Abstract
Multifunctional nano-materials that can be used to monitor the expression of specific biomarkers and serve as vehicles for controlled drug delivery are highly desirable. Herein, we report a new DNA-hybrid-gated core–shell upconversion nanoprobe (UCNP@MOF/DOX) for fluorescence analysis of microRNA-21 (miR-21), which also triggers the release of drug loaded in the probes for on-demand anti-cancer treatment. The nanoprobe is built on the merits of ultraviolet-visible light of upconversion nanoparticles (UCNPs) excited by near-infrared (NIR) and extraordinary loading capability of metal–organic frameworks (MOFs) for drug delivery. Controlled release of doxorubicin (DOX) from the nanoprobe by miR-21 underwent the following two-stage kinetics: a fast release stage specifically triggered by miR-21 and proportional to miR-21 concentration and a slow stage observed in both gated and ungated nanoprobes due to collapse of the UIO-66-NH2 coatings via ligand exchange with phosphates. In addition, the nanoprobe showed good selectivity, a linear response towards miR-21 ranging from 4 nM to 500 nM, and a limit of detection in 4 nM, which precluded unintended payload leakage due to low-abundance endogenous miR-21 expression in normal cells. Moreover, based on a dual-targeted delivery system constituted by AS1411-mediated recognition and responsive release of DOX, a specific cytotoxic efficacy was observed in MCF-7 cells. The present work provides a smart and robust nanoprobe for real-time detection of miRNA and dual-responsive drug delivery in tumor cells.
A DNA-hybrid-gated core–shell upconversion nanoprobe is prepared for both fluorescent monitoring of miR-21 and on-demand delivery of DOX. It showed good selectivity towards miR-21 and demonstrated specific cytotoxic efficacy towards MCF-7 cells.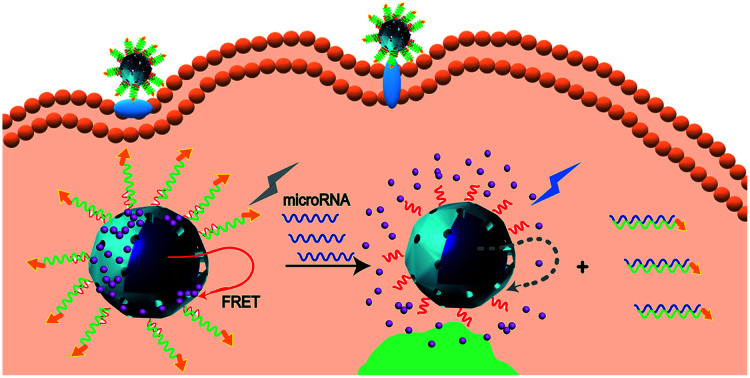
Introduction
The initiation and progression of cancers are closely related to the abnormal expression of non-coding RNAs, especially microRNAs (miRNAs).1 A growing number of studies have highlighted the importance of miRNAs as potential biomarkers for molecular diagnostics and therapeutic agents of cancers.2,3 Recently, nanotheranostics, the combination of diagnostic and therapy in a single nanoplatform, has emerged as a new and efficient strategy to precisely visualize tumor tissues and deliver drugs.4,5 Accordingly, miRNA-targeted fluorescent nanoprobes formed on nucleic acid probe-modified organic fluorophores, quantum dots or inorganic nanoparticles have been exploited as dominant theragnostic patterns.6 Our team previously presented real-time detection of microRNA-21 (miR-21) in HepG2 cells, by utilizing a 6-carboxy-fluorescein (6-FAM) labeled DNA tetrahedral modified AuNPs fluorescent probe.7 However, ultraviolet (UV) or visible light-excited fluorescence signals are confronted with drawbacks like slight penetration depth, photodamage and interference of auto-fluorescence. Considerable research shows that near-infrared nanomaterials excited by long-wavelength light have obvious advantages in bioanalysis.8,9
Rare-earth-doped upconversion nanoparticles (UCNPs) have been promising candidates for NIR nanoprobes owing to their distinctive properties, including superior stability, tunable multicolor emission and efficient upconversion from NIR to UV or visible emission.10 Currently, the construction of core–shell upconversion nanoprobes based on UCNPs is a classical strategy in applications. For instance, mesoporous silica-coated UCNPs (UCNPs@mSiO2), a typical upconversion nanocomposite, could act for optical imaging and drug delivery by loading chemotherapeutic drugs or photosensitizers.11 However, in most upconversion nanoprobes, UCNPs are used simply for NIR imaging of tumor tissue, which cannot reflect the levels of specific tumor-related biomarkers. Furthermore, drug release is mostly triggered by temperature or pH, which is susceptible to changes in physiological and pathological conditions, resulting in significant side effects.12,13 In view of the above problems, researchers proposed an upconversion nanobeacon UCNPs-MB/doxorubicin (DOX) for fluorescence detection of mRNA and controlled drug delivery. BHQ-1-modified stem-loop hairpin DNA was used to load DOX and quench the fluorescence of UCNPs due to fluorescence resonance energy transfer (FRET), which UCNP is used as the energy donor and BHQ-1 is employed as the energy acceptor.14 However, the poor loading capacity of hairpin DNA and the instability of BHQ-1 may weaken the ability of drug delivery and intracellular imaging. It is worth mentioning that a recent study made use of pH-sensitive UCNPs@mSiO2 to deliver DOX and monitor the drug release process by fluorescence signal changes due to the FRET composed of UCNPs and DOX.15 Inspired by this, we assumed that provided the release of DOX is controlled by target miRNA, the miRNA could be detected according to the change of fluorescence signal and the specific pathological cells with exclusive expression of miRNA could be specifically killed.
