Abstract
Posidonia oceanica is a sea grass belonging to the family Posidoniaceae, which stands out as a substantial reservoir of bioactive compounds. In this study, the secondary metabolites of the P. oceanica rhizome were annotated using UPLC-HRESI-MS/MS, revealing 86 compounds including simple phenolic acids, flavonoids, and their sulphated conjugates. Moreover, the P. oceanica butanol extract exhibited substantial antioxidant and antidiabetic effects in vitro. Thus, a reliable, robust drug delivery system was developed through the encapsulation of P. oceanica extract in gelatin nanoparticles to protect active constituents, control their release and enhance their therapeutic activity. To confirm these achievements, untargeted GC-MS metabolomics analysis together with biochemical evaluation was employed to investigate the in vivo anti-diabetic potential of the P. oceanica nano-extract. The results of this study demonstrated that the P. oceanica gelatin nanoparticle formulation reduced the serum fasting blood glucose level significantly (p < 0.05) in addition to improving the insulin level, together with the elevation of glucose transporter 4 levels. Besides, multivariate/univariate analyses of the GC-MS metabolomic dataset revealed several dysregulated metabolites in diabetic rats, which were restored to normalized levels after treatment with the P. oceanica gelatin nanoparticle formulation. These metabolites mainly originate from the metabolism of amino acids, fatty acids and carbohydrates, indicating that this type of delivery was more effective than the plain extract in regulating these altered metabolic processes. Overall, this study provides novel insight for the potential of P. oceanica butanol extract encapsulated in gelatin nanoparticles as a promising and effective antidiabetic therapy.
The potential of P. oceanica butanol extract encapsulated in gelatin nanoparticles as a promising and effective antidiabetic therapy has been investigated via metabolomics.
1. Introduction
Diabetes mellitus (DM) is a complex metabolic disorder, which is characterized by insulin deficiency, resistance or dysfunctional secretion from pancreatic β-cells. DM can lead to a variety of disorders including nephropathy, cardiovascular disorders and retinopathy.1 Hence, it has become a major global health concern with a projected figure of 642 million cases by 2040.2 Accordingly, plants with their complex secondary metabolites, including polyphenols, have been used for thousands of years as a conventional medicine and have emerged as a powerful source of new remedies against many life-threatening diseases.3 However, the effectiveness of polyphenols mainly depends on the stability, bioactivity and bioavailability of their active ingredients. Additionally, the low absorption, efflux mechanism in the gastrointestinal tract and unpleasant taste of most phenolic compounds also limit their application.4 Thus, incorporating polyphenols in nanoparticulate carriers, especially those based on the polymers of natural origin such as gelatin, has been investigated as an innovative and emerging technology for solving these problems.
Posidonia oceanica (L.) Delile, from the family Posidoniaceae, is a long-living, slow-growing, endemic Mediterranean seagrass.5P. oceanica has been called “the lungs of the Mediterranean” as it is one of the most important sources of oxygen in coastal waters. Interestingly, P. oceanica is also characterized by its vast content of phenolic compounds.6 These include the phenols found in P. oceanica leaves such as phenolic acids (4-hydroxybenzoic acid, gentisic acid, chicoric acid, cinnamic acid, ferulic acid, p-coumaric acid, and caffeic acid), flavonoids (kaempferol, quercetin, isorhamnetin and myricetin) and vanillin.7 The medicinal uses of P. oceanica date back to ancient Egypt, where it was used to treat skin diseases, while it has been used recently as a remedy for hypertension, respiratory infections, diabetes, colitis, and acne in the southwestern Mediterranean region.8
Metabolomics, as the metabolic complement of functional genomics, is a promising approach to explore homeostasis. The main goal of metabolomics in diabetes and other diseases is to discover metabolic biomarkers and perturbed pathways, which can be used as tools for medical practice such as the diagnosis and prognosis of therapeutic response including herbal medication.9 Recently, with the development of metabolomic technology, more reliable control of diabetes mellitus is expected to be achieved based on a patient's metabolomic profile.
The aim of the present study was to develop a metabolomic-based approach for characterizing the secondary bioactive polyphenolic metabolic pool of P. oceanica using UPLC-HRESI-MS/MS. Moreover, a reliable, robust drug delivery system was developed by encapsulating P. oceanica extract in gelatin nanoparticles to increase its bioactivity and therapeutic potency. The in vitro characterization of this novel nano-extract was performed. In addition, evaluation of its in vivo antidiabetic activity compared to the plain extract was conducted in a comparative way via GC-MS-based metabolomics together with different biochemical tests.
2. Materials and methods
2.1. Chemicals
The reagents including streptozotocin (STZ) (>98%), Daonil “glibenclamide” (Sanofi-Aventis Co.), methoxyamine HCl, pyridine, MSTFA (N-methyl-N-(trimethylsilyl)trifluoroacetamide) with 1% (v/v) trimethylchlorosilane, acetonitrile (99.8%) and n-alkane C8–C40 standards, Folin–Ciocalteu Reagent (FCR), gelatin, Kolliphor® P 188 (Poloxamer 188) and Kolliphor® P 407 (Poloxamer 407) were purchased from Sigma Aldrich Co. (St Louis, Mo, USA). Gallic acid, 98% was obtained from Acros Organics, Belgium. Glutaraldehyde (25% solution) was obtained from Bio Basic Canada Inc., Ontario, Canada. All other chemicals and solvents were of high analytical grade.
2.2. Plant materials
P. oceanica vertical rhizomes were obtained from Mersa Matruh, Egypt. A voucher specimen was identified by Dr Mona Marzouk (Professor of plant chemosystematics) and deposited at the herbarium of National Research Center, Egypt. The air-dried powdered rhizome (2 kg) was extracted exhaustively with methanol (3 × 3.5 L) at room temperature. The extract was filtered and concentrated under reduced pressure at 45 °C using a rotary evaporator. The methanolic extract was suspended in water, left overnight, and was successively partitioned with methylene chloride, followed by ethyl acetate then n-butanol.
2.3. Identification and quantification of P. oceanica secondary metabolites
Secondary metabolites of P. oceanica were identified using a UPLC (Acquity system, Waters, Milford, USA) attached to a Q Exactive hybrid MS/MS quadrupole-orbitrap mass spectrometer. The separation was performed using water (0.1% formic acid, solvent A) and acetonitrile (solvent B) at a flow rate of 0.4 mL min−1 as follows: 0–15 min from 50% to 50% B, 15–22 min to 98% B with the BEH shield C18 column (150 × 2.1 mm, 1.7 μm). The Q-Exactive MS was operated as previously reported.10
For absolute quantification of some secondary metabolites according to the availability of their analytical standards, an HPLC system (HP 1100 chromatograph, Agilent Technologies, Palo Alto, CA, USA) equipped with an auto-sampler (G1329B), quaternary pump and a diode array detector was employed. Separation was achieved using a ZORBAX Eclipse XDB C18 column (15 cm × 4.6 mm I.D., 5 μm, USA) under the following conditions: mobile phase A, 2% acetic acid; mobile phase B, acetonitrile; flow rate, 0.8 mL min−1; fixed wavelength, 280 and 360 nm; injected quantity, 10 μL; elution program, A (%)/B (%): 0 min 90/10; 15 min 50/50; 17 min 20/80; 19 min 90/10; and 20 min 90/10. Identification of phenolic compounds was performed by comparison with the retention times of standard substances.
2.4. Determination of total flavonoids, phenols, DPPH and in vitro antidiabetic activity
Total flavonoids and phenols were estimated in different extracts using the methods reported by Hamed et al.11 and the DPPH (1,1-diphenyl-2-picrylhydrazyl, 250 mM) radical scavenging assay was performed as described by Shimada et al.12 Briefly, the percentage inhibition of the DPPH radical was calculated according to the following formula: % inhibition = [(Acontrol − Asample)/Acontrol] × 100, where A is the absorbance at 517 nm. α-Glucosidase inhibitory activity was measured according to the method described by Elya et al.13
2.5. Preparation of gelatin nanoparticles
Gelatin nanoparticles were prepared via the nano-precipitation method previously reported by Lee et al.14 with some modifications. Briefly, 25 mg of P. oceanica butanol extract was dissolved in a 10 mL solvent mixture of de-ionized water: DMSO (8 : 2, v/v) via sonication. Then, 200 mg of bovine gelatin powder (type B) was added and dissolved using a magnetic stirrer at a temperature of 45–50 °C for 30 min to attain complete solubilization of the gelatin. The resultant solution was added dropwise to a solution of stabilizer dissolved in ethanol with continuous stirring by a magnetic stirrer. Ethanol was used as a non-solvent for gelatin and the volume of ethanol was varied to provide different solvent to non-solvent ratios. Also, the type of stabilizer and its corresponding amounts were varied to provide different stabilizer to gelatin mass ratios. Finally, glutaraldehyde solution was added as a cross-linking agent such that the glutaraldehyde to gelatin mass ratio of 0.125 was satisfied. The preparation was allowed to crosslink for 12 h with continuous stirring.
2.6. Characterization of the prepared gelatin nanoparticulate formulations
2.6.1. Entrapment efficiency (EE%) estimation
The unentrapped, free polyphenols were separated from the nanoparticles by cooling and centrifugation at 8000 rpm and −4 °C for 45 minutes using a cooling centrifuge (Union 32R, Hanil Science Industrial, Korea). The collected supernatants were diluted before being analyzed for unentrapped polyphenols using the previously mentioned spectrophotometric method. The entrapment efficiency (EE%) was calculated according to the following equation:15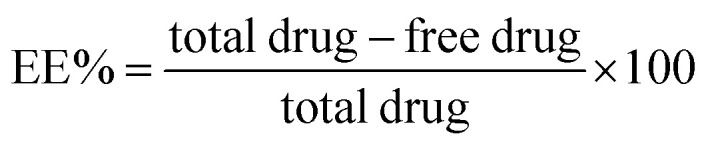
2.6.2. Estimation of particle size (PS) and zeta potential (ZP)
The particle size and zeta potential of the prepared nanoparticles were estimated via dynamic light scattering (DLS) using a Zetasizer (Malvern Instruments, Ltd, Nano Series ZS90, UK).
