Abstract
Particulate matter (PM) causes several diseases, including cardiovascular diseases (CVDs). Previous studies compared the gene expression patterns in airway epithelial cells and keratinocytes exposed to PM. However, analysis of differentially expressed gene (DEGs) in endothelial cells exposed to PM2.5 (diameter less than 2.5 μm) from fossil fuel combustion has been limited. Here, we exposed human umbilical vein endothelial cells (HUVECs) to PM2.5 from combustion of gasoline, performed RNA-seq analysis, and identified DEGs. Exposure to the IC50 concentrations of gasoline engine exhaust PM2.5 (GPM) for 24 h yielded 1081 (up-regulation: 446, down-regulation: 635) DEGs. The most highly up-regulated gene is NGFR followed by ADM2 and NUPR1. The most highly down-regulated gene is TNFSF10 followed by GDF3 and EDN1. Gene Ontology enrichment analysis revealed that GPM regulated genes involved in cardiovascular system development, tube development and circulatory system development. Kyoto Encyclopedia of Genes and Genomes and Reactome pathway analyses showed that genes related to cytokine–cytokine receptor interactions and cytokine signaling in the immune system were significantly affected by GPM. We confirmed the RNA-seq data of some highly altered genes by qRT-PCR and showed the induction of NGFR, ADM2 and IL-11 at a protein level, indicating that the observed gene expression patterns were reliable. Given the adverse effects of PM2.5 on CVDs, our findings provide new insight into the importance of several DEGs and pathways in GPM-induced CVDs.
Keywords: Gasoline engine exhaust particulate matter 2.5, HUVECs, RNA sequencing, Transcriptomics, Cardiovascular diseases
1. Introduction
Air pollution is a growing environmental health risk factor that caused an estimated 4.2 million deaths worldwide in 2015 [1]. Air pollutant is a heterogeneous complex mixture of solid particles and gases, and its composition depends on many factors, including sources and atmospheric conditions [2]. Particulate matter (PM) is classified as PM10 (diameter <10 μm), PM2.5 (diameter <2.5 μm), or PM0.1 (diameter <0.1 μm) according to particle size [2]. Combustion of fossil fuels, such as gasoline and diesel, is a major source of PM2.5 [3]. These particles are small enough to penetrate into the human respiratory system; from there, it gets into the circulatory system, where it causes inflammation and oxidative stress [4]. Accordingly, several cohort studies have reported a positive correlation between PM and cardiovascular mortality [5,6], emphasizing the importance of performing research on cardiovascular diseases (CVDs) caused by PM.
Endothelial cells form the inner side of blood vessel and regulate the vascular tone [7]. Because PM2.5 that penetrates the respiratory system circulates throughout the body in blood vessels and causes CVDs, it is important to investigate how PM2.5 leads to the endothelial cell dysfunction and to find the genes which regulate this phenomenon. In this study, we produced PM2.5 by combusting gasoline and analyzed its chemical constituents. We exposed human umbilical vein endothelial cells (HUVECs) to gasoline engine exhaust PM2.5 (GPM) and determined its IC50 concentration. We selected HUVECs as an in vitro model of GPM-induced endothelial cell toxicity because these cells recapitulate phenomena that occur in blood vessels in vivo [8] and are a good model for the study of nanoparticle-induced toxicity in endothelium [9]. We exposed HUVECs to the IC50 concentration of GPM for 24 h, performed gene expression profiling by RNA-seq, and analyzed the differentially expressed genes (DEGs). We then performed Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG), and Reactome analyses to identify the mechanisms associated with endothelial cell dysfunction. We confirmed the results of RNA-seq by quantitative real time (qRT)-PCR, western blotting, and enzyme-linked immunosorbent assay (ELISA). Finally, we discuss the potential roles of some highly altered DEGs in GPM-induced CVDs.
2. Materials and methods
2.1. Generation of PM2.5 and chemical characterization
Generation of fine particles from gasoline engine exhaust and analysis of chemical characterization were performed as previously described [10]. Briefly, the engine exhaust particles were produced from a gasoline engine (439 cc, LH8500, ZongShen General Power Machine Co., Ltd., Chongqing, China) and collected on to filters by using a PM2.5 low-volume sampler (URG-2000-30EH, URG, Chapel Hill, NC, USA) at a flow rate of 16.7 L/min for 30 min. The mass of PM2.5 was determined based on the weight of filter, which was equilibrated at 21 ± 2 °C and relative humidity of 35 ± 5% for 24 h before and after collection. The mass concentration (μg/m3) was calculated by dividing the collected PM2.5 mass (μg) by the volume of collected air (m3). We used a polytetrafluoroethylene (PTFE) filter (Zeflour, Pall Corporation, Port Washington, NY, USA) for analysis of ions and elements, and a quartz filter (Tissuquartz, Pall Corporation) for analysis of carbonaceous species (OC; organic carbon, EC; elemental carbon). Ions, elements, and carbonaceous species in PM2.5 were characterized by ion chromatography [850 Professional IC, MagIC Net professional (ver. 3.2), Metrohm, Switzerland], inductively coupled plasma-mass spectrometry [7500ce, MassHunter 4.4 Workstation (ver. C.01.04), Agilent Technologies, Santa Clara, CA, USA] and an OC-EC carbon analyzer [5L, Carbon Analysis (ver. OCEC1029), Sunset Laboratory, Tigard, OR, USA], respectively. To study cellular toxicity, we collected GPM on to a glass fiber filter (Pall Corporation) and extracted them with dimethylsulfoxide (DMSO, Sigma-Aldrich, St. Louis, MO, USA). Extracted GPM was filtered through a PTFE syringe filter (Sartorius AG, Germany) before exposed to HUVECs.
