Abstract
The incidence of central nervous system (CNS) diseases is increasing with the aging population. However, it remains challenging to deliver drugs into the CNS because of the existence of a blood–brain barrier (BBB). Notably, rabies virus glycoprotein (RVG) peptides have been developed as delivery ligands for CNS diseases. So far, massive RVG peptide modified carriers have been reported, such as liposomes, micelles, polymers, exosomes, dendrimers, and proteins. Moreover, these drug delivery systems can encapsulate almost all small molecules and macromolecule drugs, including siRNA, microRNAs, DNA, proteins, and other nanoparticles, to treat various CNS diseases with efficient and safe drugs. In this review, targeted delivery systems with RVG peptide modified carriers possessing favorable biocompatibility and delivery efficiency are summarized.
Rabies virus glycoprotein (RVG) peptides have been developed to deliver drugs for CNS diseases. In the present review, RVG-mediated drug delivery systems are summarised, which can deliver almost all small molecules and macromolecule agents.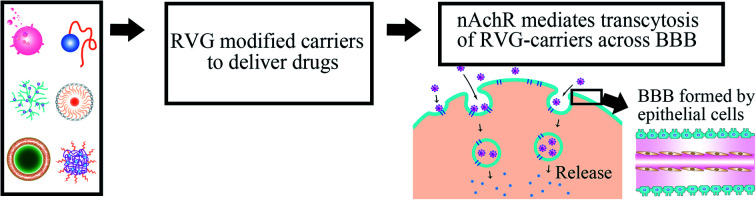
1. Introduction
As the incidence of CNS diseases is increasing with the aging population, it is a significant challenge to deliver therapeutic agents to the CNS in drug development.1 Besides, the lack of effective treatment can result in high-cost therapies compared to a quarter of all other disease burdens based on real-world data obtained from a healthcare organization in the Basque Country from 2016 to 2018.2 Therefore, drug delivery systems developed for the CNS improve many patients' well-being and significantly reduce health costs. However, a challenging obstacle must be solved to enable bioactive molecules to reach the CNS with therapeutic quantities of the BBB.3
BBB is composed of brain endothelial cells, astrocytes, and pericytes with absent fenestrations and more restrictive tight junctions to prevent mostly large-molecule and small-molecule agents from entering the CNS,4 except when many brain diseases (such as glioblastoma and spinal cord injury) destroy its integrity.5 BBB regulates the passage of solutes between the CNS and blood due to tight connections and the lack of fenestration in the epithelial layer (Fig. 1).6,7 BBB is naturally formed to prevent large molecular substances from entering the CNS except for some small lipid-soluble molecules with molecular weight (MW) <111 Da,8,9 such as proteins, nucleic acids, and other therapeutic agents with unique properties.10,11 To solve this problem, researchers have focused on cell-penetrating RVG peptides, which can be served as a suitable candidate cell-penetrating peptide with the ability to deliver therapeutic agents to the brain in a non-invasive manner.12 There are two kinds of CPPs, natural and synthetic, such as TAT13 and RVG.14 Although they have great differences in chemical structure, they still have some common properties, such as high efficiency of transmembrane transport and no obvious cytotoxicity. However, these CPPs have poor cell selectivity and can be introduced into almost all cells. Fortunately, RVG avoids this deficiency with the characteristics of the target central nervous system because RVG can travel through a very long nerve bundle from peripheral nerve endings to the CNS along the spinal cord without retarding the signal strength.15
Fig. 1. A diagrammatic sketch of the BBB structure. The fenestration of the endothelial layer does not form a barrier (arrow), and epithelial cells form BBB (green) by tight junctions (arrowhead).
2. Delivery mechanism of RVG peptides
RVG with a 505 amino acid glycoprotein is located in the lipid layer of the rabies virus surface, which enables RVG with neurotropism.16,17 Among those 189–214 amino acid sequences of rabies virus are currently the most common critical binding regions for nerve cell entry and viral fusion.18,19 Many studies showed that 29 amino acid peptides derived from 189–214 amino acid sequences (RVG29-d9R, YTIWMPENPRPGTPCDIFTNSRGKRASNGGGG (d) RRRRRR RRR) of RVG specifically deliver siRNA and cationic polymers into nerve cells by neuronal nicotinic acetylcholine receptor (nAChR)-mediated cellular transduction.20,21 Moreover, 330–357 amino acid sequences are important nerve-binding regions of rabies virus-derived peptides.22,23 Sufficient shreds of evidence proved that the derived peptide of rabies virus (KSVRTWNEIIPSKGCLRVGGRCHPHVNGGGRRRRRRRRRRRRRRRRRRRRRRRRR) precisely and effectively deliver its cargos to the CNS after in vitro administration.24,25
Receptor-mediated endocytosis is the mechanism for RVG-carrier to penetrate through the BBB modified with RVG nanocarriers and primarily bind to nAChR26 or γ-aminobutyric acid (GABA) receptor,16,27 which is widely located on the extracellular surface of target cells, such as microvascular endothelial cells and neurons.28 Then transcytosis processes were initiated, and therapeutic agents were finally released in the CNS. RVG can specifically recognize receptors on the surface BBB-specific receptor (such as nAChR29,30 or γ-aminobutyric acid receptor27) to deliver carriers into the CNS in a non-invasive way. The detection results showed that nAChR exists not only in the neuron but also in microglia,31 such as endothelial cells32 and Schwann cell.33 However, RVG only infects neurons, not glial cells.34 RVG29 sequence with the transmembrane domain (440–461 amino acid sequence) can specifically recognize nAChR.24 It was also reported to bind with neurotransmitter γ-aminobutyric acid to facilitate drug delivery into the CNS.35 RVG peptides usually move and enrich along neurons that extend from the nasal cavity to the brain by the olfactory pathway and trigeminal pathway.16 Otherwise, the whole-brain or whole-spinal cord targeting should be operated by systemic delivery.
3. Binding modes of RVG peptides and carriers for treating CNS diseases
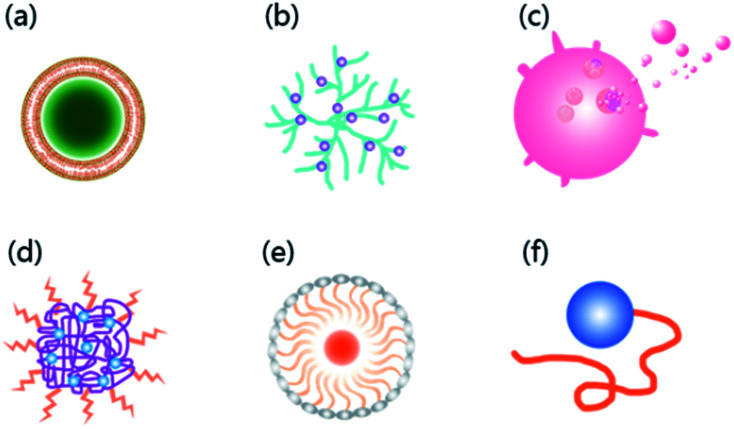
With the characteristics of neurotropic, BBB permeable, and biologically safe,36 RVG has been successfully used to transport a variety of nanocarriers into the CNS,37 such as liposomes,38 polymers,39 dendrimers,40 exosomes,25 micelles,41 bioactive proteins,41 and moieties42 with efficiency and fast permeability43,44 (Fig. 2). Also, when RVG-carriers provide biological macromolecules to the CNS, RVG modified carriers exhibit high load capacity.20,39
Fig. 2. Six common carriers with RVG as the ligand currently developed to deliver drugs across the BBB. (a) Liposomes; (b) dendrimers; (c) exosomes; (d) polymers; (e) micelles; (f) bioactive proteins.
