methylation landscape. Nature 2019, 573:281–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Izzo F, Lee SC, Poran A, Chaligne R, Gaiti F, Gross B, Murali RR, Deochand SD, Ang C, Jones PW, et al. : DNA methylation disruption reshapes the hematopoietic differentiation landscape. Nat Genet 2020, 52:378–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yang L, Chen Z, Stout ES, Delerue F, Ittner LM, Wilkins MR, Quinlan KGR, Crossley M: Methylation of a CGATA element inhibits binding and regulation by GATA-1. Nat Commun 2020, 11:2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Villasenor R, Pfaendler R, Ambrosi C, Butz S, Giuliani S, Bryan E, Sheahan TW, Gable AL, Schmolka N, Manzo M, et al. : ChromID identifies the protein interactome at chromatin marks. Nat Biotechnol 2020, 38:728–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kang M, Lee S, Lee D, Kim S: Learning Cell-Type-Specific Gene Regulation Mechanisms by Multi-Attention Based Deep Learning With Regulatory Latent Space. Front Genet 2020, 11:869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mir M, Bickmore W, Furlong EEM, Narlikar G: Chromatin topology, condensates and gene regulation: shifting paradigms or just a phase? Development 2019, 146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tsui C, Inouye C, Levy M, Lu A, Florens L, Washburn MP, Tjian R: dCas9-targeted locus-specific protein isolation method identifies histone gene regulators. Proc Natl Acad Sci U S A 2018, 115:E2734–E2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sartorelli V, Lauberth SM: Enhancer RNAs are an important regulatory layer of the epigenome. Nat Struct Mol Biol 2020, 27:521–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tauchmann S, Almosailleakh M, Schwaller J: NSD1 in erythroid differentiation and leukemogenesis. Mol Cell Oncol 2020, 7:1809919. [DOI] [PMC free article] [PubMed] [Google Scholar]