Abstract
Inoculation against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is ongoing worldwide. However, the emergence of SARS-CoV-2 variants could cause immune evasion. We developed a bivalent nanoparticle vaccine that displays the receptor binding domains (RBDs) of the D614G and B.1.351 strains. With a prime-boost or a single-dose strategy, this vaccine elicits a robust neutralizing antibody and full protection against infection with the authentic D614G or B.1.351 strain in human angiotensin-converting enzyme 2 transgene mice. Interestingly, 8 months after inoculation with the D614G-specific vaccine, a new boost with this bivalent vaccine potently elicits cross-neutralizing antibodies for SARS-CoV-2 variants in rhesus macaques. We suggest that the D614G/B.1.351 bivalent vaccine could be used as an initial single dose or a sequential enforcement dose to prevent infection with SARS-CoV-2 and its variants.
Keywords: SARS-CoV-2 variants, B.1.351 variants, bivalent nanoparticle vaccine
Graphical abstract
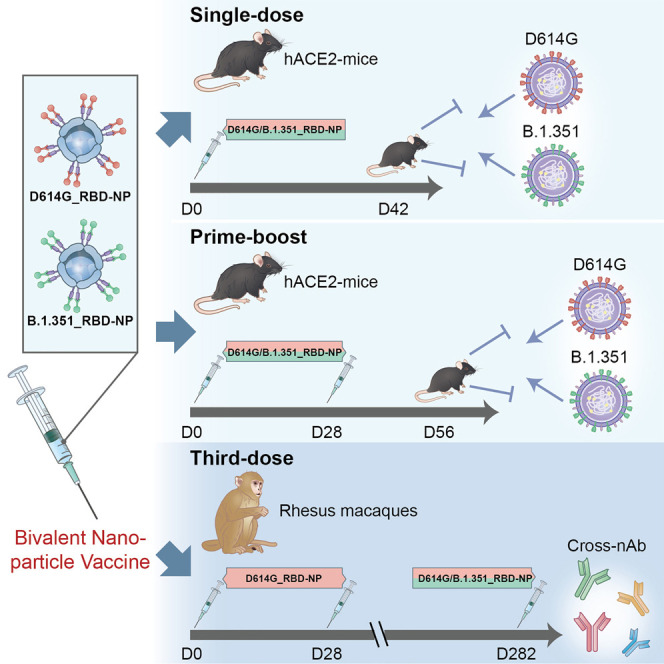
Yuan et al. construct a bivalent vaccine based on the RBD sequence of two SARS-CoV-2 variants, D614G and B.1.351. Vaccination with a single dose or a prime-boost regimen is protective against SARS-CoV-2 challenge in mice. The bivalent vaccine elicits nAbs against SARS-CoV-2 variants in rhesus macaques with a third-dose regimen
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has infected more than 245 million people, causing more than 4.9 million deaths worldwide (Wang et al., 2020a; Zhou et al., 2020). Since the outbreak started, various coronavirus disease 2019 (COVID-19) vaccines have been authorized or approved for emergency use, followed by many more at different phases in their development pipelines, including nanoparticle vaccines (Arunachalam et al., 2021; Brouwer et al., 2021; Keech et al., 2020; Ma et al., 2020; Saunders et al., 2021; Walls et al., 2020a). However, the subsequent emergence of SARS-CoV-2 variants, especially those with mutations in the receptor binding domain (RBD) of the spike protein, has caused great concern (Plante et al., 2021; Wang et al., 2021b). The RBD of the spike protein interacts with the major viral receptor angiotensin-converting enzyme 2 (ACE2) to mediate viral entry, and mutations in the RBD cause evasion from neutralizing antibodies (nAbs) or enhanced binding affinity to ACE2 (Letko et al., 2020; Walls et al., 2020b). Since late 2020, numerous variants have been reported and eventually spread worldwide, notably the B.1.1.7 (alpha, variant of concern [VOC] 202012/01), B.1.351 (known as the beta variant), P.1 (also called gamma), B.1.429, B.1.526, B.1.617.1 (kappa), and B.1.617.2 (delta) lineages, which carry various mutations in many regions of the SARS-CoV-2 genome, especially in the spike (S) region (Figure S4C; Galloway et al., 2021; Hodcroft et al., 2021; Hoffmann et al., 2021; Li et al., 2021b; Resende et al., 2021; Tegally et al., 2020; Zhang et al., 2021b). Importantly, compared with the D614 and D614G lineages, most of these variants, especially those harboring E484K/Q mutants, confer elevated resistance to neutralization from convalescent COVID-19 sera as well as many therapeutic monoclonal antibodies (Garcia-Beltran et al., 2021; Greaney et al., 2021; Hoffmann et al., 2021; Li et al., 2021a, 2021c; Planas et al., 2021; Sun et al., 2021; Wang et al., 2021a; Zhou et al., 2021). Recent clinical studies in South Africa, in parallel, confirmed reduced efficacy against symptomatic COVID-19 disease for various vaccines based on the original D614/D614G, including the NVX-CoV2373 (Novavax), BNT162b2 (Pfizer-BioNTech), AZD1222 (University of Oxford/AstraZeneca), and Ad26.COV2.S (Janssen/Johnson & Johnson) vaccines (Abu-Raddad et al., 2021; Moyo-Gwete et al., 2021; Shen et al., 2021; Shinde et al., 2021). Some recent studies also show that the protection efficiency of these vaccines against other variants harboring E484K/Q mutations, such as B1.617.1 harboring L452R/E484Q, is decreased (Cherian et al., 2021; Edara et al., 2021; Kumar et al., 2021). Therefore, strategies for developing updated vaccines against B.1.351 and other variants harboring E484K/Q mutations are urgently needed to avoid potential loss of clinical efficacy. Further, because the magnitude and duration of vaccine protection remains to be determined, an additional boost for the current vaccines could be necessary.
Given that the B.1.351 and P.1 strains harbor K417N/T, E484K, and N501Y mutations in the RBD domain, B.1.526 harbors E484K, and B.1.617.1 harbors L452R and E484Q (Figure S4C), we wanted to develop a D614G- and E484K/Q-specific bivalent vaccine. Because the mutants of B.1.351 exert the highest possibility of immune evasion so far, its RBD was chosen as the representative immunogen for E484K/Q mutant lineages (Diamond et al., 2021; Skelly et al., 2021). To this end, a bivalent D614G/B.1.351 RBD nanoparticle vaccine was developed that comprises a 1:1 mix of D614G_RBD-NP (D614G RBD nanoparticle vaccine) and B.1.351_RBD-NP (B.1.351 RBD nanoparticle vaccine). With a prime-boost or single-dose strategy, this bivalent vaccine elicited robust nAbs and full protection against infection with the authentic D614G or B.1.351 strains in human ACE2 (hACE2) transgene mice. Furthermore, 8 months after inoculation with the D614G-specific vaccine, a new boost with this bivalent vaccine potently elicited cross-nAbs for SARS-CoV-2 variants in rhesus macaques.
Results
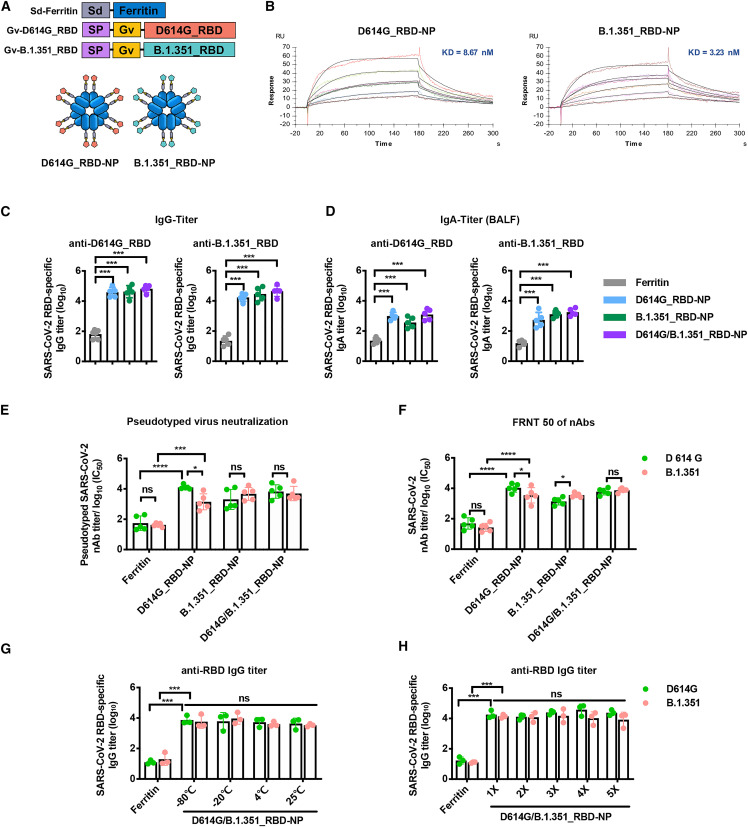
As described previously, we expressed the ferritin and RBD protein separately and then allow them to covalently conjugate through an isopeptide bond (Ma et al., 2020). To increase conjugation efficiency, we recently developed a GvTagOpti/SdCatcher (Gv/Sd) system derived from Gardnerella vaginalis and Streptococcus dysgalactiae, respectively. This conjugation system presents immunogen on nanoparticles with much higher efficiency (Zhang et al., 2021c). The Sd-coding sequence was genetically fused at the N terminus of ferritin (Sd-ferritin), whereas the Gv-coding sequence was fused at the N-terminus of the D614G_RBD or B.1.351_RBD sequence (Figure 1A). The purified Gv-D614G_RBD and Gv-B.1.351_RBD were irreversibly covalently conjugated to the Sd-ferritin to generate D614G_RBD-nanoparticle (NP) and B.1.351_RBD-NP (Figure 1A). The NP conjugates were purified by size exclusion chromatography (SEC) and analyzed by SDS-PAGE (Figures S1A and S1B). Transmission electron microscopy (TEM) of the pooled higher-molecular-weight fractions from SEC revealed well-ordered NPs (Figure S1C). The RBD-conjugated NPs showed spikes protruding from the spherical core after conjugation (Figure S1C). We characterized the antigenicity of NP conjugates by detecting their binding affinity and kinetics with the receptor hACE2 (Ramanathan et al., 2021). The measured binding dissociation constants (KD) of the D614G_RBD-NP and B.1.351_RBD-NP with the hACE2 receptor were 8.67 × 10−9 and 3.23 × 10−9 M, respectively, indicating that the epitopes on the NPs are exposed and correctly folded and that the B.1.351_RBD-NP binds to human ACE2 with increased affinity (Figures 1B and S1D). SEC, TEM, and surface plasmon resonance (SPR) confirmed successful generation of NP vaccines presenting multiple copies of the SARS-CoV-2 RBD proteins.
