Abstract
Postweaning multisystemic wasting syndrome (PMWS) is an emerging disease in swine. Increasing evidence indicates that a variant strain of porcine circovirus (PCV), designated type 2 PCV (PCV-2), is responsible for PMWS. To determine the extent of genetic heterogeneity of PCV-2 isolates, the complete genomes of six PCV-2 isolates from different regions of North America were amplified by PCR and sequenced. Sequence and phylogenetic analyses confirmed that two distinct genotypes of PCV exist: nonpathogenic genotype PCV-1 and PMWS-associated genotype PCV-2. However, within the PCV-2 genotype, several minor branches that have been identified appear to be associated with geographic origins. The genomic sequences of two French PCV-2 isolates diverge the most from those of other PCV-2 isolates and form a distinct branch. Other minor but distinguishable branches have also been identified for a Taiwan PCV-2 isolate and two of the Canadian PCV-2 isolates. All the U.S. PCV-2 isolates are closely related, but the Canadian isolates vary, to some extent, in their genomic sequences. The data from this study indicate that although the genome of PCV-2 is generally stable among different isolates, PCV-2 isolates from different geographic regions vary in their genomic sequences. This variation may have important implications for PCV-2 diagnosis and research. On the basis of genetic analyses of available PCV strains, a universal PCR-restriction fragment length polymorphism (PCR-RFLP) assay was developed to detect and differentiate between infections with PCV-1 and PCV-2. This PCR-RFLP assay should be useful for studying the pathogenesis of PCV-2, for detecting PCV-2 infection in pigs from different geographic regions, and for screening donor pigs for use in xenotransplantation.
Porcine circovirus (PCV) was originally isolated as a noncytopathic contaminant of the porcine kidney cell line PK15 (50). PCV is a small nonenveloped virus that contains a single-stranded circular DNA genome of about 1.76 kb (50). On the basis of its morphology and genomic organization, PCV was classified as a member of Circoviridae family (30, 36), which consists of two other animal circoviruses, chicken anemia virus (CAV) and psittacine beak-and-feather disease virus, and three plant circoviruses, banana bunchy top virus, coconut foliar decay virus, and subterranean clover stunt virus. Members of the three recognized animal circoviruses, PCV, CAV, and psittacine beak-and-feather disease virus, do not share nucleotide sequence homology or antigenic determinants with each other (8, 54). More recently, a human circovirus, designated TT virus (TTV), was identified from individuals with posttransfusion hepatitis (38, 43). The human TTV is similar to the circovirus CAV in its genomic organization (38). Although antibodies to PCV were found in various animal species including humans, mice, cattle, and pigs (11, 12, 22, 23, 41, 52, 53), little is known regarding the pathogenesis of PCV in these animal species. Experimental infection of pigs with the PK15-derived PCV did not produce clinical disease, and thus, this virus is not considered pathogenic for pigs (1, 51).
Postweaning multisystemic wasting syndrome (PMWS) is an emerging disease in pigs first described in 1991 (10, 20). The disease occurs in swine herds that are usually in good health and has a low rate of morbidity but causes a relatively high case fatality rate among 5- to 12-week-old pigs (6, 10). Clinically, PMWS is characterized by progressive weight loss, dyspnea, tachypnea, anemia, diarrhea, and jaundice. In an acute outbreak, the mortality rate associated with PMWS may peak at about 10% and can reach up to 50% in some cases (6, 20, 21). The microscopic lesions of PMWS include those associated with granulomatous interstitial pneumonia, lymphadenopathy, hepatitis, nephritis, and pancreatitis (20, 21). PMWS has now been recognized in pigs in Canada and most of the United States (2, 3, 6, 13, 18, 26, 27, 37, 39, 40), many European countries (4, 6, 24, 32, 48, 49), and some countries in Asia (6, 44) and has the potential to have a serious economic impact on the swine industry worldwide.
The etiology of PMWS is rather complicated, but it is believed that a variant strain of PCV, designated type 2 PCV (PCV-2), is responsible for PMWS in pigs (2, 3, 4, 6, 13, 18, 37, 39). The nonpathogenic PK15-derived PCV has been designated PCV-1 to distinguish it from the PMWS-associated PCV, PCV-2 (2, 3). PCV-2 was isolated from pigs with clinical and pathological findings consistent with PMWS (2, 3, 4, 6, 13, 19, 27, 39, 40). PCV-2 DNA and antigen were detected in various tissues and organs from pigs with natural cases of PMWS (19, 27, 32, 37, 39, 40, 46). The complete genome of PCV-2 has been determined, and surprisingly, nonpathogenic PCV-1 and PMWS-associated PCV-2 are found to share only about 75% nucleotide sequence identity (18, 37, 39). Seven open reading frames (ORFs) have been identified for PCV-1 (31, 33, 36), whereas 6 or 11 ORFs have been identified for PCV-2 (18, 37, 39). Although PMWS has been reported in most of the United States, only a few PCV-2 isolates from the United States have been genetically characterized (37, 39). On the basis of the nucleotide sequences of the U.S. and other PCV-2 isolates sequenced thus far, it appears that there exists only one genotype of PCV-2 worldwide (19, 37, 39). Nevertheless, the value of diagnosing PCV-2 infections and studying the pathogenesis of PCV-2 by PCR and other molecular approaches will depend on knowledge of the extent of genetic variation among PCV-2 isolates from different geographic regions. In addition, the development of an effective vaccine against PMWS also requires a better understanding of the extent of genetic variation among PCV-2 isolates. In this study, we genetically characterized six PCV-2 isolates from pigs with confirmed PMWS in different geographic regions of North America. The extent of genetic variation among these six PCV-2 isolates and all other known PCV isolates (both PCV-1 and PCV-2) was analyzed. On the basis of the data generated from this study, we further developed a universal PCR-restriction fragment length polymorphism (PCR-RFLP) assay to detect and differentiate between infections with PCV-1 and PCV-2 in pigs from different geographic regions.
MATERIALS AND METHODS
Sample sources.
Various tissue samples (liver, spleen, tonsil, lymph nodes, etc.) were collected from pigs with PMWS as confirmed by immunohistochemistry (IHC) (data not shown). The tissues were stored at −80°C until use. The complete PCV-2 genome was amplified, sequenced, and characterized from tissue samples from six selected pigs with PMWS that originated from different geographic regions of North America: two from Utah, one from Missouri, one from Iowa, one from Illinois, and one from Canada (Table 1). The isolates from these six pigs with PMWS, along with those from four more pigs with PMWS (Table 1) from Iowa, were also characterized by PCR-RFLP analyses.
TABLE 1.
PCV isolates used in this study as well as those reported previously
| Type | Isolate no. | Geographic location | IHC
|
ISHa of PCV | Clinical sign(s)b | Histopathological lesionsc | Reference or source | |
|---|---|---|---|---|---|---|---|---|
| PRRSV | PCV | |||||||
| PCV-2 | 26606d | Utah | + | NDe | + | Resp | Pneumonia, lymphoid depletion, enteritis | This study |
| 26607d | Utah | + | ND | + | Resp, diarrhea | Pneumonia, enteritis, hepatitis, nephritis | This study | |
| 40856d | Missouri | + | + | ND | Resp, wasting | Pneumonia, lymphoid depletion, hepatitis, nephritis | This study | |
| 40895d | Iowa | − | + | ND | Resp, wasting | Pneumonia, lymphoid depletion | This study | |
| 34464d | Canada | + | + | ND | Resp | Pneumonia | This study | |
| 10489d | Illinois | − | + | ND | Resp, wasting | Pneumonia, lymphoid depletion | This study | |
| 38835 | Iowa | + | + | ND | Resp | Pneumonia, lymphoid depletion | This study | |
| 36688 | Iowa | + | + | ND | Resp, wasting, dermatitis | Pneumonia, lymphoid depletion, nephritis, dermatitis | This study | |
| 40860 | Iowa | + | + | ND | Resp | Pneumonia | This study | |
| 40887 | Iowa | + | + | ND | Resp | Pneumonia | This study | |
| AF055391 | California | 37 | ||||||
| AF027217 | California | 19 | ||||||
| AF109397 | Francef | 29, GenBank | ||||||
| AJ223185 | Iowa | 39 | ||||||
| AF055394 | France | 37 | ||||||
| AF085695 | Canada | GenBank | ||||||
| AF086836 | Canada | GenBank | ||||||
| AF086835 | Canada | GenBank | ||||||
| AF086834 | Canada | GenBank | ||||||
| AF112862 | Canada | GenBank | ||||||
| AF166528 | Taiwan | GenBank | ||||||
| AF109399 | Canada | GenBank | ||||||
| AF109398 | Canada | GenBank | ||||||
| AF117753 | Canada | GenBank | ||||||
| AF055393 | France | 37 | ||||||
| AF055392 | Canada | 37 | ||||||
| PCV-1 | AF071879 | PK15 cell | 42 | |||||
| Y09921 | Germany | 31 | ||||||
| U49186 | Ireland | 36 | ||||||
| AF012107 | France | 31 | ||||||
ISH, in situ hybridization.