2.6.3. Transmission electron microscopy (TEM)
The selected nanoparticle formulation morphology was characterized using TEM (Jeol, JEM-1230, Tokyo, Japan).
2.6.4. In vitro release study of P. oceanica encapsulated gelatin nanoparticles
The in vitro release profile for polyphenols from the optimized gelatin nanoparticle formulation was evaluated using gradual pH-changing buffer systems.16 The dialysis bag diffusion method was employed. The pre-separated P. oceanica butanol extract-loaded gelatin nanoparticles were re-suspended and placed in presoaked cellulose dialysis bags (Dialysis Tubing Cellulose Membrane, Sigma Co., USA; molecular weight cutoff 12 000–14 000). The bags were sealed and then immersed in glass bottles filled with 50 mL receptor release medium. To mimic the gastrointestinal transit time and pH, dissolution media were altered at selected intervals. Initially, the release was conducted for 2 h in 0.1 N HCl release medium (pH = 1.2), mimicking the stomach. Then, the dialysis bags were transferred to pH 6.8 phosphate buffer solution for 4 h to mimic the proximal intestine. Finally, the pH of the release medium was increased to pH = 7.4 to mimic the ileocecal region, up to 24 h.17 The bottles were placed in a thermostated shaking water bath maintained at 37 °C ± 0.5 °C with a shaking speed of 100 rpm (Memmert, SV 1422, Schwabach, Germany).18 The released polyphenols were analyzed and the cumulative percentage of released polyphenols was calculated and plotted versus time. Different mathematical models were employed to elucidate the release mechanism. These models included: zero order, first order, Higuchi model and Peppas exponential equation. The release exponent “n” of the Peppas model indicates the drug release mechanism.19,20
2.7. Evaluation of antidiabetic effect
2.7.1. Ethics and permissions
The procedures of the study protocol were accepted ethically by the National Research Center's Ethics Review Committee in Cairo, Egypt, which ensured that the animals would not struggle at any point of the experiment (approval no. 19092) according to ARRIVE guidelines.21
2.7.2. Experimental animals
Thirty male Wistar albino rats, weighing 150–170 kg, were obtained from the National Research Centre's animal house. Animal care was consistent with the ethical guidelines of the National Research Centre's Medical Ethical Committee in Egypt. The animals were housed individually, and feed and water were provided ad libitum.
2.7.3. Experimental design in rat model
The rats were divided into two main groups. The first group (group 1, Gp1: six rats) served as the healthy control, whereas the second group (Gp2: 24 rats) was subjected to diabetes induction22 as follows: streptozotocin was induced by a single intraperitoneal dose of 40 mg kg−1 body weight dissolved in 0.05 mol L−1 citrate buffer at pH 4.5. All groups that were injected with STZ were administered 5 mL of 40% glucose orally after 2 h of STZ injection and 20% glucose in drinking water overnight. Fasting blood glucose measurement was performed on tail-vein blood with a glucometer (PRECICHECK, FIA Biomed GmbH, Germany) after 1 week of diabetes induction. Rats were considered diabetic at blood glucose levels >300 mg dL−1.23 The diabetic rats were then subdivided into four subgroups (six rats each), where sub Gp 1 was considered a positive control, and sub Gp2, Gp3, and Gp4 were treated orally for 28 days with the free butanol extract of P. oceanica (100 mg kg−1 body weight), the selected gelatin nanoparticles formulation (G3) comprising the same equivalent amount of P. oceanica butanol extract, and the reference antidiabetic drug glibenclamide (5 mg kg−1 body weight), respectively.
2.7.4. Collection of blood samples from rats
The blood samples were collected from the eyes of the rate at the end of the experimental phase. Blood samples were immediately placed on ice for 3 h and then centrifuged at 3000 rpm for 15 min. Subsequently, the serum was separated and stored at −80 °C for further analysis.
2.7.5. Biochemical analysis
After 28 days of treatment, the rats were fasted for 12 h. The fasting blood glucose (FBG) levels were measured using a glucometer. The level of insulin (INS) was determined through enzyme linked immunosorbent assay (ELISA) using the rat insulin ELISA kit (ELR-Insulin, RayBio®, Norcross, GA, USA). The level of glucose transporter 4 (GLUT 4) was evaluated by a commercially available ELISA kit (E-ElR0811, Elabscience, USA). The homeostasis model assessment of insulin resistance (HOMA-IR) was also calculated using the following formula:24,25 HOMA-IR = INS (mIU L−1) × FBG (mmol L−1)/22.5.
2.7.6. Sample preparation for GC-MS analysis
The serum samples were thawed on ice and metabolites were extracted as follows: 200 μL of cold acetonitrile was combined with 100 μL of serum and centrifuged for precipitation of proteins at 7000 rpm for 15 min. The supernatant was evaporated after centrifugation using nitrogen gas and a freeze dryer until dryness. Then 50 μL methoxyamine HCl/pyridine solution (20 mg mL−1) was first added to the dried pellet for metabolite derivatization, and then incubated at 60 °C for 1 h. 100 μL MSTFA containing 1% TMS was applied to the mixture, and then incubated at 60 °C for 1 h as the second derivatization step. The quality control (QC) sample was prepared by combining aliquots of all the samples to form a pooled sample.
2.7.7. GC-MS and multivariate chemometric analysis
The serum metabolites of the studied groups were profiled using a GC instrument (Thermo Scientific Corp., USA), coupled with a thermo mass spectrometer detector. Metabolite separation was achieved under the conditions previously described in our previous work.26 The GC-MS data was cleaned, deconvoluted and aligned using the MS-DIAL interface. QC pooled samples were used to reduce the systematic error caused by instrumental fluctuations by removing the features with high relative standard deviation (RSD). Normalization, log transformation and Pareto scaling were applied to the filtered features prior to chemometric analysis. Principal component analysis (PCA), projection of latent structure discriminate analysis (PLS-DA) and pairwise comparison by orthogonal projection to latent structure discriminate analysis (OPLS-DA) were applied using the “ropls” package27 under an R 3.3.2 environment for better understanding of the subtle common point and discrepancies of the complicated data. Univariate analysis was also implemented by analyzing the differences between the normalized quantities of metabolites using the t-test adjusted by calculating the false discovery rate (q values) and metabolite fold changes. The selected potential metabolites were searched and identified by comparing their retention indices (RI) and fragmentation patterns by those available in the Golm, Fiehn BinBase and RIKEN databases.
3. Results
3.1. Phytochemical profiling, antioxidant and in vitro antidiabetic activity of P. oceanica extracts
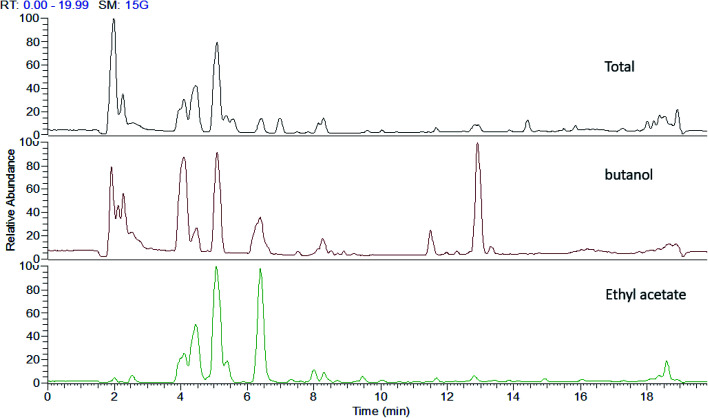
Metabolomics based on high throughput and sensitive UPLC-HRESI-MS/MS enabled in-depth investigation of the various secondary metabolites in several fractions from the P. oceanica rhizome revealing 86 different compounds, which were mainly phenolics and flavonoids. The secondary metabolites were annotated based on their exact molecular masses with Δ less than 5 ppm, fragmentation pattern and retention times. The metabolites were compared to available online databases (PubChem, ChEBI, Metlin and KNApSAck), authentic compounds and literature data (Table 1). The acquired UHPLC-HRESI-MS traces for the P. oceanica extracts are depicted in Fig. 1. Additionally, the concentrations of the metabolites with the available standards were determined quantitatively using HPLC-DAD with the highest concentrations found to be in the butanol fraction such as: p-hydroxybenzoic acid (64.04 μg g−1), protocatechuic acid (12.01 μg g−1), epi-catechin (10.06 μg g−1), syringic acid (9.83 μg g−1) and catechin (8.20 μg g−1) (Table 1).
