Highlights
-
•
Clinical –The studies are the first to identify CSPG4 as an oncoantigen which is associated with poor outcome in patients with ovarian cancer.
-
•
CSPG4 expression by tumor cells significantly enhances tumor expansion in vivo and tumor invasion, cisplatin resistance and spheroid formation.
-
•
CSPG expression stimulates cell adhesion related pathways (e.g. Focal Adhesion Kinase) which stimulates expression of ZEB1 which stimulates invasion and spheroid formation.
-
•
Antibody targeting a specific domain of CSPG4 limits invasion and promotes apoptosis of cells within spheroids.
-
•
The studies identify CSPG4 as a targetable oncoantigen that could improve ovarian cancer patient management by limiting metastasis and improving standard of care chemotherapy response.
Abstract
Epithelial ovarian cancer (EOC) is a highly heterogeneous disease encompassing several distinct molecular subtypes and clinical entities. Despite the initial success of surgical debulking and adjuvant chemotherapy, recurrence with chemotherapy resistant tumors is common in patients with EOC and leads to poor overall survival. The extensive genetic and phenotypic heterogeneity associated with ovarian cancers has hindered the identification of effective prognostic and predictive biomarkers in EOC patients. In the current studies, we identify a tumor cell surface oncoantigen, chondroitin sulfate proteoglycan 4 (CSPG4), as an independent risk factor for decreased survival of patients with EOC. Our results show that CSPG4 promotes EOC cell invasion, cisplatin resistance and spheroid formation in vitro and tumor expansion in vivo. Mechanistically, spheroid formation and tumor cell invasion are due to CSPG4-stimulated expression of the mesenchymal transcription factor ZEB1. Furthermore, we have developed a novel monoclonal anti-CSGP4 antibody against the juxtamembrane domain of the core protein that limits CSPG4-stimulated ZEB1 expression, tumor cell invasion and promotes EOC apoptosis within spheroid cultures. We therefore propose that CSPG4 expression drives phenotypic heterogeneity and malignant progression in EOC tumors. These studies further demonstrate that CSPG4 expression levels are a potential diagnostic biomarker in EOC and indicate that targeting cells which express this oncoantigen could limit recurrence and improve outcomes in patients with EOC.
Significance The studies are the first to identify CSPG4 expression as an independent risk factor of poor outcome in patients with epithelial ovarian cancer. Mechanistically, these studies show that CSPG4 promotes tumor growth, invasion, cisplatin resistance, mesenchymal transition and spheroid formation. A novel anti-CSPG4 monoclonal antibody has been identified that inhibits CSPG4-induced invasion and promotes apoptosis of EOC spheroid cultures. The data indicate that CSPG4 may be an ideal target for limiting therapy resistant recurrence and metastasis of EOC.
Introduction
Epithelial ovarian cancer (EOC) is a highly heterogeneous disease that consists of a wide spectrum of distinct molecular subtypes and clinical entities [1], [2], [3], [4]. There is a complex basis for interpatient and intrapatient genetic heterogeneity in EOC which is reflected by the distinct genetic signatures associated with different histologic subtypes or genetic/epigenetic changes induced by external stressors such as chemotherapies [5]. Although most EOC patients initially respond well to surgical debulking and adjuvant chemotherapy, the occurrence of chemoresistance is a major hurdle, with 75% of patients experiencing a relapse within five years [2,3]. Malignant progression involves extensive intra-tumoral phenotypic heterogeneity related to dynamic biological requirements at different stages in progression [3], [4], [5]. These dynamics include localized changes in growth factors, an actively remodeling tumor-associated extracellular matrix and the presence of therapy-resistant cancer stem cells, [3,[6], [7], [8]].
Ovarian carcinoma metastasis largely occurs via an intraperitoneal (IP) route and is thus distinct from other common carcinomas such as breast and prostate cancers [2,3,6]. In EOC, individual cells or cell aggregates dissociate from primary tumors to form multicellular spheroids responsible for peritoneal spread, metastasis, and recurrence [6,9]. The survival of individual cells that give rise to spheroids is facilitated by their anchorage independence and initial resistance to anoikis [6,9]. Increased compaction of cells within spheroids can lead to increased therapy resistance, in part by limiting penetration of chemotherapies into more centrally located cells within these spheroids [6,9]. Their subsequent invasion into the sub-mesothelial tissues involves stimulation by growth factors and chemokines within the microenvironment and activation of tumor associated matrix metalloproteinases which degrade the underlying extracellular matrices [9]. Malignant progression in EOC is also associated with a tumor cell phenotypic shift from an epithelial to a mesenchymal phenotype (EMT). EMT programs are impacted by complex mechanisms, which include multiple signaling pathways (e.g. growth factors, Wnt/β-catenin, Notch) and changes in expression/function of adhesion receptors (E-cadherin/N-cadherin, claudins, integrins) [10,11]. Tumor cell detachment from the primary tumor and subsequent spheroid formation has been linked to increased expression of specific mesenchymal transcription factors such as ZEB1 and Slug (Snail2) which are linked to cell ‘stemness’, resistance to apoptosis and therapy [10,11].
We have evaluated CSPG4 as a cell surface EOC biomarker and its impact on facilitating phenotypic heterogeneity and malignant progression in patients with EOC. CSPG4 is a type I transmembrane glycoprotein with a large extracellular domain and a relatively short intracellular domain [12,13]. CSPG4 binds one or more components of the extracellular matrix and promotes activation of multiple oncogenic pathways related to integrin function, growth factor signaling, and mesenchymal transition [12], [13], [14], [15], [16]. While CSPG4 is expressed at low levels on immature progenitor cell types in normal adult tissues [12,14,17], levels are increased on multiple tumor types and thus it is considered a tumor associated ‘oncoantigen’ which can be targeted therapeutically [12,14,[17], [18], [19]].
The current studies are the first to demonstrate that elevated levels of CSPG4 are linked to poor overall survival in patients with multiple subtypes of EOC. Using CRISPR/Cas9 deletion of CSPG4 in multiple ovarian cancer cell lines, we demonstrate that CSPG4 functions to promote invasion, cisplatin resistance, spheroid formation and mesenchymal transition. Loss of CSPG4 also significantly reduces tumor expansion in vivo compared to cells that express the proteoglycan. A novel antibody generated against the juxtamembrane domain of the core protein blocks invasion, ZEB1 expression and promotes apoptosis of CSPG4 stimulated spheroids. The results indicate CSPG4 may be an ideal target for limiting recurrence and improving outcome in patients with EOC.
Materials and methods
Ovarian cancer patient cohort
The cohort consists of 126 epithelial ovarian cancer patients with long‐term clinical follow‐up, who have undergone initial surgery and treatment at the Hunan Cancer Hospital, affiliated to Xiangya School of Medicine of Central South University of China, a specialized cancer hospital certified by the Joint Commission International (JCI). Inclusion criteria for the ovarian cancer patient cohort were histologically confirmed EOC including three major histopathologic subtypes (serous, mucinous, and other adenocarcinoma); treatment with platinum/taxane based chemotherapy after debulking surgery; no radiotherapy or biological therapy before surgery; and Karnofsky Performance Status score ≥80 prior to surgery. Patients were staged according to the International Federation of Gynecology and Obstetrics (FIGO) surgical staging system. Another 16 patients with benign ovarian lesions and 26 hysterectomy patients with normal ovarian tissues were also recruited. Protocols are approved by the Ethics Committee of Hunan Cancer Hospital (Changsha, China) and all patients provided written informed consent on file with the hospital.
Immunohistochemistry
The specimens were paraffin embedded and the tissue sections (4 μm) were dewaxed, rehydrated, blocked with 3% BSA, and subjected to antigen retrieval. After washing, the sections were incubated with antibody 9.2.27 against CSPG4 (1:1000) at 4°C overnight. Mouse IgG (cat#A7028, Beyotime, Shanghai, China) was used as negative control. The bound antibodies were detected using horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG (Beyotime cat#A0216) and visualized by DAB (DAB-2031, Maixin Biotech. Co., Fuzhou, China), followed by counterstaining with hematoxylin (CTS-1090, Maixin Biotech. Co.). The results were evaluated under a microscope by two pathologists in a blinded manner.
CSPG4 staining intensity and fraction of tumor cells positive were scored by pathologists blinded to patient ID. Semiquantitative immunohistochemistry scoring was based on a modified method of Allred [20] where specific percent positive thresholds were adjusted empirically based on validation of CSPG4 staining performance blinded to outcomes or other clinical features prior to execution of the full study analysis.
Cell lines
Human ovarian cancer cell lines used in these studies; HEY (RRID: CVCL_0297), A2780 (RRID: CVCL_0134) and ES-2 (RRID: CVCL_3509). Cell lines were authenticated via STR profiling by the ATCC Cell Line Authentication Service (Manassas, Virginia) in June 2019 and screened for mycoplasma upon receipt using the Universal Mycoplasma Detection Kit (ATCC cat#30–1012 K). ES-2 and A2780 cells were cultured in DMEM medium (Mediatech cat#10–013-cv), HEY cells in RPMI 1640 medium (Gibco cat#11875–093), supplemented with 10% fetal bovine serum (Atlanta Biologicals cat#SS11150H, Lot#H1810S) and 1% penicillin/ streptomycin (Gibco cat#15140–122) at 37°C/5% CO2. CSPG4-CRISPR knockout and mock stable transfected variants were maintained in medium supplemented with 0.6 μg/ml puromycin (Sigma cat#P8833).
