Abstract
The Musashi RNA-binding proteins (RBPs) regulate translation of target mRNAs and maintenance of cell stemness and tumorigenesis. Musashi-1 (MSI1), long considered as an intestinal and neural stem cell marker, has been more recently found to be over expressed in many cancers. It has served as an important drug target for treating acute myeloid leukemia and solid tumors such as ovarian, colorectal and bladder cancer. One of the reported binding targets of MSI1 is Numb, a negative regulator of the Notch signaling. However, the dynamic mechanism of Numb RNA binding to MSI1 remains unknown, largely hindering effective drug design targeting this critical interaction. Here, we have performed extensive all-atom microsecond-timescale simulations using a robust Gaussian accelerated molecular dynamics (GaMD) method, which successfully captured multiple times of spontaneous and highly accurate binding of the Numb RNA from bulk solvent to the MSI1 protein target site. GaMD simulations revealed that Numb RNA binding to MSI1 involved largely induced fit in both the RNA and protein. The simulations also identified important low-energy intermediate conformational states during RNA binding, in which Numb interacted mainly with the β2-β3 loop and C terminus of MSI1. The mechanistic understanding of RNA binding obtained from our GaMD simulations is expected to facilitate rational structure-based drug design targeting MSI1 and other RBPs.
Keywords: Musashi RNA-binding proteins, Protein-RNA binding, Binding pathway and mechanism, Free energy landscape, Gaussian accelerated molecular dynamics
Graphical abstract
Abstract graphic All-atom microsecond-timescale simulations using a robust Gaussian accelerated molecular dynamics (GaMD) method successfully captured spontaneous binding of the Numb RNA to the Musashi-1 (MSI1) protein, which involved largely induced fit in both the RNA and protein. GaMD simulations also identified important low-energy intermediate conformational states of RNA binding. The simulation findings are expected to facilitate rational structure-based drug design targeting MSI1 and other RNA-binding proteins.
1. Introduction
Protein-RNA interactions play crucial roles in various cellular activities and their dysfunction leads to a wide range of human diseases (Wu, 2020; Hentze et al., 2018; Conlon and Manley, 2017). Identification of small molecules that modulate interactions between RNA-binding proteins (RBPs) and RNA is progressing rapidly. It represents a novel strategy for discovery of drugs with new mechanisms (Wu, 2020). The Musashi (MSI) RBPs have been shown to regulate translation of target mRNAs and participate in the maintenance of cell stemness and tumorigenesis. They have been suggested as potential drug targets for treating many types of human cancer, including acute myeloid leukemia, ovarian cancer, colorectal cancer and bladder cancer (Kudinov et al., 2017). The MSI protein family has two members: MSI1 and MSI2. Each MSI protein contains two N-terminal RNA recognition motifs (RRM1 and RRM2) that mediate the binding to their target mRNAs (Sakakibara et al., 2001). MSI1 binds to the 3′-untranslated region of Numb mRNA and represses its translation, which confers to the upregulation of Notch signaling. This leads to increased cell proliferation and survival, and decreased apoptosis of cancer cells (Kudinov et al., 2017). Understanding the molecular mechanism of MSI1-Numb RNA interaction is important in both basic biology and applied medical research.
Rational design of small molecules targeting protein-RNA interactions requires structural characterizations of the RBP-RNA complexes. Due to high flexibility of MSI proteins and lack of potent ligands, only a few MSI structures have been resolved so far, including the apo structure of MSI1/2-RRM1 (Lan et al., 2020; Lan et al., 2017; Nagata et al., 1999; Miyanoiri et al., 2003; Minuesa et al., 2019) and RNA-bound structure of MSI1 (Ohyama et al., 2011; Iwaoka et al., 2017). These structures have greatly facilitated structure-based modeling and drug design targeting the MSI-RNA interactions (Lan et al., 2015; Clingman et al., 2014; Lan et al., 2018; Lan et al., 2020). For example, we have recently identified one potent compound Aza-9 by combining fluorescence polarization (FP) assay, surface plasmon resonance (SPR), nuclear magnetic resonance (NMR) spectroscopy and molecular docking (Lan et al., 2020a, Lan et al., 2020b). However, experimental structures are rather static images and the dynamic mechanism of MSI-RNA interactions remains unknown, which has largely hindered the development of potent inhibitors of MSI proteins.
Molecular dynamics (MD) is a powerful technique that enables all-atom simulations of biomolecules. MD simulations are able to fully account for the flexibility of the RBP and RNA during their interactions (Cheatham III and Case, 2013; Šponer et al., 2017; Borišek et al., 2019). In 2015, Krepl et al.(Krepl et al., 2015) provided systematic benchmarking data by simulating six structurally diverse protein/RNA complexes over multiple microsecond timescale MD runs and evaluating the simulations’ stability. Their results suggested that current force fields are able to handle microsecond MD simulations of protein/RNA complexes in many cases. For most systems, MD was possible to achieve a good but imperfect agreement with the experimental structure. However, MD could not maintain the initial experimental structure in one among six cases (3K5Y). The same group further presented a joint MD and NMR study to interpret and expand the available structural data of two RBPs bound with their single-stranded target RNAs (Krepl et al., 2016). They collected more than 50 μs simulations and showed that the MD simulation was robust enough to reliably describe structural dynamics of RBP-RNA complexes (Krepl et al., 2016). However, due to the slow dynamics and limited simulation timescale, it is rather challenging for conventional MD (cMD) simulations to sufficiently sample RBP-RNA interactions and obtain proper free energy profiles to quantitatively characterize RBP-RNA interactions.
To overcome limitations of cMD, enhanced sampling methods have been developed to improve biomolecular simulations (Mlýnský and Bussi, 2018; Abrams and Bussi, 2014; Spiwok et al., 2015; Gao et al., 2008; Liwo et al., 2008; Christen and van Gunsteren, 2008; Miao and McCammon, 2016a, Miao and McCammon, 2016b). Enhanced sampling methods have also been applied in studies of RBP-RNA interactions, including the steered MD (Wu et al., 2018), umbrella sampling (Wu et al., 2018) and Metadynamics (Laio and Parrinello, 2002; Alessandro and Francesco, 2008; Borkar et al., 2016). Nevertheless, these enhanced sampling methods require predefined collective variables and may introduce constraints on the conformational space of the proteins. In this context, Gaussian accelerated MD (GaMD) has been developed to allow for unconstrained enhanced sampling and free energy calculations of large biomolecules (Miao and McCammon, 2016a, Miao and McCammon, 2016b; Pang et al., 2017; Miao and McCammon, 2017; Wang et al., 2021). GaMD has been applied to successfully simulate protein folding and conformational changes (Pang et al., 2017; Miao and McCammon, 2017; Miao et al., 2015; Peng et al., 2019; Brown et al., 2019; Salawu, 2018), ligand binding (Pang et al., 2017; Miao and McCammon, 2016, 2017; Miao et al., 2015; Wang and Chan, 2017; Liao and Wang, 2019; Miao et al., 2018a, Miao et al., 2018b; Pawnikar and Miao, 2020; Chuang et al., 2018), protein-protein/membrane/nucleic acid interactions (Sibener et al., 2018; Park et al., 2018; Miao and McCammon, 2018; Ricci et al., 2019; Palermo et al., 2017; Bhattarai et al., 2020). More recently, GaMD simulations have successfully captured spontaneous binding of RNA to the human respiratory syncytial virus M2-1 protein (Gao et al., 2020).
In this study, we have performed all-atom enhanced sampling simulations using GaMD on MSI1-Numb RNA interactions started from the NMR structure in the bound state (Ohyama et al., 2011), as well as the unbound state with the Numb RNA moved far away from the MSI1 protein surface (Table 1). While the NMR structure was found to maintain the bound conformation of MSI1-Numb during the GaMD simulations, further simulations captured complete binding of the Numb RNA to the MSI1 protein. The simulations thus allowed us to characterize structural flexibility and free energy landscapes of the MSI1-Numb RNA interactions, which provided important insights into the mechanism of RNA recognition by the MSI1 RBP.
Table 1.
Summary of GaMD simulations performed on the Numb RNA (apo), MSI1 protein without RNA Numb (apo) and with RNA Numb started from the Bound and Unbound states.
| System | Natoms | Length(ns) | Boost potential ( kcal/mol) |
|---|---|---|---|
| Numb RNA (apo) | 14,616 | 500 × 3 | 9.84 ± 3.02 |
| MSI1 protein (apo) | 28,742 | 500 × 3 | 12.28 ± 3.54 |
| MSI1-Numb (Bound) | 29,368 | 500 × 3 | 14.08 ± 3.79 |
| MSI1-Numb (Unbound) | 29,569 | 1,200 × 19 | 11.49 ± 3.36 |
2. Methods
2.1. Gaussian accelerated molecular dynamics (GaMD)
GaMD enhances the conformational sampling of biomolecules by adding a harmonic boost potential to reduce the system energy barriers (Miao et al., 2015). When the system potential is lower than a reference energy E, the modified potential of the system is calculated as:
| (1) |
Where k is the harmonic force constant. The two adjustable parameters E and k are automatically determined on three enhanced sampling principles. First, for any two arbitrary potential values and found on the original energy surface, if , should be a monotonic function that does not change the relative order of the biased potential values; i.e., . Second, if , the potential difference observed on the smoothened energy surface should be smaller than that of the original; i.e., . By combining the first two criteria and plugging in the formula of and , we obtain
| (2) |
Where and are the system minimum and maximum potential energies. To ensure that Eq. (2) is valid, k has to satisfy: . Let us define: , then . Third, the standard deviation (SD) of needs to be small enough (i.e. narrow distribution) to ensure accurate reweighting using cumulant expansion to the second order: , where and are the average and SD of with as a user-specified upper limit (e.g., ) for accurate reweighting. When E is set to the lower bound according to Eq. (2), can be calculated as
| (3) |
Alternatively, when the threshold energy E is set to its upper bound , is set to:
| (4) |
If is calculated between 0 and 1. Otherwise, is calculated using Eq. (3).