Mass spectral data for tentative identification and quantification of phenolic acids and flavonoids in P. oceanica in the different extracts.
| No. | RT [min] | Metabolite identification | Chemical formula | [M − H]− | Relative abundance (%) | |||
|---|---|---|---|---|---|---|---|---|
| Measured, calculated | Fragmentation | MeOH | BuOH | EtOAc | ||||
| 1 | 1.55 | Citric acid | C6H8O7 | 190.9278, 191.0270 | 162.89240, 146.93741, 111.00736 | * | * | |
| 2 | 2.00 | 4-Hydroxybenzoylcholinea | C12H17O3N | 224.1281c, 224.1281 | 165.049, 155.9746, 125.2647 | * | ||
| 3 | 2.22 | Monogalloyl glucose | C13H16O10 | 331.0671, 331.0744 | 169.0132 | * | * | |
| 4 | 2.2 | Succinic acid | C4H6O4 | 177.0179, 117.0182 | 199.0072, 86.1084, 73.0279 | * | * | * |
| 5 | 2.28 | Malic acid | C4H6O5 | 133.0129, 133.0215 | 115.0022, 89.0229 | * | ||
| 6 | 2.29 | Quinic acid | C7H12O6 | 191.0553, 191.0634 | 133.0129, 111.0073 | * | * | |
| 7 | 2.36 | 5-Methoxypsoralen | C12 H8 O4 | 215.0230, 215.0339 | 113.0224, 89.0228, 71.0123 | * | * | |
| 8 | 2.47 | Phloracetophenone | C8H8O4 | 167.0201, 167.0200 | 124.0138, 118.2005, 96.086 | * | ||
| 9 | 2.47 | Hordenine | C10H15NO | 166.1226c, 166.226 | 148.0484, 121.0651, 107.0495 | * | ||
| 10 | 2.94 | Danshensu | C9H10O5 | 197.8073, 197.0449 | 162.8379, 103.9188, 122.8925 | * | * | |
| 11 | 3.35 | Benzoylcholine | C12H17NO2 | 208.1333c, 208.1305 | 149.0596, 131.0497, 105.0336 | * | ||
| 12 | 3.86 | Phloroglucinol | C6H6O3 | 125.0231, 125.0233 | 97.0280, 81.0331, 69.0330 | * | ||
| 13 | 3.93 | P-Hydroxyl benzoic acid | C13H16O8 | 299.0775 | 137.0231 | * | * | |
| 14 | 3.98 | Protocatechuic acida,b | C7H6O4 | 153.0182, 153.0266 | 109.0281 | *11.02 | *12.01 | *8.51 |
| 15 | 3.99 | Catechola | C6H6O2 | 109.0281, 109.0284 | 92.8299, 81.0330, 65.0380 | * | * | * |
| 16 | 4.00 | Protocatechuic acid hexoside | C13H16O9 | 315.0724, 315.0711 | 279.8815, 153.0183, 123.0437 | * | ||
| 17 | 4.04 | Methyl salicylatea | C8H8O3 | 151.0023, 151.0026 | 136.0155, 123.0074, 107.0124 | * | ||
| 18 | 4.48 | Vanillina | C8H8O3 | 151.0391, 151.0473 | 123.0074, 107.0123 | * | * | * |
| 19 | 4.06 | Gallic acida,b | C7H6O5 | 169.0133, 170.0215 | 125.0231 | *3.77 | *3.70 | |
| 20 | 4.41 | Vanillic acid hexoside | C14H18O9 | 329.0883, 329.0951 | 167.0341, 119.0338 | * | * | |
| 21 | 4.45 | Homovanillic acid hexoside | C15H20O9 | 343.1039, 343.1107 | 181.0495 | |||
| 22 | 4.46 | Galloyl shikimic acid | C14H14O9 | 325.0569 | 169.0134, 125.0230 | * | * | |
| 23 | 5.03 | Hydroxybenzoic acid pentoside | C12H14O7 | 269.0671, 269.0656 | 137.0232 | * | ||
| 24 | 5.05 | Hydroxyferulic acid | C10H10O5 | 209.0450, 209.0528 | * | |||
| 25 | 5.06 | P-Hydroxybenzoic acidb | C7H6O3 | 137.0231, 137.0317 | 93.0331 | *56.52 | *64.04 | *30.26 |
| 26 | 5.08 | P-Coumaric acidb | C9H8O3 | 162.838 | 119.0486 | *3.27 | *0.42 | *5.71 |
| 27 | 5.08 | Salicylic acida | C7H6O3 | 137.0231, 137.0233 | 107.0029, 93.0331, 65.0381 | * | ||
| 28 | 5.09 | 4,6-Dihydroxy-3-(1-hydroxyethyl)-5-methoxy-2-benzofuran-1(3H)-one | C11H12O6 | 239.0546, 239.0546 | 221.0453, 192.9584, 165.0183 | * | ||
| 29 | 5.12 | Protocatechuic acid sulphate | C7H6O7S | 232.976, 232.9756 | 153.0183 | * | * | |
| 30 | 5.14 | Caffeic acid hexoside | C15H18O9 | 341.1107, 341.1957 | 179.0551, 161.0442, 119.0335, 89.02283 | * | ||
| 31 | 5.26 | Benzoic acida | C7H6O2 | 121.0281, 121.0284 | 108.0204, 93.0331, 75.7648 | * | * | |
| 32 | 5.56 | 4-Deoxyphloridzin | C21H24O9 | 419.1400, 419.1395 | 257.0854, 271.1304, 196.0937 | * | ||
| 33 | 5.35 | Catechinb | C15H14O6 | 289.0716, 289.0707 | 245.0130, 179.0342, 125.0229 | *0.01 | *8.20 | *0.1 |
| 34 | 5.2 | Daphnetin | C9H6O4 | 177.0184, 177.0182 | 162.0320, 133.0281, 105.0330 | * | ||
| 35 | 5.56 | Syringaldehydea | C9H10O4 | 181.0134, 181.0131 | 166.0264, 137.0232, 125.0178, 98.0331 | * | * | |
| 36 | 5.63 | 3-Methylcatechol | C7H8O2 | 123.0436, 123.0441 | 108.0202, 95.0125, 72.9686, 68.3135 | * | ||
| 37 | 5.64 | Procyanidin B2a | C30H26O12 | 577.1420, 577.1420 | 557.2190, 407.0811, 289.0718 | * | ||
| 38 | 5.7 | Ethyl cinnamate | C11H12O2 | 174.9552, | 160.9757, 146.9598, 130.9424, 118.946, | * | ||
| 39 | 5.67 | Caffeic acid dimethyl ester | C11 H12O4 | 207.0656, 207.0652 | 179.9351, 159.858, 127.868, 103.918, 87.9237 | * | ||
| 40 | 5.79 | Gentisic acidb | C7H7O4 | 154.0248, 154.0261 | 122.8927, 109.0280, 110.031396.9586, 59.5949 | * | ||
| 41 | 5.85 | Kynurenic acid | C10H8NO3 | 188.0343, 188.0342 | 144.0443, 109.0278 | * | ||
| 42 | 5.89 | Homovanillic acida | C9H10 O4 | 181.0493, 181.0495 | 153.0181, 137.0231, 123.0437, 109.0280 | * | ||
| 43 | 6.16 | Homo gentisic acida | C8H8O4 | 167.0339, 167.0339 | 123.0072, 108.0199, 95.0124 | * | ||
| 44 | 6.19 | Epi-catechina | C15H14O6 | 289.0392 | 245.082 | *0.54 | *10.06 | |
| 45 | 6.25 | Vanillic acid 4-sulfate | C8H8O7S | 246.9914, 246.9262 | 167.0340, 159.0441 | * | ||
| 46 | 6.30 | Dihydroluteolin-O-hexoside | C21H22O11 | 449.1089, 449.1162 | 287.0563, 269.0458, 178.9978 | * | * | * |
| 47 | 6.38 | Ferulic acid hexoside | C16H20O9 | 355.1040, 355.1107 | 193.0496 | * | ||
| 48 | 6.39 | Protocatechuic aldehyde | C7H6O6S | 216.9805, 216.9801 | 199.8505, 159.8586, 172.99701, 137.02315, 119.9765 | * | ||
| 49 | 6.58 | Catechin gallate | C22H18O10 | 441.1247, 441.1239 | 169.6788, 171.9455, 160.8407, 123.2177 | * | * | |
| 50 | 6.66 | Ferulic acidb | C10 H10O4 | 193.0497, 193.0495 | 177.0545, 149.5339, 90.9320 | *0.47 | *4.42 | *0.38 |
| 51 | 6.7 | Coumaroyl quinic acid | C16 H18O8 | 337.0918, 337.0923 | 322.0846, 191.0705, 147.0801, 119.6231 | * | ||
| 52 | 6.76 | Coniferyl aldehydea | C10H10O3 | 177.1633, 177.1638 | 167.0339, 152.0103, 124.0152, 111.0074 | * | ||
| 53 | 6.69 | Sinapic acida,b | C11H12O5 | 223.0610, 223.0685 | 179.0706, 123.438 | *0.11 | *2.76 | |
| 54 | 6.92 | Vanillic acidb | C8 H8O4 | 167.0339, 167.0339 | 152.0104, 123.0074, 111.0073, 66.463 | *6.43 | *1.38 | *12.51 |
| 55 | 6.96 | Salvaianolic acid G | C18H12O7 | 399.2004 | 321.15760, 66.8679 | * | ||
| 56 | 7.06 | Quercetin-O-pentoside-O-rhamnoside | 597.2015 | 417.1549, 327.0145, 213.4272, 181.0498 | * | |||
| 57 | 7.34 | Cinnamic acida,b | C9H8O2 | 147.0440, 147.0441 | 123.9449, 102.9473, 87.9238, 61.9867 | *0.50 | *2.98 | *0.68 |
| 58 | 7.35 | Syringic acida,b | C9H10O5 | 197.8072, 197.8073 | 162.8380, 123.9005, 103.9186 | *1.49 | *9.83 | *15.01 |
| 59 | 7.37 | Gallocatechin | C15H14O7 | 305.0699, 305.0715 | 225.1128, 169.7713, 138.7856 | * | ||
| 60 | 7.52 | p-Coumaraldehydea | C9H8O2 | 147.0439, 147.0441 | 129.0334, 119.0490, 102.9473 | * | ||
| 61 | 8.09 | Gallocatechin gallate | C22H18O11 | 457.0765, 457.0771 | 137.0958, 123.0438, 114.9323 | * | * | |
| 62 | 8.19 | Scopoletinb | C10H8O4 | 191.0338, 191.0339 | 162.8923, 146.9375, 111.0073 | *0.52 | *2.61 | |
| 63 | 8.27 | Procyanidin A2a | C30H24O12 | 575.1262, 575.1300 | 407.0793, 161.8318, 125.0228 | * | ||
| 64 | 8.61 | Caffeic acida,b | C9H8O4 | 179.0343, 179.0339 | 150.9530, 134.9868, 90.9966 | *1.34 | *1.01 | *0.56 |
| 65 | 8.64 | Ellagic acida | C14H6O8 | 300.9992, 300.9979 | 257.0097, 185.0247 | * | ||
