Highlights
-
•
Osteosarcoma (OS) chemoresistance and distant metastasis are directly associated with OS recurrence and dismal patient prognosis, which are serious concerns for the medical community.
-
•
However, current knowledge on OS pathogenesis and treatment remains limited. We found that kinesin superfamily protein 4A (KIF4A) acts as a potential OS biomarker.
-
•
KIF4A promoted OS cell proliferation and anti-apoptotic in vitro and enhanced tumor growth in vivo. Our results indicate that miR-195 inhibits the expression of KIF4A by directly targeting its 3’-untranslated region
-
•
Hence, targeting KIF4A could be a novel therapeutic strategy for OS and miR-195 may be a potential KIF4A-targeting drug. Furthermore, this study demonstrates that normal human chondrocytes can be used to produce miR-195-carrying exosomes to successfully deliver miR-195 into OS cells.
-
•
Thus, our results suggest that chondrocyte-derived exosomal miR-195 may be developed into a potential adjuvant chemotherapeutic drug.
Keywords: Osteosarcoma, KIF4A, Exosomes, miR-195, Chondrocytes, Anti-apoptotic
Abstract
Background
Osteosarcoma (OS) is a primary malignant tumor of the bone that occurs in adolescents and is characterized by a young age at onset, high malignancy, high rate of metastasis, and poor prognosis. However, the factors influencing disease progression and prognosis remain unclear.
Methods
In this study, we aimed to investigate the role of chondrocyte-derived exosomal miR-195 in OS. We used normal human chondrocytes to form miR-195-carrying exosomes to deliver miR-195 into OS cells. Xenograft tumor experiments were performed in mice intratumorally injected with exosomal miR-195. We found that kinesin superfamily protein 4A (KIF4A) promoted OS tumor progression and anti-apoptotic.
Resules
We demonstrated that miR-195 inhibited the expression of KIF4A by directly targeting its 3’-untranslated region. Moreover, we observed that exosomal miR-195 successfully inhibited OS cell tumor growth and antiapoptotic in vitro and suppressed tumor growth in vivo.
Conclusion
Collectively, these results demonstrate that normal human chondrocyte-derived exosomal miR-195 can be internalized by OS cells and inhibit tumor growth and antiapoptotic by targeting KIF4A, providing a new direction for clarifying the molecular mechanism underlying OS development.
Introduction
Osteosarcoma (OS) is a malignant tumor of mesenchymal tissue origin, characterized by malignant spindle cells that produce abnormal osteoid tissue [1]. The most common clinical symptoms include pain and localized masses, and the rates of clinical misdiagnosis and missed diagnosis are relatively high. Approximately 30% of patients have poor prognosis and may develop lung metastases within one year [2]. However, the etiology of OS is not fully understood, and the mechanism of abnormal cell proliferation remains unclear. The kinesin superfamily is a class of conserved microtubule-dependent molecular motor proteins with adenosine triphosphatase (ATPase) activity and motor properties [3]. Studies have shown that abnormal expression of kinesin superfamily proteins (KIFs) is related to the development and progression of various human cancers [4,5]. Among these, KIF2A,KIF18B, KIF26B expression correlates with tumorigenicity and poor OS prognosis [5], [6], [7]. KIF4A expression has been found to be significantly up-regulated in multiple tumor tissues, and high expression of KIF4A in cancers is reported to promote tumor progression and metastasis [8], [9], [10], [11]. Moreover, Huang et al found that KIF4A enhanced cell proliferation via activation of AKT/Bax/Bcl-2 signaling axis and predicted a poor prognosis in hepatocellular carcinoma [11]. In this study, we found that KIF4A expression was up-regulated in OS tissues compared to that in normal bone tissues and that high KIF4A expression predicted adverse prognosis and drug resistance in OS by bioinformatics analysis. More importantly, we demonstrated that KIF4A promoted OS cells growth and inhibited cell apoptosis through AKT activation in vivo and in vitro.
Exosomes, a broadly investigated subgroup of extracellular vesicles, have a cup-shaped appearance when observed using electron microscopy with diameters ranging from 50 nm to 150 nm [12]. Accumulating evidence indicates that exosomes are important mediators of cell communication with several regulatory roles in various cellular functions, including proliferation, metastasis, apoptosis, and drug resistance. MicroRNAs (miRNAs) are important non-coding RNAs that are packaged in exosomes and are widely involved in the regulation of cellular functions by post-transcriptionally regulating target gene expression [13]. When cells become cancerous, exosomal miRNA secretion disorders cause aberrant expression of oncogenes and tumor suppressor genes [14]. The results of relevant studies showed that exosomal miRNAs can enhance OS development by targeting cancer suppressor genes or affecting the tumor microenvironment [15,16]. By contrast, a study has shown that a bone marrow mesenchymal stem cell-derived exosomal miRNA inhibits OS progression by targeting TRA2B [17]. However, knowledge regarding the effect of normal human chondrocyte-derived exosomal miRNAs on OS cells remains limited.
Chondrocytes synthesize and secrete the extracellular matrix, cytokines, and fibers required for cartilage development. Moreover, abnormal secretion of cytokines by chondrocytes is directly linked to inflammation and cell carcinogenesis [18]. Research has shown that chondrocyte exosomes mediate osteoarthritis progression by regulating mitochondrion and immune reactivity [19] and miRNAs can also regulate chondrocyte development [20]. Zhang et al. obtained exosomes carrying miR-206 by transfecting miR-206 mimic into BMSC cells, and then observed the therapeutic effect of exosomes on osteosarcoma in vitro and in vivo [17]. Moreover, Liang et al. conformed that Chondrocyte-exosomes also deliver miR-140 to deep cartilage regions through the dense mesochondrium, inhibit cartilage-degrading proteases, and alleviate OA progression in a rat model [21]. In this study, we aimed to investigate the role of chondrocyte-derived exosomal miR-195 in OS. We used the secretory capacity of chondrocytes in an in vitro model to produce exosomal miRNAs with oncogenic effects. Furthermore, we illustrated the molecular mechanisms underlying the progression of OS by exosomal miRNA. Our findings demonstrated that normal human chondrocyte-derived exosomal miR-195 inhibited OS cell proliferation and reduced tolerance to chemotherapeutic drugs by targeting KIF4A.
Methods
Materials
McCOY's 5A Medium, Roswell Park Memorial Institute (RPMI) 1640 medium, fetal bovine serum (FBS), trypsin, cisplatin, KIF4A shRNA, and DAPI were purchased from Sigma-Aldrich (St. Louis, MO, USA). miR-26b, miR-126, miR-195, miR-223, miR-671-5p, and miR-negative control (NC) mimics (Table S1) and primers were synthesized at GenePharma (Shanghai, China). Antibodies against KIF4A, AKT, p-AKT, Bcl-2, Bax, CD63, and CD81 were purchased from Abcam, whereas the β-tubulin antibody was purchased from MULTISCIENCES (Hangzhou, China). The RNA extraction kit was purchased from Invitrogen (CA, USA) and the cDNA Reverse Transcription and SYBR PrimeScript RT Kits were obtained from Takara Bio Inc. (Tokyo, Japan). The Annexin V/FITC and Annexin V-APC/7-AAD apoptosis kits were purchased from MULTISCIENCES (Hangzhou, China), while the PKH26 Red Fluorescent Cell Linker Kit was obtained from Sigma-Aldrich.
Methods
Cell culture and transfection
Human OS (U2OS and MG-63), chondrocyte (CHON-001), and embryonic kidney 293 (HEK293) cell lines were purchased from the American Type Culture Collection. U2OS and MG-63 cells were grown in McCOY's 5A medium and RPMI 1640 medium supplemented with 10% FBS, respectively. CHON-001 and HEK293 cells were cultured in DMEM supplemented with 10% FBS. To establish stable KIF4A-knockdown cell lines, U2OS and MG-63 cells were transfected with pLVX-puro-KIF4A. Stable clones were selected using puromycin (200 ng/ml) for 4 weeks. MG-63 cells were transiently transfected with hsa-miR-26b, hsa-miR-126, hsa-miR-195, hsa-miR-223, hsa-miR-671-5p, or the miRNA mimic NC using the Lipofectamine 3000 kit according to the manufacturer's instructions (Thermo Fisher Scientific, Waltham, MA, USA).