2.2. Energetic reweighting of GaMD simulations
For energetic reweighting of GaMD simulations to calculate potential of mean force (PMF), the probability distribution along a reaction coordinate is written as . Given the boost potential of each frame, can be reweighted to recover the canonical ensemble distribution , as:
| (5) |
where M is the number of bins, and is the ensemble-averaged Boltzmann factor of for simulation frames found in the jth bin. The ensemble-averaged reweighting factor can be approximated using cumulant expansion:
| (6) |
where the first two cumulants are given by:
| (7) |
The boost potential obtained from GaMD simulations usually follows near-Gaussian distribution (Miao and McCammon, 2017). Cumulant expansion to the second order thus provides a good approximation for computing the reweighting factor (Miao et al., 2015; Miao et al., 2014). The reweighted free energy is calculated as:
| (8) |
where is the modified free energy obtained from GaMD simulation and is a constant.
2.3. System setup
Two models were prepared for simulations of MSI1-RNA interactions. One was obtained from the first model of NMR structure of the Numb RNA-bound MSI1 protein (PDB: 2RS2, denoted as the “Bound” state) (Ohyama et al., 2011). Another one was generated with the first model by moving the Numb RNA ∼30 Å away from its binding site in MSI1 (denoted as the “Unbound” state). The models for only the MSI1 protein and only the Numb RNA (denoted as the “apo” state) were obtained from the Numb-MSI1 complex by deleting the corresponding binding partner. The four systems were solvated in explicit water using tleap in the AMBER 20 package (D.A. Case, D.S. Cerutti, T.E. Cheatham, III, T.A. Darden, R.E. Duke et al., 2020. AMBER, 2020, University of California, San Francisco). The system charges were then neutralized at 0.15M NaCl using tleap. The AMBER ff14SBonlysc force field parameters (Maier et al., 2015), RNA.LJbb (Zgarbová et al., 2011) and TIP3P model (Jorgensen et al., 1983) were applied for the protein, RNA and water molecules, respectively.
2.4. Simulation protocol
GaMD simulations were performed by using the GPU-accelerating program pmemd.cuda in AMBER 20 (D.A. Case, D.S. Cerutti, T.E. Cheatham, III, T.A. Darden, R.E. Duke, et al. 2020. AMBER, 2020, University of California, San Francisco). The hydrogen-heavy atom bonds were constrained using the SHAKE algorithm and the simulation time step was set to 2.0 fs. The particle mesh Ewald (PME) method was employed to compute the long-range electrostatic interactions and a cutoff value of 9.0 Å was applied to treat the non-bonded atomic interactions. The temperature was controlled using the Langevin thermostat with a collision frequency of 1.0 ps−1.
Each system was minimized using steepest descent for 50,000 steps and conjugate gradient for another 50,000 steps. After minimization, the system was heated from 0 to 300 K in 1 ns simulation by applying 1 kcal/(mol•Å2) harmonic position restraints to the protein and RNA heavy atoms with a constant number, volume and temperature (NVT) ensemble. Each system was further equilibrated using a constant number, pressure and temperature (NPT) ensemble at 1 atm and 300 K for 1ns with same restraints as in the NVT run. Another 1.2 ns cMD simulations were performed to collect potential energy statistics (including the maximum, minimum, average and standard deviation). Then 24 ns GaMD equilibration after applying the boost potential was performed. Previous studies showed that hundreds-of-nanosecond GaMD simulations with multiple replicas could provide good sampling of native biomolecular complexes(Miao and McCammon, 2016a, Miao and McCammon, 2016b; Bhattarai et al., 2020; Draper-Joyce et al., 2021; Wang and Miao, 2019) and microsecond GaMD simulations were able to capture ligand binding from solvent to target proteins(Miao and McCammon, 2016a, Miao and McCammon, 2016b; Pawnikar and Miao, 2020; Miao et al., 2018). Therefore, 3 independent 500 ns GaMD simulations with randomized initial atomic velocities were performed on the MSI1-Numb bound system (Table 1). While 19 independent 1,200 ns GaMD simulations were performed on the unbound system with the aim to capture at least 5 events of successful RNA binding to the MSI1 protein (Table 1). In addition, three independent 500 ns GaMD production runs with randomized initial atomic velocities were performed on the apo states of the MSI1 protein and RNA, respectively. Simulation frames were saved every 0.4 ps for analysis. The boost potential in GaMD simulation was calculated using potential energies of the present frame, as well as potential statistics including the minimum, maximum, average and standard deviation that are collected through short cMD and GaMD equilibration runs in the same simulation. The AMBER input files of GaMD equilibration and production simulations are provided in Supporting Information.
2.5. Simulation analysis
CPPTRAJ (Roe and Cheatham, 2013) and VMD (Humphrey et al., 1996) were used to analyze the GaMD simulations. Important reaction coordinates were identified from the simulation trajectories such that they involved dynamic regions (e.g., the β2-β3 loop of MSI1) and could be used to differentiate conformational states of the MSI1-Numb system. Therefore, root-mean-square deviations (RMSDs) of the backbone of core RNA (central three nucleotides UAG in Numb) and the β2-β3 loop of MSI1 relative to the first NMR conformation in the PDB with alignment of the MSI1 protein core (excluding the highly flexible C-terminus residues 96 to 103), the number of native contacts between MSI1 and Numb RNA (Ncontacts), the radius of gyration (Rg) and end-to-end distance of the Numb RNA were selected as reaction coordinates. Root-mean-square fluctuations (RMSFs) were calculated for the protein residues and RNA nucleotides, averaged over the GaMD production simulations and color coded for schematic representation of each system. Since only the Sim1 to Sim6 GaMD trajectories successfully captured complete binding of the Numb RNA to MSI1, these trajectories were used separately for structural clustering to identify the RNA binding pathways using the hierarchical agglomerative algorithm in CPPTRAJ (Roe and Cheatham, 2013). The RMSD cutoff was set to 3.0 Å for the core RNA backbone to form a cluster.
The PyReweighting (Miao et al., 2014) toolkit was applied to reweight GaMD simulations to recover the original free energy or PMF profiles of the two MSI1-Numb systems. Three 500 ns GaMD production simulations were combined for calculating the PMF profiles of the Bound MSI1-Numb system. Six of the 1200 ns GaMD production simulations that successfully captured spontaneously binding of Numb RNA to MSI1 started from the Unbound state were combined to calculate the corresponding PMF profiles. A bin size of 1.0 Å was used for the core RNA backbone RMSD, the MSI1 β2-β3 loop backbone RMSD, the Numb Rg and the end-to-end distance of Numb. A bin size of 100 was used for Ncontacts. The cutoff was set to 500 frames for all 2D PMF calculations.
3. Results
3.1. GaMD simulations captured complete binding of the Numb RNA to the MSI1 protein
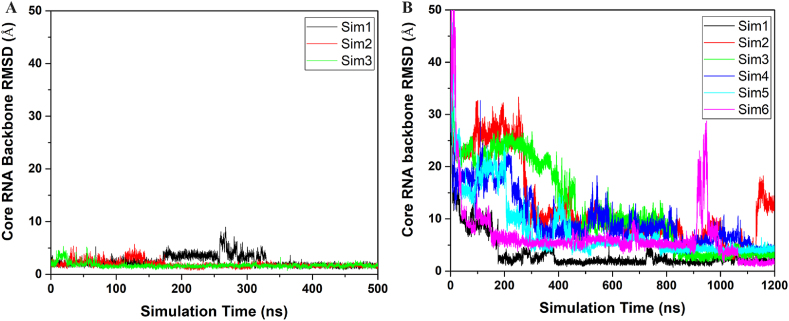
Extensive GaMD production simulations were performed on the MSI1-Numb system, including three independent 500 ns runs started from the Bound state and 19 independent 1,200 ns runs started from the Unbound state (Table 1). The GaMD simulations started from the Bound state recorded average and SD of the boost potential as 14.08 kcal/mol and 3.79 kcal/mol, respectively (Table 1). The GaMD simulations started from the Unbound state showed similar average and SD of boost potential with 11.49 kcal/mol and 3.36 kcal/mol, respectively (Table 1). The GaMD simulations of “apo” state of MSI protein showed similar average and SD of boost potential with 12.28 kcal/mol and 3.54 kcal/mol, respectively (Table 1). In contrast, the GaMD simulations of “apo” state of Numb RNA with fewer atoms recorded lower average and SD of boost potential with 9.84 kcal/mol and 3.02 kcal/mol, respectively (Table 1). The Bound MSI1-Numb complex was found to maintain the NMR structure with <3 Å RMSD of the core RNA backbone during most of the GaMD simulations (Fig. 1A). In the GaMD simulations started from the Unbound state, the core RNA backbone RMSD relative to the NMR structure in 6 of 19 simulations (∼31.5%) decreased to < 3.0 Å, suggesting that complete binding of the Numb RNA from free diffusion in the solvent to the MSI1 target site was successfully captured (Fig. 1B &S1). Spontaneous binding of RNA was observed in the Sim1 after ∼100 ns with the RNA backbone RMSD decreased to ∼3.0 Å relative to the first NMR structure (Fig. 1B). In Sim2, the Numb RNA bound to MSI1 during ∼1,010–1,130 ns and then dissociated to the solvent (Fig. 1B). The Numb RNA bound to MSI1 after ∼800 ns in Sim3, Sim4 and Sim5 (Fig. 1B). In Sim6, spontaneous binding of RNA was observed after ∼1000 ns (Fig. 1B). Multiple RNA binding events captured in the present GaMD simulations allowed us to characterize the dynamic interactions between the MSI1 protein and Numb RNA.