| 66 | 8.68 | 2′-Hydroxygenistein-7-O-glucoside | C21 H19O11 | 447.0944 | 317.1527, 285.0407, 134.4610, 103.1963 | * | ||
| 67 | 8.85 | Rutinb | C27H30O16 | 609.1588, 609.1589 | 301.0715, 173.603, 75.0797 | *9.33 | *0.68 | *1.39 |
| 68 | 8.99 | Kaempferol-3-glucuronide | C21 H18 O12 | 461.0737, 461.0715 | 403.9498, 285.0404, 148.9581 | * | ||
| 69 | 9.09 | Secoisolariciresinol | C20H27O6 | 361.1659, 361.1646 | 346.1428, 331.0833, 179.0741, 137.0230 | * | ||
| 70 | 9.39 | Rosmarinic acidb | C18H16O8 | 359.1136, 359.1125 | 257.0820, 197.0451, 179.036, 161.0233 | *0.08 | *0.02 | *1.14 |
| 71 | 9.46 | Pyridinesulfonamides | C16H20N4O3S | 347.1172, 347.1175 | 274.4805, 137.0233, 195.0652, 162.0544 | * | ||
| 72 | 9.51 | Apigenin-7-O-glucosideb | C21H20O10 | 431.0973, 431.0975 | 321.6589, 268.038, 79.8678 | *0.55 | *2.30 | *0.01 |
| 73 | 9.72 | Luteolin-5-glucoside | C21H20O11 | 447.1045, 447.1028 | 314.0438, 271.0258, 151.0024, 89.0907 | * | ||
| 74 | 9.86 | Baicalein-7-O-glucuronide | C21H18O11 | 445.0786, 445.0765 | 269.0458, 151.0388, 113.0232 | * | ||
| 75 | 10.54 | P-Anisic acida | C8H8O3 | 151.0388, 151.0390, | 136.0153, 123.0074, 107.0488, 93.0331 | |||
| 76 | 10.95 | Gibberellin A42 | C20H30O6 | 365.1975, 365.1959 | 335.0222, 267.0664, 255.0665, 166.6308 | * | ||
| 77 | 11.53 | Enterolactone | C18H18O4 | 297.1132, 297.1211 | 253.1231, 189.0550, 107.0488 | |||
| 78 | 11.71 | Luteolin b | C15H10O6 | 285.0406, 285.0394 | 250.4250, 151.0027, 137.0230, 93.0329 | *3.72 | *0.01 | *0.03 |
| 79 | 11.85 | Naringeninb | C15H12O5 | 271.0613, 271.0601 | 253.1438, 151.0023, 119.0487 | *0.23 | *3.13 | *2.49 |
| 80 | 12.95 | Gibberellic acid A44 | C20H26O5 | 345.1709, 345.1780 | 301.1812, 283.1712 | * | ||
| 81 | 13.15 | Diosmetin | C16H12O6 | 299.0565, 299.0550 | 284.0327, 237.1862, 125.0957 | * | * | |
| 82 | 13.36 | Kaempferolb | C15H10O6 | 285.0406, 285.0394 | 254.9923, 183.9123, 137.0231 | |||
| 83 | 14.13 | Butylparaben | C11H14O3 | 193.0862, 193.0859 | 178.0257, 137.0231, 106.0755 | * | ||
| 84 | 17.51 | Apigeninb | C15H10O5 | 269.0457, 269.0444 | 251.2011, 189.0912, 155.1069 | *0.67 | *0.01 | *0.91 |
| 85 | 17.62 | Methylated (−)-gallo catechin gallate | 471.0768 | 431.8876, 385.5272, 341.1092, 325.1846, 245.9779, 169.9367, | * | |||
| 86 | 18.92 | Bilobalide | 325.1844 | 250.1201, 183.0110, 108.1780 | * | |||
Compounds isolated before from the plant.
Quantitative estimation of some identified compounds (μg g−1) using/HPLC-DAD.
Data was collected in the positive-ion mode.
Fig. 1. UHPLC-MS traces of P. oceanica extracts showing different qualitative differences according to their metabolic profiles.
Interestingly, the highest concentrations of total phenols and flavonoids were also found in the butanol fraction, which is consistent with the HPLC-DAD quantification results. Also, the in vitro antidiabetic activity agreed with the total phenol and flavonoid content and the antioxidant activities using the DPPH assay, as shown in Table 2.
Total phenolic and flavonoid contents and in vitro antioxidant and anti-diabetic activities of P. oceanica extract and its fractions against α-glucosidase enzyme.
| Extract | Phenolic content (mg gallic acid/g extract) | Flavonoid content (mg rutin/g extract) | DPPH (IC50, μg mL−1) | α-Glucosidase (IC50, μg mL−1) |
|---|---|---|---|---|
| BuOH | 200.20 ± 1.09 | 40.17 ± 0.11 | 10.5 | 4.8 ± 0.3 |
| EtOAc | 140.25 ± 1.01 | 20.34 ± 0.27 | 30.4 | 8.9 ± 0.4 |
| Total alcohol | 120.37 ± 1.17 | 20.35 ± 0.03 | 76.3 | 24.8 ± 2 |
| Acarbose (SD) | — | — | — | 4.5 ± 0.27 |
3.2. Preparation, characterization and in vitro release study of P. oceanica-encapsulated gelatin nanoparticles
Based on the in vitro activity, the butanol fraction was chosen for further investigation. Initially, twelve gelatin nanoparticle preparations were examined, and various formulation parameters were varied, aiming to obtain P. oceanica-encapsulated gelatin nanoparticles with the optimum characteristics. The investigated parameters were the type of stabilizer, gelatin to stabilizer mass ratio and solvent to non-solvent ratio. The data listed in Table 3 shows the attainment of nanoparticles in eight formulations. It was observed that well dispersed nanoparticles were successful formed when employing a 1 : 8 v/v water/ethyl alcohol ratio. Conversely, visible aggregates were observed in the formulations prepared using water/ethyl alcohol at a ratio of 1 : 4, v/v (G4, G10, G11 and G12). Accordingly, the nanoparticle systems prepared employing the water/ethyl alcohol ratio of 1 : 8, v/v were subjected to further characterization.
Composition, encapsulation efficiencies, particle size and zeta potential values of gelatin nanoparticulate formulations.
| Code | Gelatin (g) | Stabilizer | Gelatin: stabilizer ratio (w/w) | Solvent, water (mL) | Non-solvent, ethanol (mL) | NP formed | EE% ± SD | PS (nm) ± SD | ZP (mV) | |
|---|---|---|---|---|---|---|---|---|---|---|
| Poloxamer 407 (g) | Poloxamer 188 (g) | |||||||||
| G1 | 0.2 | 1.6 | 0 | 1 : 8 | 10 | 80 | + | 51.68 ± 2.34 | 274.7 ± 30.5 | −10.7 |
| G2 | 0.2 | 3.2 | 0 | 1 : 16 | 10 | 80 | + | 63.76 ± 2.38 | 290.0 ± 13.66 | −12.3 |
| G3 | 0.2 | 6.4 | 0 | 1 : 32 | 10 | 80 | + | 51.68 ± 2.34 | 274.7 ± 30.5 | −10.7 |
| G4 | 0.2 | 1.6 | 0 | 1 : 8 | 10 | 40 | − | — | — | — |
| G5 | 0.2 | 3.2 | 0 | 1 : 16 | 10 | 40 | + | — | — | — |
| G6 | 0.2 | 6.4 | 0 | 1 : 32 | 10 | 40 | + | — | — | — |
| G7 | 0.2 | 0 | 1.6 | 1 : 8 | 10 | 80 | + | 32.65 ± 5.39 | 591.1 ± 37.97 | −10.0 |
| G8 | 0.2 | 0 | 3.2 | 1 : 16 | 10 | 80 | + | 41.80 ± 2.12 | 461.1 ± 47.83 | −10.5 |
| G9 | 0.2 | 0 | 6.4 | 1 : 32 | 10 | 80 | + | 49.48 ± 3.59 | 458.7 ± 53.95 | −14.8 |
| G10 | 0.2 | 0 | 1.6 | 1 : 8 | 10 | 40 | − | — | — | — |
| G11 | 0.2 | 0 | 3.2 | 1 : 16 | 10 | 40 | − | — | — | — |
| G12 | 0.2 | 0 | 6.4 | 1 : 32 | 10 | 40 | − | — | — | — |
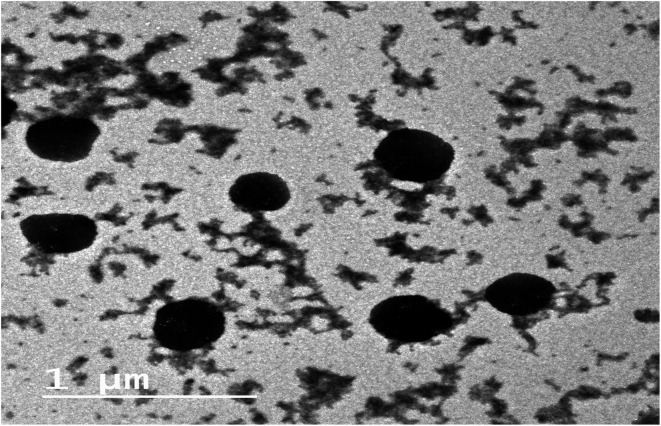
The EE% of the prepared gelatin nanoparticle formulations is reported in Table 3. It was obvious that higher EE% values were observed when Poloxamer 407 was used as a stabilizer in comparison to Poloxamer 188. These results also revealed that the increment in EE% values was directly proportional to the increase in stabilizer ratio in all the investigated formulations. This finding was more pronounced when employing high stabilizer to gelatin ratios (G2 and G3 in comparison to G8 and G9). Moreover, all the investigated formulations showed particle size values in the nanosize range (242–591 nm) (Table 3). The zeta potential of the gelatin nanoparticles exhibited negative values in the range of −10.0 to −14.8 mV, reflecting moderate physical stability for the nanoparticulate formulations. These results are in accordance with previous reports on gelatin nanoparticles.28,29 Based on the results of EE%, PS and ZP, formulation G3 was found to exhibit the highest EE% and suitable PS and ZP values, and thus was selected for further investigations. Besides, TEM was employed to elucidate the shape and morphology of the selected nanoparticle formulation, G3. Fig. 2 shows that the nanoparticles are homogeneous and appear as dark stained spheroid shapes with no signs of aggregation.