Western blotting
Total cellular protein expression was assessed using western blotting. Briefly, U2OS and MG-63 cells were rinsed twice with 1 × phosphate-buffered saline (PBS), lysed using RIPA Lysis Buffer (Biyuntian Bio-Technology Co., Ltd.) supplemented with Halt Protease Inhibitor Cocktail and Halt Phosphatase Inhibitor Cocktail (Thermo Fisher Scientific), and the protein concentration was determined using the BCA method (Thermo Fisher Scientific). The membranes were scanned using a Biorad ChemiDoc™ touch imaging system (BIO-RAD, Hercules, California, USA).
Exosome purification from human chondrocytes
The differential ultracentrifugation method was used for exosome extraction and purification from the chondrocyte culture supernatant as follows: (a) the supernatant was centrifuged at 300 g for 15 min at 4°C and supernatant A was collected; (b) supernatant A was centrifuged at 2,000 g for 30 min at 4°C, following which supernatant B was collected; (c) finally, supernatant B was centrifuged at 10,000 g for 30 min at 4°C and the supernatant was collected. The supernatant from this step was centrifuged at 100,000 g at 4°C for 1 h, and the precipitate was retained and resuspended in PBS, followed by centrifugation at 100,000 g for 1 h. Subsequently, the exosomes were collected from the bottom of the tube and resuspended in PBS. Exosomal protein levels were measured using a BCA protein assay kit (Thermo Scientific). The purified exosomes were fluorescently labeled using the PKH26 Red Fluorescent Cell Linker Kit according to the manufacturer's instructions.
Cell proliferation assay
Cell proliferation was evaluated using the Cell Counting Kit-8 (MULTISCIENCES, Hangzhou, China) according to the manufacturer's instructions. Briefly, 3 × 103 cells were seeded into 96-well plates for 96 h. The CCK-8 solution was added to each well and the cells were incubated at 37°C for 4 h. Optical density (OD) values were measured at 450 nm using a microplate reader (DNM-9602G,PERLONG Co., Ltd., Beijing, China.).
Cytotoxicity assay
MG63 and U2OS cells were treated with different concentrations of cisplatin (0, 1, 10, 25, 50, 100, 150, and 200 μM). The cell density was adjusted to 2 × 104 cells/mL, and the cells were inoculated into 96-well plates (200 μL of cell suspension/well). CCK-8 (10 μL) was added to each well, and the OD 450 value was recorded 2 h later. The half maximal inhibitory concentration (IC50) was determined as the concentration that resulted in 50% cell growth reduction compared to that of untreated cells.
Flow cytometry assay
Cell apoptosis was measured using the Annexin V/FITC and Annexin V-APC/7-AAD apoptosis kits according to the manufacturer's instructions. Briefly, cells were seeded in 60-mm dishes and incubated overnight, followed by 10 μM cisplatin induction. After the indicated treatment, cells were collected and stained with Annexin V-FITC or Annexin V-APC for 15 min followed by 5 μL PI or 7-AAD staining for 5 min at 4°C. The samples were analyzed using a flow cytometer (Beckman-Counter Electronics, Brea, CA, USA).
qRT-PCR analysis
OS cells were transfected with the microRNA mimics for 48 h, and total RNA was extracted using RNA extraction kits (Invitrogen) according to the manufacturer's instructions. Reverse transcription for cDNA synthesis and qRT-PCR was performed using a SYBR PrimeScript RT Kit following the manufacturer's instructions. The primer sequences used are listed in Table S2. qRT-PCR was performed using an ABI 7500 Real-Time PCR detection system (Thermo Fisher Scientific). At least three independent experiments were performed per sample. Actin was used as the internal reference, and the comparative cycle threshold method (2−ΔΔCt) was used to analyze differences in each transcript [22].
Transmission electron microscopy (TEM) analysis
The isolated exosomes were characterized using a transmission electron microscope (JEOL Ltd, Akishima, Japan). The exosome (30 µl) was dripped, laid flat on a 10-cm2 parafilm, and capped with a copper mesh for 5 min. Then, uranyl acetate (30 µl) was dripped on the parafilm, and the droplet was capped with a copper mesh for 5 min. The copper mesh was rinsed several times in ultrapure water with ophthalmic tweezers and examined under the transmission electron microscope at 100 kV.
Nanoparticle tracking analysis (NTA)
The size and trajectory of particles were determined using NTA (ZetaView, Particle Metrix Co., Ltd., Ammersee, Germany).
Colony formation assay
Colony formation was analyzed using a double-layer soft agar assay in 24-well plates, as previously described [23]. After 2–3 weeks, 0.3 ml P-iodonitrotetrazolium violet (1 mg/ml) was added to the top layer for staining. Colonies with a diameter of >125 μm were scored as positive.
Exosome uptake assay
The purified exosomes were fluorescently labeled using the PKH26 Red Fluorescent Cell Linker Kit. PKH26 dye solution (1:1000) was added to 20 μg exosomes for 30 min, washed with PBS, then centrifuged at 100,000g for 80 min. Then, PKH26-labeled exosomes were added to MG63 cells and cocultured for 48 h. Uptake of exosomes was observed under a confocal fluorescence microscope.
Animal experiments
Female BALB/c nude mice (3–4-week-old) were purchased from Beijing Charles River (Beijing, China). To examine the effect of KIF4A on tumorigenesis, mice were injected with 5 × 106 exogenous KIF4A-knockdown MG63 cells and vector control MG63 cells on the right flank. In addition, MG63 cells (5 × 106 cells in PBS) were injected into the right flank of nude mice to construct the xenograft model. When the volume of tumors reached 80–130 mm3, the mice were randomly divided into two groups. miR-NC-overexpressing CHON-001-derived exosomes (EXO-NC) , and miR-195-overexpressing CHON-001-derived exosomes (EXO-miR-195) were injected into tumor separately (20 μg in 100 μl PBS). The injections were performed twice per week for two weeks. After cell injection, tumor growth was measured using a Vernier caliper every 3 days. At the end of the treatments, mice were sacrificed using rapid cervical dislocation, and xenograft tumors were removed and weighed. The tumor volume was calculated according the formula: V (mm3) = length (mm) × width (mm)2/2.
Luciferase activity assay
The pmirGLO dual-luciferase miRNA target expression vector (Promega, USA) was used to assess miRNA activity by inserting the putative miRNA target site 3’untranslated region (UTR) of the firefly luciferase gene. HEK293 cells were seeded into 24-well culture plates at a density of 2 × 105 cells/well in DMEM containing 10% FBS and incubated overnight. Cells were co-transfected with the dual-luciferase reporter constructs and corresponding miRNA mimics or NC using the Lipofectamine 3000 transfection reagent. After transfection for 36 h, luciferase activity assays were performed using a dual-luciferase reporter system, according to the manufacturer's protocol [23]. All experiments were independently conducted three times in triplicate.
Bioinformatics web tools
Differentially expressed genes (DEGs) were identified using the GSE94805, GSE28425, and GSE30934 datasets (https://www.ncbi.nlm.nih.gov/geo/). A protein–protein interaction (PPI) network was constructed using the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING)(http://string-db.org/). Furthermore, the following tools were used: UALCAN (http://ualcan.path.uab.edu/), Gene Expression Profiling Interactive Analysis (GEPIA) (http://gepia.cancer-pku.cn/detail.php), SangerBox (http://www.sangerbox.com/), Cytoscape (https://cytoscape.org/), ONCOMINE (https://www.oncomine.org/resource/login.html), TargetScan (http://www.targetscan.org/vert_72/), MIRDB (http://www.mirdb.org/cgi-bin/search.cgi), and BiBiServ (https://bibiserv.cebitec.uni-bielefeld.de/index.html), DIANA TOOLS (http://diana.imis.athena-innovation.gr/DianaTools/index.php).
