Abstract
The tablet manufacturing process is a complex system, especially in continuous manufacturing (CM). It includes multiple unit operations, such as mixing, granulation, and tableting. In tablet manufacturing, critical quality attributes are influenced by multiple factorial relationships between material properties, process variables, and interactions. Moreover, the variation in raw material attributes and manufacturing processes is an inherent characteristic and seriously affects the quality of pharmaceutical products. To deepen our understanding of the tablet manufacturing process, multivariable modeling techniques can replace univariate analysis to investigate tablet manufacturing. In this review, the roles of the most prominent multivariate modeling techniques in the tablet manufacturing process are discussed. The review mainly focuses on applying multivariate modeling techniques to process understanding, optimization, process monitoring, and process control within multiple unit operations. To minimize the errors in the process of modeling, good modeling practice (GMoP) was introduced into the pharmaceutical process. Furthermore, current progress in the continuous manufacturing of tablets and the role of multivariate modeling techniques in continuous manufacturing are introduced. In this review, information is provided to both researchers and manufacturers to improve tablet quality.
Multivariate modeling techniques play important roles in the tablet manufacturing process.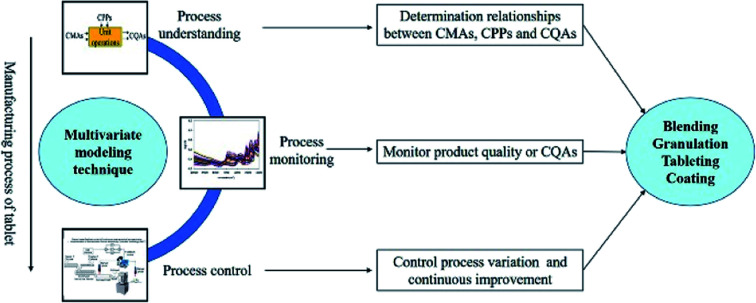
1. Introduction
Tablets are the prevalent dosage forms, which have many advantages, including high-precision dosing, good physical and chemical stability, manufacturing efficiency, and low costs.1 In the global pharmaceutical market, it has been speculated that tablets are the preferred dosage form and tablet sales are expected to exceed US$500 billion by the end of 2027.2
Due to the enormous market share of tablets, higher requirements have been put forward to improve product quality and supervision. As mentioned by Yu and Kopcha, there is still a long way to go to improve the quality of pharmaceutical products.3 One of the major challenges is determining the relationship between raw material attributes, process conditions, and critical quality attributes.4 The tablet manufacturing process involves powder blending, granulation, tablet compression, and coating operation units. This process makes it difficult for univariate methods to comprehensively study the tablet manufacturing process.
Mathematical models, which could provide a scientific understanding of the manufacturing process and predict the state of the pharmaceutical system, play an important role in the tablet manufacturing process. According to an investigational report by Kourti and Davis in 2012,5 the application of mathematical modeling can achieve lots of benefits, including improving product quality, and enhancing product and process understanding. For mechanism models, the discrete element method (DEM)6,7 and finite element method (FEM)8,9 have been used less often than the multivariable modeling technique in tablet manufacturing, due to the complexity of raw material properties and the continuum duality of particles.10 For empirical models, multivariate modeling techniques are one of the most commonly used statistical models. In the pharmaceutical field, with the application of process analytical technology (PAT) and with the emergence of continuous manufacturing (CM), multivariate modeling techniques are feasible options for mathematically describing the pharmaceutical process.11 For example, regression models and latent variable models (LVMs) can extract useful information related to product quality from a large amount of data.
In this paper, different multivariate model techniques are introduced for process understanding, optimization, quality monitoring, and control in the powder blending process, granulation, tableting and the coating process. To comprehensively investigate the roles of multivariate modeling techniques in the tablet manufacturing process, relevant studies published in the past 15–20 years (mostly within 15 years) were searched on the Web of Science, by using the following search terms: QbD, empirical model, statistics model, pharmaceutical tablet, blending, granulation, coating, continuous manufacture, near-infrared (NIR), Raman, chemometrics, process model, multivariate data analysis, and partial least squares (PLS). The review is organized as follows: Section 2 briefly exhibits the theoretical background of commonly-used multivariate modeling techniques. Section 3 describes good modeling practice (GMoP) in the pharmaceutical process. Sections 4 and 5 provide an overview of multivariate modeling techniques in the basic unit operations and continuous manufacturing of tablets.
2. Brief theoretical background of commonly-used multivariate modeling techniques
In the pharmaceutical process, multivariate methods can be used to maximize speed and minimize costs.12 Multivariate analysis (MVA) models, also called data-driven models, are usually available for analyzing data obtained from the pharmaceutical process. In this review, we aimed to illustrate the applications of multivariate modeling techniques within the tablet manufacturing process, and, therefore, little effort has been expended on the theoretical aspects of the multivariate modeling techniques. Brief explanations of the theoretical background are given below.
2.1. Brief theory of simple regression
Various regression models can be used to investigate the relationships between independent and response variables. Multiple linear regression (MLR) and polynomial regression are often used to analyze data from the design of experiment (DoE). These models quantify the relationship between several explanatory variables and one or more response variables.
The MLR model is fitted by minimizing the sum of squared residuals based on least squares. According to the principle, the regression coefficient can be calculated with eqn (1)
| B = (XTX)−1XTY | 1 |
In eqn (1), Y, X, B, and XT represent the response variable matrix, independent variables, regression coefficient matrix and transpose matrix of the independent variables, respectively. However, the inverse of XTX no longer exists due to collinearity between the independent variables. Except for cases where X-variables are controlled in designed experimentation, measured data in pharmaceutical applications are typically multivariate and collinear and MLR cannot be used.13 As a special linear regression model, polynomial regression describes the nonlinear relationship between independent and dependent variables in pharmaceutical manufacturing.
2.2. Basic theoretical concepts in latent variable modeling
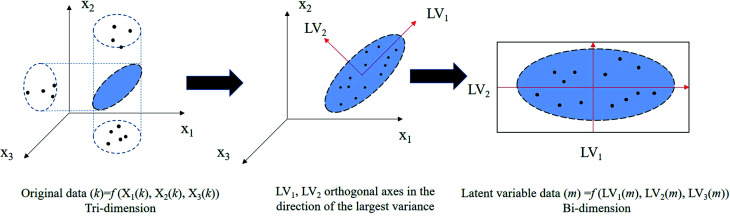
The latent variable model (LVM) is a statistical model that can be used to process large amounts of correlated data. LVMs were used for experimental data as early as the 1970s.14 By extracting latent variables (LVs), an LVM projects original high-dimensional data into a low-dimensional space, also called the latent space. The new LVs still retain as much original information as possible. Next, in the latent space, the relationships among samples, independent variables, and response variables can be analyzed. Fig. 1 displays a straightforward interpretation of LVMs.15 For a detailed introduction to LVMs, refer to related studies.10,13,16,17
Fig. 1. Simple interpretation of the LVM.
In the last decade, principal component analysis (PCA), partial least squares (PLS) and orthogonal partial least squares (OPLS) were commonly used in the pharmaceutical manufacturing process. However, PCA and PLS models are not suitable for dealing with more complex datasets including more than two matrices. Multi-block PLS (MB-PLS),18 L-shape PLS19 and joint-Y PLS (JY-PLS)20 can analyze complex datasets, and quantify the relationship between input matrices and output matrices.
2.3. Basic theoretical concepts about artificial neural networks
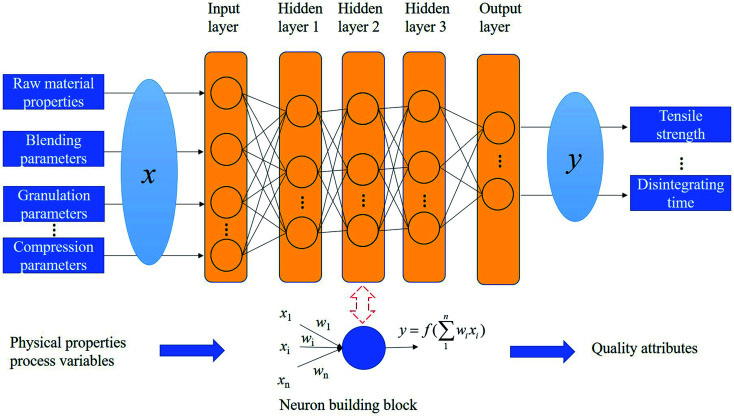
The artificial neural network (ANN) model is a type of machine learning tool, which attempts to imitate the neural structures of the biological brain. In general, ANN models can be roughly divided into two types: static ANN and dynamic ANN.21 Although many neural networks have been continually invented, all ANN models can be described by neurons, also called processing elements (PE), learning algorithms, and connection formulas.22 In the neuronal system, a PE receives one or more signals, and input data is multiplied by its weight. Finally, an activation function can generate an output.23 For the detailed principle of ANN, refer to previous publications.23–25Fig. 2 shows a feedforward multi-layer perceptron (MLP) network, which is the most frequently and successfully used ANN model in pharmaceutical processes.21 MLP can develop a non-linear mapping between independent variables and response variables by using many parallel neurons.
Fig. 2. Structure of a multilayer artificial neural network in pharmaceutical manufacturing.
However, building an ANN model involves lots of parameters and activation functions. Hence, it is not easy to obtain any insight into the structure of approximate functions. In other words, because of intermediate equations, the relationship between the input and output variables is not a direct path.26 Consequently, it is difficult to gain the mathematical equation between independent and response variables.21 Although the ANN model is a black-box model, it is a powerful tool to solve non-linear problems and multi-response systems. One of its main advantages is that it does not require a rule-based experimental design. Moreover, ANN can use historical data or incomplete data to map functions. Because the ANN model can be used to efficiently develop different response surfaces, it plays a crucial role in drug delivery and pharmaceutical processes.22
3. Brief introduction to good modeling practice in the pharmaceutical process
To minimize errors in the process of modeling, or in analysis and subsequent applications,27 it is necessary to implement GMoP to build multivariate models.
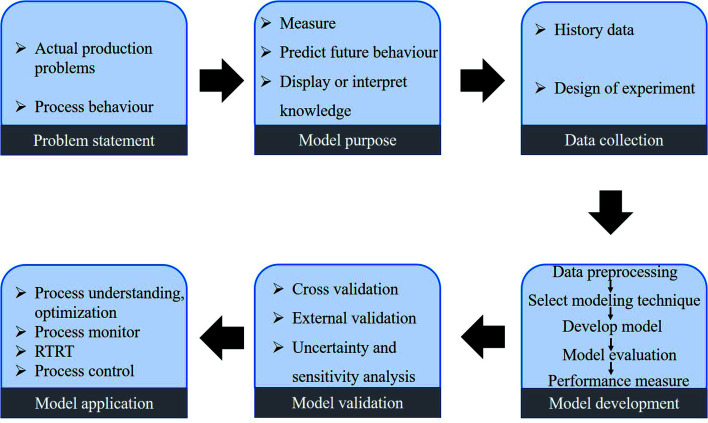
According to the practice guidelines for building a model, the key components of GMoP should include model purpose, model evaluation, and performance measures.28,29 To standardize the modeling process in the GMoP, a model development framework is proposed (shown in Fig. 3). In the pharmaceutical process, the first step in developing the model process is to put forward the issues according to the actual situation and the modeling purpose. In the first step, it is important to choose an appropriate modeling technique. For the second step, one needs to acquire data according to the DoE or historical data in the manufacturing process. Subsequently, the data should be organized and preprocessed before model development. In this stage, it is suggested that exploratory analysis of the data should be implemented to select variables. Next, the model can be built, and the performance of the model can be evaluated. In this step, especially the development of multivariate models, the interpretability of the model needs to be considered. Finally, after validation, the developed model can be used in the actual manufacturing process.
Fig. 3. Model development framework of pharmaceutical process.
In the GMoP, model verification and validation are essential parts of the modeling process before using the model for any pharmaceutical unit operation, and are directly related to the accuracy and applicability of the model. Consequently, it is necessary to perform model evaluation. For the model validation of the ANN model and LVMs, chemometric indicators, such as correlation coefficient and root mean square error (RMSE), are in general used to evaluate the model. To evaluate an MLR or a polynomial model, in addition to the above-mentioned indexes, the model should be further studied for statistical analysis, such as analysis of variance (ANOVA). The premise for further application of these developed models is that the P-value of the model is less than 0.05, and the P-value of the lack of fit is greater than 0.05. When the model meets these requirements, the relationship between response variables and independent variables is significant, and the equation is well fitted.
Model validation does not indicate model verification. Nevertheless, in the actual modeling process, model validation is always combined with model verification. The purpose of model validation is to prove that the model recreates the behavior of the system with high fidelity to meet the analysis goal. The commonly-used validation method compares the predictive model results to experimental data by using the internal or external data set. According to ICH Points to Consider (R2) guidelines,30 the model can be divided into three categories: high impact models, medium impact models, and low impact models. The developed model should be validated at different levels based on the category used in practice. For a high impact model, validation by the internal data set is often inadequate. It is necessary to use the external data set to validate model performance. In addition, data sets used for validation should take the expected variability into account in a future pharmaceutical process. Model accuracy, especially for statistically-based models, should be validated by uncertainty analysis.31,32 However, a low impact model may not implement rigorous verification. Finally, according to the pharmaceutical quality system of the company, the validated model can be used in the manufacturing process, and should be continuously updated and maintained.
4. Application of multivariate modeling techniques in tablet manufacturing
The tablet manufacturing process generally includes three pathways: i.e., direct compaction (DC), dry granulation, and wet granulation. This section will summarize and discuss the application of multivariate modeling techniques in the tablet manufacturing process.
4.1. Pharmaceutical process understanding and optimization based on multivariate modeling techniques
The main role of understanding the pharmaceutical process is identifying and controlling critical sources of variation that affect product quality. The objective is to understand how raw material properties and process variables can affect quality attributes. Based on process knowledge, pharmaceutical processes can be optimized.
4.1.1. Understanding and optimizing the blend process
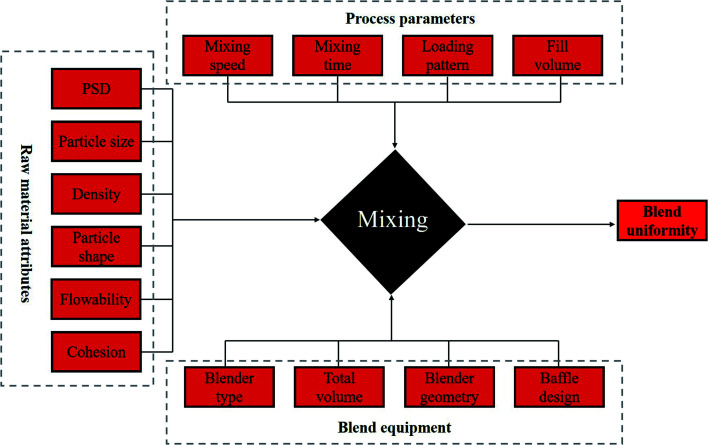
The blending unit operation is one of the basic processes for preparing solid dosage forms. In addition, it is a critical unit operation for ensuring that pharmaceutical powder mixing is homogeneous. Only homogeneous powders can ensure the content uniformity (CU) of active pharmaceutical ingredients (APIs).33,34 Blending uniformity (BU) is a critical quality attribute (CQA) of the intermediate and final product because it relates significantly to drug quality, safety, and therapeutic efficacy. However, during the blending process, BU is strongly impacted by several factors, including raw material attributes, mixing equipment, process parameters and environmental conditions (Fig. 4).35–41
Fig. 4. Critical variability for blend uniformity.
To explore the relationship between the above factors and BU, the effects of raw material properties and process parameters on the mixing process can be explored by multivariate modeling techniques. Table 1 shows the application of multivariate models in the powder mixing process. It can be seen from Table 1 that multiple regression models and LVMs were widely used. DoE-integrated multivariate modeling techniques were often utilized to quantify the impact of various factors on the mixing process, which helps to identify CMAs (critical material attributes) or CPPs (critical process parameters). Based on established models, a design space can be developed to control the mixing process, and this is beneficial for guiding and optimizing the mixing unit operation.