2.2. Cell culture
HUVECs were purchased from Lonza (Walkersville, MD, USA) and cultured in endothelial basal medium-2 (EBM-2; Lonza) supplemented with endothelial growth medium-2 (EGM-2) SingleQuots (Lonza). Vascular endothelial growth factor (VEGF) was provided as a components of the EGM-2 SingleQuots; however, to rule out an effect of VEGF on endothelial cell survival, we did not add it to the culture medium [11]. HUVECs were cultured in cell culture dishes precoated with 0.2% gelatin (Sigma-Aldrich) and used at passages 4 to 6.
2.3. Cell proliferation assay
HUVECs were seeded in 96-well cell culture plates precoated with 0.2% gelatin at a density of 1 × 104 cells/well. The next day, the cells were exposed to various concentrations of GPM or the correspondingly same volume of DMSO for 24 h. After the incubation with WST-1 reagent (Premix WST-1 Cell Proliferation Assay System, Takara Bio Inc., Shiga, Japan) for an additional 4 h, absorbance at 420 nm was measured on a SpectraMax Plus 384 microplate reader (Molecular Devices, San Jose, CA, USA). After subtracting the absorbance of culture medium plus WST-1 reagent in the absence of cells, cell viability was calculated by dividing the absorbance of each concentration GPM-treated group with the correspondingly same volume of DMSO-treated group. Cell viability in the corresponding DMSO-treated group was considered to be 100%. We incubated the cells in the complete media during this experiment.
2.4. RNA sequencing (RNA-seq)
RNA-seq was performed by Ebiogen (Seoul, Republic of Korea). Total RNA was isolated using Trizol reagent (Invitrogen, Carlsbad, CA, USA) and RNA quality was assessed on an Agilent 2100 bioanalyzer using the RNA 6000 Nano Chip (Agilent Technologies). Libraries were constructed using the QuantSeq 3′ mRNA-Seq Library Prep Kit (Lexogen Inc., Vienna, Austria). Single-end 75-nt sequencing was performed on a NextSeq 500 (Illumina, San Diego, CA, USA).
2.5. RNA-seq data analysis
QuantSeq 3′ mRNA-Seq reads were aligned using Bowtie2 [12]. For alignment to the genome and transcriptome, Bowtie2 indices were generated from the genome assembly sequence or the representative transcript sequences. The alignment file was used to assemble transcripts, estimate their abundances, and detect the differential expression of genes. DEGs were determined based on counts from unique and multiple alignments using coverage in Bedtools [13]. The RC (Read Count) data were processed by the quantile normalization method using EdgeR within R using Bioconductor [14]. Gene classification was based on searches done by DAVID (http://david.abcc.ncifcrf.gov) and Medline databases (http://www.ncbi.nlm.nih.gov).
2.6. Gene ontology and canonical pathway analysis
Gene Ontology (GO) and canonical pathways of DEGs were analyzed in Molecular Signature Data Base (MSigDB version 7.1) of the Gene Set Enrichment Analysis (GSEA) website (http://www.gsea-msigdb.org) [15]. Selected DEGs were input into MsigDB and their overlap with genes in GO, KEGG, and Reactome gene sets were computed. Enrichment of GO terms, KEGG pathways, and Reactome pathways was considered significant when the FDR q value was less than 0.05.