Statistical analyses
Statistical analyses were performed using GraphPad Prism 7 (GraphPad Software, La Jolla, CA, USA) and IBM SPSS Statistics for Windows (version 23.0; IBM Corp., Armonk, NY, USA). Data are expressed as the mean±standard deviation. The F-test was used to analyze the homogeneity of variance among quantitative data groups, the Student's t-test and non-parametric test were used to compare the data between groups, and a p-value of <0.05 was considered statistically significant.
Results
KIF4A is highly expressed in OS and associated with poor prognosis
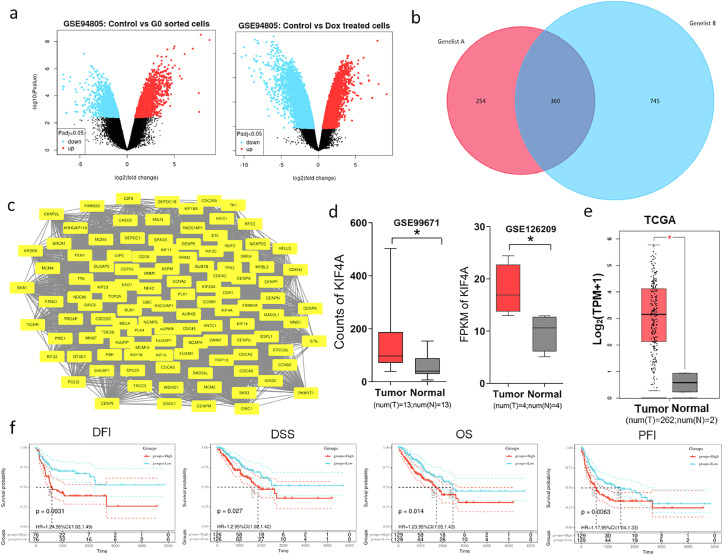
According to the GSE94805 dataset (https://www.ncbi.nlm.nih.gov/geo/), 614 and 1,105 upregulated DEGs were identified in the control group (cells undergoing normal mitosis, n=4) vs. doxorubicin (DOX)-treated OS group (cells entering senescence after DOX treatment, n=4) and in the control vs. G0-sorted OS group (cells in the G0 phase, n=4), respectively, (Figure 1a). We considered the biological processes involved in the up-regulated genes in normal OS cells were strongly related to cell proliferation, mitosis, apoptosis. So, the above 614 genes upregulated in control vs. Dox treated group were compared with the above 1,105 genes upregulated in control vs. G0 sorted group, and we found there were 360 common genes. (Figure 1b). Then, the PPI network corresponding to the 360 intersecting DEGs was analyzed using Cytoscape, and analysis revealed 113 upregulated core genes in the network (Figure 1c). It is of great significance to understand the working principle of proteins in biological systems, the reaction mechanism of biological signals and energy metabolism under special physiological states such as diseases, and the functional relationship between proteins. As a next step, we verified the expression of these genes in clinical samples by using GEPIA and GEO analysis, and kaplan-meier software was used to analyze the relationship between their expression and prognostic overall survival (OS). Overall, a total of 13 genes(AURKA, AURKB, BIRC5, CCNA2, MCM4, CDCA8, CENPA, EXO1, KIF4A, KIF15A, MYBL2, NCAPH, PLK1) met the criteria and will be further investigated. We further conformed the expression of 13 genes in the clinical samples (GSE99671; GSE126209; TCGA) by bioinformatics analysis. We also evaluated the prognostic value of 13 genes in osteosarcoma by using the SangerBox, including overall survival (OS), disease-specific survival (DSS), disease-free interval (DFI), and progression-free interval (PFI). The results showed that KIF4A is up-regulated in OS samples (Figure 1d, 1e), and that high levels of KIF4A predict a poor prognosis in patients with OS (Figure 1f, S1a, S1b). Furthermore, by analyzing the online database ONCOMINE, we showed that high-KIF4A expression is associated with chemotherapy resistance (Figure S1c). We also detected KIF4A protein and mRNA levels in normal chondrocyte (CHON-001) and OS (Saos-2, U2OS, MG63) cell lines, western blotting results revealed KIF4A was high expressed in three OS cell lines (Figure S2a). Likewise, qPCR indicated the similar result (Figure S2b).
Figure 1.
Kinesin superfamily protein 4A (KIF4A) is highly expressed in osteosarcoma (OS).
(a) Volcano plot of mRNA differential expression in the control vs. G0-sorted OS cells (left) and in the control vs. doxorubicin (DOX)-treated OS cells (right). The blue and red dots lying in the top sectors represent significantly down- and up-regulated mRNAs, respectively (p<0.05, log FC ≤-2 or ≥2). (b) Venn diagram of upregulated genes between control vs. G0 sorted cells and control vs. Dox treated cells. (c) For 360 up-regulated genes, protein-protein interaction (PPI) network analysis was constructed by using the STRING database and Cytoscape. (d, e) KIF4A was significantly overexpressed in OS cancer samples compared to its expression in normal bone cell samples. These date included 13 normal samples and 13 OS samples in GSE99671; 4 normal samples and 4 OS samples in GSE126209; 2 normal samples and 262 OS samples in TCGA; (*p<0.05). Red and grey colors represent tumor and normal tissues, respectively. (f) The expression of KIF4A was related with poor prognosis (overall survival [OS] (p=0.014, groups for KIF4A high expression n=214; groups for KIF4A low expression n=235), progression-free interval [PFI] (p=0.0063, groups for KIF4A high expression n=172; groups for KIF4A low expression n=200), disease-free interval [DFI] (p=0.0031, groups for KIF4A high expression n=108; groups for KIF4A low expression n=142), and disease-specific survival [DSS] (p=0.027, groups for KIF4A high expression n=211; groups for KIF4A low expression n=230)) as demonstrated by analyzing OS clinical samples using the Sangerbox software. Red and blue colors represent high expression groups and low expression groups, respectively. Data were obtained from TCGA database.
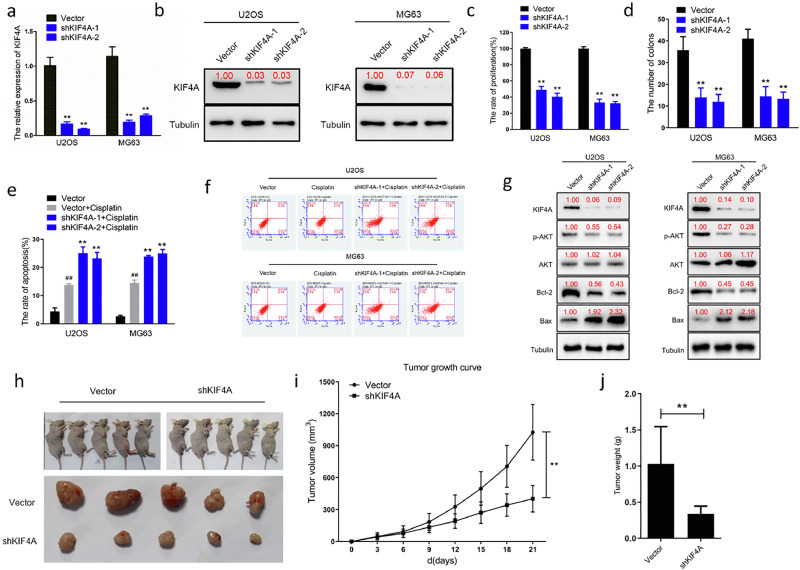
KIF4A expression promotes cell proliferation and anti-apoptotic in vitro and in vivo
To further characterize the role of KIF4A, we knocked down its expression in two OS cell lines, MG63 and U2OS, using shRNA transfections (Figure 2a, 2b). We found that low expression of KIF4A inhibited OS cell hyperproliferation and colony formation (Figure 2c, 2d). This finding demonstrates that KIF4A plays a critical role in regulating OS cell malignant proliferation. In addition, we evaluated the IC50 levels after cisplatin treatment using the CCK8 kit (Figure S2c) and selected low doses of cisplatin (0.5 µM) to treat the different OS cell groups. Flow cytometry results showed that compared with the vector group, apoptosis was higher in the cisplatin group (p<0.01). However, apoptosis in the shKIF4A groups following cisplatin treatment was more pronounced than that in the groups treated with cisplatin alone. This indicated that decreased KIF4A expression could remarkably enhance OS cell sensitivity to cisplatin (Figure 2e, 2f). Furthermore, we detected apoptosis-related proteins in OS cell lines using western blotting, and the results showed that Bcl-2 and p-AKT expression was decreased in shRNA groups compared to that in the control group, and that Bax expression was significantly higher in the shRNA group compared to that in the control group (Figure 2g). Establishing an OS xenograft model contributes to evaluating the role of KIF4A in tumor formation and progression. The in vivo experiments showed that the tumor growth rate was significantly slower in the MG63-shKIF4A group than that in the MG63-control group (p<0.01; Figure 2h, 2i). In addition, the tumor weight was markedly lower in the MG63-shKIF4A group compared to that in the MG63-control group (p<0.01; Figure 2j). These in vivo results suggest that the maintenance of rapid tumor proliferation is an important function of KIF4A. Furthermore, we detected apoptosis-related proteins using western blotting in tumor samples (Vector group n=5; shKIF4A group n=5), and the results showed that Bcl2 and p-AKT expression was decreased in shRNA groups compared to that in the control group, and that Bax expression was significantly higher in the shRNA group compared to that in the control group (Figure S3a). These findings indicated that KIF4A modulated OS cell proliferation and apoptosis by regulating the AKT and Bcl-2/Bax pathways.