The applications of multivariate methods in the mixing process.
| Models | Blender type | Application | References |
|---|---|---|---|
| MLR | KG-5 blender | The model was used to correlate the critical formulation and CPPs with the response variables. A design space for the powder mixing process was built | 42 |
| MLR | Continuous mixer | The relationship between residence time and total particle length was explored | 43 |
| Polynomial | V-blender | The effects of CPPs on CQAs were quantified to identify their relationship, and the design space was established | 44 |
| Quadratic model | Cone shape tank | Based on the regression model, the optimal mixing conditions, including the impeller speed and eccentricity, were found | 45 |
| PLS | Twin-screw blender | In twin-screw blend feeding, the relationship between blend material properties and feeding capacity was developed | 46 |
| PLS | Square cone mixer | The relationship between raw material variability and mixing time was quantified. The CMAs affecting the mixing process were identified according to VIP | 47 |
| PLS and MLR | Continuous mixer | The effect of material properties on the mean residence time was studied. The relationship between bulk density and mean residence time at different flow rates was determined | 48 |
4.1.2. Understanding and optimizing the granulation process
Granulation, as a process of powder agglomeration and size enlargement, is a key pharmaceutical unit operation. Prior to tablet compression, granulation can significantly improve the physical properties of the powder, including flowability and density.49–52 To obtain optimal characteristics of granules, statistical or mechanistic models have been used to study the physical properties of granules produced by various granulation methods. In this section, the application of multivariate modeling techniques to commonly-used granulation methods is summarized, such as high shear wet granulation (HSWG), roller compaction and twin-screw granulation processes.
HSWG is an important granulation method because of its inherent advantages. It includes several operational steps, such as mixing and wet mass. Therefore, this process is complex, and influenced by many factors, including agitator speed, massing time, liquid addition rate, and interactions between them. Based on the QbD approach, the CMAs and CPPs can be identified in the HSWG process.53 In order to comprehensively understand and study the HSWG process, multivariate modeling techniques were applied combined with QbD or DoE to explore the impact of the CMAs and CPPs on granule characteristics.54Table 2 summarizes the application of multivariate modeling techniques in HSWG. For example, Zhang et al.55 used QbD principles to improve their understanding of the HSWG process. Firstly, the CPPs were screened through a Plackett–Burman experimental design. Then, based on the Box–Behnken experimental design, a multivariate analysis (MVA) model was developed to investigate the relationship between granule size and CPPs. Finally, a design space was developed to optimize the HSWG process. Han et al.56 applied risk assessment to determine three CPPs in HSWG based on prior knowledge. In this study, the effects of agitator speed, spray rate, and massing time on the CQAs of granules were quantified by the MVA model.
Applications of multivariate models in HSWG.
| Models | Application | References |
|---|---|---|
| Polynomial regression | The effects of operational parameters, such as impeller speed, dosing speed, chopper speed and wet massing time, on granule size were quantified | 57 |
| The effects of process parameters, including granulation time, impeller, and formulation variables, on packing coefficient and strength of granules were investigated | 58 | |
| The relationship between granulation variables and the specific energy of the granules was determined | 59 | |
| The best-fit equation was used to accurately predict the Carr's index for granules under different formulation factors | 60 | |
| Polynomial, MLR | The impact of formulation variables on granule properties like flowability and size was assessed, which was beneficial for selecting the desired formulation | 61 |
| Combined with DoE, models which correlated the process parameters with granule properties, were developed. This provided the basis for adjusting process parameters according to the product quality attributes | 62 | |
| Using DoE techniques, the effects of amount of water and massing time on the key quality attributes of granules were investigated | 63 | |
| PLS | The relationship between impeller speed and total power spectral densities (TPSDs) was developed. The research demonstrated that audible acoustic emissions could monitor process changes in real time | 64 |
| Gene expression programing model | Impeller power can be predicted according to the impeller diameter, impeller speed, the percentage of the liquid and mean torque | 65 |
| PCA, MLR | The relationship between process variables on granule hardness and Carr's index was developed. Based on the PCA model, it was shown that there was a strong correlation between the impeller speed and wet massing time with the granule attributes | 66 |
| Polynomial, MLR, PLS, ANNs | Based on various MVA models, the relationship between three process parameters and CQAs of granules such as mean size and flowability was quantified | 67 |
| PLS, MBPLS, OPLS | Various MVA models were developed to investigate the effects of HSWG process variables and granule properties on tablet quality | 68 |
For dry granulation, roller compaction is the most commonly-used type of dry granulation in the manufacturing of tablets. The roller compaction process does not use any liquid binders, which are available for water or heat-sensitive pharmaceutical materials.69 In the roller compaction process, mixed powders are first fed from the hopper to the compaction region by a screw. Then, the powder in the compaction region is pressed by rotating rollers to form a compacted ribbon.69,70 Finally, the ribbon is transported to a size reduction region, where the ribbon is milled to the desired particle size.
The traditional trial and error approach is very time-consuming and costly for developing or optimizing process parameters. Therefore, several mathematical models were established, including one-dimensional (1D), 2D, and 3D models to study the roller compaction process.70–73 However, due to the diversity of pharmaceutical powders and the complexity of manufacturing solid dosage forms, these mathematical models are only suitable for particular powders and roller compactors,74 making it challenging to build a “one size fits all” model.75
The ribbon properties can significantly affect the attributes of granules, such as flowability, granule size distribution (GSD), and compressibility.52,76–78 They may influence the tablet compression process. Therefore, it is necessary to develop multivariate models to investigate the roller compaction process. In many studies, the MVA model was developed to study the complex roller compaction process.79–81 For example, Yu et al.74 developed a PLS model to explore the effect of raw material properties on ribbon properties using 81 pharmaceutical powders. Next, the CMAs and CPPs of roller compaction were identified based on the VIP of the PLS model. Finally, feasible material properties and operating regions were found by developing a multi-objective design space. The applications of the multivariate modeling techniques in the roller compaction process are presented in Table 3.
The applications of multivariate models in roller compaction.
| Multivariate model | Application | References |
|---|---|---|
| Polynomial model | The influence of process variables on ribbon properties like density and granule size was determined, which was helpful for obtaining optimal process parameters according to the target quality | 82 |
| The model was developed to explore the relationship between process variables like roll force, gap and bypass, and bypass potency | 83 | |
| Using DoE, the effects of the particle size of the raw material and fraction of API on the ribbon attributes were investigated | 84 | |
| Polynomial, MLR | The quantitative relationship, which correlated process variables with ribbon properties such as normal stress and density, was investigated. It can be used to predict the required process parameters | 85 |
| The quantitative relationship between operating process variables and pre-blend properties on normal stress was determined. The effects of normal stress and roller gap on the ribbon density were investigated | 86 | |
| The impact of process operating conditions on the nip angle and ribbon density were quantified through developing various mathematical models in the scale-up process | 73 | |
| PCA, PLS | Based on the PCA and PLS models, the effects of raw material attributes and process parameters on the ribbon properties were explored | 87 |
| PLS | Several models were developed to investigate relationships between raw material properties, process variables and the properties of ribbon and tablet | 88 |
| JY-PLS | The JY-PLS model was used for scale-up from laboratory roller compactor to full-scale roller compactor, which effectively reduced the risk of the scale-up process | 89 |
| ANNs | In roller compaction, the models were used to investigate the relationships between the formulation variables and tablet properties. Furthermore, the formulation was optimized according to the genetic model | 90 |
With the background of continuous manufacturing (CM) in the pharmaceutical industry, twin-screw granulation (TSG) is an alternative technique for transforming a batch pharmaceutical process to continuous manufacturing.91 It can granulate powders to granules in a short time and efficiently mix API and excipient.92 TSG includes three main components, respectively conveying elements, kneading blocks and comb mixer elements. To enhance the manufacturing efficiency of TSG, it is important to comprehensively study the impact of formulation and process parameters on the granule attributes. Many studies have reported on the relationship between CMAs, CPPs and CQAs.93–98 For example, Keleb et al.99 found that granulation yield decreased when increasing the screw speed. For the application of multivariate modeling techniques in TSG, Stauffer et al.100 developed an MLR model to investigate the relationships between raw material variability, process variables, and CQAs, such as GSD and granule friability. In their study, PCA was used to analyze the batch-to-batch variability of physical properties. According to the process variables in TSG, Ismail et al. built an ANN model to predict mean residence time distribution based on the screw speed and L/S ratio in the process of twin-screw granulation.101
In addition, a few studies have reported the effect of mill process parameters on the granule properties in drying granulation. Most of these studies focus on the properties of the ribbon. Consequently, a comprehensive study of all the granulation processes combined with multivariable modeling techniques is warranted.
4.1.3. Process understanding and optimization of tablet compaction
In general, the tablet compression process includes three consecutive steps: powder or granule die filling first, then compression, and finally ejection of the tablet.102 In this process, tablet formation may involve rearrangement and elastic and plastic deformation, thereby forming inter-particulate bonds.103 In brief, the mechanism of forming tablets is more complicated, leading to a complex tablet compaction process. Even a small change in the compression process has a serious impact.104 The multivariate modeling technique is a useful tool to understand the tableting process.
4.1.3.1. Die filling in tablet compaction
The weight variation between tablets, content uniformity, tensile strength, disintegration time, friability, and dissolution were defined as CQAs.105 However, these CQAs mainly depend on die filling during tablet compaction. Non-uniformity in die filling results directly in variation in the weight and content uniformity of the tablets and may critically affect the compression force.106 Hence, die filling is a critical process for obtaining the required quality of the tablet. Although a DEM could explain underlying physical phenomena by first principles107 and is used in the die filling process,108–111 the computational costs were high. At present, multivariate modeling techniques still play a key role in investigating die filling. Studies performed at Ghent University used multivariate modeling techniques to systemically study the die filling process of high-speed rotary tablet compactors.112,113 In this study, a PCA model was used to analyze powder physical properties or classify relevant data obtained by DoE experiments. A PLS model was developed to investigate the relationship between attributes of blend powder, process conditions, and responses of product and process. This revealed that powder flowability, density, turret speed, paddle speed, feed frame, etc. significantly affect the die filling performance. According to the developed model, the process conditions can be optimized to meet target CQAs. In addition, multivariate models combined with a DEM were applied to study the die filling process. The main function of the DEM is to simulate the die filling process and to visualize the movement of particles in the die. Multivariate models, such as PLS and polynomial models, were used to investigate the effects of potential CMAs, potential CPPs, and their interaction on tablet weight variation, segregation, tablet mass, etc.110,114
4.1.3.2. Tableting process
The tablet manufacturing process can be regarded as a whole system. The effects of powder properties, mixing parameters, granulation parameters, and tableting parameters on CQAs can be comprehensively investigated. Researchers at Wyeth Pharmaceuticals studied the effects of granulation process parameters, granule properties and tableting parameters on dissolution behavior.115 In this study, a PLS model was used to quantify the relationship between these variables and to identify the CMAs and CPPs. Sun et al.116 used a D-optimal experimental design to study tablet quality. Tablets were prepared using various raw materials under different process parameters of HSWG and compression. In their study, a MBPLS model was developed to quantify the relationship between raw material properties, various process conditions, tensile strength, and tablet disintegration time. The critical pharmaceutical unit operation, CMAs and CPPs were identified by the MBPLS model. Finally, according to the multiple linear regression model, a multi-objective design space was established to control tablet quality. Table 4 shows the many multivariate modeling techniques for studying the tableting process.
The applications of multivariate models in the tableting process.
| Model | Application | References |
|---|---|---|
| Polynomial model | The relationship between raw material properties and tablet tensile strength was quantified | 84 |
| PCA | The model was used to evaluate the relationship between various powder properties for direct compaction | 117 |
| PCA, PLS | The PCA model can investigate the relationship between various powder and compression properties. The PLS model was developed to identify key effecting factors, and to quantify the relationship between those factors and tablet tensile strength | 118 |
| The purpose of developing the PLS model was to study the effects of input variables, such as properties of raw materials or intermediates, and process conditions on tablet dissolution | 119 | |
| The PCA model can reduce the dimensionality of the original data and explore the relationship between different physical properties. The PLS model was developed to investigate the effects of raw material attributes, tableting process variables and compression behavior indices on the tablet quality attributes | 120 | |
| PCA, MBPLS | According to the PCA model, the variability in powder and granules was analyzed. MBPLS was used to identify critical factors and critical process units. It can quantify the relationship between process variables and tablet dissolution | 121 |
| PCA, MLR | The linear model was used to determine the effect of excipient properties on tablet attributes, and the PCA model was built to correlate the properties of filler and binder with tablet properties | 122 |
| PLS | The purpose of developing the PLS model was to explore the impact of process variables on the mass flow rate per unit orifice area in the die filling | 123 |
| MLR | Based on the model, the effects of roller compaction conditions and milling process variables on the attributes of granule and tablet were studied | 124 |
| ANN | The relationships between material physical properties, process parameters and tablet quality attributes were determined | 125 |
In an LVM, such as a PCA or PLS model, multivariate modeling techniques are mainly applied in the tableting compression process. The MBPLS algorithm analyzes the multiple block data from various unit operations in tablet manufacturing, which is beneficial for investigating the tablet manufacturing process.
4.1.4. Coating process
Coating of tablets is an important pharmaceutical process. In general, during the coating process, tablets are repeatedly exposed to a spray that contains several solutes and solvents. The coating process is repeated until the desired coating quality and/or uniformity are achieved.126 The purpose of coating includes masking the taste or odor to improve compliance, thereby obtaining a better appearance and easier distinguishability, changing drug release behavior, and adding a second API in the coat.127–129 The pharmaceutical coating process needs to ensure that the tablets are coated uniformly.130 To ensure uniformity of the coating, the variability of the coating is mainly controlled by the thickness of inter- and intra-coating. Although coating is a widespread pharmaceutical process and has been used for many decades, there are still some serious issues, such as a lack of comprehensive understanding of the coating process.131
To solve the above issues and promote a more in-depth understanding, the multivariate modeling technique can be used to study the coating process. Rege et al.132 applied the Plackett–Burman experimental design to investigate the relationship between manufacturing parameters and final product quality. In their study, the effects of process parameters on the CQAs were analyzed by an MLR model. The atomization pressure, pan speed, and coating time were identified as CPPs affecting content uniformity. For the film coating process, Tanabe et al.133 generated a PLS model according to the data derived from the production process. Moreover, Cahyadi et al.134 used a multivariate model combined with DoE to optimize process parameters in the quasi-continuous tablet coating process. Table 5 shows other studies involved in the application of multivariate modeling techniques in the coating process.
The applications of multivariate modeling techniques in the coating process.
| Models | Application | References |
|---|---|---|
| MLR, polynomial | The MLR model identifies critical factors affecting CQAs. The polynomial model quantifies the relationship between process variables and CQA in the coating process. Based on the model, the optimal region for process variables was defined | 135 |
| According to a full factorial design, MLR models were developed to quantify the relationship between the coating process variable and loss on drying, coating process efficiency, and coating uniformity | 136 | |
| Polynomial | Using a full factorial design, polynomial models were developed to correlate coating process conditions with response variables, such as coating uniformity and surface roughness | 137 |
| According to DoE, based on the polynomial model, the effects of critical process variables on coating uniformity were investigated at lab and pilot scales | 138 | |
| Based on the central composite design, the polynomial model was developed to study the relationship between key process parameters and CQAs, such as weight gain and surface roughness | 139 | |
| Multivariant model | According to a central composite – face-centered response surface design, the relationship between five process parameters and CQAs like tablet appearance were correlated by a multivariant model. The optimal process variables could be determined based on the model | 140 |
| Quadratic polynomial | A multivariate model, which links four operating variables, such as spray rate, rotation speed of pan, and spray temperature to the weight variability index, was established | 141 |
In summary, the above-mentioned tables (from Tables 1–5) show that PLS, MLR, and polynomial models are the most widely used in the pharmaceutical process, while OPLS, MBPLS, and ANN models only have a few applications. However, manufacturing tablets that includes several operation units may separate the process variables into specific blocks. MBPLS and OPLS can be applied to investigate the effects of each block of data on the response variables.142 The most prominent shortcoming of these methods in the process analysis is that the prediction blocks are independent. In the actual manufacturing process, the various process units are usually highly correlated; therefore, the multivariate modeling technique should consider these relationships. PLS-path modeling (PLS-PM) is beneficial for understanding the inner mechanism of the manufacturing process by incorporating process knowledge into predictive modeling.143 However, PLS-PM is rarely used in the tablet manufacturing process. An ANN model is a black-box model. However, in a recent study,144 an ANN model combined with specialized software identified CMAs and CPPs. This demonstrated that an ANN model is not an absolute “black-box” model and that ANN can be used as a valuable tool to understand and optimize the pharmaceutical process. Liu et al. developed a controllable and readable ANN model (CR-ANN),145 also called a white-box model. The CR-ANN model can improve the readability of the internal structure; therefore, it is easy to overcome the shortcomings of a traditional ANN model. CR-ANN can be further widely used in process understanding and optimization. In particular, the mechanism model combined with multivariate modeling techniques should be developed for the pharmaceutical process, which will be conducive to comprehensively gaining process knowledge. Thus, in the future, it will be necessary to expand the application of this modeling technique in the manufacturing process.