Nucleic acid-functionalized porous nanoparticles display fascinating features for stimuli-controlled drug delivery. Bulky DNA gating systems are formed by bridged duplexes and two- or three-dimensional DNA structures for capping the pores. Different triggers including strand displacement, small molecules or pH stimuli could unfold the DNA gates resulting in the release of contents.16,17 Mesoporous silica nanoparticles (MSNPs) have been a common material for DNA gating systems.18 Following MSNPs, the emergence of metal–organic frameworks (MOFs) has attracted considerable attention.19,20 MOFs are porous organic–inorganic hybrid materials self-assembled by organic ligands and metal ions or clusters.21 Compared with MSNPs, the surface area of MOFs is approximately 10 times larger, thus leading to a higher loading capacity.22 Moreover, MOFs have a uniform pore size distribution and could be specifically nucleic acid-functionalized by varieties of covalent bonds, so that the stimuli-triggered release is more orderly and controllable.23,24 Therefore, MOFs are considered promising candidates for the fabrication of drug delivery systems with less leakage, responsive delivery, and spatio-temporal delivery.
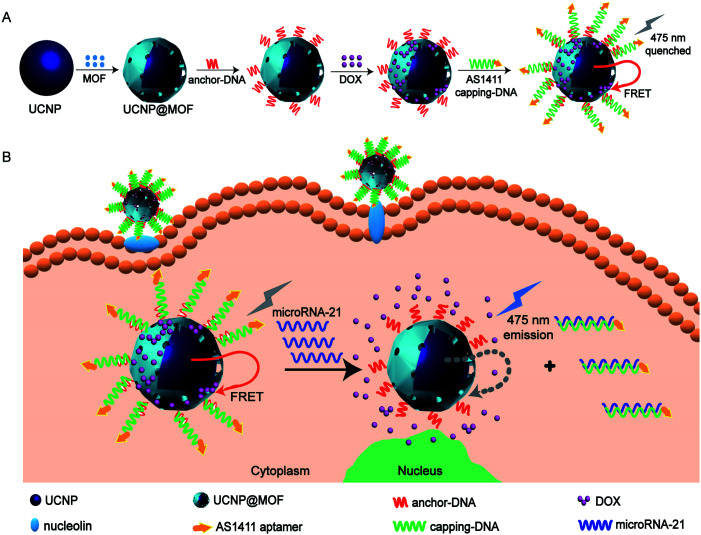
Herein, we developed a new upconversion nanoprobe based on DNA-hybrid-gated UCNPs@MOF/DOX for miRNA fluorescence detection and dual-targeted controlled drug delivery in tumor cells under NIR light (miR-21 as illustration, one of the oncogenic miRNAs overexpressed in breast cancer) (Scheme 1). The representative MOF, UiO-66-NH2, was employed as the porous shell of the core–shell composites. After loading DOX efficiently, the composites were nucleic acid-functionalized to cap the pores by forming a DNA gating system. By virtue of the FRET between UCNPs and DOX, upconversion fluorescence (UCF) at 475 nm was quenched (Scheme 1A). The nanoprobe could be targeted and efficiently endocytosed into MCF-7 cells via modified aptamer AS1411 which could target nucleolin overexpressed on the tumor cell membrane.25 Upon the nanoprobe identified with miR-21, the DNA gate collapsed by toehold-mediated strand displacement reaction, simultaneously initiating the responsive release of DOX. Owing to the weakening of FRET between UCNPs and DOX, the UCF at 475 nm gradually recovered. By analyzing the change of fluorescence signal, we ingeniously realized the detection of miR-21 (Scheme 1B).
Scheme 1. Schematic illustration of (A) synthesis of the DNA-hybrid-gated UCNP@UIO-66-NH2/DOX nanoprobe and (B) the process of miRNA detection and target-responsive drug delivery.
Results and discussion
Structural characterization of UCNP@UIO-66-NH2 nanocomposites
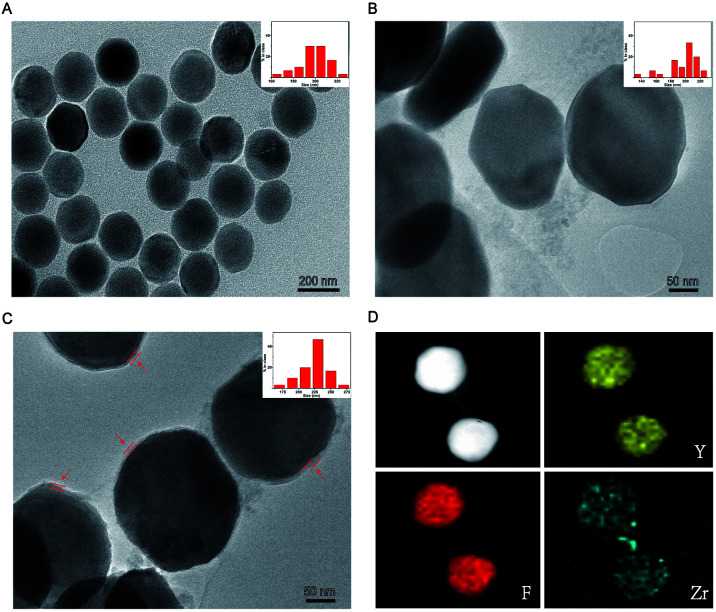
The general synthetic route used for the preparation of UCNP@UIO-66-NH2 is illustrated in Scheme 1A. The originally prepared oleic acid (OA)-coated NaYF4:Yb/Tm displayed an obvious hexagonal plate, which matches well with the characteristic morphology of NaYF4 crystal in the β-phase (Fig. 1A and S1A†).26 Compared with X-ray diffraction (XRD) spectrum of cubic pattern (JCPDS: 77-2042), UCNPs show unique characteristic peaks consistent with hexagonal plate pattern (JCPDS: 16-0334) (Fig. S1B†). The water-dispersed UCNPs were synthesized by ligand exchange approach that OA was displaced by PEG-phosphate ligand (Fig. 1B).27 During the synthesis of the nanocomposites, the PEG-phosphate layer could serve as nucleation sites for the MOF growth, because Zr4+ ions were absorbed by PEG-phosphate layer and then nucleated on the polymer upon BDC-NH2 addition.28 Once a thin MOF layer formed, the MOF shell grew continuously in a layer-by-layer manner, which prevented the formation of free MOF particles. Different thicknesses of MOF shell were formed through repetitive addition of Zr4+ and BDC-NH2 linkers. As such, after three repeats, the thickness of the coated MOF shell was approximately 10.7 nm, which could achieve an excellent FRET efficiency (Fig. 1C). The hydrodynamic size of PEG-phosphate coated UCNPs is around 237 nm. After MOF coating, it increases to 256 nm (Fig. S2†). Elemental mapping techniques and energy dispersive X-ray spectrum were subsequently applied to confirm the compositions of the prepared nanocomposites. As suggested by the results (Fig. 1D and S3†), elemental Y, F, Tm and Zr were uniformly distributed on the particles, which apparently revealed that the nanocomposites were consisted of NaYF4:Yb/Tm cores and UIO-66-NH2 shells.