Figure 2.
Kinesin superfamily protein 4A (KIF4A) regulates osteosarcoma (OS) cell proliferation and apoptosis in vitro and in vivo.
(a) qPCR demonstrating successful shRNA-mediated knockdown of KIF4A in U2OS and MG63 cells. (b) Western blotting demonstrating successful shRNA-mediated knockdown of KIF4A in U2OS and MG63 cells. (c) Proliferation of KIF4A-knockdown cells (shKIF4A-1, shKIF4A-2) were significantly lower than that of the control cells (Vector) at 96 h (**p<0.01). (d) The number of clones of U2OS and MG63 cells with KIF4A-knockdown were significantly fewer than that of the control group in vitro (**p<0.01). (e, f) Significant apoptosis were detected in cisplatin (0.5 µM) -treated U2OS and MG63 cells(Vector vs. Vector+Cisplatin: ##p<0.01). KIF4A knockdown enhanced cellular sensitivity to cisplatin (0.5 µM) -induced apoptosis in U2OS and MG63 cells (Vector+Cisplatin vs shKIF4A+Cisplatin: **p<0.01). (g) The expression levels of apoptosis-related proteins were measured by western blotting. Knockdown of KIF4A reduced phosphorylated AKT, BCL-2 expression and increased Bax expression in vitro. (h) Nude mice were inoculated subcutaneously with MG63 cells (Groups: Vector, shKIF4A; 5 × 106 cells/animal; n=5; Time: 21 days) (i) Tumor growth curve showing that low expression of KIF4A inhibits tumor growth in vivo (**p<0.01). (j) Tumor weight measurements showing that the xenograft tumor weight in the group with KIF4A-knockdown (shKIF4A) was lower than the control group (Vector) in vivo(**p<0.01).
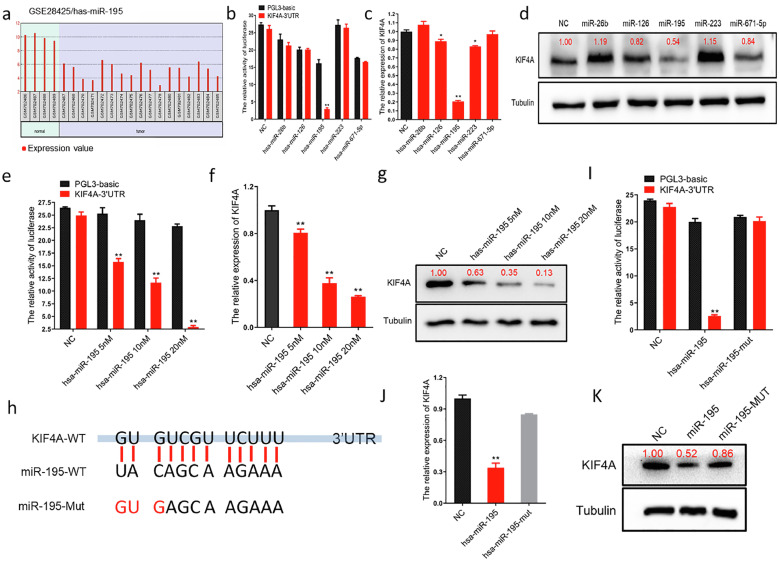
mir-195 inhibits the expression of KIF4A in OS cells by targeting the KIF4A mRNA 3′UTR
By analyzing the NCBI GEO database-derived dataset GSE28425, we found that the expression of miRNAs was significantly lower in OS cell lines compared to that in normal bone cells (Figure 3a). The five miRNAs identified (miR-26b, miR-126, miR-195, miR-223, and miR-671-5p), which potentially bind to KIF4A, were predicted using Target Scan Human, DIANA TOOLS, and BiBiServ2. We constructed a luciferase reporter plasmid containing the 3’UTR of the KIF4A transcript (pGL3-KIF4A-3’UTR). This plasmid (including the control plasmid), miRNA mimics, and the Renilla luciferase plasmid were co-transfected into HEK293 cells. The results showed that, relative to other miRNAs, only the miR-195 mimic effectively inhibited luciferase activity (Figure 3b). After transfection of the five miRNAs, the KIF4A mRNA and protein expression levels were investigated using qRT-PCR and western blotting, respectively. The results showed that miR-195 could inhibit the expression of KIF4A in OS cells more effectively than the other miRNAs (Figure 3c, 3d and Figure S4a, S4b). The pGL3-KIF4A-3’UTR plasmid (including the control plasmid) and different concentrations of the miR-195 mimic (5 nM, 10 nM, and 20 nM) were co-transfected into HEK293 cells. The results showed that there was a dose-dependent decline in luciferase activity with increasing doses of the miR-195 mimic (Figure 3e). Next, we transfected OS cells with different concentrations of the miR-195 mimic (5 nM, 10 nM, and 20 nM) and KIF4A mRNA and protein expression levels were detected using qRT-PCR and western blotting, respectively (Figure 3f, 3g and Figure S4c, S4d). The results showed that there was a dose-dependent decline in the expression of KIF4A with increasing doses of the miR-195 mimic. To further verify KIF4A as a target of miR-195, we analyzed the miRNA binding sites using the BiBiServ2 software (Figure S3b). We mutated three miR-195 seed sequences, and then again performed the luciferase, qRT-PCR, and western blotting experiments (Figure 3h). Dual-luciferase reporter assay results displayed significant reduction of luciferase activity in the wild-type group (miR-195 mimic) and restored luciferase activity in the mutant group (miR-195 mut mimic) (Figure 3i). The qRT-PCR results revealed that KIF4A expression was significantly reduced in OS cells transfected with the wild-type miR-195 and that its expression was restored in the group transfected with the mutant miR-195 (Figure 3j and Figure S4e), consistent with the western blotting results (Figure 3k and Figure S4f). All the above observations unambiguously demonstrated that miR-195 inhibits the expression of KIF4A in OS cells by targeting the 3′UTR of KIF4A mRNA.
Figure 3.
miR-195 inhibits the expression of kinesin superfamily protein 4A (KIF4A) in osteosarcoma (OS) cells by targeting the KIF4A 3’ untranslated region (UTR).