Figure 1.
The bivalent D614G/B.1.351_RBD-NP vaccine elicits a robust immune responses in BALB/c mice and is thermostable and resilient
(A) Schematic of the bivalent D614G/B.1.351_RBD-NP. The bivalent D614G/B.1.351_RBD-NP consists of Sd-ferritin, Gv-D614G_RBD, and Gv-B.1.351_RBD. The ratio is 50/50 of D614G_RBD-NP and B.1.351_RBD-NP in bivalent D614G/B.1.351_RBD-NP. Sd, SdCatcher; SP, secretory signal peptide; Gv, GvTagOpti.
(B) Representative BIAcore plots of D614G_RBD-NP and B.1.351_RBD-NP bound to hACE2. The KD values were calculated by the software BIAevaluation. The KD value shown was a mean of three independent experiments.
(C and D) D614G_RBD- and B.1.351_RBD-specific IgG/IgA titers of immunized BALB/c mice were detected by ELISA. Antibody titers of serum and BALF, which were collected at week 6, were determined by ELISA, and the data are represented as the reciprocal of the endpoint serum dilution.
(E) Groups of serially diluted serum were examined for nAbs against pseudotyped SARS-CoV-2 (D614G/B.1.351). Data represent the 50% neutralizing titers (NT50) of nAbs in each group. Experiments were conducted independently in triplicate.
(F) The nAbs titer of each vaccine group for the authentic SARS-CoV-2 (D614G/B.1.351) was determined by FRNT and is represented as half-maximal inhibitory concentration (IC50).
(G and H) Immunoreactivity of bivalent D614G/B.1.351_RBD-NP for D614G_RBD and B.1.351_RBD, determined by ELISA after storage at various temperatures for 2 weeks (G) or after one to five cycles of freezing and thawing (H). Antibody titers of serum collected at week 2 were determined by ELISA, and the data are represented as the reciprocal of the endpoint serum dilution. Each dot represents serum from one animal.
Experiments were conducted independently in triplicate. Data are represented as mean ± SD. Adjusted p values were calculated by one-way ANOVA with Tukey’s multiple comparisons test. Significant differences between groups linked by horizontal lines are indicated by asterisks. ns, not significant; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
To evaluate the immunogenicity of these bivalent NP vaccines against the ancestral D614G and B.1.351 variants, BALB/c mice were immunized with these vaccines. We compared two different aluminum adjuvants with the Sigma adjuvant system (SAS) used in our previous study and found that there was no difference in immunogenicity between each group (Figure S1E). Therefore, we chose alhydrogel (InvivoGen) as the adjuvant for this study. BALB/c mice were immunized subcutaneously with 10 μg of B.1.351_RBD-NP or D614G/B.1.351_RBD-NP adjuvant with alhydrogel in a prime-boost manner. Serum was collected, and mice were euthanized 2 weeks after boost. The B.1.351_RBD-NP and D614G/B.1.351_RBD-NP vaccines induced RBD-specific immunoglobulin G (IgG) in serum at approximately 105 titer and RBD-specific IgA secretion in bronchoalveolar lavage fluid (BALF) specific for the D614G and B.1.351 variants (Figures 1C and 1D). By utilizing pseudovirus neutralization assays, the nAbs induced by D614G/B.1.351_RBD-NP strongly inhibited D614G and B.1.351 pseudotyped variants (Figure 1E). In line with the previous findings that serum from convalescent COVID-19 individuals showed reduced neutralization against the B.1.351 variant, the D614G_RBD-NP vaccine showed a reduction of nAbs to the B.1.351 variant in BALB/c mice (Figure 1E; Li et al., 2021c). To study whether these nAbs could inhibit infection with authentic D614G and B.1.351 strains, a focus reduction neutralizing test (FRNT) was conducted (Ma et al., 2020). The nAbs in all NP-vaccinated mice strongly inhibited replication of the authentic D614G and B.1.351 strains. The B.1.351_RBD-NP vaccine elicited higher neutralization titers against B.1.351 compared with that of D614G, whereas the bivalent D614G/B.1.351_RBD-NP induced a similar robust neutralization response against the authentic D614G and B.1.351 strains, with no significant difference in neutralization titers (Figure 1F). We further determined the cellular immune response of the bivalent NP vaccines. 2 weeks after boost vaccination, splenocytes were obtained, and intracellular cytokine staining (ICCS) was conducted (Figure S2A). All NP-vaccinated mice showed strong polyfunctional CD8+ T cells expressing interferon γ (IFN-γ) and interleukin-2 (IL-2) and Th1-biased CD4+ T cells expressing IFN-γ (Figure S2B). There was no difference across different groups for Th2-biased IL-4+-expressing CD4+ T cells (Figure S2B).
To determine the stability of NP vaccines, we stored the D614G/B.1.351_RBD-NP vaccine at −80°C, −20°C, 4°C, and 25°C for 2 weeks. SDS-PAGE profiles indicated that the protein remained highly stable after 2 weeks of storage at all tested temperatures (Figure S3A), with no loss of immunogenicity in BALB/c mice (Figure 1G). We next assessed the resilience of the D614G/B.1.351_RBD-NP vaccine by challenging it with multiple rounds of freezing and thawing. Even after five rounds of freezing and thawing, no degradation was observed, based on SDS-PAGE analysis (Figure S3B), and no loss of immunogenicity in BALB/c mice (Figure 1H). The B.1.351_RBD-NP vaccine and the bivalent D614G/B.1.351_RBD-NP vaccine induced robust humoral and cellular immune responses with a high level of stability.
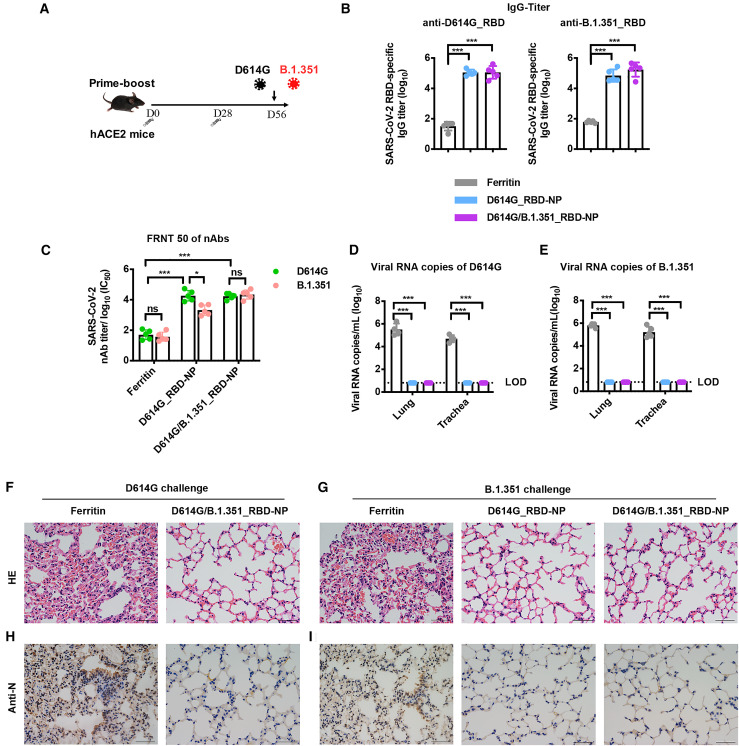
To determine bivalent D614G/B.1.351_RBD-NP vaccine protection against D614G and B.1.351 variant infection, we immunized transgenic hACE2 mice, which expressed humanized ACE2, with NP vaccines (Figure 2A). The RBD-specific IgG against the D614G and B.1.351 strains was quite high in all NP-immunized hACE2 mice 2 weeks after boost (Figure 2B). In the 50% focus reduction neutralizing test (FRNT50) assay, the D614G/B.1.351_RBD-NP vaccine elicited a similar robust neutralization response against the authentic D614G and B.1.351 strains, whereas a 9-fold decrease of neutralization against the B.1.351 strain was observed in D614G_RBD-NP-immunized hACE2 mice (Figure 2C). All immunized hACE2 mice were challenged intranasally with 4 × 104 plaque-forming units (PFUs) of the D614G or B.1.351 strains, respectively, and euthanized 5 days after challenge. The SARS-CoV-2 viral RNA copies in the lungs and trachea were used to quantify virus replication. Control hACE2 mice had an average of 5.84 × 105 and 6.10 × 104 copies/mL for D614G and 6.75 × 105 and 2.69 × 105 copies/mL for the B.1.351 strain in the lungs and trachea, respectively, whereas D614G/B.1.351_RBD-NP-immunized hACE2 mice had undetectable levels of viral RNA (Figures 2D and 2E). The D614G_NP vaccine remained overall efficacious and delivered notable cross-protection against the B.1.351 variant (Figure 2E). Moreover, the SARS-CoV-2 nucleocapsid (N) antigen was undetectable in the lung tissue of all NP-vaccinated hACE2 mice but was detected in control mice (Figures 2F–2I). Hematoxylin and eosin (H&E) staining of lung tissue showed a reduction of inflammation in immunized hACE2 mice compared with control mice (Figures 2F–2I).
Figure 2.
The bivalent D614G/B.1.351_RBD-NP vaccine protects against D614G and (B)1.351 variant infection in hACE2 mice
(A) Schematic of hACE2 mouse vaccination. Five mice in each group were primer-boost-vaccinated with the bivalent D614G/B.1.351_RBD-NP on day 0 and day 28. Mice were challenged with authentic SARS-CoV-2 (D614G/B.1.351) on day 56. All mice were bled and euthanized 5 days after challenge.
(B) D614G_RBD-specific and B.1.351_RBD-specific IgG antibody titers of serum were determined using ELISA by serial dilution and are represented as the reciprocal of the endpoint serum dilution.
(C) The serum of each mouse was 10-fold serially diluted and incubated with 500 focus-forming units (ffu) of authentic SARS-CoV-2 (D614G/B.1.351), followed by incubation with Vero E6 cells. The FRNT spots of each well were counted. FRNT50 of nAbs of each vaccine group was determined by FRNT and is represented as IC50.
(D and E) Viral RNA copies in the lungs and trachea of each mouse were determined by qRT-PCR and plotted as log10 copies per milliliter. A dotted line indicates the limit of detection (LOD).
(F and G) H&E staining of the lungs of each mouse.
(H and I) Immunohistochemistry (IHC) against N proteins was evaluated in the lungs of each mouse. Scale bars (F–I) represent 50 μm. Each dot represents serum from one animal.