Fig. 1. Characterization of UCNP@UIO-66-NH2. TEM images and size distributions of (A) OA-coated UCNPs, (B) PEG-phosphate coated UCNPs, (C) UCNP@UIO-66-NH2 NPs (the UIO-66-NH2 layers were indicated by red arrows) and (D) elemental mappings of UCNP@UIO-66-NH2 NPs.
Characterization of the upconversion nanoprobe
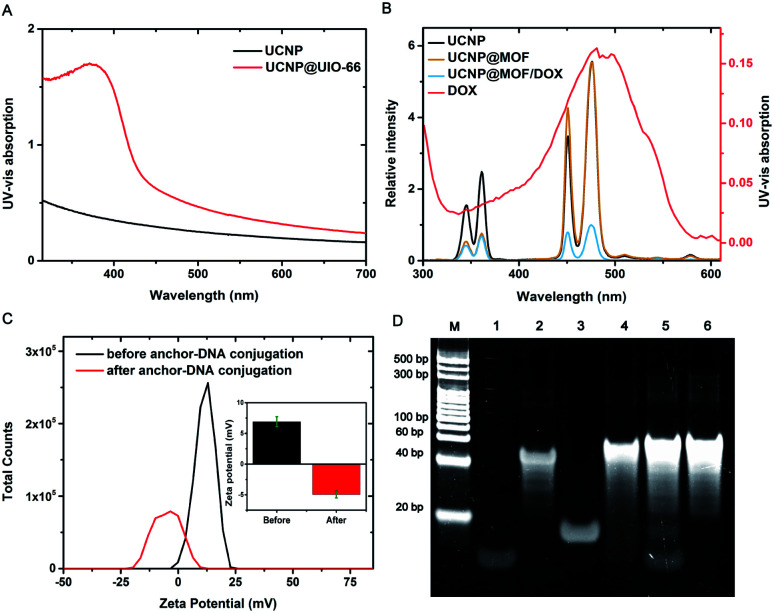
In the process of nanoprobe construction, the UV-vis-NIR absorption spectrum of UCNPs displayed a significant change after subsequent MOF growth (Fig. 2A). Upon coating with UiO-66-NH2, a strong absorption peak mainly located at 300–400 nm raised due to MOF absorption. This absorption feature convincingly demonstrates the successful coating of MOF shell. The upconversion luminescence spectra were further recorded (Fig. 2B). UCNPs gave two obvious emission peaks located at 360 nm and 475 nm under 980 nm excitation, which originated from the transit ions of Tm3+ ions. Significantly, upon coating with UiO-66-NH2, 360 nm emission from UCNPs decreased to nearly fully quenched due to the spectral overlap of MOF absorption. The 475 nm emission from UCNPs can also be quenched through DOX loading due to the UV absorption of DOX. These spectral changes unambiguously reflect the energy transfer between UCNPs and DOX as well as MOFs, which indicated that the assembly of the nanoprobe was successfully realized.
Fig. 2. Characterization of DNA-hybrid-gated UCNPs@UIO-66-NH2/DOX. (A) UV-visible absorption spectra, (B) upconversion luminescence spectra under excitation at 980 nM of different samples and UV-visible absorption of DOX, (C) zeta potentials of the nanoparticles, and (D) polyacrylamide gel electrophoresis for verification of strand displacement reaction (M: marker; lane 1: anchor-DNA; 2: capping-DNA; 3: miR-21; 4: DNA duplexes hybridized by anchor-DNA and capping-DNA; 5: mixed system of DNA duplexes and miR-21; 6: DNA duplexes hybridized by capping-DNA and miR-21).