Generation of CSPG4 CRISPR cell lines
The guide RNA target sequences used to make the CSPG4-CRISPR knockout cells are 5′ CGAGCGCGGCTCTGCTCCTG 3′ and 5′AGAGACCTGGAGACACCAGG 3′. Guide RNAs were inserted into plasmid pCR4-TOPO-U6-HPRT-gRNA. Guide RNA plasmids were co-transfected with plasmid expressing the CAS9 enzyme (pT3.5 Caggs-FLAG-hCas9) as well as plasmids for puromycin and GFP selection, pcDNA-PB7 and pPB SB-CG-LUC-GFP (Puro)(+CRE), using Lipofectamine 2000 reagent (Invitrogen cat#11668–019) following the manufacturer's suggested protocol. Mock cell lines were transfected with selection plasmids only and selected as a pool by culture in puromycin containing medium (0.6 µg/ml). CSPG4-CRISPR knockout cell lines were selected by clonal plating in 96 well plates in puromycin containing media (0.6 μg/ml). Single cell derived colonies were expanded and screened by genomic PCR for the deletion of the CSPG4 gene using primers 5′ GGGCCCTTTAAGAAGGTTGA 3′ and 5′ GTTTTGACAGCCCAAACCAG 3′, and by immunoblot and flow cytometry (Supplementary Figs. 2 and 5) to verify the loss of CSPG4 protein.
Other antibodies and reagents
See Supplementary Table 1.
Transfection of siRNA
Small interfering RNA (siRNA) specific for human ZEB1 (cat# sc-38643) was purchased from Santa Cruz Biotechnology (Dallas, TX), and negative control siRNA was purchased from Qiagen, Inc. (cat# 1027281, Germantown, MD). Cells were transfected with siRNA at 60–70% confluence using the Lipofectamine RNAimax transfection reagent (cat# 13778075, ThermoFisher Scientific) following the manufacturer's suggested protocol. Cells were harvested 48h post-transfection for plating in growth, invasion and spheroid formation assays.
Anchorage independent growth assay
Soft agar growth assays were performed as previously described [21] with the following modification: cells were plated in the upper agarose layer at a final concentration of 0.6% agarose. Colonies were counted in five random fields/well from replicate wells at the indicated time point (see figure legends) for each cell line. Experiments were performed a minimum of 3 times.
Cell invasion assays
Cells (2.5–5.0 × 104) in normal growth medium were added to the top chamber of triplicate wells of matrigel invasion chambers (8 μm, Corning, NY), the bottom chambers filled with complete medium (ES-2, A2780 cell lines) or serum free medium (HEY cell line) and cultured for 16–24 h at 37 °C, 5% CO2 atmosphere. Remaining cells in the upper chamber were removed with a cotton swab and the invaded cells fixed and stained using Differential Quick Staining Kit (Electron Microscopy Sciences, Hatfield, PA). Invaded cells were enumerated under a microscope at 100X magnification from five random fields/well. Each experiment was repeated a minimum of three times.
Spheroid formation assays and survival analysis
2 × 105 cells were suspended in 1% high viscosity methylcellulose (Sigma cat# M0512) diluted in complete growth media, plated in 6-well plates coated with Poly-HEMA and cultured for 7 days. Spheroids with a diameter over 100 μm were enumerated under a microscope at 100x magnification from five random fields/well. Spheroids grown in methylcellulose culture were harvested by dilution-dispersion in PBS, centrifugation at 400x g for 15 min, and washed twice in PBS for subsequent analysis. Each experiment was repeated a minimum of three times. For quantification of cell growth/survival in spheroids, spheroids were collected from methylcellulose cultures and re-plated overnight in normal growth media on tissue culture plates. The following day all cells from each well were collected and counted with trypan blue exclusion.
Cisplatin cytotoxicity assay
Cisplatin stock (3.3 mM in 0.9% saline solution) was diluted in growth medium to the required concentrations before each experiment. Cells were seeded into 96-well plates at 1.0 × 103 cells/well in 100μl of growth media and allowed to adhere overnight. The following day media was removed from wells and replaced with 100 µl media containing the indicated treatment concentration or media alone (baseline) in triplicate wells. After 96h of treatment, 20 µl of MTS reagent (Promega, cat#G3580) was added to each well and plates incubated in the dark for 2 h at 37°C, 5% CO2. Absorbance at 570 nm was collected on a Tecan 200 plate reader. Each experiment was repeated a minimum of three times.
Xenograft intraperitoneal injection mouse model
All animal studies were approved by the University of Minnesota Institutional Animal Care and Use Committee (IACUC 1908–37330A). NOD/SCID/γc−/− (NSG) 12-week-old female mice (Jackson Laboratories, Bar Harbor, ME) were used for this experiment. Mice were sub-lethally irradiated (225 cGy) and the following day 4 mice were injected intraperitoneally with 2 × 105 A2780 Mock luciferase expressing tumor cells and 5 mice injected intraperitoneally with 2 × 105 A2780 CSPG4-CRISPR luciferase expressing tumor cells. Tumor burden was monitored post luciferin (Goldbio, St. Louis, MO) injection by bioluminescent imaging (BLI) using the IVIS Spectrum in vivo Imaging system (PerkinElmer) on days 6, 13, and 27 post cell injection. Image analysis was performed using Living Image 4.5 software (PerkinElmer). Negative control mice were injected with luciferin only.
RNA seq analysis
Total RNA was isolated in two technical replicates from the ES-2 parent, Mock, and CSPG4-CRISPR cell lines using the Rneasy RNA isolation kit (Qiagen, cat# 74104) following the manufacturer's suggested protocol. RNA samples were submitted to the University of Minnesota Genomics Center for quality control assessment on an Agilent Bioanalyzer and quantification using a fluorimetric RiboGreen assay. A strand-specific RNA-seq library was generated and sequenced on an IlluminaiSeq 2500 in high output mode, ∼20 million reads/sample (duplicate samples) with 2 × 125 bp paired end reads.
Bulk RNAseq samples were processed and aligned using the CHURP version 0.2.2 command line interface framework. A full description of the CHURP pipeline can be found in [22]. Briefly, trimmomatic version 0.33 [23] was used to clean reads for adapter contamination and low-quality sequence. FastQC [24] was used to generate sequence quality reports for raw and trimmed reads. HISAT2 version 2.1.0 [25] was used to align samples to the genome reference consortium H. sapiens build 38 reference genome. FeatureCounts v1.6.2 [26] was used to count mapped reads to genes.
Gene expression and pathway analysis
All differential gene expression and pathway analyses were done in R v 3.6.3 (R Core Team, 2020). Differential gene expression analysis was done using EdgeR v 3.28.1 [27]. Differentially expressed genes were identified between the ES-2 CSPG4-CRISPR/Cas9 knock-out cell line and the average of the ES-2 parent and mock cell lines. Counts were normalized using the relative log expression normalization method and only genes with counts per million greater than one in two or more samples were kept. A general linear model approach was used to test for differentially expressed genes. A gene was categorized as differentially expressed if the p-value was less than 0.01 after p-value adjustment and log2 fold change was greater than one. P-values were adjusted using the Benjamini & Hochberg method. GO term enrichment analysis and gene set enrichment analysis (GSEA) were done using the ClusterProfler R package [28]. The hallmark gene set from the Molecular Signatures Database v 7.1 (https://www.gsea-msigdb.org/gsea/msigdb/index.jsp) was used in the GSEA.
Survival analysis was performed using the Kaplan-Meier Plotter web-based informatics tool [29,30]. Fifteen ovarian cancer patient cohorts were included in the combined analysis, including the ovarian cancer TCGA cohort and 14 additional cohorts from the Gene Expression Omnibus (GEO) database; most cases in these datasets are serous histology, with a small proportion of endometrioid type ovarian cancers. JetSet optimal probes [31] were selected for CSPG4 (MCSPG) and ZEB1 expression analysis; 1656 patients had available data for CSPG4, while 355 patients had available data for ZEB1. The cohort was separated by the median for either normalized single gene expression or the mean of normalized combined expression of both genes. Outcomes were censored at 5 years to harmonize with the duration of follow up for separately performed immunohistochemical studies.
Analysis of EMT signature enrichment was performed using the web based Xena informatics tool [32] on the ovarian cancer TCGA dataset. The cohort was separated by mean CSPG4 gene expression, and EMT signature score was calculated using Xena genomic signatures feature [33].
Western blot
Western blots were performed using standard methods as described previously [21].
Confocal microscopy
Cells were plated on coverslips for 48h, fixed with 4% paraformaldehyde for 15 min, permeabilized with 0.05% Triton X-100 for 5 min at room temperature, and blocked with 1% donkey serum for 1h. Coverslips were incubated with the indicated primary anti-CSPG4 antibodies at 1 µg/ml overnight at 4°C, washed twice with PBS+1% BSA for 10 min at room temperature, and incubated with Cy3-conjugated anti-mouse secondary antibody (1:5000) for 1h at room temperature. After washing cells twice with PBS+1% BSA, images were captured as described previously [15].
Flow cytometry
Cells were released in PBS/5 mM EDTA solution and washed 2 times with FACS buffer (RPMI media supplemented with 1% goat serum and 5 mM HEPES). Cells were incubated with the indicated primary antibody for 45 min at 4°C, washed 3 times with FACS buffer, and then incubated with goat anti-mouse phycoerythrin-conjugated secondary antibody for 30 min at 4°C. Antibody staining was analyzed on a BD Biosciences Accuri C6 flow cytometry system and data graphed using the Accuri C6 software (BD Biosciences).