There are usually two linking modes between the RVG peptides and carrier components. One is the stable linking mode of covalent connection,45 and the other is electrostatic interaction on the surface of the carrier.46 For example, polylactic acid-glycolic acid (PLGA) was modified by RVG with a covalent connection to promote the CNS-specific delivery of encapsulated molecules.47 PLGA is a block copolymer made of polylactic acid (PLA)48 and polyglycolic acid (PGA)49 with biodegradable, biocompatible, and easy functionalization properties, which can easily be hydrolyzed and metabolized.50 Furthermore, it has been approved for extensive disease treatment by the FDA and European Medicine Agency.51 In a non-covalent strategy, electrostatic interaction has been applied extensively. Cationic RVG peptides and negatively charged therapeutic molecules (such as negatively charged nucleic acids) usually self-assemble into nanocomposites, which enable carriers to deliver the drug into the CNS.52 Also, RVG assembles with the oxidative degradable arginine-grafted polymer (PAM-ABP) and acquired condensed long-chain DNA by electrostatic interaction.53 The dendrimers–reagent complex completes the transport function perfectly. For example, dendrimers, such as polyamide amine (PAMAM), protect the agent from damage and the net positive charge of dendrimers–agent complex determines the transfection efficiency.54
4. Application progress of RVG-modified carriers for CNS disease treatment
4.1. RVG peptides modified liposomes for treating CNS diseases
As common drug delivery nanocarriers, liposomes are popular agent delivery systems with good biocompatibility, biosafety, and tissue specificity.55,56 Many liposome delivery systems consist of almost all hydrophilic and lipophilic small molecule therapeutic agents to the CNS. For example, liposome-siRNA-peptide complexes (LSPCs) were carried to the CNS to reduce the normal cellular prion protein expression level, PrPC. Our research team first transplanted U251MG cells into the striatum to prepare a mouse glioma model. Then RVG-derived peptide (RDP)-modified liposome loaded curcumin was used to treat glioma-bearing mice. Moreover, the experimental results showed that RDP could deliver curcumin into the CNS to prolong glioma-bearing mice's life span.57 Similarly, the liposome encapsulating polypyridyl ruthenium complex was led by RDP to reduce the inflammation of meningeal encephalitis.58
Also, RVG peptide modified liposomes can effectively silence target genes. RVG combined to siRNA/trimethylated chitosan (TMC) via bifunctional polyethylene glycol (PEG) showed binding serum stability, very low toxicity, and high intake rate of cells, compared with unmodified siRNA/TMC-mPEG complexes. In addition, experimental results showed that significantly enriched therapeutic agents in the brain of mice were injected with RVG-peptide complexes, which proved that the BACE1 gene was knocked out by the RVG-chitosan complex.59 Conceição's studies showed that stable nucleic acid-lipid particles were prepared by linking RVG27-9R with liposome loaded siRNA mutant for ataxin-3. Further animal diseases models in mice indicated that RVG27-9R modified liposome loaded stable nucleic acid-lipid particles (SNLPs) can effectively silence ataxin-3, then reduce neuropathological and behavioral abnormal in Machado–Joseph disease, which provides a new way for neurodegenerative therapy.60
4.2. RVG peptide modified dendrimers for treating CNS diseases
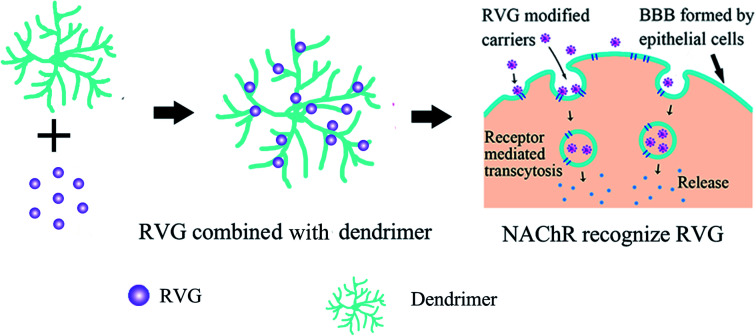
Multifunctional dendrimers (such as PLGA,61 PAMAM62) have attracted researchers' attention as drug delivery agents because of their unique properties, such as multifunction, biocompatibility, and adjustable size.63 RVG is usually first linked to dendrimer molecules on its surface. Then RVG peptides bind receptors (nAchR or γ-aminobutyric acid) on the epithelial cells, which are the BBB's main structure to mediate the transcytosis of carriers across the BBB. At last, the internalization of carriers allows drug release into the CNS, as shown in Fig. 3.
Fig. 3. Delivery of RVG modified dendrimer.
There was no effective treatment of brain injury after circulatory arrest for neuritis and excitotoxicity regulation following hypothermic circulatory arrest (HCA). The dendrimers assisted enormously to treat brain injury after circulatory arrest. For example, dendrimer-N-acetylcysteine (D-NAC) and dendrimer-valproic acid (D-VPA) conjugates were produced for the canine brain injury model following circulatory arrest. Mishra found that PAMAM dendrites with hydroxyl end groups were absorbed in the injured brain after systemic administration and localized in injured neurons and microglia.64 Moreover, compared with the combination of high-dose VPA, NAC, or free VPA, the low-dose group's 24 hour neurological deficit score was significantly improved.
As a highly ordered and well-defined macromolecule, PAMAM has been widely utilized in various fields, including gene therapy,65 biomedical imaging,63 and sensing of dendrimers.66 PAMAM dendrimers are multibranched monomers of polymer molecules that are derived from a single core with open, flat, asymmetrical shapes in the next generation.67 After the core of dendrimers was removed, some uniform fragments were maintained. The number of dendrons depends on the diversity of the central core (2, 3, 4, or more).