As depicted in Scheme 1A, the DNA functionalization of nanocomposites is a key step. Firstly, the phosphate modified anchor DNA was firmly and specifically bound on the surface of MOF shell, through the Zr–O–P bond. Owing to the negative charge of DNA skeleton, the zeta potential was significantly transformed after the successful modification of anchor-DNA (Fig. 2C). As a supplement, we also tested the fluorescence intensity of the supernatant after the reaction by using 6-FAM labeled anchor DNA, which confirms the DNA functionalization of the nanocomposites (Fig. S4†). Furthermore, the capping DNA and the anchor DNA were hybridized to form a stable DNA gate. Compared with UCNP@MOF, the hydrodynamic size reached to 265 nm upon DNA conjugation (Fig. S2†). We tested the hybridization ability of two DNA strands and the efficient occurrence of toehold-medicated strand replacement. By means of PAGE (polyacrylamide gel electrophoresis) (Fig. 2D), the anchor DNA (15 bp) was successfully hybridized with the capping DNA (48 bp) (lane 4). Subsequently, the target miR-21 was added to the solution of DNA duplexes. miR-21 hybridized with the capping DNA specifically and replaced the anchor DNA, resulting in the collapse of DNA gate (lane 5). In addition, the ratio of DNA strands was further optimized (Fig. S5†). Besides, the loading capacity of DOX in the nanoprobe was evaluated. The loading capacity is calculated to be ∼24 μg DOX for 1 mg nanoprobe using linear fitting (Fig. S6†). Considering the DOX-DNA interaction and the excellent loading capacity of UIO-66-NH2, we assumed that the loading of DOX was mainly attribute to the UIO-66-NH2 shell, supplemented by double-strand DNA loading.14
Analytical performance and target-responsive drug delivery of the upconversion nanoprobe
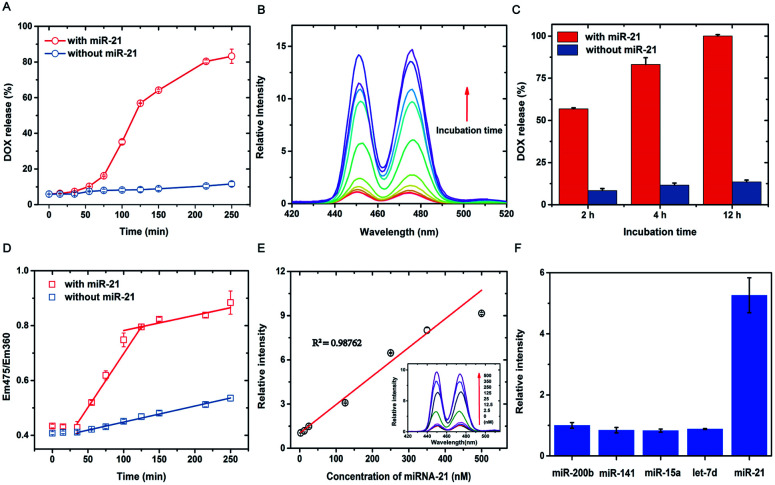
After successful construction of the DNA-hybrid-gated nanoprobe UCNPs@UIO-66-NH2/DOX, the performance of miRNA detection and target-responsive drug delivery were evaluated in vitro. Specifically, the emission at 475 nm was used to characterize the release of DOX as it was quenched by loaded DOX. As shown in Fig. 3A, the 475 nm emission of nanoprobe was efficiently suppressed under 980 nm excitation caused by FRET-induced quenching. After addition of miR-21, a gradual growth of fluorescence intensity at 475 nm was observed until it reached a plateau after about 4 h (Fig. S7†). The relative fluorescence intensity, which was calculated by dividing the original intensity before miR-21 addition, increased approximately 15 times (Fig. 3B). In comparison, the miR-21-free trial showed no clear increase in overall emission at 475 nm, except for a slight gain of intensity due to minimal leakage of DOX. After incubation for 2 h, over 55% DOX was released from the nano-carrier (Fig. 3C). The release percentage reached more than 80% in 4 h and further extension of incubation time to 12 h only ended in no more than 20% release. We further analyzed the release kinetics using the ratio of emission at 475 nm and that at 360 nm, which should remain constant with or without DOX loading. Notably, when plotting the Em475/Em360 value against the incubation time, a clear two-phase increase was observed for trials with miR-21 addition whereas those without miR-21 suggested only monotonically slow growth over time. In addition, the ratio of Em475/Em360 is well correlated with the incubation time through linear fitting for miR-21 positivity in both growth courses (R2 > 0.95), during which the first phase indicated a dramatic release of DOX approximately 30 min after the introduction of miR-21 and lasted for 1 h. After that, DOX release turned to ease down, and the unloading rate, estimated by the slope of fitting, was close to that without miR-21. These results jointly supported that the triggered release of DOX from the DNA-hybrid-gated nanoprobe by miR-21 underwent a two-phase kinetic rather than a single kinetic process, as observed in most drug delivery nano-carriers. Specifically, the fast release period was due to the opening of the DNA gate via strand displacement by miR-21, which directly exposes the nanopores of the UIO-66-NH2 coatings to the surrounding buffer and immediately spurs the discharge of DOX. After releasing the majority of the payload, the nanoprobe went to the second period, in which the release of DOX is ascribed to the gradual collapse of the UIO-66-NH2 coatings due to ligand exchange with phosphates in the buffer. As the release of DOX induced by the target is much faster than that by buffer, it would lead to a burst of drug concentration as long as the target miRNA is overexpressed or accumulated in specific cells.
Fig. 3. Evaluation of target-responsive drug delivery and quantitative detection in vitro. Data are means ± SD (n = 3 experimental replicates). (A) The kinetic study of DOX released from the nanoprobe (0.75 mg mL−1) with and without miR-21 (100 nM). (B) The relative fluorescence spectra of the nanoprobe (0.75 mg mL−1) incubated with miR-21 (100 nM) at different time intervals. (C) Comparative study of the DOX released from the nanoprobe (0.75 mg mL−1) with and without miR-21 (100 nM) at different time intervals (2 h, 4 h, 8 h). (D) The kinetic study of Em475/Em360 with and without miR-21 (100 nM). (E) Linear relationship of relative fluorescence intensity and the concentrations of miR-21 from 2.5 nM-500 nM. (F) Specificity of the nanoprobe for the detection of different miRNAs (100 nM).
Furthermore, a quantitative analysis of nanoprobes to detect target miR-21 was subsequently investigated. As shown in Fig. 3E, after consecutive addition of miR-21 at various concentrations, the intensity of 475 nm emission increased proportionally in the range from 4 nM to 500 nM. Linear fitting between the fluorescence intensity and miR-21 concentration suggested a good correlation coefficient over 0.98. The lowest detection limit (LOD) was calculated to be ∼4 nM (LOD = 3σ/slope, where σ is the standard deviation of blank samples), and incubation of the probe with miR-21 lower than LOD hardly resulted in significant gain of fluorescence even after extending the time course. Therefore, although the detection limit of the nanoprobe renders it not sensitive enough to detect miR-21 in ultralow concentrations, it would be a merit for controlled drug release, as miR-21 is also present in low abundance for many noncancerous cells. Moreover, the selectivity of the nanoprobe was investigated in the presence of four miRNA counterparts, let-7d, miR-141, miR-15a, and miR-200b, under identical conditions. As listed in Fig. 3F, none of the potential homologues other than target miR-21 generated significant enhancement of the UCF signal, thus demonstrating that UCF enhancement was selectively induced by miR-21. We thus anticipate that the present DNA-hybrid-gated nanoprobe would serve as an efficient, controlled drug release system and nano-carrier.