Fig. 2. TEM micrograph of a selected gelatin nanoparticle formulation, G3.
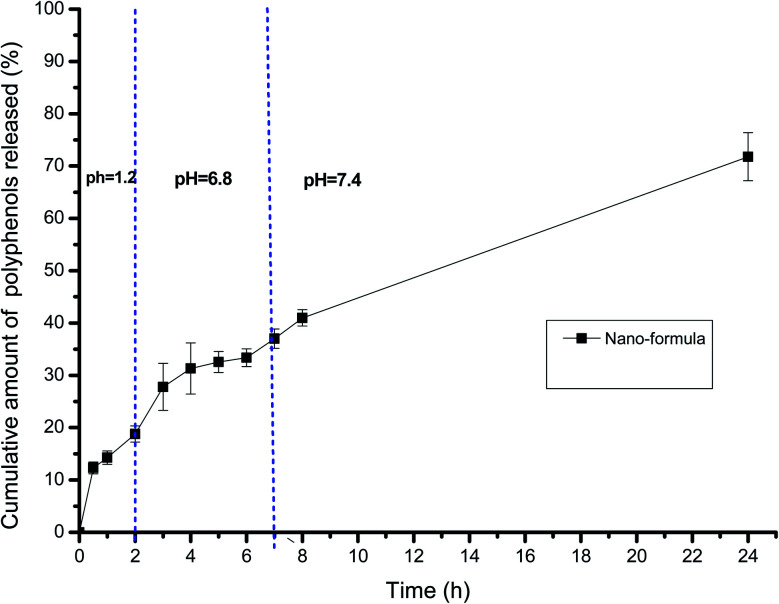
Fig. 3 illustrates the release profile of the polyphenols from the selected gelatin nanoparticle formulation, G3. The depicted release profile was gradual, which extended over 24 h, indicating that encapsulating the extract within the gelatin nanoparticle formula was successful in affording a controlled release pattern in the GIT pH mimetic condition. There was no detectable abrupt burst release for the polyphenols. The cumulative percentage release amount was satisfactory, reaching about 20% within 2 h at pH 1.2. This was followed by a slower gradual rate of release at the intestinal pH of pH 6.8, attaining about 36% after 6 h. Subsequently, further amounts of the encapsulated polyphenols were released within the subsequent 18 h at pH 7.4, where the total cumulative amount of released phenolics reached nearly 70% within 24 h.
Fig. 3. Release pattern of polyphenols from the selected gelatin nanoparticle formulation, G3.
The kinetic study of the release data disclosed that both the diffusion (Higuchi) model and first-order model depicted high regression coefficient values (0.98 and 0.97, respectively). Further, fitting 60% of the release data to the Peppas model revealed that the “n” value was 0.46, thus lying in the range between 0.43 and 0.85, indicating an anomalous non-Fickian release pattern.20 This confirmed that different contributing factors dominated the release of the polyphenols from the gelatin nanoparticles.30
3.3. Evaluation of the in vivo anti-diabetic potential of P. oceanica based on biochemical parameters and metabolomic analysis
The effects of P. oceanica and its nano-extract on the serum FBG, INS, HOMA-IR and GLUT 4 levels in type 2 diabetic rats are shown in Table 4. The STZ-induced diabetic rats showed a significant (p < 0.05) increase in insulin concentration and HOMA-IR level with a highly significant (p < 0.05) rise in fasting blood glucose level compared to the normal control rats. In addition, the level of GLUT 4 was significantly lower (P < 0.05) in the serum of the diabetic rats than that in the normal control rats. Oral administration of P. oceanica gelatin nanoparticles considerably (P < 0.05) recovered the hyperglycemic state and improved the insulin, HOMA-IR and GLUT-4 levels towards normal compared to the diabetic group.
In vivo biochemical evaluation of fasting blood glucose (FBG), insulin (INS), homeostasis model assessment of insulin resistance (HOMA-IR) and glucose transporter 4 (Glut 4) among the different groups.
| Groups | FBG (mmol L−1) | INS (μIU mL−1) | HOMA-IR | GLUT 4 (ng mL−1) |
|---|---|---|---|---|
| Normal control | 4.79 ± 0.12a | 11.23 ± 0.58a | 2.45 ± 0.14a | 11.66 ± 0.12a |
| Diabetic rats | 21.41 ± 0.45b | 43.13 ± 4.67b | 41.38 ± 3.67b | 8.61 ± 0.44b |
| Plain extract treated group | 12.10 ± 1.25a,b | 35.03 ± 4.48b | 20.38 ± 3.78a,b | 8.42 ± 0.38b |
| Standard drug treated group | 11.86 ± 0.92a,b | 26.30 ± 4.05a | 13.10 ± 2.07a | 9.17 ± 0.66b |
| Nano-extract treated group | 7.88 ± 0.44a | 13.50 ± 2.14a | 4.82 ± 0.96a | 11.27 ± 0.42a |
P < 0.05 versus diabetic rats.
P < 0.05 versus normal control.
A metabolomics approach based on GC-MS was also developed to investigate the antidiabetic activity of P. oceanica in rats. A total of 207 characteristic m/z features were detected in this study, among which 72 annotated metabolites survived the QC-based filtering procedures. Data normalization by sum of total intensities for each sample was carried out to make the metabolite intensity more comparable. Besides, a combination of log transformation and Pareto scaling yielded a Gaussian distribution for the data. This normality of the data enabled the application of different multivariate and univariate tests to examine the trends among the studied groups to reveal the significant metabolites associated with the antidiabetic activity of P. oceanica and its nano-formulation. Twenty-five differential metabolites were revealed from the statistical analysis of the GC-MS data, which belonged to different classes, including organic acids, amino acids, sugars and fatty acids. The identities, chromatographic and mass spectrometry data of these metabolites are listed in Table 1S.†
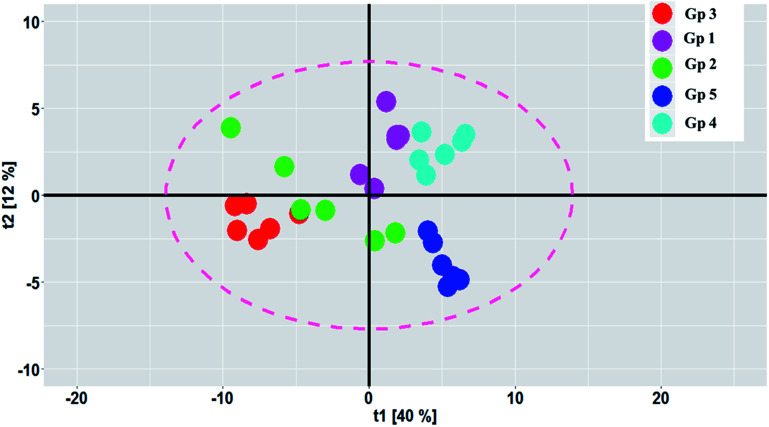
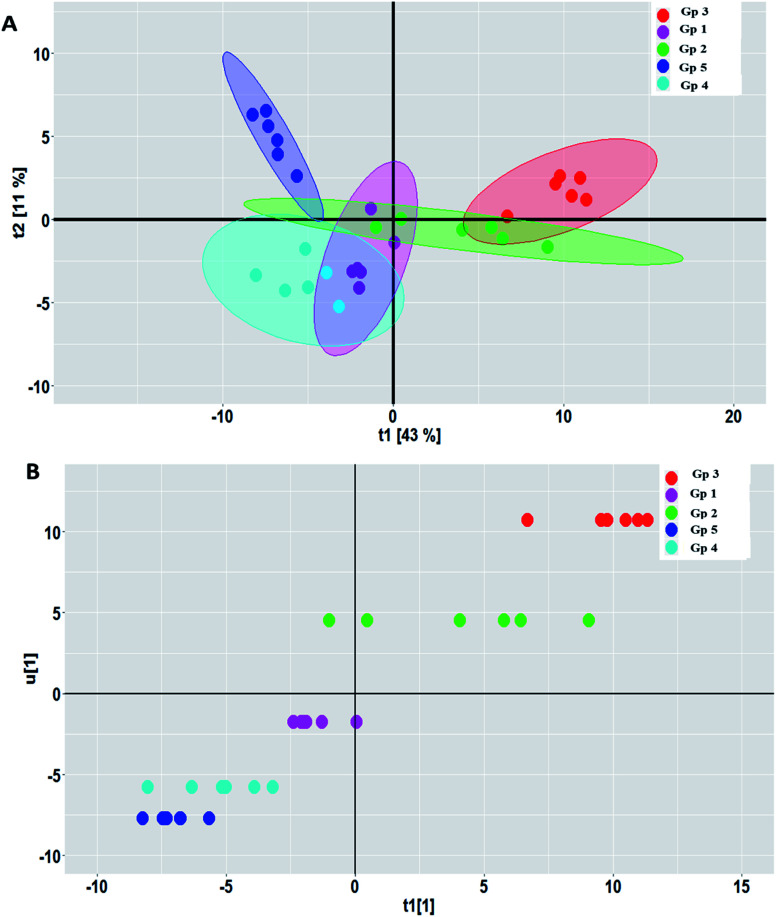
PCA was carried out to visualize the patterns among the different studied groups in an unsupervised manner. The scores of the control group and groups treated with P. oceanica gelatin nanoparticles and glibenclamide showed a separation trend along the first projection with positive score values from the diabetic and the butanol-treated groups with negative score values, but further differentiation was not obvious (Fig. 4). Hence, PLS-DA as a supervised chemometric approach was employed to derive better sample classification owing to its capability to identify correlation with the phenotypic variable of interest. The PLS-DA score plot showed clearer discrimination among sample groups compared to PCA, with an R2 value (0.66) and Q2 value (0.52) using three latent component models (Fig. 5a). The PLS-DA class inner relationship model of all the groups revealed that the diabetic and P. oceanica butanol-treated group rats were located the furthest from normal rats and other treatment groups (Fig. 5b). Remarkably, the P. oceanica gelatin nanoparticle-treated group was the closest to the control normal rats, suggesting that this line was the most effective treatment approach to restore the metabolite profile in the diabetic rats, mirroring that in the normal rats. The validity of the developed PLS-DA model was confirmed by performing a permutation test (1000 times) with pR2Y and pQ2 (>0.001) for the multi-group comparison. The metabolites contributing this group segregation were revealed by calculating the variable influence of projection (VIP) together with their regression coefficients (Table 2S†). They include beta-hydroxybutyric acid, glucosamine, butane-2,3-diol, glucose, 1,5-anhydro-d-glucitol, aminobutyric acid, myo-inositol and GABA.