Experiments were conducted independently in triplicate. Data are represented as mean ± SD. Adjusted p values were calculated by one-way ANOVA with Tukey’s multiple comparisons test. Significant differences between groups linked by horizontal lines are indicated by asterisks. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
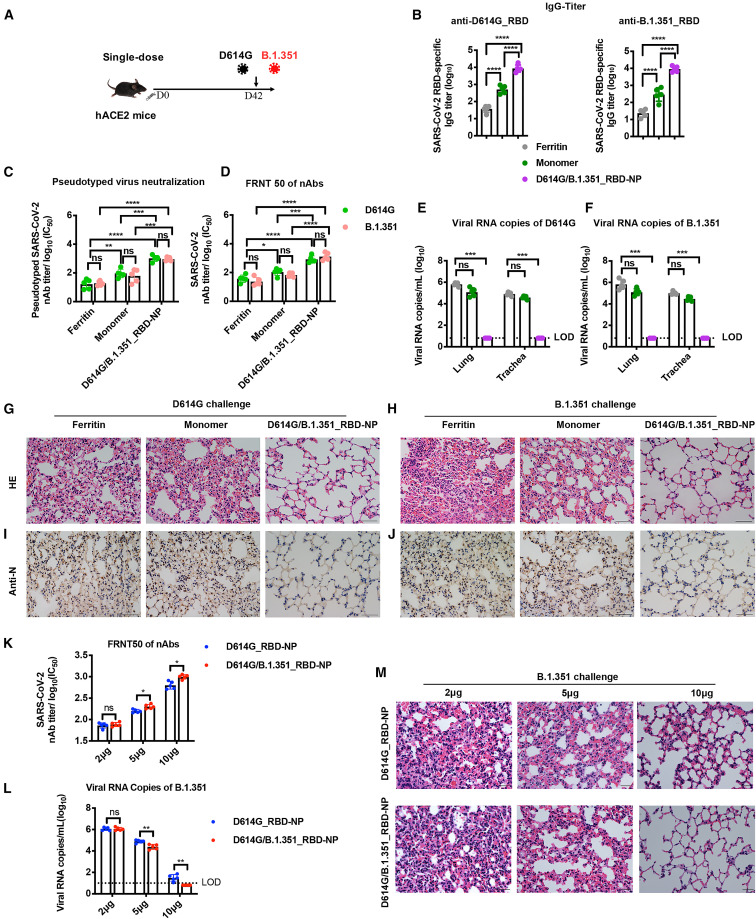
In our previous study, we reported that a significant amount of nAbs had been induced by our NP vaccines 4 weeks after priming but before boost vaccination, indicating that single-dose vaccination could be enough to elicit sufficient nAbs against SARS-CoV-2 (Ma et al., 2020). Here we evaluate the immunogenicity and in vivo protection ability of a single dose of the bivalent D614G/B.1.351_RBD-NP vaccine (Figure 3A). Five hACE2 mice were immunized with 10 μg of the D614G/B.1.351_RBD-NP vaccine or an equimolar amount of D614G/B.1.351_RBD-monomer as a control. Because the prime-boost strategy of the NP vaccine has been carefully evaluated using a monomer as a control in our previous study, here we also set the D614G/B.1.351_RBD-monomer group as a control for the single-dose strategy (Ma et al., 2020). Serum was collected 6 weeks after vaccination (Figure 3A). RBD-specific IgG against the D614G and B.1.351 strains was detectable in all NP-immunized hACE2 mice (Figure 3B). With the FRNT50 assay and pseudovirus neutralization assays, a single dose of the D614G/B.1.351_RBD-NP vaccine induced significantly higher neutralization titers against the pseudovirus and authentic D614G and B.1.351 variants compared with D614G/B.1.351_RBD-monomer (Figures 3C, 3D and, S4A). All immunized hACE2 mice were challenged intranasally with 4 × 104 PFUs of the D614G or B.1.351 strains, respectively, and euthanized 5 days after challenge. Although the single dose of D614G/B.1.351_RBD-monomer had an average of 1.62 × 105 and 3.88 × 104 copies/mL for the D614G strain and 1.54 × 105 and 3.17 × 104 copies/mL for the B.1.351 strain in the lungs and trachea, respectively, all single-dose D614G/B.1.351_RBD-NP-immunized hACE2 mice had undetectable levels of viral RNA (Figures 3E and 3F). The lungs were analyzed for histopathology and immunohistochemistry. Histopathology examination indicated severe bronchopneumonia and interstitial pneumonia in the D614G/B.1.351_RBD-monomer group, with edema and bronchial epithelial cell desquamation and infiltration of lymphocytes within alveolar spaces. In contrast, only very mild bronchopneumonia was observed in the D614G/B.1.351_RBD-NP vaccine group (Figures 3G and 3H). Similarly, immunohistochemistry assays detected the SARS-CoV-2 N antigen in the D614G/B.1.351_RBD-monomer group, but the SARS-CoV-2 N antigen was undetectable in lung tissue of the single-dose-immunized D614G/B.1.351_RBD-NP group (Figures 3I and 3J). These data demonstrated that single-dose vaccination of the D614G/B.1.351_RBD-NP vaccine caused significant prevention of replication of the authentic D614G and B.1.351 strains in the lungs.
Figure 3.
Protection efficacy of a single dose of the bivalent D614G/B.1.351_RBD-NP vaccine against D614G and (B)1.351 variant infection in hACE2 mice
(A) Schematic of hACE2 mouse vaccination. Five mice in each group were vaccinated with a single dose of the bivalent D614G/B.1.351_RBD-NP on day 0. Mice were challenged with authentic SARS-CoV-2 on day 42. All mice were bled and euthanized 5 days after challenge.
(B) D614G_RBD-specific and B1.351_RBD-specific IgG antibody titers of serum were determined using ELISA by serial dilution and are represented as the reciprocal of the endpoint serum dilution.
(C) The nAbs titer for SARS-CoV-2 pseudovirus (D614G/B.1.351) of vaccinated hACE2 mice by pseudotyped virus neutralization assay, represented as IC50.
(D) The serum of each mouse was 10-fold serially diluted and incubated with 500 ffu of authentic SARS-CoV-2 (D614G/B.1.351), followed by incubation with Vero E6 cells. The FRNT spots of each well were counted. FRNT50 of nAbs of each vaccine group was determined by FRNT and is represented as IC50.
(E and F) Viral RNA copies in the lungs and trachea of each mouse were determined by qRT-PCR and plotted as log10 copies per milliliter. A dotted line indicates the LOD.
(G and H) H&E staining in the lungs of each mouse.
(I and J) IHC against N proteins was evaluated in the lungs of each mouse.
(K) The serum of each mouse was 10-fold serially diluted and incubated with 500 ffu of the authentic B).1.351 strain, followed by incubation with Vero E6 cells. The FRNT spots of each well were counted. FRNT50 of nAbs of each vaccine group was determined by FRNT and is represented as IC50.
(L) Viral RNA copies in the lungs of each mouse were determined by qRT-PCR and plotted as log10 copies per milliliter. A dotted line indicates the LOD.
(M) H&E staining in the lungs of each mouse.
Scale bars in (G)–(J) and (M) represent 50 μm. Each dot represents serum from one animal. Experiments were conducted independently in triplicate. Data are represented as mean ± SD. Adjusted p values were calculated by one-way ANOVA with Tukey’s multiple comparisons test. Significant differences between groups linked by horizontal lines are indicated by asterisks. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
Further, we compared the protection efficiency of the bivalent vaccine strategy and the original D614G_RBD-NP vaccine from the B.1.351 strain. hACE2 mice were immunized with 2 μg, 5 μg, and 10 μg of the bivalent vaccine and D614G_RBD-NP vaccine in a single-dose regimen. Six weeks after vaccination, serum RBD-specific IgG titers against the B.1.351 strain were roughly comparable for the two groups at three different doses (Figure S4B). In the FRNT50 assay, the D614G/B.1.351_RBD-NP vaccine produced higher nAb responses against the authentic B.1.351 strain in mice vaccinated with a dose of 5 μg and 10 μg (Figure 3K). All immunized hACE2 mice were challenged intranasally with 4 × 104 PFUs of the B.1.351 strain and euthanized 5 days after challenge. Compared with the D614G_RBD-NP vaccine group, the bivalent D614G/B.1.351_RBD-NP vaccine showed better protection, as demonstrated by lower levels of viral RNA in the lungs and a reduction of inflammation, as seen by histopathological examination of lung tissue (Figure 3L and 3M). These data demonstrated the superiority and necessity of the bivalent vaccine strategy, particularly in a single-dose regimen.
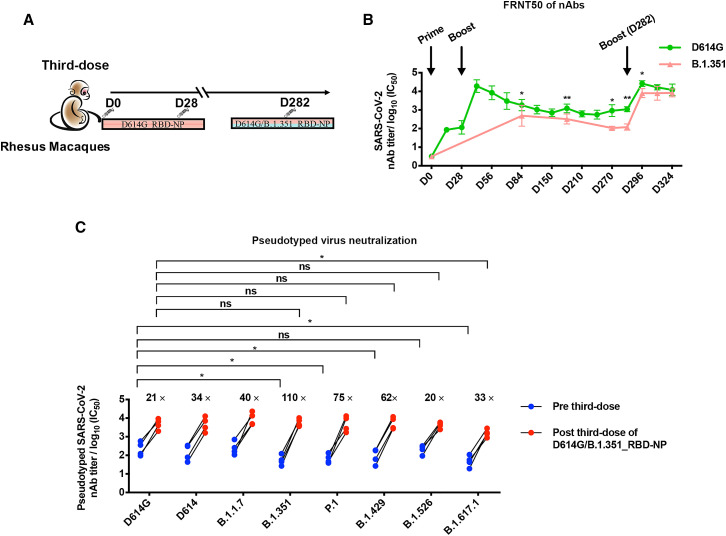
Finally, we assess the cross-protection of the updated bivalent D614G/B.1.351_RBD-NP vaccine as a third dose on previously immunized rhesus macaques (Wu et al., 2021). Rhesus macaques were immunized with 50 μg of the D614G_RBD-NP vaccine on days 1 and 28, and the robustness of the nAbs against the authentic D614G and B.1.351 strains was monitored over the course of more than 8 months (Figure 4A; Ma et al., 2020). As measured by FRNT50 assay against the D614G authentic virus, nAb titers in serum from immunized rhesus macaques were elicited 14 days after priming by the D614G_RBD-NP vaccine and peaked following another 14 days after the first boost. The nAb titers thereafter remained significant but eventually waned over a course of 8 months. Interestingly, serum from immunized animals showed consistently higher nAb titers against the D614G strain than the B.1.351 strain (Figure 4B).
Figure 4.
A third dose of the bivalent D614G/B.1.351_RBD-NP vaccine in rhesus macaques previously vaccinated with two doses of D614G_RBD-NP induces cross-neutralization of viral variants
(A) Schematic of rhesus macaque vaccination. Four rhesus macaques were first vaccinated with primer-boost D614G_RBD-NP on day 0 and day 28 and then vaccinated with bivalent D614G/B.1.351_RBD-NP on day 282. Serum was collected at the indicated times.