Intracellular upconversion imaging of miRNA
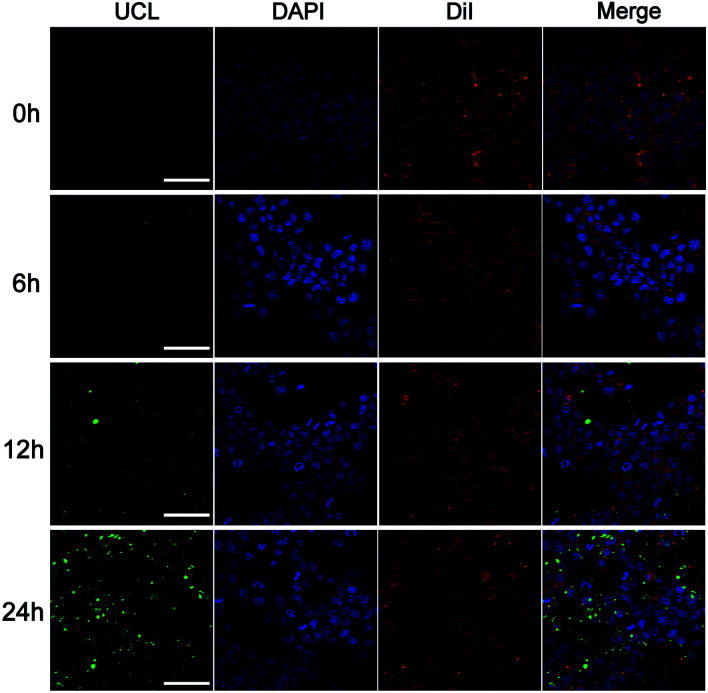
To assess the potential application of the upconversion nanoprobe as a miRNA-targeted nano-carrier, the in situ imaging of miR-21 in living cells was further investigated. The uptake efficiency and reaction dynamics of the nanoprobes were firstly investigated by incubating human breast cancer cells (MCF-7) with this nanoprobe (50 μg mL−1) for different time periods (1–24 h) (Fig. 4). The UCF reached an appropriate intensity at 12 h, which was thus selected as the incubation time in the subsequent experiments. It's worth noting that although the fluorescence intensity increased with incubation time, the fluorescence at 24 h appeared in the area without cells. It is likely that the nanoprobe was effused from damaged cells caused by DOX. Furthermore, the upconversion nanoprobe was used to analyze miR-21 levels in different cell lines, including MCF-7 cells and HEK-293T cells. As shown in Fig. 5, distinct UCF intensities were observed in particular cell lines, suggesting differences in miR-21 expression levels. Consistent with the miR-21 expression level in previous studies, the brightest UCF signal was observed in MCF-7 cells and a weaker UCF signal was displayed in HEK-293T cells.29 As it is critical to release drugs in consonance with the relative expression level of target miRNA during different states in living cells, we then incubated the nanoprobe with MCF-7 cells treated with curcumin which is a universal miR-21 inhibitor for down-regulation of the expression.30,31 As expected, a significant decrease of UCF intensity was captured in curcumin treated MCF-7 cells. These results indicated that this nanoprobe possessed practicality in miRNA imaging and dynamic monitoring of living cells.
Fig. 4. Two-photon excited confocal laser scanning microscopy (CLSM) images of MCF-7 cells after incubation with DNA-hybrid-gated UCNP@UIO-66-NH2/DOX (50 μg mL−1) at different times. Scale bar: 75 μm.
Fig. 5. Intracellular upconversion detection and imaging of miR-21. Fluorescence confocal microscopy image of MCF-7 cells, MCF-7 cells treated with curcumin and normal HEK-293T cells after incubation with DNA-hybrid-gated UCNP@UIO-66-NH2/DOX (50 μg mL−1) for 12 h under 980 nm excitation. Scale bar: 75 μm.
Intracellular miRNA-triggered drug delivery
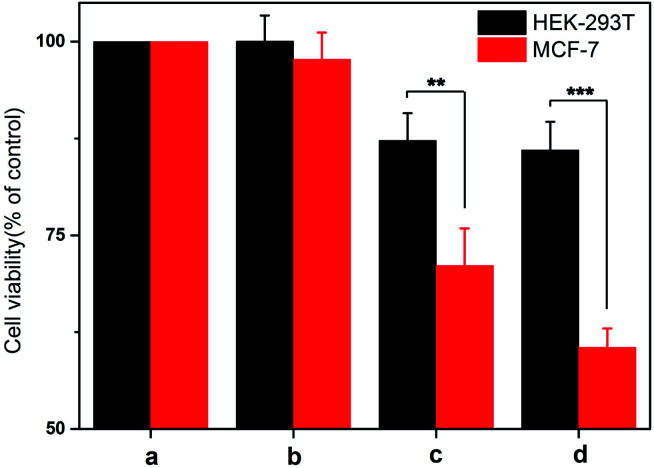
The drug delivery performance was evaluated by the cytotoxicity of nanoprobe on different cells. In this experiment, different cells were treated with the probe for a time interval of 12 h to allow the permeation of nanoprobe and the cell viability was examined after 24 h by Cell Counting Kit-8 (CCK-8) assay. As displayed in Fig. 6, the viability of HEK-293T cells decreased by approximately 10% and there was no significant difference between the nanoprobe with AS1411 and that without AS1411. In turn, the viability of MCF-7 cells decreased by approximately 25% and 35% under the treatment of nanoprobe with AS1411 and that without AS1411 respectively, which suggested that AS1411 played an indispensable role in cell permeation and specific killing. It should be noted that treatment of the HEK-293T and MCF-7 cells using the nanoprobe without DOX loading did not have any effect on cell viability. This result implied that the nanoprobe had excellent biocompatibility. These results were consistent with the intensity of UCF signal in cell imaging and proved that this nanoprobe possessed impressive selectivity and efficacy on cell viability.