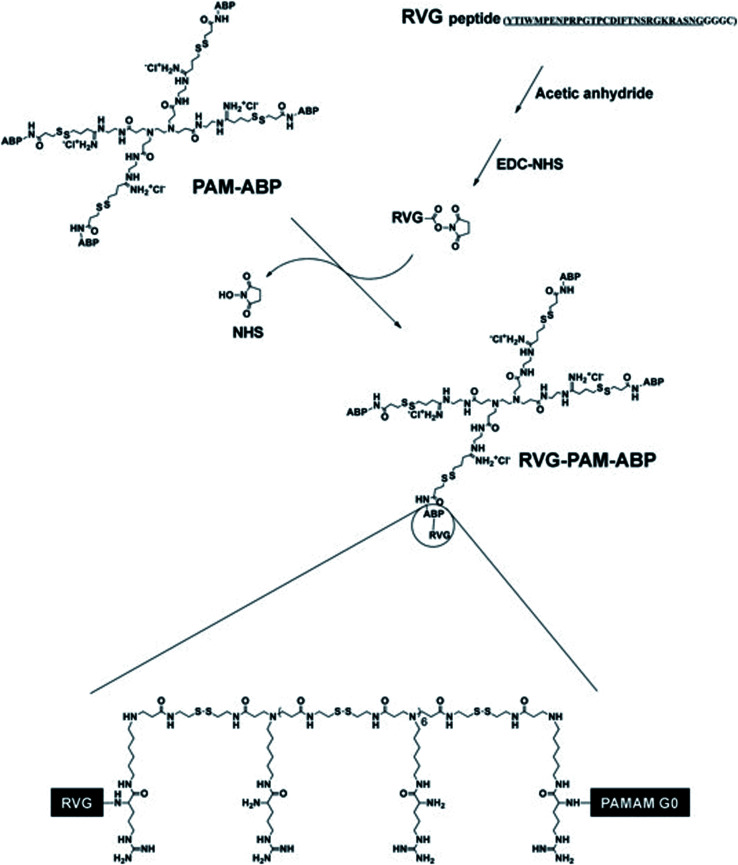
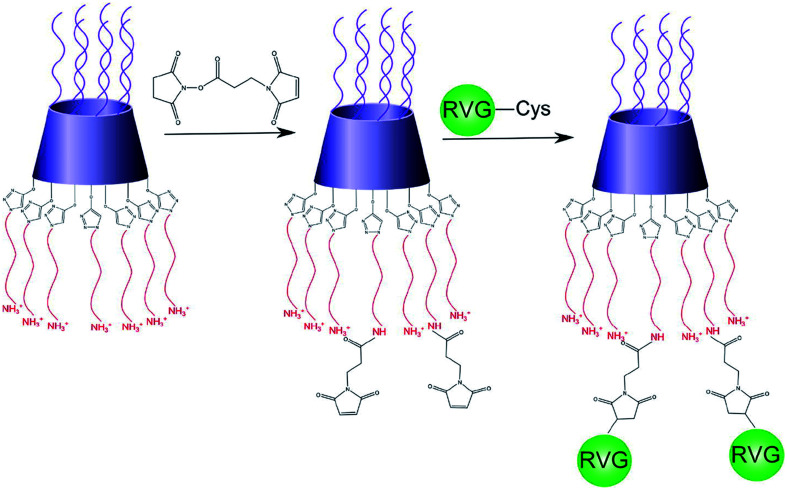
Studies found that PAMAM may be used as a nano-scale spherical polymer for efficient gene encapsulation to improve the transfection efficiency of nerve cells, and cell-penetrating peptides (CPPs) modified PAMAM can facilitate its delivery.65 For example, RVG29 was covalently bonded with PAMAM with the help of bifunctional polyethylene glycol (PEG) to prepare PAMAM-PEG-RVG29, which combined with DNA to form PAMAM-PEG-RVG/DNA composite. Then the targeting efficiency of PAMAM-PEG-RVG29 encapsulating DNA was evaluated in vitro and in vivo. The experimental results showed that RVG-modified PAMAM-PEG exhibited higher BBB crossover efficiency compared with PAMAM/DNA in vitro. Besides, in vivo imaging showed that PAMAM-PEG-RVG29 could preferably accumulate in the CNS with higher reporter gene expression efficiency than that of RVG unmodified PAMAM.68 Also, to further increase the transfection efficiency, arginine-grafted bio-reducible poly(disulfide amine) (ABP) was combined with PAMAM to form a PAM-ABP complex (Scheme 1). By combining RVG with redox-sensitive biodegradable arginine-grafted polymer (PAM-ABP), RVG can produce particles with plasmid DNA without changing the releasing characteristics of environmentally sensitive DNA and the toxicity distribution of PAM-ABP, which showed that RVG-PAM-ABP quantitatively enhanced the cell transfection efficiency.53
Scheme 1. Synthesis progress of RVG-PAM-ABP (this figure has been reproduced from ref. 53 from its author, copyright 2014).
4.3. RVG peptide modified exosomes for CNS disease treatment
As extracellular vesicles (40–120 nm) are released by many cell types, exosomes can transfer mRNA,69 microRNA,70 and protein71 for therapeutic purposes.
Exosomes can effectively deliver therapeutic RNA or nucleoprotein for gene treatment.70 Lydia combined RDP with exosomes to express Lamp2b and found that the complexes selectively bind to nAChR expressed on neurons, which may enable exosomes to enter brain cells effectively. In addition, intravenous RVG-targeted exosomes specifically deliver glyceraldehyde-3-phosphate dehydrogenase (GAPDH) siRNA to the CNS, leading to specific gene knockout.72 Also, the exosomes of mesenchymal stem cells (MSCs) can prevent memory impairment in Alzheimer's disease (AD) animal models with normalized inflammatory cytokine levels.73
4.3.1. Targeted delivery of RVG-modified exosome loaded siRNA to treat Alzheimer's disease or drug addiction (such as morphine, fentanyl, and methadone)
Alzheimer's disease is the most common cause of dementia among the aging population. Brains of patients with this condition have a marked inflammatory response.74 Fortunately, RVG-exosomes can deliver therapeutic agents to the CNS for regulating inflammatory responses73 and aggregation of amyloid-beta (A-beta) peptides75 in Alzheimer's disease. Besides, RVG-modified exosomes were prepared from human embryonic kidney 293T (HEK 293T) cells and expressed targeting peptides on exosomes' surface for the delivery of MOR siRNA to treat morphine addiction. The experimental results show that the RVG-modified exosome loaded siRNA were safe and efficiently delivered into the CNS to reduce efficient inhibition of morphine recurrence.76
4.3.2. Targeted delivery of RVG-exosomes for stroke in the juvenile and adult nervous system
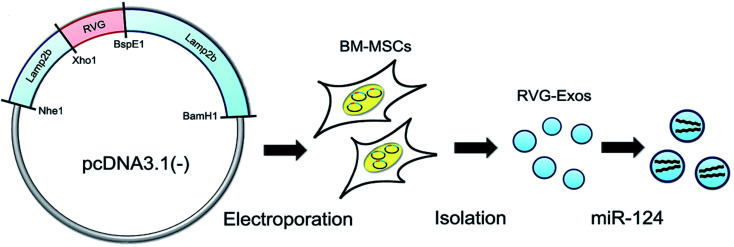
Stroke is a group of diseases with cerebral ischemia and hemorrhagic injury as primary clinical manifestations, also known as stroke or cerebrovascular accident, with high mortality and disability rate, such as hemorrhagic stroke (cerebral hemorrhage or subarachnoid hemorrhage) and ischemic stroke (cerebral infarction, cerebral thrombosis). High Mobility Group Box 1 (HMGB1) was delivered into the CNS by rabies virus glycoprotein (RVG) peptide-decorated exosomes for the ischemic brain, and the results showed good treatment results with infarct size decrease.77 To produce RVG-exosomes for ischemic injury, Yang et al. fused RVG peptide with lysosome-related membrane glycoprotein 2b (Lamp2b) and led pcDNA3.1(−)-RVG-Lamp2b plasmid into bone marrow mesenchymal stem cell (BM-MSC). Then RVG-exosomes loaded with mimic or disordered microRNAs by electroporation purified the exosomes from the culture supernatant of BM-MSC (Fig. 4). The experimental results showed that the systematic application of RVG-exosomes containing miR-124 could promote cortical neural progenitor cells' neural function and protect ischemic damage through strong cortical neurogenesis, indicating that RVG-exosomes can be used for neuromodulation after stroke.78
Fig. 4. RVG-modified exosomes, including plasmid recombination, BM-MSC electroporation, exosome separation, and cargo loading (this figure has been reproduced from ref. 78 from Elsevier Science, copyright 2017).