(B) The serum of each rhesus macaque at different times was 10-fold serially diluted and incubated with 500 ffu of authentic SARS-CoV-2 (D614G/B.1.351), followed by incubation with Vero E6 cells. The FRNT spots of each well were counted. FRNT50 of nAbs of authentic SARS-CoV-2 (D614G/B.1.351) was determined by FRNT and plotted as a time-course curve.
(C) The nAbs titer for the SARS-CoV-2 pseudovirus (D614G/D614/B.1.1.7/B.1.351/P.1/B.1.429/B.1.526/B.1.617.1) of rhesus macaques before and after the third boost with D614G/B.1.351_RBD-NP was determined by pseudotyped virus neutralization assay and is represented as IC50. Each dot represents serum from one animal.
Experiments were conducted independently in triplicates. Data are represented as mean ± SD. Adjusted p values were calculated by one-way ANOVA with Tukey’s multiple comparisons test. Significant differences between groups linked by horizontal lines are indicated by asterisks. ∗p < 0.05, ∗∗p < 0.01.
To evaluate the ability of the bivalent D614G/B.1.351_RBD-NP vaccine to boost pre-existing immunity and increase neutralization of the D614G and B.1.351 strains, a third dose of 50 μg of the bivalent D614G/B.1.351_RBD-NP vaccine was administered on day 282 (Figure 4A). The RBD-specific IgG titers against the D614G and B.1.351 strains were quite high after a third dose of the vaccine, reaching approximately 105 titers (Figure S4D). Following the bivalent D614G/B.1.351_RBD-NP vaccine booster injection, the nAb titers against the authentic D614G virus increased significantly, exceeding the previously measured peak for the D614G_RBD-NP level. The third dose of the D614G/B.1.351_RBD-NP vaccine increased the neutralization titers against the D614G and B.1.351 strains 15-fold and 65-fold, respectively (Figures 4B and S4E). Then we determined whether the D614G/B.1.351_RBD-NP vaccine booster elicited nAbs against other SARS-CoV-2 variants. 4 weeks after the third dose of the D614G/B.1.351_RBD-NP vaccine, the rhesus macaque sera potently neutralized the pseudotyped viruses of current major SARS-CoV-2 variants, including B.1.1.7, B.1.351, P.1, B.1.429, B.1.526, and B.1.617.1 (Figure 4C). Fifteen convalescent sera, which were collected in Guangzhou and Zhuhai in South China before April 15, 2020 and therefore excluded existence of the epidemic variants of SARS-CoV-2 except for the D614 and D614G strains, were used as controls for the pseudovirus neutralizing assay against SARS-CoV-2 variants (Figure S4F; Liu et al., 2020). The convalescent sera were 10-fold less effective at neutralizing B.1.351 and 23-fold less effective for B.1.617.1 in comparison with its neutralization of the D614G strain (Figure S4F). Similarly, the sera from rhesus macaques on day 282 (before the third dose) showed a significant reduction of neutralizing capacity of the B.1.351 and P.1 strains and especially the B.1.617.1 strain. Interestingly, the third dose of D614G/B.1.351_RBD-NP immunization for rhesus macaques potently elicited nAbs against all viral variants we tested (Figure 4C). This new boost especially enhances the nAb titer for B.1.617.1 33-fold, which is still slightly lower than that of the D614G strain (Figure 4C).
Discussion
In this study, we developed a bivalent D614G/B.1.351_RBD-NP vaccine to prevent infection with the early strain and viral variants harboring E484K/Q mutants. The nAbs induced by this bivalent vaccine are quite high, and the protection from the authentic D614G and B.1.351 strains in hACE2 mice is complete. Protection not only occurs with the initial prime-boost strategy but also with the initial single-dose strategy. Therefore, this bivalent NP vaccine could be used for initial prime-boost or single-dose vaccination for those who never received any COVID-19 vaccine. In addition, we proposed a third-dose (second booster) strategy, which is particularly important given the accelerating rollout of global vaccination (Kreier, 2021). Notably, with the single-dose strategy, a robust immune response against B.1.351 viruses depends on at least a 5-μg dose in our experiment settings. This result suggests that a certain amount of vaccine is required for initiating the immune response. In rhesus macaques receiving initial prime-boost inoculation of the D614G-specific NP vaccine, the nAbs for the D614G and viral variants eventually decreased. A third-dose inoculation with a bivalent NP vaccine significantly boosted the nAb titers against the D614G and B.1.351 strains at almost the same level. Notably, the deterioration rate of the D614G nAb titer is slower after the third dose compared with that after the second dose, suggesting involvement of a stronger memory immune response. Importantly, the third dose of immunization with the bivalent NP vaccine also potently elicited nAbs against almost all variants we tested, albeit with a slight deficiency for B.1.617.1.
A recent work indicates that a NP vaccine based on an early strain is enough to fully protect an animal from infection with viral variants, including B.1.351 (Saunders et al., 2021). Although our data support this claim, we show that, after initial prime-boost with the NP vaccine targeting D614G, the nAb titers for the D614G strain will eventually decrease, and the decrease of nAb titers for B.1.351 is even more significant. This causes concern regarding the long persistence of immunity against viral variants after NP vaccine administration targeting the early strain. Given that SARS-CoV-2 variants with the E484K/Q mutant dominate in many regions of the world and that the bivalent vaccine has a potent capability to boost nAb titers against viral variants, we suggest that it is necessary to develop a bivalent NP vaccine targeting the early strain and B.1.351 strain, which could be sufficient for the current COVID-19 pandemic with the threat of 484K/Q mutants. Given that SARS-CoV-2 exhibits a significant capability to evolve after an almost 2-year epidemic, a vaccine for SARS-CoV-2 variants could be developed.
To deal with the pandemic caused by SARS-CoV-2 variants, in addition to updating the design of immunogen, the inoculation strategy should also be optimized (Huang et al., 2021b; Powell et al., 2021; Yahalom-Ronen et al., 2020). Because the bivalent NP vaccine has potent protection efficiency in hACE2 mice with a single dose and is stable at ambient temperature and resistant to freezing and thawing with minimal loss of immunogenicity, long-distance distribution of this bivalent NP vaccine could become quite easy, especially to countries where cold-chain resources are incomplete.
Limitations of the study
Our study has potential limitations. Only the D614G/B.1.351_RBD-NP vaccine was evaluated as a third dose. The ability of D614G_RBD-NP or B.1.351_RBD-NP to boost immunity against the D614G and B.1.351 strains has not yet been assessed, although cross-protecting nAbs against the viral variants are expected because of the high levels of titers.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Goat anti-Mouse IgG (H+L) Secondary Antibody, HRP | Invitrogen | Cat#31430; RRID: AB_228307 |
| Goat anti-Monkey IgG (H+L) Secondary Antibody, HRP | Invitrogen | Cat#PA1-84631; RRID: AB_933605 |
| HRP∗Polyclonal Goat Anti Mouse IgA | Immunoway | Cat#RS030211 |
| Rabbit Polyclonal anti-SARS-CoV-2 Nucleoprotein (N) Antibody | Sino Biological | Cat#40143-T62; RRID: AB_2892769 |
| Goat Anti-Rabbit IgG Secondary Antibody (HRP) | Sino Biological | Cat#SSA004 |
| Ultra-LEAF Purified anti-mouse CD28 Antibody | Biolegend | Cat#102116; RRID: AB_11147170 |
| CD3e Monoclonal Antibody (145-2C11), PE-Cyanine7 | eBioscience | Cat#25-0031-82; RRID: AB_469572 |
| Alexa Fluor 700 anti-mouse CD4 Antibody | Biolegend | Cat#100429; RRID: AB_493698 |
| CD8a Monoclonal Antibody (53-6.7), FITC | eBioscience | Cat# 11-0081-86; RRID: AB_464917 |
| APC anti-mouse IFN-γ Antibody | Biolegend | Cat#505810; RRID: AB_315404 |
| PerCP/Cyanine5.5 anti-mouse IL-2 Antibody | Biolegend | Cat#503822; RRID: AB_2123676 |
| PE anti-mouse IL-4 Antibody | Biolegend | Cat#504103; RRID: AB_315317 |
| Bacterial and virus strains | ||
| E.coli BL21 | Takara | Cat#9126 |
| SARS-CoV-2 (D614G) | This paper | hCoV-19/CHN/SYSU-IHV/2020 (D614G); GISAID: EPI_ISL_444969 |
| SARS-CoV-2 (B.1.351) | Guangdong Provincial Center for Disease Control and Prevention, Guangzhou | 19nCoV-CDC-Tan-GDPCC (B.1.351) |
| Biological samples | ||
| Serum samples from BALB/c mice | This paper | N/A |
| Serum samples from hACE2 mice | This paper | N/A |
| Serum samples from rhesus macaques | This paper | N/A |
| Lung samples from hACE2 mice | This paper | N/A |
| Trachea samples from hACE2 mice | This paper | N/A |
| Serum samples from convalescent COVID-19 patients | Guangzhou 8th People’s Hospital and Fifth Affiliated Hospital of Sun Yat-sen University |
N/A |
| Chemicals, peptides, and recombinant proteins | ||
| eBioscience Fixable Viability Dye eFluor 780 | Invitrogen | Cat#65-0865 |
| Isopropyl β-D-1 thiogalactopyranoside (IPTG) | Takara | Cat#9030 |
| Aluminium hydroxide gel | InvivoGen | Cat#vac-alu-250 |
| Sigma adjuvant System (SAS) | Sigma-Aldrich | Cat#S6322 |
| eBioscience Intracellular Fixation & Permeabilization Buffer Set | Invitrogen | Cat#88-8824 |
| eBioscience™ Brefeldin A Solution (1000X) | Thermo Scientific | Cat#00-4506-51 |
| eBioscience™ Monensin Solution (1000X) | Thermo Scientific | Cat#00-4505-51 |
| ELISA Stop Solution | Solarbio | C1058 |
| eBioscience TMB Solution | eBioscience | Cat#00-4201 |