Fig. 1.
Time courses of the backbone RMSDs of core RNA (central three nucleotides GUA of the Numb RNA) relative to the first NMR conformation (PDB: 2RS2) are calculated from the (A) three independent 500 ns GaMD simulations of the MSI1-Numb system started from the Bound state and (B) six of the 19 independent 1200 ns GaMD simulations that successfully captured binding of Numb to MSI1 started from the Unbound state. The remaining 13 GaMD trajectories (Sim7-Sim19) that started from the Unbound state and did not capture RNA binding are plotted in in Fig. S1.
3.2. Variations of structural flexibility upon MSI1-Numb RNA binding
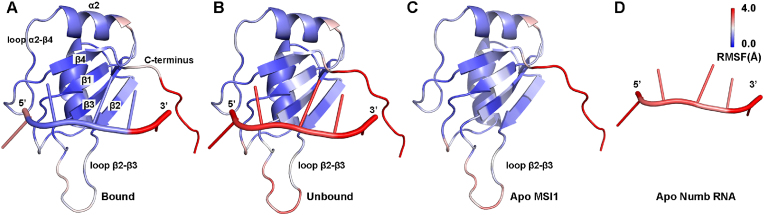
We analyzed structural flexibility of both the MSI1 protein and Numb RNA in the GaMD simulations. During GaMD simulations started from the Bound NMR structure, the MSI1 protein underwent small fluctuations except the loop connecting β2 and β3 strands (the β2-β3 loop) and the C terminus (Fig. S2A). The fifth nucleotide in the Numb RNA exhibited significantly higher flexibility than the other nucleotides, especially the central three ones UAG (denoted as the core RNA). Similar findings were observed in the simulations of the apo Numb RNA (Fig. 2D). This suggested that interactions between the core RNA and the MSI1 were strong. Thus, the core RNA might play an important role in the interactions between the MSI1 protein and Numb RNA. Furthermore, the MSI1 β2-β3 loop and C-terminus and the Numb RNA exhibited significantly higher fluctuations in the GaMD simulations started from the Unbound state than those in simulations started from the Bound state (Fig. S2B). Note that both Bound and Unbound conformations of MSI1-Numb were observed in the GaMD simulations (Sim1-Sim6) started from the Unbound state (Fig. 1B). In this regard, trajectories of Sim7 to Sim19 started from the Unbound state that did not capture the Numb RNA binding were used for RMSF calculation of the system in the Unbound state (Fig. 1B). The three GaMD trajectories started from the Bound state (Fig. 1A) plus the 100–1200 ns trajectory of Sim1, 850–1200 ns trajectory of Sim3, 800–1200 trajectory of Sim5 and 1000–1200 ns trajectory of Sim6 started from the Unbound state (Fig. 1B) were used for RMSF calculation of the system in the Bound state (Fig. 2A). Results showed that the flexibility of loop β2-β3 and C-terminus of MSI1 in the Bound state was significantly lower than in the Unbound and apo state (Fig. 2), being similar to the findings observed in Fig. S2. These motifs were suggested to be important for binding of the Numb RNA (Ohyama et al., 2011) and small molecules (Clingman et al., 2014) to the MSI1 protein.
Fig. 2.
Structural flexibility of MSI1-RNA obtained from GaMD simulations: root-mean-square fluctuations (RMSFs) of the MSI1-RNA complex when the Numb RNA was in the (A) Bound or (B) Unbound states, and RMSF of the (C) MSI1 protein and (D) Numb RNA in the apo state. A color scale of 0.0 Å (blue) to 4.0 Å (red) is used. Note that the three GaMD trajectories started from the Bound state (Fig. 1A) plus the 100–1200 ns trajectory of Sim1, 850–1200 ns trajectory of Sim3, 800–1200 ns trajectory of Sim5 and 1100–1200 ns trajectory of Sim6 started from the Unbound state (Fig. 1B) were used for RMSF calculation of the system in the Bound state. The 13 GaMD trajectories that started from the Unbound state and did not capture RNA binding (Sim7-Sim19 in Fig. S1) were used for RMSF calculation of the system in the Unbound state (Fig. 1B).
3.3. Free energy profiles of RNA binding to the MSI1 protein
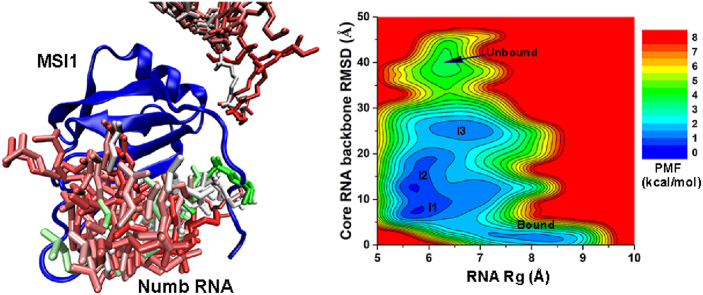
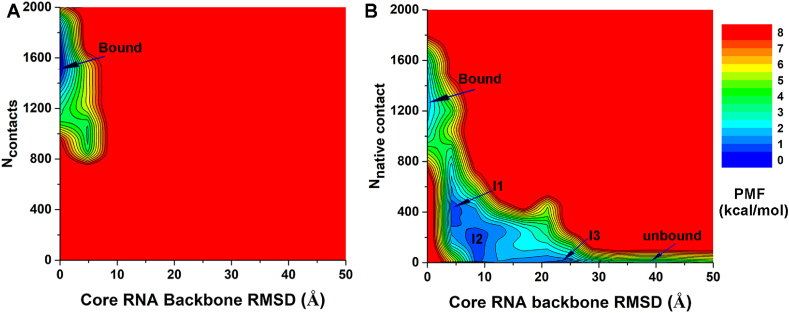
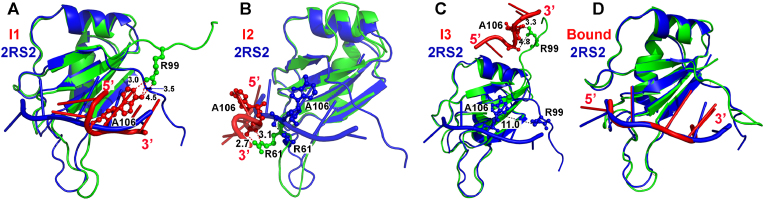
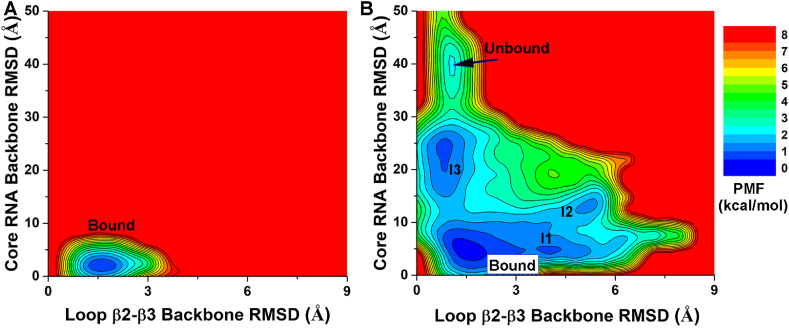
Free energy profiles were calculated from the GaMD simulations using the core RNA backbone RMSD relative to the NMR structure and the number of native contacts between MSI1 and Numb RNA (Ncontacts) as reaction coordinates. Only one low-energy minimum of the “Bound” conformation was identified from the GaMD simulations on the NMR structure, in which the core RNA backbone RMSD and Ncontacts centered around (1.2 Å, 1600) (Fig. 3A). Five low-energy minima were identified from GaMD simulations started from the Unbound state, including the “Bound”, “Intermediate I1”, “Intermediate I2”, “Intermediate I3” and “Unbound” states, in which the core RNA backbone RMSD and Ncontacts centered around (2.0 Å, 1500), (5.2 Å, 480), (9.5 Å, 200), (25.0 Å, 10) and (40 Å, 0), respectively (Fig. 3B). The intermediate I1, I2 and I3 conformational states are shown in Fig. 4. The Numb RNA binding to MSI1 involved large conformational changes in both the RNA and protein (Figs. 4 and 5B, Movies S1 and S2). In the Unbound state, RNA diffused far away from the protein with ∼40 Å RMSD in the core RNA backbone relative to the NMR complex structure, while the protein loop β2-β3 could adopt a conformation close to the NMR structure with ∼1.0 Å RMSD (Fig. 5B). As the RNA moved towards the MSI protein in the I2 and I3 intermediate states, the Numb RNA exhibited RMSDs of 10.8 Å and 22.3 Å from the target binding conformation and interacted with the protein loop β2-β3 and C-terminus, respectively (Fig. 4). Such interactions induced significant conformational changes of the corresponding regions, which appeared to pull the Numb RNA to the protein target site ( Movies S1 and S2). Then, the Numb RNA moved closer to the target site in the I1 intermediate state with reduced RMSD of ∼5.2 Å. Meanwhile, the protein loop β2-β3 and C-terminus in the I1 intermediate state showed RMSDs of ∼4.0 Å and ∼21.2 Å, respectively, relative to the NMR structure (Fig. 4, Fig. 5B). Finally, the protein (especially the β2-β3 loop and C-terminus) and RNA rearranged their conformations and formed complex in the “Bound” state that was similar to the NMR experimental structure (Fig. 4D). Remarkably, positively charged residues (Arg61 and Arg99) in the β2-β3 loop and C-terminal region of the MSI1 protein formed favorable salt-bridge and hydrogen bond interactions with the central nucleotide A106 of Numb RNA. In the intermediate I1 state, the Numb RNA formed interactions with both the β2-β3 loop and C terminus of MSI1, leading to large conformational changes of these two regions (Fig. 4A). Notably, the sidechain of residue Arg99 in the C terminus of the MSI1 protein formed three hydrogen bonds with the sidechain of nucleotide A106 in the Numb RNA (Fig. 4A). In the intermediate I2 state, the Numb RNA formed interactions with the β2-β3 loop of MSI1, leading to a large conformational change of this loop (Fig. 4B). The sidechain of residue Arg61 in MSI1 could flip out to the solvent, forming salt-bridge with the backbone (oxygen atom in the phosphate group) of the nucleotide A106 in Numb RNA (Fig. 4B). In the intermediate I3 state, Arg99 in the C terminus of MSI1 formed a hydrogen bond and a salt-bridge with sidechain and backbone of the nucleotide A106 in Numb RNA, respectively, for which a large conformational change of the protein C terminus was observed (Fig. 4C). No contact between MSI1 Arg99 and Numb A106 was found in the NMR experimental structure (Fig. 4C). The distance between MSI1 Arg99 and Numb A106 and the Numb Core RMSD were chosen as reaction coordinates to calculate 2D free energy profiles (Fig. S3). Indeed, a salt-bridge interaction between MSI1 Arg99 and Numb A106 was identified in low-energy intermediate states (I1 and I3) during RNA binding to MSI1 (Fig. S3B). Thus, the electrostatic interaction between MSI1 Arg99 and Numb A106 played a significant role in the recognition and binding of the Numb RNA to MSI1 protein.