Generation of novel CSPG4 antibody
CSPG4-specific mouse monoclonal antibody 7H5A2 (IgG1) was generated by Promab Biotechnologies Inc (Richmond, CA) by injection of a recombinant CSPG4 protein immunogen corresponding to aa 1538–2221 of the CSPG4 core protein extracellular domain, expressed and purified from a eukaryotic expression system (sequence in Supplementary Fig. 5). The specificity of the antibody was determined by screening against CSPG4 wild type and knockout cell lysates via western blot and cell staining via immunofluorescence (Supplementary Fig. 5).
Statistical analysis
All statistical analysis was performed with Graphpad PRISM 6 (Graphpad Software, San Diego, CA) unless otherwise indicated. Comparison of two independent samples was done utilizing two-tailed Student's t-test with Welch's correction. Differences in tumor burden over time within the xenograft model were analyzed by ordinary two-way ANOVA with Sidak's multiple comparison test. Values of p<0.05 are considered statistically significant. Overall survival and disease-free survival curves for patients with low and high CSPG4 expression in tumor specimens was analyzed using the Kaplan–Meier method and compared using the log-rank test (SPSS 15.0 software, Chicago, IL, USA). Univariate and multivariate analysis are performed using Cox regression model after adjusting for baseline characteristics. p<0.05 is considered statistically significant.
Results
CSPG4 is a protein biomarker of poor survival in ovarian cancer patients
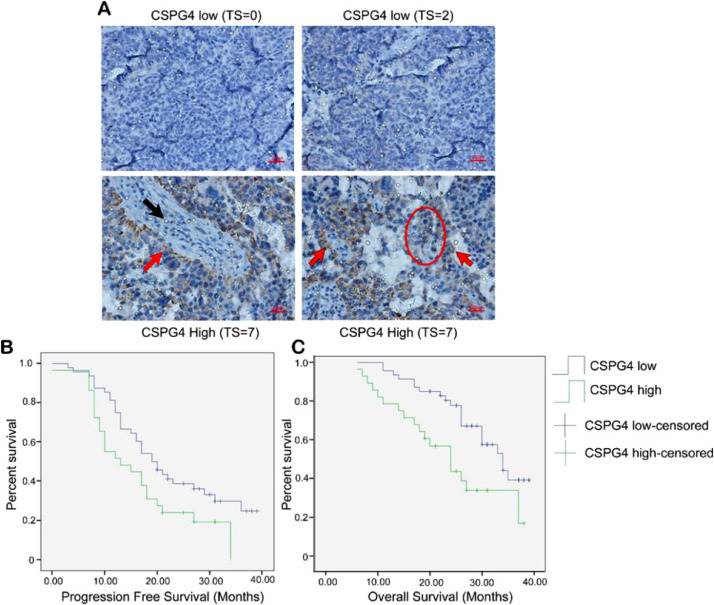
We first evaluated CSPG4 protein expression by IHC in a cohort of 126 ovarian cancer patients. CSPG4 protein expression is significantly higher in ovarian cancer (39.68%, 50/126) compared to benign (12.5%, 2/16) or normal ovarian tissue (11.54%, 3/26) (X2=11.04, p=0.004) (Table 1). CSPG4 was detected in uniform patterns in tumor cells in contact with tumor associated stroma, however, CSPG4 positivity was more heterogeneous in areas distant from stroma (Fig. 1A). Patients with low expression of CSPG4 protein have significantly longer progression free survival (Kaplan-Meier PFS: 22.615±1.754 vs 16.559±1.940, X2=4.316, p=0.038) and improved overall survival (Kaplan-Meier OS: 31.027±1.353 vs 24.046±2.177, X2=7.366, p=0.007) compared to patients with high expression of CSPG4 (Fig. 1B, 1C). The poor prognosis associated with elevated CSPG4 was independent of patient age, tumor subtype (1 or 2), clinical stage (I/II vs. III/IV), degree of differentiation, the presence of omental or lymphatic metastasis or the volume of ascites (Table 2). Hazard ratio analysis demonstrated that high CSPG4 levels are an independent indicator of poor overall survival in both univariate (HR=2.33; 95% CI 1.230 to 4.427; p=0.009) and multivariate (HR=2.54; 95% CI 1.255–5.140; p=0.010) analyses (Table 3).
Table 1.
CSPG4 expression in ovarian cancer patient cohort.
| Group | N | CSPG4 |
High expression rate (%) | Χ2 | P-value | |
|---|---|---|---|---|---|---|
| Low | High | |||||
| Malignant | 126 | 76 | 50 | 39.68 | ||
| Normal | 26 | 23 | 3 | 11.54 | 11.042 | 0.004 |
| Benign | 16 | 14 | 2 | 12.5 | ||
CSPG4 expression in ovarian cancer, benign ovarian lesions and normal ovarian tissues. CSPG4 expression is enriched in malignant tumor tissue vs. normal and benign ovarian tissue. CSPG4 high vs. low expression is defined in Fig. 1.
Fig. 1.
CSPG4 protein expression characterized in 126 patient ovarian cancer cohort. (A) Representative images from IHC staining for CSPG4 in the ovarian cancer patient cohort. Tumor tissue staining indicated by red arrows; stromal staining indicated by black arrow. An area of staining heterogeneity in the tumor parenchyma is indicated by the red circle. Staining intensity was scored on a scale from 0 to 3 [0 (negative), 1 (weak), 2 (moderate) and 3 (strong)]. The fraction of CSPG4 staining was scored from 0 to 4, reflecting the percentage of positively stained tumor cells in the sample [0 (0%), 1 (1–25%), 2 (25–50%), 3 (50–75%) and 4 (75–100%)]. The intensity and fraction positive scores were added together to generate the total score (TS). TS ≥ 4 was considered high expression of CSPG4, while TS ≤ 3 was considered low expression. Red bar = 20μm. (B + C) Kaplan-Meier curves over 40 months for censored data from this 126-patient ovarian cancer cohort. (B) CSPG4 high expression (green line; 24/29) vs. low CSPG4 blue line (33/48) correlates with reduced progression free survival (PFS: 22.615 ± 1.754 vs 16.559 ± 1.940, X2 = 4.316, p = 0.038). (C) High CSPG4 expression (green line 19/29) vs low CSPG4 (blue line 22/48) correlates with reduced overall survival (OS: 31.027 ± 1.353 vs 24.046 ± 2.177, X2 = 7.366, p = 0.007).
Table 2.
CSPG4 as an independent prognostic factor of patient outcome.
| Variable | N | CSPG4 |
P-value | |
|---|---|---|---|---|
| Low | High | |||
| Age | 0.394 | |||
| <50 years | 46 | 30 | 16 | |
| ≥50 years | 80 | 46 | 34 | |
| Type | 0.995 | |||
| I | 41 | 25 | 16 | |
| II | 75 | 45 | 30 | |
| Non-Type I or II | 10 | 6 | 4 | |
| Clinical Stage | 0.190 | |||
| I + II | 22 | 16 | 6 | |
| III + IV | 104 | 60 | 44 | |
| Differentiation Degree | 0.143 | |||
| Low | 109 | 63 | 46 | |
| Middle and High | 17 | 13 | 4 | |
| Omental Metastasis | 0.116 | |||
| No | 51 | 35 | 16 | |
| Yes | 75 | 41 | 34 | |
| Lymph Metastasis | 0.524 | |||
| No | 42 | 27 | 15 | |
| Yes | 62 | 36 | 26 | |
| Not Cleaned | 22 | 13 | 9 | |
| Ascites Volume | 0.562 | |||
| <500ml | 59 | 34 | 25 | |
CSPG4 expression level is an independent prognostic factor in ovarian cancer. The poor prognosis associated with elevated CSPG4 is independent of patient age, tumor subtype (I or II), clinical stage (I/II vs. III/IV), degree of differentiation, the presence of omental or lymphatic metastasis or the volume of ascites. CSPG4 high vs. low expression is defined in Fig. 1.
Table 3.
Univariate and multivariate analysis of patient cohort data.
| Variables | Univariate analysis |
Multivariate analysis |
||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P | HR | 95% CI | P | |
| Age | 1.726 | 0.839–3.549 | 0.138 | 1.244 | 0.586–2.638 | 0.570 |
| Clinical Stage | 4.216 | 1.016–17.506 | 0.048 | 3.634 | 0.657–20.203 | 0.139 |
| Differentiation Degree | 0.578 | 0.225–1.483 | 0.254 | 0.727 | 0.269–1.965 | 0.529 |
| Omental Metastasis | 2.298 | 1.345–6.376 | 0.007 | 1.870 | 0.761–4.597 | 0.172 |
| Lymph Metastasis | 0.972 | 0.612–1.544 | 0.905 | 0.603 | 0.336–1.083 | 0.090 |
| Ascites Volume | 1.499 | 0.776–2.895 | 0.228 | 1.681 | 0.845–3.343 | 0.139 |
| CSPG4 (High/Low) | 2.334 | 1.230–4.427 | 0.009 | 2.540 | 1.255–5.140 | 0.010 |
CSPG4 expression level (High vs. Low) is a significant indicator of overall patient survival (OS) in both univariate and multivariate analysis. CSPG4 expression level (high vs. low) was determined by IHC (see Fig. 1 for definition of high vs. low CSPG4).