Clinical signs: Resp, respiratory disease; wasting, anorexia and weight loss.
Histopathological lesions: pneumonia, interstitial pneumonia.
PCV-2 isolates sequenced in this study.
ND, not determined.
Bovine isolate.
Isolation of DNA from tissues.
DNA was extracted from various tissue samples with the QIAamp DNA Mini kit (Qiagen, Inc., Valencia, Calif.) according to the protocol supplied by the manufacturer. For each DNA extraction, 25 mg of tissue sample was used. The resulting DNA was eluted in DNase-, RNase-, and proteinase-free water (Eppendorf 5 Primer, Inc., Boulder, Colo.).
PCR amplification of the complete genome of PCV-2.
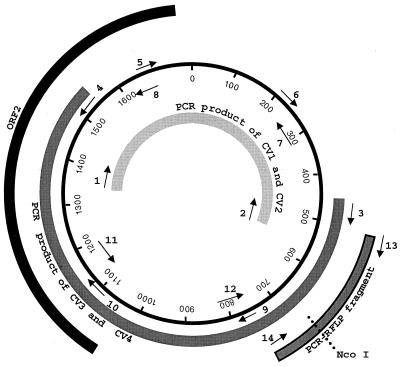
Two sets of PCR primers were designed on the basis of the published PCV-2 sequence. These primers amplify two overlapping fragments that represent the entire genome of PCV-2 (Fig. 1). The first set of primers, CV1 and CV2 (Table 2), amplifies a 989-bp fragment, and the second set of primers, CV3 and CV4 (Table 2), amplifies a 1,092-bp fragment. The extracted DNA was amplified by PCR with AmpliTaq Gold polymerase (Perkin-Elmer, Norwalk, Conn.). The PCR consisted of 35 cycles of denaturation at 94°C for 1 min, annealing at 55°C for 1 min, and extension at 72°C for 3 min, followed by a terminal extension at 72°C for 7 min.
FIG. 1.
Genome organization of PCV-2. The origin of replication (0), the putative capsid gene (ORF2), the PCR-RFLP fragment, and the two overlapping PCR fragments used to determine the complete genome of PCV-2 are indicated in the circular map. The relative positions of the oligonucleotide primers used in this study are indicated by arrows with respective numbers: 1, CV1; 2, CV2; 3, CV3; 4, CV4; 5, CV1-1; 6, CV1-2; 7, CV2-1; 8, CV2-2; 9, CV3-1; 10, CV3-2; 11, CV4-1; 12, CV4-2; 13, MCV1; 14, MCV2. The sequences and designations of these primers are listed in Table 2.
TABLE 2.
Oligonucleotide primers used in the study
| Primer | Primer sequence | Application | Positiona |
|---|---|---|---|
| CV1 | 5′-AGGGCTGTGGCCTTTGTTAC-3′ | PCR, Seqb | 1336–1355 |
| CV2 | 5′-TCTTCCAATCACGCTTCTGC-3′ | PCR, Seq | 536–556 |
| CV3 | 5′-TGGTGACCGTTGCAGAGCAG-3′ | PCR, Seq | 452–471 |
| CV4 | 5′-TGGGCGGTGGACATGATGAG-3′ | PCR, Seq | 1525–1544 |
| CV1-1 | 5′-GAGGATCTGGCCAAGATGGCTG-3′ | Seq | 1674–1695 |
| CV1-2 | 5′-AGGACGAACACCTCACCTCCAG-3′ | Seq | 213–234 |
| CV2-1 | 5′-GCAGCGGGCACCCAAATACCAC-3′ | Seq | 279–300 |
| CV2-2 | 5′-ACGTATCCAAGGAGGCGTTACC-3′ | Seq | 1718–1739 |
| CV3-1 | 5′-AGACTAAAGGTGGAACTGTACC-3′ | Seq | 770–791 |
| CV3-2 | 5′-TTGTACATACATGGTTACACGG-3′ | Seq | 1083–1104 |
| CV4-1 | 5′-TGTGGACCACGTAGGCCTCGGC-3′ | Seq | 1146–1167 |
| CV4-2 | 5′-TGGTAATCAGAATACTGCGGGC-3′ | Seq | 799–820 |
| MCV1 | 5′-GCTGAACTTTTGAAAGTGAGCGGG-3′ | PCR-RFLP | 508–531 |
| MCV2 | 5′-TCACACAGTCTCAGTAGATCATCCCA-3′ | PCR-RFLP | 724–749 |
The relative positions of these oligonucleotide primers are indicated in Fig. 1.
Seq, primers used for DNA sequencing to determine the complete genome of PCV-2.
Nucleotide sequencing and sequence and phylogenetic analyses.
The PCR products of the expected sizes were purified by electrophoresis on a 1% agarose gel, followed by extraction with a Geneclean Kit (Bio 101, Inc., La Jolla, Calif.). Both strands were sequenced with a variety of sequencing primers (Table 2) with an ABI automated DNA sequencer at the Virginia Polytechnic Institute and State University DNA Sequencing Facility. The sequences of the primers used to sequence the complete genome of PCV-2 are listed in Table 2, and their relative positions in the circular genome are indicated in Fig. 1. The sequences were compiled and analyzed with the MacVector program (Oxford Molecular Ltd., Beaverton, Oreg.). The percentages of sequence identity among different PCV isolates were determined with the Clustal alignment program in the MacVector package. Sequence alignments were performed with the ALIGN program in the MacVector package. Phylogenetic analyses were conducted with the aid of the PAUP program (from David L. Swofford, Smithsonian Institution, Washington, D.C., and distributed by Sinauer Associates, Inc., Sunderland, Mass.). The branch-and-bound searching and midpoint rooting options were used to produce a consensus tree.
Development of a PCR-RFLP assay.