Fig. 6. The cytotoxicity of normal HEK293T and malignant MCF-7 cells treated with the various nanoprobes by Cell Counting Kit-8 assay. Data are means ± SD (n = 3 experimental replicates). **P < 0.01 versus normal cells, ***P < 0.001 versus normal cells by t-test. (a) Untreated cells, (b) cells treated with unloaded DNA-hybrid-gated UCNP@UIO-66-NH2, (c) cells treated with DNA-hybrid-gated UCNP@UIO-66-NH2/DOX without modified AS1411, (d) cells treated with DNA-hybrid-gated UCNP@UIO-66-NH2/DOX with modified AS1411.
Conclusions
In conclusion, a novel DNA-hybrid-gated core–shell upconversion nanoprobe has been developed as an intelligent nano-carrier for miR-21 fluorescence detection and dual-targeted controlled drug delivery in living cells. Specifically, the UCNP@UIO-66-NH2 NPs were loaded with DOX efficiently and subsequently functionalized by a DNA gating system modified with AS1411. Once the nanoprobes were delivered into the cytoplasm of MCF-7 cells by AS1411-mediated endocytosis, endogenous miR-21 acted as a trigger to initiate the collapse of the DNA gating system and release DOX by a strand replacement reaction. In light of this strategy, the nanoprobe realized the detection of miR-21 by FRET between UCNPs and DOX with low interferences and delivered drugs efficiently via a dual-targeted controlled mode. Moreover, considering the functional and regulatory mechanisms of miRNAs, this nanoprobe, which could hybridize with endogenous miR-21, has certain potential in combination with chemotherapy and gene therapy. Evaluation of the nanoprobe capability in in situ monitoring of miR-21 and drug delivery in xenograft tumor mode and related biological safety as well as its clearance are currently being examined in our lab. These results reveal that the present novel nanoprobe, which achieved sensitive and selective detection of miRNA and excellent efficacy of drug delivery, is an intelligent nano-carrier and provides potential applications in precise diagnosis and personalized treatment.
Experimental
Materials
All commercial chemicals were analytical grade and used without further purification. Rare earth chlorides, NH4F and DOX were purchased from Aladdin Reagent, Ltd. (Shanghai, China). Oligonucleotides were synthesized by Sangon biotechnology Co. Ltd. (Shanghai, China) and sequences are listed in Table S1.† Breast cancer cells MCF-7 (ATCC, HTB-22) and normal epithelial cells HEK-293T (ATCC, CRL-11268) were purchased from American Type Culture Collection (Manassas, VA, USA). Dulbecco's modified Eagles medium (DMEM), fetal bovine serum, penicillin and streptomycin were gotten from GE Hyclone (Pittsburgh, USA). Cell Counting Kit-8 (CCK-8) was obtained from MCE (New Jersey, USA). The apparatuses and measurements are shown in ESI.†
Synthesis of β-NaYF4:Yb (20%), Tm (0.5%) UCNPs
The β-NaYF4:Yb (20%), Tm (0.5%) UCNPs were prepared according to the previously reported method.15 YCl3·6H2O (482 mg), YbCl3·6H2O (155 mg), and TmCl3·6H2O (2.8 mg) in deionized water (2 mL) were added to a 100 mL flask containing oleic acid (15 mL) and 1-octadecene (30 mL). The resulting solution was stirred at room temperature and exclude O2 with argon for 1 h. Then the mixture was slowly heated to 120 °C to get rid of water and maintained at 156 °C for about 1 h until a homogeneous transparent yellow solution was obtained. The system was then cooled down to room temperature with the flowing of argon. Then a methanol solution (10 mL) containing NH4F (296 mg) and NaOH (200 mg) was added. Then the mixture was heated to 70 °C and kept for 20 min to evaporate methanol. After that, the solution was heated to 290 °C and kept for 1.5 h before it was cooled down to room temperature. The mixture were first precipitated by the addition of ethanol (20 mL), and collected by centrifugation at 9000 rpm for 10 min. After four times washing, the final product was re-dispersed in cyclohexane (20 mL) and lyophilized for further use.
Synthesis of PEG-phosphate stabilized UCNPs
UCNPs (30 mg) were added to absolute ethanol (2 mL) with PEG (1000)-phosphate ligand (1 g). Then the resulting solution was heated slowly to 70 °C in water bath. The reaction mixture was maintained at 70 °C for a minimum of 8 h. It was then cooled down to room temperature. Subsequently, the PEG-phosphate stabilized UCNPs were precipitated via centrifugation at 7000 rpm. Then the product was firstly washed with cyclohexane (2 mL) for two times to remove free oleate from the sample. Then the pellets were washed with absolute ethanol (2 mL) for four times to remove excessive PEG (1000)-phosphate from the system. The final product was re-dispersed in N,N-dimethylformamide (DMF) (2 mL) for the later experiment of UIO-66-NH2 shell coating.