CSPG4 promotes EOC growth, invasion, cisplatin resistance and spheroid formation
We next explored the functional significance of the CSPG4 expression in EOC cells using multiple in vitro correlate analyses of malignant phenotypes. For these studies we selected 3 ovarian cell lines that had high levels of CSPG4 expression out of 11 cell lines (listed in Supplementary Fig. 1) initially screened by western blot analysis (not shown). The selected ovarian cancer cell lines (ES-2, HEY, A2780) originated from patients harboring different subtypes of EOC [34]. These three cell lines were subsequently screened by confocal analysis (Supplementary Fig. 1) to verify cell surface staining of CSPG4.We deleted the entire CSPG4 locus in each of these cell lines using CRISPR/Cas9. Deletion of the CSPG4 locus was first confirmed by genomic PCR (not shown). Clones that were positive by PCR were subsequently screened for loss of CSPG4 protein by flow cytometry (Supplementary Fig. 2) and western blot (Supplementary Fig. 5) and were used for subsequent studies.
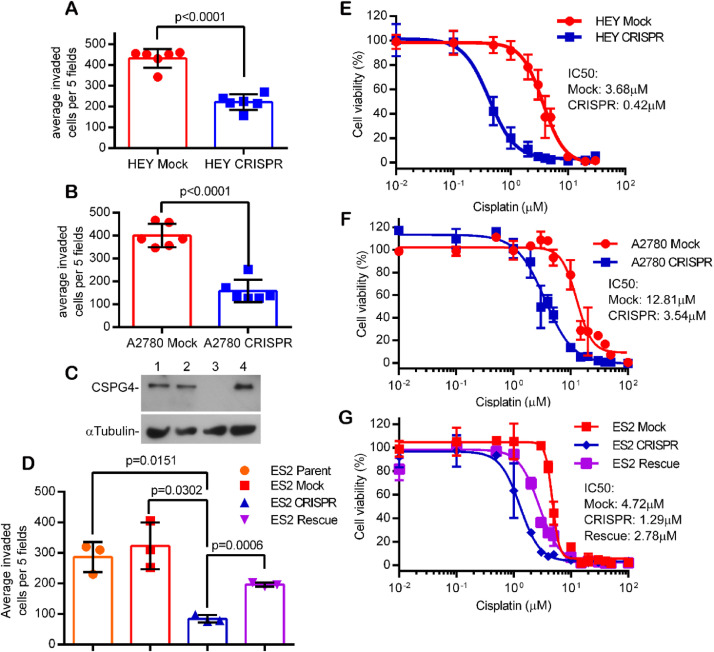
Using a standard matrigel invasion assay we determined that loss of CSPG4 expression significantly decreased the invasive phenotype of all three cell lines (Fig. 2A–D). Importantly, we could rescue the invasive phenotype by re-expression of CSPG4 in the ES-2 CRISPR (CSPG4-deleted) cells (Fig. 2C, D), thus indicating that loss of an invasive phenotype was not due to spurious off targeting of our CRISPR/Cas9 deletion.
Fig. 2.
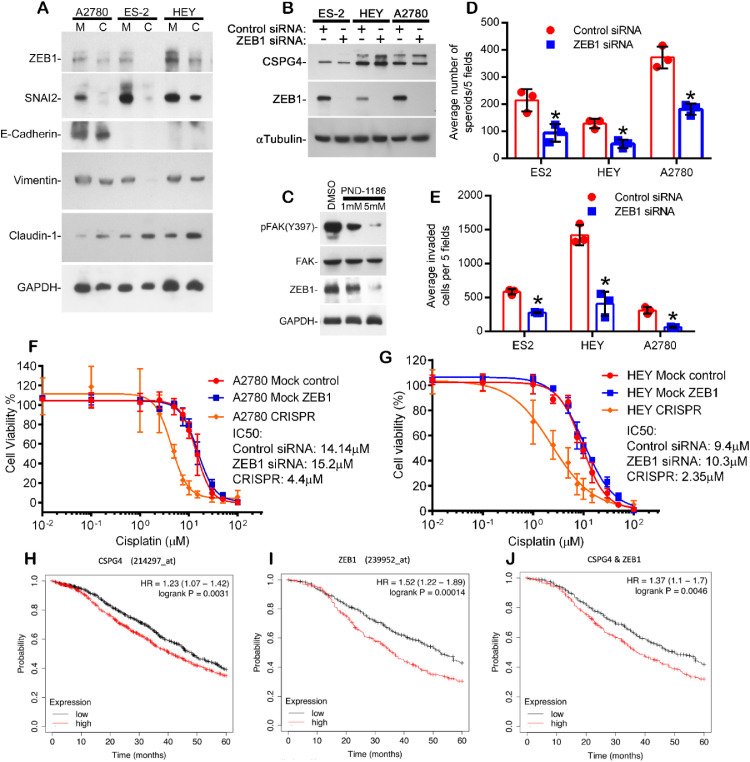
CSPG4 knockout results in significant loss of invasive capacity and cisplatin resistance in multiple ovarian tumor cell lines. Invasion assays using control (Mock) and CSPG4 knockout (CRISPR) HEY cells (A) or A2780 cells (B). Bars represent the total number of invading cells from five random fields/well from triplicate wells, +/- S.D., from three replicate experiments, n=6. P-values determined by student's t-test with Welch's correction. C) Western blot for CSPG4 in ES2 cell lines. Lanes: 1- ES-2 Parent, 2- ES-2 Mock, 3- ES-2 CSPG4 knockout, 4- ES-2 CSPG4 Rescue. (Note that CSPG4 can be expressed with various levels of CS modification and in this blot the primary band represents predominantly the core protein) D) Invasion assay using indicated ES-2 cell lines. Invasive capacity is rescued in the ES-2 CRISPR knockout line with CSPG4 re-expression (Rescue, purple bar). P-values determined by student's t-test with Welch's correction. (E-G) Cell viability of mock and knockout (CRISPR) HEY cells (E), mock and knockout A2780 cells (F), and mock, knockout and rescue ES-2 cells (G) treated with increasing concentrations of cisplatin. Dose response curves were plotted as the percent of MTS staining vs. untreated cells for each cell line +/- s.e.m. from three replicate experiments (n=9). Relative resistance to cisplatin in the ES-2 CRISPR cells was restored by re-expression of CSPG4 in the ES-2 CRISPR cells (ES-2 Rescue, purple line in G).
While platinum-based therapy is widely used as a first-line adjuvant therapy for patients with EOC, recurrence with resistant disease is a common complication that reduces overall survival. To investigate whether CSPG4 expression impacts cisplatin resistance in EOC, we evaluated the cisplatin sensitivity of our cell lines using an MTS assay. Loss of CSPG4 resulted in increased cisplatin sensitivity in all three cell lines, as evidenced by a 3.6-to-8.8-fold reduction in the IC50 for the knockout cell lines (Fig. 2E–G). As was observed for the invasion phenotype, re-expression of CSPG4 in edited cells resulted in reversal of cisplatin sensitivity (Fig. 2G purple curve).
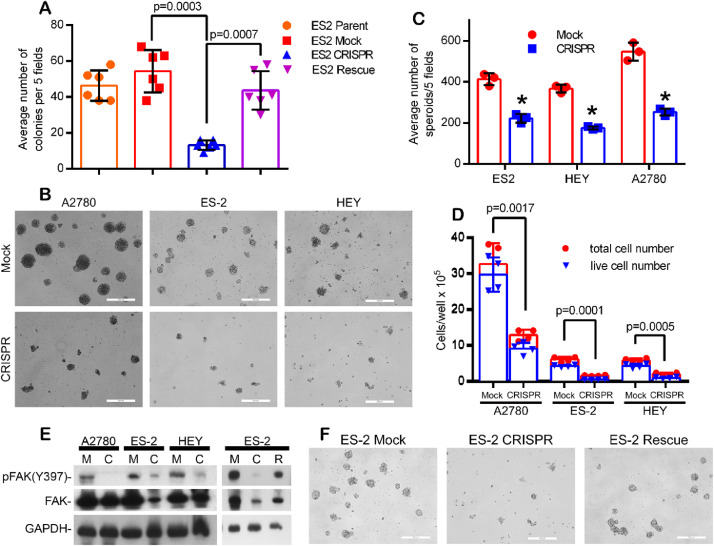
Like the decreased invasive phenotype and decreased cisplatin resistance, loss of CSPG4 resulted in the reduced ability to form anchorage independent colonies (Fig. 3A) which is an in vitro correlate of tumorigenicity. The loss of anchorage independence could also be reversed by CSPG4 re-expression (Fig. 3A). EOC metastasis involves the shedding of cellular aggregates (termed spheroids) from the primary tumor into the abdominal cavity [3,9]. To determine the effect of CSPG4 on spheroid formation, we performed a spheroid formation assay by culturing cells in methylcellulose. Loss of CSPG4 resulted in a highly significant reduction in the ability of all three cell lines to form spheroids (Fig. 3B–D). .
Fig. 3.