Fig. 4. PCA score plot showing the partial segregation of the samples along the first two principal components. The samples are coded as follow: Gp 1, healthy control group; Gp 2, non-treated diabetic group; Gp 3, diabetic group treated with plain butanol extract of P. oceanica; Gp 4, diabetic group treated with the nano-extract of P. oceanica; and Gp 5, diabetic group treated with glibenclamide.
Fig. 5. (A) PLS-DA score plots and (B) class inner relation obtained from modelling the serum metabolites in the different rat groups. The rat groups with diabetes (Gp 2) and P. oceanica butanol treatment group (Gp 3) showing separation from the normal control group (Gp1) and P. oceanica nano-extract treatment group (Gp 4).
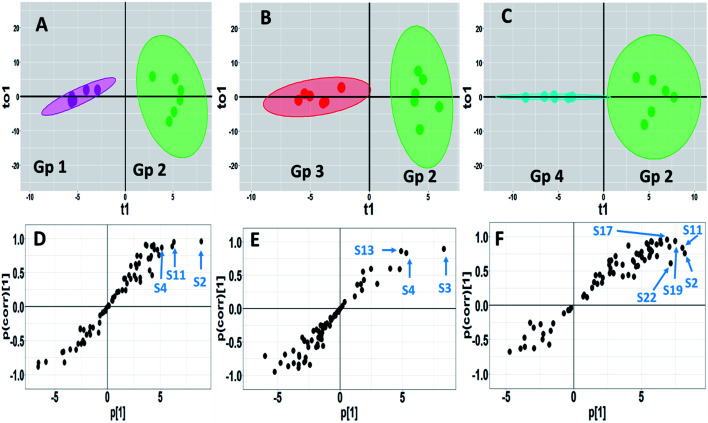
OPLS-DA is another supervised approach that is superior to other multivariate methods in terms of discriminating ability and biomarker discovery owing to its capability to remove the non-predictive variations through orthogonal signal correction. However, only pairwise comparison can be implemented using this model, and consequently, different OPLS-DA models were constructed to reveal the pharmacometabolomic effect of P. oceanica and its nano-extract. The R2Y and Q2Y values were used to determine the quality of each OPLS-DA model and evaluate its prediction power. For the control group versus the streptozotocin-induced diabetic group, the model validation parameters were: R2 = 0.958 and Q2 = 0.674 with clear separation along the predictive component, as shown in Fig. 6a. Another separation trend was depicted in Fig. 6b from modelling the P. oceanica butanol-treated group versus the diabetic samples with R2 = 0.952 and Q2 = 0.721. Moreover, a final model was built comparing the P. oceanica (L.) extract encapsulated in gelatin nanoparticle-treatment group versus their diabetic counterparts. The score plot of this model as depicted in Fig. 6c shows a clear separation with R2 = 0.928 and Q2 = 0.774. Additionally, all the OPLS-DA models were strictly validated using permutation tests (1000), where all the Q2 values of the permuted data set show a significant model with pQ2 (0.001 each) for pairwise comparison of the diabetic rats versus the control, P. oceanica butanol- and nano-extract treatment groups. Besides, the S plot loadings, a useful tool generated for each OPLS-DA model by plotting the covariance (p) against correlation p(corr), was implemented to decipher the relevant biomarkers contributing to the detected differences, as shown in Fig. 6d–f. The results revealed some metabolites with higher p(corr) values, accounting for the higher expression in diabetic rats compared to the control and P. oceanica encapsulated in gelatin nanoparticles treatment groups including glucose, alanine, leucine, isoleucine, proline, tyrosine, phenylalanine, oleic acid and linoleic acid.
Fig. 6. GC-MS-based OPLS-DA score plots derived from modelling diabetic rats versus other groups (A–C). The scores of the samples are coded as follows: Gp 1, healthy control group; Gp 2, non-treated diabetic group; Gp 3, diabetic group treated with plain butanol extract of P. oceanica; Gp 4, diabetic group treated with the nano-extract of P. oceanica. Derived S-plots (D–F) showing the covariance p[1] against the correlation p(cor)[1]. Selected variables follow that listed in Table S2† for metabolite identification: S2; butane-2,3-diol, S3; lactic acid, S4; alanine, S11; proline, S13; serine, S17; phenylalanine, S19; glucose, and S22; tyrosine.
However, for a better selection of the differential metabolites, a combined univariate and multivariate strategy was developed considering the VIP of the OPLS-DA models together with the univariate t-test statistical analysis and metabolite fold change (Table 3S†). The variables that influence the projection (VIP) >1.3 with a q value <0.05 and normalized fold changes >2 or <0.5 were selected. In agreement with the OPLS-DA-derived S plots, the nano P. oceanica treatment group has the highest number of metabolites, among which, their relative concentrations were statistically different from the diabetic group [13 metabolites belonged mainly to amino acids, fatty acids and sugars] followed by the control group [8 metabolites] and butanol P. oceanica treatment group [5 metabolites]. The fold change of the diabetic rats divided by the normal control and nano P. oceanica treatment groups suggested a significant increase in butane-2,3-diol, alanine, leucine, isoleucine, proline, tyrosine, phenylalanine, methionine, 2-hydroxyhexanoic acid, oleic acid and linoleic acid (Table 3S†). However, the butanol P. oceanica treatment group failed to decrease the relative concentration levels of these metabolites.
4. Discussion
In this study, we comprehensively characterized the secondary metabolites in the hydroalcoholic extract together with its two fractions (ethyl acetate and butanol) using UPLC-HRESI-MS/MS. The analysis revealed 86 secondary metabolites belonging to simple phenols, lignans, phenolic acids, sulphated phenolic acid conjugates, cinnamic acid derivatives and flavonoids in addition to plant hormones and alkaloids. The total flavonoid and phenolic contents were more pronounced in the butanol extract. The latter also exhibited potent antidiabetic activity in vitro against α-glucosidase, an enzyme that is mainly involved in diabetes. This bioactivity of the P. oceanica butanol fraction is owing to its richness with known antidiabetic metabolites such as p-hydroxybenzoic acid,31 protocatechuic acid,32 epi-catechin,33 syringic acid34 and catechin.35
To provide a gradual controlled release that provides protection of these polyphenols against degradation in the harsh GIT condition especially, the butanol fraction was formulated in a nano form. Gelatin nanoparticulate formulations were prepared employing the nanoprecipitation technique. In this process, nanoparticle formation is based on the interfacial turbulence generated by solvent displacement from the internal phase.36 Successful preparation was observed when a higher solvent/non solvent ratio was used (1 : 8 v/v). The solvent/non-solvent ratio is an important parameter for preparing stable nanoparticles,37 where higher ratios lead to the production of small stable nanoparticles. It is also worth noting that unlike Poloxamer 188, Poloxamer 407 had the ability to form stable nanoparticles when higher concentrations were employed.
The increase in EE% indicates that the stabilizing effect of poloxamers on the gelatin nanoparticles is concentration dependent.37 Higher EE% values and smaller particle sizes were observed when Poloxamer 407 was used as the stabilizer compared to Poloxamer 188. This observation was more pronounced when higher concentrations of the stabilizer were used and can be attributed to the higher molecular weight of Poloxamer 407 (12 000 g mol−1) compared to Poloxamer 188 (8350 g mol−1). It was previously reported that stabilizers having a lower molecular weight failed to produce the required stabilizing effect.37
The in vitro release profile of the selected formulation, G3, showed that the cumulative polyphenol amount released at the intestinal region pH (6.8–7.4) was larger than that released at the acidic stomach pH. This can be attributed to the gelatin type used in the nanoparticle preparation (type B), which will become negatively charged at near neutral pH values, thus causing greater swelling and permitting a larger amount of the encapsulated drug to be released in comparison to acidic pH.38 This gradual controlled release provides protection to the polyphenols against degradation in the harsh environment in the stomach region. Also, it will permit a larger amount of polyphenols to reach their target site considering that it has been reported that the intestinal region is the major site of polyphenol absorption.39,40
This investigated formulation (G3) was further subjected to in vivo evaluation in diabetic rats to estimate the implication of encapsulation in the gelatin nanoparticles on promoting the therapeutic activity of P. oceanica extract. Therefore, induction of diabetes was first performed and assessed by measuring the levels of fasting blood glucose, which showed a significant increase in the STZ hyperglycemic rats due to the destruction of glucose homeostasis.41 The muscle and fat cells are “resistant” to insulin action in type 2 diabetes (T2DM) and compensatory mechanisms are triggered in the β-cells to secrete more insulin.42 Therefore, the HOMA-IR levels in diabetic rats were found to be significantly higher than that in normal control rats. However, these levels decreased significantly after treatment with P. oceanica extract encapsulated in gelatin nanoparticles for 28 days, suggesting that the P. oceanica extract encapsulated in the gelatin nanoparticles is capable of reducing insulin resistance and improving the sensitivity of the body to insulin.
Additionally, there was a significant reduction in the level of Glut 4 in the diabetic rats compared to the normal control, in agreement with Sukanya et al.,43 who reported that the level of Glut 4 was significantly reduced in diabetic rats by mediating glucose uptake and regulating the transport of glucose into muscle cells. Interestingly, the P. oceanica extract encapsulated in gelatin nanoparticles also showed substantial improvement in the level of Glut 4 and glucose homeostasis.