2.7. Quantitative real time-polymerase chain reaction (qRT-PCR)
Total cellular RNA was extracted using the Trizol reagent. RNA (2 μg) was reverse transcribed using the RNA to cDNA EcoDry Premix (Clontech, Mountain View, CA, USA), and each gene of interest was amplified using the Power SYBR PCR Master Mix (Applied Biosystems, Foster City, CA, USA) in the QuantStudio 6 Flex Real Time PCR System (Applied Biosystems). The expression level of each gene was calculated in triplicate and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) mRNA was used for normalization. The primers sequences were as follows: for nerve growth factor receptor (NGFR), 5′-CGACAACCTCATCCCTGTCT-3′ (forward) and 5′-GCTGTTCCACCTCTTGAAGG-3′ (reverse); for adrenomedullin 2 (ADM2), 5′-CTGCCAGGTGCAGAATCTC-3′ (forward) and 5′-GGGTCCACAGGAGCTGAGT-3′ (reverse); for nuclear protein 1 (NUPR1), 5′-AGAGAGAAGCTGCTGCCAAC-3′ (forward) and 5′-CCTCGCTTCTTCCTCTCTGA-3′ (reverse); for IL-11, 5′-ACAGCTGAGGGACAAATTCC-3′ (forward) and 5′-AGCTGTAGAGCTCCCAGTGC-3′ (reverse); for matrix metallopeptidase-1 (MMP-1), 5′-GGTCTCTGAGGGTCAAGCAG-3′ (forward) and 5′-CCAGGTCCATCAAAAGGAGA-3′ (reverse); for tumor necrosis factor (ligand) superfamily, member 10 (TNFSF10), 5′-TTCACAGTGCTCCTGCAGTC-3′ (forward) and 5′-ATCTGCTTCAGCTCGTTGGT-3′ (reverse); for growth differentiation factor 3 (GDF3), 5′-CCGGAAAAATTTCGGGTTAT-3′ (forward) and 5′-TCTGGCACAGGTGTCTTCAG-3′ (reverse); for aquaporin 1 (AQP1), 5′-GGACACCTCCTGGCTATTGA-3′ (forward) and 5′-TCCAGTGGTTGCTGAAGTTG-3′ (reverse); for IL-33, 5′-CAAAGAAGTTTGCCCCATGT-3′ (forward) and 5′-AAGGCCTTTTGGTGGTTTCT-3′ (reverse); for tissue inhibitor of metalloproteinase 3 (TIMP3), 5′-ACCTGCCTTGCTTTGTGACT-3′ (forward) and 5′-GGCGTAGTGTTTGGACTGGT-3′ (reverse); for GAPDH, 5′- ACGGATTTGGTCGTATTGGG-3′ (forward) and 5′- TGATTTTGGAGGGATCTCGC-3′ (reverse).
2.8. Western blotting
HUVECs exposed to DMSO or GPM for 24 h were washed with cold PBS and lysed in cell lysis buffer (Cell Signaling Technology, Beverly, MA, USA) supplemented with protease inhibitor cocktail (Sigma-Aldrich). Equal amounts of cell lysates were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Proteins were transferred onto polyvinylidene difluoride membranes (Millipore, Billerica, MA) and subsequently blocked in 5% nonfat milk (Santa Cruz Biotechnology, Santa Cruz, CA) in 0.1% Tween 20 containing Tris-buffered saline (TBS) for 1 h. Membranes were incubated overnight at 4 °C with appropriate primary antibodies. After being washed 3 times with 0.1% Tween 20 containing TBS, membranes were incubated with horseradish peroxidase (HRP)-conjugated secondary antibody (1:5000 dilution, Santa Cruz Biotechnology) for 1 h. After washing with 0.1% Tween 20 containing TBS, the signals were visualized with ImageQuant LAS4000 mini system (GE Healthcare, Chicago, IL, USA) using the Western Blotting Luminol Reagent (Santa Cruz Biotechnology). Densitometric analysis was performed using Image J software (ver. 1.53). NGFR antibody was purchased from Cell Signaling Technology. ADM2 antibody was from Invitrogen and α-tubulin antibody was from Sigma-Aldrich.
2.9. ELISA
Medium of HUVECs exposed to DMSO or GPM for 24 h were harvested and centrifuged at 13,000 g for 10 min at 4 °C. After collecting the supernatant, the level of interleukin (IL)-11 in the supernatant was measured using the IL-11 ELISA kit (R&D Systems, Minneapolis, MN, USA), according to manufacturer's instruction.
2.10. Statistics
Results of the western blotting and ELISA are presented as means ± SD. Statistical analyses were performed using Student's t-test. We used the Microsoft Excel (ver. 2013) in Windows 10 operating system for statistical analysis. P < 0.05 was considered significant.
3. Results
3.1. Chemical composition analysis of GPM
GPM was subjected to analysis of mass fractions of chemical components (elements, ions, and carbonaceous species). The GPM had the highest mass fraction of organic carbon (OC; 78.10%) followed by ions (2.27%), elemental carbon (EC; 1.30%), and elements (0.10%), including others (unidentified chemical compounds; 18.23%) (Table S1). The Al was the most abundant element (34.27%) in the GPM elements, followed by Se (24.38%) and Na (21.17%) (Table S2). The NO3− was the most abundant ion (60.42%) in ionic species, followed by NH4+(22.38%) and SO42−(7.06%) (Table S3).
3.2. Cytotoxic effect of GPM on HUVECs
We treated HUVECs with various concentrations of GPM or DMSO (solute control) and measured cell viability after incubation for 24 h. Relative to DMSO, GPM induced cell death in a dose-dependent manner (Fig. 1A). The IC50 concentrations was 59.0 μg/ml. To study the change in gene expression profile in GPM-exposed HUVECs, we used the IC50 concentration. Exposure of HUVECs to the IC50 concentration of GPM resulted in cytotoxicity (Fig. 1B).
Fig. 1.
