Abstract
Genomic fingerprints from 92 capsulated and noncapsulated strains of Haemophilus influenzae from Mexican children with different diseases and healthy carriers were generated by PCR using the enterobacterial repetitive intergenic consensus (ERIC) sequences. A cluster analysis by the unweighted pair-group method with arithmetic averages based on the overall similarity as estimated from the characteristics of the genomic fingerprints, was conducted to group the strains. A total of 69 fingerprint patterns were detected in the H. influenzae strains. Isolates from patients with different diseases were represented by a variety of patterns, which clustered into two major groups. Of the 37 strains isolated from cases of meningitis, 24 shared patterns and were clustered into five groups within a similarity level of 1.0. One fragment of 1.25 kb was common to all meningitis strains. H. influenzae strains from healthy carriers presented fingerprint patterns different from those found in strains from sick children. Isolates from healthy individuals were more variable and were distributed differently from those from patients. The results show that ERIC-PCR provides a powerful tool for the determination of the distinctive pathogenicity potentials of H. influenzae strains and encourage its use for molecular epidemiology investigations.
Haemophilus influenzae is an important cause of human disease worldwide, with serotype b (Hib) capsulated strains causing invasive bacteremic infections such as meningitis, epiglottitis, septicemia, and septic arthritis, particularly in infants. Strains lacking a capsule (HiNT) are well established as etiologic agents of otitis media and lower respiratory tract infections in children and account for millions of deaths among children in developing countries (20). The availability of Hib conjugate vaccines has dramatically reduced the incidence of invasive disease in Western Europe and North America (2, 5, 30). As the incidence of Hib decreases, focus in this public health problem has turned to other capsular types (a and c to f) and noncapsulate strains (8, 36).
H. influenzae strains have been traditionally classified by determination of the biotype and capsular serotype. Such methods are subject to phenotypic variations and do not give information on the clonal origin of the strains. More discriminatory methods, such as outer membrane protein analysis, lipopolysaccharide profiles, and multilocus enzyme electrophoresis, have been used to study the epidemiology and pathogenesis of H. influenzae infections (1, 11, 14, 23). Studies of the pathobiology of Haemophilus indicate that marked differences in virulence occur among strains (31). However, associations between specific diseases and virulence determinants are sometimes difficult to establish.
Genome variation of H. influenzae has been evaluated by applying several molecular biology techniques, including analysis of DNA restriction fragment length polymorphisms, PCR with arbitrary primers (randomly amplified polymorphic DNA), and rRNA gene restriction patterns (4, 28). The last method has a low discriminatory capability, and the first two give results in complex patterns. Binary data output and computer-assisted cluster analysis are of additional value.
Interspersed repetitive DNA sequences have been described for eubacteria. In 1992, de Bruijn (7) examined the distribution of the enterobacterial repetitive intergenic consensus (ERIC) sequences in the genomes of a number of gram-negative isolates. ERIC sequences are highly conserved at the nucleotide sequence level, but their chromosomal locations differ between species or strains (7). ERIC sequences are 126 bp long and appear to be restricted to transcribed regions of the genome, either in intergenic regions of polycistronic operons or in untranslated regions upstream or downstream of open reading frames (18). These elements have been successfully used for molecular typing purposes. By use of PCR differences in band sizes which represent polymorphisms in the distances between repetitive elements of different genomes, ERIC-PCR allows for the identification of interstrain genotypic diversity and has the potential to differentiate pathovars (17, 18, 34, 35).
In H. influenzae and other bacterial species, some of the genes encoding pathogenicity determinants have been shown to contain contiguous repetitive DNA that appeared to be related to adaptive virulence (12, 13, 25, 33).
This study reports an analysis of ERIC variability distribution in genomic DNAs from H. influenzae strains isolated from sick Mexican children and from healthy children who were carriers. The aim of the study was to ascertain whether restricted variability among isolates was related to the clinical origin. The data on levels of similarity shown here reflect the extent of variability among H. influenzae strains. The results revealed little variability among clinical H. influenzae strains, providing additional evidence of clonality. The full richness of diversity of nontypeable H. influenzae is discussed in relation to some possible genetic mechanisms.
MATERIALS AND METHODS
Bacterial strains.
This study was based on a collection of 92 H. influenzae strains isolated from Mexican children between 1990 and 1997. These included 24 nontypeable and 48 serotype b strains recovered during episodes of different diseases. A further 20 nontypeable strains which had been isolated from throat swabs obtained from healthy child carriers were also included. Clinical isolates were part of collections from the Instituto Nacional de Diagnóstico, Referencia y Estudios Epidemiológicos (INDRE), and the Hospital Infantil de México “Federico Gómez” (HIM), both located in Mexico City. With the exception of seven strains which were obtained from children in the Mexican states of Michoacan and Morelos, all strains were recovered in the metropolitan area of Mexico City.
The specific characteristics of the isolates are given in Table 1. Strains were assigned to biotypes on the basis of standard biochemical assays of indole, urease, and ornithine decarboxylase production. Nontypeable H. influenzae strains were identified by their lack of reaction with monovalent antisera against the capsular antigens a, b, c, d, e, and f, the typeable b character was determined by agglutination with monovalent antiserum against b capsule (Difco Laboratories, Detroit, Mich.). Enteropathogenic Escherichia coli strain E234869 (serotype 0127:H6) was used as a reference strain for positive ERIC sequences.
TABLE 1.