(a) Analysis of the Gene Expression Omnibus (GEO) database-derived dataset GSE28425. Bar charts showed that the miR-195 expression level was significantly lower in OS tissue (n=4) than in normal tissue (n=17) (p<0.01). (b) The targeted binding effect between the KIF4A 3′ untranslated region (UTR) and miRNAs determined using a luciferase assay in HEK293 cells (Black columns represent the luciferase activity co-transfected with PGL-3 basic; Red columns represent the luciferase activity co-transfected with KIF4A 3′UTR, **p<0.01). (c) The inhibitory effect of miRNAs on KIF4A detected using qPCR in MG63 cells. miR-195 mimic inhibited the expression level of KIF4A in MG63 cells (**p<0.01). (d) The inhibitory effect of miRNAs on KIF4A detected using western blotting in MG63 cells. (e) Luciferase dose-dependent assay showing significant binding of miR-195 to the 3’UTR region of KIF4A in HEK293 cells (Black columns represent the luciferase activity co-transfected with PGL-3 basic; Red columns represent the luciferase activity co-transfected with KIF4A 3′UTR, **p<0.01). (f) qPCR dose-dependent assay showing that miR-195 inhibited the expression of KIF4A in MG63 cells (**p<0.01). (g) western blotting dose-dependent assay showing that miR-195 inhibited the expression of KIF4A in MG63 cells. (h) Potential binding sites between miR-195 and KIF4A were predicted with miRNA target prediction software including BiBiServ, MIRDB, and DIANA.. (i) The effect of miR-195 point mutations on KIF4A binding determined using a luciferase assay in HEK293 cells (**p<0.01). (j) The effect of miR-195 point mutations on KIF4A binding detected using qPCR in MG63 cells. (k) The effect of miR-195 point mutations on KIF4A binding detected using western blotting in MG63 cells.
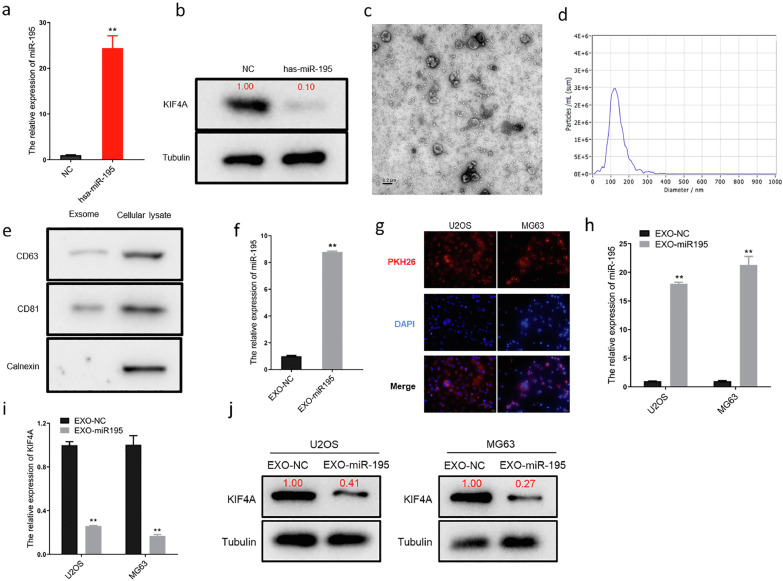
Normal human chondrocyte-derived exosomal miR-195 can be internalized by osteosarcoma cells
It has recently become clear that exosome-mediated miRNA transfer is an important strategy for cancer-targeting therapy. Therefore, we cultured normal human chondrocytes (CHON001) and transfected them with a miR-195 mimic. After 24 h of transfection, the cells were harvested for qRT-PCR and western blotting analyses. The results revealed that the level of miR-195 was markedly increased in the transfected cells (Figure 4a), whereas the expression of KIF4A was significantly reduced (Figure 4b). Normal human chondrocyte-derived exosomes were extracted using ultracentrifugation and identified using TEM (Figure 4c) and NTA (Figure 4d). Normal human chondrocyte-derived exosomes showed a distinct membrane structure with a diameter of approximately 100 nm. Moreover, the expression of exosome-specific markers CD63 and CD81, but not that of calnexin, was observed using western blotting (Figure 4e). The qRT-PCR results revealed that the expression of miR-195 was significantly higher in the exosome group (transfected with miR-195, EXO-miR-195) compared to that in the control group (transfected with NC, EXO-NC) (Figure 4f). Confocal laser microscopy images displayed the penetration of fluorescent PKH26-labeled exosomes into U2OS and MG63 cells after 24 h of co-culturing (Figure 4g). Furthermore, the level of miR-195 was markedly elevated in U2OS and MG63 cells as detected using qRT-PCR after the exosome intervention (Figure 4h). In addition, western blotting and qRT-PCR assays showed that the exosome intervention noticeably decreased the expression levels of both KIF4A mRNA (Figure 4i) and protein (Figure 4j) in U2OS and MG63 cells. Therefore, we established that normal human chondrocyte-derived exosomal miR-195 could be internalized by OS cells and downregulated the expression of KIF4A.
Figure 4.
Chondrocyte-derived exosomal miR-195 is internalized by OS cells.
(a) The expression of miR195 in chondrocytes after transfection detected using qPCR. The expression of miR-195 was significantly high in chondrocytes (**p<0.01). (b) The level of KIF4A in chondrocytes after transfection detected using western blotting. (c) The morphology of exosomes showed a bowl-shaped bilayer membrane structure using transmission electron microscopy.(d) Size distribution of exosomes measured using nanoparticle tracking analysis. (e) Exosome marker proteins CD63, CD81, and Calnexin detected using western blotting. (f) The expression of miR195 in exosomes detected using qPCR. (g) Confocal microscopy images showing that exosomal miR-195 is internalized by MG63cells (PKH26 marked exsomes in red and DAPI marked nucleus in blue, magnification 600 × ). (h) The level of miR-195 detected in U2OS and MG63 cells using qPCR. (i, j) The mRNA and protein levels of miR-195 detected in U2OS and MG63 cells using qPCR and western blotting. *p<0.05 and **p<0.01.
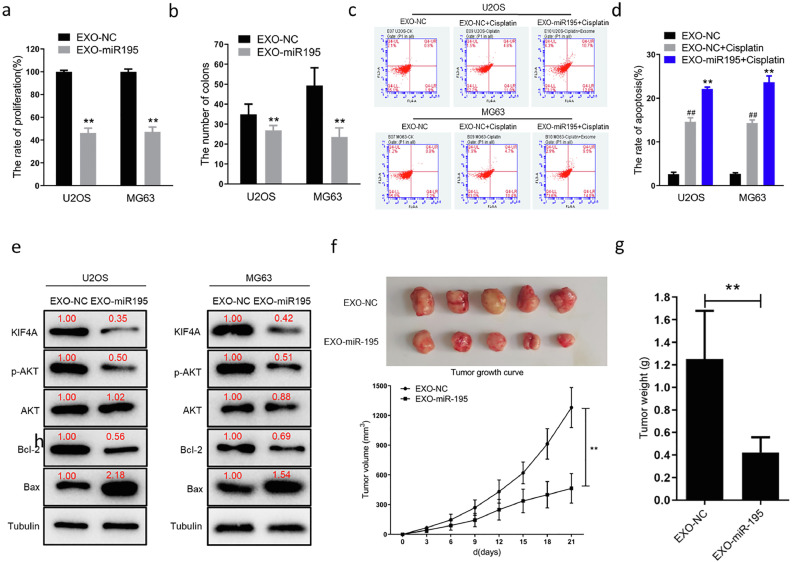
Normal human chondrocyte-derived exosomal miR-195 suppresses cell proliferation and anti-apoptotic in vitro and vivo
To further investigate the functional role of miR-195 delivery via normal human chondrocyte-derived exosomes in U2OS and MG63 cells, after 48 h of treatment with exosomes, OS cell hyperproliferation and colony formation were assessed. These phenotypes were suppressed in the EXO-miR-195 group compared to those in the EXO-NC group (Figure 5a, 5b). By analyzing the Gene Expression Omnibus (GEO) database-derived dataset GSE30934, OS patients with high miR-195 expression were found to be more drug-sensitive than those with low miR-195 expression (Figure S3c). To validate this result, we used a low dose of cisplatin (0.5 µM) in the EXO-NC and EXO-miR-195 groups. Flow cytometry results suggested that OS cell apoptosis was remarkably increased in the EXO-miR-195 group compared to that in the EXO-NC group (Figure 5c, 5d). Based on these data, we concluded that normal human chondrocyte-derived exosomal miR-195 suppressed cell proliferation and anti-apoptotic in OS cells. Furthermore, we detected the expression of some proliferation and apoptosis-related proteins in OS cell lines using western blotting. The results showed that KIF4A, Bcl2, and p-AKT expression was decreased in the EXO-miR-195 group compared to that in the EXO-NC group and that Bax expression was significantly higher in the EXO-miR-195 group than in the EXO-NC group (Figure 5e). An OS xenograft model was established to evaluate the role of normal human chondrocyte-derived exosomal miR-195 in tumor formation and progression. The results of the xenograft model showed that the EXO-miR-195 group had a significantly lower tumor growth rate than the EXO-NC group (Figure 5f), indicating that tumor growth was suppressed. In addition, tumor weight was markedly lower in the EXO-miR-195 group than in the EXO-NC group (Figure 5g). These in vivo results suggested that normal human chondrocyte-derived exosomal miR-195 inhibited OS cell growth in vivo. Subsequently, we detected the expression of some proliferation and apoptosis-related proteins in tumor samples using western blotting. The results showed that KIF4A, Bcl2, and p-AKT expression was decreased in the EXO-miR-195 group (n=5) compared to that in the EXO-NC group (n=5) and that Bax expression was significantly higher in the EXO-miR-195 group than in the EXO-NC group (Figure S3d). These findings indicate that human chondrocyte-derived exosomal miR-195 modulates OS cell proliferation and apoptosis via the regulation of the AKT and Bcl-2/Bax pathways.