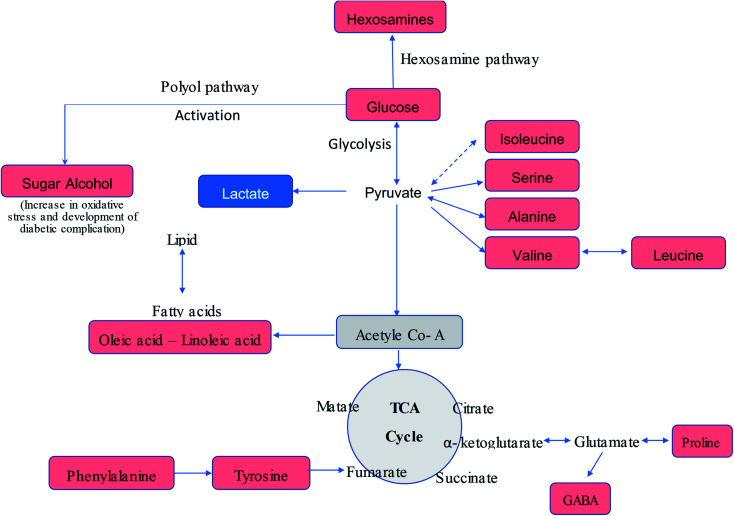
The serum metabolic profiling of the control and T2DM treatment groups was also investigated to explore the potential serum biomarkers and changes in the metabolic pathways involved in the treatment of diabetic rats with the P. oceanica butanol fraction in both its free form and encapsulated in gelatin nanoparticles (Fig. 7). As predicted, the hexose level in our present study was strongly correlated with T2DM, where hexose involves not only glucose, but all six-carbon monosaccharides. Pancreatic beta-cell dysfunction and insulin resistance can be implied by an elevated hexose level. Our result is consistent with the results of other studies,44,45 which confirmed that hexose metabolites are relevant for the assessment of T2DM associations. The present results indicated a significant reduction in the blood glucose level in the diabetic rats pretreated with P. oceanica extract and that encapsulated in the gelatin nanoparticle formula was more effective. This finding suggests that encapsulation in the form of nanoparticles is highly effective in reducing the endogenous glucose production (EGP) metabolite level in diabetic rats. This study also demonstrated that many saccharide species related to glucose metabolism were notably decreased in the group treated with the P. oceanica nanoparticle formulation (G3). The reduced saccharide species in this group implied the improvement of glucose metabolism, leading to depletion in the risk of disease complications.
Fig. 7. Schematic representation of the altered metabolic pathway in T2DM as detected by GC-MS metabolomic analysis. Metabolites coded in red color show increase in their levels in the diabetic group compared with the control samples, while lactic acid, coded in blue color, shows a decrease in its level in the diabetic group compared with the control samples.
Furthermore, the diabetic group depicted a significant increase in the concentration of BCAAs (valine, leucine, and isoleucine), proline, methionine, phenylalanine, alanine and tyrosine (Table 3S† and Fig. 7) in comparison to the normal control group. These results are in agreement with that of Zhang et al.,1 who reported that an elevation in the level of branched-chain amino acids actually leads to insulin resistance by declining the activity of AMP-activated protein kinase, which finally leads to T2DM.46 Moreover, increased BCAAs elicit catabolic substances (propionyl CoA and succinyl CoA), resulting in the aggregation of incompletely beta-oxidized fatty acids and glucose, decreased insulin effect, and glucose control disorder.45 It was observed that this increase was significantly reversed by the P. oceanica extract encapsulated in gelatin nanoparticles. The beneficial effects of P. oceanica butanol extract on the enhanced metabolism of amino acids can be clarified by its beneficial impact on circulating insulin concentrations and insulin sensitivity by activating the cAMP/PKA-dependent ERK1/2 signaling pathway.47
It was also found that aromatic amino acids such as phenylalanine and tyrosine were perturbed in the diabetic group, which is consistent with previous metabolomic studies of T2DM.48,49 Asserted pathways that relate this association include inhibiting the transport/phosphorylation of glucose50 and enhancing of insulin resistance by phosphorylation of insulin receptor substrate 1.51 Phenylalanine and tyrosine were reported to be related to insulin resistance, which confirms their roles in the pathogenesis of T2DM.52
The elevation of the sugar alcohol butane-2,3-diol concentrations suggests dysregulation of glucose homeostasis and stimulation of specific pathways such as polyol metabolism (Fig. 7). Indeed, sugar alcohols can also be derived from microbiota and food metabolism in the gut due to impaired intestinal barriers,53 and thus the attenuated level of butane-2,3-diol in the group treated with the P. oceanica extract encapsulated in gelatin nanoparticles suggests the regulation of glucose homeostasis.
Increased oleic and linoleic acids levels were also observed, which displayed lipotoxicity in the rats treated with STZ. This is consistent with the findings of Lu et al.,54 who reported that fatty acids inhibit insulin action via the Randle cycle, intracellular lipid derivative accumulation, inflammation, oxidative stress and mitochondrial dysfunction. The ameliorated level of these fatty acids reflects the improvement of lipid metabolism due to treatment with P. oceanica extract encapsulated in gelatin nanoparticles. Glucosamine is a naturally occurring amino monosaccharide and well known as a precursor for glycosaminoglycans and mucopolysaccharides. The elevations of glucosamine level in the current study are in agreement with Omori et al.,55 who reported that an increase in hexosamines may play a role in hyperglycemia-induced damage of vascular endothelial cells, and thus may contribute to the development of insulin resistance and hypertension. Thus, these findings confirm the ability of P. oceanica extract encapsulated in gelatin nanoparticles as a potent antidiabetic agent.
5. Conclusions
In the current study, the secondary metabolites of P. oceanica rhizome were annotated via UHPLC-MS/MS, where the different polyphenol classes found included major polyphenols, i.e., 32 phenolic acids, 17 cinnamic acids and 29 flavonoids. Thus, a gelatin nanoparticle formulation, G3, was formulated, which showed the highest EE%, suitable PS and moderate physical stability as exhibited by its ZP values. Furthermore, this formulation exhibited a sustained in vitro release profile with slower release in the intestinal pH range (pH 6.8 and 7.4). The study of the release kinetics revealed an anomalous, non-Fickian release pattern. Accordingly, this formulation was used for therapeutic evaluation as an antidiabetic agent. Metabolomic analysis complemented with biochemical assays revealed major metabolic changes in T2DM. The P. oceanica extract encapsulated in gelatin nanoparticles demonstrated capability to ameliorate T2DM, as indicated by the improvement in the pathological metabolic state and biochemical parameters of the diabetic rats to an almost normal stage.
Conflicts of interest
The authors have no conflicts of interest to declare.
Supplementary Material
Acknowledgments
We would like to express our great thanks and appreciation to National Research Centre, Egypt for providing facilities and financing this work project, reference number: (11010314). We would like also to express our deep gratitude and grateful thanks to Prof. Dr Hab. Piotr Kachlicki head of metabolomics team at Institute of Plant Genetics PAS Poznan for his support and assistance for using UPLC/HRMS.
Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra09606g
References
- Zhang Y. Wang P. Xu Y. Meng X. Zhang Y. J. Evidence-Based Complementary Altern. Med. 2016;2016:1–12. [Google Scholar]
- Wu T. Qiao S. Shi C. Wang S. Ji G. J. Diabetes Invest. 2018;9:244–255. doi: 10.1111/jdi.12723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salem M. A. Perez de Souza L. Serag A. Fernie A. R. Farag M. A. Ezzat S. M. Alseekh S. Metabolites. 2020;10:37. doi: 10.3390/metabo10010037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Z. Bhandari B. Trends Food Sci. Technol. 2010;21:510–523. doi: 10.1016/j.tifs.2010.08.003. [DOI] [Google Scholar]
- Vacchi M. De Falco G. Simeone S. Montefalcone M. Morri C. Ferrari M. Bianchi C. N. Earth Surf. Processes Landforms. 2017;42:42–54. doi: 10.1002/esp.3932. [DOI] [Google Scholar]
- Grignon-Dubois M. Rezzonico B. Bot. Mar. 2015;58:379. [Google Scholar]
- Haznedaroglu M. Z. Zeybek U. Pharm. Biol. 2007;45(10):745–748. doi: 10.1080/13880200701585717. [DOI] [Google Scholar]
- Cornara L. Pastorino G. Borghesi B. Salis A. Clericuzio M. Marchetti C. Damonte G. Burlando B. Mar. Drugs. 2018;16:21. doi: 10.3390/md16010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serkova N. J. Standiford T. J. Stringer K. A. Am. J. Respir. Crit. Care Med. 2011;184:647–655. doi: 10.1164/rccm.201103-0474CI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piasecka A. Sawikowska A. Krajewski P. Kachlicki P. J. Mass Spectrom. 2015;50(3):513–532. doi: 10.1002/jms.3557. doi: 10.1002/jms.3557. [DOI] [PubMed] [Google Scholar]
- Hamed M. A. Mohammed M. A. Aboul Naser A. F. Matloub A. A. Fayed D. B. Ali S. A. Khalil W. K. B. J. Biol. Act. Prod. Nat. 2019;9:335–351. [Google Scholar]
- Shimada K. Fujikawa K. Yahara K. Nakamura T. J. Agric. Food Chem. 1992;40:945–948. doi: 10.1021/jf00018a005. [DOI] [Google Scholar]
- Elya B. Basah K. Mun'im A. Yuliastuti W. Bangun A. Septiana E. K. J. Biomed. Biotechnol. 2012;2012:1–6. doi: 10.1155/2012/281078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E. J. Khan S. A. Park J. K. Lim K. H. Bioprocess Biosyst. Eng. 2012;35:297–307. doi: 10.1007/s00449-011-0591-2. [DOI] [PubMed] [Google Scholar]
- Kassem A. A. Abd El-Alim S. H. Asfour M. H. Int. J. Pharm. 2017;517:256–268. doi: 10.1016/j.ijpharm.2016.12.018. [DOI] [PubMed] [Google Scholar]
- Makhlof A. Tozuka Y. Takeuchi H. Eur. J. Pharm. Biopharm. 2009;72:1–8. doi: 10.1016/j.ejpb.2008.12.013. [DOI] [PubMed] [Google Scholar]
- Asfour M. H. Mohsen A. M. J. Adv. Res. 2018;9:17–26. doi: 10.1016/j.jare.2017.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmotasem H. Farag H. K. Salama A. A. A. Int. J. Pharm. 2018;547:83–96. doi: 10.1016/j.ijpharm.2018.05.038. [DOI] [PubMed] [Google Scholar]
- Dash S. Murthy P. N. Nath L. Chowdhury P. Acta Pol. Pharm. 2010;67:217–223. [PubMed] [Google Scholar]
- Peppas N. A. Sahlin J. J. Int. J. Pharm. 1989;57:169–172. doi: 10.1016/0378-5173(89)90306-2. [DOI] [Google Scholar]
- Kilkenny C. Browne W. J. Cuthill I. C. Emerson M. Altman D. G. PLoS Biol. 2010;8:e1000412. doi: 10.1371/journal.pbio.1000412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhutada P. Mundhada Y. Bansod K. Bhutada C. Tawari S. Dixit P. Mundhada D. Neurobiol. Learn. Mem. 2010;94(3):293–302. doi: 10.1016/j.nlm.2010.06.008. doi: 10.1016/j.nlm.2010.06.008. [DOI] [PubMed] [Google Scholar]
- Mendes J. J. Leandro C. I. Bonaparte D. P. Pinto A. L. Comp. Med. 2012;62:37–48. [PMC free article] [PubMed] [Google Scholar]
- Chen M. Liao Z. Lu B. Wang M. Lin L. Zhang S. Li Y. Liu D. Liao Q. Xie Z. Front. Microbiol. 2018;9:2380. doi: 10.3389/fmicb.2018.02380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. P. Yuan Y. Zhang W. Y. Jiang Z. Hu T. J. Feng Y. T. Liu M. X. J. Pharm. Pharmacol. 2019;71:220–229. doi: 10.1111/jphp.13023. [DOI] [PubMed] [Google Scholar]
- Hassan H. A. Ammar N. M. Serag A. Shaker O. G. El Gendy A. N. Abdel-Hamid A.-H. Z. Microchem. J. 2020;155:104742. doi: 10.1016/j.microc.2020.104742. [DOI] [Google Scholar]
- Thévenot E. A. Roux A. Xu Y. Ezan E. Junot C. J. Proteome Res. 2015;14:3322–3335. doi: 10.1021/acs.jproteome.5b00354. [DOI] [PubMed] [Google Scholar]
- Vandervoort J. Ludwig A. Eur. J. Pharm. Biopharm. 2004;57:251–261. doi: 10.1016/S0939-6411(03)00187-5. [DOI] [PubMed] [Google Scholar]
- Tseng C.-L. Wang T.-W. Dong G.-C. Yueh-Hsiu Wu S. Young T.-H. Shieh M.-J. Lou P.-J. Lin F.-H. Biomaterials. 2007;28:3996–4005. doi: 10.1016/j.biomaterials.2007.05.006. [DOI] [PubMed] [Google Scholar]
- Salama A. H. Elmotasem H. Salama A. A. A. Int. J. Pharm. 2020;584:119411. doi: 10.1016/j.ijpharm.2020.119411. [DOI] [PubMed] [Google Scholar]
- Peungvicha P. Temsiririrkkul R. Prasain J. K. Tezuka Y. Kadota S. Thirawarapan S. S. Watanabe H. J. Ethnopharmacol. 1998;62:79–84. doi: 10.1016/S0378-8741(98)00061-0. [DOI] [PubMed] [Google Scholar]
- Harini R. Pugalendi K. V. J. Basic Clin. Physiol. Pharmacol. 2010;21:79–91. doi: 10.1515/jbcpp.2010.21.1.79. [DOI] [PubMed] [Google Scholar]
- Bone A. J. Hii C. S. Brown D. Smith W. Howell S. L. Biosci. Rep. 1985;5:215–221. doi: 10.1007/BF01119590. [DOI] [PubMed] [Google Scholar]
- Muthukumaran J. Srinivasan S. Venkatesan R. S. Ramachandran V. Muruganathan U. J. Acute Dis. 2013;2:304–309. doi: 10.1016/S2221-6189(13)60149-3. [DOI] [Google Scholar]
- Mrabti H. N. Jaradat N. Fichtali I. Ouedrhiri W. Jodeh S. Ayesh S. Cherrah Y. Faouzi M. E. A. Plants. 2018;7:31. doi: 10.3390/plants7020031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E. J. Khan S. A. Lim K. H. J. Biomater. Sci., Polym. Ed. 2011;22:753–771. doi: 10.1163/092050610X492093. [DOI] [PubMed] [Google Scholar]
- Khan Saeed A. Schneider M. Macromol. Biosci. 2013;13:455–463. doi: 10.1002/mabi.201200382. [DOI] [PubMed] [Google Scholar]
- Kuan Y. H. Nafchi A. M. Huda N. Ariffin F. Karim A. A. J. Sci. Food Agric. 2017;97:1663–1671. doi: 10.1002/jsfa.7970. [DOI] [PubMed] [Google Scholar]
- Faridi Esfanjani A. Assadpour E. Jafari S. M. Trends Food Sci. Technol. 2018;76:56–66. doi: 10.1016/j.tifs.2018.04.002. [DOI] [Google Scholar]
- Jafari S. M., McClements D. J. and Toldrá F., in Advances in Food and Nutrition Research, Academic Press, 2017, vol. 81, pp. 1–30 [DOI] [PubMed] [Google Scholar]
- Iftikhar A. Aslam B. Iftikhar M. Majeed W. Batool M. Zahoor B. Amna N. Gohar H. Latif I. Oxid. Med. Cell. Longevity. 2020;2020:9020219. doi: 10.1155/2020/9020219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell G. I. Polonsky K. S. Nature. 2001;414:788–791. doi: 10.1038/414788a. [DOI] [PubMed] [Google Scholar]
- Sukanya V. Pandiyan V. Vijayarani K. Padmanath K. Indian J. Clin. Biochem. 2020;35:488–496. doi: 10.1007/s12291-019-00852-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiehn O. Garvey W. T. Newman J. W. Lok K. H. Hoppel C. L. Adams S. H. PLoS One. 2010;5:e15234. doi: 10.1371/journal.pone.0015234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S. J. Kwak S.-Y. Jo G. Song T.-J. Shin M.-J. Sci. Rep. 2018;8:8207. doi: 10.1038/s41598-018-26320-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newgard C. B. An J. Bain J. R. Muehlbauer M. J. Stevens R. D. Lien L. F. Haqq A. M. Shah S. H. Arlotto M. Slentz C. A. Rochon J. Gallup D. Ilkayeva O. Wenner B. R. Yancy W. S. Eisenson H. Musante G. Surwit R. S. Millington D. S. Butler M. D. Svetkey L. P. Cell Metab. 2009;9:311–326. doi: 10.1016/j.cmet.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasain J. K. Peng N. Rajbhandari R. Wyss J. M. Phytomedicine. 2012;20:17–23. doi: 10.1016/j.phymed.2012.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu G. Zheng Y. Wang H. Sun J. Ma H. Xiao Y. Li Y. Yuan Y. Yang H. Li X. Min X. Zhang C. Xu C. Jiang Y. Zhang X. He M. Yang M. Hu Z. Tang H. Shen H. Hu F. B. Pan A. Wu T. Int. J. Epidemiol. 2016;45:1507–1516. doi: 10.1093/ije/dyw221. [DOI] [PubMed] [Google Scholar]
- Tam Z. Y. Ng S. P. Tan L. Q. Lin C.-H. Rothenbacher D. Klenk J. Boehm B. O. Sci. Rep. 2017;7:4392. doi: 10.1038/s41598-017-01735-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs M. Krssak M. Bernroider E. Anderwald C. Brehm A. Meyerspeer M. Nowotny P. Roth E. Waldhausl W. Roden M. Diabetes. 2002;51:599–605. doi: 10.2337/diabetes.51.3.599. [DOI] [PubMed] [Google Scholar]
- Stančáková A. Civelek M. Saleem N. K. Soininen P. Kangas A. J. Cederberg H. Paananen J. Pihlajamäki J. Bonnycastle L. L. Morken M. A. Boehnke M. Pajukanta P. Lusis A. J. Collins F. S. Kuusisto J. Ala-Korpela M. Laakso M. Diabetes. 2012;61(7):1895–1902. doi: 10.2337/db11-1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillin T. Hughes A. D. Wang Q. Würtz P. Ala-Korpela M. Sattar N. Forouhi N. G. Godsland I. F. Eastwood S. V. McKeigue P. M. Chaturvedi N. Diabetologia. 2015;58:968–979. doi: 10.1007/s00125-015-3517-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta H. Youn G. S. Shin M. J. Suk K. T. Microorganisms. 2019;7(5):121. doi: 10.3390/microorganisms7050121. doi: 10.3390/microorganisms7050121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y. Wang Y. Ong C. N. Subramaniam T. Choi H. W. Yuan J. M. Koh W. P. Pan A. Diabetologia. 2016;59(11):2349–2359. doi: 10.1007/s00125-016-4069-2. doi: 10.1007/s00125-016-4069-2. [DOI] [PubMed] [Google Scholar]
- Omori K. Katakami N. Yamamoto Y. Ninomiya H. Takahara M. Matsuoka T. A. Bamba T. Fukusaki E. Shimomura I. J. Atheroscler. Thromb. 2019;26(3):233–245. doi: 10.5551/jat.42945. doi: 10.5551/jat.42945. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.