Characteristics of H. influenzae isolates analyzed in the study
| Epa | Isolateb
|
Presencec of fragments with the following sizes (kb):
|
Sourced | Serotype | Biotype | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Name | No. | 2.11 | 1.75 | 1.5 | 1.25 | 1.1 | 0.97 | 0.84 | 0.72 | 0.65 | 0.59 | 0.51 | 0.45 | 0.43 | ||||
| 1 | In 30 | 1 | – | – | – | – | – | – | – | CSF | b | IV | ||||||
| In 19 | 14 | – | – | – | – | – | – | – | CSF1 | b | VI | |||||||
| In 29 | 23 | – | – | – | – | – | – | – | CSF2 | b | IV | |||||||
| Lc144 | 78 | – | – | – | – | – | – | – | CSF | b | VI | |||||||
| Lc175 | 79 | – | – | – | – | – | – | – | CSF2 | b | IV | |||||||
| 2 | In 8 | 6 | – | – | – | – | – | – | – | – | CSF | b | IV | |||||
| In 26 | 21 | – | – | – | – | – | – | – | – | CSF | b | IV | ||||||
| Gb392 | 80 | – | – | – | – | – | – | – | – | SAR | b | IV | ||||||
| Gb638 | 81 | – | – | – | – | – | – | – | – | BS | b | IV | ||||||
| 3 | In 10 | 8 | – | – | – | – | – | – | – | – | – | BS | NTe | II | ||||
| LC755 | 71 | – | – | – | – | – | – | – | – | – | CSF1 | b | VI | |||||
| LC849 | 72 | – | – | – | – | – | – | – | – | – | CSF | b | IV | |||||
| LC795 | 73 | – | – | – | – | – | – | – | – | – | CSF | b | IV | |||||
| 4 | In 18 | 13 | – | – | – | – | – | – | – | – | CSF1 | b | IV | |||||
| In 23 | 18 | – | – | – | – | – | – | – | – | CSF | b | VI | ||||||
| Le 8 | 53 | – | – | – | – | – | – | – | – | SAR | b | IV | ||||||
| Le 16 | 58 | – | – | – | – | – | – | – | – | CSF | b | IV | ||||||
| Gb567 | 83 | – | – | – | – | – | – | – | – | CSF | b | VI | ||||||
| Gb513 | 74 | – | – | – | – | – | – | – | – | CSF | b | VI | ||||||
| 5 | In 21 | 16 | – | – | – | – | – | – | – | – | – | CSF | b | IV | ||||
| In 27 | 22 | – | – | – | – | – | – | – | – | – | CSF2 | b | VI | |||||
| Le 1 | 45 | – | – | – | – | – | – | – | – | – | CSF | b | IV | |||||
| Le 2 | 46 | – | – | – | – | – | – | – | – | – | B S | NT | VI | |||||
| LC323 | 70 | – | – | – | – | – | – | – | – | – | CSF | b | VI | |||||
| 6 | In 9 | 7 | – | – | – | – | – | – | – | NF | NT | IV | ||||||
| 7 | Le34 | 69 | – | – | – | – | – | – | – | – | – | – | NF | NT | IV | |||
| 8 | Hinf | 29 | – | – | – | – | – | – | – | CSF | b | I | ||||||
| 9 | Hinf6 | 30 | – | – | – | – | – | – | – | – | CSF | b | VI | |||||
| 10 | Nez 6 | 85 | – | – | – | – | – | – | – | AC | NT | IV | ||||||
| 11 | 646 | 86 | – | – | – | – | – | – | – | – | CSF | b | IV | |||||
| 12 | In 7 | 5 | – | – | – | – | – | – | – | CSF | b | I | ||||||
| 13 | GO | 27 | – | – | – | – | – | – | – | BS | b | IV | ||||||
| 14 | LC132 | 26 | – | – | – | – | – | – | – | – | CSF | b | IV | |||||
| 15 | 140gb | 82 | – | – | – | – | – | – | – | CSF2 | b | IV | ||||||
| 16 | 08ii | 35 | – | – | – | – | – | – | – | AC | NT | IV | ||||||
| 17 | Le 19 | 61 | – | – | – | – | – | – | – | – | NF | NT | I | |||||
| 18 | In 22 | 17 | – | – | – | – | – | – | – | BS | b | II | ||||||
| 19 | Le 26 | 65 | – | – | – | – | – | – | BS | NT | V | |||||||
| 20 | Le 11 | 56 | – | – | – | – | – | – | SP | NT | IV | |||||||
| 21 | In 2 | 24 | – | – | – | – | – | – | – | – | – | SP | NT | IV | ||||
| 22 | Le 27 | 66 | – | – | – | – | – | – | – | – | – | GT | NT | VI | ||||
| 23 | Le 4 | 48 | – | – | – | – | – | – | – | CSF | b | VI | ||||||
| 24 | LC390 | 76 | – | – | – | – | – | – | CSF | b | IV | |||||||
| 25 | LC861 | 75 | – | – | – | – | – | – | – | – | – | CSF | b | IV | ||||
| 26 | In 3 | 2 | – | – | – | – | – | – | – | – | – | NF | NT | IV | ||||
| 27 | Le 18 | 60 | – | – | – | – | – | – | – | NF | NT | I | ||||||
| 28 | In 25 | 20 | – | – | – | – | – | – | – | – | – | – | BS | b | VI | |||
| 29 | LE 22 | 64 | – | – | – | – | – | – | – | – | ME | NT | IV | |||||
| 30 | In 6 | 4 | – | – | – | – | – | – | – | CSF | b | IV | ||||||
| 31 | 15II | 10 | – | – | – | – | – | – | – | – | AC | NT | I | |||||
| 32 | 39ii | 43 | – | – | – | – | – | – | AC | NT | I | |||||||
| 33 | Le 33 | 77 | – | – | – | – | – | NF | NT | II | ||||||||
| 34 | C7196 | 36 | – | – | – | – | – | – | – | – | BS | NT | III | |||||
| 35 | Le13 | 47 | – | – | – | – | – | – | BS | NT | IV | |||||||
| 36 | In 16 | 11 | – | – | – | – | – | – | – | – | CSF | b | II | |||||
| In 17 | 12 | – | – | – | – | – | – | – | – | BS | b | IV | ||||||
| In 32 | 25 | – | – | – | – | – | – | – | – | CSF | b | IV | ||||||
| 37 | In 20 | 15 | – | – | – | – | – | – | – | – | – | CSF | b | IV | ||||
| 38 | Le 6 | 50 | – | – | – | – | – | – | – | – | EY | NT | IV | |||||
| 39 | Le 20 | 62 | – | – | – | – | – | – | – | – | NF | b | II | |||||
| 40 | In 24 | 19 | – | – | – | – | – | – | – | – | – | – | CSF | b | I | |||
| 41 | Le 10 | 51 | – | – | – | – | – | – | – | – | BS | NT | IV | |||||
