Abstract
Proposing efficient prophylactic and therapeutic strategies for coronavirus 2019 (COVID-19) requires precise knowledge of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pathogenesis. An array of platforms, including organoids and microfluidic devices, have provided a basis for studies of SARS-CoV-2. Here, we summarize available models as well as novel drug screening approaches, from simple to more advanced platforms. Notably, organoids and microfluidic devices offer promising perspectives for the clinical translation of basic science, such as screening therapeutics candidates. Overall, modifying these advanced micro and macro 3D platforms for disease modeling and combining them with recent advances in drug screening has significant potential for the discovery of novel potent drugs against COVID-19.
Keywords: SARS-CoV-2, Drug screening, Organoid technology, Microfluidic technology, COVID-19 modeling
COVID-19 pathophysiology
The global pandemic caused by the novel SARS-CoV-2 and its associated morbidity and mortality are prompting researchers to develop reliable COVID-19 models. Indeed, more comprehensive knowledge of the pathogenesis of SARS-CoV-2, and disease model platforms for drug screening, should enable scientists to identify preventive and therapeutic strategies.1
The SARS-CoV-2 genome comprises a 5′ terminal open reading frame (ORF1/ab) that encodes the replicase polyprotein 1a (pp1a) and pp1ab. These polyproteins are broken by papain-like cysteine protease (PLpro) and 3C-like serine protease (3CLpro) to generate nonstructural proteins, including RNA-dependent RNA polymerase (RdRp) and helicase (Hel), the key enzymes engaged in transcription and replication of CoVs. The structural proteins of the virus, including spike (S), envelope (E), membrane (M) and nucleocapsid (N), are encoded from the 3′ one-third of the CoV genome. The S-glycoprotein mediates entry of the virus by binding to the angiotensin-converting enzyme 2 (ACE2) receptor, or other alternative receptors on host cells.2, 3 Upon virus entry, its genomic RNA is replicated and new viral particles are released. The damage-associated molecular patterns (DAMPs) produced by infected cells initiate production of proinflammatory cytokines, such as interleukin (IL)-6, which leads to the recruitment of immune cells, including macrophages and T cells, to the site of infection. Overproduction of these proinflammatory cytokines, a phenomenon called cytokine release syndrome (CRS), causes severe lung damage, acute respiratory distress syndrome (ARDS), and, ultimately, multiorgan damage.4
Given the heavy burden that COVID-19 pandemic has imposed on all aspects of life worldwide, there is a significant need to determine the crucial mechanisms involved in the pathogenesis of the virus and to identify efficient drugs. To this end, introducing reliable models to mimic pathological events would be helpful for understanding disease pathogenesis as well as for drug screening. Of note, the principal concern in drug development for new emerging diseases is unknown side effects and possible toxicity, which make the in vitro and in vivo evaluation of new candidates vital.5
Genetic background and gene editing in the clinical course of COVID-19
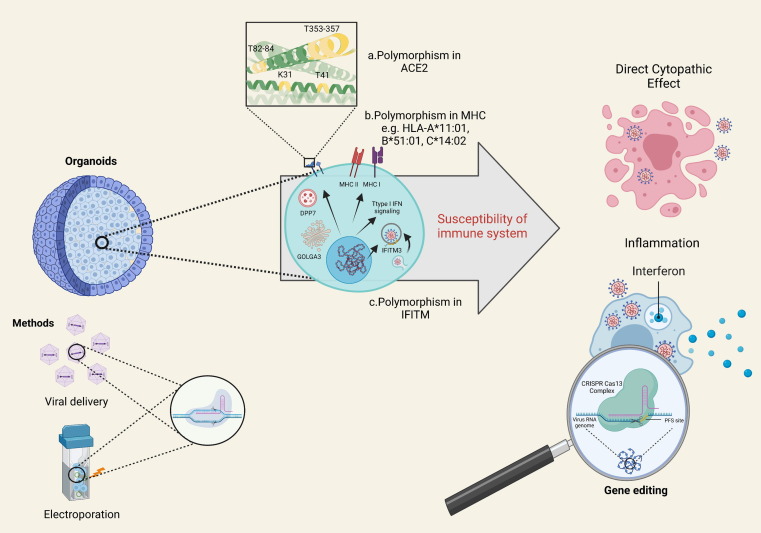
Beside risk factors, such as age and pre-existing conditions, genetic background can also affect COVID-19 susceptibility. This is more evident in familial cases of COVID-19.6 Genetic variations in three main classes of gene, encoding receptors, HLA, and ‘other’ immune-related genes, are of important in this regard (Fig. 1 ).7 For instance, determining the allele frequencies in different areas of the ACE gene that encode the S-protein binding domain, such as residues near lysine 31, and tyrosine 41, 82–84, and 353–357 in ACE2, especially in different geographic populations, would be informative.8 Although not all polymorphisms in ACE2 were shown to be correlated significantly with SARS-CoV-1 susceptibility, some might have a role in COVID-19 susceptibility (Table S1 in the supplemental information online).7, 8 Moreover, based on computational models, it appears that the intermolecular interactions of two alleles of this gene, rs73635825 and rs143936283, with the S protein exhibit significant differences.9 Research demonstrated the association between ACE2 expression and specific conditions, such as essential hypertension, diabetic heart disease, and dyslipidemia.10 Although further investigation is required, it is possible that the level of ACE2 expression might be higher in Asian populations compared with Caucasian and Black populations.11 This highlights the importance