4.2. Pharmaceutical process monitoring based on multivariate modeling techniques
To ensure consistency of product quality and stability of therapeutics, it is necessary to monitor, analyze, and control each unit operation. With American Food and Drug Administration (FDA) guidance on PAT, most scholars and the pharmaceutical industry applied the new process analysis methods to research the pharmaceutical process combined with multivariate modeling techniques. Most of these studies used multivariate modeling techniques to establish a multivariate calibration model; then the calibration model was implemented to predict the quality of the product in real time.
4.2.1. Monitoring blending processes
To effectively detect the CQAs and gain a deeper understanding of the change in dynamics of the mixing process, near-infrared (NIR) instruments are widely used PAT tools to monitor the blending process.47,146–151 The application of Raman spectroscopy in a study of the mixing process has also been reported.152–154 However, in order to obtain meaningful results, spectral data must be transformed by chemometrics.47 Multivariate modeling approaches, which play an important role in translating spectral signals to BU, can be applied to monitor the mixing process.
For qualitative methods, PCA, as one LVM, is one of the most preeminent approaches that provides reliable results.49 PCA has been used in many studies to detect and monitor mixing homogeneity.49,155 For PCA, one method plotted the score graph of the first principal component (PC1) as a function of mixing time. It assumes that mixed uniformity is achieved when the score level is stable and close to a point.156,157 In another method, the first two principal components of homogeneous powders (the target mixture) were analyzed. If the powders (the test mixture) approach or overlap the target mixture cluster in the score plot, this would indicate that the test mixture had achieved a state of uniformity.157,158 For the PLS model, the content of constituents can be predicted during the mixing process. Firstly, it is necessary to develop an off-line PLS calibration model.159 Then, ingredient contents could be predicted in real time during the mixing process. When the deviation of component concentration reaches a pre-defined limit within a specific time, it can be considered that the test mixture had reached a state of mixed uniformity.160 Another multivariate model is the ANN algorithm. An ANN model detects blend uniformity in a similar manner to a PLS model. Compared with a PLS model, the performance of an ANN model is excellent.146 However, in practice, considering the complexity and time-consuming nature of developing an ANN model, ANN models were applied relatively less often. This is shown in Table 6.
Pharmaceutical applications of multivariate modeling techniques in monitoring the powder mixing process.
| Models | Characterization methods | Application | References |
|---|---|---|---|
| PCR | NIR | Detection of blending homogeneity | 156, 158, 161 and 162 |
| NIR-CI | Detection of blending end-point | 163 | |
| NIR-CI | Determination of mixture homogeneity | 164 | |
| NIR | Confirming the end-point of blending | 165 | |
| Raman | Confirming the end-point of blending | 165 | |
| NIR | The endpoint of the start-up phase | 166 | |
| NIR | Process spectral data | 167 | |
| NIR | Determining the blend uniformity | 146 | |
| PLS | NIR | Detection of blending end-point | 147, 158, 159, 165, 168 and 169 |
| NIR-CI | Verifying the NIRS analyzer response and assessing homogeneity | 167 | |
| NIR | Drug concentration | 169 | |
| NIR | Confirmation of blend uniformity | 170 | |
| NIR | Monitoring the drug level at the outlet of the continuous blender | 166 | |
| NIR | Measurement of contents | 171 and 172 | |
| NIR | Assessment of powder blend uniformity | 146 and 173–176 | |
| NIR | Evaluation of degree of homogeneity | 177 | |
| PCA-ANN | NIR | Measurement of blend uniformity | 175 |
| ANN | Image analysis | Prediction of mixing time | 178 |
| ANN | NIR | Determining the blend uniformity | 146 |
4.2.2. Monitoring the granulation process
The multivariate modeling technique is not only used to understand the granulation process, but also to monitor the granule properties or content of API based on PAT. For example, Khorasani et al.179 applied a PCA model to map the ribbon porosity distribution based on near-infrared chemical imaging (NIR-CI) technology. The API concentrations in the ribbon were quantified by a PLS model. In many studies, NIR-CI or NIR were used, as shown in Table 7. NIR or microwaves can measure key properties of the ribbon online during the roller compaction process in real time. For instance, Gupta et al.180 used a PLS calibration model to simultaneously predict relative density, moisture content, and tensile strength in real time. Gupta et al.181 used microwave online real-time monitoring density, API content and moisture content of ribbon based on ANN or PLS models. In this study, the performance of the different multivariate models was compared. The prediction accuracy of microwave and NIR sensors was also compared. In addition, the multivariate modeling technique has been widely used to predict or detect CQAs or API contents in the twin screw granulation process.182–186
Applications of multivariate models in monitoring or detecting the granulation process.
| Model | Application | References |
|---|---|---|
| PCA, PLS | The PCA model was used to detect changes in ribbon density qualitatively. However, the PLS model can quantitatively monitor ribbon density during the roller compaction process | 187 |
| The envelope density and moisture content of the ribbon were monitored | 188 | |
| The PCA model can be used to analyze textural descriptors of the ribbon. The density can be predicted by the PLS model | 189 | |
| The PCA model can obtain the variation between various ribbons, and the PLS model can detect the porosity of the ribbon | 190 | |
| The water contents in the wet mass can be measured by in-line NIR based on the PLS model | 191 | |
| PLS | Granule properties like granule size can be detected in real time according to the PLS model | 192 |
| It can be used to monitor the hardness of the ribbon online | 193 | |
| The content uniformity and ribbon properties, like moisture content or relative density, could be determined in real time | 180 and 194 | |
| The key ribbon properties, like relative density and tensile strength, could be predicted by the model | 195 and 196 | |
| The model was used to predict the solid fraction of ribbon | 197 | |
| The attributes of flakes, such as tensile strength, Young's modulus, and relative density, can be monitored in real time | 194 | |
| PLS, ANN | Based on the models, the APIs in traditional Chinese medicine (TCM) granules can be detected | 198 |
4.2.3. Monitoring or prediction of tablet quality
The pharmacopoeia stipulates that items to be inspected for tablet quality include content uniformity, hardness, dissolution, and disintegration time. Content uniformity is a critical quality parameter, which is directly related to drug efficacy. The distribution of API in the tablet, which may affect texture and dissolution, is another CQA. The disintegration time affects the release of the drug in vivo. Mechanical properties, such as fragility and hardness, affect the transport and storage of the tablets. Hence, non-destructive and quick detection of the quality of tablets is an important task. For example, Blanco et al.199 used NIR spectroscopy to simultaneously detect the chemical composition and physical properties of tablets. In many reports, the component content in the tablet can be predicted by multivariate modeling techniques200–203 in real-time monitoring of the tableting process. In many studies, real-time release test (RTRT) models were developed, and aimed to monitor or predict the CQAs of the product during the process, without requiring lengthy off-line assays.204 For example, based on a PLS or MLR model, the content uniformity, hardness, disintegration time, friability, and dissolution of tablets can be tested in the manufacturing process,204,205 and can significantly reduce the release time. The applications of multivariate modeling techniques combined with the PAT tool to predict or monitor tablet CQAs are shown in Table 8.
The applications of multivariate models in monitoring or detecting CQAs for tablets.
| Model | Tool | Application | References |
|---|---|---|---|
| MLR | NIR | Detection of tablet hardness | 199 and 206 |
| NIR | Measurement of drug content | 207 | |
| PCA | NIR | Measurement of drug content | 208 |
| PCR | NIR | Detection of tablet hardness | 209–212 |
| NIR | Measurement of tablet weight variation | 212 | |
| NIR | Prediction of tablet porosity | 209 | |
| PLS | NIR | Detection of tablet hardness | 199, 206, 207, 213 and 214 |
| NIR | Prediction of tablet porosity | 213 | |
| NIR | Measurement of tensile strength | 215–217 | |
| Multispectral UV imaging | Measurement of tensile strength | 218 | |
| NIR | Measurement of moisture | 216 | |
| NIR | Prediction of dissolution behavior | 199 and 219–227 | |
| NIR-CI | Prediction of dissolution behavior | 228 | |
| Raman | Prediction of dissolution behavior | 224 | |
| NIR | Measurement of drug content | 199, 201, 203, 214, 216 and 229–249 | |
| NIR-CI | Measurement of drug content | 250 and 251 | |
| Raman | Measurement of drug content | 235, 248, 252 and 253 | |
| MIR | Measurement of drug content | 241 | |
| Multispectral UV imaging | Measurement of drug content | 218 | |
| NIR | Measurement of disintegration time | 254 | |
| NIR | Measurement of tablet weight | 237 | |
| ANN | NIR | Prediction of dissolution behavior | 224 |
| Raman | Prediction of dissolution behavior | 224 | |
| NIR | Measurement of drug content | 214, 229 and 255 | |
| NIR | Detection of tablet hardness | 214 |
4.2.4. Monitoring or prediction of the tablet coating process
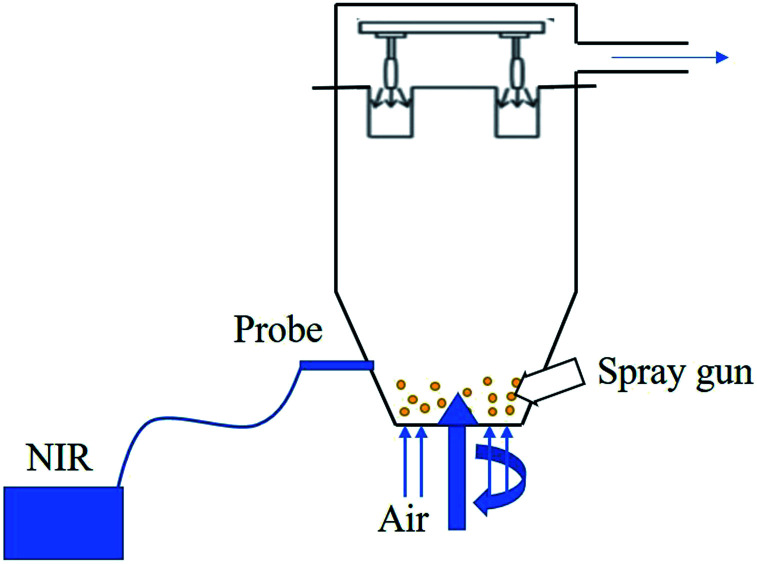
Based on the multivariate model, there are many reports of using the PAT tool to monitor the coating process. As the most commonly used PAT tool, NIR can measure and monitor any CQAs during the coating process. Romer et al. developed an off-line PLS model to in-line predict the coating thickness in the film process.256 Lee et al.257 used the following equipment (Fig. 5) to in-line monitor the fluid bed coating process based on a PCA model and a PLS model. There are many studies on NIR testing of the coating process based on a multivariate model, as shown in Table 9, which shows that NIR combined with the multivariate modeling technique is widely applied in the pharmaceutical coating process.
Fig. 5. Diagram for NIR in-line monitoring coating process.
The applications of multivariate models in monitoring or detecting for the coating process.
| Model | Application | Analysis tool | References |
|---|---|---|---|
| PCA | Clustering of spectral data | NIR | 257 and 262 |
| PCA | Visualization of the coating process | NIR | 263 |
| PLS | Prediction of coating thickness | NIR | 256, 257, 262 and 263 |
| End-point detection of a coating | NIR | 264 | |
| Prediction of weight gain | NIR | 262 and 263 | |
| Prediction of moisture content | NIR | 263, 265 and 266 | |
| Measurement of curing degree | NIR | 267 | |
| Prediction of API content | Raman | 259, 260 and 268–270 | |
| Prediction of weight gain | Raman | 259 | |
| Prediction of coating thickness | Raman | 271 | |
| Prediction of moisture content | Raman | 266 |
As another commonly used PAT tool, Raman spectroscopy is also used in the pharmaceutical coating process. Hagrasy et al. first demonstrated that real-time monitoring of the coating process could be achieved by using in-line Raman spectroscopy.258 Muller et al.259–261 systematically investigated the application of Raman spectroscopy in the coating process. In their study, a calibration PLS model was built to link Raman spectral data to diprophylline contents in the coating layer. According to the PLS model, the diprophylline contents could be determined in real time. The developed method was validated in accordance with ICH guidelines. This proved the feasibility of in-line monitoring by Raman spectroscopy of the coating process combined with a multivariable modeling technique. In the following study, Raman spectroscopy could measure other CQAs, such as thickness based on the PLS model. Table 9 summarizes the applications of NIR and Raman spectroscopy in the coating process, and shows that PLS models are widely used in the coating process.
In summary, in pharmaceutical process monitoring, a PLS model is one of the standard tools in the arsenal of making predictions based on the large amounts of data collected during production processes.143 A PLS model can extract the most predictive and correlative information from a large number of spectral and high-dimensional data. Consequently, a PLS model is powerful in monitoring and predicting the tablet manufacturing process. However, the disadvantage is that the model handles all metrics without hierarchy or conditions. Therefore, a PLS model is limited in understanding the pharmaceutical process that produces variation in the measurement data.272 Therefore, much of the process knowledge is still untouched, because information about the pharmaceutical process is not comprehensively included in predictive modeling. The performance of an ANN model was proven to be superior to conventional multivariate modeling techniques.273 For example, according to a study by Nagy et al.,224 it was found that the prediction error provided by an ANN model was lower than that of a PLS model in detecting dissolution behavior. However, ANN models in pharmaceutical process monitoring are still used relatively less often, which is in agreement with the findings presented in a previous report.224 For an ANN model, since the spectra contain thousands of variables, directly inputting them as independent variables into the ANN model will greatly increase the complexity of the model and significantly reduce the efficiency of modeling. This may be one of the reasons why ANN model are used less often in pharmaceutical process monitoring. Dimensionality reduction of the spectral data is an effective way to improve the efficiency of ANN modeling, while maintaining a strong prediction performance. In the pharmaceutical process, PCA is always used to reduce the dimensionality of the data. Hence, an ANN model combined with PCA is utilized to monitor CQAs in the tablet manufacturing process.175
4.3. Pharmaceutical process control based on multivariate modeling techniques
Pharmaceutical product quality is multivariate in nature, and all measured properties must meet requirements simultaneously. Process control depends on an understanding of the pharmaceutical process and the accumulation of relevant knowledge. Multivariate modeling techniques, especially LVMs, are important tools for pharmaceutical process control.
LVMs can be used to analyze the historical data from the pharmaceutical process, and thus corresponding feedback or feedforward control strategies could be developed. For example, combined with the NIR tool, Hattori274 developed a robust feed-forward control model based on a PLS model in the tableting process. The process parameters predicted by the control model could produce tablets that met the required quality. Westerhuis et al.275 established a step-by-step control method for the wet granulation and tableting process based on PLS and MBPLS. First, the properties of the granules were determined by the formulation variables and granulation process parameters. Then, the tableting process parameters were adjusted according to the properties of the granules. Consequently, this control scheme for manufacturing tablets can adjust process conditions to better meet the specifications. Based on a PLS model, García et al.276 developed a feed-forward control method to deal with the variation in initial raw material quality in high-speed shear wet granulation. A comprehensive design space was developed, considering a complex network of relationships between process conditions and product quality. Muteki et al.277 developed a feed-forward control method for the process of dry granulation and tableting. For different batches of raw materials, this method can simulate and calculate the best operating parameters, and reduce the differences in tablet quality.
In addition, based on the multivariate modeling technique, the multivariate statistical process control (MSPC) tool deals with large sets of data derived from pharmaceutical processes. It is an effective tool for process control. For example, Burggraeve et al.278 developed a real-time online monitoring method for fluidized bed granulation based on LVMs. This method monitors the quality of particles in real time, and products that do not meet the expected quality requirements can be identified. Table 10 shows the applications of these models in manufacturing tablets.