| 42 | Le 32 | 68 | – | – | – | – | – | – | – | ME | NT | I | ||||||
| 43 | In 5 | 3 | – | – | – | – | – | SP | NT | IV | ||||||||
| 44 | Le 17 | 59 | – | – | – | – | – | NF | b | V | ||||||||
| 45 | Le | 55 | – | – | – | – | – | CSF | b | I | ||||||||
| 46 | In 11 | 9 | – | – | – | – | – | BS | b | III | ||||||||
| 47 | UAM | 31 | – | – | – | – | CSF | b | I | |||||||||
| Le 7 | 32 | – | – | – | – | SAR | NT | IV | ||||||||||
| 48 | H2 | 28 | – | – | – | CSF | b | II | ||||||||||
| 49 | 13ii | 39 | – | – | – | – | – | AC | NT | V | ||||||||
| 50 | Le 5 | 49 | – | – | – | – | NF | NT | IV | |||||||||
| 51 | 06ii | 33 | – | – | – | – | AC | NT | VI | |||||||||
| 52 | 07ii | 34 | – | – | – | – | – | AC | NT | I | ||||||||
| 53 | Nez 8 | 84 | – | – | – | – | – | – | AC | NT | VI | |||||||
| 54 | 12ii | 38 | – | – | – | – | AC | NT | I | |||||||||
| 55 | 19ii | 41 | – | – | – | AC | NT | IV | ||||||||||
| 56 | Nez15 | 92 | – | – | – | – | AC | NT | IV | |||||||||
| 57 | Nez12 | 88 | – | – | – | – | – | AC | NT | IV | ||||||||
| 58 | Nez13 | 90 | – | – | – | AC | NT | IV | ||||||||||
| 59 | 14ii | 37 | – | – | – | – | AC | NT | IV | |||||||||
| In 12 | 40 | – | – | – | BS | b | IV | |||||||||||
| 60 | 49ii | 44 | – | – | – | AC | NT | IV | ||||||||||
| 61 | 01ii | 52 | – | – | – | – | AC | NT | IV | |||||||||
| 62 | 18ii | 42 | – | – | – | – | AC | NT | I | |||||||||
| 63 | Nez10 | 87 | – | – | AC | NT | IV | |||||||||||
| 64 | Le 9 | 54 | – | – | – | – | – | – | BS | NT | IV | |||||||
| 65 | Le 21 | 63 | – | – | – | – | – | – | NF | NT | I | |||||||
| 66 | Nez14 | 91 | – | – | – | – | AC | NT | IV | |||||||||
| 67 | Le 15 | 57 | – | – | – | – | – | – | – | – | BS | NT | III | |||||
| 68 | Nez11 | 89 | – | – | – | – | – | – | – | AC | NT | IV | ||||||
| 69 | Le 31 | 67 | – | – | – | – | – | ME | NT | V | ||||||||
Eps are arbitrarily numbered as per the phenogram in Fig. 1.
Clinical strains and strains from healthy child carriers shipped to our institution from the INDRE and HIM, SSA, Mexico City, Mexico. Isolates are numbered arbitrarily as in the phenogram.
–, fragment present.
CSF, (meningitis); SAR, supurative arthritis; BS, bronchial secretion; SP, sputum; NF, nasopharynx; GT, genital tract; ME, middle ear; EY, eye; AC, asymptomatic carrier; CSF1 and CSF2, CSF (meningitis) strains from the Mexican states of Michoacan and Morelos, respectively.
DNA extraction.
All Haemophilus strains were incubated overnight at 37°C on brain heart infusion agar plates supplemented with 2.5% Fildes fluid (Difco Laboratories). For DNA extraction, bacterial growth was scraped from the plates and suspended in 1 ml of phosphate-buffered saline (pH 7.0). DNA was isolated from this bacterial suspension using the guanidium isothiocyanate protocol as described by Boom et al. (3). DNA extracts were suspended in 100 μl of Tris-EDTA (10 mM Tris-HCl, 0.01 mM EDTA [pH 8.0]). The concentration was adjusted fluorometrically at excitation and emission wavelengths of 365 and 460 nm, respectively, using the Hoechst 33258 dye and a DyNA Quant 200-115 v minifluorometer (Hoefer Pharmacia Biotech Inc., San Francisco, Calif.). Aliquots were stored at 4°C for further analysis.
ERIC-PCR and amplification conditions.
The ERIC-PCR was optimized for template, deoxyribonucleoside triphosphates, magnesium ion concentrations, and primers by a modified Taguchi method, based on orthogonal arrays as described by Cobb and Clarkson (6). The optimized amplification reactions were performed in 50-μl volumes containing 10 mM Tris-HCl (pH 9.0), 50 mM KCl, and 2.5 mM MgCl (Sigma Chemical Co., St. Louis, Mo.). Each reaction mixture included 35 pmol of each primer (ERIC 1 and ERIC 2; 3′CACTTAGGGGTCCTCGAATGTA5′ and 5′AAGTAAGTGACTGGGGTGAGCG3′, respectively) (Lakeside, Mannheim Boeringer Biochemicals, San Francisco, Calif.), deoxyribonucleoside triphosphates to a final concentration of 0.2 mM each (Ultrapure dNTP Set; Pharmacia, Biotech, Piscataway, N.J.), 50 ng of DNA sample, and 0.5 U of Taq DNA polymerase (Gibco, BRL Life Technologies, Inc., Gaithersburg, Md.). A negative control, consisting of the same reaction mixture but with no DNA template, was included in each amplification procedure. E. coli genomic DNA obtained using the same extraction protocol was used as a positive control. The PCR was initiated with a 5-min denaturation period at 94°C, followed by 35 cycles of denaturation (1 min at 94°C), annealing (1 min at 40°C), and enzymatic chain extension (4 min at 74°C) with a final extension at 74°C for 10 min. Amplifications were performed in an automated thermal cycler (DNA Thermal Cycler 480; Perkin-Elmer).