CSPG4 stimulates EOC anchorage independent growth, spheroid formation and FAK activation. (A) ES-2 parent, Mock, CRISPR and stable CSPG4-rescue cells were grown in a soft agar colony formation assay for seven days. Bars represent the total number colonies counted from five random fields/well from replicate wells, +/- S.E., n=6. P-values determined by student's t-test with Welch's correction. (B and C) CSPG4-CRISPR cells form fewer/smaller spheroids when cultured in methylcellulose media. (B) Representative images showing spheroids from Mock and CSPG4-CRISPR cultures for three EOC cell lines. Bar=200μm. (C) Spheroid counts from five random fields/well from three replicate experiments +/- S.D. *p<0.001 by students t-test with Welch's correction. Spheroids were quantified 7 days post plating. n=3 from 3 individual experiments (D) Quantification of cell growth/survival in spheroids. Spheroids were collected from methylcellulose culture and re-plated overnight in normal growth media on tissue culture plates. The following day all cells from each well were collected and counted. P-values determined by student's t-test with Welch's correction, comparing live cell numbers (based on trypan blue exclusion) between Mock and CRISPR cells for each cell line, n=4. (E) Cells (Mock, M; CRISPR, C; CRISPR rescued with CSPG4 re-expression, R) were cultured in 1.0% methylcellulose/complete media for 7 days, harvested and lysates analyzed by western blot for FAK expression/phosphorylation. (F) Representative images showing rescue of ES-2 CSPG4 CRISPR spheroid formation after CSPG4 re-expression. Bar=200μm.
CSPG4 expression in spheroids was also linked to activation of FAK (Fig. 3E), and loss of FAK activation and decreased spheroid formation in the CSPG4 CRISPR/Cas9 cells could be restored by CSPG4 re-expression (Fig. 3E and 3F). Gene ontology (GO) term enrichment analysis of our RNAseq dataset of differentially expressed genes in the ES-2 control and CSPG4 knockout cell lines identifies a significant enrichment (adjusted p-value <0.005) for GO terms associated with regulation of cell adhesion, associated signaling pathways and extracellular matrix collagen/organization (Supplementary Fig. 3 and attached Excel Spreadsheet). This association supports multiple studies functionally linking CSPG4 to these processes in numerous other tumor cell model systems [[12], [13], [14],17]. The impact of CSPG4 on FAK activation may partially explain the impact of CSPG4 expression on promoting anchorage independent growth and spheroid formation by EOC tumor cells.
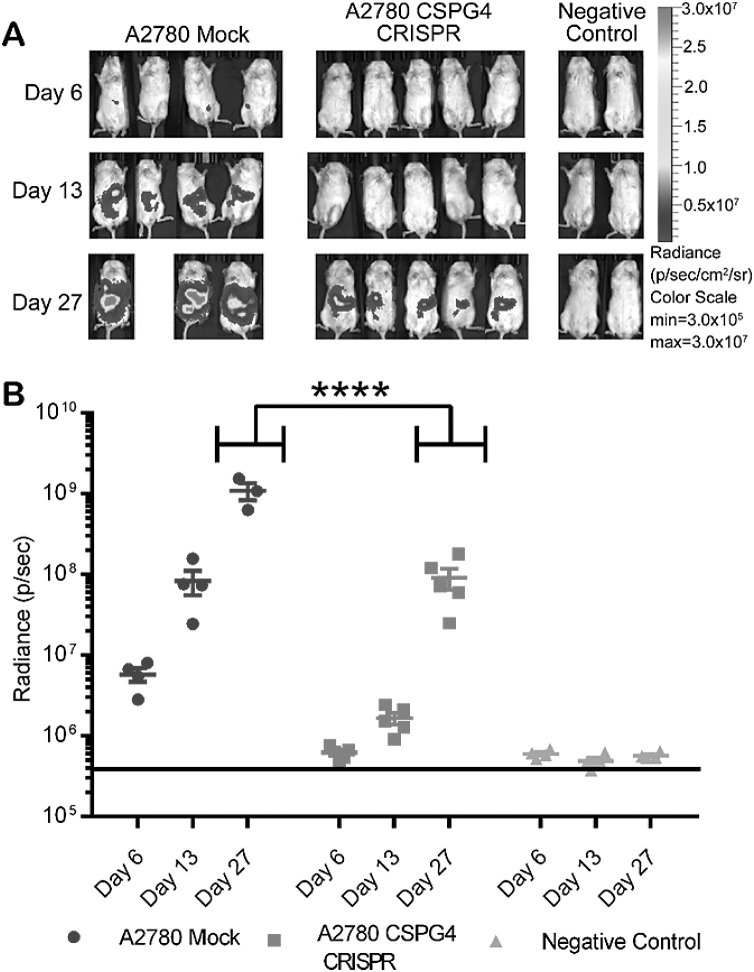
Finally, to determine if CSPG4 expression impacts tumor growth in vivo, we utilized an intraperitoneal xenograft injection model [35], [36], [37]. Mice were injected with 2 × 105 mock transfected or CSPG4 knockout A2780 cells. Tumor growth, as monitored by bioluminescence, was significantly reduced in mice receiving CSPG4 knockout cells (Fig. 4). By 27 days post injection, CSPG4 expression promoted almost an order of magnitude growth advantage compared to CSPG4 knockout counterparts. The results collectively lead to the conclusion that CSPG4 could negatively impact EOC patient outcome by promoting multiple aspects of malignant progression which could include enhanced tumor expansion in patients.
Fig. 4.
CRISPR knockout of CSPG4 in A2780 ovarian carcinoma cells results in reduced tumor growth in vivo. Mice were injected I.P. with 2.0 × 105 luc+ A2780 Mock or A2780 CSPG4-CRISPR knockout cells. (A) Tumor growth was monitored by bioluminescent imaging (BLI) on day 6, 13, and 27. Color scale bar indicates photon/s/cm2/sr. (B) Quantification of tumor burden based on BLI total flux (photons/sec). Data are shown as mean ± SD. ****p<0.0001 by ordinary two-way ANOVA with Sidak's multiple comparison test. Mock: n=4, CRISPR, n=5, Negative control, n=5.
CSPG4 expression is associated with epithelial to mesenchymal changes in ovarian cancer cells
Gene set enrichment analysis (GSEA) of the ES-2 RNA seq dataset indicated that CSPG4 deletion impacts the differential expression of several genes within the hallmark epithelial to mesenchymal transition gene set from the Molecular Signatures Database v 7.1 (https://www.gsea-msigdb.org/gsea/msigdb/index.jsp) (Supplementary Fig. 4A). Analysis of a large array of gene expression data from the ovarian cancer TCGA also demonstrates that high CSPG4 expression is associated with an EMT signature, leading us to conclude that high CSPG4 portends a more mesenchymal phenotype in EOC tumors [33] (Supplementary Fig. 4B).
To explore this further, we initially analyzed the impact of CSPG4 expression on several mesenchymal/epithelial markers by western blots of all three EOC cell lines (Fig. 5A). CRISPR/Cas9 driven loss of CSPG4 expression decreased the expression of multiple mesenchymal biomarkers, including vimentin and the mesenchymal transcription factors SNAI2 and ZEB1, while promoting increased expression of the epithelial biomarker Claudin-1 (Fig. 5A). Not surprisingly, given the context dependent/cell line variations in EMT biomarker expression [11], the initial western blot results indicated that loss of CSPG4 caused no consistent change in levels of the epithelial biomarker E-cadherin (Fig. 5A).
Fig. 5.
CSPG4 expression is associated with epithelial-to-mesenchymal plasticity, mediated by CSPG4 associated changes in ZEB1 expression. (A) The indicated cell lines both Mock(M) and CRISPR(C) were cultured separately in spheroid formation assays for 7 days, collected and analyzed by western blot for various EMT markers. (B) Western blot for ZEB1 and CSPG4 in cancer cell lines treated with siRNA for ZEB1 or control for 48h. (C) Western blot for pFAK, FAK and ZEB1 in HEY cells treated with either DMSO (control) or FAK inhibitor PND-1186 at the indicated concentrations in normal growth medium for 24h. (D) Spheroid formation in methylcellulose of cancer cell lines treated with siRNA to ZEB1 or control for 7 days. Bars represent spheroid counts from five random fields/well from triplicate wells +/- S.D. *p<0.02 by students t-test with Welch's correction. n=3 from 3 individual experiments. (E) Invasion assay of cancer cells treated with ZEB1 or control siRNA (methods). *p<0.002 by student's t-test with Welch's correction. n=3 from 3 individual experiments. (F and G) Cells were transfected with control or ZEB1 siRNA overnight and plated in the cisplatin cytotoxicity assay the following day. MTS data was collected after 4 days of cisplatin treatment. Untransfected CSPG4-CRISPR cells for A2780 (F) and HEY (G) were included as a control. Dose response curves were plotted as the percent of MTS staining vs. untreated cells for each group. Combined data from three independent experiments are shown (n=9). (H–J) Kaplan-Meier curves of ovarian cancer patients in a combined cohort of TCGA and 14 GEO datasets demonstrate that tumors that express CSPG4 (H), ZEB1 (I), and the mean combined expression of both (J) are associated with decreased 5-year survival (red lines) when compared to data from tumors that are negative for these markers (black lines).
For subsequent studies we further focused on the relationship between CSPG4 and ZEB1 levels, since ZEB1 is a transcriptional factor previously associated with the development of a mesenchymal phenotype and poor outcome in EOC patients [38,39]. Independently inhibiting ZEB1 expression using RNAi (Fig. 5B) had no inhibitory impact on CSPG4 levels, indicating that ZEB1 expression is downstream of CSPG4. Furthermore, CSPG4 stimulated ZEB1 expression is mediated by FAK activation, since ZEB1 expression is inhibited by a small molecule inhibitor of FAK activation, PND-1186 (Fig. 5C).