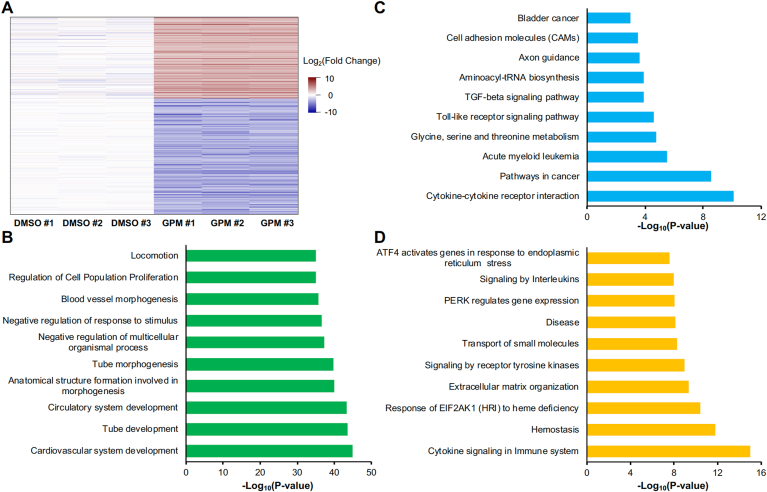
IC50 concentration of GPM induces 1081 DEGs in HUVECs. (A) HUVECs were exposed to various concentrations of GPM for 24 h. Cell viability was measured by WST-1 assay. (B) Images were acquired after 24 h exposure to DMSO (solute control) and GPM IC50. (C) Volcano plot of DEGs between GPM-exposed and DMSO-exposed HUVECs. |Fold change| > 3 and P < 0.05 were used as the threshold for significant differences in gene expression. Red dots: significantly up-regulated genes; green dots: significantly down-regulated genes (D) Numbers of up- or down-regulated genes in HUVECs exposed to GPM. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.3. Gene expression profile of HUVECs exposed to GPM
We exposed HUVECs to IC50 concentration of GPM or DMSO for 24 h, and then subjected the samples to RNA-seq. Using criteria of |fold change| > 3 and P value < 0.05, we identified 1081 genes (out of 25,737 genes detected) that were differentially expressed following exposure to GPM. In GPM-exposed HUVECs, 446 genes were up-regulated and 635 genes were down-regulated (Fig. 1C and D).
3.4. DEGs analysis of HUVECs exposed to GPM
The top 10 up-regulated and down-regulated genes in GPM-exposed cells are listed in Table 1. The most highly up-regulated gene is NGFR followed by ADM2 and NUPR1 and the most highly down-regulated gene is TNFSF10 followed by GDF3 and EDN1. A heatmap analysis of 3 DMSO-exposed HUVECs and 3 GPM-exposed HUVECs illustrates the change in gene expression pattern (Fig. 2A). To identify GO enrichment and canonical pathways affected, we mapped the 1081 GPM-induced DEGs to GO terms, KEGG pathways, and Reactome pathways. The most highly enriched GO term was “cardiovascular system development”, followed by “tube development” and “circulatory system development” (Fig. 2B). KEGG pathway analysis revealed that many DEGs were involved in cytokine–cytokine receptor interactions, cancer pathways, and acute myeloid leukemia (Fig. 2C). Reactome pathway analysis revealed that many DEGs were involved in cytokine signaling in the immune system, hemostasis, and response of EIF2AK1 (HRI) to heme deficiency (Fig. 2D).
Table 1.
Top 10 up-regulated and down-regulated genes.
| Category | Gene | Log2(FC) | Description |
|---|---|---|---|
| Up-regulated genes | NGFR | 9.552 | nerve growth factor receptor |
| ADM2 | 8.034 | adrenomedullin 2 | |
| NUPR1 | 6.604 | nuclear protein 1, transcriptional regulator | |
| FAM129A | 6.12 | family with sequence similarity 129 member A | |
| PCLO | 6.002 | piccolo presynaptic cytomatrix protein | |
| BEX2 | 5.978 | brain expressed X-linked 2 | |
| STK32A | 5.836 | serine/threonine kinase 32A | |
| EPGN | 5.821 | epithelial mitogen | |
| SGCG | 5.792 | sarcoglycan gamma | |
|
IL11 |
5.575 |
interleukin 11 |
|
| Down-regulated genes | TNFSF10 | −6.716 | tumor necrosis factor superfamily member 10 |
| GDF3 | −6.147 | growth differentiation factor 3 | |
| EDN1 | −6.018 | endothelin 1 | |
| AQP1 | −5.721 | aquaporin 1 (Colton blood group) | |
| TIMP3 | −5.299 | TIMP metallopeptidase inhibitor 3 | |
| ZNF367 | −5.285 | zinc finger protein 367 | |
| SGSM1 | −5.186 | small G protein signaling modulator 1 | |
| CCL2 | −5.151 | C–C motif chemokine ligand 2 | |
| GIMAP8 | −5.143 | GTPase, IMAP family member 8 | |
| SLCO2A1 | −5.079 | solute carrier organic anion transporter family member 2A1 |
Fig. 2.