Electrophoretic patterns (Eps).
An 8-μl portion of each amplified PCR product was loaded in gel slots and separated by electrophoresis at 10°C and 5 V/cm for 6 to 7 h on 1.4% agarose gels (Ultrapure; Gibco, BRL Life Technologies, Inc.) of 24 by 20 cm in 1× TBE buffer (10 mM Tris base, 50 mM boric acid, 2 mM EDTA [pH 8.0]). Gels were stained with 0.5 μg of ethidium bromide per ml in 1× TBE for 20 to 35 min and were visualized and photographed under UV transillumination for 45 to 60 s on Polaroid type 55 film.
To assess whether reproducible banding patterns were generated, a protocol involving eight selected isolates was tested. Fingerprints generated from independent DNA preparations extracted from 10 colonies of the same isolate were run side by side on an agarose gel. This procedure was repeated three times over three 3-month periods.
Cluster analysis and statistics.
The cluster analysis of the 92 strains was conducted on the basis of the characteristics of the fingerprints generated. Based on the data for presence or absence of 13 different DNA fragments in the fingerprints of the 92 strains of H. influenzae, a binary data matrix was created. Overall similarity between pairs of strains was calculated from the binary data matrix using the simple matching coefficient (29). The resulting similarity matrix was used as the input data for a cluster analysis by the unweighted pair-group method with arithmetic averages (UPGMA) (27). The goodness of the clustering method was assessed by calculating the cophenetic correlation coefficient (r). The Numerical Taxonomy and Multivariate Analysis System version 2.0 was used to carry out these analyses (26). The comparison of the distributions in the phenogram of Eps associated with groups of diseases and carriers was analyzed by the Wilcoxon sum rank test.
In appropriate cases, the chi-square test with Yates correction or the two-tailed Fisher exact test was done by using the EpiInfo software, version 6.04.
RESULTS
General characteristics and diversity of the Eps.
PCR with primers ERIC I and ERIC II produced multiple fragments of DNA in sizes ranging between 0.43 and 2.11 kb. A total of 69 Eps were found among the 92 H. influenzae strains studied. Only eight of the Eps were found more than once, indicating a high variability among the H. influenzae strains. Sixty-one of the 69 Eps (88%) were represented by a single strain. Table 1 shows the data obtained in terms of Ep, assigned reference number, fragment sizes, anatomic source, serotype, and biotype for each analyzed strain.
In a few cases, Eps were shared between isolates recovered from different disease types; for example, patterns Ep 2, Ep 3, Ep 4, Ep 5, Ep 36, and Ep 47 were found in isolates recovered from cerebrospinal fluid (CSF) and occasionally were also found in strains from suppurative arthritis and bronchial secretions. Most of the identical patterns were typically shared among strains from patients with meningitis. The 48 serotype b pathogenic strains were represented by 29 of the 69 Eps obtained. The 40 remaining patterns were found in strains that failed to react with antisera for all capsular H. influenzae polysaccharides. None of the noncapsulated isolates from carriers or sick children shared patterns, and these were significantly more variable than the capsule b strains isolated from disease cases. The only pattern shared between isolates from a particular disease episode (pneumonia) and an asymptomatic carrier was Ep 59 (Table 1).
With the exception of strains associated with meningitis, there were no specific bands related to particular clinical entities. Patterns Ep 1, Ep 2, Ep 3, Ep 4, and Ep 5 were typically shared between isolates recovered from CSF (19 of 37) and showed the presence of seven analogous fragments of 1.75, 1.5, 1.25, 1.1, 0.97, 0.72, and 0.59 kb (Table 2). One common band of 1.25 kb was found in all (37 of 37) meningitis strains examined. Of strains from this clinical condition, 85% exhibited one fragment of 1.5 kb, which was found in only 37 and 45% of isolates from other disease types (P < 0.0001) and carriers (P < 0.0001), respectively. In comparison with Eps of healthy carriers, meningitis and other disease isolates had significant differences (P < 0.05) in the frequencies of bands of 1.75, 1.25, 0.84, 0.72, 0.65, and 0.59 kb. There were no observations of any common fragments appearing in all H. influenzae strains studied (Tables 1 and 2).
TABLE 2.
Distribution and frequency of amplification products among strains of H. influenzae determined by ERIC-PCR
| Fragment size (kb) | No. (%) of strains from:
|
|||
|---|---|---|---|---|
| Hib meningitisa | Meningitisb | Other disease typesc | Carriers | |
| 2.1 | 5 (26) | 12 (32) | 8 (23) | 8 (40) |
| 1.75 | 19 (100) | 34 (92) | 30 (86) | 13 (65) |
| 1.5 | 19 (100) | 31 (85) | 13 (37) | 9 (45) |
| 1.25 | 19 (100) | 37 (100) | 27 (77) | 11 (55) |
| 1.1 | 19 (100) | 33 (89) | 31 (88) | 16 (80) |
| 0.97 | 19 (100) | 33 (89) | 26 (74) | 18 (90) |
| 0.84 | 12 (60) | 24 (65) | 26 (74) | 7 (35) |
| 0.72 | 19 (100) | 32 (87) | 27 (77) | 5 (25) |
| 0.65 | 0 | 8 (22) | 13 (37) | 2 (10) |
| 0.59 | 19 (100) | 27 (73) | 19 (54) | 3 (15) |
| 0.51 | 4 (22) | 11 (30) | 11 (28) | 2 (10) |
| 0.45 | 0 | 1 (3) | 9 (26) | 1 (5) |
| 0.43 | 0 | 0 | 4 (11) | 1 (5) |
Hib strains recovered from patients with meningitis which shared Eps and were distributed within Eps 1 to 5 (see Table 1).
Total meningitis strains.
Hib and noncapsulated isolates from clinical origins other than meningitis.