Fig. 3.
2D potential of mean force (PMF) free energy profiles of the core RNA backbone RMSD relative to the first NMR conformation (PDB: 2RS2) and number of native contacts between MSI1 and Numb RNA are calculated from GaMD simulations started from the (A) Bound and (B) Unbound states of the MSI1-Numb system.
Fig. 4.
Low-energy intermediate conformational states (I1, I2 and I3) and “Bound” state as identified from the 2D PMF profile of the MSI1-RNA simulation system started from the Unbound state. The MSI1 protein and Numb RNA are shown in green and red, respectively. The NMR structure of the MSI1-Numb complex is shown in blue for comparison. MSI1 protein residues Arg99 and Arg61, and nucleotide A106 of Numb RNA are highlighted in balls and sticks. The hydrogen bonds between the side-chain of residues in the MSI1 protein and Numb RNA are shown in red. The salt-bridge interactions between the side-chain of residues in the MSI1 protein and backbone (oxygen atom) of the Numb RNA are shown in black.
Fig. 5.
2D PMF profiles of the MSI1 β2-β3 loop backbone RMSD and core RNA backbone RMSD relative to the first NMR conformation (PDB: 2RS2) are calculated from GaMD simulations started from the (A) Bound and (B) Unbound states of the MSI1-Numb system.
As described above, binding of the Numb RNA induced higher flexibility of the MSI1 β2-β3 loop (Fig. 2B) and large conformational change of the same region was observed in the intermediate I1 and I2 states (Fig. 4A and B). Therefore, the MSI1 β2-β3 loop backbone RMSD and core Numb RNA backbone RMSD relative to the experimental structure were used as reaction coordinates to further compute 2D free energy profiles (Fig. 5). The MSI1 β2-β3 loop was highly flexible, sampling a large conformational space with the backbone RMSD ranging from ∼0 Å to ∼8.0 Å (Fig. 5B&S4). This loop sampled two distinct low-energy conformations, including the “Closed” (bound) (RMSD <1 Å) and “Open” (free) states (RMSD ∼3–5 Å) (Fig. 5B). Compared to the “Open” state, the MSI1 β2-β3 loop moved closer to the core domain in the “Closed” state (Fig. 4A). Five low-energy states were identified from GaMD simulations starting with the Unbound state, including the “Unbound/Open”, “Intermediate I3/Open”, “Intermediate I2/Closed”, “Intermediate I1/Closed” and “Bound/Open” states, in which the MSI1 β2-β3 loop backbone RMSD and core RNA backbone RMSD were located around (1.0 Å, 40 Å), (1.2 Å, 25.0 Å), (5.0 Å, 11.5 Å), (4.2 Å, 5.5 Å) and (1.5 Å, 2.0 Å), respectively (Fig. 5B). The Numb RNA and MSI1 β2-β3 loop accommodated to each other to form the final bound conformation (Fig. 5B).
3.4. Pathways of RNA binding to the MSI1 protein
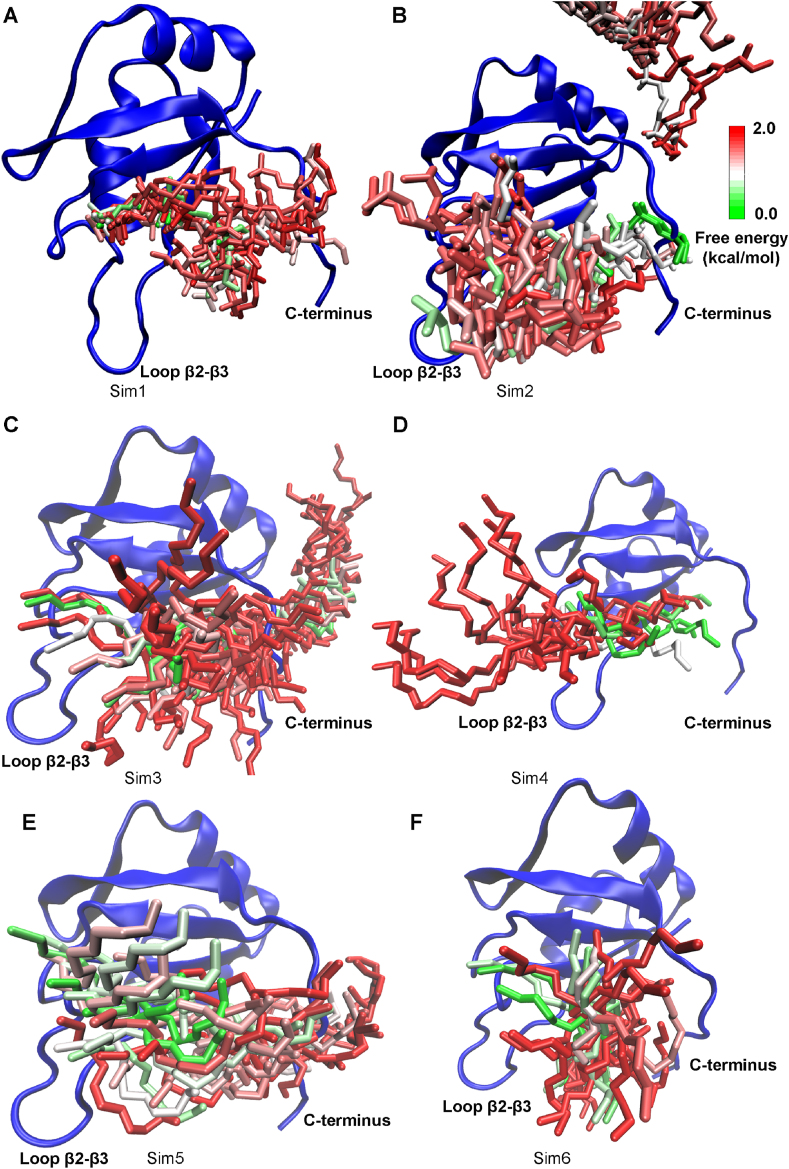
Here, we focused on exploring the pathway and mechanism of RNA binding, for which only six out of 19 GaMD simulations of the unbound system successfully captured the RNA binding process. Therefore, all these six trajectories were analyzed in detail to determine the RNA binding pathways. Thus, structural clustering was performed separately on the GaMD trajectories of Sim1-Sim6 to identify the representative binding pathways of the Numb RNA to the MSI1 protein. Movies S1 and S2 show GaMD trajectories of Sim1 and Sim2 started from the Unbound state in Supporting Information, respectively. The structural clusters were reweighted to obtain their original free energy values, which ranged from 0.0 kcal/mol to ∼4.5 kcal/mol. The top reweighted clusters with PMF ≤2.0 kcal/mol were selected to represent the pathways of the Numb RNA binding to MSI1 (Fig. 6). In Sim1, Sim3, Sim5 and Sim6, the Numb RNA bound to MSI1 via interactions with the protein C terminus (Fig. 6A, C, 6E and 6F). In Sim4, the Numb RNA bound to MSI1 via interactions with the β2-β3 loop of MSI1 (Fig. 6D). In Sim2, both the β2-β3 loop and C terminus of MSI1 contributed important interactions with the Numb RNA during its binding to the protein target site (Fig. 6B). These findings revealed the important roles of the β2-β3 loop and C terminus of MSI1 in binding of the Numb RNA, especially for their large conformational changes in the three intermediate states of RNA binding (Fig. 4). It is worth to noting that no strong interactions were formed between the Numb RNA and the MSI1 β2-β3 loop and C terminus in the final “Bound” state. Rather these two regions formed important interactions with RNA in the intermediate states during the RNA binding process. This was consistent with the above RMSF analysis that higher flexibilities were observed in these two dynamic regions of MSI1 (Fig. 2).