Inhibiting ZEB1 limits both spheroid formation (Fig. 5D) and invasion (Fig. 5E), which is consistent with studies demonstrating that spheroid formation is associated with a mesenchymal transition [9,40]. However, limiting ZEB1 expression had no detectable impact on cisplatin sensitivity (Fig. 5F and 5G), indicating that the impact of CSPG4 on cisplatin sensitivity is independent of increased ZEB1 expression. Aggregated survival analysis of multiple ovarian cancer patient datasets shows that elevated transcript expression of both CSPG4 and ZEB1, either individually or in combination, is significantly associated with decreased overall survival at 5 years (Fig. 5H–J), supporting the conclusion that both biomarkers can function in concert to promote recurrence and relapse.
Anti-CSPG4 antibody decreases EOC invasion and spheroid formation by inhibiting CSPG4 activated FAK-ZEB1 pathway
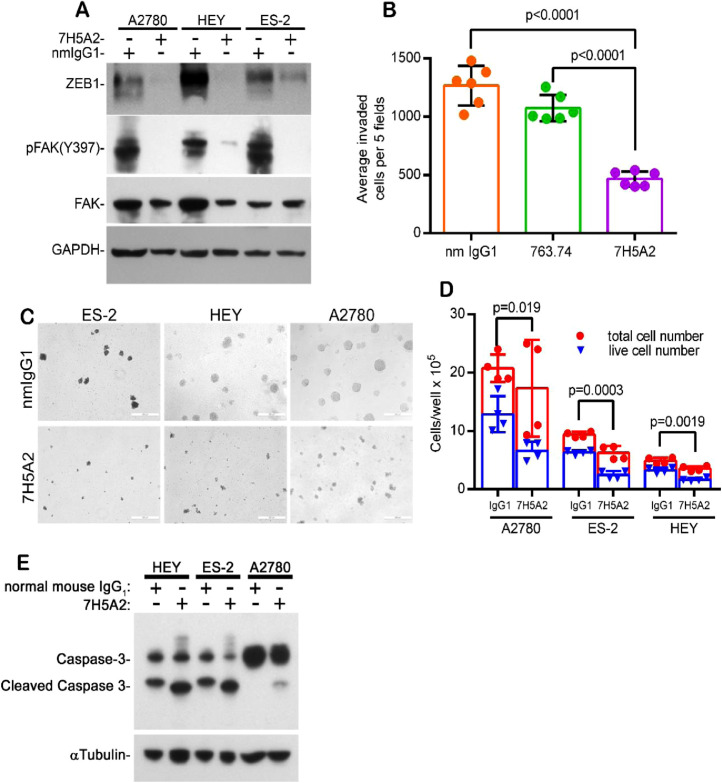
While multiple structural/functional domains of the extracellular portion of the CSPG4 core protein have been identified, many of these sites (e.g. collagen and growth factor binding sites, chondroitin sulfate attachment sites, laminin G-domain) map to membrane distal regions of the core protein [12,13]. By contrast there is less known of the potential functional importance of domains in the core protein that are membrane proximal. To examine the importance of this core protein region in CSPG4 on EOC cells, a recombinant fragment of the CSPG4 core protein containing this region (Q1538-N2221, Supplemental Fig. 5A) was purified from a eukaryotic expression system and used in the production of mouse monoclonal antibodies. We subsequently identified an antibody clone (7H5A2) that specifically recognizes CSPG4 on the cell surface (Supplementary Fig. 5B and 5C). The antibody is antagonistic to CSPG4 function and similar to CSPG4 deletion, inhibited activation of FAK and ZEB1 expression in spheroid cultures of all three cell lines (Fig. 6A). The antibody, which does not appreciably decrease the level of CSPG4, also significantly inhibited EOC invasion (Fig. 6B) to a greater extent than the anti-CSPG4 antibody 763.74. Antibody 763.74 has previously been shown to impact CSPG4 mediated invasion of other tumor types [41], but in contrast to 7H5A2, it recognizes a distinct membrane distal epitope on the core protein [42]. The 7H5A2 antibody also significantly inhibited spheroid formation (Fig. 6C and 6D) and enhanced caspase 3 activation (Fig. 6E), indicating the antibody promoted apoptosis of CSPG4 positive EOC cells within the spheroids.
Fig. 6.
Anti-CSPG4 antibody 7H5A2 inhibits ZEB1 expression and FAK activation, cell invasion and spheroid formation. (A) Western blot for pFAK, FAK, and ZEB1 in cells plated in 1% methylcellulose in the presence of 50μg/ml normal mouse IgG1 (nmIgG1) or anti-CSPG4 antibody 7H5A2 cultured in suspension for 24h, (B) Invasion assay using HEY cells pre-incubated with control (nm IgG1) or anti-CSPG4 (763.74 and 7H5A2) antibodies at 50 μg/ml for 2h and then plated in invasion chambers in the presence of the antibody in the top chamber only for 24h. Bars represent the total number of invading cells counted in five random fields/well from duplicate wells, +/- S.D. P-values determined by student's t-test with Welch's correction. n=6 from 3 separate experiments. (C, D) Spheroid formation assay plated in the presence of the indicated antibodies at 50 μg/ml in 1% methylcellulose for 7 days (HEY) or 14 days (A2780 and ES-2). (C) Representative images showing spheroid formation, bar=200μm. (D) Quantification of cell growth/survival in spheroids. Spheroids were collected from methylcellulose culture at 7 (HEY) or 14 days (ES-2 and A2780), and re-plated overnight in normal growth media in tissue culture plates. The following day all cells are collected from each well and counted. P-values determined by student's t-test with Welch's correction, comparing live cell numbers (trypan blue excluding) vs. total cell number isolated from each condition, n=4. (E) Cells were pre-treated with normal mouse IgG1 or antibody 7H5A2 (50 μg/ml) for 1h and then plated in 1% methylcellulose supplemented with 50 μg/ml of the indicated antibody. Spheroids were harvested after 72h and assayed for caspase-3 activation by western blot.
Discussion
While it is well known that EOC tumors are genetically heterogeneous, there is also extensive evidence that malignant progression is driven by phenotypic heterogeneity which is related to the dynamic biology of malignant progression [1], [2], [3], [4]. IHC data from this EOC patient cohort demonstrates that high levels of CSPG4 within tumors are an independent risk factor for poor overall survival. CSPG4 expression promotes tumor cell invasion, cisplatin resistance and spheroid formation in vitro and tumor expansion in vivo.
CSPG4 does not signal directly on its own, but rather functions as a co-receptor/plasma membrane scaffold that enhances the intensity and duration of multiple stimulated oncogenic pathways. A longer duration of signal transduction activation can lead to nuclear changes that impact on the transcriptome, as we have shown for CSPG4-mediated prolonged activation of Erk 1,2 which causes nuclear localization of activated Erk 1,2 and a shift to a mesenchymal transcriptome in melanoma cells, which promotes their tumorigenic potential [12,21]. Thus, localized elevations of CSPG4 levels in subpopulations of EOC cells may sustain tumor cell subpopulations that have enhanced oncogenic signaling leading to increased growth, survival and/or invasive potential. This is consistent with the tumor growth data in vivo, which link CSPG4 expression to significantly enhanced tumor expansion compared to CRISPR/Cas9 deleted counterparts. It is also important to note that although high CSPG4 expression levels in EOC tumors negatively impact patient survival, the CSPG4 staining pattern in the tumor parenchyma is heterogeneous. Since CSPG4 expression is stimulated by microenvironmental changes in hypoxia or inflammatory mediators such as TNFα, we predict that heterogeneous CSPG4 expression may be related to these, or additional microenvironmental factors in the expanding tumor [43].
One consequence of CSPG4 expression is that it stimulates a mesenchymal shift in the phenotype of EOC cells, which is associated with spheroid formation and subsequent intraperitoneal metastasis to other organs such as omentum [3,9]. These spheroids, which are associated with the transition to a mesenchymal phenotype [9,40] originate from individual cells that have acquired an anchorage independent phenotype, can adhere to mesothelial-lined surfaces and form metastases by invading into sub-mesothelial tissues [[1], [2], [3],6,9,44]. Thus, current data implicate CSPG4 expression in promoting intraperitoneal metastasis, which is a major cause of therapy failure in patients with EOC.
While TCGA gene expression data link elevated levels of CSPG4 to a generalized mesenchymal transition, the current data indicate that CSPG4 may be mechanistically linked to elevated expression of the mesenchymal transcription factor ZEB1 which has been linked to malignant progression in EOC and other tumors [9,38,45,46]. As a transcriptional regulator, ZEB1 represses the expression of multiple epithelial genes while it stimulates the expression of genes that are associated with an invasive, mesenchymal phenotype [45,46]. Since TCGA analysis indicates co-expression of CSPG4 and ZEB1 is associated with poor overall survival, the two may function in concert to promote metastasis as evidenced by their impact on spheroid formation and invasion. Furthermore, CSPG4 expressing tumor cells are also more resistant to cisplatin, suggesting that CSPG4 expressing tumor cells may form a therapy-resistant tumor cell reservoir that promotes relapse following initial standard of treatment.
CSPG4 promotes spheroid formation and invasion by activating FAK to enhance ZEB1 expression. This is consistent with data from other cell model systems (including fibroblasts isolated from FAK-null animals) which link FAK activation to ZEB1 expression [47,48]. The current study shows that a well characterized inhibitor of FAK activation limits ZEB1 expression and our novel anti-CSPG4 specific antibody also inhibits FAK activation, ZEB1 expression, tumor cell invasion and spheroid formation. A link between CSPG4, activated β1 integrins and FAK activation has previously been shown by our laboratory and others [12,15,16,49,50]. Thus, these data support a model in which EOC cell surface CSPG4 functions to promote cancer progression by interacting with components of the tumor microenvironment (specific ECM components or various growth factors) to enhance cell adhesion, motility and mesenchymal transition.