Pathway analysis of DEGs from GPM-exposed HUVECs. (A) Heatmap analysis of 446 genes up-regulated and 635 genes down-regulated in response to GPM exposure. Color indicates the relative expression level of each gene. (B–D) Pathways associated with DEGs, based on GO analysis (B), KEGG analysis (C) and Reactome analysis (D). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.5. Confirmation of DEGs by qRT-PCR, western blotting, and ELISA
To validate the data from RNA-seq analysis, we performed qRT-PCR for the some of top 10 up- or down-regulated genes. These genes were chosen because they regulate the cardiovascular cell dysfunction and are related to CVDs [[16], [17], [18], [19], [20], [21], [22], [23]]; we will discuss these functions below. The results of qRT-PCR in GPM-exposed HUVECs were comparable to those of RNA-seq (Fig. 3A). We also explored the expression of some DEGs at a protein level and found that GPM increased the expression of NGFR, ADM2 and IL-11 (Fig. 3B–D). Together, the results of these validation experiments confirm that the results of this study are reliable.
Fig. 3.
Confirmation of DEGs by qRT-PCR, western blotting, and ELISA. (A) Fold change values in RNA-seq data of selected GPM-regulated genes were compared with those in qRT-PCR data. (B) Expression of NGFR, ADM2 and α-tubulin in HUVECs exposed to GPM or DMSO for 24 h. (C) The expressions of NGFR and ADM2 in Fig. 3B were quantified by being normalized to the corresponding level of α-tubulin. *P < 0.02 versus DMSO. (D) The amount of IL-11 in the medium of HUVECs exposed to GPM or DMSO for 24 h was determined by ELISA. *P < 0.02 versus DMSO.
4. Discussion
Several epidemiological studies have shown that PM contributes to various human diseases, including CVDs [5,6] and suggested that it is necessary to extend our understanding into the candidate genes responsible for the initiation or exacerbation of diseases. Recent studies reported changes in gene expression patterns in PM-exposed bronchial epithelial cells [24] and keratinocytes [25]. One recent study reported the changes in gene expression pattern in Beijing-collected PM2.5-exposed HUVECs [26]; however, the analysis of DEGs in HUVECs exposed to GPM has been limited. Accordingly, we performed gene expression analysis in GPM-exposed HUVECs to elucidate the candidate genes involved in blood vessel dysfunction and incidence of CVDs.
Our DEG analyses identified the genes which are up- or down-regulated by GPM exposure. Among them, the gene encoding nerve growth factor receptor (NGFR) was the most up-regulated. Expression of NGFR (neurotrophin p75 receptor; p75NTR) in endothelial cell (EC) is up-regulated in type-1 diabetes and hindlimb ischemia under conditions where EC apoptosis is induced [16]. p75NTR is up-regulated by ischemia–reperfusion injury and decreases the expression of tight junction proteins (ZO-1, claudin-5) in EC, leading to apoptosis [27]. Furthermore, microRNA-503 produced in a p75NTR-dependent manner in EC is transferred to recipient pericytes, leading to reduced production of vascular endothelial growth factor and EC dysfunction [28]. Taken together, these observations suggest that NGFR may play a critical role in PM2.5-induced EC apoptosis and vascular dysfunction. The second up-regulated gene is adrenomedullin 2 (ADM2; also known as intermedin). The level of plasma ADM2 was higher in patients with major adverse cardiovascular events than in healthy controls and was suggested as a prognostic marker of ST-segment elevation acute myocardial infarction (STEMI) [29]. However, ADM2 exerts cardiovascular protective effects in a congestive heart failure model [17] and an ischemia/reperfusion model [30]. In addition, ADM2 ameliorates atherosclerosis in ApoE−/- mouse [31]; thus, ADM2 counters the detrimental consequences of CVD-inducing stimuli [32]. These reports suggest that EC may produce ADM2 as a counter-peptide in response to PM2.5-induced EC death. We found that nuclear protein 1 (NUPR1) was highly up-regulated. It was reported that NUPR1 deficiency resulted in the downregulation of several endoplasmic reticulum (ER) stress-regulating genes [33], suggesting the possible involvement of ER stress in GPM-induced endothelial cell dysfunction. Supportively, our Reactome pathway analysis showed that GPM regulates the ER stress-related signaling pathway (Fig. 2D). Furthermore, NUPR1 mediates methamphetamine-induced ER stress by upregulating the expression of CHOP and activates p53-p53 upregulated modulator of apoptosis (PUMA) pathway to lead to EC apoptosis [18]. In light of previous studies on PM-induced ER stress in EC [34] and keratinocyte [35], NUPR1 is likely to play an important role in ER stress–mediated EC dysfunction in response to GPM. We focused on the up-regulation of interleukin-11 (IL-11). Previously, IL-11 was identified as a critical determinant of cardiovascular fibrosis; thus, it has been proposed as a potential therapeutic target of cardiac fibrosis [36]. Specifically, fibroblast-specific IL-11 transgenic mice develop cardiac fibrosis [19]. Endogenous production of IL-11 increases in the heart of both an angiotensin II infusion mouse model and a transverse aortic constriction mouse model and when Schafer et al. applied these models to Il11ra1−/− mice, less cardiac fibrosis developed [19]. Thus, we believe that GPM-stimulated IL-11 up-regulation in EC could contribute to the incidence of heart fibrosis. Taken together, these findings indicate that up-regulation of those genes could be one mechanism by which GPM induces CVDs.