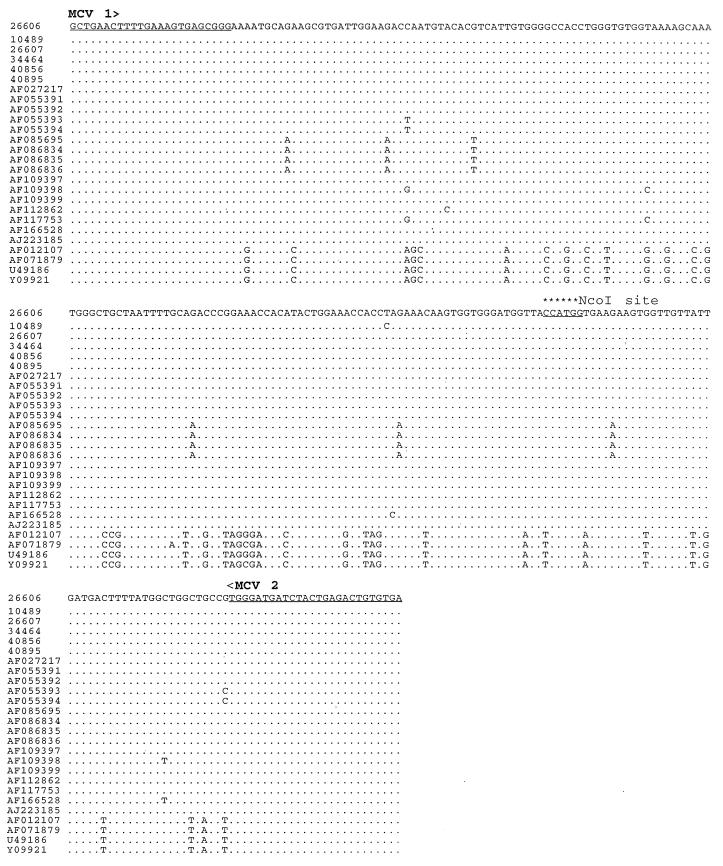
A PCR-RFLP assay was developed to differentiate between strains of PCV-1 and PCV-2 infecting the pigs. Briefly, the complete sequences of the six PCV-2 isolates from this study and the complete sequences of all other PCV sequences available in GenBank (those of both PCV-1 and PCV-2) were aligned with the Clustal program (data not shown). On the basis of this alignment, a set of conserved PCR primers (primers MCV1 and MCV2; Table 2) was designed to amplify a fragment of 243 bp from samples that contained either PCV-1 or PCV-2, or both. The sequences of the two PCR primers chosen from the sequences of PCV-1 and PCV-2 isolates are identical for all known PCV-1 and PCV-2 isolates including the six PCV-2 isolates sequenced in this study (Fig. 2). The PCR consisted of 37 cycles of denaturation at 94°C for 1 min, annealing at 56°C for 1 min, and extension at 72°C for 1.5 min. The amplified PCR products were subsequently digested with a unique restriction enzyme, NcoI, which is present in all PCV-2 isolates but not in PCV-1 isolates (Fig. 2). The digested PCR products are separated on a 2% agarose gel for RFLP analysis.
FIG. 2.
Nucleotide sequence alignment of the region amplified in the PCR-RFLP assay. The regions from which the consensus PCR primers (MCV1 and MCV2) were chosen are underlined. The unique NcoI restriction enzyme site that is present in all PCV-2 isolates is indicated by asterisks. The sequence of PCV-2 isolate 26606 from this study is shown on top, and only differences from that sequence are indicated for the other isolates. The sequences used in the alignment are cited in the text.
Nucleotide sequence accession numbers.
The complete genomic sequences of the six PCV-2 isolates reported in this paper have been deposited with the GenBank database under accession numbers AF264038, AF264039, AF264040, AF264041, AF264042, and AF264043.
RESULTS
Genetic characterization of PCV-2 isolates from pigs with PMWS in different geographic regions.
To determine the extent of genetic heterogeneity among PCV-2 isolates, the complete genome of PCV-2 was amplified and sequenced from one pig with PMWS in Canada (isolate 34464) and five pigs with PMWS in the United States: two from Utah (isolates 26606 and 26607), one from Missouri (isolate 40856), one from Iowa (isolate 40895), and one from Illinois (isolate 10489). The pigs with PMWS included in this study possessed clinical signs consistent with PMWS (Table 1) and were confirmed to be positive for PCV-2 antigen by IHC (data not shown). All six pigs with PMWS included in the study were negative for swine influenza virus, but four of the six pigs were found to be positive for porcine reproductive and respiratory syndrome virus (PRRSV) antigen (Table 1).
The lengths of the genomic DNAs of the PCV-1 isolates ranged from 1,758 to 1,760 bp. Sequence analyses of the complete genomes of six PCV-2 isolates from this study showed that, like all other PCV-2 isolates, the complete genome of each of the six PCV-2 isolates is 1,768 bp in length. All the PCV-2 isolates sequenced are closely related to each other, displaying 95 to 99% nucleotide sequence identities (Table 3). Two French PCV-2 isolates, isolates AF055393 and AF055394, displayed the most sequence divergence from the other PCV-2 isolates, with the identities ranging from 95 to 96%. Similarly, the four PCV-1 isolates sequenced thus far (isolates AF071879, Y09921, U49186, and AF012107) are closely related to each other, and their entire genomes share 98 to 99% nucleotide sequence identity (Table 3). Moreover, the nucleotide sequence identity between the entire genomes of PCV-1 and PCV-2 is only about 75 to 77%.
TABLE 3.
Pairwise comparison of the complete genomic and putative capsid gene (ORF2) sequences of PCV-1 and PCV-2
| Isolate | % Sequence identitya
|
|||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 26606 | 10489 | 26607 | 40895 | 34464 | 40856 | AF085695 | AF086834 | AF086835 | AF086836 | AF109398 | AF109399 | AF112862 | AF117753 | AF027217 | AF055391 | AF055392 | AF055393 | AF055394 | AF109397 | AF166528 | AJ223185 | AF012107 | AF071879 | U49186 | YO9921 | |
| 26606 | 98/96 | 99/99 | 98/95 | 96/95 | 99/98 | 98/97 | 98/96 | 98/97 | 98/97 | 93/93 | 94/93 | 97/95 | 93/93 | 98/95 | 98/95 | 98/96 | 92/92 | 92/92 | 97/95 | 95/95 | 98/96 | 66/66 | 65/64 | 65/65 | 65/65 | |
| 10489 | 99 | 98/97 | 99/98 | 97/97 | 98/96 | 98/98 | 99/100 | 99/98 | 98/98 | 94/94 | 95/95 | 98/98 | 93/93 | 99/98 | 99/98 | 98/98 | 92/93 | 92/93 | 99/98 | 96/98 | 99/99 | 67/68 | 66/66 | 66/66 | 66/66 | |
| 26607 | 99 | 99 | 98/96 | 96/95 | 99/98 | 98/97 | 98/97 | 99/97 | 98/97 | 93/93 | 94/93 | 97/95 | 93/93 | 98/96 | 98/96 | 98/97 | 92/92 | 92/92 | 98/96 | 95/95 | 98/96 | 66/66 | 65/64 | 66/65 | 66/65 | |
| 40895 | 99 | 99 | 99 | 97/97 | 98/95 | 98/96 | 99/98 | 98/97 | 98/96 | 94/95 | 95/95 | 98/96 | 93/93 | 99/99 | 99/99 | 98/96 | 92/93 | 92/93 | 99/98 | 96/97 | 99/99 | 66/67 | 66/65 | 66/66 | 66/66 | |
| 34464 | 98 | 98 | 98 | 98 | 96/95 | 96/95 | 97/97 | 97/96 | 96/95 | 94/93 | 97/97 | 96/95 | 93/92 | 97/96 | 97/96 | 96/95 | 92/91 | 92/91 | 97/96 | 96/96 | 97/97 | 66/67 | 65/65 | 65/66 | 65/66 | |