The applications of multivariate models in process control for the tablet manufacturing process.
| Model | Application | References |
|---|---|---|
| MSPC | Based on the PCA model, the MSPC tool can be developed to control the granulation and drying process. After the deviation has been corrected, the process system can return to a stable state | 279 |
| An MSPC tool was built based on PLS and PCA models. The control chart was used to monitor humidity and temperature in the granulation process and to detect process abnormalities | 280 | |
| PLS | According to the feed-forward PLS model, it can be used to determine the process conditions based on the preceding conditions of unit operation | 254 |
| According to the PLS model, the feed-forward process can construct a control plan to determine the optimal process conditions. Then a release profile meeting the required quality was obtained | 281 | |
| The feed-forward model for tablet compaction was developed based on NIR. Based on this model, the process parameters that meet the final product quality can be predicted | 282 | |
| Based on NIR, the PLS model can measure the bulk density in real time. The measured signals can be used for forward feed control to ensure small density variation | 283 | |
| Latent projection model | According to the latent projection model, the variations in raw material and from batch-to-batch can be controlled by adjusting some process parameters | 284 |
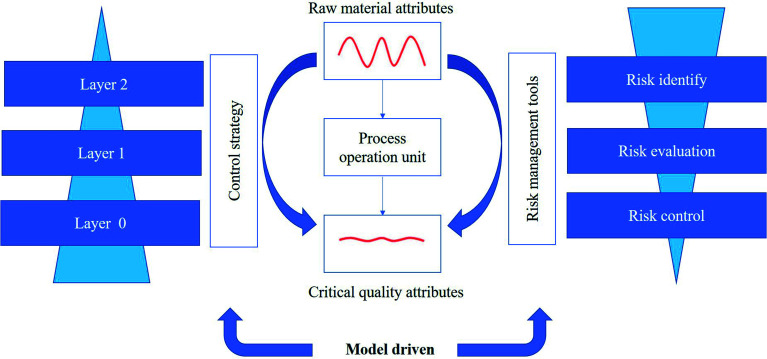
In the tablet manufacturing process, multivariate modeling techniques play an important role in process control. The core of using the multivariate model technique for process quality control is model inversion. According to the quality goals for the final product, optimal process operating conditions can be solved by the developed model, which can accurately determine the specific plan and autonomously control it. Consequently, the risk of manufacturing a product outside the acceptance criteria would be decreased. Based on the hierarchical three-layer control design proposed by Su et al.,285 the above-mentioned control strategies are layer 1 control approaches. At this control level, an efficient feedback or feed-forward control method is used to diminish the effect of variation that may propagate in downstream unit operations.286 Another feature of layer 1 control is that automatic adjustment usually spans several pharmaceutical operation units. Real-time monitoring of CQAs is one of the key components in layer 1 control. However, this remains challenging due to the complexity of PAT tool calibration and model validation in real time.287 In particular, PAT sensor positions, sampling problems, and contamination can cause measurement drift and deviations, which can affect the accuracy of real-time process data. Therefore, it is necessary to systematically study the PAT tool network design to develop approaches for maintaining reliable measurements of CQA. In the future, layer 2 control design should be developed in the tablet manufacturing process based on dynamic models. At this control level, many advanced process control techniques, such as predictive models and real-time optimization, can be integrated to achieve multi-input multi-output control. Even smart manufacturing (SM)288,289 can be developed for the pharmaceutical process, which can rapidly and in real time optimize manufacturing pharmaceutical products and the dynamic response to the demand for pharmaceutical product quality.290 Combined with risk management tools (Fig. 6), the consistency of the quality of the final product can be ensured.
Fig. 6. Overview of the control strategy combined with risk management tools.
5. Application of multivariate modeling techniques in a continuous manufacturing line
For manufacturing tablets, continuous manufacturing (CM), as an approach for modernizing the production system, can produce pharmaceutical products of higher quality.291 Because CM has many advantages, such as flexibility, speeding up the supply chain, lower risk of stock-outs, and reduced space and investment costs,292,293 it has been widely used. Nowadays, seven drug products from four different pharmaceutical companies, Vertex, Johnson & Johnson, Eli Lilly and Pfizer, have been approved by different regulatory agencies for continuous manufacturing.294 Drug products from CM have brought significant economic benefits to these four pharmaceutical companies. A study from Transparency Market Research forecast that from 2016 to 2025, the market values of pharmaceutical continuous manufacturing technology might increase from US$1.74 billion to US$3.69 billion.295 Therefore, the CM of pharmaceutical products has vast market prospects.
Using the quality by control (QbC) approach,287 multivariate modeling techniques are a useful tool for studying continuous manufacturing. For example, as an integrated continuous tablet manufacturing line, the PROMIS-line was developed at the University of Eastern Finland.296 Furthermore, in the process of powder feeding, the PCA model was applied to analyze the process data and the time evolution of powder feeding can be visualized. In another study at the University of Eastern Finland an end-to-end continuous direct compression (CDC) line was built.201,297 In these studies, multivariate modeling techniques, including PLS and OPLS models, were used to correlate spectral data with API contents. Hattori et al.282 developed a feed-forward control method to control a tablet continuous compression process based on a PLS model. Pauli et al.298 applied LVM to monitor the continuous manufacturing tablet process. In their study, a PLS model was used to predict the API content to evaluate blending uniformity and content uniformity in the tablets. The PCA model qualitatively describes the process monitoring. Stauffer et al.299 investigated the relationships between the variability of raw materials, CPPs and CQAs based on empirical models, such as PLS and MLR models. A design space of the CM line was developed, which can guarantee the consistency of the quality of the final product and process stability. Reports on the CM of tablets are presented in Table 11.
The applications of multivariate modeling techniques in continuous tablet manufacturing.
| Model | Application | References |
|---|---|---|
| PLS, PCA | In the continuous powder blending and tableting process, the PCA model, as an exploratory data analysis tool, was used to explore the effects of experimental variables on PAT spectra. The PLS model was applied to predict the CQAs of the tablet | 300 |
| In the CDC manufacturing process, the PCA model was applied to identify possible outliers or abnormalities. Transmission NIR spectroscopy, combined with the PLS model, was used to measure blending uniformity and detect tablet content uniformity | 301 | |
| In the CM line, the PLS model was performed to detect CQAs. The multivariate analysis model could be applied for process monitoring | 302 | |
| The PCA model was performed to analyze spectral data. The PLS model, which links spectral data to response variables of interest, was established. The developed multivariate models can be integrated into the online prediction tool | 303 | |
| Based on the PCA and PLS model, multivariate monitoring charts could monitor various units in the continuous manufacturing process | 304 | |
| PLS/PCA, multiblock PCA/PLS | Various latent models were used to describe and monitor the time variables in the continuous twin-screw granulation and drying process. It can detect and diagnose deviations in the continuous manufacturing process | 305 |
| PCA, MLR | In the RTRT of the tablet, the multivariate model was developed to predict dissolution profiles in a CDC system. The NIR data were processed by the PCA model. Based on the NIR spectra, MLR could be applied to predict tablet dissolution behavior | 204 |
| PLS | Based on the calibration model, the powder density in a continuous line can be predicted | 306 |
| The PLS model was developed to predict blending powder bulk density in the CDC manufacturing process based on the NIR data | 283 | |
| Using the NIR tool, the developed off-line PLS calibration model could monitor the continuous pharmaceutical manufacturing process's API concentration | 307 | |
| NIR spectroscopy as a PAT tool was used to measure API content combined with the PLS model | 308 | |
| MLR | MLR models were built to explore the relationship between process conditions and response variables, such as flowability, ejection force, and tablet strength. Based on the model, a design space was developed for high-dose tablets in CM | 309 |
| PCA | The PCA model can extract concentration-related information from NIR spectral data | 310 |
Although continuous manufacturing has many advantages and CM lines, such as that of GEA Pharma Systems, have been built,294 the progress towards CM is still slow.292 Furthermore, CM is facing many challenges, such as developmental challenges, and supervisory and quality control challenges.291 In the CM process, there is limited hold-up in each pharmaceutical operation unit. Thus, the variability of raw materials in upstream unit operations can rapidly and directly affect downstream pharmaceutical processes, which can significantly impact final pharmaceutical product quality.285 Furthermore, control engineering theories for pharmaceutical continuous manufacturing process have not yet achieved a common understanding or wider application in the pharmaceutical process. The material library, which was proposed to gain sufficient knowledge about raw material attributes,74,311,312 has not been studied for the CM process in solid oral dosages. In the future, based on the material library, multivariate modeling techniques or hybrid models should be established to study the relationship between critical equipment settings (CEEs), CMAs, CPPs and CQAs. Continuous improvement is another important task in the CM process. However, relevant studies are rarely reported by the pharmaceutical industry. Based on the QbC concept, continuous improvement in pharmaceutical CM can easily be achieved at multiple levels, such as process performance monitoring, and predictive maintenance.287 To ensure the consistent quality of the product at every pharmaceutical operation unit, comprehensive control strategies and risk analysis should be proposed and implemented in the pharmaceutical process.
6. Conclusion
The tablet manufacturing process is inherently multivariate because it involves several basic pharmaceutical unit operations and produces multivariate data. However, the multivariate modeling technique is an appropriate tool to analyze these data. Consequently, this article provides a review of the applications of multivariate modeling techniques in tablet manufacturing. Multivariate modeling techniques can be used to understand the complex pharmaceutical phenomenon by investigating the relationships between measured variables. It can, to some extent, reveal the causes of process behavior, and plays an irreplaceable role in process optimization, monitoring, control, and continuous manufacturing processes. Through this review, we have aimed to provide more information to both researchers and manufacturers to improve tablet quality.
Conflicts of interest
The authors declare no conflict of interest.
Supplementary Material
Acknowledgments
This review was supported by the National Major Scientific and Technological Special Project for “Significant New Drugs Development” (2019ZX09301160, 2019ZX09721001-005-002).
References
- Patel S. Kaushal A. M. Bansal A. K. Crit. Rev. Ther. Drug Carrier Syst. 2006;23:1–66. doi: 10.1615/CritRevTherDrugCarrierSyst.v23.i1.10. [DOI] [PubMed] [Google Scholar]
- Solid dose: under-hyped but not under-represented, https://www.pharmamanufacturing.com/articles/2019/solid-dose-under-hyped-but-not-under-represented/
- Yu L. X. Kopcha M. Int. J. Pharm. 2017;528:354–359. doi: 10.1016/j.ijpharm.2017.06.039. [DOI] [PubMed] [Google Scholar]
- Ojha V. K. Schiano S. Wu C. Snasel V. Abraham A. Neural. Comput. Appl. 2018;29:467–481. doi: 10.1007/s00521-016-2545-8. [DOI] [Google Scholar]