We identified TNFSF10 (tumor necrosis factor–related apoptosis-inducing ligand; TRAIL) as the first down-regulated gene. Although originally identified as a tumor necrosis–inducing cytokine, it also has a non-apoptotic function in the vascular system. Trail−/−ApoE−/− double-knockout mice develop larger and more frequent plaques than ApoE−/- mice in response to a high-fat diet [20]. In addition, TRAIL phosphorylates endothelial nitric oxide synthase at Ser1177 to produce nitric oxide, a molecule that plays an important role in vascular homeostasis [37]. These studies suggest a protective role for TRAIL, consistent with previous report describing an inverse association with the advanced coronary artery disease [38]. Based on these reports, we consider TRAIL a candidate biomarker for GPM-induced CVDs. The second down-regulated gene was growth differentiation factor 3 (GDF3). GDF3 plays a cardioprotective role by promoting macrophage differentiation into an anti-inflammatory phenotype and decreasing inflammation [21]. We found that aquaporin 1 (AQP1) was the fourth down-regulated gene. AQP1 is a water-transporting membrane protein expressed in cardiac ECs [22]. Ischemia and hypoxia stimulation down-regulates expression of AQP1 in cardiac ECs, but the exact function of this change remains unclear [22]. However, in light of a previous study showing that AQP1 transports nitric oxide from EC to vascular smooth muscle cells [39], down-regulation of AQP1 is likely to impair vascular relaxation. We observed that TIMP metallopeptidase inhibitor 3 (TIMP3) was down-regulated by GPM. Expression of TIMP3 is reduced in atherosclerotic plaques [40], and overexpression in macrophages decreases atherosclerosis [23]. In addition, TIMP3 deficiency leads to abdominal aortic aneurysms [41], suggesting that it plays a protective role against CVDs. Taken together, these observations suggest that down-regulation of these genes could be one mechanism by which GPM induces CVDs.
Surprisingly, we found that C–C motif chemokine ligand 2 (CCL2, monocyte chemoattractant protein 1; MCP1) was down-regulated. This is interesting because CCL2 recruits monocytes/macrophages and increases inflammation [42]. Consistent with our results, microarray data using HUVECs exposed to Beijing-collected PM2.5 showed down-regulation of CCL2 [26]. In addition, another study showed that in patients with coronary artery disease, both lower and higher serum levels of CCL2 are associated with all-cause and cardiovascular mortality [43]. Nevertheless, the interpretation of this result remains unclear. One possible explanation is provided by Nahrendorf et al. [44], who reported the existence of a biphasic monocyte response after a CVD-causing event such as myocardial infarction. Ly-6Chigh monocytes are initially recruited to the infarction site in response to CCL2. Those monocytes participate in the infarct wound healing process, including the digestion of infarcted tissue and removal of necrotic debris (phase 1), followed by resolution of inflammation and propagation of repair by Ly-6Clow monocytes (phase 2) [44,45]. When CCL2 is absent or present at low levels, fewer Ly-6Chigh monocytes are recruited to infarcted tissue and wound healing is not initiated, potentially resulting in severe CVD. However, more studies are required to fully explain the down-regulation of CCL2 in GPM-exposed HUVECs.
In conclusion, we identified several candidate genes and biomechanical pathways that could contribute to CVDs induced by PM2.5. Future studies should attempt to determine the precise role of up- or down-regulated genes such as NGFR, ADM2, NUPR1, IL11, TNFSF10, GDF3, APQ1 and TIMP3.
Author contribution
Inkyo Jung: Investigation, Resources. Minhan Park: Investigation, Resources. Myong-Ho Jeong: Data Curation. Kihong Park: Writing - Original Draft. Won-Ho Kim: Conceptualization. Geun-Young Kim: Conceptualization, Formal analysis, Writing - Original Draft, Writing - Review & Editing, Visualization, Supervision.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
This work was supported by an intramural research grants from the Korea National Institute of Health (2019-NI-101-01 and 2021-NI-024-00).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbrep.2021.101190.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Cohen A.J., Brauer M., Burnett R., et al. Estimates and 25-year trends of the global burden of disease attributable to ambient air pollution: an analysis of data from the Global Burden of Diseases Study 2015. Lancet. 2017;389:1907–1918. doi: 10.1016/S0140-6736(17)30505-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hamanaka R.B., Mutlu G.M. Particulate matter air pollution: effects on the cardiovascular system. Front. Endocrinol. 2018;9:680. doi: 10.3389/fendo.2018.00680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vohra K., Vodonos A., Schwartz J., et al. Global mortality from outdoor fine particle pollution generated by fossil fuel combustion: results from GEOS-Chem. Environ. Res. 2021;195:110754. doi: 10.1016/j.envres.2021.110754. [DOI] [PubMed] [Google Scholar]