4.3.3. RVG modified exosome loaded siRNA for treating neurodegenerative diseases
Parkinson's disease is a neurodegenerative disease and one of the common nervous system diseases with abnormal alpha-synuclein (α-Syn) aggregates caused by misfolded proteins.
To achieve widespread delivery of siRNA to the brain, Cooper injected RVG-modified exosomes that express the rabies virus glycoprotein. In S129Dα-Syn transgenic mice, they found that the levels of α-SynmRNA and protein in the whole brain decreased 7 days after injection, which resulted in a large decrease in protein aggregates in the nerves, including a reduction of dopaminergic neurons in the substantia nigra.79 Cooper's research highlights the therapeutic potential of RVG-modified exosomes, which means that RVG-exosome delivery targeting siRNA technology could treat protein-misfolding diseases.
4.4. RVG peptide modified polymers for CNS disease treatment
Polymer delivery systems have been widely promoted in CNS disease treatment.80 As a biodegradable polymer, GLIADEL wafer can load multiple therapeutic agents for the treatment of malignant glioma in an animal model.81 The critical property of the biological activity of polymers (such as cyclodextrin,82 pluronic 85,83 pluronic 123,84 and pluronic F127 (ref. 85)) is that they can bind to RVG peptides and then deliver agents into the CNS to achieve therapeutic purposes. On this basis, polymers have stealth characteristics, which can avoid reticular endothelial system (RES) and other scavenging. To make full use of different polymers, the mixed use of polymers with complementary properties can improve their delivery efficiency.86 For example, RVG-amphiphilic cyclodextrin was successfully prepared, optimally at a molar ratio of 1 : 1.5 : 0.5 (cationic cyclodextrin : PEGylated cyclodextrin : RVG-tagged PEGylated cyclodextrin) with a size of 281 ± 39.72 nm (Scheme 2), which indicates a promising polymer delivery system for CNS disease treatment.
Scheme 2. Schematic of RVG peptide combining with an amphiphilic cyclodextrin molecule (reproduced with permission from ref. 87 from Elsevier Science, copyright 2015).
You et al. report a kind of RVG combined monomethoxy polyethylene glycol (mPEG)-polylactic acid glycolic acid (PLGA) nanoparticles (RNP-DFO), which can not only effectively transmit to the CNS but also extend the cycle time of deferoxamine. This RVG delivery system can significantly enhance the concentration of deferoxamine and greatly reduce their cerebral iron content and ROS level in the CNS.28 Zhang's results showed that the micelles composed of pluronic P123 and pluronic F127 combined the multidrug resistance (MDR) sensitization characteristics of P123 with the long cycle effect of pluronic F127, and enhanced the activity of PTX in overcoming MDR in lung cancer.88 Liu's research shows that tryptophan derivatives functionalized pluronic P123/F127 should be developed to deliver antiepileptic agents to the CNS by transport-mediated endocytosis.89
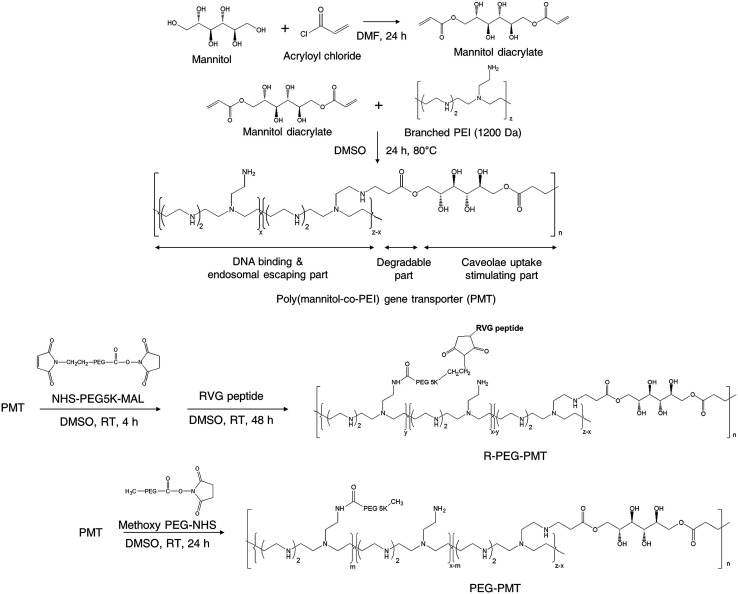
Also, nucleic acids combine with cationic polymers via ionic interaction to efficiently deliver genes and escape from them using proton buffering capacity. Therefore, RNA interference (RNAi) became one of the powerful treatment tools for neurodegenerative diseases by directly blocking pathogenic genes. For example, RVG-modified poly(mannitol-co-PEI) gene transporter (PMT) delivered PMT/siRNA complexes to the CNS by binding nAChR expressed on BBB in vitro and in vivo. RVG-modified PMT can transport β-site amyloid precursor protein-cleaving enzyme 1 (BACE1, a therapeutic target in Alzheimer's disease) siRNA into the CNS, and the progress of Alzheimer's disease (AD) was blocked after the injection of RVG-modified PMT/siRNA complexes,90 as shown in Scheme 3. Besides, RVG and chitosan modified purron-based nanocarriers had been used to deliver reporter protein β-galactosidase (β-Gal) to the CNS. The results indicated that the nanocarriers, including chitosan and RVG, were very influential in facilitating drug delivery into the CNS, suggesting that RVG-chitosan nanocarriers are available for delivering proteins to the CNS.91
Scheme 3. Synthetic route of RVG-PEG-PMT (this scheme has been reproduced from ref. 90 with permission from Elsevier Science, copyright 2014).
4.5. RVG peptide modified micelles for CNS diseases treatment
The drug delivery system based on micelles has achieved significant solubility, metabolic stability, and cycle time.92,93 RVG can help the therapeutic drug delivery to the CNS despite the BBB.94 Besides, many other factors were affecting micelles' in vivo effects to deliver into the brain, including the physical and chemical properties of micelles, enzymatic degradation, and so on.95 For example, Zou-D designed and synthesized ganglioside GM1 micelles spontaneously encapsulating anticancer drug ganglioside DOX with slow drug release for CNS targeting effects in vitro and in vivo.96
For penicillin-sensitive and drug-resistant pneumococcal meningitis, PEG-based nano-BA12K was prepared with RVG29 and P-glycoprotein (P-gp) inhibitor (pluronic P85 monomer) to make up an integrated micelle system (RVG29-nano-BAP85) for resisting penicillin-sensitive and drug-resistant pneumococcal meningitis. The results showed that RVG29-nano-BAP85 had higher BBB traversal ability than single formulation micelles. Excellent CNS targeting performance of the mixed micelles have the characteristics of higher receptor binding affinity, lower P-gp function, and negligible systemic toxicity to penicillin-sensitive and drug-resistant pneumococcal meningitis.19
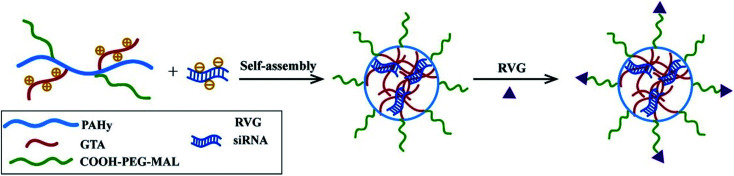
Besides, RVG peptides can modify micelles to provide CNS-targeting nucleic acids by electrostatic interaction.97 Hong Hu connected RVG and micelles under heterobifunctional PEG to enhance the stability and biocompatibility (Fig. 5). The experimental results showed that micelles modified by RVG could effectively deliver siRNA to the CNS.98
Fig. 5. The preparation of RVG-PEG-g-PAHy-GTA/siRNA micelles (reproduced with permission from ref. 98 with permission from Elsevier Science, copyright 2015).