Fig. 6.
Binding pathways of the Numb RNA to the MSI1 protein revealed from GaMD simulations: Starting from free diffusion in the solvent, the Numb RNA spontaneously bound to the target site of the MSI1 via intermediate conformations in the (A) “Sim1”, (B) “Sim2”, (C) “Sim3”, (D) “Sim4”, (E) “Sim5” and (F) “Sim6” GaMD trajectories. In the intermediate conformations, the Numb RNA interacted with the C-terminus (A, C, E and F), the β2-β3 loop (D) or both the C-terminus and β2-β3 loop (B) of MSI1. The MSI1 protein is shown in blue ribbons. The Numb RNA structural clusters (sticks) are colored by the reweighted PMF free energy values in a green (0.0 kcal/mol)-white-red (2.0 kcal/mol) color scale.
3.5. The Numb RNA bound to the MSI1 protein via an induced fit mechanism
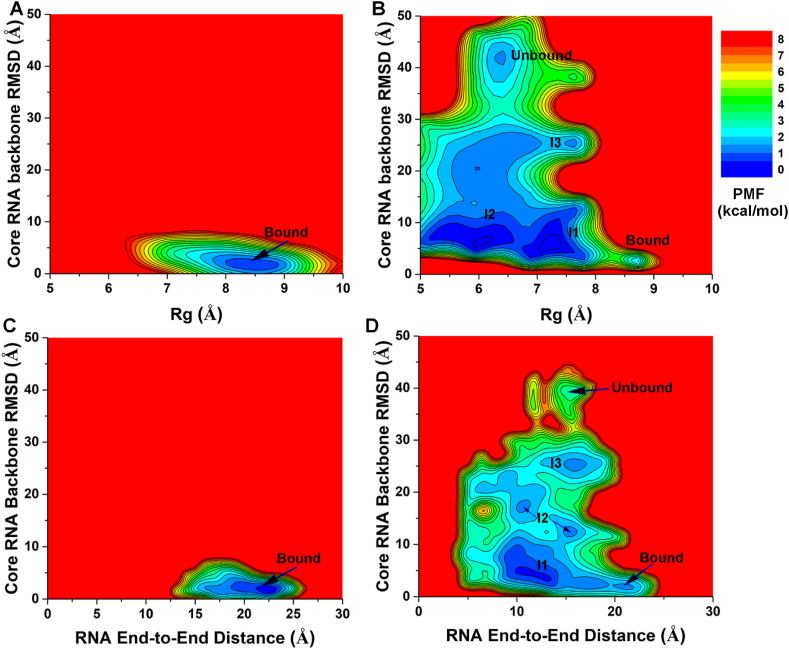
In order to further explore the mechanism of RNA binding to the MSI1 protein, we computed free energy profiles to characterize conformational changes of the Numb RNA upon binding to MSI1. In this regard, the radius of gyration (Rg) and the end-to-end distance of Numb were calculated to monitor its possible conformational changes. We used the Rg and end-to-end distance of the Numb RNA and the core RNA backbone RMSDs as reaction coordinates to calculate 2D PMF profiles (Fig. 7). Notably, the Numb RNA sampled a large conformational space during binding to the MSI1 protein in the GaMD simulations started from the Unbound state (Fig. 7B). From the reweighted 2D PMF profiles, we identified a similar “Bound” low-energy well in simulations started from both the Bound and Unbound states, for which the Numb RNA Rg and core RNA backbone RMSD centered around (8.5 Å, 2.0 Å) and (8.5 Å, 2.5 Å), respectively (Fig. 7A-B). Another four low-energy states were identified in GaMD simulations started from Unbound conformation, including the “Unbound”, “Intermediate I1”, “Intermediate I2” and “Intermediate I3”, for which the core RNA backbone RMSD and Rg of Numb centered around (40.0 Å, 6.2 Å), (5.0 Å, 7.2 Å), (6.9 Å, 6.2 Å) and (25.0 Å, 7.5 Å), respectively (Fig. 7B).
Fig. 7.
(A–B) 2D PMF profiles of the radius of gyration (Rg) of the Numb RNA and core RNA backbone RMSD relative to the first NMR conformation (PDB: 2RS2) are calculated from GaMD simulations started from the (A) Bound and (B) Unbound states of the MSI1-Numb system. (C–D) 2D PMF profiles of the end-to-end distance of the Numb RNA and core RNA backbone RMSD relative to the first NMR conformation (PDB: 2RS2) are calculated from GaMD simulations started from the (C) Bound and (D) Unbound states of the MSI1-Numb system.
Furthermore, we calculated 2D PMF profiles regarding the core RNA backbone RMSD and the end-to-end distance of Numb RNA (Fig. 7C-D). Only one low-energy state (“Bound”) was identified in the 2D PMF profile calculated from the GaMD simulations of the bound NMR structure, in which the Numb RNA adopted primarily the “Extended” conformation (Fig. 7C). The Numb end-to-end distance and core RNA backbone RMSD centered around (22.5 Å, 1.8 Å) (Fig. 7C). In contrast, six distinct low-energy states were identified from the 2D PMF profile in the GaMD simulations started from the Unbound conformation, including the “Unbound”, “Intermediate I1”, “Intermediate I2”, “Intermediate I3” and “Bound”, in which the Numb RNA adopted primarily the “Curled”, “Curled”, “Curled”, “Curled”, and “Extended” conformations, respectively (Fig. 4, Fig. 7). The end-to-end distance of Numb RNA and core RNA backbone RMSD centered around (15.0 Å, 40.0 Å) in the “Unbound/Curled” state, (15.2 Å, 25.0 Å) in the “Intermediate I3/Curled” state, (∼15–10 Å, 10–17 Å) in the “Intermediate I2/Curled” state, (12.5 Å, 5.2 Å) in the “Intermediate I1/Curled” state and finally (22.0 Å, 2.5 Å) in the “Bound/Extended” state (Fig. 7D).
In comparison, the Numb RNA sampled a larger conformational space with a wider range of Rg or end-to-end distance in the “Bound” state than in the “Unbound” and even the three Intermediate conformational states (Fig. 7B and D), suggesting binding of the Numb RNA to the MSI1 protein involved largely induced fit. To further support the “induced-fit” mechanism of RNA binding to MSI1, we have added 500 ns x 3 GaMD simulations on the apo form of both MSI1 protein and Numb RNA (Table 1). In the RNA-bound MSI1, the Numb RNA sampled the “Bound” low-energy state with Rg and end-to-end distance centered around (8.5 Å, 22.0 Å) (Fig. S5D). In contrast, the apo Numb RNA could not sampled the “Bound” state (Fig. S5F). In this context, Numb RNA in GaMD simulations of the RNA unbound system sampled a large conformational space covering that of both the bound and apo systems (Fig. S5E). Similar findings were observed for the MSI1 protein. The MSI1 β2-β3 loop backbone RMSD and MSI1 C-terminus backbone RMSD were used to calculate the free energy landscapes (Figs. S5A–S5C). In the RNA-MSI1 complex, the protein sampled the “Bound” low-energy state with the β2-β3 loop backbone RMSD and C-terminus backbone RMSD centered around (3.0 Å, 20 Å) (Fig. S5A). It is worth noting that the large RMSD of MSI1 C-terminus observed in the “Bound” state (∼20 Å) was reasonable because similar values were obtained among the 20 conformations in the NMR experimental structure (PDB: 2RS2). MSI1 in GaMD simulations of the RNA unbound system sampled a large conformational space covering that of both the bound and apo systems (Fig. S5B). In comparison, the apo MSI1 could rarely sample these conformations (Fig. S5C). Together, these results suggested that the RNA binding to MSI1 protein adopted an “induced-fit” mechanism.
4. Discussion
In this study, we have applied extensive all-atom GaMD simulations with a total length of 24,300 ns to investigate dynamic interactions between the Numb RNA and MSI1 protein. The GaMD simulations unprecedentedly captured multiple times of spontaneous and highly accurate binding of the Numb RNA from bulk solvent to the MSI1 protein with <2 Å RMSD in the core RNA backbone compared with the experimental structure. Proper energetic reweighting of the GaMD simulations allowed us to calculate free energy profiles to characterize the MSI1-Numb binding process.