- 4.Bourdrel T., Bind M.A., Bejot Y., et al. Cardiovascular effects of air pollution. Arch. Cardiovasc. Dis. 2017;110:634–642. doi: 10.1016/j.acvd.2017.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miller K.A., Siscovick D.S., Sheppard L., et al. Long-term exposure to air pollution and incidence of cardiovascular events in women. N. Engl. J. Med. 2007;356:447–458. doi: 10.1056/NEJMoa054409. [DOI] [PubMed] [Google Scholar]
- 6.Hayes R.B., Lim C., Zhang Y., et al. PM2.5 air pollution and cause-specific cardiovascular disease mortality. Int. J. Epidemiol. 2020;49:25–35. doi: 10.1093/ije/dyz114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tousoulis D., Kampoli A.M., Tentolouris C., et al. The role of nitric oxide on endothelial function. Curr. Vasc. Pharmacol. 2012;10:4–18. doi: 10.2174/157016112798829760. [DOI] [PubMed] [Google Scholar]
- 8.Le N.T., Heo K.S., Takei Y., et al. A crucial role for p90RSK-mediated reduction of ERK5 transcriptional activity in endothelial dysfunction and atherosclerosis. Circulation. 2013;127:486–499. doi: 10.1161/CIRCULATIONAHA.112.116988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cao Y., Gong Y., Liu L., et al. The use of human umbilical vein endothelial cells (HUVECs) as an in vitro model to assess the toxicity of nanoparticles to endothelium: a review. J. Appl. Toxicol. 2017;37:1359–1369. doi: 10.1002/jat.3470. [DOI] [PubMed] [Google Scholar]
- 10.Park M., Joo H.S., Lee K., et al. Differential toxicities of fine particulate matters from various sources. Sci. Rep. 2018;8:17007. doi: 10.1038/s41598-018-35398-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim G.Y., Kim H., Lim H.J., et al. Coronin 1A depletion protects endothelial cells from TNFalpha-induced apoptosis by modulating p38beta expression and activation. Cell. Signal. 2015;27:1688–1693. doi: 10.1016/j.cellsig.2015.04.012. [DOI] [PubMed] [Google Scholar]
- 12.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gentleman R.C., Carey V.J., Bates D.M., et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Subramanian A., Tamayo P., Mootha V.K., et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Caporali A., Pani E., Horrevoets A.J., et al. Neurotrophin p75 receptor (p75NTR) promotes endothelial cell apoptosis and inhibits angiogenesis: implications for diabetes-induced impaired neovascularization in ischemic limb muscles. Circ. Res. 2008;103:e15–26. doi: 10.1161/CIRCRESAHA.108.177386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hirose T., Totsune K., Mori N., et al. Increased expression of adrenomedullin 2/intermedin in rat hearts with congestive heart failure. Eur. J. Heart Fail. 2008;10:840–849. doi: 10.1016/j.ejheart.2008.06.020. [DOI] [PubMed] [Google Scholar]
- 18.Cai D., Huang E., Luo B., et al. Nupr1/Chop signal axis is involved in mitochondrion-related endothelial cell apoptosis induced by methamphetamine. Cell Death Dis. 2016;7:e2161. doi: 10.1038/cddis.2016.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schafer S., Viswanathan S., Widjaja A.A., et al. IL-11 is a crucial determinant of cardiovascular fibrosis. Nature. 2017;552:110–115. doi: 10.1038/nature24676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Di Bartolo B.A., Chan J., Bennett M.R., et al. TNF-related apoptosis-inducing ligand (TRAIL) protects against diabetes and atherosclerosis in Apoe (-)/(-) mice. Diabetologia. 2011;54:3157–3167. doi: 10.1007/s00125-011-2308-0. [DOI] [PubMed] [Google Scholar]
- 21.Wang L., Li Y., Wang X., et al. GDF3 protects mice against sepsis-induced cardiac dysfunction and mortality by suppression of macrophage pro-inflammatory phenotype. Cells. 2020;9 doi: 10.3390/cells9010120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rutkovskiy A., Bliksoen M., Hillestad V., et al. Aquaporin-1 in cardiac endothelial cells is downregulated in ischemia, hypoxia and cardioplegia. J. Mol. Cell. Cardiol. 2013;56:22–33. doi: 10.1016/j.yjmcc.2012.12.002. [DOI] [PubMed] [Google Scholar]
- 23.Casagrande V., Menghini R., Menini S., et al. Overexpression of tissue inhibitor of metalloproteinase 3 in macrophages reduces atherosclerosis in low-density lipoprotein receptor knockout mice. Arterioscler. Thromb. Vasc. Biol. 2012;32:74–81. doi: 10.1161/ATVBAHA.111.238402. [DOI] [PubMed] [Google Scholar]
- 24.Zhou Z., Liu Y., Duan F., et al. Transcriptomic analyses of the biological effects of airborne PM2.5 exposure on human bronchial epithelial cells. PLoS One. 2015;10 doi: 10.1371/journal.pone.0138267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim H.J., Bae I.H., Son E.D., et al. Transcriptome analysis of airborne PM2.5-induced detrimental effects on human keratinocytes. Toxicol. Lett. 2017;273:26–35. doi: 10.1016/j.toxlet.2017.03.010. [DOI] [PubMed] [Google Scholar]