Eighteen of the 48 biotype IV strains (37.5%) shared ERIC-PCR genotypes.
The reproducibility of the ERIC-PCR fingerprinting method was examined. Eight strains that yielded some of the most complex fingerprint patterns were selected for this purpose. Smaller, unstable fragments tended to appear in the DNA banding patterns when a constant concentration of genomic DNA (50 ng) was used with increasing primer concentrations. It is likely that the higher the primer concentration, the more primers may anneal to less-specific target sequences, causing smaller “ghost” PCR fragments to be generated. The best stability of bands in the DNA fingerprints was achieved with 50 ng of DNA and a total amount of 70 pmol of primers (35 pmol each of ERIC I and ERIC II). Identical resulting bands were obtained when 10 colonies of the same strain were tested at different times (data not shown).
Relationships among genotypes.
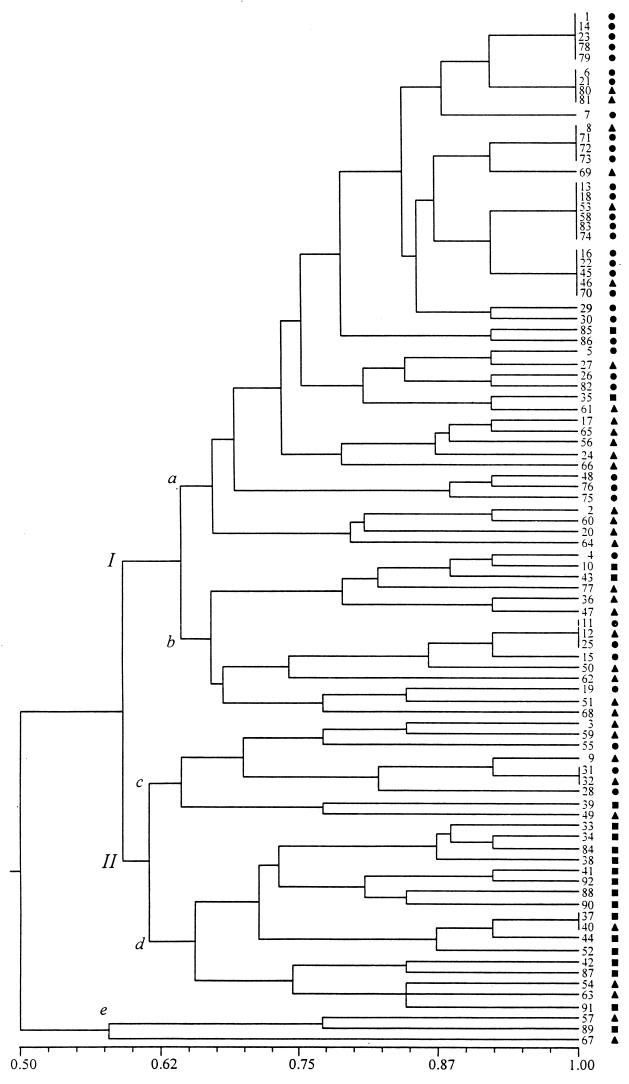
The phenogram shown in Fig. 1 represents the similarity relationships between the 69 Eps found. As can be seen from this phenogram, two major groups were formed at a similarity level of 0.59. They are identified in Fig. 1 as I and II. Two outliers were discriminated from the rest of the strains at the lowest value of similarity, namely, 0.57. Cluster I includes two smaller clusters, a and b, which are formed at a level of similarity of 0.64. Cluster II also includes two groups, d and c, which were formed at a similarity level of 0.61. Forty-eight strains were found within the large cluster, designated subgroup Ia. Interestingly, 95% of these (46 of 48) were Hib and HiNT strains isolated from sick children. This was the subgroup in which the most similar pairs of strains were found; many of them were grouped into five clusters, each one including between four and six strains which were identical (they grouped at a similarity level of 1.0). Seventy-nine percent (19 of 24) of the strains distributed within these five clusters were isolated from patients with meningitis.
FIG. 1.
Similarity relationships between 92 H. influenzae isolates. The phenogram of fingerprints was generated from a simple-matching similarity matrix by means of the unweighted pair-group average clustering method. A similarity of 1.0 indicates 100% identity between strains. The cophenetic correlation coefficient (r) was 7.33. ●, Hib strains recovered from meningitis; ▴, Hib and noncapsulated isolates from diseases other than meningitis; ■, noncapsulated isolates from healthy carriers.
Eighty-two percent of clinical strains (59 of 72) were grouped into subgroups Ia and Ib at moderate and high similarity levels, respectively (Table 3).
TABLE 3.
Number and frequency of isolates of H. influenzae from various diseases in groups I and II of the phenogram shown in Fig. 1
| Anatomic site of culture | Group Ia
|
Group IIb
|
|||||
|---|---|---|---|---|---|---|---|
| No. in subgroup:
|
% | No. in subgroup:
|
% | ||||
| a | b | c | d | e | |||
| CSF (meningitis) | 29 | 5 | 92 | 3 | 0 | 0 | 8 |
| Bronchial secretionc | 7 | 4 | 73 | 1 | 2 | 1 | 27 |
| Nasopharynx | 4 | 2 | 66 | 2 | 1 | 0 | 34 |
| Sputumd | 2 | 0 | 1 | 0 | 0 | ||
| Middle ear | 1 | 1 | 0 | 0 | 1 | ||
| Conjunctiva | 0 | 1 | 0 | 0 | 0 | ||
| Joint | 2 | 0 | 1 | 0 | 0 | ||
| Genital tract | 1 | 0 | 0 | 0 | 0 | ||
| Total | 46 | 13 | 82 | 8 | 3 | 2 | 18 |
Strains clustered in subgroups a and b at moderate to high similarity levels.
Subgroups c and d comprised most of the distinct genotypes, and subgroup e strains were very poorly related to other isolates.
Specimens were obtained from bronchial secretions recovered by bronchial aspiration.
Sputa were expectorated under the supervision of a physiotherapist.