Relatively low-energy conformational states of RNA binding to the MSI1 protein were identified from the GaMD simulations, including the Unbound, Intermediate I1, Intermediate I2, Intermediate I3, and Bound states (Fig. 4). In the intermediate states, the β2-β3 loop and C terminus of the MSI1 protein were found to be essential for recognition and binding of the Numb RNA (Fig. 4A–C & 6). The charged residues Arg61 and Arg99 of MSI1 formed critical hydrogen bond and salt-bridge interactions with the Numb RNA during the GaMD simulations, particularly in the Intermediate and Bound states. The important role of Arg99 was consistent with the previous finding that mutation of the corresponding residue in MSI2 (Arg100Ala) decreased the binding affinity of Numb RNA (Minuesa et al., 2019). The salt bridge between Arg61 (MSI1 protein) and A106 (Numb RNA) was also observed in the bound NMR structure (Fig. 4B). Furthermore, Arg61 was characterized as a key residue for inhibitor binding as the MSI1 Arg61Glu mutant exhibited ∼5 fold decrease in the inhibitor binding affinity (Clingman et al., 2014). Additionally, strong binding between the RNA core and MSI1 protein was observed in the GaMD simulations started from the Bound state. This agreed well with the previous finding obtained by Zearfoss et al.(Zearfoss et al., 2014) that the central UAG RNA nucleotides form the MSI recognition element and make major contributions to the binding affinity. Therefore, our GaMD simulations revealed that long-range electrostatic interactions played an important role in the Numb RNA binding to the MSI1 protein and identified two critical protein residues (Arg61 and Arg99) for RNA recognition, being highly consistent with previous experimental findings.
Conformation selection (Tsai et al., 1999; Tsai et al., 1999; Kumar et al., 2000) and induce fit (Okazaki and Takada, 2008; Williamson, 2000) are two common models for describing biomolecular recognition. In this context, our GaMD simulations have revealed that binding of the Numb RNA to the MSI1 protein involved predominantly an induced fit mechanism, in which both the RNA and protein underwent significant conformational changes during binding (Figs. S4 and S5). This is consistent with previous studies of other protein-RNA interactions (Williamson, 2000; Leulliot and Varani, 2001; Suryadi et al., 2005,Pitici et al., 2002), including ribosomal protein S15-rRNA and U1A-RNA complexes. A major conformational change of the rRNA was found upon binding to the S15 protein through comparison of the free and bound structures of S15 and rRNA, suggesting induced fit of the protein and RNA (Williamson, 2000). MD simulations combined with available structure analysis also indicated that binding of the U1A protein and RNA followed an induced fit mechanism (Pitici et al., 2002). For the U1A protein, MD simulations indicated that induced fit upon binding involved a non-native thermodynamic substate, in which the structure is preorganized for binding. In contrast, induced fit of the RNA involved a distortion of the native structure to an unstable form in solution.
It is important to note that the presented GaMD simulations captured six binding events of Numb RNA to MSI1 protein. In addition to free energy profiles calculated by combining all six successfully RNA binding simulations (Fig. 3B), we calculated free energy profiles from each of the six successful GaMD simulations of RNA binding to as shown in Fig. S6. While the Unbound, Bound and Intermediate low-energy states could be sampled in the individual simulations, these free energy profiles showed differences in terms of the positions and values of the free energy minima and barrier heights. Results suggested that our current GaMD simulations were still not converged. Nevertheless, Numb RNA could bind to the MSI1 protein in two main pathways via the protein β2-β3 loop and C-terminus interaction sites, which were observed for 4 and 2 times during the six successful GaMD simulations, respectively. For future studies, more events of RNA binding and unbinding would still need to be simulated in order to calculate the RNA binding free energies and kinetic rates quantitatively. Nevertheless, the focus of this study was to uncover the dynamic pathways and mechanism of RNA binding to the MSI1 protein. The binding of RNA to MSI1 protein via interacting with the two main binding sites (β2-β3 loop and C-terminus) were observed multiple times during GaMD simulations. Therefore, the six GaMD trajectories should reveal accurate Numb RNA binding pathways. More binding and unbinding events would still need to be simulated in order to calculate the RNA binding free energies and kinetic rates quantitatively. In this regard, our recently developed selective GaMD algorithms (Miao et al., 2020; Wang and Miao, 2020) could be useful to address the challenge. In particular, the peptide GaMD (Pep-GaMD) method (Wang and Miao, 2020), which works by selectively boosting the essential peptide potential energy, has been demonstrated to capture repetitive binding and unbinding of highly flexible peptides to the target protein within microsecond simulations (Wang and Miao, 2020). Apart from enhanced conformational sampling, accurate force fields are also needed especially for the RNA (Šponer et al., 2018; Tan et al., 2018; Kührová et al., 2019; Cesari et al., 2019) in order to simulate repetitive RNA dissociation and binding to RBPs. Even the force field works well individually for the protein and RNA, combination of protein and RNA force fields in the MD simulations could introduce additional challenges (Šponer et al., 2017; Krepl et al., 2015; Šponer et al., 2018). Nevertheless, we have observed multiple complete binding events of the Numb RNA to the MSI1 protein with our current force field settings and GaMD simulations, which shall guide future studies of RNA-protein interactions.
5. Conclusions
In summary, all-atom GaMD simulations with unconstrained enhanced sampling and free energy calculations have provided important insights into the mechanism of the Numb RNA binding to the MSI1 protein. The results and the methods used in this study would help in simulating binding process of RNA to the RBPs in general, to accurately predict the binding mechanism of protein−RNA interactions. For future studies, the effects of small molecule binding in the MSI1-Numb interactions still need to be determined and our simulation findings await validation in the wet-lab experiments. Further studies are planned to simulate both dissociation and binding of RNA to the RBPs and accurately predict the thermodynamics and kinetics of protein−RNA interactions. These efforts are expected to greatly facilitate rational drug design targeting the MSI1 and other RBPs.
CRediT authorship contribution statement
Jinan Wang: performed the research, analyzed the data, wrote the paper. Lan Lan: analyzed the data. Xiaoqing Wu: analyzed the data. Liang Xu: analyzed the data. Yinglong Miao: designed research, wrote the paper.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported in part by the National Institutes of Health (R01GM132572) and the startup funding in the College of Liberal Arts and Sciences at the University of Kansas (to Y.M.), NIH R01 CA191785 and KU Cancer Center Pilot Project Award (to L.X.). This work used supercomputing resources with allocation Award No. TG-MCB180049 through the Extreme Science and Engineering Discovery Environment (XSEDE), which is supported by the National Science Foundation, Grant No. ACI-1548562, and Project No. M2874 through the National Energy Research Scientific Computing Center (NERSC), which is a U.S. Department of Energy Office of ScienceUser Facility operated under Contract No. DE-AC02-05CH11231, and the Research Computing Cluster at the University of Kansas.
Handling Editor: Nagasuma Chandra
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.crstbi.2021.12.002.
Appendix B. Supplementary data
The following are the Supplementary data to this article:
The Supporting Information is available free of charge on the CRSB Publications website. Movies S1 and S2 show GaMD trajectories of Sim1 and Sim2 started from the Unbound state, respectively. AMBER input files of GaMD equilibration and production simulations and Figs. S1–S6 are also included.