Figure 5.
Chondrocyte-derived exosomal miR-195 inhibits osteosarcoma (OS) cell proliferation and promotes OS cell apoptosis in vitro and in vivo.
(a) Proliferation of exosome treated cells (EXO-miR195) were significantly lower than that of the control cells (EXO-NC) at 96 h (**p<0.01). (b) The number of clones of U2OS and MG63 cells with exosome treated (EXO-miR195) were significantly fewer than that of the control group (EXO-NC) in vitro (**p<0.01). (c, d) Cell apoptosis detected using flow cytometry. Significant apoptosis were detected in cisplatin (0.5 µM) -treated U2OS and MG63 cells (EXO- NC vs. EXO-NC+Cisplatin: ##p<0.01). The treatment of exosomal miR-195 enhanced cellular sensitivity to cisplatin (0.5 µM) -induced apoptosis in U2OS and MG63 cells (EXO-NC+Cisplatin vs EXO-miR195+Cisplatin: **p<0.01). (e) Treatment of exosomal miR-195 reduced KIF4A, phosphorylated AKT, BCL-2 expression and increased Bax expression in vitro. (f) Nude mice were inoculated subcutaneously with MG63 cells (5 × 106 cells/animal). The two groups were treated with intratumoral injection of EXO-miR195 or EXO-NC (20 μg per treatment, twice per week and continued for 2 weeks.). Tumor growth curve showing that chondrocyte-derived exosomal miR-195 inhibits tumor growth in vivo (**p<0.01). (g) Tumor weight measurements showing that the xenograft tumor weight in the group with chondrocyte-derived exosomal miR-195 treated (EXO-miR195) was lower than the control group (EXO-NC) in vivo (**p<0.01).
Discussion
OS is the most common primary malignant tumor that develops in the bone. Because of the limitations in diagnosis and treatment, approximately 30% of patients are still prone to lung metastases with poor prognosis and high mortality [24]. Currently, a combination of surgery and chemotherapy is the primary means of treating OS [25]. However, the prognostic treatment outcome has not improved greatly in recent years, and the emergence of resistance is an important contributing factor [26]. In this study, using bioinformatic analyses, we demonstrated that the levels of KIF4A were higher in OS tissues than in normal tissues and that its high expression was also inversely associated with patient OS, PFI, DFI, DSS, and drug resistance. Cell experiments showed that KIF4A promotes cell proliferation and inhibits apoptosis by regulating the apoptotic signaling pathways in vitro and in vivo. KIF4A is an important member of the kinesin superfamily. KIFs play important roles in various biological processes, such as mitosis, meiosis, membrane transport, mRNA and protein transport, signal transduction, and microtubule transport [3,27,28]. KIF2A and KIF18B have been reported to act as oncogenes that promote OS cell growth and metastasis both in vitro and in vivo [6,7]. These are particularly interesting findings, and at least these genes can act as poor prognostic markers. Furthermore, their conserved regions may be candidates for targeted gene therapies, such as the miRNA approach.
It is established that miRNAs, found in different cells and body fluids, such as blood, saliva and urine, are important genomic regulators of mRNA expression [29]. Using bioinformatic analysis, we identified and demonstrated that miR-195 regulates the expression of KIF4A in OS. Therefore, we believe that this is of particular importance in the diagnosis of OS. Moreover, it provides new strategies for OS therapeutic targets. Interestingly, miR-195 also functions as a tumor suppressor in multiple cancer cell lines [30], [31], [32]. It is well known that miRNAs can target multiple genes. In the present study, however, we focused mainly on the tumor-suppressor function of miR-195. There are limited reports on the negative impact of miR-195 on normal cell function. Therefore, in the future, we aim to investigate functional changes in chondrocytes after transfection with the miR-195 mimic. If there are no adverse effects or if adverse effects are controllable, miR-195 can be used for local drug administration after surgery to reduce OS recurrence. Furthermore, the mechanisms underlying the function of miR-195 function in OS cells need to be elucidated.
In recent years, scientists have demonstrated that exosomes play an important role in tumor diagnosis and treatment [33]. An increasing number of studies indicate that miRNAs packaged in exosomes establish intercellular communication, in which exosomes transport and transduce miRNAs into cancer cells, either in close proximity or distant from the cells of exosome origin [34]. Therefore, researchers have attempted to use exosomes as nanocarriers to accurately transfer miRNAs, siRNAs, and even chemotherapy drugs into target cells [35,36]. For example, exosomes derived from normal mesenchymal cells were designed to carry oncogenic KRAS shRNA into pancreatic cancer cells. Treatment with exosomes suppressed cancer in mouse models of pancreatic cancer and significantly prolonged survival [37]. Another promising area for exosomes is anti-cancer vaccine development, as exosomes can deliver or present tumor-derived antigens. Exosomes isolated from the ascites of patients with colon cancer were injected into patients as a vaccine and showed safe, well-tolerated performance, and cytotoxic T-cell activation [38]. In addition to their therapeutic use, exosomes have been employed as biomarkers because their abnormal expression is closely associated with cancer progression and metastasis [39]. Studies have shown that the abnormal expression of miR-17-5p and miR-92a-3p in exosomes can be used to predict the pathological staging and grading of colon cancer [40]. We used normal human chondrocytes to secrete exosomal miR-195 to treat OS cells. Our results showed that normal human chondrocytes could release sufficient amounts of exosomal miR-195 and that exosomal miR-195 could efficiently enter OS cells. Finally, miR-195 completely inhibited the expression of KIF4A in OS cells and suppressed cell proliferation and the anti-apoptotic effects of KIF4A by regulating the apoptotic signaling pathways in vitro and in vivo. In this study, we confirmed that exosomal miR-195 inhibited the level of phosphorylation AKT and Bcl-2. Some studies have indicated phosphorylated AKT can promote the expression of Bcl-2 by binding to its promoter region directly and Bcl-2 can suppresses apoptosis by inhibiting Ca2+ signaling [41]. The O subfamily of FOX transcription factors, including FOXO1, FOXO3, FOXO4 and FOXO6, are key downstream transcription factors of the Akt pathway. The activated Akt inactivates FoxO by inducing its translocation from the nucleus to the cytoplasm, thereby blocking the expression of effector genes (Bim, p21, p27) for anti-apoptotic effects [42]. Unphosphorylated BAD binds to and inhibits the anti-apoptotic protein Bcl-xL, BCL-2, thereby promoting the release of cytochrome C from mitochondria, activating apoptotic proteins and triggering apoptosis. After BAD at the ser99 is phosphorylated by Akt, BAD dissociates from mitochondria, thus inhibiting apoptosis [43]. The ATP-binding cassette (ABC) protein family, including P-glycoprotein (P-gp) and multidrug resistance-associated protein (MRP1), increase the drug efflux, thereby reducing its accumulation in tumor cells, and Nath et al. found that activation of Akt pathway induces drug resistance by up-regulating MRP1 expression in pancreatic cancer cells [44]. According to the studies above, we think that exosomal miR-195 may increase the drug sensitivity of OS cells by inhibiting the activity of AKT signaling pathway and it is necessary to conduct further exploration on the these signals in the future. In mouse studies, we selected intratumoral injection of exosomal miR-195 over tail vein injection [45]. We believe that this method can allow exosomal miR-195 to directly target OS cells while decreasing the adverse effects on other cells. Naturally, this may also contribute to other problems, such as frequent tumor punctures that increase the risk of tumor cell metastasis. Further studies considering these variables need to be undertaken. We will establish an orthotopic OS model by directly implanting OS cells or tumor tissue into the distal femur of mice, and the sample size needs to be expanded further. In this way, we will treat models with medication alone, medication+adjuvant exosomes, surgery, surgery+adjuvant exosomes and puncture exosomes respectively. A comparison of the result will help us to assess accurately the effect of exosomes on the proliferation, metastasis, drug sensitivity of OS cells in vivo.