| Carboxymethylcellulose sodium salt (CMC) | Sigma-Aldrich | 21902; CAS9004-32-4 |
| TrueBlue HRP Substrate | KPL | 50-78-02 |
| Paraformaldehyde | Sigma-Aldrich | P6148; CAS30525-89-4 |
| Penicillin-Streptomycin, Liquid | ThermoFisher | Cat#15140122 |
| Fetal Bovine Serum (FBS) | ThermoFisher | Cat#10270-106 |
| SARS-CoV-2 Spike Glycoprotein Peptides Pool | GenScript | Cat#RP30020 |
| synthetic B.1.351_RBD peptide (B.1.351_RBD 405-424 and B.1.351_RBD 495-514) |
GenScript (This paper) | N/A |
| Recombinant Sd-Ferritin protein | This paper | N/A |
| Recombinant Gv-D614G_RBD | This paper | N/A |
| Recombinant Gv-B.1.351_RBD | This paper | N/A |
| Critical commercial assays | ||
| RNeasy Mini Kit | QIAGEN | Cat#74104 |
| SARS-CoV-2 RNA detection kit (PCR-Fluorescence Probing) | Da An Gene Co. | DA0931 |
| Pierce Rapid Gold BCA Protein Assay Kit | ThermoFisher | Cat#A53225 |
| Luciferase Assay System | Promega | Cat#E4550 |
| Experimental models: Cell lines | ||
| HEK293T | ATCC | CRL-3216; RRID: CVCL_0063 |
| Vero E6 | ATCC | CRL-1586; RRID:CVCL_0574 |
| CHO-K1 | ATCC | CCL-61 |
| Experimental models: Organisms/strains | ||
| BALB/c mice | Guangdong Medical Laboratory Animal Center | N/A |
| Transgenic hACE2 mice (C57BL/6) | GemPharmatech Co, Ltd | N/A |
| Rhesus macaques | Guangdong Landau Biotechnology Co, Ltd | N/A |
| Oligonucleotides | ||
| SARS-CoV-2 nucleocapsid (N) qPCR Forward Primer:5’- CAGTAGGGGAACTTCTCCTGCT-3’ | Da An Gene Co. | DA0931 |
| SARS-CoV-2 nucleocapsid (N) qPCR Reverse Primer:5’-CTTTGCTGCTGCTTGACAGA-3’ | Da An Gene Co. | DA0931 |
| SARS-CoV-2 nucleocapsid (N) Probe: 5’-FAM-CTGGCAATGGCGGTGATGCTGC-BHQ1-3’ | Da An Gene Co. | DA0931 |
| Recombinant DNA | ||
| pET28a-Sd-Ferritin | This paper | N/A |
| pLVX-SP-Gv-D614G_RBD | This paper | N/A |
| pLVX-SP-Gv-B.1.351_RBD | This paper | N/A |
| psPAX2 | Dr. Didier Trono | Addgene Plasmid #12260 |
| pHIV-Luciferase | Dr. Bryan Welm | Addgene Plasmid #21375 |
| SARS-CoV-2 (D614G) Spike Gene | This paper | hCoV-19/CHN/SYSU-IHV/2020 strain; GISAID: EPI_ISL_444969 |
| SARS-CoV-2 (D614) Spike Gene | This paper | Wuhan-Hu-1; GISAID: EPI_ISL_402125 |
| SARS-CoV-2 (B.1.1.7) Spike Gene | This paper | B.1.1.7 (GISAID: EPI_ISL_581117) |
| SARS-CoV-2 (B.1.351) Spike Gene | This paper | B.1.351 (GISAID: EPI_ISL_678597) |
| SARS-CoV-2 (P.1) Spike Gene | This paper | P.1 (GISAID: EPI_ISL_792683) |
| SARS-CoV-2 (B.1.429) Spike Gene | This paper | B.1.429 (GISAID: EPI_ISL_1675148) |
| SARS-CoV-2 (B.1.526) Spike Gene | This paper | B.1.526 (GISAID: EPI_ISL_1098596) |
| SARS-CoV-2 (B.1.617.1) Spike Gene | This paper | B.1.525 (GISAID: EPI_ISL_1093465) |
| Software and algorithms | ||
| GraphPad Prism v8.0 software | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| BD LSRFortessa cell analyzer | BD Biosciences | http://www.bdbiosciences.com/in/instruments/lsr/index.jsp |
| FlowJo v10 | Tree Star | https://www.flowjo.com/ |
| Image Studio Lite v4.0 | LI-COR Biosciences | https://www.licor.com/bio/products/software/image_studio_lite/ |
| QuantStudio 7 Flex System | ThermoFisher | https://www.thermofisher.com/order/catalog/product/4485701#/4485701 |
| GloMax 96 Microplate Luminometer Software v1.9.3 | Promega | https://www.promega.com/resources/software-firmware/detection-instruments-software/promega-branded-instruments/glomax-96-microplate-luminometer/ |
| SkanIt SW for Microplate Readers | ThermoFisher | https://www.thermofisher.com/order/catalog/product/5187139?SID=srch-srp-5187139 |
| ImmunoSpot software v5.1.34 | Cellular Technology Ltd | http://www.immunospot.com/ImmunoSpot-analyzers |
| Other | ||
| Sepharose 6 FF | GE Healthcare | Cat#90100367 |
| KR2i TangentialFlow Filtration system | Repligen | Cat#SYR2-U20 |
| Capto SP ImpRes | GE Healthcare | Cat#17546815 |
| Amicon Ultra-15 Centrifugal Filter Unit | Millipore | UFC900396 |
| Olympus BX63 | Olympus | https://www.olympus-lifescience.com.cn/zh/microscopes/upright/fluorescence/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Dr. Hui Zhang (zhangh92@mail.sysu.edu.cn).
Materials availability
Plasmids sequences for vaccine components will be made available upon request. Purified proteins for in vitro experiments can be generated upon execution of a material transfer agreement (MTA) with inquiries directed to Dr. Hui Zhang.
Experimental model and subject details
Ethics statements
The Ethics Review Board of Sun Yat-sen University approved this study. Animal experiments were carried out in compliance with the guidelines and regulations of the Laboratory Monitoring Committee of Guangdong Province of China. The animal experiments were also approved by the Ethics Committee of Zhongshan School of Medicine (ZSSOM) of Sun Yat-sen University on Laboratory Animal Care (Assurance Number: 2017-061). Authentic SARS-CoV-2 challenge studies were approved by the Ethics Committee of ZSSOM of Sun Yat-sen University on Laboratory Animal Care (Assurance Number: 2017-061) as well. Non-human primates experiments were approved by the Institutional Animal Care and Use Committee (IACUC) of Guangdong Landau Biotechnology Co, Ltd. (IACUC Approval No: LDACU 20200216-01). The Ethics Review Boards of Sun Yat-sen University, Guangzhou 8th People’s Hospital and Fifth Affiliated Hospital of Sun Yat-sen University approved this study. Fifteen serum samples from convalescent COVID-19 patients were obtained from Guangzhou 8th People’s Hospital and Fifth Affiliated Hospital of Sun Yat-sen University. All the convalescent sera were positive for RBD-specific antibodies (Liu et al., 2020).
Cells and viruses
HEK293T, CHO-K1 and Vero E6 cells were obtained from ATCC. These adherent cells were cultured in Dulbecco's modified Eagle medium (DMEM) supplemented with 1% penicillin-streptomycin (ThermoFisher) and 10% FBS (ThermoFisher). HEK293T expressing hACE2 (hACE2/HEK293T) was constructed in home. All cells were cultured in the sterile incubator at 37°C and 5% CO2. All cells have been confirmed to be mycoplasma free.
SARS-CoV-2 strains, named as hCoV-19/CHN/SYSU-IHV/2020 (D614G) (Accession ID on GISAID: EPI_ISL_444969) and 19nCoV-CDC-Tan-GDPCC (B.1.351) were propagated in Vero E6 cells as published before (Huang et al., 2021a; Ma et al., 2020). The D614G strain was isolated from the sputum of a female COVID-19 patient who was infected by a UK traveler in April 2020 by us, and the B.1.351 strain was isolated from a South Africa traveler by the Guangdong Center for Disease Control in January 2021. The sequence of this B.1.351 strain was performed by Guangdong Center for Disease Control and confirmed by us. The concentrations of D614G and B.1.351 were determined by FRNT as described below. All the authentic SARS-CoV-2-related experiments were conducted in the BSL-3 facility of Sun Yat-sen University.
Animal models
Transgenic hACE2 mice (C57BL/6) were purchased from GemPharmatech Co, Ltd. The generation procedure was described as published before (Zhang et al., 2021d). Specific-pathogen-free (SPF) 5-6-week old female BALB/c mice were purchased from Guangdong Medical Laboratory Animal Center. All mice were housed and vaccinated in SPF facilities at the Laboratory Animal Center of Sun Yat-sen University. Four adult rhesus macaques (2 male and 2 female) between 2-4-year old were purchased from Guangdong Landau Biotechnology Co, Ltd. Monkeys experiments were conducted according to the guidelines and regulations of the Laboratory Monitoring Committee of Guangdong Province of China.
Method details
Protein expression and purification
The RBD nanoparticle vaccine was constructed as described previously (Ma et al., 2020). To further improve the conjugation efficiency of the NP vaccine, we screened and modified a variety of natural CnaB2 proteins with isopeptide bonds from various bacterial strains. Additionally, we optimized the conjugation efficiency by generating several mutants based upon the structural prediction. Finally, we identified a potent combination of GvTagOpti (Gv) and SdCatcher (Sd) from Gardnerella vaginalis and Streptococcus dysgalactiae respectively, which significantly enhanced the assemble efficiency and the production of the SARS-CoV-2 NP vaccine (Zhang et al., 2021c). The Sd-Ferritin was expressed and purified from Escherichia coli (E.coli). Briefly, the Sd was genetically fused at the N terminus of Ferritin (Sd-Ferritin), DNA sequences of Sd-Ferritin were cloned to the pET28a vector. The construct was transformed into BL21 (Takara). A single clone was amplified in LB with kanamycin while shaking at 37°C. The isopropyl b-D-1 thiogalactopyranoside (IPTG) (Takara) was added to the bacterial solution to induce protein expression. Eighteen hours after induction, the bacteria expressing the protein were harvested and lysed by sonication. After centrifugation, the supernatants were loaded onto a Sepharose 6 FF (GE Healthcare) size exclusion column that was pre-equilibrated with 20 mM Tris 50 mM NaCl buffer (pH 7.5) and eluted with the same buffer at a rate of 10 mL min−1. The total volume of the column (Vt) was 53 mL, and the elution volume (Ve) of Sd-ferritin NP was 26 mL. The concentration of Sd-Ferritin was determined by BCA assay. The bacterial endotoxins in nanoparticles were quantified by the Tachypleus amebocyte lysate test (≤10 EU/dose). Coomassie blue staining, size-exclusion chromatography (SEC), and transmission electron microscopy (TEM) were executed to confirm the purity and homogeneity (Wang et al., 2020b).