- Kourti T. Davis B. Pharmaceut. Eng. 2012;32:52–62. [Google Scholar]
- Behjani M. A. Motlagh Y. G. Bayly A. E. Hassanpour A. Powder Technol. 2020;366:73–81. doi: 10.1016/j.powtec.2019.10.102. [DOI] [Google Scholar]
- Hildebrandt C. Gopireddy S. R. Scherlies R. Urbanetz N. A. Adv. Powder Technol. 2020;31:755–769. doi: 10.1016/j.apt.2019.11.030. [DOI] [Google Scholar]
- Takayama K. Sato T. Sato K. Todo H. Obata Y. Sugibayashi K. J. Drug Delivery Sci. Technol. 2019;52:1021–1031. doi: 10.1016/j.jddst.2019.06.017. [DOI] [Google Scholar]
- Kumar A., Dhondt J., Bertels J., Klingeleers D., Gernaey K. V., Nopens I. and De Beer T., 2017, https://biblio.ugent.be/publication/8542476
- Ierapetritou M. G. and Ramachandran R., Process simulation and data modeling in solid oral drug development and manufacture, Springer, 2016 [Google Scholar]
- Troup G. M. Georgakis C. Comput. Chem. Eng. 2013;51:157–171. doi: 10.1016/j.compchemeng.2012.06.014. [DOI] [Google Scholar]
- Gabrielsson J. Lindberg N. O. Lundstedt T. J. Chemom. 2002;16:141–160. doi: 10.1002/cem.697. [DOI] [Google Scholar]
- Rajalahti T. Kvalheim O. M. Int. J. Pharm. 2011;417:280–290. doi: 10.1016/j.ijpharm.2011.02.019. [DOI] [PubMed] [Google Scholar]
- Kowalski B. R. J. Chem. Inf. Comput. Sci. 1975;15:201–203. doi: 10.1021/ci60004a002. [DOI] [Google Scholar]
- Tomba E. Facco P. Bezzo F. Barolo M. Int. J. Pharm. 2013;457:283–297. doi: 10.1016/j.ijpharm.2013.08.074. [DOI] [PubMed] [Google Scholar]
- Geladi P. Kowalski B. R. Anal. Chim. Acta. 1986;185:1–17. doi: 10.1016/0003-2670(86)80028-9. [DOI] [Google Scholar]
- Ferreira A. P. Tobyn M. Pharm. Dev. Technol. 2015;20:513–527. doi: 10.3109/10837450.2014.898656. [DOI] [PubMed] [Google Scholar]
- Westerhuis J. A. Kourti T. Macgregor J. F. J. Chemom. 1998;12:301–321. doi: 10.1002/(SICI)1099-128X(199809/10)12:5<301::AID-CEM515>3.0.CO;2-S. [DOI] [Google Scholar]
- Kettanehwold N. Chemom. Intell. Lab. Syst. 1992;14:57–69. doi: 10.1016/0169-7439(92)80092-I. [DOI] [Google Scholar]
- Munoz S. G. Macgregor J. F. Kourti T. Chemom. Intell. Lab. Syst. 2005;79:101–114. doi: 10.1016/j.chemolab.2005.04.009. [DOI] [Google Scholar]
- Ibrić S. Djuriš J. Parojčić J. Djurić Z. Pharmaceutics. 2012;4:531–550. doi: 10.3390/pharmaceutics4040531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutariya V. Groshev A. Sadana P. Bhatia D. Pathak Y. Open Bioinf. J. 2013;7:49–62. doi: 10.2174/1875036201307010049. [DOI] [Google Scholar]
- Bourquin J. Schmidli H. Van Hoogevest P. Leuenberger H. Pharm. Dev. Technol. 1997;2:95–109. doi: 10.3109/10837459709022615. [DOI] [PubMed] [Google Scholar]
- Bourquin J. Schmidli H. van Hoogevest P. Leuenberger H. Pharm. Dev. Technol. 1997;2:111–121. doi: 10.3109/10837459709022616. [DOI] [PubMed] [Google Scholar]
- Jain A. K. Mao J. Mohiuddin K. Computer. 1996;29(3):31–44. doi: 10.1109/2.485891. [DOI] [Google Scholar]
- Simões M. F. Silva G. Pinto A. C. Fonseca M. Silva N. E. Pinto R. M. Simões S. Eur. J. Pharm. Biopharm. 2020;152:282–295. doi: 10.1016/j.ejpb.2020.05.012. [DOI] [PubMed] [Google Scholar]
- Sin G. Gernaey K. V. Lantz A. E. Biotechnol. Prog. 2009;25:1043–1053. doi: 10.1002/btpr.166. [DOI] [PubMed] [Google Scholar]
- Good modelling practice, https://digitalcommons.unl.edu/cgi/viewcontent.cgi?article=1072&context=usepapapers
- Crout N., Kokkonen T., Jakeman A. J., Norton J. P. and Whitfield P., Good Modelling Practice, 2008, vol. 3, pp. 15–31 [Google Scholar]
- ICH-Endorsed Guide for ICH Q8/Q9/Q10 Implementation, https://database.ich.org/sites/default/files/Q8_Q9_Q10_Q%26As_R4_Points_to_Consider_2.pdf
- Saffaj T. Ihssane B. Jhilal F. Bouchafra H. Laslami S. Sosse S. A. Analyst. 2013;138:4677–4691. doi: 10.1039/C3AN00519D. [DOI] [PubMed] [Google Scholar]
- Wu Z. Xu B. Du M. Sui C. Shi X. Qiao Y. J. Pharm. Biomed. Anal. 2012;62:1–6. doi: 10.1016/j.jpba.2011.12.005. [DOI] [PubMed] [Google Scholar]
- Scheibelhofer O. Balak N. Wahl P. Koller D. M. Glasser B. J. Khinast J. AAPS PharmSciTech. 2013;14:234–244. doi: 10.1208/s12249-012-9910-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Maesschalck R. Sanchez F. C. Massart D. L. Doherty P. Hailey P. A. Appl. Spectrosc. 1998;52:725–731. doi: 10.1366/0003702981944148. [DOI] [Google Scholar]
- Florianalgarin M. Mendez R. Powder Technol. 2015;276:156–165. doi: 10.1016/j.powtec.2015.02.024. [DOI] [Google Scholar]
- Escotetespinoza M. S. Moghtadernejad S. Oka S. Wang Y. Romanospino A. D. Schafer E. Cappuyns P. Van Assche I. Futran M. Ierapetritou M. G. Powder Technol. 2019;342:744–763. doi: 10.1016/j.powtec.2018.10.040. [DOI] [Google Scholar]
- Chaudhuri B. Mehrotra A. Muzzio F. J. Tomassone M. S. Powder Technol. 2006;165:105–114. doi: 10.1016/j.powtec.2006.04.001. [DOI] [Google Scholar]
- Flament M. P. Leterme P. Gayot A. Int. J. Pharm. 2004;275:201–209. doi: 10.1016/j.ijpharm.2004.02.002. [DOI] [PubMed] [Google Scholar]
- Jallo L. J. Ghoroi C. Gurumurthy L. Patel U. Dave R. N. Int. J. Pharm. 2012;423:213–225. doi: 10.1016/j.ijpharm.2011.12.012. [DOI] [PubMed] [Google Scholar]
- Alyami H. Dahmash E. Z. Bowen J. Mohammed A. PLoS One. 2017;12 doi: 10.1371/journal.pone.0178772. doi: 10.1371/journal.pone.0178772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenoy P. Viau M. Tammel K. Innings F. Fitzpatrick J. J. Ahrne L. Powder Technol. 2015;272:165–172. doi: 10.1016/j.powtec.2014.11.023. [DOI] [Google Scholar]
- Wu H. White M. Khan M. A. Org. Process Res. Dev. 2015;19:215–226. doi: 10.1021/op500085m. [DOI] [Google Scholar]
- Portillo P. M. Vanarase A. U. Ingram A. Seville J. Ierapetritou M. G. Muzzio F. J. Chem. Eng. Sci. 2010;65:5658–5668. doi: 10.1016/j.ces.2010.06.036. [DOI] [Google Scholar]
- Yeom S. B. Choi D. H. Pharmaceutics. 2019;11:264. doi: 10.3390/pharmaceutics11060264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hormann T. Suzzi D. Adam S. Khinast J. J. Pharm. Innov. 2012;7:181–194. doi: 10.1007/s12247-012-9142-x. [DOI] [Google Scholar]
- Stauffer F. Vanhoorne V. Pilcer G. Chavez P. Schubert M. A. Vervaet C. De Beer T. Eur. J. Pharm. Biopharm. 2019;135:49–60. doi: 10.1016/j.ejpb.2018.12.012. [DOI] [PubMed] [Google Scholar]
- Shi G. Xu B. Zhang Z. Yang C. Dai S. Lin Z. Shi X. Fu J. Qiao Y. Processes. 2019;7:568. doi: 10.3390/pr7090568. [DOI] [Google Scholar]
- Vanarase A. U. Osorio J. G. Muzzio F. J. Powder Technol. 2013;246:63–72. doi: 10.1016/j.powtec.2013.05.002. [DOI] [Google Scholar]
- Matero S. Den Berg F. V. Poutiainen S. Rantanen J. Pajander J. J. Pharm. Sci. 2013;102:1385–1403. doi: 10.1002/jps.23472. [DOI] [PubMed] [Google Scholar]
- Giry K. Genty M. Viana M. Wuthrich P. Chulia D. Drug Dev. Ind. Pharm. 2006;32:509–530. doi: 10.1080/03639040500529119. [DOI] [PubMed] [Google Scholar]
- Shanmugam S. BioImpacts. 2015;5:55. doi: 10.15171/bi.2015.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inghelbrecht S. Remon J. P. Int. J. Pharm. 1998;171:195–206. doi: 10.1016/S0378-5173(98)00195-1. [DOI] [Google Scholar]
- Pandey P. Badawy S. Drug Dev. Ind. Pharm. 2016;42:175–189. doi: 10.3109/03639045.2015.1100199. [DOI] [PubMed] [Google Scholar]
- Kumar A. Gernaey K. V. De Beer T. Nopens I. Eur. J. Pharm. Biopharm. 2013;85:814–832. doi: 10.1016/j.ejpb.2013.09.013. [DOI] [PubMed] [Google Scholar]
- Zhang Y. Cheng B. C. Zhou W. Xu B. Gao X. Qiao Y. Luo G. Pharmaceutics. 2019;11:519. doi: 10.3390/pharmaceutics11100519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J. K. Shin B. S. Choi D. H. Pharmaceutics. 2019;11:252. doi: 10.3390/pharmaceutics11060252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajdik J. Baki G. Szentkirallyi Z. Knop K. Kleinebudde P. Pintyehodi K. J. Pharm. Biomed. Anal. 2008;48:694–701. doi: 10.1016/j.jpba.2008.06.023. [DOI] [PubMed] [Google Scholar]
- Mangwandi C. Adams M. J. Hounslow M. J. Salman A. D. Int. J. Pharm. 2012;427:328–336. doi: 10.1016/j.ijpharm.2012.02.029. [DOI] [PubMed] [Google Scholar]
- Gabbott I. Husban F. A. Reynolds G. K. Eur. J. Pharm. Biopharm. 2016;106:70–78. doi: 10.1016/j.ejpb.2016.03.022. [DOI] [PubMed] [Google Scholar]
- Lee A. R. Kwon S. Y. Choi D. H. Park E. Int. J. Pharm. 2017;534:144–158. doi: 10.1016/j.ijpharm.2017.10.004. [DOI] [PubMed] [Google Scholar]
- Fayed M. H. Abdelrahman S. I. Alanazi F. K. Ahmed M. O. Tawfeek H. M. Ali B. E. Acta Pol. Pharm. 2017;74:551. [PubMed] [Google Scholar]
- Fayed M. H. Abdelrahman S. I. Alanazi F. K. Ahmed M. O. Tawfeek H. M. Alshdefat R. Drug Dev. Ind. Pharm. 2017;43:1584–1600. doi: 10.1080/03639045.2017.1326930. [DOI] [PubMed] [Google Scholar]
- Fayed M. H. Abdel-Rahman S. I. Alanazi F. K. Ahmed M. O. Tawfeek H. M. Al-Shedfat R. I. Acta Pol. Pharm. 2017;74:235–248. [PubMed] [Google Scholar]
- Hansuld E. M. Briens L. Sayani A. Mccann J. A. B. Drug Dev. Ind. Pharm. 2013;39:331–341. doi: 10.3109/03639045.2012.681055. [DOI] [PubMed] [Google Scholar]
- Landin M. J. Pharm. Sci. 2017;106:273–277. doi: 10.1016/j.xphs.2016.09.022. [DOI] [PubMed] [Google Scholar]
- Thapa P. Choi D. H. Kim M. Jeong S. H. Asian J. Pharm. Sci. 2019;14:287–304. doi: 10.1016/j.ajps.2018.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z. Gong Y. Ming L. Xie Y. Chin. Herb. Med. 2018;10:396–404. doi: 10.1016/j.chmed.2018.07.005. [DOI] [Google Scholar]
- Sun F. Xu B. Zhang Y. Dai S. Yang C. Cui X. Shi X. Qiao Y. Drug Des., Dev. Ther. 2016;10:3909–3924. doi: 10.2147/DDDT.S119122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng Y. Qiu Z. Wen H. Eur. J. Pharm. Biopharm. 2009;73:219–229. doi: 10.1016/j.ejpb.2009.04.008. [DOI] [PubMed] [Google Scholar]
- Mazor A. Orefice L. Michrafy A. De Ryck A. Khinast J. Powder Technol. 2017;337:3–16. doi: 10.1016/j.powtec.2017.04.053. [DOI] [Google Scholar]
- Bindhumadhavan G. Seville J. P. K. Adams M. J. Greenwood R. Fitzpatrick S. Chem. Eng. Sci. 2005;60:3891–3897. doi: 10.1016/j.ces.2005.02.022. [DOI] [Google Scholar]
- Liu Y. Wassgren C. Powder Technol. 2016;297:294–302. doi: 10.1016/j.powtec.2016.04.017. [DOI] [Google Scholar]
- Nesarikar V. V. Patel C. Early W. Vatsaraj N. Sprockel O. L. Jerzweski R. Int. J. Pharm. 2012;436:486–507. doi: 10.1016/j.ijpharm.2012.06.027. [DOI] [PubMed] [Google Scholar]
- Yu J. Xu B. Zhang K. Shi C. Zhang Z. Fu J. Qiao Y. Pharmaceutics. 2019;11:662. doi: 10.3390/pharmaceutics11120662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boersen N. Carvajal M. T. Morris K. R. Peck G. E. Pinal R. Drug Dev. Ind. Pharm. 2015;41:1470–1478. doi: 10.3109/03639045.2014.958754. [DOI] [PubMed] [Google Scholar]
- Wiedey R. Sibanc R. Wilms A. Kleinebudde P. Eur. J. Pharm. Biopharm. 2018;133:232–239. doi: 10.1016/j.ejpb.2018.10.021. [DOI] [PubMed] [Google Scholar]
- Inghelbrecht S. Remon J. P. Int. J. Pharm. 1998;161:215–224. doi: 10.1016/S0378-5173(97)00356-6. [DOI] [Google Scholar]
- Sun C. C. Kleinebudde P. Eur. J. Pharm. Biopharm. 2016;106:9–14. doi: 10.1016/j.ejpb.2016.04.003. [DOI] [PubMed] [Google Scholar]
- Shi W. Sprockel O. L. Eur. J. Pharm. Biopharm. 2016;106:15–19. doi: 10.1016/j.ejpb.2016.02.005. [DOI] [PubMed] [Google Scholar]
- Dumarey M. Wikstrom H. Fransson M. Sparen A. Tajarobi P. Josefson M. Trygg J. Int. J. Pharm. 2011;416:110–119. doi: 10.1016/j.ijpharm.2011.06.018. [DOI] [PubMed] [Google Scholar]
- Gupte A. Dehart M. Stagner W. C. Haware R. V. Drug Dev. Ind. Pharm. 2017;43:574–583. doi: 10.1080/03639045.2016.1272118. [DOI] [PubMed] [Google Scholar]
- Sajjia M. Albadarin A. B. Walker G. Int. J. Pharm. 2016;513:453–463. doi: 10.1016/j.ijpharm.2016.09.052. [DOI] [PubMed] [Google Scholar]
- Ende M. T. A. Moses S. K. Carella A. J. Gadkari R. A. Graul T. W. Otano A. L. Timpano R. J. Pharm. Dev. Technol. 2007;12:391–404. doi: 10.1080/10837450701369253. [DOI] [PubMed] [Google Scholar]
- Herting M. G. Kleinebudde P. Int. J. Pharm. 2007;338:110–118. doi: 10.1016/j.ijpharm.2007.01.035. [DOI] [PubMed] [Google Scholar]
- Reddy J. P. Phanse R. Nesarikar V. V. Pharm. Dev. Technol. 2019;24:1250–1257. doi: 10.1080/10837450.2019.1658775. [DOI] [PubMed] [Google Scholar]
- Nesarikar V. V. Vatsaraj N. Patel C. Early W. Pandey P. Sprockel O. L. Gao Z. Jerzewski R. L. Miller R. Levin M. Int. J. Pharm. 2012;426:116–131. doi: 10.1016/j.ijpharm.2012.01.032. [DOI] [PubMed] [Google Scholar]
- Soh J. L. P. Wang F. Boersen N. Pinal R. Peck G. E. Carvajal M. T. Cheney J. Valthorsson H. Pazdan J. Drug Dev. Ind. Pharm. 2008;34:1022–1035. doi: 10.1080/03639040801925990. [DOI] [PubMed] [Google Scholar]