In contrast to clinical isolates of Hib and HiNT, most of the noncapsulated H. influenzae carrier isolates did not group together, with 80% (16 of 20) being clustered throughout group II (Table 4). Clusters within group II had a tendency to distribute at the lower simmilarity values, indicating that they comprised the most genotypically different isolates by ERIC-PCR (Fig. 1).
TABLE 4.
Number and frequency of Hib and noncapsulated isolates in groups I and II of the phenogram
The goodness of the clustering method was quantitied by calculating the cophenetic correlation coefficient (r = 7.33). In addition, the distribution in the phenogram of isolates from various clinical conditions and carriers was statistically analyzed; differences between groups were assessed by use of the two-tailed Wilcoxon ranking test. Carrier isolates differed significantly from meningitis (CSF) isolates (P < 0.0001) and other clinical isolates (P < 0.001). Thus, in addition to the fact that no Eps were shared between carrier isolates and clinical isolates, the ranking of carrier isolates is significantly different from that of clinical isolates.
DISCUSSION
This study is the first practical application of ERIC-PCR and clustering analysis to assess the variability between clinical Hib isolates from children. Clinical Hib strains were included in this study due to their interest from any perspective, since serotype b is very seldom found in North America and Western Europe following the introduction of conjugate vaccines into the Expanded Programme of Immunization.
Noncapsulated strains of H. influenzae from cases of chronic obstructive pulmonary disease and cystic fibrosis were analyzed previously by van Belkum et al. (32) using ERIC-PCR, with the data revealing marked differences between isolates. Those results are positively correlated with the data in this study, which also shows large genetic differences at this DNA level between most of the nontypeable isolates. Differences in the molecular sizes ranging from 430 to 2,110 bp in the present study (using a total primer amount of 70 pmol), in comparison with those previously published by van Belkum et al., where fragments of 100 to 2,000 bp (using a total primer amount of 100 pmol) were obtained, could be due to differences in technical procedures (see Results).
The results of the present study show that some H. influenzae strains grouped according to the type of infection (meningitis strains from group Ia versus other diseases isolates from groups Ia and Ib) and, to some extent, the typeable b character (Tables 3 and 4). By using ERIC-PCR, many of the type b strains causing meningitis, isolated from unrelated hosts, were shown to be identical. The clone concept postulates that bacterial populations are arrays of lineages that maintain their genetic identity for extended periods of time (24). Bacteria of the same (or nearly the same) genotype can be isolated from samples from different geographic regions or at different times. Our data showed the recovery of Ep 1 to Ep 5 in the Mexico City area and the occurrence of Ep 1, Ep 3, Ep 4, and Ep 5 in two different Mexican states (Michoacan and Morelos). Although a reduced number of strains isolated in localities other than the Mexico City area were included, these strains were recovered during a 7-year period. The observations for this data set agree with the hypothesis of clonal persistence for Hib strains, in concordance with other lines of evidence obtained by using multilocus enzyme electrophoresis (22, 23). Examination by ERIC-PCR of additional isolates from disease episodes on different continents or intracontinentaly and for more than 7 years will provide additional insights.
Musser et al. (23) reported that type b strains of electrophoretic types (ETs) 12.5 and 12.8, commonly associated with outer membrane protein (OMP) subtype 1, and ET 21.8, which occurred in 83% of strains of OMP subtype 1c, were nonranodmly associated with different types of invasive disease. Strains of ET 21.8 OMP subtype 1c caused proportionally more meningitis and less of other invasive disease types than did those of ET 12.5 and ET 12.8. This observation was interpreted as showing a true difference in the virulence of isolates expressing ET 21.8 OMP subtype 1c. Considering that properties analyzed by multilocus enzyme electrophoresis, OMP typing, and ERIC-PCR tend to be distributed in restricted sets of strains, we hypothesized that ET 21.8 OMP subtype 1c are marking clones especially successful in invading the CSF. However, the high occurrence of Ep 1 to Ep 5 in strains recovered from cases of meningitis does not mean that these, per se, have a role in virulence or organotropism.
Capsule-deficient mutants of type b strains arise with high frequency. It would be interesting to see where clinical capsulated strains other than b strains and capsule-deficient mutants fall in the phenogram. A small number of nontypeable isolates expressed the same Ep as type b strains. The analyzed nontypeable isolates may represent spontaneous b mutants.
In clonal populations, such as those of Hib, strong nonrandom associations between genetic loci (linkage disequilibrium) are generated (15, 22, 23). The results obtained for Hib meningitis strains are probably showing some linkage disequilibrium between some virulence factors and ERIC elements. Further theoretical and experimental bases are necessary to determine whether meningitis isolates present specific characteristics related to ERIC motifs and if these in turn may be recognized as having some involvement in the pathogenesis of Hib meningitis strains. Additional analysis may contribute to better understanding of the study results. Work on direct sequencing of ERIC-PCR-generated amplification fragments is in progress in this laboratory. It is important to mention that other type b meningitis strains were also spread within other groups (Ib and IIc).
Studies of noncapsulated H. influenzae isolates by OMP and lipopolysaccharide (LPS) profiles revealed that nontypeable strains isolated from patients with different disease types are, as a group, significantly more variable than those Hib strains recovered from invasive disease sources. The latter were represented by a restricted number of OMP and LPS Eps (21). The distribution of noncapsulated isolates from bronchial secretions, nasopharynx, sputum, genital tract, middle ear, eye, and suppurative arthritis, as shown in the phenogram, was represented by a variety of single Eps. In this way, any restricted distribution related to clinical origin was observed among noncapsulated strains. In addition, these patterns were not shared with patterns of carrier isolates. The repeated findings on the strong genetic diversity of noncapsulated H. influenzae, which are consistent with the results described here using ERIC-PCR, allowed some authors to make hypothetical predictions suggesting the possibility of a nonclonal population for nontypeable H. influenzae (22). However, Fusté et al. (9), who recently surveyed the extent of clonality within nontypeable strains, showed a basic clonal structure with little possibility of recombination. Mutations in the H. influenzae genome probably contribute to the strong variability of noncapsulated isolates, as observed using ERIC-PCR. It is possible that the error rate of bacterial DNA synthesis is increased during persistent infections, such as cystic fibrosis and chronic bronchitis, and during carriage in healthy individuals.