Importantly, CSPG4 expression also causes a significant increase in platinum IC50s, suggesting a potential increase in the therapeutic window for EOC cells expressing this oncoantigen. While a CSPG4/ZEB1 axis is linked to mesenchymal transition, independently inhibiting ZEB1 expression has no impact on cisplatin sensitivity, leading us to conclude that CSPG4 impacts cisplatin sensitivity by mechanisms that are ZEB1-independent. As a multifunctional transmembrane signaling node, CSPG4 functions to alter the activation initiated by multiple extracellular stimuli (e.g., TGFβ, FGF, HGF) and depending on the cellular context, it can activate multiple oncogenic pathways (e.g. FAK, MAPK, PI3K, NF-kB) in tumor cells [12,14,[17], [18], [19]]. Since reduced cisplatin sensitivity in standard of care therapy may impact the survival of resistant clones of EOC tumor cells, it will be important to further define the mechanisms by which CSPG4 alters the response to this therapy. One approach is to rescue CSPG4 null cells using several well-defined CSPG4 structural mutants to identify domains that fail to reverse the loss of cisplatin sensitivity [12,13,[15], [16], [17]]. This approach may lead to enhanced targeting by identifying CSPG4 domains that limit cisplatin sensitivity by mechanism(s) that are coincident with, or independent of, regulating ZEB1 expression.
While the current data indicate that CSPG4 may directly reduce tumor cell sensitivity to cisplatin, we propose that CSPG4 in the larger context of complex tumor tissues may also impact poor outcome in EOC patients by contributing to cell adhesion-related mechanisms associated with environmental mediated drug resistance (EMDR) [7]. The concept that underlies EMDR is that adherent tumor cell subpopulations, which initially resist therapy, can form a reservoir of resistant cells which may undergo additional mutations that are responsible for therapy resistant relapse following standard of care [7]. This is analogous to, but distinct from, the hypothesis that therapy resistant cancer initiating stem cells are responsible for therapy failure [51]. Numerous cell adhesion related mechanisms (e.g. related to integrin and growth factor/cytokine mediated pathways) can function to promote survival in the absence of transcriptomic profiles that regulate cancer initiating stem cells [7]. Mesenchymal shifts in EOC, driven by factors such as TGF-β, are associated with a collagen remodeling fibrotic gene signature that correlates with metastasis and poor overall survival [52]. The fibrotic signature associated with mesenchymal EOC includes elevated type VI collagen, a major ECM ligand for CSPG4 [52] and elevations in type VI collagen in the tumor parenchyma are associated with decreased EOC patient survival [53]. Those studies demonstrated that EOC cells adherent on type VI collagen coated surfaces exhibited increased resistance to cisplatin in vitro. The potential clinical impact is that localized CSPG4/ECM interactions may cause the formation of therapy-resistant adherent ‘niches’ consisting of deeply embedded EOC populations that may evade detection following standard of care surgical debulking. Thus, anti-CSPG4 antibodies that either directly inhibit CSPG4 function or can promote immune mediated toxicity of EOC cells may be effective therapeutically to improve patient outcome.
Author contributions
J.Y. - originally conceived of these studies and led the execution of experiments throughout the study. Q.L and J.W- responsible for the IHC analysis of the EOC patient cohort. M.P.- worked with J.Y. to design and interpret the experiments. B.M. and N.W. -Designed guide RNAs for CRISPR/Cas 9 deletion of the entire locus of CSPG4. M.F and J.S.M – Advised on generating and characterizing novel CSPG4 antibody. M.A.G, L. B. R.H. – Provided advice into clinical aspects and biology of epithelial ovarian cancer and designed the animal study. T.K.S – Performed and interpreted RNA seq analysis for the EOC cell lines. C.H.O'C – Performed and aided in the interpretation of gene ontology and gene set expression analysis. S. T. and A.C.N, - Performed bioinformatic analysis using TCGA database for CSPG4 in EOC patients. E.T. – Advised on signal transduction mechanisms related to CSPG4 and critically read the manuscript. J.B.M. – Integrated efforts in the study, oversaw the writing and assumes responsible for the integrity of the studies.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by NCI grants P01 CA111412 and R35 CA197292 awarded to J.S.M, grants from the National Natural Science Foundation of China (81972836) and the Science and Technology Innovation Program of Hunan Province (2020SK2120) awarded to J. W. and Q.L. Additional funding was provided by the Atwater Fund, Elsa Pardee and Chairman's Fund Professor in Cancer Research to J.B.M. T.K.S was supported by grants from Minnesota Ovarian Cancer Alliance, the Randy Shaver Cancer Research and Community Fund, and the University of Minnesota Grand Challenges. A.C.N is supported by the American Cancer Society 132574-CSDG-18-139-01-CSM. The authors also acknowledge the Imaging Core and the Flow Cytometry Core at the University of Minnesota for their excellent service. The authors have declared that no conflict of interest exists.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2021.101318.
Contributor Information
Jing Wang, Email: wangjing0081@hnszlyy.com.
James B. McCarthy, Email: mccar001@umn.edu.
Appendix. Supplementary materials
References
- 1.Kim J., Park E.Y., Kim O., Schilder J.M., Coffey D.M., Cho C.H., et al. Cell origins of high-grade serous ovarian cancer. Cancers. 2018;10 doi: 10.3390/cancers10110433. (Basel) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kroeger P.T., Drapkin R. Pathogenesis and heterogeneity of ovarian cancer. Curr. Opin. Obstet. Gynecol. 2017;29:26–34. doi: 10.1097/GCO.0000000000000340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lengyel E. Ovarian cancer development and metastasis. Am. J. Pathol. 2010;177:1053–1064. doi: 10.2353/ajpath.2010.100105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Testa U., Petrucci E., Pasquini L., Castelli G., Pelosi E. Ovarian cancers: genetic abnormalities, tumor heterogeneity and progression, clonal evolution and cancer stem cells. Medicines. 2018;5:1–74. doi: 10.3390/medicines5010016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moffitt L., Karimnia N., Stephens A., Bilandzic M. Therapeutic targeting of collective invasion in ovarian cancer. Int. J. Mol. Sci. 2019;20 doi: 10.3390/ijms20061466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Al Habyan S., Kalos C., Szymborski J., McCaffrey L. Multicellular detachment generates metastatic spheroids during intra-abdominal dissemination in epithelial ovarian cancer. Oncogene. 2018;37:5127–5135. doi: 10.1038/s41388-018-0317-x. Oncogene. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meads M.B., Gatenby R.A., Dalton W.S. Environment-mediated drug resistance: a major contributor to minimal residual disease. Nat. Rev. Cancer. 2009;9:665–674. doi: 10.1038/nrc2714. [DOI] [PubMed] [Google Scholar]
- 8.Paullin T., Powell C., Menzie C., Hill R., Cheng F., Martyniuk C.J., et al. Spheroid growth in ovarian cancer alters transcriptome responses for stress pathways and epigenetic responses. PLoS ONE. 2017;12 doi: 10.1371/journal.pone.0182930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shield K., Ackland M.L., Ahmed N., Rice G.E. Multicellular spheroids in ovarian cancer metastases: biology and pathology. Gynecol. Oncol. 2009;113:143–148. doi: 10.1016/j.ygyno.2008.11.032. [DOI] [PubMed] [Google Scholar]
- 10.Deng J., Wang L., Chen H., Hao J., Ni J., Chang L., et al. Targeting epithelial-mesenchymal transition and cancer stem cells for chemoresistant ovarian cancer. Oncotarget. 2016;7:55771–55788. doi: 10.18632/oncotarget.9908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang J., Antin P., Berx G., Blanpain C., Brabletz T., Bronner M., et al. Guidelines and definitions for research on epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2020;21:341–352. doi: 10.1038/s41580-020-0237-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Price M.A., Colvin Wanshura L.E., Yang J., Carlson J., Xiang B., Li G., et al. CSPG4, a potential therapeutic target, facilitates malignant progression of melanoma. Pigment Cell Melanoma Res. 2011;24:1148–1157. doi: 10.1111/j.1755-148X.2011.00929.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tamburini E., Dallatomasina A., Quartararo J., Cortelazzi B., Mangieri D., Lazzaretti M., et al. Structural deciphering of the NG2/CSPG4 proteoglycan multifunctionality. FASEB J. 2019;33:3112–3128. doi: 10.1096/fj.201801670R. [DOI] [PubMed] [Google Scholar]
- 14.Wang X., Wang Y., Yu L., Sakakura K., Visus C., Schwab J.H., et al. CSPG4 in cancer: multiple roles. Curr. Mol. Med. 2010;10:419–429. doi: 10.2174/156652410791316977. [DOI] [PubMed] [Google Scholar]