- Soh J. L. P. Boersen N. Carvajal M. T. Morris K. R. Peck G. E. Pinal R. J. Pharm. Innov. 2007;2:106–124. doi: 10.1007/s12247-007-9013-z. [DOI] [Google Scholar]
- Liu Z. Bruwer M. Macgregor J. F. Rathore S. S. S. Reed D. E. Champagne M. J. R. Ind. Eng. Chem. Res. 2011;50:10696–10706. doi: 10.1021/ie102316b. [DOI] [Google Scholar]
- Turkoglu M. Aydin I. Murray M. Sakr A. Eur. J. Pharm. Biopharm. 1999;48:239–245. doi: 10.1016/S0939-6411(99)00054-5. [DOI] [PubMed] [Google Scholar]
- Bandari S. Nyavanandi D. Kallakunta V. R. Janga K. Y. Sarabu S. Butreddy A. Repka M. A. Int. J. Pharm. 2020;580:119215. doi: 10.1016/j.ijpharm.2020.119215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail H. Y. Shirazian S. Singh M. Whitaker D. Albadarin A. B. Walker G. Int. J. Pharm. 2020;576:118737. doi: 10.1016/j.ijpharm.2019.118737. [DOI] [PubMed] [Google Scholar]
- Liu H. Galbraith S. C. Ricart B. Stanton C. Smithgoettler B. Verdi L. Oconnor T. F. Lee S. Yoon S. Int. J. Pharm. 2017;525:249–263. doi: 10.1016/j.ijpharm.2017.04.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portier C. Pandelaere K. Delaet U. Vigh T. Pretoro G. D. De Beer T. Vervaet C. Vanhoorne V. Int. J. Pharm. 2020;576:119004. doi: 10.1016/j.ijpharm.2019.119004. [DOI] [PubMed] [Google Scholar]
- Vercruysse J. Diaz D. C. Peeters E. Fonteyne M. Delaet U. Van Assche I. De Beer T. Remon J. P. Vervaet C. Eur. J. Pharm. Biopharm. 2012;82:205–211. doi: 10.1016/j.ejpb.2012.05.010. [DOI] [PubMed] [Google Scholar]
- Ye X. Kallakunta V. Kim D. W. Patil H. Tiwari R. V. Upadhye S. B. Vladyka R. S. Repka M. A. J. Pharm. Sci. 2019;108:2895–2904. doi: 10.1016/j.xphs.2019.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A. Alakarjula M. Vanhoorne V. Toiviainen M. De Leersnyder F. Vercruysse J. Juuti M. Ketolainen J. Vervaet C. Remon J. P. Eur. J. Pharm. Sci. 2016;90:25–37. doi: 10.1016/j.ejps.2015.12.021. [DOI] [PubMed] [Google Scholar]
- Fonteyne M. Correia A. De Plecker S. Vercruysse J. Ilic I. Zhou Q. Vervaet C. Remon J. P. Onofre F. Bulone V. Int. J. Pharm. 2015;478:705–717. doi: 10.1016/j.ijpharm.2014.11.070. [DOI] [PubMed] [Google Scholar]
- Keleb E. Vermeire A. Vervaet C. Remon J. P. Int. J. Pharm. 2004;273:183–194. doi: 10.1016/j.ijpharm.2004.01.001. [DOI] [PubMed] [Google Scholar]
- Stauffer F. Vanhoorne V. Pilcer G. Chavez P. Vervaet C. De Beer T. Int. J. Pharm. 2019;561:265–273. doi: 10.1016/j.ijpharm.2019.03.012. [DOI] [PubMed] [Google Scholar]
- Ismail H. Y. Singh M. Darwish S. Kuhs M. Shirazian S. Croker D. M. Khraisheh M. Albadarin A. B. Walker G. Powder Technol. 2019;343:568–577. doi: 10.1016/j.powtec.2018.11.060. [DOI] [Google Scholar]
- Sierravega N. O. Romanach R. J. Mendez R. Int. J. Pharm. 2019;572:118728. doi: 10.1016/j.ijpharm.2019.118728. [DOI] [PubMed] [Google Scholar]
- Nyström C. and Karehill P.-G., Drugs and the Pharmaceutical Sciences, Marcel Dekker, New York, 1996, vol. 71, pp. 17–54 [Google Scholar]
- Steendam R. Eissens A. C. E. Frijlink H. W. Lerk C. F. Int. J. Pharm. 2000;204:23–33. doi: 10.1016/S0378-5173(00)00456-7. [DOI] [PubMed] [Google Scholar]
- Escotetespinoza M. S. Vadodaria S. Singh R. Muzzio F. J. Ierapetritou M. G. Int. J. Pharm. 2018;543:274–287. doi: 10.1016/j.ijpharm.2018.03.036. [DOI] [PubMed] [Google Scholar]
- Schomberg A. K. Kwade A. Finke J. H. Pharmaceutics. 2020;12:248. doi: 10.3390/pharmaceutics12030248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeom S. B. Ha E. Kim M. Jeong S. H. Hwang S. Choi D. H. Pharmaceutics. 2019;11:414. doi: 10.3390/pharmaceutics11080414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts K. J., Docherty R. and Tamura R., Engineering Crystallography: From Molecule to Crystal to Functional Form, Springer Netherlands, 2017 [Google Scholar]
- Guo Y. Wu C. Kafui K. D. Thornton C. Powder Technol. 2010;197:111–119. doi: 10.1016/j.powtec.2009.09.003. [DOI] [Google Scholar]
- Furukawa R. Shiosaka Y. Kadota K. Takagaki K. Noguchi T. Shimosaka A. Shirakawa Y. J. Drug Delivery Sci. Technol. 2016;35:284–293. doi: 10.1016/j.jddst.2016.08.004. [DOI] [Google Scholar]
- Mehrotra A. Chaudhuri B. Faqih A. Tomassone M. S. Muzzio F. J. Powder Technol. 2009;188:295–300. doi: 10.1016/j.powtec.2008.05.016. [DOI] [Google Scholar]
- Van Snick B. Grymonpre W. Dhondt J. Pandelaere K. Pretoro G. D. Remon J. P. De Beer T. Vervaet C. Vanhoorne V. Int. J. Pharm. 2018;549:476–488. doi: 10.1016/j.ijpharm.2018.08.015. [DOI] [PubMed] [Google Scholar]
- Grymonpre W. Vanhoorne V. Van Snick B. Prudilova B. B. Detobel F. Remon J. P. De Beer T. Vervaet C. Int. J. Pharm. 2018;548:54–61. doi: 10.1016/j.ijpharm.2018.06.047. [DOI] [PubMed] [Google Scholar]
- Hildebrandt C. Gopireddy S. R. Scherlies R. Urbanetz N. A. Powder Technol. 2019;345:390–404. doi: 10.1016/j.powtec.2019.01.015. [DOI] [Google Scholar]
- Huang J. Kaul G. Cai C. Chatlapalli R. Hernandezabad P. Ghosh K. Nagi A. Int. J. Pharm. 2009;382:23–32. doi: 10.1016/j.ijpharm.2009.07.031. [DOI] [PubMed] [Google Scholar]
- Sun F. Xu B. Dai S. Zhang Y. Lin Z. Qiao Y. Pharmaceutics. 2019;11:474. doi: 10.3390/pharmaceutics11090474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z. Wu F. Zhao L. Lin X. Shen L. Feng Y. Adv. Powder Technol. 2018;29:2881–2894. doi: 10.1016/j.apt.2018.08.009. [DOI] [Google Scholar]
- Haware R. V. Tho I. Bauerbrandl A. Eur. J. Pharm. Biopharm. 2009;73:424–431. doi: 10.1016/j.ejpb.2009.08.005. [DOI] [PubMed] [Google Scholar]
- Kaul G. Huang J. Chatlapalli R. Ghosh K. Nagi A. AAPS PharmSciTech. 2011;12:1064–1076. doi: 10.1208/s12249-011-9676-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai S. Xu B. Zhang Z. Yu J. Wang F. Shi X. Qiao Y. Int. J. Pharm. 2019;572:118742. doi: 10.1016/j.ijpharm.2019.118742. [DOI] [PubMed] [Google Scholar]
- Sun F. Xu B. Zhang Y. Dai S. Shi X. Qiao Y. Bioengineered. 2017;8:61–70. doi: 10.1080/21655979.2016.1227591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willecke N. Szepes A. Wunderlich M. Remon J. P. Vervaet C. De Beer T. Int. J. Pharm. 2018;545(1–2):128–143. doi: 10.1016/j.ijpharm.2018.04.017. [DOI] [PubMed] [Google Scholar]
- Goh H. P. Heng P. W. S. Liew C. V. Eur. J. Pharm. Sci. 2018;122:105–115. doi: 10.1016/j.ejps.2018.06.026. [DOI] [PubMed] [Google Scholar]
- Perez-Gandarillas L. Perez-Gago A. Mazor A. Kleinebudde P. Lecoq O. Michrafy A. Eur. J. Pharm. Biopharm. 2016;106:38–49. doi: 10.1016/j.ejpb.2016.05.020. [DOI] [PubMed] [Google Scholar]
- Zawbaa H. M. Schiano S. Perezgandarillas L. Grosan C. Michrafy A. Wu C. Adv. Powder Technol. 2018;29:2966–2977. doi: 10.1016/j.apt.2018.11.008. [DOI] [Google Scholar]
- Toschkoff G. Khinast J. Int. J. Pharm. 2013;457:407–422. doi: 10.1016/j.ijpharm.2013.08.022. [DOI] [PubMed] [Google Scholar]
- Yeom S. B. Ha E.-S. Kim M.-S. Jeong S. H. Hwang S.-J. Choi D. H. Pharmaceutics. 2019;11:414. doi: 10.3390/pharmaceutics11080414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop K. Kleinebudde P. Int. J. Pharm. 2013;457:527–536. doi: 10.1016/j.ijpharm.2013.01.062. [DOI] [PubMed] [Google Scholar]
- Korasa K. Vrečer F. Eur. J. Pharm. Sci. 2018;111:278–292. doi: 10.1016/j.ejps.2017.10.010. [DOI] [PubMed] [Google Scholar]
- Boehling P. Toschkoff G. Just S. Knop K. Kleinebudde P. Funke A. Rehbaum H. Rajniak P. Khinast J. Eur. J. Pharm. Sci. 2016;93:74–83. doi: 10.1016/j.ejps.2016.08.018. [DOI] [PubMed] [Google Scholar]
- Suzzi D. Radl S. Khinast J. Chem. Eng. Sci. 2010;65:5699–5715. doi: 10.1016/j.ces.2010.07.007. [DOI] [Google Scholar]
- Rege B. D. Gawel J. Kou J. H. Int. J. Pharm. 2002;237:87–94. doi: 10.1016/S0378-5173(02)00037-6. [DOI] [PubMed] [Google Scholar]
- Tanabe S. Nakagawa H. Watanabe T. Minami H. Kano M. Urbanetz N. A. Int. J. Pharm. 2016;511:341–350. doi: 10.1016/j.ijpharm.2016.07.023. [DOI] [PubMed] [Google Scholar]
- Cahyadi C. Heng P. W. S. Chan L. W. AAPS PharmSciTech. 2011;12:119–131. doi: 10.1208/s12249-010-9567-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinamaki J. Ruotsalainen M. Lehtola V. Antikainen O. Yliruusi J. Pharm. Dev. Technol. 1997;2:357–364. doi: 10.3109/10837459709022634. [DOI] [PubMed] [Google Scholar]
- Oza N. Khodakiya A. Sagar S. Int. J. Appl. Pharm. 2019:251–257. doi: 10.22159/ijap.2019v11i4.31905. [DOI] [Google Scholar]
- Patel J. Shah A. Sheth N. Int. J. PharmTech Res. 2009;1:235–240. [Google Scholar]
- Just S. Toschkoff G. Funke A. Djuric D. Scharrer G. Khinast J. Knop K. Kleinebudde P. Int. J. Pharm. 2013;457:1–8. doi: 10.1016/j.ijpharm.2013.09.010. [DOI] [PubMed] [Google Scholar]
- Nayak B. Elchidana P. Sahu P. Indian J. Pharm. Sci. 2017;79:345–352. [Google Scholar]
- Jason Teckoe T. M., Farrell T. and Rajabi-Siahboomi A., Use of a Quality by Design Approach to Optimize Coating Process Parameters for Opadry® 200, https://www.colorcon.com/resource-center/quality-by-design/download/362/648/34?method=view
- Dubey A. Boukouvala F. Keyvan G. Hsia R. Saranteas K. Brone D. Misra T. Ierapetritou M. G. Muzzio F. J. AAPS PharmSciTech. 2012;13:231–246. doi: 10.1208/s12249-011-9723-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F. Xu B. Zhang Y. Dai S. Yang C. Cui X. Shi X. Qiao Y. Drug Des., Dev. Ther. 2016;10:3909. doi: 10.2147/DDDT.S119122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kollenburg G. H. van Es J. Gerreten J. Lanters H. Bouman R. Koelewijn W. Davies A. N. Buydens L. van Manen H.-J. Jansen J. J. Comput. Chem. Eng. 2020:106841. doi: 10.1016/j.compchemeng.2020.106841. [DOI] [Google Scholar]
- Simões M. F. Silva G. Pinto A. C. Fonseca M. Silva N. E. Pinto R. M. A. Simões S. Eur. J. Pharm. Biopharm. 2020;152:282–295. doi: 10.1016/j.ejpb.2020.05.012. [DOI] [PubMed] [Google Scholar]
- Liu G. and Wang J., 2020, p. arXiv:2004.03955
- Tewari J. Strong R. Boulas P. Spectrochim. Acta, Part A. 2017;173:886–891. doi: 10.1016/j.saa.2016.10.013. [DOI] [PubMed] [Google Scholar]
- Berntsson O. Danielsson L.-G. Lagerholm B. Folestad S. Powder Technol. 2002;123:185–193. doi: 10.1016/S0032-5910(01)00456-9. [DOI] [Google Scholar]
- Uchiyama J. Aoki S. Uemoto Y. Chem. Pharm. Bull. 2015;63:164–179. doi: 10.1248/cpb.c14-00634. [DOI] [PubMed] [Google Scholar]
- Casian T. Iurian S. Gavan A. Revnic C. Porav S. Porfire A. Vlase L. Tomuţă I. Talanta. 2018;188:404–416. doi: 10.1016/j.talanta.2018.05.101. [DOI] [PubMed] [Google Scholar]
- Fonteyne M. Vercruysse J. De Leersnyder F. Besseling R. Gerich A. Oostra W. Remon J. P. Vervaet C. De Beer T. Anal. Chim. Acta. 2016;935:213–223. doi: 10.1016/j.aca.2016.07.020. [DOI] [PubMed] [Google Scholar]
- Romañach R. J. Méndeza R. Esbensenb K. H. Spectrosc. Eur. 2019;31:22–27. [Google Scholar]
- Riolo D. Piazza A. Cottini C. Serafini M. Lutero E. Cuoghi E. Gasparini L. Botturi D. Marino I. G. Aliatis I. J. Pharm. Biomed. Anal. 2018;149:329–334. doi: 10.1016/j.jpba.2017.11.030. [DOI] [PubMed] [Google Scholar]
- Hausman D. S. Cambron R. T. Sakr A. Int. J. Pharm. 2005;298:80–90. doi: 10.1016/j.ijpharm.2005.04.011. [DOI] [PubMed] [Google Scholar]
- De Beer T. Bodson C. Dejaegher B. Walczak B. Vercruysse P. Burggraeve A. Lemos A. Delattre L. Vander Heyden Y. Remon J. P. J. Pharm. Biomed. Anal. 2008;48:772–779. doi: 10.1016/j.jpba.2008.07.023. [DOI] [PubMed] [Google Scholar]
- Sanchez F. C. Toft J. Van den Bogaert B. Massart D. Dive S. Hailey P. Fresenius. J. Anal. Chem. 1995;352:771–778. doi: 10.1007/BF00323062. [DOI] [Google Scholar]
- Sekulic S. S. Wakeman J. Doherty P. Hailey P. A. J. Pharm. Biomed. Anal. 1998;17:1285–1309. doi: 10.1016/S0731-7085(98)00025-9. [DOI] [PubMed] [Google Scholar]
- Khorasani M. Amigo J. M. Bertelsen P. Van Den Berg F. Rantanen J. J. Pharm. Sci. 2015;104:2541–2549. doi: 10.1002/jps.24533. [DOI] [PubMed] [Google Scholar]
- El-Hagrasy A. S. Morris H. R. D'amico F. Lodder R. A. Drennen III J. K. J. Pharm. Sci. 2001;90:1298–1307. doi: 10.1002/jps.1082. [DOI] [PubMed] [Google Scholar]
- Sulub Y. Wabuyele B. Gargiulo P. Pazdan J. Cheney J. Berry J. Gupta A. Shah R. Wu H. Khan M. J. Pharm. Biomed. Anal. 2009;49:48–54. doi: 10.1016/j.jpba.2008.10.001. [DOI] [PubMed] [Google Scholar]
- Mohan S. Momose W. Katz J. M. Hossain M. N. Velez N. Drennen III J. K. Anderson C. A. J. Pharm. Biomed. Anal. 2018;148:51–57. doi: 10.1016/j.jpba.2017.09.011. [DOI] [PubMed] [Google Scholar]