Synthesis of core−shell UCNP@UIO-66-NH2 NPs
A layer-by-layer growing method was employed for the formation of UIO-66-NH2 shell on UCNPs. In a typical procedure, the above PEG-phosphate stabilized UCNPs were dispersed in DMF (10 mL), and then ZrCl4 solution (5 mL, 5 mM) was added. The solution was stirred for 40 min at 60 °C for absorbing Zr4+ on the particle surface. After that, BDC-NH2 solution (5 mL, 5 mM) was added to the solution, which was heated to 120 °C and maintained for 1 h. After a thin layer of UIO-66-NH2 was grown, the intermediate products were collected by centrifugation at 9000 rpm for 10 min. To increase the shell thickness of MOF, the above growing process was repeated according to the same protocol until a desirable thickness was obtained. Finally, the products were washed with ethanol and water alternately for 6 times (10 mL) and re-dispersed in DMF (10 mL) for stable storage.
PAGE to test the feasibility of strand displacement
The anchor-DNA (10 μL, 10 μM) and capping-DNA (10 μL, 10 μM) were added to Tris–HCl buffer (30 μL), mixed well and incubated at 37 °C for 1 h to form DNA duplexes. Subsequently, miR-21 (10 μL, 10 μM) was added to the above reaction solution, mixed well and incubated at 37 °C for 1 h. The displacement efficiency was validated by PAGE (12% gel).
Anchor-DNA functionalization of UCNP@UIO-66-NH2 NPs
UCNP@UIO-66-NH2 NPs (3 mg, 1 mL) were re-dispersed from DMF to water. Then an aqueous solution of phosphate modified anchor-DNA (2 nmol) was added to the solution and mixed on a mechanical shaker at room temperature for 14 h. During the period, NaCl was added to the solution over 2 h in three equal aliquots to a final concentration of 0.5 M. The anchor-DNA functionalized UCNP@UIO-66-NH2 NPs were washed in PBS buffer three times to remove unbound DNA and finally re-dispersed in PBS buffer (1 mL). The 6-FAM fluorescence of the supernatant at 525 nm was measured to determine the amount of DNA functionalization.
Loading DOX and capping the pores
UCNP@UIO-66-NH2 NPs (3 mg) were incubated with DOX (1.0 mg mL−1) for 24 h in PBS buffer (2 mL). Then, capping-DNA (2.5 nmol) was added to the above solution to seal DOX. Then, after sealing DOX with capping-DNA for 12 hours, the precipitates were collected by centrifugation at 9000 rpm for 10 min, and the supernatant was diluted 10 times to determine its UV visible absorption. Compared with the standard curve, the DOX loading efficiency was computed. Finally, DNA-hybrid-gated UCNP@UIO-66-NH2/DOX was re-dispersed in PBS buffer (2 mL).
Evaluation of drug release and detection ability
For the kinetic study, the above DNA-hybrid-gated UCNP@UIO-66-NH2/DOX solution (2 mL) was incubated with miR-21 (2 nmol) at 37 °C with different incubation times. The upconversion fluorescence was measured with 980 nm excitation (1.5 W). For linear research, 2 mL of the above solution was treated with variable concentrations of miR-21. After 3 h of incubation, the fluorescence of the respective sample solutions was recorded under the same conditions. For the specificity experiment, the above solution was mixed with let-7d, miR-141, miR-15a, miR-200b and miR-21 (2 nmol). After 3 h of reaction, the upconversion fluorescence was detected in the same way. All experiments were repeated three times.
Fluorescence imaging of miRNA
Breast cancer cells (MCF-7) and normal epithelial cells (HEK-293T) were cultured in Dulbecco's Modified Eagle Medium (DMEM) supplied with 10% fetal bovine serum and 100 U mL−1 antibiotics penicillin/streptomycin (1%). MCF-7 cells and HEK-293T cells were seeded in 35 mm confocal culture dishes at a density of 7 × 105 cells per well. After 24 h, the MCF-7 cells were then divided randomly into two groups. The medium of HEK-293T cells and the first group were replaced with fresh culture medium and that of the other group was replaced with fresh medium containing curcumin (5 μM). After 24 h, the cells were further incubated with DNA-hybrid-gated UCNP@UIO-66-NH2/DOX (50 μg mL−1) for 12 h, afterwards the cells were washed three times with PBS buffer intensely and immobilized with 4% paraformaldehyde. The cell membrane was stained with Dil and the nucleus was stained with DAPI. The UCF images (emission range 400–500 nm) were recorded with the excitation at 980 nm.
Cell viability experiments by Cell Counting Kit-8 assay
MCF-7 cells and HEK-293T cells were seeded in a 96-well plate at a density of 5 × 103 cells per well in culture medium (100 μL) and incubated for 24 h. After removing the medium, fresh medium (100 μL) containing nanoprobes (50 μg mL−1) with different treatments was added into the wells and incubated for 12 h. Following intensive washing, the cells were further incubated for another 24 h with fresh DMEM. Then the cell medium was replaced by fresh medium (100 μL) containing CCK-8 solution (10 μL), and the 96-well plate was incubated at 37 °C for 2 h. The absorption of samples was measured with a microplate reader at a wavelength of 450 nm.
Author contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
This research work was financially supported by the National Natural Science Foundation of China (No. 81972011, 81972025), Chongqing Technology Innovation and Application Demonstration Project (cstc2019jscx-msxmX0088), the Key Project of Education Department of Sichuan (No. 16ZA0181) and Top Talent Project of Chongqing Medical University (BJRC201817).
Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ra00154j
Notes and references
- Esteller M. Nat. Rev. Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- He L. Thomson J. M. Hemann M. T. Hernando-Monge E. Mu D. Goodson S. Powers S. Cordon-Cardo C. Lowe S. W. Hannon G. J. Hammond S. M. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J. Getz G. Miska E. A. Alvarez-Saavedra E. Lamb J. Peck D. Sweet-Cordero A. Ebet B. L. Mak R. H. Ferrando A. A. Downing J. R. Jacks T. Horvitz H. R. Golub T. R. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- Lammers T. Aime S. Hennink W. E. Storm G. Kiessling F. Acc. Chem. Res. 2011;44:1029–1038. doi: 10.1021/ar200019c. [DOI] [PubMed] [Google Scholar]
- Ma Y. Huang J. Song S. Chen H. Zhang Z. Small. 2016;12:4936–4954. doi: 10.1002/smll.201600635. [DOI] [PubMed] [Google Scholar]
- Conde J. Edelman E. R. Artzi N. Adv. Drug Delivery Rev. 2015;81:169–183. doi: 10.1016/j.addr.2014.09.003. [DOI] [PubMed] [Google Scholar]
- Bai S. Xu B. Guo Y. Qiu J. Yu W. Xie G. Theranostics. 2018;8:2424–2434. doi: 10.7150/thno.23852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong K. T. Law W. C. Hu R. Ye L. Liu L. Swihart M. T. Prasad P. N. Chem. Soc. Rev. 2013;42:1236–1250. doi: 10.1039/C2CS35392J. [DOI] [PubMed] [Google Scholar]
- Zhu S. Tian R. Antaris A. L. Chen X. Dai H. Adv. Mater. 2019;31:1900321. doi: 10.1002/adma.201900321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S. Weitemier A. Z. Zeng X. He L. McHugh T. J. Science. 2018;359:679–684. doi: 10.1126/science.aaq1144. [DOI] [PubMed] [Google Scholar]
- Idris N. M. Gnanasammandhan M. K. Zhang J. Ho P. C. Mahendran R. Zhang Y. Nat. Med. 2012;18:1580–1585. doi: 10.1038/nm.2933. [DOI] [PubMed] [Google Scholar]
- Xu C. T. Zhan Q. Liu H. Somesfalean G. Qian J. He S. Andersson-Engels S. Laser Photonics Rev. 2013;7:663–697. doi: 10.1002/lpor.201200052. [DOI] [Google Scholar]
- Ji Y. Lu F. Hu W. Zhao H. Tang Y. Li B. Hu X. Li X. Lu X. Fan Q. Huang W. Biomaterials. 2019;219:119393. doi: 10.1016/j.biomaterials.2019.119393. [DOI] [PubMed] [Google Scholar]
- Ding Q. Zhan Q. Zhou X. Zhang T. Xing D. Small. 2016;12:5944–5953. doi: 10.1002/smll.201601724. [DOI] [PubMed] [Google Scholar]
- Xu J. Han W. Cheng Z. Yang P. Bi H. Yang D. Niu N. He F. Gai S. Lin J. Chem. Sci. 2018;9:3233–3247. doi: 10.1039/C7SC05414A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C. H. Willner I. Angew. Chem., Int. Ed. 2015;54:12212–12235. doi: 10.1002/anie.201503054. [DOI] [PubMed] [Google Scholar]
- Vazquez-Gonzalez M. Willner I. Langmuir. 2018;34:14692–14710. doi: 10.1021/acs.langmuir.8b00478. [DOI] [PubMed] [Google Scholar]
- Zhang P. Cheng F. Zhou R. Cao J. Li J. Burda C. Min Q. Zhu J. J. Angew. Chem., Int. Ed. 2014;53:2371–2375. doi: 10.1002/anie.201308920. [DOI] [PubMed] [Google Scholar]
- Kahn J. S. Freage L. Enkin N. Garcia M. A. Willner I. Adv. Mater. 2017;29:1602782. doi: 10.1002/adma.201602782. [DOI] [PubMed] [Google Scholar]
- Chen W. H. Xu Y. Liao W. C. Yang S. S. Willner I. Adv. Funct. Mater. 2017;27:1702102. doi: 10.1002/adfm.201702102. [DOI] [Google Scholar]
- Furukawa H. Cordova K. E. O'Keeffe M. Yaghi O. M. Sci. 2013;341:1230444. doi: 10.1126/science.1230444. [DOI] [PubMed] [Google Scholar]
- Wu M. X. Yang Y. W. Adv. Mater. 2017;29:1606134. doi: 10.1002/adma.201606134. [DOI] [PubMed] [Google Scholar]
- Wang S. McGuirk C. M. Ross M. B. Wang S. Chen P. Xing H. Liu Y. Mirkin C. A. J. Am. Chem. Soc. 2017;139:9827–9830. doi: 10.1021/jacs.7b05633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S. Li C. Shi H. Huang Y. Li G. Anal. Chem. 2018;90:9929–9935. doi: 10.1021/acs.analchem.8b02127. [DOI] [PubMed] [Google Scholar]
- Rothlisberger P. Hollenstein M. Adv. Drug Delivery Rev. 2018;134:3–21. doi: 10.1016/j.addr.2018.04.007. [DOI] [PubMed] [Google Scholar]
- Wang M. Abbineni G. Clevenger A. Mao C. Xu S. Nanomedicine. 2011;7:710–729. doi: 10.1016/j.nano.2011.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer J. C. Manseau M. P. Murray J. I. van Veggel F. C. Langmuir. 2010;26:1157–1164. doi: 10.1021/la902260j. [DOI] [PubMed] [Google Scholar]
- Guo C. Hu P. Yu L. Yuan F. Mater. Lett. 2009;63:1013–1015. doi: 10.1016/j.matlet.2008.12.053. [DOI] [Google Scholar]
- Liang C. P. Ma P. Q. Liu H. Guo X. Yin B. C. Ye B. C. Angew. Chem., Int. Ed. 2017;56:9077–9081. doi: 10.1002/anie.201704147. [DOI] [PubMed] [Google Scholar]
- Mudduluru G. George-William J. N. Muppala S. Asangani I. A. Kumarswamy R. Nelson L. D. Allgayer H. Biosci. Rep. 2011;31:185–197. doi: 10.1042/BSR20100065. [DOI] [PubMed] [Google Scholar]
- Wu Y. Han J. Xue P. Xu R. Kang Y. Nanoscale. 2015;7:1753–1759. doi: 10.1039/C4NR05447D. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.