Conclusions
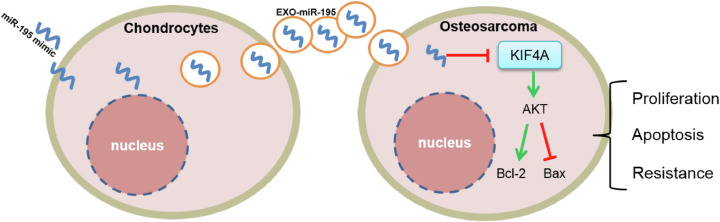
This study demonstrated that normal human chondrocyte-derived exosomal miR-195 was successfully transferred into OS cells and inhibited cell proliferation and reduced tolerance to chemotherapeutic drugs by targeting KIF4A (Figure 6), opening a new horizon for scientists to investigate OS treatment strategies. However, more studies are needed to elucidate the potential mechanism underlying OS development and identify effective treatment options.
Figure 6.
A schematic drawing of chondrocyte-derived exosomal miR-195 entering osteosarcoma (OS) cells.
miR-195 is wrapped in chondrocyte-derived exosomes and then delivered to OS cells. In OS cells, highly expressed miR-195 inhibits OS cell proliferation and promotes their apoptosis by inhibiting kinesin superfamily protein 4A (KIF4A)
Funding
Jiangxi Provincial Department of Education Fund
Author contributions
Conceptualization, P.H. X.L. and Y.L.; Methodology, G.C., Y.L., H.L., Y.H. and H.F.; Data analysis, G.C., Y.H., and H.F.; Writing – Original Draft Preparation, Y.L. and G.C.; Writing – Review & Editing, Y.L. and P.H.; All authors reviewed and approved the final manuscript.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Ethics Committee of Zhejiang XiShu Biological Co., Ltd (protocol code 2021070701).
Data Availability Statement
All data generated or analyzed during this study are included in this article and its supplementary information files.
Declaration of competing Interest
The authors declare no conflict of interest.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2021.101289.
Contributor Information
Xiaojian Liu, Email: liuxiaojian0123@sina.com.
Panpan Huang, Email: huangpp@gmu.edu.cn.
Appendix. Supplementary materials
References
- 1.Ji GR, Yu NC, Xue X, Li ZG. PERK-mediated Autophagy in Osteosarcoma Cells Resists ER Stress-induced Cell Apoptosis. International journal of biological sciences. 2015;11(7):803–812. doi: 10.7150/ijbs.11100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang J, Yang J, Wang HQ, Pan Z, Yan X, Hu C, Li Y, Lyu J. Development and validation of a nomogram for osteosarcoma-specific survival: A population-based study. Medicine. 2019;98(23):e15988. doi: 10.1097/MD.0000000000015988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miki H, Okada Y, Hirokawa N. Analysis of the kinesin superfamily: insights into structure and function. Trends in cell biology. 2005;15(9):467–476. doi: 10.1016/j.tcb.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 4.Yu Y, Feng YM. The role of kinesin family proteins in tumorigenesis and progression: potential biomarkers and molecular targets for cancer therapy. Cancer. 2010;116(22):5150–5160. doi: 10.1002/cncr.25461. [DOI] [PubMed] [Google Scholar]
- 5.Lu Y, Song T, Xue X, Cao G, Huang P. Kinesin superfamily proteins: roles in osteosarcoma. Frontiers in bioscience. 2021;26(8):370–378. doi: 10.52586/4950. [DOI] [PubMed] [Google Scholar]
- 6.Wang ZX, Ren SC, Chang ZS, Ren J. Identification of Kinesin Family Member 2A (KIF2A) as a Promising Therapeutic Target for Osteosarcoma. BioMed research international. 2020;2020 doi: 10.1155/2020/7102757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gao T, Yu L, Fang Z, Liu J, Bai C, Li S, Xue R, Zhang L, Tan Z, Fan Z. KIF18B promotes tumor progression in osteosarcoma by activating beta-catenin. Cancer biology & medicine. 2020;17(2):371–386. doi: 10.20892/j.issn.2095-3941.2019.0452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cao Q, Song Z, Ruan H, Wang C, Yang X, Bao L, Wang K, Cheng G, Xu T, Xiao W, et al. Targeting the KIF4A/AR Axis to Reverse Endocrine Therapy Resistance in Castration-resistant Prostate Cancer. Clinical cancer research: an official journal of the American Association for Cancer Research. 2020;26(6):1516–1528. doi: 10.1158/1078-0432.CCR-19-0396. [DOI] [PubMed] [Google Scholar]
- 9.Hu G, Yan Z, Zhang C, Cheng M, Yan Y, Wang Y, Deng L, Lu Q, Luo S. FOXM1 promotes hepatocellular carcinoma progression by regulating KIF4A expression. Journal of experimental & clinical cancer research: CR. 2019;38(1):188. doi: 10.1186/s13046-019-1202-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xue D, Cheng P, Han M, Liu X, Xue L, Ye C, Wang K, Huang J. An integrated bioinformatical analysis to evaluate the role of KIF4A as a prognostic biomarker for breast cancer. OncoTargets and therapy. 2018;11:4755–4768. doi: 10.2147/OTT.S164730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang Y, Wang H, Lian Y, Wu X, Zhou L, Wang J, Deng M, Huang Y. Upregulation of kinesin family member 4A enhanced cell proliferation via activation of Akt signaling and predicted a poor prognosis in hepatocellular carcinoma. Cell death & disease. 2018;9(2):141. doi: 10.1038/s41419-017-0114-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thery C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nature reviews Immunology. 2009;9(8):581–593. doi: 10.1038/nri2567. [DOI] [PubMed] [Google Scholar]
- 13.Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nature reviews Drug discovery. 2017;16(3):203–222. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- 14.Fan Q, Yang L, Zhang X, Peng X, Wei S, Su D, Zhai Z, Hua X, Li H. The emerging role of exosome-derived non-coding RNAs in cancer biology. Cancer letters. 2018;414:107–115. doi: 10.1016/j.canlet.2017.10.040. [DOI] [PubMed] [Google Scholar]
- 15.Raimondi L, De Luca A, Gallo A, Costa V, Russelli G, Cuscino N, Manno M, Raccosta S, Carina V, Bellavia D, et al. Osteosarcoma cell-derived exosomes affect tumor microenvironment by specific packaging of microRNAs. Carcinogenesis. 2020;41(5):666–677. doi: 10.1093/carcin/bgz130. [DOI] [PubMed] [Google Scholar]
- 16.Wang JW, Wu XF, Gu XJ, Jiang XH. Exosomal miR-1228 From Cancer-Associated Fibroblasts Promotes Cell Migration and Invasion of Osteosarcoma by Directly Targeting SCAI. Oncology research. 2019;27(9):979–986. doi: 10.3727/096504018X15336368805108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang H, Wang J, Ren T, Huang Y, Liang X, Yu Y, Wang W, Niu J, Guo W. Bone marrow mesenchymal stem cell-derived exosomal miR-206 inhibits osteosarcoma progression by targeting TRA2B. Cancer letters. 2020;490:54–65. doi: 10.1016/j.canlet.2020.07.008. [DOI] [PubMed] [Google Scholar]
- 18.Song H, Park KH. Regulation and function of SOX9 during cartilage development and regeneration. Seminars in cancer biology. 2020;67(1):12–23. doi: 10.1016/j.semcancer.2020.04.008. Pt. [DOI] [PubMed] [Google Scholar]
- 19.Zheng L, Wang Y, Qiu P, Xia C, Fang Y, Mei S, Fang C, Shi Y, Wu K, Chen Z, et al. Primary chondrocyte exosomes mediate osteoarthritis progression by regulating mitochondrion and immune reactivity. Nanomedicine. 2019;14(24):3193–3212. doi: 10.2217/nnm-2018-0498. [DOI] [PubMed] [Google Scholar]