4.6. RVG peptide modified bioactive proteins for CNS disease treatment
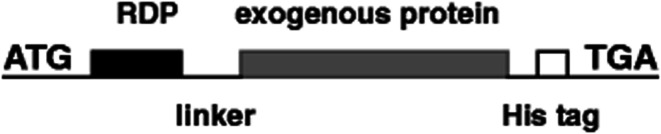
With the increasing aging population and nervous system diseases, more than 12% of disease deaths are caused by nervous system diseases.99 In recent years, targeted therapy systems have brought hope to these patients, especially for single-gene illnesses that cause protein dysfunction and follow pathological cascade.100 Many experiments proved that it is feasible to control diseases by activating or inhibiting gene transcription using bioactive proteins.101,102 For instance, CNS-derived neurotrophic factor (BDNF) was combined by our research team with RDP, and then injected into the mice to repair damaged CNS (Fig. 6). Subsequent inspections found that the BDNF protein can efficiently and specifically enter the CNS in 15 min with the therapeutic effect of reducing stroke volume and neural deficit.103 Besides, 43 amino acid peptides derived from RVG were fused to β-Gal, and it was found that the fusion protein was targeted to hippocampal neurons after systemic administration,43 which may open a new possibility for targeted delivery of small molecules and macromolecule to local brain regions using neurotrophic glycoprotein-derived peptides.
Fig. 6. The structural representation of RDP and exogenous proteins, such as BDNF, β-galactosidase (β-Gal), and luciferase (Luc) (reproduced with permission from ref. 103 with permission from Springer Nature, copyright 2012).
Gene editing technologies promote clinical practice through rectifying genetic mutation and effective gene therapy of immune cells.104 However, if its treatment can be targeted, the curative effects will significantly be improved. For example, as a 38 kD site-specific recombinase in phage P1, Cre protein had been a promising tool for genome editing to shear the plasmid and traverse the cell membrane of cultured Neuro2a cells. The studies found that RVG not only sends Cre to the targeted area in the CNS by infecting Cre-expressing neurons105 but also modifies Cre recombinase for genome editing in mouse brain.106 Furthermore, after RVG-Cre was intravenously administered to mouse lines mTmG and Rosa26lacZ, Zou-Z found that RVG-Cre can be enriched in the brain and the somatic genome was safely edited in adult mice.107
5. Conclusion and perspectives
With the widespread unhealthy lifestyles and the prolonged life expectancy of modern people, the incidence of CNS diseases worldwide has gradually increased in recent years. As a special CNS-targeted drug delivery system, RVG delivery systems are one of the most promising topics in the field of brain disease treatment. RVG-mediated drug delivery systems have been widely applied to treat CNS diseases with great advantages in delivering nanocarriers across the BBB into the brain with biological properties: stable and suitable physical and chemical properties for in vivo applications, superior targeting, and therapeutic efficiency. However, a more detailed understanding of its application progress will promote its engineering level, such as scaling up and quality control. More efforts, including new biological materials and biotechnology, should be made to achieve the translation of RVG delivery systems.
Author contributions
Q. Wang, S. Cheng, and F. Qin collected and extensively researched the literature in the field; A. Fu and C. Fu conceived and wrote the manuscript. All authors have approved the final version of the manuscript.
Funding
This research was funded by the Chongqing Science and Technology Commission (No. cstc2018jcyjAX0612, cstc2018jscx-msybX0304) and a start-up fund for the doctor of Southwestern University.
Conflicts of interest
The authors declare no conflict of interest.
Supplementary Material
References
- Dawson W. D. Bobrow K. Ibanez A. Booi L. Pintado-Caipa M. Yamamoto S. Tarnanas I. Evans T. Comas-Herrera A. Cummings J. Kaye J. Yaffe K. Miller B. L. Eyre H. A. Lancet Neurol. 2020;19:972–974. doi: 10.1016/S1474-4422(20)30358-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Inhiesto E. Acaiturri-Ayesta M. T. Ustarroz-Aguirre I. Camahuali D. Urtaran-Laresgoiti M. Basabe-Aldecoa M. Nuño-Solinís R. Urizar E. J. Parkinson's Dis. 2020;2020:9106026. doi: 10.1155/2020/9106026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang Z. Shi J. Zhou Y. Wang K. Zhao Y. Li Q. Qiao Z. Wu A. Tan Z. Liu W. Bioorg. Chem. 2021;107:104602. doi: 10.1016/j.bioorg.2020.104602. [DOI] [PubMed] [Google Scholar]
- Cai Q. Wang L. Deng G. Liu J. Chen Q. Chen Z. Am. J. Transl. Res. 2016;8:749–764. [PMC free article] [PubMed] [Google Scholar]
- Hsu J. F. Chu S. M. Liao C. C. Wang C. J. Wang Y. S. Lai M. Y. Wang H. C. Huang H. R. Tsai M. H. Cancers. 2021;13:195. doi: 10.3390/cancers13020195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H. Gallet S. Klemm P. Scholl P. Folz-Donahue K. Altmüller J. Alber J. Heilinger C. Kukat C. Loyens A. Müller-Fielitz H. Sundaram S. Schwaninger M. Prevot V. Brüning J. C. Neuron. 2020;107:306–319.e9. doi: 10.1016/j.neuron.2020.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick Z. Frazer G. Ferro A. Clare S. Bouladoux N. Ferdinand J. Tuong Z. K. Negro-Demontel M. L. Kumar N. Suchanek O. Tajsic T. Harcourt K. Scott K. Bashford-Rogers R. Helmy A. Reich D. S. Belkaid Y. Lawley T. D. McGavern D. B. Clatworthy M. R. Nature. 2020;587:472–476. doi: 10.1038/s41586-020-2886-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manrique-Suarez V. Vispo N. S. Ramos O. S. Curr. Pharm. Biotechnol. 2021;22:1–11. doi: 10.2174/1389201022666210104115224. [DOI] [PubMed] [Google Scholar]