| 40856 | 99 | 99 | 99 | 98 | 98 | 98/97 | 98/96 | 98/97 | 98/97 | 93/93 | 94/93 | 97/95 | 92/92 | 98/95 | 98/95 | 98/96 | 92/92 | 92/92 | 97/95 | 95/95 | 98/96 | 66/66 | 65/63 | 65/64 | 65/64 | |
| AF085695 | 98 | 98 | 98 | 98 | 97 | 98 | 99/98 | 99/99 | 100/100 | 94/94 | 94/93 | 97/96 | 93/93 | 98/96 | 98/96 | 99/99 | 92/93 | 92/93 | 98/96 | 96/96 | 98/97 | 66/67 | 66/65 | 66/66 | 66/66 | |
| AF086834 | 98 | 98 | 98 | 98 | 97 | 98 | 99 | 99/98 | 99/98 | 94/94 | 95/95 | 98/98 | 93/93 | 99/98 | 99/98 | 99/98 | 92/93 | 93/93 | 99/98 | 96/98 | 99/99 | 67/68 | 66/66 | 66/66 | 66/66 | |
| AF086835 | 98 | 98 | 98 | 98 | 97 | 98 | 99 | 99 | 99/99 | 94/95 | 94/93 | 98/96 | 93/93 | 98/97 | 98/97 | 99/99 | 93/93 | 93/93 | 98/97 | 96/97 | 98/98 | 67/67 | 66/65 | 66/66 | 66/66 | |
| AF086836 | 98 | 98 | 98 | 98 | 97 | 98 | 99 | 98 | 99 | 94/94 | 94/93 | 97/96 | 93/93 | 98/96 | 98/96 | 99/99 | 92/93 | 92/93 | 98/96 | 96/96 | 98/97 | 66/67 | 66/65 | 66/66 | 66/66 | |
| AF109398 | 96 | 96 | 96 | 96 | 96 | 96 | 95 | 95 | 95 | 95 | 93/92 | 94/94 | 97/96 | 94/95 | 94/95 | 94/94 | 93/93 | 93/93 | 95/96 | 93/94 | 94/95 | 67/67 | 66/65 | 66/66 | 66/66 | |
| AF109399 | 97 | 97 | 97 | 97 | 98 | 97 | 96 | 96 | 96 | 96 | 96 | 94/93 | 93/91 | 95/94 | 95/94 | 94/93 | 91/90 | 91/90 | 95/94 | 94/94 | 95/95 | 66/65 | 65/63 | 65/63 | 65/63 | |
| AF112862 | 98 | 99 | 98 | 99 | 98 | 98 | 98 | 98 | 98 | 97 | 96 | 97 | 93/92 | 98/96 | 98/96 | 98/96 | 92/92 | 92/92 | 98/97 | 95/96 | 98/97 | 67/67 | 66/65 | 66/66 | 66/66 | |
| AF117753 | 96 | 96 | 96 | 96 | 96 | 95 | 95 | 95 | 95 | 95 | 97 | 96 | 96 | 93/93 | 93/93 | 93/93 | 91/91 | 92/91 | 93/93 | 93/93 | 93/93 | 66/66 | 65/64 | 65/65 | 65/65 | |
| AF027217 | 99 | 99 | 99 | 99 | 98 | 99 | 98 | 98 | 98 | 98 | 96 | 97 | 99 | 96 | 99/99 | 98/96 | 93/93 | 93/93 | 99/98 | 96/97 | 99/99 | 66/67 | 66/65 | 66/66 | 66/66 | |
| AF055391 | 99 | 99 | 99 | 99 | 98 | 99 | 98 | 98 | 98 | 98 | 96 | 97 | 99 | 96 | 99 | 98/96 | 93/93 | 93/93 | 99/98 | 96/97 | 99/99 | 66/67 | 66/65 | 66/66 | 66/66 | |
| AF055392 | 99 | 99 | 99 | 99 | 98 | 99 | 99 | 98 | 98 | 98 | 96 | 97 | 99 | 96 | 99 | 99 | 93/93 | 93/93 | 98/96 | 96/96 | 98/97 | 66/67 | 66/65 | 66/66 | 66/66 | |
| AF055393 | 95 | 95 | 95 | 95 | 95 | 95 | 95 | 95 | 95 | 95 | 95 | 95 | 95 | 95 | 96 | 96 | 96 | 99/99 | 92/93 | 93/93 | 92/93 | 67/68 | 66/66 | 66/66 | 66/66 | |
| AF055394 | 95 | 96 | 95 | 96 | 95 | 95 | 95 | 95 | 95 | 95 | 95 | 95 | 95 | 95 | 96 | 96 | 96 | 99 | 92/92 | 93/93 | 92/93 | 67/68 | 66/66 | 66/66 | 66/66 | |
| AF109397 | 99 | 99 | 99 | 99 | 98 | 99 | 98 | 98 | 98 | 98 | 97 | 97 | 99 | 96 | 99 | 99 | 99 | 95 | 95 | 96/97 | 99/99 | 67/68 | 66/66 | 66/66 | 66/66 | |
| AF166528 | 97 | 97 | 97 | 97 | 97 | 97 | 96 | 97 | 96 | 97 | 96 | 96 | 97 | 95 | 97 | 97 | 97 | 96 | 96 | 97 | 96/98 | 66/67 | 65/65 | 65/66 | 65/66 | |
| AJ223185 | 99 | 99 | 99 | 99 | 98 | 98 | 98 | 98 | 98 | 98 | 96 | 97 | 99 | 96 | 99 | 99 | 99 | 95 | 95 | 99 | 97 | 66/68 | 66/66 | 66/66 | 66/66 | |
| AF012107 | 76 | 77 | 76 | 76 | 76 | 76 | 76 | 76 | 76 | 76 | 76 | 76 | 76 | 76 | 76 | 76 | 76 | 77 | 77 | 77 | 76 | 76 | 97/95 | 97/96 | 97/94 | |
| AF071879 | 76 | 76 | 76 | 76 | 76 | 76 | 76 | 75 | 76 | 76 | 76 | 76 | 76 | 75 | 76 | 76 | 76 | 76 | 76 | 76 | 76 | 76 | 98 | 99/98 | 98/95 | |
| U49186 | 76 | 76 | 76 | 76 | 76 | 76 | 76 | 76 | 76 | 76 | 76 | 76 | 76 | 75 | 76 | 76 | 76 | 76 | 77 | 76 | 76 | 76 | 98 | 99 | 98/96 | |
| YO9921 | 76 | 76 | 76 | 76 | 76 | 76 | 76 | 76 | 76 | 76 | 76 | 76 | 76 | 75 | 76 | 76 | 76 | 76 | 76 | 76 | 76 | 76 | 98 | 99 | 99 | |
The values in the table are percent identity of amino acid or nucleotide sequences. The percent nucleotide sequence identities of the complete genomes are presented in the lower left half. The four PCV-1 isolates (AF012107, AF071879, U49186, YO9921) are highlighted in boldface type. The nucleotide/amino acid sequence identities of the putative capsid (ORF2) gene are shown at the upper right.
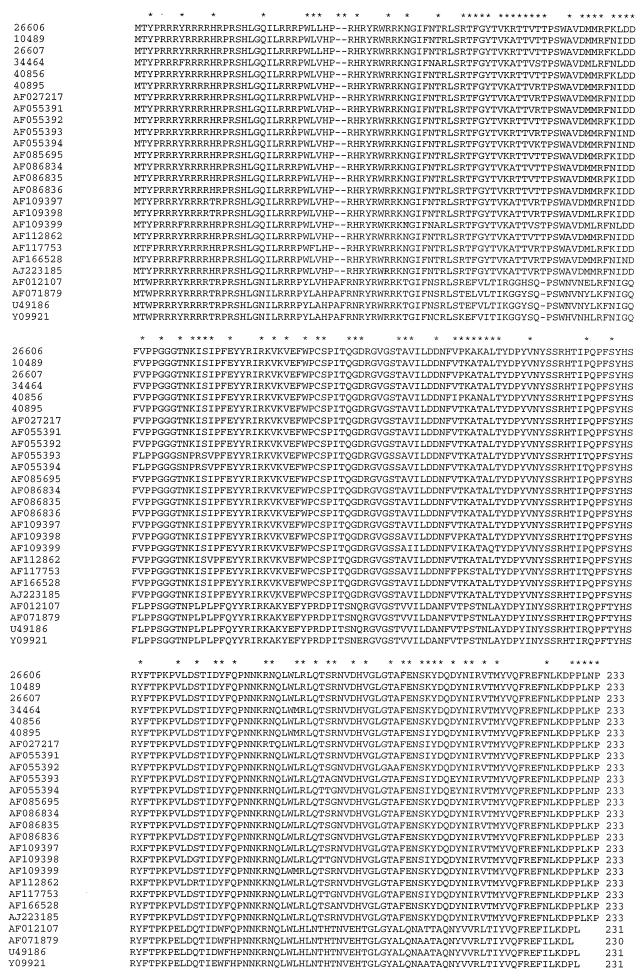
ORF2 of PCV is believed to code for the putative capsid protein (31, 33, 42). Sequence analysis indicated that the ORF2 genes of PCV-1 isolates encode a protein of 230 to 231 amino acid residues, whereas the ORF2 genes of PCV-2 isolates encodes a protein of 233 amino acid residues (Fig. 3). Pairwise sequence comparisons revealed that the ORF2 genes of all PCV-2 isolates shared 91 to 100% nucleotide sequence identity and 90 to 100% amino acid sequence identity (Table 3). The two French isolates, isolates AF055393 and AF055394, have only about 90 to 93% nucleotide sequence identity with the other PCV-2 isolates (Table 3). The ORF2 genes of the four PCV-1 isolates share 97 to 99% nucleotide sequence identity and 94 to 98% amino acid sequence identity. Between the ORF2 genes of PCV-1 and PCV-2 isolates, there exists only 65 to 67% nucleotide sequence identity and 63 to 68% amino acid sequence identity (Table 3). However, sequence analysis revealed that the N-terminal region of ORF2 is very rich in basic amino acid residues (arginine and lysine) and is highly conserved among PCV isolates, both PCV-1 and PCV-2 isolates (Fig. 3).