- 20.Shvedova M, Kobayashi T. MicroRNAs in cartilage development and dysplasia. Bone. 2020;140 doi: 10.1016/j.bone.2020.115564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liang Y, Xu X, Li X, Xiong J, Li B, Duan L, Wang D, Xia J. Chondrocyte-Targeted MicroRNA Delivery by Engineered Exosomes toward a Cell-Free Osteoarthritis Therapy. ACS applied materials & interfaces. 2020;12(33):36938–36947. doi: 10.1021/acsami.0c10458. [DOI] [PubMed] [Google Scholar]
- 22.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 23.Huang P, Liao R, Chen X, Wu X, Li X, Wang Y, Cao Q, Dong C. Nuclear translocation of PLSCR1 activates STAT1 signaling in basal-like breast cancer. Theranostics. 2020;10(10):4644–4658. doi: 10.7150/thno.43150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chou AJ, Geller DS, Gorlick R. Therapy for osteosarcoma: where do we go from here? Paediatric drugs. 2008;10(5):315–327. doi: 10.2165/00148581-200810050-00005. [DOI] [PubMed] [Google Scholar]
- 25.Wang L, Huang X, You X, Yi T, Lu B, Liu J, Lu G, Ma M, Zou C, Wu J, et al. Nanoparticle enhanced combination therapy for stem-like progenitors defined by single-cell transcriptomics in chemotherapy-resistant osteosarcoma. Signal transduction and targeted therapy. 2020;5(1):196. doi: 10.1038/s41392-020-00248-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheng C, Ding Q, Zhang Z, Wang S, Zhong B, Huang X, Shao Z. PTBP1 modulates osteosarcoma chemoresistance to cisplatin by regulating the expression of the copper transporter SLC31A1. Journal of cellular and molecular medicine. 2020;24(9):5274–5289. doi: 10.1111/jcmm.15183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goldstein LS, Philp AV. The road less traveled: emerging principles of kinesin motor utilization. Annual review of cell and developmental biology. 1999;15:141–183. doi: 10.1146/annurev.cellbio.15.1.141. [DOI] [PubMed] [Google Scholar]
- 28.Hirokawa N, Noda Y, Tanaka Y, Niwa S. Kinesin superfamily motor proteins and intracellular transport. Nature reviews Molecular cell biology. 2009;10(10):682–696. doi: 10.1038/nrm2774. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Y, Huang B, Wang HY, Chang A, Zheng XFS. Emerging Role of MicroRNAs in mTOR Signaling. Cellular and molecular life sciences: CMLS. 2017;74(14):2613–2625. doi: 10.1007/s00018-017-2485-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pidikova P, Reis R, Herichova I. miRNA Clusters with Down-Regulated Expression in Human Colorectal Cancer and Their Regulation. International journal of molecular sciences. 2020;21(13) doi: 10.3390/ijms21134633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang JY, Li YN, Mu X, Pan ZL, Liu WB. Targeted regulation of miR-195 on MAP2K1 for suppressing ADM drug resistance in prostate cancer cells. European review for medical and pharmacological sciences. 2020;24(15):7911. doi: 10.26355/eurrev_202008_22445. [DOI] [PubMed] [Google Scholar]
- 32.Rao G, Dwivedi SKD, Zhang Y, Dey A, Shameer K, Karthik R, Srikantan S, Hossen MN, Wren JD, Madesh M, et al. MicroRNA-195 controls MICU1 expression and tumor growth in ovarian cancer. EMBO reports. 2020;21(10):e48483. doi: 10.15252/embr.201948483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnsen KB, Gudbergsson JM, Skov MN, Pilgaard L, Moos T, Duroux M. A comprehensive overview of exosomes as drug delivery vehicles - endogenous nanocarriers for targeted cancer therapy. Biochimica et biophysica acta. 2014;1846(1):75–87. doi: 10.1016/j.bbcan.2014.04.005. [DOI] [PubMed] [Google Scholar]
- 34.Tkach M, Thery C. Communication by Extracellular Vesicles: Where We Are and Where We Need to Go. Cell. 2016;164(6):1226–1232. doi: 10.1016/j.cell.2016.01.043. [DOI] [PubMed] [Google Scholar]
- 35.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nature cell biology. 2007;9(6):654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 36.Ren J, He W, Zheng L, Duan H. From structures to functions: insights into exosomes as promising drug delivery vehicles. Biomaterials science. 2016;4(6):910–921. doi: 10.1039/c5bm00583c. [DOI] [PubMed] [Google Scholar]
- 37.Kamerkar S, LeBleu VS, Sugimoto H, Yang S, Ruivo CF, Melo SA, Lee JJ, Kalluri R. Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature. 2017;546(7659):498–503. doi: 10.1038/nature22341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dai S, Wei D, Wu Z, Zhou X, Wei X, Huang H, Li G. Phase I clinical trial of autologous ascites-derived exosomes combined with GM-CSF for colorectal cancer. Molecular therapy: the journal of the American Society of Gene Therapy. 2008;16(4):782–790. doi: 10.1038/mt.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thakur BK, Zhang H, Becker A, Matei I, Huang Y, Costa-Silva B, Zheng Y, Hoshino A, Brazier H, Xiang J, et al. Double-stranded DNA in exosomes: a novel biomarker in cancer detection. Cell research. 2014;24(6):766–769. doi: 10.1038/cr.2014.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fu F, Jiang W, Zhou L, Chen Z. Circulating Exosomal miR-17-5p and miR-92a-3p Predict Pathologic Stage and Grade of Colorectal Cancer. Translational oncology. 2018;11(2):221–232. doi: 10.1016/j.tranon.2017.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chou CH, Lai SL, Chen CN, Lee PH, Peng FC, Kuo ML, Lai HS. IL-6 regulates Mcl-1L expression through the JAK/PI3K/Akt/CREB signaling pathway in hepatocytes: implication of an anti-apoptotic role during liver regeneration. PloS one. 2013;8(6):e66268. doi: 10.1371/journal.pone.0066268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boreddy SR, Pramanik KC, Srivastava SK. Pancreatic tumor suppression by benzyl isothiocyanate is associated with inhibition of PI3K/AKT/FOXO pathway. Clinical cancer research: an official journal of the American Association for Cancer Research. 2011;17(7):1784–1795. doi: 10.1158/1078-0432.CCR-10-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sakamaki J, Daitoku H, Ueno K, Hagiwara A, Yamagata K, Fukamizu A. Arginine methylation of BCL-2 antagonist of cell death (BAD) counteracts its phosphorylation and inactivation by Akt. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(15):6085–6090. doi: 10.1073/pnas.1015328108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nath S, Daneshvar K, Roy LD, Grover P, Kidiyoor A, Mosley L, Sahraei M, Mukherjee P. MUC1 induces drug resistance in pancreatic cancer cells via upregulation of multidrug resistance genes. Oncogenesis. 2013;2:e51. doi: 10.1038/oncsis.2013.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Naseri Z, Oskuee RK, Jaafari MR. Forouzandeh Moghadam M: Exosome-mediated delivery of functionally active miRNA-142-3p inhibitor reduces tumorigenicity of breast cancer in vitro and in vivo. International journal of nanomedicine. 2018;13:7727–7747. doi: 10.2147/IJN.S182384. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this article and its supplementary information files.