- Xie B.-S. Wang X. Pan Y.-H. Jiang G. Feng J.-F. Lin Y. Theranostics. 2021;11:1177–1191. doi: 10.7150/thno.46992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohmwald K. Soto J. A. Andrade-Parra C. Fernandez-Fierro A. Espinoza J. A. Rios M. Eugenin E. A. Gonzalez P. A. Opazo M. C. Riedel C. A. Kalergis A. M. Brain, Behav., Immun. 2021;91:159–171. doi: 10.1016/j.bbi.2020.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nwafor D. Brown C. Neural Regener. Res. 2021;16:99–100. doi: 10.4103/1673-5374.286958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardani K. Milani A. Shabani S. H. Bolhassani A. Expert Opin. Drug Delivery. 2019;16:1227–1258. doi: 10.1080/17425247.2019.1676720. [DOI] [PubMed] [Google Scholar]
- Vale N. Duarte D. Silva S. Correia A. S. Costa B. Gouveia M. J. Ferreira A. Pharmacol. Res. 2020;162:105231. doi: 10.1016/j.phrs.2020.105231. [DOI] [PubMed] [Google Scholar]
- Rodrigues B. D. Arora S. Kanekiyo T. Singh J. Brain Res. 2020;1734:10. doi: 10.1016/j.brainres.2020.146738. [DOI] [PubMed] [Google Scholar]
- Potratz M. Zaeck L. M. Weigel C. Klein A. Freuling C. M. Mueller T. Finke S. Acta Neuropathol. Commun. 2020;8:1–15. doi: 10.1186/s40478-019-0875-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung E. P. Cotter J. D. Prakapenka A. V. Cook R. L. DiPerna D. M. Sirianni R. W. Pharmaceutics. 2020;12:93. doi: 10.3390/pharmaceutics12020093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debnath A. Pathak D. C. D'Silva A. L. Batheja R. Ramamurthy N. Vakharia V. N. Chellappa M. M. Dey S. Vet. Microbiol. 2020;251:108890. doi: 10.1016/j.vetmic.2020.108890. [DOI] [PubMed] [Google Scholar]
- Dietzschold B. Li J. Faber M. Schnell M. Future Virol. 2008;3:481–490. doi: 10.2217/17460794.3.5.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mebatsion T. J. Virol. 2001;75:11496–11502. doi: 10.1128/JVI.75.23.11496-11502.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y. Gao C. Wang H. Sun J. Liang M. Feng Y. Liu Q. Fu S. Cui L. Gao C. Li Y. Yang Y. Sun B. Bioact. Mater. 2021;6:529–542. doi: 10.1016/j.bioactmat.2020.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han H. Zhang Y. Jin S. Chen P. Liu S. Xie Z. Jing X. Wang Z. New J. Chem. 2020;44:5692–5701. doi: 10.1039/C9NJ05366B. [DOI] [Google Scholar]
- Benmansour A. Leblois H. Coulon P. Tuffereau C. Gaudin Y. Flamand A. Lafay F. J. Virol. 1991;65:4198–4203. doi: 10.1128/JVI.65.8.4198-4203.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuffereau C. Leblois H. Benejean J. Coulon P. Lafay F. Flamand A. Virology. 1989;172:206–212. doi: 10.1016/0042-6822(89)90122-0. [DOI] [PubMed] [Google Scholar]
- Fu C. Xiang Y. Li X. Fu A. Mater. Sci. Eng., C. 2018;87:155–166. doi: 10.1016/j.msec.2017.12.029. [DOI] [PubMed] [Google Scholar]
- Liu L. Li Y. Peng H. Liu R. Ji W. Shi Z. Shen J. Ma G. Zhang X. Sci. Adv. 2020;6:eaba3967. doi: 10.1126/sciadv.aba3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huey R. Rathbone D. McCarron P. Hawthorne S. J. Drug Targeting. 2019;27:959–970. doi: 10.1080/1061186X.2019.1567737. [DOI] [PubMed] [Google Scholar]
- Fu A. Zhao Z. Gao F. Zhang M. Pharm. Res. 2013;30:2108–2117. doi: 10.1007/s11095-013-1068-6. [DOI] [PubMed] [Google Scholar]
- You L. H. Wang J. Liu T. Q. Zhang Y. L. Han X. X. Wang T. Guo S. S. Dong T. Y. Xu J. C. Anderson G. J. Liu Q. Chang Y. Z. Lou X. Nie G. J. ACS Nano. 2018;12:4123–4139. doi: 10.1021/acsnano.7b08172. [DOI] [PubMed] [Google Scholar]
- Bu X. Yin C. Zhang X. Zhang A. Shao X. Zhang Y. Yan Y. Med. Sci. Monit. 2019;25:5482–5492. doi: 10.12659/MSM.915251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu X. Zhang A. Chen Z. Zhang X. Zhang R. Yin C. Zhang J. Zhang Y. Yan Y. BMC Cancer. 2019;19:976. doi: 10.1186/s12885-019-6225-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbas M. Alzarea S. Papke R. L. Rahman S. CNS Neurol. Disord.: Drug Targets. 2020;19:1–13. doi: 10.2174/1871527319666201230102616. [DOI] [PubMed] [Google Scholar]
- Alwazzan A. Mehboob R. Gilani S. A. Hassan A. Perveen S. Tanvir I. Waseem H. Ehsan K. Ahmad F. J. Akram J. Front. Physiol. 2020;11:607239. doi: 10.3389/fphys.2020.607239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro R. Taetzsch T. Vaughan S. K. Godbe K. Chappell J. Settlage R. E. Valdez G. eLife. 2020;9:e56935. doi: 10.7554/eLife.56935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. Liu T. Dong Y. Kondoh K. Lu Z. Neurosci. Bull. 2019;35:909–920. doi: 10.1007/s12264-019-00374-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan L. Li Z. Y. Lv Q. K. Huang W. Int. J. Pharm. 2019;567:118449. doi: 10.1016/j.ijpharm.2019.118449. [DOI] [PubMed] [Google Scholar]
- Chen Y. X. Wei C. X. Lyu Y. Q. Chen H. Z. Jiang G. Gao X. L. Biomater. Sci. 2020;8:1073–1088. doi: 10.1039/C9BM01395D. [DOI] [PubMed] [Google Scholar]
- Pinheiro R. G. R. Granja A. Loureiro J. A. Pereira M. C. Pinheiro M. Neves A. R. Reis S. Pharm. Res. 2020;37:139. doi: 10.1007/s11095-020-02865-1. [DOI] [PubMed] [Google Scholar]
- Grafals-Ruiz N. Rios-Vicil C. I. Lozada-Delgado E. L. Quiñones-Díaz B. I. Noriega-Rivera R. A. Martínez-Zayas G. Santana-Rivera Y. Santiago-Sánchez G. S. Valiyeva F. Vivas-Mejía P. E. Int. J. Nanomed. 2020;15:2809–2828. doi: 10.2147/IJN.S241055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barani M. Mukhtar M. Rahdar A. Sargazi G. Thysiadou A. Kyzas G. Z. Molecules. 2021;26:186. doi: 10.3390/molecules26010186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y. L. Kuo L. W. Hsu C. H. Chiang C. S. Lu Y. J. Chang S. J. Hu S. H. J. Controlled Release. 2020;321:159–173. doi: 10.1016/j.jconrel.2020.02.018. [DOI] [PubMed] [Google Scholar]
- Mostafavi E. Medina-Cruz D. Vernet-Crua A. Cheng J. Cholula-Díaz J. L. Guisbiers G. Webster T. J. Expert Opin. Drug Delivery. 2020 doi: 10.1080/17425247.2021.1865306. [DOI] [PubMed] [Google Scholar]