- El-Hagrasy A. S. D'Amico F. Drennen III J. K. J. Pharm. Sci. 2006;95:392–406. doi: 10.1002/jps.20467. [DOI] [PubMed] [Google Scholar]
- Wargo D. J. Drennen J. K. J. Pharm. Biomed. Anal. 1996;14:1415–1423. doi: 10.1016/0731-7085(96)01739-6. [DOI] [PubMed] [Google Scholar]
- Ma H. Anderson C. A. J. Pharm. Sci. 2008;97:3305–3320. doi: 10.1002/jps.21230. [DOI] [PubMed] [Google Scholar]
- Amigo J. M. Cruz J. Bautista M. Maspoch S. Coello J. Blanco M. TrAC, Trends Anal. Chem. 2008;27:696–713. doi: 10.1016/j.trac.2008.05.010. [DOI] [Google Scholar]
- Lee S.-H. Lee J.-H. Cho S. Do S.-H. Woo Y.-A. Arch. Pharmacal Res. 2012;35:1599–1607. doi: 10.1007/s12272-012-0911-3. [DOI] [PubMed] [Google Scholar]
- Martinez L. Peinado A. Liesum L. Betz G. Eur. J. Pharm. Biopharm. 2013;84:606–615. doi: 10.1016/j.ejpb.2013.01.016. [DOI] [PubMed] [Google Scholar]
- Puchert T. Holzhauer C. V. Menezes J. C. Lochmann D. Reich G. Eur. J. Pharm. Biopharm. 2011;78:173–182. doi: 10.1016/j.ejpb.2010.12.015. [DOI] [PubMed] [Google Scholar]
- Shi Z. Cogdill R. P. Short S. M. Anderson C. A. J. Pharm. Biomed. Anal. 2008;47:738–745. doi: 10.1016/j.jpba.2008.03.013. [DOI] [PubMed] [Google Scholar]
- Elhagrasy A. S. Drennen J. K. J. Pharm. Sci. 2006;95:422–434. doi: 10.1002/jps.20465. [DOI] [PubMed] [Google Scholar]
- Momose W. Imai K. Yokota S. Yonemochi E. Terada K. Powder Technol. 2011;210:122–131. doi: 10.1016/j.powtec.2011.03.005. [DOI] [Google Scholar]
- Liew C. V. Karande A. D. Heng P. W. S. Int. J. Pharm. 2010;386:138–148. doi: 10.1016/j.ijpharm.2009.11.011. [DOI] [PubMed] [Google Scholar]
- Karande A. D. Liew C. V. Heng P. W. S. Int. J. Pharm. 2010;395:91–97. doi: 10.1016/j.ijpharm.2010.05.011. [DOI] [PubMed] [Google Scholar]
- Bakri B. Weimer M. Hauck G. Reich G. Eur. J. Pharm. Biopharm. 2015;97:78–89. doi: 10.1016/j.ejpb.2015.10.002. [DOI] [PubMed] [Google Scholar]
- Sulub Y. Konigsberger M. Cheney J. J. Pharm. Biomed. Anal. 2011;55:429–434. doi: 10.1016/j.jpba.2011.02.017. [DOI] [PubMed] [Google Scholar]
- Xiao Jie X. U. Song L. L. Zhu X. K. Fan B. Y. Guo H. Xiang B. R. Chin. J. Mod. Appl. Pharm. 2006;S1:644–646. [Google Scholar]
- Alvaradohernandez B. B. Scicolone J. V. Ortegazuniga C. Romanospino A. D. Colonlugo Y. M. Aymat E. Sanchez E. Muzzio F. J. Romanach R. J. J. Pharm. Biomed. Anal. 2020;180:113054. doi: 10.1016/j.jpba.2019.113054. [DOI] [PubMed] [Google Scholar]
- Virtanen S. Antikainen O. Yliruusi J. Int. J. Pharm. 2007;345:108–115. doi: 10.1016/j.ijpharm.2007.05.044. [DOI] [PubMed] [Google Scholar]
- Mahdi Y. Mouhi L. Guemras N. Daoud K. J. Fundam. Appl. Sci. 2018;8:655–670. doi: 10.4314/jfas.v8i3.1. [DOI] [Google Scholar]
- Khorasani M. Amigo J. M. Sun C. C. Bertelsen P. Rantanen J. Eur. J. Pharm. Biopharm. 2015;93:293–302. doi: 10.1016/j.ejpb.2015.04.008. [DOI] [PubMed] [Google Scholar]
- Gupta A. Peck G. E. Miller R. W. Morris K. R. J. Pharm. Sci. 2005;94:1589–1597. doi: 10.1002/jps.20375. [DOI] [PubMed] [Google Scholar]
- Gupta A. Austin J. Davis S. Harris M. T. Reklaitis G. V. J. Pharm. Sci. 2015;104:1787–1794. doi: 10.1002/jps.24409. [DOI] [PubMed] [Google Scholar]
- Mundozah A. L. Yang J. Tridon C. C. Cartwright J. J. Omar C. S. Salman A. D. Int. J. Pharm. 2019;568:118541. doi: 10.1016/j.ijpharm.2019.118541. [DOI] [PubMed] [Google Scholar]
- Harting J. Kleinebudde P. Eur. J. Pharm. Biopharm. 2019;137:77–85. doi: 10.1016/j.ejpb.2019.02.015. [DOI] [PubMed] [Google Scholar]
- Meng W. Romanospino A. D. Panikar S. S. Ocallaghan C. Gilliam S. J. Ramachandran R. Muzzio F. J. Adv. Powder Technol. 2019;30:879–894. doi: 10.1016/j.apt.2019.01.017. [DOI] [Google Scholar]
- Harting J. Kleinebudde P. Eur. J. Pharm. Biopharm. 2018;125:169–181. doi: 10.1016/j.ejpb.2018.01.015. [DOI] [PubMed] [Google Scholar]
- Romanospino A. D. Tamrakar A. Igne B. Dimaso E. T. Airiau C. Clancy D. Pereira G. Muzzio F. J. Singh R. Ramachandran R. Int. J. Pharm. 2020;574:118848. doi: 10.1016/j.ijpharm.2019.118848. [DOI] [PubMed] [Google Scholar]
- Acevedo D. Muliadi A. R. Giridhar A. Litster J. D. Romanach R. J. AAPS PharmSciTech. 2012;13:1005–1012. doi: 10.1208/s12249-012-9825-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin J. Gupta A. Mcdonnell R. Reklaitis G. V. Harris M. T. J. Pharm. Sci. 2013;102:1895–1904. doi: 10.1002/jps.23536. [DOI] [PubMed] [Google Scholar]
- Souihi N. Nilsson D. Josefson M. Trygg J. Int. J. Pharm. 2015;483:200–211. doi: 10.1016/j.ijpharm.2015.02.006. [DOI] [PubMed] [Google Scholar]
- Khorasani M. Amigo J. M. Sonnergaard J. Olsen P. Bertelsen P. Rantanen J. J. Pharm. Biomed. Anal. 2015;109:11–17. doi: 10.1016/j.jpba.2015.02.008. [DOI] [PubMed] [Google Scholar]
- Rantanen J. Wikstrom H. Turner R. Taylor L. S. Anal. Chem. 2005;77:556–563. doi: 10.1021/ac048842u. [DOI] [PubMed] [Google Scholar]
- Luukkonen P. Fransson M. Bjorn I. N. Hautala J. Lagerholm B. Folestad S. J. Pharm. Sci. 2008;97:950–959. doi: 10.1002/jps.20998. [DOI] [PubMed] [Google Scholar]
- Bijlani V. Delgadolopez M. Adeyeye C. M. Drennen J. K. NIR News. 2002;13:8–10. doi: 10.1255/nirn.680. [DOI] [Google Scholar]
- Samanta A. K. Karande A. D. Ng K. Heng P. W. S. AAPS PharmSciTech. 2013;14:86–100. doi: 10.1208/s12249-012-9890-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A. Peck G. E. Miller R. W. Morris K. R. J. Pharm. Sci. 2005;94:2301–2313. doi: 10.1002/jps.20430. [DOI] [PubMed] [Google Scholar]
- Van Quyet P. Samanta A. K. Liew C. V. Chan L. W. Heng P. W. S. J. Pharm. Sci. 2013;102:2667–2678. doi: 10.1002/jps.23635. [DOI] [PubMed] [Google Scholar]
- Talwar S. Nunes C. Stevens T. Nesarikar V. Timmins P. Anderson C. A. Drennen J. K. Appl. Spectrosc. 2017;71:1209–1221. doi: 10.1177/0003702816671960. [DOI] [PubMed] [Google Scholar]
- Zhao J. Tian G. Qiu Y. Qu H. Spectrochim. Acta, Part A. 2020;245:118878. doi: 10.1016/j.saa.2020.118878. [DOI] [PubMed] [Google Scholar]
- Blanco M. Alcala M. Gonzalez J. M. Torras E. J. Pharm. Sci. 2006;95:2137–2144. doi: 10.1002/jps.20653. [DOI] [PubMed] [Google Scholar]
- Chavez P. Sacre P. De Bleye C. Netchacovitch L. Mantanus J. Motte H. Schubert M. A. Hubert P. Ziemons E. Talanta. 2015;144:1352–1359. doi: 10.1016/j.talanta.2015.08.018. [DOI] [PubMed] [Google Scholar]
- Jarvinen K. Hoehe W. Jarvinen M. Poutiainen S. Juuti M. Borchert S. Eur. J. Pharm. Sci. 2013;48:680–688. doi: 10.1016/j.ejps.2012.12.032. [DOI] [PubMed] [Google Scholar]
- Karande A. D. Heng P. W. S. Liew C. V. Int. J. Pharm. 2010;396:63–74. doi: 10.1016/j.ijpharm.2010.06.011. [DOI] [PubMed] [Google Scholar]
- Dalvi H. Langlet A. Colbert M. Cournoyer A. Guay J. Abatzoglou N. Gosselin R. Talanta. 2019;195:87–96. doi: 10.1016/j.talanta.2018.11.034. [DOI] [PubMed] [Google Scholar]
- Pawar P. Wang Y. Keyvan G. Callegari G. Cuitino A. M. Muzzio F. J. Int. J. Pharm. 2016;512:96–107. doi: 10.1016/j.ijpharm.2016.08.033. [DOI] [PubMed] [Google Scholar]
- Pestieau A. Krier F. Thoorens G. Dupont A. Chavez P. Ziemons E. Hubert P. Evrard B. J. Pharm. Biomed. Anal. 2014;98:60–67. doi: 10.1016/j.jpba.2014.05.002. [DOI] [PubMed] [Google Scholar]
- Morisseau K. M. Rhodes C. T. Pharm. Res. 1997;14:108–111. doi: 10.1023/A:1012071904673. [DOI] [PubMed] [Google Scholar]
- Ebube N. K. Thosar S. S. Roberts R. A. Kemper M. S. Rubinovitz R. L. Martin D. L. Reier G. E. Wheatley T. A. Shukla A. J. Pharm. Dev. Technol. 1999;4:19–26. doi: 10.1080/10837459908984220. [DOI] [PubMed] [Google Scholar]
- Li W. Bagnol L. Berman M. Chiarella R. Gerber M. Int. J. Pharm. 2009;380:49–54. doi: 10.1016/j.ijpharm.2009.06.032. [DOI] [PubMed] [Google Scholar]
- Otsuka M. Chemom. Intell. Lab. Syst. 2006;82:109–114. doi: 10.1016/j.chemolab.2005.04.015. [DOI] [Google Scholar]
- Otsuka M. Yamane I. J. Pharm. Sci. 2006;95:1425–1433. doi: 10.1002/jps.20514. [DOI] [PubMed] [Google Scholar]
- Tanabe H. Otsuka K. Otsuka M. Anal. Sci. 2007;23:857–862. doi: 10.2116/analsci.23.857. [DOI] [PubMed] [Google Scholar]
- Otsuka M. Yamane I. J. Pharm. Sci. 2009;98:4296–4305. doi: 10.1002/jps.21748. [DOI] [PubMed] [Google Scholar]
- Donoso M. Kildsig D. O. Ghaly E. S. Pharm. Dev. Technol. 2003;8:357–366. doi: 10.1081/PDT-120024689. [DOI] [PubMed] [Google Scholar]
- Wu J. Luo W. Wang X. Cheng Q. Sun C. Li H. J. Pharm. Biomed. Anal. 2013;80:186–191. doi: 10.1016/j.jpba.2013.03.004. [DOI] [PubMed] [Google Scholar]
- Li J. Jiang Y. Fan Q. Chen Y. Wu R. Spectrochim. Acta, Part A. 2014;125:278–284. doi: 10.1016/j.saa.2014.01.097. [DOI] [PubMed] [Google Scholar]
- Yang H. Liao X. Peng F. Wang W. Liu Y. Yan J. Li H. Drug Dev. Ind. Pharm. 2015;41:1877–1887. doi: 10.3109/03639045.2015.1019354. [DOI] [PubMed] [Google Scholar]
- Casian T. Reznek A. Vonicagligor A. L. Van Renterghem J. De Beer T. Tomuţă I. Talanta. 2017;167:333–343. doi: 10.1016/j.talanta.2017.01.092. [DOI] [PubMed] [Google Scholar]
- Klukkert M. Wu J. X. Rantanen J. Carstensen J. M. Rades T. Leopold C. S. Eur. J. Pharm. Sci. 2016;90:85–95. doi: 10.1016/j.ejps.2015.12.004. [DOI] [PubMed] [Google Scholar]
- Freitas M. P. Sabadin A. Silva L. M. Giannotti F. M. Couto D. A. D. Tonhi E. Medeiros R. S. Coco G. L. Russo V. Martins J. A. J. Pharm. Biomed. Anal. 2005;39:17–21. doi: 10.1016/j.jpba.2005.03.023. [DOI] [PubMed] [Google Scholar]
- Tabasi S. H. Moolchandani V. Fahmy R. Hoag S. W. Int. J. Pharm. 2009;382:1–6. doi: 10.1016/j.ijpharm.2009.07.029. [DOI] [PubMed] [Google Scholar]
- Hernandez E. Pawar P. Keyvan G. Wang Y. Velez N. Callegari G. Cuitino A. M. Michniakkohn B. Muzzio F. J. Romanach R. J. J. Pharm. Biomed. Anal. 2016;117:568–576. doi: 10.1016/j.jpba.2015.10.012. [DOI] [PubMed] [Google Scholar]
- Baranwal Y. Romanospino A. D. Keyvan G. Ha J. M. Hong E. P. Muzzio F. J. Ramachandran R. Int. J. Pharm. 2019;565:419–436. doi: 10.1016/j.ijpharm.2019.05.022. [DOI] [PubMed] [Google Scholar]
- Ojala K. Myrskyranta M. Liimatainen A. Kortejärvi H. Juppo A. Int. J. Pharm. 2020:119028. doi: 10.1016/j.ijpharm.2020.119028. [DOI] [PubMed] [Google Scholar]
- Nagy B. Petra D. Galata D. L. Demuth B. Borbas E. Marosi G. Nagy Z. K. Farkas A. Int. J. Pharm. 2019;567:118464. doi: 10.1016/j.ijpharm.2019.118464. [DOI] [PubMed] [Google Scholar]
- Smetisko J. Miljanic S. J. Pharm. Biomed. Anal. 2017;145:322–330. doi: 10.1016/j.jpba.2017.06.055. [DOI] [PubMed] [Google Scholar]
- Neves A. C. O. Soares G. M. De Morais S. C. Costa F. S. L. Porto D. L. Lima K. M. G. J. Pharm. Biomed. Anal. 2012;57:115–119. doi: 10.1016/j.jpba.2011.08.029. [DOI] [PubMed] [Google Scholar]
- Ojala K. Myrskyranta M. Liimatainen A. Kortejärvi H. Juppo A. Int. J. Pharm. 2020;577:119028. doi: 10.1016/j.ijpharm.2020.119028. [DOI] [PubMed] [Google Scholar]
- Yekpe K. Abatzoglou N. Bataille B. Gosselin R. Sharkawi T. Simard J. Cournoyer A. Int. J. Pharm. 2015;486:242–251. doi: 10.1016/j.ijpharm.2015.03.060. [DOI] [PubMed] [Google Scholar]
- Chalus P. Walter S. Ulmschneider M. Anal. Chim. Acta. 2007;591:219–224. doi: 10.1016/j.aca.2007.03.076. [DOI] [PubMed] [Google Scholar]
- Ito M. Suzuki T. Yada S. Kusai A. Nakagami H. Yonemochi E. Terada K. J. Pharm. Biomed. Anal. 2008;47:819–827. doi: 10.1016/j.jpba.2008.03.033. [DOI] [PubMed] [Google Scholar]
- Tabasi S. H. Fahmy R. Bensley D. Obrien C. Hoag S. W. J. Pharm. Sci. 2008;97:4040–4051. doi: 10.1002/jps.21303. [DOI] [PubMed] [Google Scholar]
- Blanco M. Cuevamestanza R. Peguero A. J. Pharm. Biomed. Anal. 2010;51:797–804. doi: 10.1016/j.jpba.2009.09.038. [DOI] [PubMed] [Google Scholar]
- Ito M. Suzuki T. Yada S. Nakagami H. Teramoto H. Yonemochi E. Terada K. J. Pharm. Biomed. Anal. 2010;53:396–402. doi: 10.1016/j.jpba.2010.04.029. [DOI] [PubMed] [Google Scholar]
- Bu D. Wan B. Mcgeorge G. Chemom. Intell. Lab. Syst. 2013;120:84–91. doi: 10.1016/j.chemolab.2012.11.005. [DOI] [Google Scholar]
- Hennigan M. C. Ryder A. G. J. Pharm. Biomed. Anal. 2013;72:163–171. doi: 10.1016/j.jpba.2012.10.002. [DOI] [PubMed] [Google Scholar]
- Pieters S. Saeys W. Den Kerkhof T. V. Goodarzi M. Hellings M. De Beer T. Heyden Y. V. Anal. Chim. Acta. 2013;761:62–70. doi: 10.1016/j.aca.2012.11.034. [DOI] [PubMed] [Google Scholar]
- Dong Y. Li J. Zhong X. Cao L. Luo Y. Fan Q. Spectrochim. Acta, Part A. 2016;159:78–82. doi: 10.1016/j.saa.2016.01.030. [DOI] [PubMed] [Google Scholar]
- Alam A. Drennen J. K. Anderson C. A. J. Pharm. Biomed. Anal. 2017;145:230–239. doi: 10.1016/j.jpba.2017.06.012. [DOI] [PubMed] [Google Scholar]