- 15.Yang J., Price M.A., Neudauer C.L., Wilson C., Ferrone S., Xia H., et al. Melanoma chondroitin sulfate proteoglycan enhances FAK and ERK activation by distinct mechanisms. J. Cell Biol. 2004;165:881–891. doi: 10.1083/jcb.200403174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang J., Price M.A., Wanshura L.E.C., He J., Yi M., Welch D.R., et al. Chondroitin sulfate proteoglycan 4 enhanced melanoma motility and growth requires a cysteine in the core protein transmembrane domain. Melanoma Res. 2019;29:365–375. doi: 10.1097/CMR.0000000000000574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nicolosi P.A., Dallatomasina A., Perris R. Theranostic impact of NG2/CSPG4 proteoglycan in cancer. Theranostics. 2015;5:530–544. doi: 10.7150/thno.10824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rolih V., Barutello G., Iussich S., De Maria R., Quaglino E., Buracco P., et al. CSPG4: a prototype oncoantigen for translational immunotherapy studies. J. Transl. Med. 2017;15:151. doi: 10.1186/s12967-017-1250-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang X., Osada T., Wang Y., Yu L., Sakakura K., Katayama A., et al. CSPG4 protein as a new target for the antibody-based immunotherapy of triple-negative breast cancer. J. Natl. Cancer Inst. 2010;102:1496–1512. doi: 10.1093/jnci/djq343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Allred D.C., Harvey J.M., Berardo M., Clark G.M. Prognostic and predictive factors in breast cancer by immunohistochemical analysis. Mod. Pathol. 1998;11:155–168. [PubMed] [Google Scholar]
- 21.Yang J., Price M.A., Li G.Y., Bar-Eli M., Salgia R., Jagedeeswaran R., et al. Melanoma proteoglycan modifies gene expression to stimulate tumor cell motility, growth, and epithelial-to-mesenchymal transition. Cancer Res. 2009;69:7538–7547. doi: 10.1158/0008-5472.CAN-08-4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baller J., Kono T., Herman A., Zhang Y. Proceedings of the Practice and Experience in Advanced Research Computing on Rise of the Machines (learning) 2019. CHURP: a Lightweight CLI Framework to Enable Novice Users to Analyze Sequencing Datasets in Parallel. Article No.: 96, 1–5. < https://dl.acm.org/doi/10.1145/3332186.3333156>. [Google Scholar]
- 23.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Andrews S. 2010. FastQC: a Quality Control Tool for High Throughput Sequence.https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ <. >. [Google Scholar]
- 25.Kim D., Paggi J.M., Park C., Bennett C., Salzberg S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019;37:907–915. doi: 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 27.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu G., Wang L.G., Han Y., He Q.Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gyorffy B., Lanczky A., Szallasi Z. Implementing an online tool for genome-wide validation of survival-associated biomarkers in ovarian-cancer using microarray data from 1287 patients. Endocr. Relat. Cancer. 2012;19:197–208. doi: 10.1530/ERC-11-0329. [DOI] [PubMed] [Google Scholar]
- 30.Nagy A., Lanczky A., Menyhart O., Gyorffy B. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci. Rep. 2018;8:9227. doi: 10.1038/s41598-018-27521-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gyorffy B., Lanczky A., Eklund A.C., Denkert C., Budczies J., Li Q., et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res. Treat. 2010;123:725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- 32.Goldman M.J., Craft B., Hastie M., Repecka K., McDade F., Kamath A., et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat. Biotechnol. 2020;38:675–678. doi: 10.1038/s41587-020-0546-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salt M.B., Bandyopadhyay S., McCormick F. Epithelial-to-mesenchymal transition rewires the molecular path to PI3K-dependent proliferation. Cancer Discov. 2014;4:186–199. doi: 10.1158/2159-8290.CD-13-0520. [DOI] [PubMed] [Google Scholar]
- 34.Domcke S., Sinha R., Levine D.A., Sander C., Schultz N. Evaluating cell lines as tumour models by comparison of genomic profiles. Nat. Commun. 2013;4:2126. doi: 10.1038/ncomms3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hernandez L., Kim M.K., Lyle L.T., Bunch K.P., House C.D., Ning F., et al. Characterization of ovarian cancer cell lines as in vivo models for preclinical studies. Gynecol. Oncol. 2016;142:332–340. doi: 10.1016/j.ygyno.2016.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hijaz M., Chhina J., Mert I., Taylor M., Dar S., Al-Wahab Z., et al. Preclinical evaluation of olaparib and metformin combination in BRCA1 wildtype ovarian cancer. Gynecol. Oncol. 2016;142:323–331. doi: 10.1016/j.ygyno.2016.06.005. [DOI] [PubMed] [Google Scholar]
- 37.Wiechert A., Saygin C., Thiagarajan P.S., Rao V.S., Hale J.S., Gupta N., et al. Cisplatin induces stemness in ovarian cancer. Oncotarget. 2016;7:30511–30522. doi: 10.18632/oncotarget.8852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen D., Wang J., Zhang Y., Chen J., Yang C., Cao W., et al. Effect of down-regulated transcriptional repressor ZEB1 on the epithelial-mesenchymal transition of ovarian cancer cells. Int. J. Gynecol. Cancer. 2013;23:1357–1366. doi: 10.1097/IGC.0b013e3182a5e760. [DOI] [PubMed] [Google Scholar]
- 39.Wei L., He Y., Bi S., Li X., Zhang J., Zhang S. miRNA199b3p suppresses growth and progression of ovarian cancer via the CHK1/Ecadherin/EMT signaling pathway by targeting ZEB1. Oncol. Rep. 2020;45:570–581. doi: 10.3892/or.2020.7895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rafehi S., Ramos Valdes Y., Bertrand M., McGee J., Prefontaine M., Sugimoto A., et al. TGFbeta signaling regulates epithelial-mesenchymal plasticity in ovarian cancer ascites-derived spheroids. Endocr. Relat. Cancer. 2016;23:147–159. doi: 10.1530/ERC-15-0383. [DOI] [PubMed] [Google Scholar]
- 41.Luo W., Ko E., Hsu J.C., Wang X., Ferrone S. Targeting melanoma cells with human high molecular weight-melanoma associated antigen-specific antibodies elicited by a peptide mimotope: functional effects. J. Immunol. 2006;176:6046–6054. doi: 10.4049/jimmunol.176.10.6046. [DOI] [PubMed] [Google Scholar]
- 42.Geiser M., Schultz D., Le Cardinal A., Voshol H., Garcia-Echeverria C. Identification of the human melanoma-associated chondroitin sulfate proteoglycan antigen epitope recognized by the antitumor monoclonal antibody 763.74 from a peptide phage library. Cancer Res. 1999;59:905–910. [PubMed] [Google Scholar]
- 43.Ampofo E., Schmitt B.M., Menger M.D., Laschke M.W. The regulatory mechanisms of NG2/CSPG4 expression. Cell Mol. Biol. Lett. 2017;22:4. doi: 10.1186/s11658-017-0035-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Burleson K.M., Hansen L.K., Skubitz A.P. Ovarian carcinoma spheroids disaggregate on type I collagen and invade live human mesothelial cell monolayers. Clin. Exp. Metastasis. 2004;21:685–697. doi: 10.1007/s10585-004-5768-5. [DOI] [PubMed] [Google Scholar]
- 45.Caramel J., Ligier M., Puisieux A. Pleiotropic roles for ZEB1 in cancer. Cancer Res. 2018;78:30–35. doi: 10.1158/0008-5472.CAN-17-2476. [DOI] [PubMed] [Google Scholar]
- 46.Sanchez-Tillo E., Siles L., de Barrios O., Cuatrecasas M., Vaquero E.C., Castells A., et al. Expanding roles of ZEB factors in tumorigenesis and tumor progression. Am. J. Cancer Res. 2011;1:897–912. [PMC free article] [PubMed] [Google Scholar]
- 47.Li X.Y., Zhou X., Rowe R.G., Hu Y., Schlaepfer D.D., Ilic D., et al. Snail1 controls epithelial-mesenchymal lineage commitment in focal adhesion kinase-null embryonic cells. J. Cell Biol. 2011;195:729–738. doi: 10.1083/jcb.201105103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ungewiss C., Rizvi Z.H., Roybal J.D., Peng D.H., Gold K.A., Shin D.H., et al. The microRNA-200/Zeb1 axis regulates ECM-dependent beta1-integrin/FAK signaling, cancer cell invasion and metastasis through CRKL. Sci. Rep. 2016;6:18652. doi: 10.1038/srep18652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fukushi J., Makagiansar I.T., Stallcup W.B. NG2 proteoglycan promotes endothelial cell motility and angiogenesis via engagement of galectin-3 and alpha3beta1 integrin. Mol. Biol. Cell. 2004;15:3580–3590. doi: 10.1091/mbc.E04-03-0236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tillet E., Gential B., Garrone R., Stallcup W.B. NG2 proteoglycan mediates beta1 integrin-independent cell adhesion and spreading on collagen VI. J. Cell. Biochem. 2002;86:726–736. doi: 10.1002/jcb.10268. [DOI] [PubMed] [Google Scholar]
- 51.Chefetz I., Grimley E., Yang K., Hong L., Vinogradova E.V., Suciu R., et al. A Pan-ALDH1a inhibitor induces necroptosis in ovarian cancer stem-like cells. Cell Rep. 2019;26:3061–3075. doi: 10.1016/j.celrep.2019.02.032. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cheon D.J., Tong Y., Sim M.S., Dering J., Berel D., Cui X., et al. A collagen-remodeling gene signature regulated by TGF-beta signaling is associated with metastasis and poor survival in serous ovarian cancer. Clin. Cancer Res. 2014;20:711–723. doi: 10.1158/1078-0432.CCR-13-1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sherman-Baust C.A., Weeraratna A.T., Rangel L.B., Pizer E.S., Cho K.R., Schwartz D.R., et al. Remodeling of the extracellular matrix through overexpression of collagen VI contributes to cisplatin resistance in ovarian cancer cells. Cancer Cell. 2003;3:377–386. doi: 10.1016/s1535-6108(03)00058-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.