- Kim H. Kim E. H. Kwak G. Chi S. G. Kim S. H. Yang Y. Int. J. Mol. Sci. 2021;22:14. doi: 10.3390/ijms22010014. [DOI] [Google Scholar]
- Xiang L. Zhou R. Fu A. Xu X. Huang Y. Hu C. J. Drug Targeting. 2011;19:632–636. doi: 10.3109/1061186X.2010.523788. [DOI] [PubMed] [Google Scholar]
- Kumar P. Wu H. McBride J. L. Jung K. E. Kim M. H. Davidson B. L. Lee S. K. Shankar P. Manjunath N. Nature. 2007;448:39–43. doi: 10.1038/nature05901. [DOI] [PubMed] [Google Scholar]
- Mo X., Liu E. and Huang Y., The intra-brain distribution of brain targeting delivery systems, Elsevier, Deutsch, 2019 [Google Scholar]
- Alfagih I. M. Aldosari B. AlQuadeib B. Almurshedi A. Alfagih M. M. Pharmaceutics. 2020;13:45. doi: 10.3390/pharmaceutics13010045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung E. P. Cotter J. D. Prakapenka A. V. Cook R. L. DiPerna D. M. Sirianni R. W. J. P. Pharmaceutics. 2020;12:93. doi: 10.3390/pharmaceutics12020093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosi G. Duskey J. T. Kreuter J. Expert Opin. Drug Delivery. 2020;17:23–32. doi: 10.1080/17425247.2020.1698544. [DOI] [PubMed] [Google Scholar]
- Gupta P. K. Tripathi S. K. Pappuru S. Chabattula S. C. Govarthanan K. Gupta S. Biswal B. K. Chakraborty D. Verma R. S. Mater. Sci. Eng., C. 2020;107:110285. doi: 10.1016/j.msec.2019.110285. [DOI] [PubMed] [Google Scholar]
- Mehta T. A., Shah N., Parekh K., Dhas N. and Patel J. K., in Surface Modification of Nanoparticles for Targeted Drug Delivery, Springer, 2019, pp. 33–71 [Google Scholar]
- Zhang Y. García-Gabilondo M. Rosell A. Roig A. Pharmaceutics. 2019;12:16. doi: 10.3390/pharmaceutics12010016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian Z. Ji T. J. Mater. Chem. B. 2020;8:6517–6529. doi: 10.1039/D0TB00713G. [DOI] [PubMed] [Google Scholar]
- Beloor J. Ramakrishna S. Nam K. Seon Choi C. Kim J. Kim S. H. Cho H. J. Shin H. Kim H. Kim S. W. Lee S. K. Kumar P. Small. 2015;11:2069–2079. doi: 10.1002/smll.201402933. [DOI] [PubMed] [Google Scholar]
- Rai D. B. Pooja D. Kulhari H. Microsyst. Technol. 2020:211–231. [Google Scholar]
- Hosseinpour S. Xu C. Walsh L. J. J. Photochem. Photobiol., B. 2021;215:112108. doi: 10.1016/j.jphotobiol.2020.112108. [DOI] [PubMed] [Google Scholar]
- Chen H. Liu L. Ma A. Yin T. Chen Z. Liang R. Qiu Y. Zheng M. Cai L. Biomaterials. 2021;269:120639. doi: 10.1016/j.biomaterials.2020.120639. [DOI] [PubMed] [Google Scholar]
- Zhao M. Zhao M. Fu C. Yu Y. Fu A. Int. J. Nanomed. 2018;13:1601–1610. doi: 10.2147/IJN.S157019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q. Fu C. Zhao Z. Fu A. Mol. Pharm. 2020;17:145–154. doi: 10.1021/acs.molpharmaceut.9b00848. [DOI] [PubMed] [Google Scholar]
- Gao Y. Wang Z. Y. Zhang J. Zhang Y. Huo H. Wang T. Jiang T. Wang S. Biomacromolecules. 2014;15:1010–1018. doi: 10.1021/bm401906p. [DOI] [PubMed] [Google Scholar]
- Conceicao M. Mendonca L. Nobrega C. Gomes C. Costa P. Hirai H. Moreira J. N. Lima M. C. Manjunath N. Pereira de Almeida L. Biomaterials. 2016;82:124–137. doi: 10.1016/j.biomaterials.2015.12.021. [DOI] [PubMed] [Google Scholar]
- Gao X. Li L. Cai X. Huang Q. Xiao J. Cheng Y. Biomaterials. 2021;265:120404. doi: 10.1016/j.biomaterials.2020.120404. [DOI] [PubMed] [Google Scholar]
- Giarola J. F. Souto D. E. P. Kubota L. T. Anal. Sci. 2021:20P394. doi: 10.2116/analsci.20P394. [DOI] [PubMed] [Google Scholar]
- Nagai K. Sato T. Kojima C. Bioorg. Med. Chem. Lett. 2020;33:127726. doi: 10.1016/j.bmcl.2020.127726. [DOI] [PubMed] [Google Scholar]
- Mishra M. K. Beaty C. A. Lesniak W. G. Kambhampati S. P. Zhang F. Wilson M. A. Blue M. E. Troncoso J. C. Kannan S. Johnston M. V. Baumgartner W. A. Kannan R. M. ACS Nano. 2014;8:2134–2147. doi: 10.1021/nn404872e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai Z. Ma J. Ding J. Pan H. Chai R. Zhu C. Cui Z. Chen Z. Zhu Q. Int. J. Nanomed. 2020;15:10305–10320. doi: 10.2147/IJN.S282107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sęk J. P. Kaczmarczyk S. Guńka K. Kowalczyk A. Borys K. M. Kasprzak A. Nowicka A. M. Dalton Trans. 2020;50:880–889. doi: 10.1039/D0DT03776A. [DOI] [PubMed] [Google Scholar]
- Ali I. Mukhtar S. D. Ali H. S. Scotti M. T. Scotti L. Curr. Pharm. Des. 2020;26:1637–1649. doi: 10.2174/1381612826666200203124330. [DOI] [PubMed] [Google Scholar]
- Liu Y. Huang R. Han L. Ke W. Shao K. Ye L. Lou J. Jiang C. Biomaterials. 2009;30:4195–4202. doi: 10.1016/j.biomaterials.2009.02.051. [DOI] [PubMed] [Google Scholar]
- Ying W. Gao H. Dos Reis F. C. G. Bandyopadhyay G. Ofrecio J. M. Luo Z. Ji Y. Jin Z. Ly C. Olefsky J. M. Cell Metab. 2021;33:1–10. doi: 10.1016/j.cmet.2020.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzenbach H. Gahan P. B. Noncoding RNAs. 2021;7:4. [Google Scholar]
- Rezaie J. Aslan C. Ahmadi M. Zolbanin N. M. Kashanchi F. Jafari R. Cell Biosci. 2021;11:19. doi: 10.1186/s13578-021-00537-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Erviti L. Seow Y. Yin H. Betts C. Lakhal S. Wood M. J. A. Nat. Cell Biol. 2011;29:341. doi: 10.1038/nbt.1807. [DOI] [PubMed] [Google Scholar]
- Cui G. H. Guo H. D. Li H. Zhai Y. Gong Z. B. Wu J. Liu J. S. Dong Y. R. Hou S. X. Liu J. R. Immun. Ageing. 2019;16:10. doi: 10.1186/s12979-019-0150-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chard D. T. Alahmadi A. A. S. Audoin B. Charalambous T. Enzinger C. Hulst H. E. Rocca M. A. Rovira À. Sastre-Garriga J. Schoonheim M. M. Tijms B. Tur C. Gandini Wheeler-Kingshott C. A. M. Wink A. M. Ciccarelli O. Barkhof F. Nat. Rev. Neurol. 2021:1–12. doi: 10.1038/s41582-020-00439-8. [DOI] [PubMed] [Google Scholar]
- Wirths O. Zampar S. Expert Opin. Ther. Targets. 2019;23:991–1004. doi: 10.1080/14728222.2019.1702972. [DOI] [PubMed] [Google Scholar]