The D614G_RBD and B.1.351_RBD were expressed and purified from Chinese hamster ovary K (CHO-K1) cells. Briefly, The Gv coding sequence was fused at the N-terminus of the D614G_RBD or B.1.351_RBD sequence. DNA sequences of SP-Gv-D614G_RBD and SP-Gv-B.1.351_RBD were codon-optimized for mammalian cell expression and cloned into vector plasmid pLVX. The constructed pLVX, lentiviral packaging plasmids psPAX2, and pLP/VSVG were transfected into HEK293T cells cultured in DMEM conditioned medium. Lentiviruses released in the supernatant were collected 60 h after transfection and then infected anchorage-dependent CHO-K1 cells cultured in F12K medium. The supernatant was removed 8 h later and changed with new a F12K medium for another day of culture. The F12K medium was then replaced with CHO S4 medium, which was used for cell suspension and expansion to a density of 3×106 cells/mL. Seven days later, supernatants were collected and centrifuged to discard cellular debris. The cleared supernatants were passed through the KR2i TangentialFlow Filtration system equipped with filters (Repligen) with 10-kDa and 100-kDa molecular weight cutoffs (MWCO) to obtain the 10-100kDa molecular weight proteins. The concentrates were purified by AKTA pure system using Capto SP Impres (GE Healthcare) columns running phosphate-buffered (PH= 7) and eluted with the 150 mM NaCl phosphate-buffered (PH= 7) buffer at a rate of 5 mL min−1. The purified proteins were concentrated and buffer-replaced with conventional Tris buffer. The concentrations of D614G_RBD and B.1.351_RBD were determined by BCA assay. Coomassie blue staining was executed to confirm the purity.
The purified Gv-D614G_RBD and Gv-B.1.351_RBD were irreversibly covalently conjugated to the Sd-Ferritin to generate the D614G_RBD-nanoparticle (NP) and B.1.351_RBD-NP. The bivalent D614G/B.1.351_RBD-NP vaccine comprises a 1:1 mix of D614G_RBD-NP and B.1.351_RBD-NP.
Surface plasmon resonance (SPR)
The measurements of Ferritin, D614G_RBD, B.1.351_RBD, D614G_RBD-NP, and B.1.351_RBD-NP binding to hACE2 were carried out with a BIAcore T100 instrument (GE Healthcare). A BIAcore CM5 Sensor Chip and an amine coupling kit were purchased from GE Healthcare. hACE2 was immobilized on a CM5 Sensor Chip (carboxymethylated dextran covalently attached to a gold surface) with the amine coupling kit (GE Healthcare). Ferritin, D614G_RBD-NP, and B.1.351_RBD-NP were diluted into different concentrations and then injected (30 mL min−1). hACE2-bound protein was monitored for about 120 s for each protein. The dissociation time was 200 s with running buffer per cycle. The signals were recorded by the Biacore T100 instrument with the standard protocols.
Size-exclusion chromatography (SEC)
Gv-D614G_RBD and Gv-B.1.351_RBD were incubated with Sd-Ferritin in the Tris-HCl buffer. After 8 h, the bound protein was subjected to Size-Exclusion Chromatography (SEC). D614G_RBD-NP and B.1.351_RBD-NP were eluted in retention of retaining 11 mL to 14 mL. The elution of nanoparticles was concentrated, and the concentration was measured by BCA assay. Coomassie blue staining, size-exclusion chromatography (SEC), and transmission electron microscopy (TEM) were executed to confirm the purity and homogeneity (Ma et al., 2020).
Transmission electron microscopy (TEM)
Transmission electron microscopy (TEM) Grids of SD-Ferritin, D614G_RBD-NP, and B.1.351_RBD-NP have proceeded to negative-stain electron microscopy in Shuimu BioSciences Ltd. Briefly, the Talos L120C (ThermoFisher) was operated at an accelerating voltage 120 kV to collect data on the sample. Samples were inspected with magnifications of 36 kx and 57 kx, respectively, with a pixel size of 0.245 nm and an electron dose of 15 e-/A2, and a total of 164 data were collected. Using cisTEM software to process the collected data, 45244 particles were picked out, and two-dimensional classification was performed with EMAN2.
Animal vaccination
For BALB/c mice vaccination, five BALB/c mice were subcutaneously immunized with 10 μg of D614G/B.1.351_RBD-NP formulated with Alum (InvivoGen) adjuvant. Mice in D614G_RBD-NP and B.1.351_RBD-NP groups were immunized with equal moles of D614G_RBD-NP and B.1.351_RBD-NP. The moles of D614G/B.1.351 RBD in the monomers group were the same as D614G_RBD-NP and B.1.351_RBD-NP in the D614G/B.1.351_RBD-NP group, respectively. Mice in the Ferritin group were immunized with equal moles of ferritin, which were the same as the D614G/B.1.351_RBD-NP group. All the mice were vaccinated with the above vaccines in a prime/boost manner, which vaccinated mice at week 0 and week 4. Serum was collected every two weeks. Mice were euthanized at week 6.
For hACE2 mice vaccination, for prime-boost strategy, all the mice were vaccinated as in BALB/c mice. Mice were challenged with authentic SARS-CoV-2 four weeks post-boost vaccination and euthanized 5 days post-challenge. Serum was collected at weeks 0, 2, 6, and 8. For single-dose strategy, mice were subcutaneously immunized with 2μg, 5μg, and 10 μg of vaccine formulated with Alum (InvivoGen) adjuvant. Mice were challenged with authentic SARS-CoV-2 six weeks post-vaccination and euthanized 5 days post-challenge. Serum was collected at weeks 0, 2, and 6.
For rhesus macaques vaccination, four monkeys were immunized with 50 μg of D614G_RBD-NP vaccine via intramuscular injection in a prime/boost manner, which was vaccinated at week 0 and week 4. Sera from monkeys were collected every two weeks. On Day 282, four monkeys were immunized with 50 μg of D614G/B.1.351_RBD-NP vaccine. Serum was collected every two weeks after the third dose immunization. All vaccines were formulated with Sigma Adjuvant System (SAS) adjuvant.
Quantitative reverse transcription polymerase chain reaction (qRT-PCR)
Different tissues of each mouse, including lung and trachea, were collected and homogenized with gentleMACS M tubes (Miltenyi Biotec, 130-093-236) in a gentleMACS dissociator (Miltenyi Biotec, 130-093-235). RNAs of homogenized tissues were extracted with RNeasy Mini Kit (QIAGEN,74104) according to the manufacturer’s instruction, followed by qRT-PCR to determine viral RNA copies of different tissues utilizing a one-step SARS-CoV-2 RNA detection kit (PCR-Fluorescence Probing) (Da An Gene Co., DA0931). The SARS-CoV-2 nucleocapsid (N) gene was cloned into a pcDNA3.1 expression plasmid and in vitro transcribed to obtain RNAs as standards to generate a standard curve. The indicating copies of N standards were 10-fold serially diluted and proceeded with qRT-PCR utilizing the same one-step SARS-CoV-2 RNA detection kit to obtain standard curves. The reactions were carried out in a QuantStudio 7 Flex System (Applied Biosystems). The N-specific primers, probes and reaction conditions were the same as published before (Li et al., 2021a). The viral RNA copies of each tissue were calculated into copies per mL and presented as a log10 scale. In each qRT-PCR experiment, both positive control and negative control of simulated RNA virus particles were included to monitor the entire experimental process and ensure the reliability of the test results.
Enzyme linked immunosorbent assay (ELISA)
Recombinant D614G_RBD and B.1.351_RBD protein was diluted to a concentration of 2 μg/mL concentration in coating buffer to coat on high-binding 96-well plates (Corning) respectively, overnight at 4 °C. After washed three times with PBS, the plates were blocked with 5% non-fat milk/PBS for 1 h. After another round of washing, the immunized animal serum was serially diluted and added into each well in duplicate, followed by incubating at room temperature for 1 h. The detection of antigen-specific IgG antibody in serum of BALB/c mice, hACE2 mice, or rhesus macaques was conducted through adding HRP-conjugated goat anti-mouse or goat anti-monkey secondary antibody (Invitrogen) respectively at dilution of 1:10000, and incubating for another 1 h after washing with PBS/T (containing 1% Tween-20) three times. The plates were washed four times before 100 μL TMB solution (eBioscience) was added to each well. After 5 min of chromogenic progress at room temperature, 100 μL stop solution (Solarbio) was added to quench reaction followed by absorption measure at 450 nm. The IgA antibodies in BALF of BALB/c mice were measured similarly using an HRP-conjugated polyclonal goat anti-mouse IgA antibody (1:10000 diluted in PBS; Immunoway). The data were analyzed using GraphPad Prism 8.0 software for non-linear regression to calculate endpoint titers.
SARS-CoV-2 infection
Specific-pathogen-free (SPF), transgenic hACE2 mice (C57BL/6), which have been immunized with different vaccines, were challenged with authentic SARS-CoV-2 in the BSL-3 facility (Zhang et al., 2021a; Zhang et al., 2021d). Littermates of the same sex were randomly assigned to un-infection or infection groups. D614G and B.1.351 were used to challenge mice. Mice were anesthetized with isoflurane and inoculated intranasally with 4×104 FFU (Focus-forming unit) of SARS-CoV-2 viruses. The lungs were collected at 5 days post-infection (d.p.i.). The virus stocks were obtained from the supernatant of Vero E6 after inoculation for 48 h, and the titers were determined by FRNT assay targeting nucleocapsid (N) protein.
Histopathology and immunohistochemistry
SARS-CoV-2-challenged hACE2 mice were euthanized in the BSL-3 facility. Lungs were collected and fixed in 4% paraformaldehyde buffer for 48 h, followed by embedding with paraffin. Longitudinal sections were performed on these tissues. The sections (3-4 μm) were stained with hematoxylin and eosin (H&E). For immunohistochemistry, lung sections of each mouse were incubated with rabbit anti-SARS-CoV-2 Nucleoprotein (N) at 1:200 dilution and the IHC were conducted as published before (Zhang et al., 2021d).
Focus reduction neutralizing test (FRNT)
The method was consistent as described previously (Li et al., 2021c; Ma et al., 2020). Briefly, the Vero E6 cells were seeded in 96-well plates at a density of 2×104 cells per well and incubated the plates until cells reached 90-100% confluent. The serum of BALB/c mice, hACE2 mice, and rhesus macaques at each time point was 10-fold serially diluted. Five hundred FFU of authentic SARS-CoV-2 viruses were mixed with the diluted serum in a ratio of 1:1 for 1 h incubation at 37°C. Cell culture medium was removed from the 96-well plate, followed by the incubating with virus/serum mixture. Plates were then incubated for 1 h at 37°C. The DMEM containing 1.6% CMC was added to each well incubated for 24 hours after the supernatant was removed. On the next day, released the supernatant and cells were fixed with 200 μl of 4% paraformaldehyde in each well. After further incubation at 4°C for 12 h, removed the fixative, and plates were washed with 200 μl PBS each well for 3 times. And then, 100 μl PBS containing 0.2% Triton X-100 and 1% BSA was added to each well. After reaction for 30 mins at room temperature, each well was washed with 200 μl PBS for 3 times and incubated 50μl of the diluted primary antibody (Anti-SARS-N; 40143-T62-100), which was diluted to 1:1000 with PBS solution containing 1% BSA at 37°C for 1 h. After primary antibody incubation, the cell within each well was washed 3 times with 200 μl PBS/T (0.1% Tween). The secondary antibody (Goat anti-rabbit IgG HRP; SSA004-1) was diluted to 1:2000 with PBS solution containing 1% BSA. 50 μl of the diluted secondary antibody was added to each well and incubated at 37°C for 1 h, followed by washing with PBS/T three times. 50 μl TrueBlue (KPL) was added to each well and set for 5 mins shaking at room temperature. Plates were washed with ddH2O, and the liquid was eradicated, followed by spot counting using with ImmunoSpot Microanalyzer. The reduction rates of the serial dilution assay were analyzed by GraphPad Prism 8.0 using non-linear regression to measure the FRNT50 titer (Zhang et al., 2021b).