- Porfire A. Filip C. Tomuta I. J. Pharm. Biomed. Anal. 2017;138:1–13. doi: 10.1016/j.jpba.2017.01.030. [DOI] [PubMed] [Google Scholar]
- Nagy B. Farkas A. Gyurkes M. Komaromyhiller S. Demuth B. Szabo B. Nusser D. Borbas E. Marosi G. Nagy Z. K. Int. J. Pharm. 2017;530:21–29. doi: 10.1016/j.ijpharm.2017.07.041. [DOI] [PubMed] [Google Scholar]
- Antonio M. Maggio R. M. J. Pharm. Biomed. Anal. 2018;149:603–611. doi: 10.1016/j.jpba.2017.11.053. [DOI] [PubMed] [Google Scholar]
- De Leersnyder F. Peeters E. Djalabi H. Vanhoorne V. Van Snick B. Hong K. Hammond S. V. Liu A. Y. Ziemons E. Vervaet C. J. Pharm. Biomed. Anal. 2018;151:274–283. doi: 10.1016/j.jpba.2018.01.032. [DOI] [PubMed] [Google Scholar]
- Sierravega N. O. Sanchezpaternina A. Maldonado N. Cardenas V. Romanach R. J. Mendez R. J. Pharm. Biomed. Anal. 2018;154:384–396. doi: 10.1016/j.jpba.2018.03.017. [DOI] [PubMed] [Google Scholar]
- Goodwin D. J. Den Ban S. V. Denham M. Barylski I. Int. J. Pharm. 2018;537:183–192. doi: 10.1016/j.ijpharm.2017.12.011. [DOI] [PubMed] [Google Scholar]
- Teixeira K. S. S. Fonseca S. G. D. C. De Moura L. C. B. De Moura M. L. R. Borges M. H. P. Barbosa E. G. Moura T. F. A. D. L. E. J. Pharm. Biomed. Anal. 2018;149:557–563. doi: 10.1016/j.jpba.2017.11.001. [DOI] [PubMed] [Google Scholar]
- De Leersnyder F. Vanhoorne V. Kumar A. Vervaet C. De Beer T. Int. J. Pharm. 2019;565:358–366. doi: 10.1016/j.ijpharm.2019.05.006. [DOI] [PubMed] [Google Scholar]
- Wahl P. Fruhmann G. Sacher S. Straka G. Sowinski S. Khinast J. Eur. J. Pharm. Biopharm. 2014;87:271–278. doi: 10.1016/j.ejpb.2014.03.021. [DOI] [PubMed] [Google Scholar]
- Pino-Torres C. Maspoch S. Castillo-Felices R. Pérez-Rivera M. Aranda-Bustos M. Peña-Farfal C. Microchem. J. 2020;154:104576. doi: 10.1016/j.microc.2019.104576. [DOI] [Google Scholar]
- Tomuta I. Rus L. Iovanov R. Rus L. L. J. Pharm. Biomed. Anal. 2013;84:285–292. doi: 10.1016/j.jpba.2012.12.020. [DOI] [PubMed] [Google Scholar]
- Palou A. Cruz J. Blanco M. Tomas J. Rios J. D. L. Alcala M. J. Pharm. Anal. 2012;2:90–97. doi: 10.1016/j.jpha.2011.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl P. Pucher I. Scheibelhofer O. Kerschhaggl M. Sacher S. Khinast J. Int. J. Pharm. 2017;518:130–137. doi: 10.1016/j.ijpharm.2016.12.003. [DOI] [PubMed] [Google Scholar]
- Wabuyele B. Sotthivirat S. Zhou G. Ash J. Dhareshwar S. S. J. Pharm. Sci. 2017;106:579–588. doi: 10.1016/j.xphs.2016.10.014. [DOI] [PubMed] [Google Scholar]
- Gomez D. A. Coello J. Maspoch S. J. Pharm. Biomed. Anal. 2016;124:207–215. doi: 10.1016/j.jpba.2016.02.055. [DOI] [PubMed] [Google Scholar]
- Skibsted E. Westerhuis J. A. Smilde A. K. Witte D. T. J. Pharm. Biomed. Anal. 2007;43:1297–1305. doi: 10.1016/j.jpba.2006.10.037. [DOI] [PubMed] [Google Scholar]
- Dou Y. Qu N. Wang B. Chi Y. Z. Ren Y. Eur. J. Pharm. Sci. 2007;32:193–199. doi: 10.1016/j.ejps.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Romer M. Heinamaki J. Strachan C. J. Sandler N. Yliruusi J. AAPS PharmSciTech. 2008;9:1047–1053. doi: 10.1208/s12249-008-9142-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. Park C. Kim A. Kwon B. Bang K. Cho Y. Jeong M. Y. Choi G. J. Pharm. Sci. 2010;99:325–335. doi: 10.1002/jps.21795. [DOI] [PubMed] [Google Scholar]
- El Hagrasy A. Chang S.-Y. Desai D. Kiang S. Am. Pharm. Rev. 2006;9:40–45. [Google Scholar]
- Muller J. Knop K. Thies J. Uerpmann C. Kleinebudde P. Drug Dev. Ind. Pharm. 2009;36:234–243. doi: 10.3109/03639040903225109. [DOI] [PubMed] [Google Scholar]
- Muller J. Knop K. Wirges M. Kleinebudde P. J. Pharm. Biomed. Anal. 2010;53:884–894. doi: 10.1016/j.jpba.2010.06.016. [DOI] [PubMed] [Google Scholar]
- Muller J. Brock D. Knop K. Zeitler J. A. Kleinebudde P. Eur. J. Pharm. Biopharm. 2012;80:690–697. doi: 10.1016/j.ejpb.2011.12.003. [DOI] [PubMed] [Google Scholar]
- Gendre C. Genty M. Boiret M. Julien M. Meunier L. Lecoq O. Baron M. Chaminade P. Pean J. M. Eur. J. Pharm. Sci. 2011;43:244–250. doi: 10.1016/j.ejps.2011.04.017. [DOI] [PubMed] [Google Scholar]
- Moltgen C. V. Puchert T. Menezes J. C. Lochmann D. Reich G. Talanta. 2012;92:26–37. doi: 10.1016/j.talanta.2011.12.034. [DOI] [PubMed] [Google Scholar]
- Gendre C. Boiret M. Genty M. Chaminade P. Pean J. M. Int. J. Pharm. 2011;421:237–243. doi: 10.1016/j.ijpharm.2011.09.036. [DOI] [PubMed] [Google Scholar]
- Hudovornik G. Korasa K. Vrecer F. Eur. J. Pharm. Sci. 2015;75:160–168. doi: 10.1016/j.ejps.2015.04.007. [DOI] [PubMed] [Google Scholar]
- Bogomolov A. Engler M. Melichar M. Wigmore A. J. Chemom. 2010;24:544–557. doi: 10.1002/cem.1329. [DOI] [Google Scholar]
- Tabasi S. H. Fahmy R. Bensley D. Obrien C. Hoag S. W. J. Pharm. Sci. 2008;97:4067–4086. doi: 10.1002/jps.21420. [DOI] [PubMed] [Google Scholar]
- Wirges M. Funke A. Serno P. Knop K. Kleinebudde P. J. Pharm. Sci. 2013;102:556–564. doi: 10.1002/jps.23383. [DOI] [PubMed] [Google Scholar]
- Wirges M. Muller J. Kasa P. Regdon G. Pintyehodi K. Knop K. Kleinebudde P. Pharmaceutics. 2011;3:723–730. doi: 10.3390/pharmaceutics3040723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirges M. Funke A. Serno P. Knop K. Kleinebudde P. J. Pharm. Biomed. Anal. 2013;78:57–64. doi: 10.1016/j.jpba.2013.01.037. [DOI] [PubMed] [Google Scholar]
- Romerotorres S. Perezramos J. D. Morris K. R. Grant E. R. J. Pharm. Biomed. Anal. 2006;41:811–819. doi: 10.1016/j.jpba.2006.01.033. [DOI] [PubMed] [Google Scholar]
- Wold S., Martens H. and Wold H., in Matrix pencils, Springer, 1983, pp. 286–293 [Google Scholar]
- Yang Y. Ye Z. Su Y. Zhao Q. Li X. Ouyang D. Acta Pharm. Sin. B. 2019;9:177–185. doi: 10.1016/j.apsb.2018.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori Y. Naganuma M. Otsuka M. Pharmaceutics. 2020;12:85. doi: 10.3390/pharmaceutics12010085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerhuis J. A. Coenegracht P. M. J. Lerk C. F. Int. J. Pharm. 1997;156:109–117. doi: 10.1016/S0378-5173(97)00191-9. [DOI] [Google Scholar]
- Garcíamunoz S. Dolph S. Ward H. W. Comput. Chem. Eng. 2010;34:1098–1107. doi: 10.1016/j.compchemeng.2010.02.027. [DOI] [Google Scholar]
- Muteki K. Swaminathan V. Sekulic S. S. Reid G. L. IFAC Proceedings Volumes. 2012;45:51–56. doi: 10.3182/20120710-4-SG-2026.00076. [DOI] [Google Scholar]
- Burggraeve A. Den Kerkhof T. V. Hellings M. Remon J. P. Vervaet C. De Beer T. Eur. J. Pharm. Sci. 2011;42:584–592. doi: 10.1016/j.ejps.2011.03.002. [DOI] [PubMed] [Google Scholar]
- Silva A. F. Sarraguca M. C. Fonteyne M. Vercruysse J. De Leersnyder F. Vanhoorne V. Bostijn N. Verstraeten M. Vervaet C. Remon J. P. Int. J. Pharm. 2017;528:242–252. doi: 10.1016/j.ijpharm.2017.05.075. [DOI] [PubMed] [Google Scholar]
- Kona R. Qu H. Mattes R. Jancsik B. Fahmy R. M. Hoag S. W. Int. J. Pharm. 2013;452:63–72. doi: 10.1016/j.ijpharm.2013.04.039. [DOI] [PubMed] [Google Scholar]
- Matero S. Reinikainen S.-P. Lahtela-Kakkonen M. Korhonen O. Ketolainen J. Poso A. J. Chemom. 2008;22:653–660. doi: 10.1002/cem.1148. [DOI] [Google Scholar]
- Hattori Y. Otsuka M. Int. J. Pharm. 2017;524:407–413. doi: 10.1016/j.ijpharm.2017.04.004. [DOI] [PubMed] [Google Scholar]
- Singh R. Romanospino A. D. Romanach R. J. Ierapetritou M. G. Ramachandran R. Int. J. Pharm. 2015;495:612–625. doi: 10.1016/j.ijpharm.2015.09.029. [DOI] [PubMed] [Google Scholar]
- Kourti T. Am. Pharm. Rev. 2009;12:118. [Google Scholar]
- Su Q. Moreno M. Giridhar A. Reklaitis G. V. Nagy Z. K. J. Pharm. Innov. 2017;12:327–346. doi: 10.1007/s12247-017-9297-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R. Muzzio F. J. Ierapetritou M. Ramachandran R. Processes. 2015;3:339–356. doi: 10.3390/pr3020339. [DOI] [Google Scholar]
- Su Q. Ganesh S. Moreno M. Bommireddy Y. Gonzalez M. Reklaitis G. V. Nagy Z. K. Comput. Chem. Eng. 2019;125:216–231. doi: 10.1016/j.compchemeng.2019.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- S. M. L. Coalition, Implementing 21st Century Smart Manufacturing, www.academia.edu/1984950/Implementing_21st_Century_Smart_Manufacturing
- Zuehlke D. Annu. Rev. Contr. 2010;34:129–138. doi: 10.1016/j.arcontrol.2010.02.008. [DOI] [Google Scholar]
- Woo J. Shin S.-J. Seo W. Meilanitasari P. Int. J. Adv. Manuf. Technol. 2018;99:2193–2217. doi: 10.1007/s00170-018-2416-9. [DOI] [Google Scholar]
- Helal N. A. Elnoweam O. Eassa H. A. Amer A. M. Eltokhy M. A. Helal M. A. Fayyaz H. A. Nounou M. I. Pharm. Pat. Anal. 2019;8:139–161. doi: 10.4155/ppa-2019-0011. [DOI] [PubMed] [Google Scholar]
- Badman C. Cooney C. L. Florence A. J. Konstantinov K. Krumme M. Mascia S. Nasr M. Trout B. L. J. Pharm. Sci. 2019;108:3521–3523. doi: 10.1016/j.xphs.2019.07.016. [DOI] [PubMed] [Google Scholar]
- Laske S. Paudel A. Scheibelhofer O. Sacher S. Hoermann T. Khinast J. Kelly A. L. Rantannen J. Korhonen O. Stauffer F. J. Pharm. Sci. 2017;106:667–712. doi: 10.1016/j.xphs.2016.11.011. [DOI] [PubMed] [Google Scholar]
- Vanhoorne V. Vervaet C. Int. J. Pharm. 2020;579:119194. doi: 10.1016/j.ijpharm.2020.119194. [DOI] [PubMed] [Google Scholar]
- Global Pharmaceutical Continuous Manufacturing Technology Market: Snapshot, https://www.transparencymarketresearch.com/pharmaceutical-continuous-manufacturing-technology-market.html
- Simonaho S. P. Ketolainen J. Ervasti T. Toiviainen M. Korhonen O. Eur. J. Pharm. Sci. 2016;90:38–46. doi: 10.1016/j.ejps.2016.02.006. [DOI] [PubMed] [Google Scholar]
- Lakio S. Ervasti T. Tajarobi P. Wikstrom H. Fransson M. Karttunen A. Ketolainen J. Folestad S. Abrahmsenalami S. Korhonen O. Eur. J. Pharm. Sci. 2017;109:514–524. doi: 10.1016/j.ejps.2017.09.018. [DOI] [PubMed] [Google Scholar]
- Pauli V. Roggo Y. Pellegatti L. Trung N. Q. N. Elbaz F. Ensslin S. Kleinebudde P. Krumme M. J. Pharm. Biomed. Anal. 2019;162:101–111. doi: 10.1016/j.jpba.2018.09.016. [DOI] [PubMed] [Google Scholar]
- Stauffer F. Vanhoorne V. Pilcer G. Chavez P. Vervaet C. De Beer T. Int. J. Pharm. 2019;1:118525. doi: 10.1016/j.ijpharm.2019.118525. [DOI] [PubMed] [Google Scholar]
- Nagy B. Farkas A. Magyar K. Demuth B. Nagy Z. K. Marosi G. Eur. J. Pharm. Sci. 2018;123:10–19. doi: 10.1016/j.ejps.2018.07.025. [DOI] [PubMed] [Google Scholar]
- Sierravega N. O. Romanospino A. D. Scicolone J. V. Muzzio F. J. Romanach R. J. Mendez R. Int. J. Pharm. 2019;560:322–333. doi: 10.1016/j.ijpharm.2019.01.073. [DOI] [PubMed] [Google Scholar]
- Roggo Y. Pauli V. Jelsch M. Pellegatti L. Elbaz F. Ensslin S. Kleinebudde P. Krumme M. J. Pharm. Biomed. Anal. 2020;179:112971. doi: 10.1016/j.jpba.2019.112971. [DOI] [PubMed] [Google Scholar]
- Singh R. Sahay A. Muzzio F. Ierapetritou M. Ramachandran R. Comput. Chem. Eng. 2014;66:186–200. doi: 10.1016/j.compchemeng.2014.02.029. [DOI] [Google Scholar]
- Zomer S. Zhang J. Talwar S. Chattoraj S. Hewitt C. Int. J. Pharm. 2018;547:506–519. doi: 10.1016/j.ijpharm.2018.06.034. [DOI] [PubMed] [Google Scholar]
- Silva A. F. Vercruysse J. Vervaet C. Remon J. P. Lopes J. A. De Beer T. Sarraguça M. C. Eur. J. Pharm. Biopharm. 2018;128:36–47. doi: 10.1016/j.ejpb.2018.04.011. [DOI] [PubMed] [Google Scholar]
- Romanospino A. D. Singh R. Ierapetritou M. G. Ramachandran R. Mendez R. Ortegazuniga C. Muzzio F. J. Romanach R. J. Int. J. Pharm. 2016;512:61–74. doi: 10.1016/j.ijpharm.2016.08.029. [DOI] [PubMed] [Google Scholar]
- Hetrick E. M. Shi Z. Barnes L. E. Garrett A. W. Rupard R. G. Kramer T. T. Cooper T. M. Myers D. P. Castle B. C. Anal. Chem. 2017;89:9175–9183. doi: 10.1021/acs.analchem.7b01907. [DOI] [PubMed] [Google Scholar]
- Pauli V. Kleinebudde P. Krumme M. Eur. J. Pharm. Biopharm. 2020;153:200–210. doi: 10.1016/j.ejpb.2020.05.030. [DOI] [PubMed] [Google Scholar]
- Taipalekovalainen K. Karttunen A. Ketolainen J. Korhonen O. Eur. J. Pharm. Sci. 2018;115:1–10. doi: 10.1016/j.ejps.2017.12.021. [DOI] [PubMed] [Google Scholar]
- Manley L. Shi Z. Int. J. Pharm. 2018;551:60–66. doi: 10.1016/j.ijpharm.2018.08.059. [DOI] [PubMed] [Google Scholar]
- Van Snick B. Dhondt J. Pandelaere K. Bertels J. Mertens R. Klingeleers D. Di Pretoro G. Remon J. P. Vervaet C. De Beer T. Int. J. Pharm. 2018;549:415–435. doi: 10.1016/j.ijpharm.2018.08.014. [DOI] [PubMed] [Google Scholar]
- Wang Y. O'Connor T. Li T. Ashraf M. Cruz C. N. Int. J. Pharm. 2019;569:118551. doi: 10.1016/j.ijpharm.2019.118551. [DOI] [PubMed] [Google Scholar]