- Liu Y. Li D. Liu Z. Zhou Y. Chu D. Li X. Jiang X. Hou D. Chen X. Chen Y. Yang Z. Jin L. Jiang W. Tian C. Zhou G. Zen K. Zhang J. Zhang Y. Li J. Zhang C. Y. Sci. Rep. 2015;5:17543. doi: 10.1038/srep17543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M. Kim G. Hwang D. W. Lee M. J. Biomed. Nanotechnol. 2019;15:2401–2412. doi: 10.1166/jbn.2019.2866. [DOI] [PubMed] [Google Scholar]
- Yang J. Zhang X. Chen X. Wang L. Yang G. Mol. Ther.--Nucleic Acids. 2017;7:278–287. doi: 10.1016/j.omtn.2017.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper J. M. Wiklander P. B. Nordin J. Z. Al-Shawi R. Wood M. J. Vithlani M. Schapira A. H. Simons J. P. El-Andaloussi S. Alvarez-Erviti L. Mov. Disord. 2015;29:1476–1485. doi: 10.1002/mds.25978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q. Zhou Y. Chen J. Huang N. Wang Z. Cheng Y. Int. J. Nanomed. 2021;16:185–199. doi: 10.2147/IJN.S286221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng Y. Y. Yang T. C. Chen S. M. Yang S. T. Tang Y. L. Liu S. J. Pharmaceutics. 2020;12:479. doi: 10.3390/pharmaceutics12050479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González L. F. Acuña E. Arellano G. Morales P. Sotomayor P. Oyarzun-Ampuero F. Naves R. J. Controlled Release. 2020;331:443–459. doi: 10.1016/j.jconrel.2020.11.019. [DOI] [PubMed] [Google Scholar]
- Ansari M. T. Ramlan T. A. Jamaluddin N. N. Zamri N. Salfi R. Khan A. Sami F. Majeed S. Hasnain M. S. Curr. Pharm. Des. 2020;26:4272–4276. doi: 10.2174/1381612826666200720235752. [DOI] [PubMed] [Google Scholar]
- Zhang X. Chen W. Bai J. Jin L. Kang X. Zhang H. Wang Z. Aging. 2020;12:8289–8300. doi: 10.18632/aging.103138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsenosy F. M. Abdelbary G. A. Elshafeey A. H. Elsayed I. Fares A. R. Int. J. Nanomed. 2020;15:9517–9537. doi: 10.2147/IJN.S277352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D. C. Eldredge A. C. Hickey J. C. Muradyan H. Guan Z. Biomacromolecules. 2020;21:1613–1624. doi: 10.1021/acs.biomac.0c00211. [DOI] [PubMed] [Google Scholar]
- Gooding M. Malhotra M. McCarthy D. J. Godinho B. M. Cryan J. F. Darcy R. O'Driscoll C. M. Eur. J. Pharm. Sci. 2015;71:80–92. doi: 10.1016/j.ejps.2015.02.007. [DOI] [PubMed] [Google Scholar]
- Zhang W. Shi Y. Chen Y. Ye J. Sha X. Fang X. Biomaterials. 2011;32:2894–2906. doi: 10.1016/j.biomaterials.2010.12.039. [DOI] [PubMed] [Google Scholar]
- Liu J. He Y. Zhang J. Li J. Yu X. Cao Z. Meng F. Zhao Y. Wu X. Shen T. Hong Z. Biomaterials. 2016;74:64–76. doi: 10.1016/j.biomaterials.2015.09.041. [DOI] [PubMed] [Google Scholar]
- Park T. E. Singh B. Li H. Lee J. Y. Kang S. K. Choi Y. J. Cho C. S. Biomaterials. 2015;38:61–71. doi: 10.1016/j.biomaterials.2014.10.068. [DOI] [PubMed] [Google Scholar]
- Caprifico A. E. Foot P. J. S. Polycarpou E. Calabrese G. Pharmaceutics. 2020;12:1013. doi: 10.3390/pharmaceutics12111013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J. Yan X. Ge X. Zhang M. Dang X. Yang Y. Xu F. Luo Y. Li G. J. Mater. Chem. B. 2021;9:1297–1314. doi: 10.1039/D0TB02192J. [DOI] [PubMed] [Google Scholar]
- Sonaje K. Tyagi V. Chen Y. Kalia Y. N. Pharmaceutics. 2021;13:88. doi: 10.3390/pharmaceutics13010088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaliq N. U. Park D. Y. Yun B. M. Yang D. H. Jung Y. W. Seo J. H. Hwang C. S. Yuk S. H. Int. J. Pharm. 2019;556:30–44. doi: 10.1016/j.ijpharm.2018.11.064. [DOI] [PubMed] [Google Scholar]
- Segal M. Ozery L. Slor G. Wagle S. S. Ehm T. Beck R. Amir R. J. Biomacromolecules. 2020;21:4076–4086. doi: 10.1021/acs.biomac.0c00882. [DOI] [PubMed] [Google Scholar]
- Zou D. Wang W. Lei D. Yin Y. Ren P. Chen J. Yin T. Wang B. Wang G. Wang Y. Int. J. Nanomed. 2017;12:4879–4889. doi: 10.2147/IJN.S138257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M. Ji N. Wang Y. Dai L. Xiong L. Sun Q. Compr. Rev. Food Sci. Food Saf. 2021;20:1075–1100. doi: 10.1111/1541-4337.12677. [DOI] [PubMed] [Google Scholar]
- Huo H. Gao Y. Wang Y. Zhang J. Wang Z. Y. Jiang T. Wang S. J. Colloid Interface Sci. 2015;447:8–15. doi: 10.1016/j.jcis.2015.01.043. [DOI] [PubMed] [Google Scholar]
- Samaridou E. Walgrave H. Salta E. Alvarez D. M. Castro-Lopez V. Loza M. Alonso M. J. Biomaterials. 2020;230:119657. doi: 10.1016/j.biomaterials.2019.119657. [DOI] [PubMed] [Google Scholar]
- Dosta P. Tamargo I. Ramos V. Kumar S. Kang D. W. Borrós S. Jo H. Adv. Healthcare Mater. 2021:e2001894. doi: 10.1002/adhm.202001894. doi: 10.1002/adhm.202001894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koay T. W. Osterhof C. Orlando I. M. C. Keppner A. Andre D. Yousefian S. Alonso M. S. Correia M. Markworth R. Schödel J. Hankeln T. Hoogewijs D. J. Biol. Chem. 2021:100291. doi: 10.1016/j.jbc.2021.100291. doi: 10.1016/j.jbc.2021.100291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castan-Laurell I. Masri B. Valet P. Expert Opin. Ther. Targets. 2019;23:215–225. doi: 10.1080/14728222.2019.1561871. [DOI] [PubMed] [Google Scholar]
- Fu A. Wang Y. Zhan L. Zhou R. Pharm. Res. 2012;29:1562–1569. doi: 10.1007/s11095-012-0667-y. [DOI] [PubMed] [Google Scholar]
- Bosch J. A. Birchak G. Perrimon N. Proc. Natl. Acad. Sci. U. S. A. 2021;118:e2021996118. doi: 10.1073/pnas.2021996118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz L. A. Miyamichi K. Gao X. J. Beier K. T. Weissbourd B. DeLoach K. E. Ren J. Ibanes S. Malenka R. C. Kremer E. J. Luo L. Nature. 2015;524:88–92. doi: 10.1038/nature14600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanada M. and Gilad A. A., in Nucleic Acid Nanotheranostics, Elsevier, 2019, pp. 409–420 [Google Scholar]
- Nagy A. Genesis. 2000;26:99–109. doi: 10.1002/(SICI)1526-968X(200002)26:2<99::AID-GENE1>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]