Recently a hypothesis on association between repetitive DNA in the bacterial genome and virulence potential has been described and proved. A characteristic of H. influenzae is phase variation in its fimbriae and antigenic variation in LPS (10). Intragenomic variation mechanisms, mediated by high-frequency mutations in H. influenzae repetitive sequences, are responsible for modulated expression of such surface molecules (10, 16; E. R. Moxon, Abstr. 38th Intersci. Conf. Antimicrob. Agents Chemother., abstr. 96, 1998). Repeat variability in H. influenzae related to phenotypic switching as a consequence of host selective pressures is well documented (19, 37, 38). It would be worth analyzing the possible variation of ERIC-PCR Eps due to immunological or physiological selection imposed by the individual patient during the infectious process.
This study is the first to identify correlations between Eps determined by ERIC-PCR and distinctive pathogenicities among H. influenzae strains. The data presented here can be utilized to generate additional hypotheses regarding the pathogenicity and epidemiology of H. influenzae.
ACKNOWLEDGMENTS
We thank Juan Jose Garcia for assistance and in performing statistical analyses. We also thank Gabriel Perez, Jorge Saldivar, and Lino Mendez Franco for excellent technical assistance. We are grateful to Ian Shepherd for assistance in preparation of the manuscript.
A grant (IN220799) from Direccion General de Asuntos de Personal Academico, Universidad Nacional Autonoma de Mexico (UNAM), through its programme PAPIIT, supported this study, and the study was supported in part by the Programa de Apoyo a Investigadores Nivel PRIDE, Facultad de Medicina, UNAM.
REFERENCES
- 1.Barenkamp S J, Munson R S, Granoff D M. Outer membrane protein and biotype analysis of pathogenic Heamophilus influenzae. Infect Immun. 1982;36:535–540. doi: 10.1128/iai.36.2.535-540.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bisgard K M, Kao A, Leake J, Strebel P M, Perkins B A, Wharton M. Haemophilus influenzae invasive disease in the United States, 1994–1995: near disappearance of a vaccine-preventable childhood disease. Emerg Infect Dis. 1998;4:229–237. doi: 10.3201/eid0402.980210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boom R, Sol C J A, Salimans M M M, Jansen C L, Wertheim-van Dillen P M E, van der Noordaa J. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990;28:595–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bruce K D, Jordens J Z. Characterization of noncapsulate Haemophilus influenzae by whole-cell polypeptide profiles, restriction endonuclease analysis, and rRNA gene restriction patterns. J Clin Microbiol. 1991;29:291–296. doi: 10.1128/jcm.29.2.291-296.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cabrera L, Gómez-de-León P, Cravioto A. Vacunas: fundamentos para su desarrollo. Mexico D.F., Mexico: Universidad Nacional Automona de Mexico-El Manual Moderno Ed; 1996. [Google Scholar]
- 6.Cobb B D, Clarkson J M. A simple procedure for optimising the polymerase chain reaction (PCR) using modified Taguchi methods. Nucleic Acids Res. 1994;22:3801–3805. doi: 10.1093/nar/22.18.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Bruijn F J. Use of repetitive (extragenic palindromic and enterobacterial repetitive intergenic consensus) sequences and the polymerase chain reaction to fingerprint the genomes of Rhizobium meliloti isolates and other soil bacteria. Appl Environ Microbiol. 1992;58:2180–2185. doi: 10.1128/aem.58.7.2180-2187.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Falla T J, Crook D W M, Brophy L N, Maskell D, Kroll J S, Moxon E R. PCR for capsular typing of Haemophilus influenzae. J Clin Microbiol. 1994;32:2382–2386. doi: 10.1128/jcm.32.10.2382-2386.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fusté M C, Pineda M A, Palomar J, Viñas M, Lorén J G. Clonality of multidrug-resistant nontypeable strains of Haemophilus influenzae. J Clin Microbiol. 1996;34:2760–2765. doi: 10.1128/jcm.34.11.2760-2765.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gildsdorf J R. Antigenic diversity and gene polymorphisms in Haemophilus influenzae. Infect Immun. 1998;66:5053–5059. doi: 10.1128/iai.66.11.5053-5059.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gómez-de-León P, Molinari J L, Cabrera R, Gómez-Barreto D, Cravioto J, Cravioto A. Serum response to outer membrane proteins of Haemophilus influenzae type b in Mexican children with or without evidence of invasive disease. Immunol Infect Dis. 1995;5:238–243. [Google Scholar]
- 12.Hood D W, Deadman M E, Jennings M P, Bisercic M, Fleishmann R D, Venter J C, Moxon E R. DNA repeats identify novel virulence genes in Haemophilus influenzae. Proc Natl Acad Sci USA. 1996;93:11121–11125. doi: 10.1073/pnas.93.20.11121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hulton C S J, Higgins C F, Sharp P M. ERIC sequences: a novel family of repetitive elements in the genomes of Escherichia coli, Salmonella typhimurium and other enterobacteria. Mol Microbiol. 1991;5:525–834. doi: 10.1111/j.1365-2958.1991.tb00755.x. [DOI] [PubMed] [Google Scholar]
- 14.Inzana T J. Electrophoretic heterogeneity and interstrain variation of the lipopolysaccharide of Haemophilus influenzae. J Infect Dis. 1983;148:492–499. doi: 10.1093/infdis/148.3.492. [DOI] [PubMed] [Google Scholar]