- 26.Hu H., Asweto C.O., Wu J., et al. Gene expression profiles and bioinformatics analysis of human umbilical vein endothelial cells exposed to PM2.5. Chemosphere. 2017;183:589–598. doi: 10.1016/j.chemosphere.2017.05.153. [DOI] [PubMed] [Google Scholar]
- 27.Kuo M.H., Lee H.F., Tu Y.F., et al. Astaxanthin ameliorates ischemic-hypoxic-induced neurotrophin receptor p75 upregulation in the endothelial cells of neonatal mouse brains. Int. J. Mol. Sci. 2019;20 doi: 10.3390/ijms20246168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Caporali A., Meloni M., Nailor A., et al. p75(NTR)-dependent activation of NF-kappaB regulates microRNA-503 transcription and pericyte-endothelial crosstalk in diabetes after limb ischaemia. Nat. Commun. 2015;6:8024. doi: 10.1038/ncomms9024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang B., Zhong Z., Shen H.W., et al. Intermedin as a prognostic factor for major adverse cardiovascular events in patients with ST-segment elevation acute myocardial infarction. Peptides. 2014;58:98–102. doi: 10.1016/j.peptides.2014.06.009. [DOI] [PubMed] [Google Scholar]
- 30.Zhang H.Y., Jiang W., Liu J.Y., et al. Intermedin is upregulated and has protective roles in a mouse ischemia/reperfusion model. Hypertens. Res. 2009;32:861–868. doi: 10.1038/hr.2009.120. [DOI] [PubMed] [Google Scholar]
- 31.Zhang X., Gu L., Chen X., et al. Intermedin ameliorates atherosclerosis in ApoE null mice by modifying lipid profiles. Peptides. 2012;37:189–193. doi: 10.1016/j.peptides.2012.07.011. [DOI] [PubMed] [Google Scholar]
- 32.Bell D., McDermott B.J. Intermedin (adrenomedullin-2): a novel counter-regulatory peptide in the cardiovascular and renal systems. Br. J. Pharmacol. 2008;153(Suppl 1):S247–S262. doi: 10.1038/sj.bjp.0707494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Santofimia-Castano P., Lan W., Bintz J., et al. Inactivation of NUPR1 promotes cell death by coupling ER-stress responses with necrosis. Sci. Rep. 2018;8:16999. doi: 10.1038/s41598-018-35020-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Y., Tang M. PM2.5 induces autophagy and apoptosis through endoplasmic reticulum stress in human endothelial cells. Sci. Total Environ. 2020;710:136397. doi: 10.1016/j.scitotenv.2019.136397. [DOI] [PubMed] [Google Scholar]
- 35.Piao M.J., Ahn M.J., Kang K.A., et al. Particulate matter 2.5 damages skin cells by inducing oxidative stress, subcellular organelle dysfunction, and apoptosis. Arch. Toxicol. 2018;92:2077–2091. doi: 10.1007/s00204-018-2197-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fernandez-Ruiz I. Cardioprotection: IL-11 is a potential therapeutic target in cardiovascular fibrosis. Nat. Rev. Cardiol. 2018;15:1. doi: 10.1038/nrcardio.2017.197. [DOI] [PubMed] [Google Scholar]
- 37.Zauli G., Pandolfi A., Gonelli A., et al. Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) sequentially upregulates nitric oxide and prostanoid production in primary human endothelial cells. Circ. Res. 2003;92:732–740. doi: 10.1161/01.RES.0000067928.83455.9C. [DOI] [PubMed] [Google Scholar]
- 38.Ajala O., Zhang Y., Gupta A., et al. Decreased serum TRAIL is associated with increased mortality in smokers with comorbid emphysema and coronary artery disease. Respir. Med. 2018;145:21–27. doi: 10.1016/j.rmed.2018.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Herrera M., Hong N.J., Garvin J.L. Aquaporin-1 transports NO across cell membranes. Hypertension. 2006;48:157–164. doi: 10.1161/01.HYP.0000223652.29338.77. [DOI] [PubMed] [Google Scholar]
- 40.Cardellini M., Menghini R., Martelli E., et al. TIMP3 is reduced in atherosclerotic plaques from subjects with type 2 diabetes and increased by SirT1. Diabetes. 2009;58:2396–2401. doi: 10.2337/db09-0280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Basu R., Fan D., Kandalam V., et al. Loss of Timp3 gene leads to abdominal aortic aneurysm formation in response to angiotensin II. J. Biol. Chem. 2012;287:44083–44096. doi: 10.1074/jbc.M112.425652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ohman M.K., Wright A.P., Wickenheiser K.J., et al. Monocyte chemoattractant protein-1 deficiency protects against visceral fat-induced atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 2010;30:1151–1158. doi: 10.1161/ATVBAHA.110.205914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ding D., Su D., Li X., et al. Serum levels of monocyte chemoattractant protein-1 and all-cause and cardiovascular mortality among patients with coronary artery disease. PLoS One. 2015;10 doi: 10.1371/journal.pone.0120633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nahrendorf M., Pittet M.J., Swirski F.K. Monocytes: protagonists of infarct inflammation and repair after myocardial infarction. Circulation. 2010;121:2437–2445. doi: 10.1161/CIRCULATIONAHA.109.916346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Prabhu S.D., Frangogiannis N.G. The biological basis for cardiac repair after myocardial infarction: from inflammation to fibrosis. Circ. Res. 2016;119:91–112. doi: 10.1161/CIRCRESAHA.116.303577. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.