Pseudotyped virus neutralization assay
Briefly, HEK293T cells were co-transfected with packaging plasmid psPAX2, luciferase-expressing lentivirus plasmid, and respective variant spike protein-expressing plasmid using polyethyleneimine (PEI, Sigma). Forty-eight hours after transfection, culture supernatants were collected, clarified of cells before stored in -80°C. Virus titration was performed by serial-diluted virus infection on hACE2 cells, and infectivity was measured by detection of luminescence, details would be shown as follows. All the convalescent serum and immunized animal serum were serially diluted and incubated with pre-titrated amounts of SARS-CoV-2 pseudotyped virus at 37°C for 1 h. Then the serum/virus mixture was then added into wells containing 1×104 hACE2 cells and incubated at 37°C in 5% CO2 for 48 h. Cells were then lysed with lysis buffer (Promega), and the lysate was the measure for luciferase activity by detecting relative luminescence units (RLU) in a luminometer (Promega). Neutralizing antibody titers of serum against the pseudotyped virus were analyzed using GraphPad Prism 8.0 software (Zhang et al., 2021d).
Intracellular cytokine staining (ICCS)
The spleen was collected in PBS, homogenized through a 70 μm strainer and incubated in ACK lysis buffer to remove red blood cells (RBCs), followed by passing through a 40 μm filter to obtain single splenocytes. To quantify the percentages of antigen-specific T cells, approximately 1 million splenocytes were seeded into each well and stimulated with peptides pool, which consists of SARS-CoV-2 Spike Glycoprotein peptides pool (GenScript) and two synthetic B.1.351_RBD peptide (B.1.351_RBD 405-424 and B.1.351_RBD 495-514). Each peptide was used at a final concentration of 2 μg/mL. Cells were co-stimulated with 2 μg/mL anti-CD28 (Biolegend) at 37°C with 5% CO2 for 1 h. Cells were then incubated with 5 μg/mL brefeldin A (Thermo Scientific) and 2 μM monensin (Thermo Scientific). DMSO was used as a negative control. PMA/ionomycin was used as a positive control. After a total of 6 h, the LIVE/DEAD Fixable Viability Dyes (Thermo Scientific) were used to exclude dead cells from analysis (Zhang et al., 2021e). Subsequently, cells were performed with a fixation/permeabilization kit (BD Biosciences). The following cytokine antibodies were used: anti-IFN-γ (XMG1.2), anti-IL-2 (JES6-5H4), and anti-IL-4 (11B11). The percentages of cytokine-specific splenocytes were analyzed by flow cytometry.
Quantification and statistical analysis
All the statistical details of specific experiments, which included the statistical tests used, number of samples, mean values, standard errors of the mean (SD), and p-values derived from indicated tests, had been described in the figure legends and showed in figures. Statistical analyses were conducted utilizing Graphpad Prism 8.0 or Microsoft Excel. Triplicate, sextuplicate, and other replicative data were presented as mean ± SD. Value of p < 0.05 was considered to be statistically significant and represented as an asterisk (∗). Value of p < 0.01 was supposed to be more statistically significant and described as double asterisks (∗∗). Value of p < 0.001 was considered the most statistically significant and represented as triple asterisks (∗∗∗). Value of p < 0.0001 was supposed to be extremely statistically substantial and described as quadruple asterisks (∗∗∗∗). For comparison between two treatments, a Student’s t-test was used. For comparison between each group with the mean of every other group within a dataset containing more than two groups, one-way ANOVA with Tukey’s multiple comparisons test was used.
Acknowledgments
This work was supported by the Special 2019-nCoV Project of the National Key Research and Development Program of China (2020YFC0841400), the Emergency Key Program of Guangzhou Laboratory (EKPG21-24), the Special 2019-nCoV Program of the Natural Science Foundation of China (NSFC; 82041002), the Special Research and Development Program of Guangzhou (202008070010), and the Important Key Program of the NSFC (81730060) (to H.Z.). This work was also supported by National Natural Science Foundation of China (32000613) and the Guangdong Basic and Applied Basic Research Foundation (2019A1515010882) (to Yiwen Zhang) and the Guangzhou Basic and Applied Basic Research Foundation (202102021094) (to Xu Zhang).
Author contributions
Conceptualization, Y.Y., Yiwen Zhang, and H.Z.; methodology, Y.Y., Xiantao Zhang, R.C., Yiwen Zhang, and H.Z.; investigation, Y.Y., Xiantao Zhang, R.C., Y. Li, B.W., R.L., X.W., Q.C., J.D., Yongli Zhang, T.C., F.Z., Y. Lin, S.Y., X.M., Xu Zhang, C.L., X.B., Y.P., K.D., T.P., X.H., Yiwen Zhang, and H.Z.; writing – original draft, Y.Y., Xiantao Zhang, R.C., Yiwen Zhang, and H.Z.; writing – review & editing, Y.Y., Xiantao Zhang, R.C., Yiwen Zhang, and H.Z.; funding acquisition, Yiwen Zhang and H.Z.; resources, R.C., F.Z., C.K., K.D., T.P., X.H., Yiwen Zhang, and H.Z.; supervision, H.Z.
Declaration of interests
The authors declare no competing interests.
Published: December 23, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.110256.
Supplemental information
Data and code availability
-
•
All data supporting the findings of this study are available within the paper or from the corresponding author upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Abu-Raddad L.J., Chemaitelly H., Butt A.A., National Study Group for C.-V. Effectiveness of the BNT162b2 Covid-19 vaccine against the B.1.1.7 and B.1.351 variants. N. Engl. J. Med. 2021;385:187–189. doi: 10.1056/NEJMc2104974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arunachalam P.S., Walls A.C., Golden N., Atyeo C., Fischinger S., Li C., Aye P., Navarro M.J., Lai L., Edara V.V., et al. Adjuvanting a subunit COVID-19 vaccine to induce protective immunity. Nature. 2021;594:253–258. doi: 10.1038/s41586-021-03530-2. [DOI] [PubMed] [Google Scholar]
- Brouwer P.J.M., Brinkkemper M., Maisonnasse P., Dereuddre-Bosquet N., Grobben M., Claireaux M., de Gast M., Marlin R., Chesnais V., Diry S., et al. Two-component spike nanoparticle vaccine protects macaques from SARS-CoV-2 infection. Cell. 2021;184:1188–1200.e1119. doi: 10.1016/j.cell.2021.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherian S., Potdar V., Jadhav S., Yadav P., Gupta N., Das M., Rakshit P., Singh S., Abraham P., Panda S. Convergent evolution of SARS-CoV-2 spike mutations, L452R, E484Q and P681R, in the second wave of COVID-19 in Maharashtra, India. bioRxiv. 2021 doi: 10.1101/2021.04.22.440932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond M., Chen R., Xie X., Case J., Zhang X., VanBlargan L., Liu Y., Liu J., Errico J., Winkler E. SARS-CoV-2 variants show resistance to neutralization by many monoclonal and serum-derived polyclonal antibodies. Res. Sq. 2021 doi: 10.21203/rs.3.rs-228079/v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edara V.-V., Lai L., Sahoo M., Floyd K., Sibai M., Solis D., Flowers M.W., Hussaini L., Ciric C.R., Bechnack S. Infection and vaccine-induced neutralizing antibody responses to the SARS-CoV-2 B. 1.617. 1 variant. bioRxiv. 2021 doi: 10.1101/2021.05.09.443299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galloway S.E., Paul P., MacCannell D.R., Johansson M.A., Brooks J.T., MacNeil A., Slayton R.B., Tong S.X., Silk B.J., Armstrong G.L., et al. Emergence of SARS-CoV-2 B.1.1.7 lineage - United States, december 29, 2020-january 12, 2021. Mmwr-Morbidity Mortality Weekly Rep. 2021;70:95–99. doi: 10.15585/mmwr.mm7003e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Beltran W.F., Lam E.C., Denis K.S., Nitido A.D., Garcia Z.H., Hauser B.M., Feldman J., Pavlovic M.N., Gregory D.J., Poznansky M.C. Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell. 2021;184:2372–2383.e9. doi: 10.1016/j.cell.2021.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaney A.J., Loes A.N., Crawford K.H.D., Starr T.N., Malone K.D., Chu H.Y., Bloom J.D. Comprehensive mapping of mutations in the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human plasma antibodies. Cell Host Microbe. 2021;29:463–476.e6. doi: 10.1016/j.chom.2021.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodcroft E.B., Domman D.B., Snyder D.J., Oguntuyo K., Van Diest M., Densmore K.H., Schwalm K.C., Femling J., Carroll J.L., Scott R.S., et al. Emergence in late 2020 of multiple lineages of SARS-CoV-2 Spike protein variants affecting amino acid position 677. medRxiv. 2021 doi: 10.1101/2021.02.12.21251658. [DOI] [Google Scholar]
- Hoffmann M., Arora P., Gross R., Seidel A., Hornich B.F., Hahn A.S., Kruger N., Graichen L., Hofmann-Winkler H., Kempf A., et al. SARS-CoV-2 variants B.1.351 and P.1 escape from neutralizing antibodies. Cell. 2021;184:2384–2393.e12. doi: 10.1016/j.cell.2021.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B., Dai L., Wang H., Hu Z., Yang X., Tan W., Gao G.F. Neutralization of SARS-CoV-2 VOC 501Y.V2 by human antisera elicited by both inactivated BBIBP-CorV and recombinant dimeric RBD ZF2001 vaccines. bioRxiv. 2021 doi: 10.1101/2021.02.01.429069. [DOI] [Google Scholar]
- Huang Q., Ji K., Tian S., Wang F., Huang B., Tong Z., Tan S., Hao J., Wang Q., Tan W., et al. A single-dose mRNA vaccine provides a long-term protection for hACE2 transgenic mice from SARS-CoV-2. Nat. Commun. 2021;12:776. doi: 10.1038/s41467-021-21037-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keech C., Albert G., Cho I., Robertson A., Reed P., Neal S., Plested J.S., Zhu M., Cloney-Clark S., Zhou H., et al. Phase 1-2 trial of a SARS-CoV-2 recombinant spike protein nanoparticle vaccine. N. Engl. J. Med. 2020;383:2320–2332. doi: 10.1056/NEJMoa2026920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreier F. 'Unprecedented achievement': who received the first billion COVID vaccinations? Nature. 2021 doi: 10.1038/d41586-021-01136-2. [DOI] [PubMed] [Google Scholar]