- 15.Krawiec S, Riley M. Organization of the bacterial chromosome. Microbiol Rev. 1990;54:502–539. doi: 10.1128/mr.54.4.502-539.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lensky R, Travisano M. Dynamics of adaptation and diversification: a 10,000 generation experiment with bacterial populations. Proc Natl Acad Sci USA. 1994;91:6808–6814. doi: 10.1073/pnas.91.15.6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Louws F J, Fulbright D W, Stephens C T, de Bruijn F J. Specific genomic fingerprints of phytopathogenic Xanthomonas and Pseudomonas pathovars and strains generated by repetitive sequences and PCR. Appl Environ Microbiol. 1994;60:2286–2295. doi: 10.1128/aem.60.7.2286-2295.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lupski J R, Weinstock G M. Short interspersed repetitive DNA sequences in prokaryotic genomes. J Bacteriol. 1992;174:4525–4529. doi: 10.1128/jb.174.14.4525-4529.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moxon E R, Rainey P B, Nowak M A, Lensky R E. Adaptive evolution of highly mutable loci in pathogenic bacteria. Curr Biol. 1994;4:24–33. doi: 10.1016/s0960-9822(00)00005-1. [DOI] [PubMed] [Google Scholar]
- 20.Moxon E R. Haemophilus influenzae. In: Mandell G L, Bennett J E, Dolin R E, editors. Principles and practice of infectious diseases. New York, N.Y: Churchill Livingstone; 1995. pp. 2039–2045. [Google Scholar]
- 21.Murphy T F, Bernstein J M, Dryja D M, Campagnari A A, Apicella M A. Outer membrane protein and lipooligosaccharide analysis of paired nasopharyngeal and middle ear isolates in otitis media due to non typeable Haemophilus influenzae: pathogenic and epidemiological observations. J Infect Dis. 1987;156:723–731. doi: 10.1093/infdis/156.5.723. [DOI] [PubMed] [Google Scholar]
- 22.Musser J M, Barenkamp S J, Granoff D M, Selander R K. Genetic relationships of serologicaly nontypable and serotype b strains of Haemophilus influenzae. Infect Immun. 1986;52:183–191. doi: 10.1128/iai.52.1.183-191.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Musser J M, Barenkamp S J, Granoff D M, et al. Global genetic structure and molecular epidemiology of encapsulated Haemophilus influenzae. Rev Infect Dis. 1990;12:75–111. doi: 10.1093/clinids/12.1.75. [DOI] [PubMed] [Google Scholar]
- 24.Orskov F, Orskov T, Evans D J, Jr, Sack R B, Sack D A, Wadstrom T. Special Escherichia coli serotypes among enterotoxigenic strains from diarrhoea in adults and children. Med Microbiol Immunol. 1976;162:73–80. doi: 10.1007/BF02121318. [DOI] [PubMed] [Google Scholar]
- 25.Patti J M, Allen B L, McGavin M J, Höök M. MSCRAMM-mediated adherence of microorganisms to host tissues. Annu Rev Microbiol. 1994;48:585–617. doi: 10.1146/annurev.mi.48.100194.003101. [DOI] [PubMed] [Google Scholar]
- 26.Rohlf F. Numerical taxonomy and multivariate analysis system, version 2.00. Stauket, New York, N.Y.: Exeter software.; 1997. [Google Scholar]
- 27.Romesbrug H C. Cluster analysis for researchers. Belmont, Calif: Lifetime Learning Publications; 1984. [Google Scholar]
- 28.Smith-Vaughan H C, Sriprakash K S, Matews J D, Kemp D J. Nonencapsulated H. influenzae in aboriginal infants with otitis media: prolonged carriage of P2 porin variants and evidence for horizontal P2 gene transfer. Infect Immun. 1997;65:1468–1474. doi: 10.1128/iai.65.4.1468-1474.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sneath P H A, Sokal R R. Numerical taxonomy. San Francisco, Calif: Freeman; 1973. [Google Scholar]
- 30.St. Geme J W., III Progress towards a vaccine for nontypable H. influenzae. Ann Med. 1996;28:31–37. doi: 10.3109/07853899608999071. [DOI] [PubMed] [Google Scholar]
- 31.Turk D. The pathogenicity of Haemophilus influenzae. J Med Microbiol. 1984;18:1–16. doi: 10.1099/00222615-18-1-1. [DOI] [PubMed] [Google Scholar]
- 32.van Belkum A, Duim B, Regelink A, Moeller L, Quint W, van Alphen L. Genomic DNA fingerprinting of clinical Haemophilus influenzae isolates by polymerase chain reaction amplification: comparison with major outer membrane protein and restriction fragment length polymorphism analysis. J Med Microbiol. 1994;41:63–68. doi: 10.1099/00222615-41-1-63. [DOI] [PubMed] [Google Scholar]
- 33.van Belkum A, Scherer S, van Alphen L, Verbrugh H A. Short-sequence DNA repeats in prokaryotic genomes. Microbiol Mol Biol Rev. 1998;62:275–293. doi: 10.1128/mmbr.62.2.275-293.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Versalovic J, Koeuth T, Lupski J R. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 1991;19:406–409. doi: 10.1093/nar/19.24.6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Versalovic J, Schneieder M, de Bruijn F J, Lupsky J R. Genomic fingerprinting of bacteria using repetitive sequence based polymerase chain reaction. Methods Mol Cell Biol. 1994;5:25–40. [Google Scholar]
- 36.Villaseñor A, Santos J I. Outer membrane protein profiles of paired nasopharyngeal and middle ear isolates of nontypable Haemophilus influenzae from Mexican children with acute otitis media. Clin Infect Dis. 1999;28:267–273. doi: 10.1086/515098. [DOI] [PubMed] [Google Scholar]
- 37.Weiser J N, Love J M, Moxon E R. The molecular mechanism of phase variation of Haemophilus influenzae lipopolysaccharide. Cell. 1989;59:657–665. doi: 10.1016/0092-8674(89)90011-1. [DOI] [PubMed] [Google Scholar]
- 38.Weiser J N, Maskell D J, Butler P D, Lindberg A A, Moxon E R. Characterization of repetitive sequences controlling phase variation of Haemophilus influenzae lipopolysaccharide. J Bacteriol. 1990;172:3304–3309. doi: 10.1128/jb.172.6.3304-3309.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]