References
- Abrams C., Bussi G. Enhanced sampling in molecular dynamics using metadynamics, replica-exchange, and temperature-acceleration. Entropy. 2014;16:163–199. [Google Scholar]
- Alessandro L., Francesco L.G. Metadynamics: a method to simulate rare events and reconstruct the free energy in biophysics, chemistry and material science. Rep. Prog. Phys. 2008;71:126601. [Google Scholar]
- Bhattarai A., Wang J., Miao Y. G-protein-coupled receptor-membrane interactions depend on the receptor activation state. J. Comput. Chem. 2020;41:460–471. doi: 10.1002/jcc.26082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borišek J., Saltalamacchia A., Gallì A., Palermo G., Molteni E., Malcovati L., Magistrato A. Disclosing the impact of carcinogenic SF3b mutations on pre-mRNA recognition via all-atom simulations. Biomolecules. 2019;9:633. doi: 10.3390/biom9100633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borkar A.N., Bardaro M.F., Camilloni C., Aprile F.A., Varani G., Vendruscolo M. Structure of a low-population binding intermediate in protein-RNA recognition. Proc. Natl. Acad. Sci. U.S.A. 2016;113:7171–7176. doi: 10.1073/pnas.1521349113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown B.P., Zhang Y.-K., Westover D., Yan Y., Qiao H., Huang V., Du Z., Smith J.A., Ross J.S., Miller V.A., et al. On-target resistance to the mutant-selective EGFR inhibitor Osimertinib can develop in an allele-specific manner dependent on the original EGFR-activating mutation. Clin. Cancer Res. 2019;25:3341–3351. doi: 10.1158/1078-0432.CCR-18-3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case D.A., Cerutti D.S., Cheatham T.E., III, Darden T.A., Duke R.E., et al. University of California; San Francisco: 2020. AMBER 2020. [Google Scholar]
- Cesari A., Bottaro S., Lindorff-Larsen K., Banáš P., Šponer J., Bussi G. Fitting corrections to an RNA force field using experimental data. J. Chem. Theor. Comput. 2019;15:3425–3431. doi: 10.1021/acs.jctc.9b00206. [DOI] [PubMed] [Google Scholar]
- Cheatham T.E., III, Case D.A. Twenty-five years of nucleic acid simulations. Biopolymers. 2013;99:969–977. doi: 10.1002/bip.22331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christen M., van Gunsteren W.F. On searching in, sampling of, and dynamically moving through conformational space of biomolecular systems: a review. J. Comput. Chem. 2008;29:157–166. doi: 10.1002/jcc.20725. [DOI] [PubMed] [Google Scholar]
- Chuang C.H., Chiou S.J., Cheng T.L., Wang Y.T. A molecular dynamics simulation study decodes the Zika virus NS5 methyltransferase bound to SAH and RNA analogue. Sci. Rep. 2018;8:6336. doi: 10.1038/s41598-018-24775-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clingman C.C., Deveau L.M., Hay S.A., Genga R.M., Shandilya S.M.D., Massi F., Ryder S.P. Allosteric inhibition of a stem cell RNA-binding protein by an intermediary metabolite. eLife. 2014;3 doi: 10.7554/eLife.02848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conlon E.G., Manley J.L. RNA-binding proteins in neurodegeneration: mechanisms in aggregate. Genes Dev. 2017;31:1509–1528. doi: 10.1101/gad.304055.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper-Joyce C.J., Bhola R., Wang J., Bhattarai A., Nguyen A.T.N., Cowie-Kent I., O'Sullivan K., Chia L.Y., Venugopal H., Valant C., et al. Positive allosteric mechanisms of adenosine A1 receptor-mediated analgesia. Nature. 2021;597:571–576. doi: 10.1038/s41586-021-03897-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y.Q., Yang L.J., Fan Y.B., Shao Q. Thermodynamics and kinetics simulations of multi-time-scale processes for complex systems. Int. Rev. Phys. Chem. 2008;27:201–227. [Google Scholar]
- Gao Y., Cao D., Pawnikar S., John K.P., Ahn H.M., Hill S., Ha J.M., Parikh P., Ogilvie C., Swain A., et al. Structure of the human respiratory syncytial virus M2-1 protein in complex with a short positive-sense gene-end RNA. Structure. 2020;28:979–990. doi: 10.1016/j.str.2020.07.001. e974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentze M.W., Castello A., Schwarzl T., Preiss T. A brave new world of RNA-binding proteins. Nat. Rev. Mol. Cell Biol. 2018;19:327–341. doi: 10.1038/nrm.2017.130. [DOI] [PubMed] [Google Scholar]
- Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14(33–38):27–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Iwaoka R., Nagata T., Tsuda K., Imai T., Okano H., Kobayashi N., Katahira M. Structural insight into the recognition of r(UAG) by Musashi-1 RBD2, and construction of a model of Musashi-1 RBD1-2 bound to the minimum target RNA. Molecules. 2017;22:1207. doi: 10.3390/molecules22071207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen W.L., Chandrasekhar J., Madura J.D., Impey R.W., Klein M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- Krepl M., Havrila M., Stadlbauer P., Banas P., Otyepka M., Pasulka J., Stefl R., Sponer J. Can we execute stable microsecond-scale atomistic simulations of protein–RNA complexes? J. Chem. Theor. Comput. 2015;11:1220–1243. doi: 10.1021/ct5008108. [DOI] [PubMed] [Google Scholar]
- Krepl M., Cléry A., Blatter M., Allain F.H.T., Sponer J. Synergy between NMR measurements and MD simulations of protein/RNA complexes: application to the RRMs, the most common RNA recognition motifs. Nucleic Acids Res. 2016;44:6452–6470. doi: 10.1093/nar/gkw438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudinov A.E., Karanicolas J., Golemis E.A., Boumber Y. Musashi RNA-binding proteins as cancer drivers and novel therapeutic targets. Clin. Cancer Res. 2017;23:2143–2153. doi: 10.1158/1078-0432.CCR-16-2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kührová P., Mlýnský V., Zgarbová M., Krepl M., Bussi G., Best R.B., Otyepka M., Šponer J., Banáš P. Improving the performance of the amber RNA force field by tuning the hydrogen-bonding interactions. J. Chem. Theor. Comput. 2019;15:3288–3305. doi: 10.1021/acs.jctc.8b00955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Ma B., Tsai C.J., Sinha N., Nussinov R. Folding and binding cascades: dynamic landscapes and population shifts. Protein Sci. 2000;9:10–19. doi: 10.1110/ps.9.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laio A., Parrinello M. Escaping free-energy minima. Proc. Natl. Acad. Sci. U.S.A. 2002;99:12562–12566. doi: 10.1073/pnas.202427399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan L., Appelman C., Smith A.R., Yu J., Larsen S., Marquez R.T., Liu H., Wu X., Gao P., Roy A. Natural product (−)-gossypol inhibits colon cancer cell growth by targeting RNA-binding protein Musashi-1. Mol. Oncol. 2015;9:1406–1420. doi: 10.1016/j.molonc.2015.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan L., Xing M., Douglas J.T., Gao P., Hanzlik R.P., Xu L. Human oncoprotein Musashi-2 N-terminal RNA recognition motif backbone assignment and identification of RNA-binding pocket. Oncotarget. 2017;8:106587. doi: 10.18632/oncotarget.22540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan L., Liu H., Smith A.R., Appelman C., Yu J., Larsen S., Marquez R.T., Wu X., Liu F.Y., Gao P., et al. Natural product derivative Gossypolone inhibits Musashi family of RNA-binding proteins. BMC Cancer. 2018;18:809. doi: 10.1186/s12885-018-4704-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan L., Xing M., Kashipathy M., Douglas J., Gao P., Battaile K., Hanzlik R., Lovell S., Xu L. Crystal and solution structures of human oncoprotein Musashi-2N-terminalRNA recognition motif 1. Proteins. 2020;88:573–583. doi: 10.1002/prot.25836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan L., Liu J., Xing M., Smith A.R., Wang J., Wu X., Appelman C., Li K., Roy A., Gowthaman R., et al. Identification and validation of an Aspergillus nidulans secondary metabolite derivative as an inhibitor of the Musashi-RNA interaction. Cancers. 2020;12:2221. doi: 10.3390/cancers12082221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leulliot N., Varani G. Current topics in RNA−protein recognition:control of specificity and biological function through induced fit and conformational capture. Biochemistry. 2001;40:7947–7956. doi: 10.1021/bi010680y. [DOI] [PubMed] [Google Scholar]
- Liao J.-M., Wang Y.-T. In silico studies of conformational dynamics of Mu opioid receptor performed using Gaussian accelerated molecular dynamics. J. Biomol. Struct. Dyn. 2019;37:166–177. doi: 10.1080/07391102.2017.1422025. [DOI] [PubMed] [Google Scholar]
- Liwo A., Czaplewski C., Oldziej S., Scheraga H.A. Computational techniques for efficient conformational sampling of proteins. Curr. Opin. Struct. Biol. 2008;18:134–139. doi: 10.1016/j.sbi.2007.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier J.A., Martinez C., Kasavajhala K., Wickstrom L., Hauser K.E., Simmerling C. ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theor. Comput. 2015;11:3696–3713. doi: 10.1021/acs.jctc.5b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Y., McCammon J.A. Unconstrained enhanced sampling for free energy calculations of biomolecules: a review. Mol. Simulat. 2016;42:1046–1055. doi: 10.1080/08927022.2015.1121541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Y., McCammon J.A. Graded activation and free energy landscapes of a muscarinic G-protein–coupled receptor. Proc. Natl. Acad. Sci. U.S.A. 2016;113:12162–12167. doi: 10.1073/pnas.1614538113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Y., McCammon J.A. Gaussian accelerated molecular dynamics: theory, implementation, and applications. Annu. Rep. Comput. Chem. 2017;13:231–278. doi: 10.1016/bs.arcc.2017.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Y., McCammon J.A. Mechanism of the G-protein mimetic nanobody binding to a muscarinic G-protein-coupled receptor. Proc. Natl. Acad. Sci. U.S.A. 2018;115:3036–3041. doi: 10.1073/pnas.1800756115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Y., Sinko W., Pierce L., Bucher D., Walker R.C., McCammon J.A. Improved reweighting of accelerated molecular dynamics simulations for free energy calculation. J. Chem. Theor. Comput. 