- Kumar V., Singh J., Hasnain S.E., Sundar D. Possible link between higher transmissibility of B. 1.617 and B. 1.1. 7 variants of SARS-CoV-2 and increased structural stability of its spike protein and hACE2 affinity. bioRxiv. 2021 doi: 10.1101/2021.04.29.441933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letko M., Marzi A., Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020;5:562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Nie J., Wu J., Zhang L., Ding R., Wang H., Zhang Y., Li T., Liu S., Zhang M., et al. SARS-CoV-2 501Y.V2 variants lack higher infectivity but do have immune escape. Cell. 2021;184:2362–2371.e9. doi: 10.1016/j.cell.2021.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Liu J., Zhang H. The challenge of emerging SARS-CoV-2 mutants to vaccine development. J. Genet. Genomics. 2021;48:102–106. doi: 10.1016/j.jgg.2021.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Ma X., Deng J., Chen Q., Liu W., Peng Z., Qiao Y., Lin Y., He X., Zhang H. Differential efficiencies to neutralize the novel mutants B.1.1.7 and 501Y.V2 by collected sera from convalescent COVID-19 patients and RBD nanoparticle-vaccinated rhesus macaques. Cell Mol Immunol. 2021;18:1058–1060. doi: 10.1038/s41423-021-00641-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Shi Y., Zhang W., Li R., He Z., Yang X., Pan Y., Deng X., Tan M., Zhao L., et al. Recovered COVID-19 patients with recurrent viral RNA exhibit lower levels of anti-RBD antibodies. Cell Mol Immunol. 2020;17:1098–1100. doi: 10.1038/s41423-020-00528-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X., Zou F., Yu F., Li R., Yuan Y., Zhang Y., Zhang X., Deng J., Chen T., Song Z., et al. Nanoparticle vaccines based on the receptor binding domain (RBD) and heptad repeat (HR) of SARS-CoV-2 elicit robust protective immune responses. Immunity. 2020;53:1315–1330.e19. doi: 10.1016/j.immuni.2020.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyo-Gwete T., Madzivhandila M., Makhado Z., Ayres F., Mhlanga D., Oosthuysen B., Lambson B.E., Kgagudi P., Tegally H., Iranzadeh A., et al. Cross-reactive neutralizing antibody responses elicited by SARS-CoV-2 501Y.V2 (B.1.351) N. Engl. J. Med. 2021 doi: 10.1056/NEJMc2104192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planas D., Bruel T., Grzelak L., Guivel-Benhassine F., Staropoli I., Porrot F., Planchais C., Buchrieser J., Rajah M.M., Bishop E., et al. Sensitivity of infectious SARS-CoV-2 B.1.1.7 and B.1.351 variants to neutralizing antibodies. Nat. Med. 2021:1–8. doi: 10.1038/s41591-021-01318-5. [DOI] [PubMed] [Google Scholar]
- Plante J.A., Mitchell B.M., Plante K.S., Debbink K., Weaver S.C., Menachery V.D. The variant gambit: COVID-19's next move. Cell Host Microbe. 2021;29:508–515. doi: 10.1016/j.chom.2021.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell A.E., Zhang K., Sanyal M., Tang S., Weidenbacher P.A., Li S., Pham T.D., Pak J.E., Chiu W., Kim P.S. A single immunization with spike-functionalized ferritin vaccines elicits neutralizing antibody responses against SARS-CoV-2 in mice. ACS Cent. Sci. 2021;7:183–199. doi: 10.1021/acscentsci.0c01405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanathan M., Ferguson I., Miao W., Khavari P. SARS-CoV-2 B. 1.1. 7 and B. 1.351 Spike variants bind human ACE2 with increased affinity. bioRxiv. 2021 doi: 10.1016/S1473-3099(21)00262-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resende P.C., Bezerra J.F., Vasconcelos R., Arantes I., Appolinario L., Mendonça A.C., Paixao A.C., Rodrigues A.C.D., Silva T., Rocha A.S. Spike E484K mutation in the first SARS-CoV-2 reinfection case confirmed in Brazil, 2020. Virological. 2021;10 [Google Scholar]
- Saunders K.O., Lee E., Parks R., Martinez D.R., Li D., Chen H., Edwards R.J., Gobeil S., Barr M., Mansouri K., et al. Neutralizing antibody vaccine for pandemic and pre-emergent coronaviruses. Nature. 2021;594:553–559. doi: 10.1038/s41586-021-03594-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X., Tang H., Pajon R., Smith G., Glenn G.M., Shi W., Korber B., Montefiori D.C. Neutralization of SARS-CoV-2 variants B.1.429 and B.1.351. N. Engl. J. Med. 2021;384:2352–2354. doi: 10.1056/NEJMc2103740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinde V., Bhikha S., Hoosain Z., Archary M., Bhorat Q., Fairlie L., Lalloo U., Masilela M.S.L., Moodley D., Hanley S., et al. Efficacy of NVX-CoV2373 Covid-19 vaccine against the B.1.351 variant. N. Engl. J. Med. 2021;384:1899–1909. doi: 10.1056/NEJMoa2103055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skelly D.T., Harding A.C., Gilbert-Jaramillo J., Knight M.L., Longet S., Brown A., Adele S., Adland E., Brown H., Team M.L. 2021. Vaccine-Induced Immunity Provides More Robust Heterotypic Immunity than Natural Infection to Emerging SARS-CoV-2 Variants of Concern. [DOI] [Google Scholar]
- Sun D., Sang Z., Kim Y.J., Xiang Y., Cohen T., Belford A.K., Huet A., Conway J.F., Sun J., Taylor D.J., et al. Potent neutralizing nanobodies resist convergent circulating variants of SARS-CoV-2 by targeting novel and conserved epitopes. bioRxiv. 2021 doi: 10.1101/2021.03.09.434592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegally H., Wilkinson E., Giovanetti M., Iranzadeh A., Fonseca V., Giandhari J., Doolabh D., Pillay S., San E.J., Msomi N. Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. medRxiv. 2020 doi: 10.1101/2020.12.21.20248640. [DOI] [Google Scholar]
- Walls A.C., Fiala B., Schafer A., Wrenn S., Pham M.N., Murphy M., Tse L.V., Shehata L., O'Connor M.A., Chen C., et al. Elicitation of potent neutralizing antibody responses by designed protein nanoparticle vaccines for SARS-CoV-2. Cell. 2020;183:1367–1382.e1317. doi: 10.1016/j.cell.2020.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292.e86. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Horby P.W., Hayden F.G., Gao G.F. A novel coronavirus outbreak of global health concern. Lancet. 2020;395:470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Zhou X., Bian Y., Wang S., Chai Q., Guo Z., Wang Z., Zhu P., Peng H., Yan X., et al. Dual-targeting nanoparticle vaccine elicits a therapeutic antibody response against chronic hepatitis B. Nat. Nanotechnol. 2020;15:406–416. doi: 10.1038/s41565-020-0648-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Nair M.S., Liu L., Iketani S., Luo Y., Guo Y., Wang M., Yu J., Zhang B., Kwong P.D., et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021;593:130–135. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- Wang R., Zhang Q., Ge J., Ren W., Zhang R., Lan J., Ju B., Su B., Yu F., Chen P., et al. Spike mutations in SARS-CoV-2 variants confer resistance to antibody neutralization. bioRxiv. 2021 doi: 10.1101/2021.03.09.434497. [DOI] [Google Scholar]
- Wu K., Choi A., Koch M., Elbashir S., Ma L., Lee D., Woods A., Henry C., Palandjian C., Hill A. Variant SARS-CoV-2 mRNA vaccines confer broad neutralization as primary or booster series in mice. bioRxiv. 2021 doi: 10.1101/2021.04.13.439482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahalom-Ronen Y., Tamir H., Melamed S., Politi B., Shifman O., Achdout H., Vitner E.B., Israeli O., Milrot E., Stein D., et al. A single dose of recombinant VSV-G-spike vaccine provides protection against SARS-CoV-2 challenge. Nat. Commun. 2020;11:6402. doi: 10.1038/s41467-020-20228-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.S., Huang F., Xia B.J., Yuan Y.C., Yu F., Wang G.W., Chen Q.Y., Wang Q., Li Y.Z., Li R., et al. The interferon-stimulated exosomal hACE2 potently inhibits SARS-CoV-2 replication through competitively blocking the virus entry. Signal. Transduction Targeted Ther. 2021;6:1–11. doi: 10.1038/s41392-021-00604-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Davis B.D., Chen S.S., Sincuir Martinez J.M., Plummer J.T., Vail E. Emergence of a novel SARS-CoV-2 variant in southern California. JAMA. 2021;325:1324–1326. doi: 10.1001/jama.2021.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Yuan Y., Wu B., Wang X., Lin Y., Luo Y., Li R., Chen T., Deng J., Zhang X., et al. Improvement of a SARS-CoV-2 vaccine by enhancing the conjugation efficiency of the immunogen to self-assembled nanoparticles. Cell Mol. Immunol. 2021;18:2042–2044. doi: 10.1038/s41423-021-00736-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Chen Y., Li Y., Huang F., Luo B., Yuan Y., Xia B., Ma X., Yang T., Yu F., et al. The ORF8 protein of SARS-CoV-2 mediates immune evasion through down-regulating MHC-Iota. Proc. Natl. Acad. Sci. U S A. 2021;118 doi: 10.1073/pnas.2024202118. e2024202118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Hu H., Liu W., Yan S.M., Li Y., Tan L., Chen Y., Liu J., Peng Z., Yuan Y., et al. Amino acids and RagD potentiate mTORC1 activation in CD8(+) T cells to confer antitumor immunity. J. Immunother. Cancer. 2021;9:e002137. doi: 10.1136/jitc-2020-002137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D., Dejnirattisai W., Supasa P., Liu C., Mentzer A.J., Ginn H.M., Zhao Y., Duyvesteyn H.M.E., Tuekprakhon A., Nutalai R., et al. Evidence of escape of SARS-CoV-2 variant B.1.351 from natural and vaccine-induced sera. Cell. 2021;184:2348–2361.e2346. doi: 10.1016/j.cell.2021.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All data supporting the findings of this study are available within the paper or from the corresponding author upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.