FIG. 3.
Amino acid sequence alignment of the putative capsid protein (ORF2) of PCV-1 and PCV-2 isolates sequenced thus far. Deletions are indicated by hyphens. Amino acid sequence differences are indicated with asterisks above the alignment. The sequences used in the alignment are cited in the text.
Phylogenetic analysis of PCV-1 and PCV-2 isolates from different geographic regions worldwide.
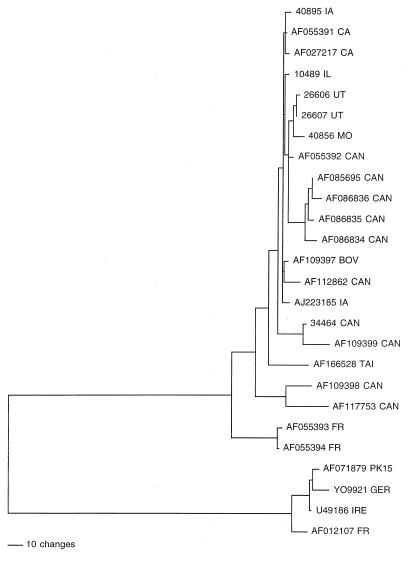
To gain a better understanding of the genetic relationship and evolution of PCV, phylogenetic analyses were performed on the basis of the complete genomic sequences of 26 PCV isolates (both PCV-1 and PCV-2) worldwide, including the six North American PCV-2 isolates sequenced in this study (Fig. 4). These sequences either were published (18, 19, 31, 32, 33, 36, 37, 39, 41, 42, 54) or are available in GenBank (Table 1). Phylogenetic analysis confirmed that two distinct genotypes of PCV exist: PCV-1 and PCV-2 (Fig. 4). All 22 PCV-2 isolates are clustered together and form one distinct branch. Similarly, all four PCV-1 isolates are closely related and form another branch. Within the major genotype of PCV-2, a few minor branches were identified, and some of these minor branches appear to be associated with the geographic origins of the isolates. All the PCV-2 isolates that were from different geographic regions of the United States and that were sequenced in this study are grouped closely with other U.S. and most of the Canadian PCV-2 isolates (Fig. 4). Canadian isolate 34464, sequenced in this study, is closely related to another Canadian isolate, 109399, but is less related to the U.S. and other Canadian isolates. Two other Canadian isolates, AF109398 and AF117753, form a distinguishable branch and are distantly related to other Canadian and U.S. isolates. An isolate of PCV-2 from Taiwan, AF166526, is clustered with the North American PCV-2 isolates but forms a single minor branch. The two French isolates of PCV-2, AF055393 and AF055394, are closely related to each other but diverge the most from North American PCV-2 isolates. Interestingly, a bovine isolate of circovirus is most closely related to the U.S. isolates of PCV-2.
FIG. 4.
Phylogenetic tree based on the complete genomic nucleotide sequences of all PCV isolates. The tree was constructed with the aid of the PAUP program. Branch-and-bound searching and midpoint rooting options were used to produce a consensus tree. A scale bar that represents the numbers of character-state changes is shown. Branch lengths are proportional to the numbers of character-state changes. The geographic locations of the isolates are also indicated with the usual state abbreviations and the following country abbreviations: CAN, Canada; TAI, Taiwan; FR, France; GER, Germany; IRE, Ireland. BOV, bovine.
Development of a PCR-RFLP assay to diagnose PCV-2 infection and to differentiate between PCV-1 and PCV-2 infections.
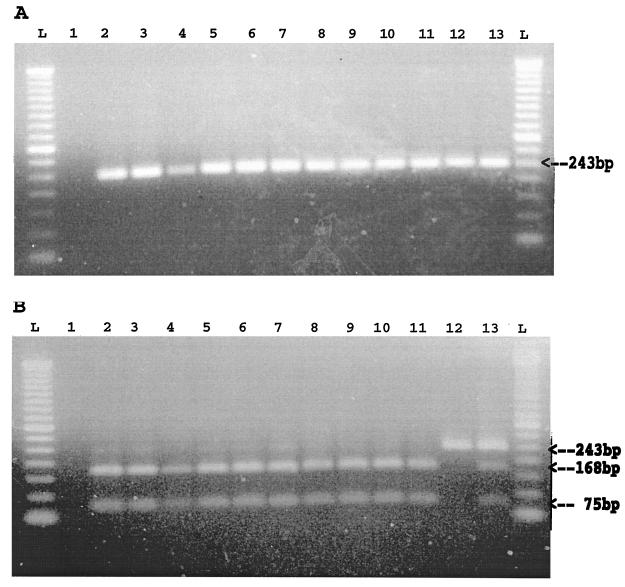
On the basis of the sequence alignments of all PCV-1 and PCV-2 isolates sequenced thus far, a set of consensus PCR primers was selected from two conserved regions of the PCV genome to amplify a fragment of 243 bp for both PCV-1 and PCV-2 isolates (Fig. 5A). To test the feasibility of using these primers for the detection of both PCV-1 and PCV-2 isolates in clinical samples, DNA was extracted from tissue samples from the six pigs with PMWS. The PCV-2 genomic sequences from these samples have been determined. DNA was also extracted from tissue samples from four additional pigs with PMWS from Iowa (Table 1), but the PCV-2 sequences in samples from these four pigs with PMWS have not been determined. DNA extracted from the PK15 cell line (ATCC CCL-33) was used as the source of PCV-1 DNA. DNA extracted from a sample of liver tissue collected from a specific-pathogen-free pig was used as a negative control. We were able to amplify an expected fragment of the PCV genome from tissue samples from all 10 pigs with PMWS as well as from the PCV-1-contaminated PK15 cells. By using an unique restriction enzyme site (NcoI) that is present only in the sequences of PCV-2 isolates (Fig. 2), a PCR-RFLP assay was developed to differentiate between infections with PCV-1 and PCV-2. After digestion of the PCR products with NcoI, the resulting RFLP patterns revealed that all products amplified from PCV-2 isolates produced two fragments of 168 and 75 bp, whereas the PCR product amplified from PCV-1 produced only the undigested fragment of 243 bp (Fig. 5). DNA extracted from a sample that contained both PCV-2 (from pig 40860) and PCV-1 (PK15 cells) was also subjected to PCR amplification. After digestion with the NcoI restriction enzyme, the PCR product amplified from the sample with both PCV-1 and PCV-2 produced three fragments of 243, 168, and 75 bp, respectively (Fig. 5B). Thus, this PCR-RFLP assay is able to detect PCV-1 and PCV-2 in clinical samples with potential dual infection with PCV-1 and PCV-2.
FIG. 5.
Detection and differentiation of PCV infections by a PCR-RFLP assay. (A) Results of PCR amplification of a 243-bp fragment from tissue samples that contained PCV isolates (both PCV-1 and PCV-2) but not from a negative control liver tissue sample (lane 1). (B) Results of RFLP analysis of the PCR products. Lane L, 50-bp DNA ladder; lane 1, a sample of liver tissue from a control specific-pathogen-free pig; lanes 2 to 11, tissue samples from 10 pigs with PMWS, respectively; lane 12, PK15 cells containing PCV-1; lane 13, a sample containing both PCV-1 and PCV-2. The expected PCR fragment (A) and three RFLP fragments of 243, 168, and 75 bp, respectively (B), are indicated with arrows.