2014;10:2677–2689. doi: 10.1021/ct500090q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Y., Feher V.A., McCammon J.A. Gaussian accelerated molecular dynamics: unconstrained enhanced sampling and free energy calculation. J. Chem. Theor. Comput. 2015;11:3584–3595. doi: 10.1021/acs.jctc.5b00436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Y., Bhattarai A., Nguyen A.T.N., Christopoulos A., May L.T. Structural basis for binding of allosteric drug leads in the adenosine A1 receptor. Sci. Rep. 2018;8:16836. doi: 10.1038/s41598-018-35266-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Y., Huang Y-mM., Walker R.C., McCammon J.A., Chang C-eA. Ligand binding pathways and conformational transitions of the HIV protease. Biochemistry. 2018;57:1533–1541. doi: 10.1021/acs.biochem.7b01248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Y., Bhattarai A., Wang J. Ligand Gaussian accelerated molecular dynamics (LiGaMD): characterization of ligand binding thermodynamics and kinetics. J. Chem. Theor. Comput. 2020;16:5526–5547. doi: 10.1021/acs.jctc.0c00395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minuesa G., Albanese S.K., Xie W., Kazansky Y., Worroll D., Chow A., Schurer A., Park S.-M., Rotsides C.Z., Taggart J., et al. Small-molecule targeting of MUSASHI RNA-binding activity in acute myeloid leukemia. Nat. Commun. 2019;10:2691. doi: 10.1038/s41467-019-10523-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyanoiri Y., Kobayashi H., Imai T., Watanabe M., Nagata T., Uesugi S., Okano H., Katahira M. Origin of higher affinity to RNA of the N-terminal RNA-binding domain than that of the C-terminal one of a mouse neural protein, Musashi1, as revealed by comparison of their structures, modes of interaction, surface electrostatic potentials, and backbone dynamics. J. Biol. Chem. 2003;278:41309–41315. doi: 10.1074/jbc.M306210200. [DOI] [PubMed] [Google Scholar]
- Mlýnský V., Bussi G. Exploring RNA structure and dynamics through enhanced sampling simulations. Curr. Opin. Struct. Biol. 2018;49:63–71. doi: 10.1016/j.sbi.2018.01.004. [DOI] [PubMed] [Google Scholar]
- Nagata T., Kanno R., Kurihara Y., Uesugi S., Imai T., Sakakibara S-i, Okano H., Katahira M. Structure, backbone dynamics and interactions with RNA of the C-terminal RNA-binding domain of a mouse neural RNA-binding protein, Musashi111Edited by P. E. Wright. J. Mol. Biol. 1999;287:315–330. doi: 10.1006/jmbi.1999.2596. [DOI] [PubMed] [Google Scholar]
- Ohyama T., Nagata T., Tsuda K., Kobayashi N., Imai T., Okano H., Yamazaki T., Katahira M. Structure of Musashi1 in a complex with target RNA: the role of aromatic stacking interactions. Nucleic Acids Res. 2011;40:3218–3231. doi: 10.1093/nar/gkr1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki K-i, Takada S. Dynamic energy landscape view of coupled binding and protein conformational change: induced-fit versus population-shift mechanisms. Proc. Natl. Acad. Sci. U.S.A. 2008;105:11182–11187. doi: 10.1073/pnas.0802524105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palermo G., Miao Y., Walker R.C., Jinek M., McCammon J.A. CRISPR-Cas9 conformational activation as elucidated from enhanced molecular simulations. Proc. Natl. Acad. Sci. U.S.A. 2017;114:7260–7265. doi: 10.1073/pnas.1707645114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang Y.T., Miao Y., Wang Y., McCammon J.A. Gaussian accelerated molecular dynamics in NAMD. J. Chem. Theor. Comput. 2017;13:9–19. doi: 10.1021/acs.jctc.6b00931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.B., Kim Y.H., Yoo Y., Kim J., Jun S.-H., Cho J.W., El Qaidi S., Walpole S., Monaco S., García-García A.A., et al. Structural basis for arginine glycosylation of host substrates by bacterial effector proteins. Nat. Commun. 2018;9:4283. doi: 10.1038/s41467-018-06680-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawnikar S., Miao Y. Pathway and mechanism of drug binding to chemokine receptors revealed by accelerated molecular simulations. Future Med. Chem. 2020;12:1213–1225. doi: 10.4155/fmc-2020-0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y., Cao S., Kiselar J., Xiao X., Du Z., Hsieh A., Ko S., Chen Y., Agrawal P., Zheng W., et al. A metastable contact and structural disorder in the estrogen receptor transactivation domain. Structure. 2019;27:229–240 e224. doi: 10.1016/j.str.2018.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitici F., Beveridge D.L., Baranger A.M. Molecular dynamics simulation studies of induced fit and conformational capture in U1A–RNA binding: do molecular substates code for specificity? Biopolymers. 2002;65:424–435. doi: 10.1002/bip.10251. [DOI] [PubMed] [Google Scholar]
- Ricci C.G., Chen J.S., Miao Y., Jinek M., Doudna J.A., McCammon J.A., Palermo G. Deciphering off-target effects in CRISPR-Cas9 through accelerated molecular dynamics. ACS Cent. Sci. 2019;5:651–662. doi: 10.1021/acscentsci.9b00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roe D.R., Cheatham T.E., 3rd PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theor. Comput. 2013;9:3084–3095. doi: 10.1021/ct400341p. [DOI] [PubMed] [Google Scholar]
- Sakakibara S., Nakamura Y., Satoh H., Okano H. Rna-binding protein Musashi2: developmentally regulated expression in neural precursor cells and subpopulations of neurons in mammalian CNS. J. Neurosci. 2001;21:8091–8107. doi: 10.1523/JNEUROSCI.21-20-08091.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salawu E.O. The impairment of torsinA's binding to and interactions with its activator: an atomistic molecular dynamics study of primary dystonia. Front. Mol. Biosci. 2018;5 doi: 10.3389/fmolb.2018.00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibener L.V., Fernandes R.A., Kolawole E.M., Carbone C.B., Liu F., McAffee D., Birnbaum M.E., Yang X., Su L.F., Yu W., et al. Isolation of a structural mechanism for uncoupling T cell receptor signaling from peptide-MHC binding. Cell. 2018;174:672–687 e627. doi: 10.1016/j.cell.2018.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiwok V., Sucur Z., Hosek P. Enhanced sampling techniques in biomolecular simulations. Biotechnol. Adv. 2015;33:1130–1140. doi: 10.1016/j.biotechadv.2014.11.011. [DOI] [PubMed] [Google Scholar]
- Šponer J., Krepl M., Banáš P., Kührová P., Zgarbová M., Jurečka P., Havrila M., Otyepka M. How to understand atomistic molecular dynamics simulations of RNA and protein–RNA complexes? WIREs RNA. 2017;8 doi: 10.1002/wrna.1405. [DOI] [PubMed] [Google Scholar]
- Šponer J., Bussi G., Krepl M., Banáš P., Bottaro S., Cunha R.A., Gil-Ley A., Pinamonti G., Poblete S., Jurečka P., et al. RNA structural dynamics as captured by molecular simulations: a comprehensive overview. Chem. Rev. 2018;118:4177–4338. doi: 10.1021/acs.chemrev.7b00427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suryadi J., Tran E.J., Maxwell E.S., Brown B.A. The crystal structure of the methanocaldococcus jannaschii multifunctional L7Ae RNA-binding protein reveals an induced-fit interaction with the Box C/D RNAs. Biochemistry. 2005;44:9657–9672. doi: 10.1021/bi050568q. [DOI] [PubMed] [Google Scholar]
- Tan D., Piana S., Dirks R.M., Shaw D.E. RNA force field with accuracy comparable to state-of-the-art protein force fields. Proc. Natl. Acad. Sci. U.S.A. 2018;115:E1346–E1355. doi: 10.1073/pnas.1713027115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai C.J., Kumar S., Ma B., Nussinov R. Folding funnels, binding funnels, and protein function. Protein Sci. 1999;8:1181–1190. doi: 10.1110/ps.8.6.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai C.J., Ma B., Nussinov R. Folding and binding cascades: shifts in energy landscapes. Proc. Natl. Acad. Sci. U.S.A. 1999;96:9970–9972. doi: 10.1073/pnas.96.18.9970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.-T., Chan Y.-H. Understanding the molecular basis of agonist/antagonist mechanism of human mu opioid receptor through Gaussian accelerated molecular dynamics method. Sci. Rep. 2017;7:7828. doi: 10.1038/s41598-017-08224-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Miao Y. Mechanistic insights into specific G protein interactions with adenosine receptors. J. Phys. Chem. B. 2019;123:6462–6473. doi: 10.1021/acs.jpcb.9b04867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Miao Y. Peptide Gaussian accelerated molecular dynamics (Pep-GaMD): enhanced sampling and free energy and kinetics calculations of peptide binding. J. Chem. Phys. 2020;153:154109. doi: 10.1063/5.0021399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Arantes P.R., Bhattarai A., Hsu R.V., Pawnikar S., Huang Y-mM., Palermo G., Miao Y. WIREs Computational Molecular Science n/a; 2021. Gaussian Accelerated Molecular Dynamics: Principles and Applications. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson J.R. Induced fit in RNA–protein recognition. Nat. Struct. Biol. 2000;7:834–837. doi: 10.1038/79575. [DOI] [PubMed] [Google Scholar]
- Wu P. Inhibition of RNA-binding proteins with small molecules. Nat. Rev. Chem. 2020;4:441–458. doi: 10.1038/s41570-020-0201-4. [DOI] [PubMed] [Google Scholar]
- Wu S., Wang J., Pu X., Li L., Li Q. T7 RNA polymerase discriminates correct and incorrect nucleoside triphosphates by free energy. Biophys. J. 2018;114:1755–1761. doi: 10.1016/j.bpj.2018.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zearfoss N.R., Deveau L.M., Clingman C.C., Schmidt E., Johnson E.S., Massi F., Ryder S.P. A conserved three-nucleotide core motif defines Musashi RNA binding specificity. J. Biol. Chem. 2014;289:35530–35541. doi: 10.1074/jbc.M114.597112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zgarbová M., Otyepka M., Šponer J., Mládek A., Banáš P., Cheatham T.E., Jurečka P. Refinement of the Cornell et al. nucleic acids force field based on reference quantum chemical calculations of glycosidic torsion profiles. J. Chem. Theor. Comput. 2011;7:2886–2902. doi: 10.1021/ct200162x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.