DISCUSSION
PMWS is a new and unique disease of swine. Since the first recognition of the disease in 1991 (10, 20), PMWS has emerged to be an economically important global disease of swine (2, 3, 4, 6, 13, 18, 24, 26, 27, 32, 37, 39, 40, 44, 48, 49). The clinical and pathological presentations and etiology of PMWS are very complicated (6, 10, 16, 20, 21, 22, 46, 47). Many known swine pathogens such as PRRSV, swine influenza virus, hemagglutinating encephalomyocarditis virus, the bacterium that causes porcine proliferative enteropathy, Mycoplasma hyopneumoniae, Haemophilus parasuis, the bacterium that causes postweaning colibacillosis, etc., could cause postweaning wasting of pigs (21). However, increasingly, data indicate that PCV-2 is the causative agent of PMWS (2, 3, 4, 6, 13, 18, 37, 39). Experimental inoculation of conventional pigs with homogenates of tissue from pigs with clinical PMWS produced PMWS-like lesions, and PCV-2 DNA and antibody to PCV-2 were detected in the inoculated pigs (7). Ellis et al. (14) experimentally inoculated neonatal gnotobiotic piglets with filtered tissue culture materials and homogenates of tissue from PMWS-affected pigs. The inoculated gnotobiotic piglets developed lesions typical of PMWS, but the study was complicated by the detection of porcine parvovirus (PPV) in inoculated piglets. In fact, coinfection with PPV and PCV in pigs with naturally acquired PMWS has been reported (16). It has also been shown that PCV-2 alone induced PMWS lesions in colostrum-deprived conventional pigs but that concurrent infection with PPV increased the severity of the lesions (5, 25), suggesting that PMWS is a complex disease syndrome and that multiple factors may be involved in the pathogenesis of PMWS. It has been suspected that some of the clinical signs and pathological lesions attributable to PRRSV may actually be induced by PCV-2 as a result of PCV-2 infection or coinfection (15, 27). Synergism between a circovirus (CAV) and a reovirus was observed following dual infection of chickens by a natural route (34). In the present study, PCV-2 was readily detectable from pigs with PMWS in different regions of the North America, but PRRSV antigen was also detected in most of the pigs with PMWS (Table 1). The etiological significance of PCV-2 in PMWS and its interrelationship with PRRSV, PPV, or other agents need to be further studied.
The extent of genetic variation of PCV-2 isolated from different geographic regions of the United States is not known since only a few PCV-2 isolates from the United States have been genetically characterized (37, 39). In this study, we genetically characterized six North American isolates of PCV-2 (one Canadian isolate and five U.S. isolates) from pigs with PMWS from different geographic regions. Sequence analysis of the complete genome and of the putative capsid gene ORF2 indicated that these six North American isolates of PCV-2 are closely related to other known PCV-2 isolates worldwide. The putative capsid gene (ORF2) of PCV is highly variable, and the ORF2 genes of certain PCV-2 isolates share as little as 90% sequence identity. Despite the overall heterogeneic nature of ORF2, the N-terminal region of ORF2 among all PCVs is highly conserved and possesses a high percentage of basic amino acids, suggesting that the amino-terminal region of the putative capsid protein may have DNA binding activity and may be in contact with the PCV DNA in the native virion (42). Phylogenetic analysis revealed that all PCV-2 isolates sequenced thus far form a major genotype, whereas all PCV-1 isolates are closely related and form another genotype. On the basis of phylogenetic analysis, it is evident that both PCV-1 and PCV-2 evolved from the same ancestor, but they may have undergone divergent evolution. Gibbs and Weiller (17) analyzed the genomes of circoviruses and plant nanoviruses and showed that circoviruses most likely evolved from a plant nanovirus. It is believed that the plant nanovirus switched hosts to infect a vertebrate and then recombined with a vertebrate-infecting virus (17). Within the major genotype of PCV-2, several minor branches were also identified. The two French PCV-2 isolates (isolates AF055393 and AF055394) diverge the most from all other PCV-2 isolates. The clinical significance of this divergence is not known. LeCann et al. (29) reported that a PCV-1-like virus was isolated from pigs with wasting disease in France, but they failed to experimentally reproduce the disease with this isolate. Phylogenetically, all the U.S. PCV-2 isolates sequenced thus far are closely related. However, genetic variation was observed among the Canadian PCV-2 isolates. Two of the Canadian isolates, isolates AF109398 and AF117753, form a minor branch that is separate from other Canadian or U.S. isolates. Two other Canadian isolates (isolate 34464, sequenced in this study, and isolate AF109399) also differ phylogenetically from the U.S. and other Canadian PCV-2 isolates. A PCV-2 isolate from Taiwan (isolate AF116528) also forms a distinguishable minor branch. The origin of the bovine circovirus isolate is not known, but its close genetic relatedness to PCV-2 suggested that the bovine circovirus may be of swine origin and that cross-species infection of PCV between bovines and swine is possible. These data suggest that although the genome of PCV-2 is relatively stable in general, minor genetic differences do exist among PCV-2 isolates from different geographic regions. This observed difference might have important implications for the diagnosis of PCV-2 infection by nucleic acid-based assays such as PCR. Genetic characterization of additional PCV-2 isolates from other geographic regions worldwide is warranted.
Since PCV-1 is nonpathogenic and widespread in pig populations (6, 11, 12, 22, 23, 51, 52, 53), a test is needed to differentiate between infections with PCV-1 and PCV-2. In addition, since PCV antibody has been detected in humans (53), a major and growing concern is the inadvertent transmission of PCV from pig organs to human recipients during xenotransplantation. In xenotransplantation, there is zero tolerance for circovirus infection, regardless of its pathogenic potential, since nonpathogenic PCV-1 may become pathogenic in immunocomprised xenograft recipients. Therefore, sensitive and easy-to-perform assays are needed to screen for both PCV-1 and PCV-2 infection in xenograft donor pigs. Several techniques such as PCR (19, 27, 28, 39, 40, 45), IHC (35, 39, 46), and in situ hybridization (9, 35, 39, 46) are available for the detection of PCV-2 infection; however, the ability of these tests to detect PCV-2 isolates from different geographic regions is not known. The data from this study suggest that the genomic sequences of PCV-2 isolates from different geographic regions vary to some extent. Thus, a universal and more sensitive PCR assay that can detect PCV isolates from various geographic regions is needed. On the basis of genetic analyses of all PCV isolates, we developed a universal PCR-RFLP assay for the diagnosis of PCV-2 infection and differentiation between infections with PCV-1 and PCV-2 in pigs. This assay uses a pair of PCR primers selected from two conserved regions of the PCV-1 and PCV-2 genomes and a unique NcoI restriction enzyme site that exists only in PCV-2 isolates. The feasibility of this PCR-RFLP assay to detect PCV-2 and to differentiate between PCV-2 and PCV-1 was validated by using clinical samples from pigs from different geographic regions with confirmed cases of PMWS and a sample that intentionally contained both PCV-1 and PCV-2. Our results indicate that this PCR-RFLP assay is accurate and fast in diagnosing PCV-2 infection in pigs with PMWS from different geographic regions, in differentiating PCV-1 and PCV-2 infections, and in detecting dual infection with PCV-1 and PCV-2. This universal PCR-RFLP assay should help clinicians diagnose cases of PMWS associated with PCV-2 infection in different geographic regions of the world and should also be useful for the screening of xenograft donor pigs to ascertain that they are free of circovirus infection.
ACKNOWLEDGMENTS
This project was supported by an Innovation Grant Award from Fort Dodge Animal Health, Inc., Fort Dodge, Iowa.
We thank Lee Weigt of Virginia Tech's DNA Sequencing Facility for assistance with sequencing and sequence analyses, Jill Sible of Department of Biology at Virginia Tech for critical review of the manuscript, and Denis Guenettee and Crystal Gilbert for editorial assistance.
REFERENCES
- 1.Allan G M, McNeilly F, Cassidy J P, Reilly G A, Adair B, Ellis W A, McNulty M S. Pathogenesis of porcine circovirus; experimental infections of colostrum deprived piglets and examination of pig foetal material. Vet Microbiol. 1995;44:49–64. doi: 10.1016/0378-1135(94)00136-k. [DOI] [PubMed] [Google Scholar]
- 2.Allan G M, Meehan B, Todd D, Kennedy S, McNeilly F, Ellis J, Clark E G, Harding J, Espuna E, Botner A, Charreyre C. Novel porcine circoviruses from pigs with wasting disease syndromes. Vet Rec. 1998;142:467–468. [PubMed] [Google Scholar]
- 3.Allan G M, McNeilly F, Kennedy S, Daft B, Clarke E G, Ellis J A, Haines D M, Meehan B M, Adair B M. Isolation of porcine circovirus-like viruses from pigs with a wasting disease in the USA and Europe. J Vet Diagn Invest. 1998;10:3–10. doi: 10.1177/104063879801000102. [DOI] [PubMed] [Google Scholar]
- 4.Allan G M, McNeilly F, Meehan B M, Kennedy S, Mackie D P, Ellis J A, Clark E G, Espuna E, Saubi N, Riera P, Botner A, Charreyre C E. Isolation and characterisation of circoviruses from pigs with wasting syndromes in Spain, Denmark and Northern Ireland. Vet Microbiol. 1999;66:115–123. doi: 10.1016/s0378-1135(99)00004-8. [DOI] [PubMed] [Google Scholar]
- 5.Allan G M, Kennedy S, McNeilly F, Foster J C, Ellis J A, Krakowka S J, Meehan B M, Adair B M. Experimental reproduction of severe wasting disease by co-infection of pigs with porcine circovirus and porcine parvovirus. J Comp Pathol. 1999;121:1–11. doi: 10.1053/jcpa.1998.0295. [DOI] [PubMed] [Google Scholar]
- 6.Allan G M, Ellis J A. Porcine circoviruses: a review. J Vet Diagn Invest. 2000;12:3–14. doi: 10.1177/104063870001200102. [DOI] [PubMed] [Google Scholar]
- 7.Balasch M, Segales J, Rosell C, Domingo M, Mankertz A, Urniza A, Plana-Duran J. Experimental inoculation of conventional pigs with tissue homogenates from pigs with post-weaning multisystemic wasting syndrome. J Comp Pathol. 1999;121:139–148. doi: 10.1053/jcpa.1999.0310. [DOI] [PubMed] [Google Scholar]
- 8.Bassami M R, Berryman D, Wilcox G E, Raidal S R. Psittacine beak and feather disease virus nucleotide sequence analysis and its relationship to porcine circovirus, plant circoviruses, and chicken anaemia virus. Virology. 1998;249:453–459. doi: 10.1006/viro.1998.9324. [DOI] [PubMed] [Google Scholar]
- 9.Choi C, Chae C. In-situ hybridization for the detection of porcine circovirus in pigs with postweaning multisystemic wasting syndrome. J Comp Pathol. 1999;121:265–270. doi: 10.1053/jcpa.1999.0315. [DOI] [PubMed] [Google Scholar]
- 10.Clark E G. Proceedings of American Association of Swine Practitioners. Quebec City, Canada: American Association of Swine Practitioners; 1997. Postweaning multisystemic wasting syndrome; pp. 499–501. [Google Scholar]
- 11.Dulac G C, Afshar A. Porcine circovirus antigens in PK-15 cell line (ATCC CCL-33) and evidence of antibodies to circovirus in Canadian pigs. Can J Vet Res. 1989;53:431–433. [PMC free article] [PubMed] [Google Scholar]
- 12.Edwards S, Sands J J. Evidence of circovirus infection in British pigs. Vet Rec. 1994;134:680–681. doi: 10.1136/vr.134.26.680. [DOI] [PubMed] [Google Scholar]
- 13.Ellis J, Hassard L, Clark E, Harding J, Allan G, Willson P, Strakappe J, Martin K, McNeilly F, Meehan B, Todd D, Haines D. Isolation of circovirus from lesions of pigs with postweaning multisystemic wasting syndrome. Can Vet J. 1998;39:44–51. [PMC free article] [PubMed] [Google Scholar]
- 14.Ellis J, Krakowka S, Lairmore M, Haines D, Bratanich A, Clark E, Allan G, Konoby C, Hassard L, Meehan B, Martin K, Harding J, Kennedy S, McNeilly F. Reproduction of lesions of postweaning multisystemic wasting syndrome in gnotobiotic piglets. J Vet Diagn Invest. 1999;11:3–14. doi: 10.1177/104063879901100101. [DOI] [PubMed] [Google Scholar]
- 15.Ellis J A. “The clinical scope of porcine reproductive and respiratory syndrome virus infection has expanded since 1987”: an alternative perspective. Vet Pathol. 1999;36:262–264. doi: 10.1354/vp.36-3-262. [DOI] [PubMed] [Google Scholar]
- 16.Ellis J A, Bratanich A, Clark E G, Allan G, Meehan B, Haines D M, Harding J, West K H, Krakowka S, Konoby C, Hassard L, Martin K, McNeilly F. Coinfection by porcine circoviruses and porcine parvovirus in pigs with naturally acquired postweaning multisystemic wasting syndrome. J Vet Diagn Invest. 2000;12:21–27. doi: 10.1177/104063870001200104. [DOI] [PubMed] [Google Scholar]
- 17.Gibbs M J, Weiller G F. Evidence that a plant virus switched hosts to infect a vertebrate and then recombined with a vertebrate-infecting virus. Proc Natl Acad Sci USA. 1999;96:8022–8027. doi: 10.1073/pnas.96.14.8022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hamel A L, Lin L L, Nayar G P. Nucleotide sequence of porcine circovirus associated with postweaning multisystemic wasting syndrome in pigs. J Virol. 1998;72:5262–5267. doi: 10.1128/jvi.72.6.5262-5267.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hamel A L, Lin L L, Sachvie C, Grudeski E, Nayar G P. PCR detection and characterization of type-2 porcine circovirus. Can J Vet Res. 2000;64:44–52. [PMC free article] [PubMed] [Google Scholar]
- 20.Harding J C. Proceedings of American Association of Swine Practitioners. Quebec City, Canada: American Association of Swine Practitioners; 1997. Postweaning multisystemic wasting syndrome (PMWS): preliminary epidemiology and clinical presentation; p. 503. [Google Scholar]
- 21.Harding J C, Clark E G. Recognizing and diagnosing postweaning multisystemic wasting syndrome (PMWS) Swine Health Prod. 1997;5:201–203. [Google Scholar]
- 22.Hines R K, Lukert P D. Porcine circovirus: a serological survey of swine in the United States. Swine Health Prod. 1995;3:71–73. [Google Scholar]
- 23.Hines R K, Lukert P D, Dau D, Case D. Some effects of porcine circovirus on performance. Swine Health Prod. 1995;3:251–255. [Google Scholar]
- 24.Kennedy S, Allan G, McNeilly F, Adair B M, Hughes A, Spillane P. Porcine circovirus infection in Northern Ireland. Vet Rec. 1998;142:495–496. [PubMed] [Google Scholar]
- 25.Kennedy S, Moffett D, McNeilly F, Meehan B, Ellis J, Krakowka S, Allan G M. Reproduction of lesions of postweaning multisystemic wasting syndrome by infection of conventional pigs with porcine circovirus type 2 alone or in combination with porcine parvovirus. J Comp Pathol. 2000;122:9–24. doi: 10.1053/jcpa.1999.0337. [DOI] [PubMed] [Google Scholar]
- 26.Kiupel M, Stevenson G W, Mittal S K, Clark E G, Haines D M. Circovirus-like viral associated disease in weaned pigs in Indiana. Vet Pathol. 1998;35:303–307. doi: 10.1177/030098589803500411. [DOI] [PubMed] [Google Scholar]
- 27.Larochelle R, Morin M, Antaya M, Magar R. Identification and incidence of porcine circovirus in routine field cases in Quebec as determined by PCR. Vet Rec. 1999;145:140–142. doi: 10.1136/vr.145.5.140. [DOI] [PubMed] [Google Scholar]
- 28.Larochelle R, Antaya M, Morin M, Magar R. Typing of porcine circovirus in clinical specimens by multiplex PCR. J Virol Methods. 1999;80:69–75. doi: 10.1016/s0166-0934(99)00032-4. [DOI] [PubMed] [Google Scholar]
- 29.LeCann P, Albina E, Madec F, Cariolet R, Jestin A. Piglet wasting disease. Vet Rec. 1997;141:660. [PubMed] [Google Scholar]
- 30.Lukert P D, de Boer G F, Dale J L, Keese P, McNulty M S, Randles J W, Tisher I. The Circoviridae. In: Murphy F A, Fauquet C M, Bishop D H L, Ghabrial S A, Jarvis A W, Martelli G P, Mayo M A, Summers M D, editors. Virus taxonomy: sixth report of the International Committee on Taxonomy of Viruses. Vienna, Austria: Springer-Verlag; 1995. pp. 166–168. [Google Scholar]
- 31.Mankertz A, Persson F, Mankertz J, Blaess G, Buhk H J. Mapping and characterization of the origin of DNA replication of porcine circovirus. J Virol. 1997;71:2562–2566. doi: 10.1128/jvi.71.3.2562-2566.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mankertz A, Domingo M, Folch J M, LeCann P, Jestin A, Segales J, Chmielewicz B, Plana-Duran J, Soike D. Characterisation of PCV-2 isolates from Spain, Germany and France. Virus Res. 2000;66:65–77. doi: 10.1016/s0168-1702(99)00122-7. [DOI] [PubMed] [Google Scholar]
- 33.Mankertz J, Buhk H J, Blaess G, Mankertz A. Transcription analysis of porcine circovirus (PCV) Virus Genes. 1998;16:267–276. doi: 10.1023/a:1008022521329. [DOI] [PubMed] [Google Scholar]
- 34.McNeilly F, Smyth J A, Adair B M, McNulty M S. Synergism between chicken anemia virus (CAV) and avian reovirus following dual infection of 1-day-old chicks by a natural route. Avian Dis. 1995;39:532–537. [PubMed] [Google Scholar]
- 35.McNeilly F, Kennedy S, Moffett D, Meehan B M, Foster J C, Clarke E G, Ellis J A, Haines D M, Adair B M, Allan G M. A comparison of in situ hybridization and immunohistochemistry for the detection of a new porcine circovirus in formalin-fixed tissues from pigs with post-weaning multisystemic wasting syndrome (PMWS) J Virol Methods. 1999;80:123–128. doi: 10.1016/s0166-0934(99)00043-9. [DOI] [PubMed] [Google Scholar]
- 36.Meehan B M, Creelan J L, McNulty M S, Todd D. Sequence of porcine circovirus DNA: affinities with plant circoviruses. J Gen Virol. 1997;78:221–227. doi: 10.1099/0022-1317-78-1-221. [DOI] [PubMed] [Google Scholar]
- 37.Meehan B M, McNeilly F, Todd D, Kennedy S, Jewhurst V A, Ellis J A, Hassard L E, Clark E G, Haines D M, Allan G M. Characterization of novel circovirus DNAs associated with wasting syndromes in pigs. J Gen Virol. 1998;79:2171–2179. doi: 10.1099/0022-1317-79-9-2171. [DOI] [PubMed] [Google Scholar]
- 38.Miyata H, Tsunoda H, Kazi A, Yamada A, Khan M A, Murakami J, Kamahora T, Shiraki K, Hino S. Identification of a novel GC-rich 113-nucleotide region to complete the circular, single-stranded DNA genome of TT virus, the first human circovirus. J Virol. 1999;73:3582–3586. doi: 10.1128/jvi.73.5.3582-3586.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morozov I, Sirinarumitr T, Sorden S D, Halbur P G, Morgan M K, Yoon K J, Paul P S. Detection of a novel strain of porcine circovirus in pigs with postweaning multisystemic wasting syndrome. J Clin Microbiol. 1998;36:2535–2541. doi: 10.1128/jcm.36.9.2535-2541.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nayar G P, Hamel A, Lin L. Detection and characterization of porcine circovirus associated with postweaning multisystemic wasting syndrome in pigs. Can Vet J. 1997;38:385–386. [PMC free article] [PubMed] [Google Scholar]
- 41.Nayar G P, Hamel A L, Lin L, Sachvie C, Grudeski E, Spearman G. Evidence for circovirus in cattle with respiratory disease and from aborted bovine fetuses. Can Vet J. 1999;40:277–278. [PMC free article] [PubMed] [Google Scholar]
- 42.Niagro F D, Forsthoefel A N, Lawther R P, Kamalanathan L, Ritchie B W, Latimer K S, Lukert P D. Beak and feather disease virus and porcine circovirus genomes: intermediates between the geminiviruses and plant circoviruses. Arch Virol. 1998;143:1723–1744. doi: 10.1007/s007050050412. [DOI] [PubMed] [Google Scholar]
- 43.Nishizawa T, Okamoto H, Konishi K, Yoshizawa H, Miyakawa Y, Mayumi M. A novel DNA virus (TTV) associated with elevated transaminase levels in posttransfusion hepatitis of unknown etiology. Biochem Biophys Res Commun. 1997;241:92–97. doi: 10.1006/bbrc.1997.7765. [DOI] [PubMed] [Google Scholar]
- 44.Onuki A, Abe K, Togashi K, Kawashima K, Taneichi A, Tsunemitsu H. Detection of porcine circovirus from lesions of a pig with wasting disease in Japan. J Vet Med Sci. 1999;61:1119–1123. doi: 10.1292/jvms.61.1119. [DOI] [PubMed] [Google Scholar]
- 45.Ouardani M, Wilson L, Jette R, Montpetit C, Dea S. Multiplex PCR for detection and typing of porcine circoviruses. J Clin Microbiol. 1999;37:3917–3924. doi: 10.1128/jcm.37.12.3917-3924.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rosell C, Segales J, Plana-Duran J, Balasch M, Rodriguez-Arrioja G M, Kennedy S, Allan G M, McNeilly F, Latimer K S, Domingo D. Pathological, immunohistochemical, and in-situ hybridization studies of natural cases of postweaning multisystemic wasting syndrome (PMWS) in pigs. J Comp Pathol. 1999;120:59–78. doi: 10.1053/jcpa.1998.0258. [DOI] [PubMed] [Google Scholar]
- 47.Rosell C, Segales J, Ramos-Vara J A, Folch J M, Rodriguez-Arrioja G M, Duran C O, Balasch M, Plana-Duran J, Domingo M. Identification of porcine circovirus in tissues of pigs with porcine dermatitis and nephropathy syndrome. Vet Rec. 2000;146:40–43. doi: 10.1136/vr.146.2.40. [DOI] [PubMed] [Google Scholar]
- 48.Segales J, Sitjar M, Domingo M, Dee S, Del Pozo M, Noval R, Sacristan C, De las Heras A, Ferro A, Latimer K S. First report of postweaning multisystemic wasting syndrome in Spain. Vet Rec. 1997;141:600–601. [PubMed] [Google Scholar]
- 49.Spillane P, Kennedy S, Meehan B, Allan G. Porcine circovirus infection in the Republic of Ireland. Vet Rec. 1998;143:511–512. [PubMed] [Google Scholar]
- 50.Tischer I, Gelderblom H, Vettermann W, Koch M A. A very small porcine virus with circular single-stranded DNA. Nature. 1982;295:64–66. doi: 10.1038/295064a0. [DOI] [PubMed] [Google Scholar]
- 51.Tischer I, Mields W, Wolff D, Vagt M, Griem W. Studies on epidemiology and pathogenicity of porcine circovirus. Arch Virol. 1986;91:271–276. doi: 10.1007/BF01314286. [DOI] [PubMed] [Google Scholar]
- 52.Tischer I, Bode L, Peters D, Pociuli S, Germann B. Distribution of antibodies to porcine circovirus in swine populations of different breeding farms. Arch Virol. 1995;140:737–743. doi: 10.1007/BF01309961. [DOI] [PubMed] [Google Scholar]
- 53.Tischer I, Bode L, Apodaca J, Timm H, Peters D, Rasch R, Pociuli S, Gerike E. Presence of antibodies reacting with porcine circovirus in sera of humans, mice, and cattle. Arch Virol. 1995;140:1427–1439. doi: 10.1007/BF01322669. [DOI] [PubMed] [Google Scholar]
- 54.Todd D, Niagro F D, Ritchie B W, Curran W, Allan G M, Lukert P D, Latimer K S, Steffens W L, McNulty M S. Comparison of three animal viruses with circular single-stranded DNA genomes. Arch Virol. 1991;117:129–135. doi: 10.1007/BF01310498. [DOI] [PubMed] [Google Scholar]