of investigating SNPs that affect the gene expression profile among different populations. Recently, the variant rs12329760 in the TMPRSS2 protein was also shown to have a lesser frequency among patients critically ill with COVID-19.12 This variant presents differential allelic frequency in different populations and might be related with decreased stability of the TMPRSS2 protein and ACE2 binding (Table S1 in the supplemental information online).12, 13 In addition, there is evidence that certain HLA haplotypes present in different ethnicity groups are associated with different risks for SARS-CoV2 infections (Fig. 1).12, 14, 15 Genetic variations in other genes involved in innate and adaptive immunity can also alter COVID-19 susceptibility.7 For example, the single nucleotide polymorphism (SNP) rs12252 in interferon (IFN)-induced transmembrane 3 (IFITM3) was demonstrated to have a significant association with the risk of hospitalization because of COVID-19,16 which might also hold true for genetic variants of genes encoding the main members of the type I IFN signaling pathway, as well as GOLGA3 and DPP7.12, 17 Although information regarding such genetic variations is invaluable for modeling COVID-19 and, consequently, drug screening, diverse environmental factors and other interacting genetic elements should also be considered, which will also affect the application of pharmacogenomics in different populations.8 Pharmacogenomics is the study of genetic variations in terms of individual drug response to reduce adverse effects and to enhance the therapeutic efficacy of a drug.18 It combines pharmacokinetics (PK) and pharmacodynamics (PD) of candidate drugs to determine drug dosing and adverse drug reactions (ADRs). For instance, individuals who are carriers of the HLA-B*5701 allele demonstrated increased sensitivity to the anti-HIV drug, abacavir.19 The significance of pharmacogenomics in a COVID-19 context has also been underscored. In this regard, the different frequencies of SNP alleles associated with toxicity or therapeutic responses to the most promising and repurposed drugs in patients with COVID-19 were recently described.20
Figure 1.
Organoids as potential platforms for investigating the role of genetic background in coronavirus 2019 (COVID-19) pathogenesis. Genetic polymorphisms in three classes of gene, including receptors (a), HLAs (b) and other immune-related genes (c), might affect the susceptibility of individuals exposed to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). For instance, a. human leukocyte antigen (HLA)-A*11:01 causes more severe symptoms in patients with COVID-19. Manipulating the organoid genome through cluster regularly interspaced palindromic repeat (CRISPR) technology could help to reveal the direct cytopathic effects of virus together with other involved mechanisms. Abbreviations: ACE, angiotensin-converting enzyme; IFITM, interferon-induced transmembrane proteins; MHC, major histocompatibility complex.
Common in vitro and in vivo models for viral disease modeling
The unprecedented challenges posed by COVID-19 necessitate fast identification of appropriate models to recapitulate different aspects of this disease.
In vitro modeling
Classically, in vitro cell (line) monolayer models are used to unravel the pathogenesis mechanism of virulent agents, including coronaviruses, because they are relatively simple. Using human airway epithelial cells (HAECs), kinetics of viral infection, alterations in tissue levels of cellular ultrastructures, as well as transcriptional immune profiles induced by SARS-CoV2 have been analyzed. This model was also used to assess the inhibitory effects of remdesivir and remdesivir combined with diltiazem on SARS-CoV-2 infection.21 Another example is the replication assessment of a synthesized recombinant chimeric BtCoV-HKU5 that encodes the S glycoprotein of SARS-CoV in Vero E6 cells.22 A high-throughput screen of ∼12 000 known compounds revealed that MDL-28170, and ONO 5334 and apilimod, members of the cysteine protease inhibitor and PIKfyve kinase inhibitor classes, respectively, show antiviral activity in Vero E6, Huh-7, HEK293T, and human induced pluripotent stem cell (hiPSC)-derived pneumocyte-like cells.23 In accordance with the importance of interactions between the DNA-damage response (DDR) pathways and RNA viruses, Garcia et al. recently identified berzosertib, an ATR kinase inhibitor, which blocks SARS-CoV-2 at post-entry level in the Vero E6 cell line.24
In vivo modeling
Given the significance of in vivo evaluations for developing prophylactic and therapeutic strategies, several animal models have been used for SARS-CoV investigations, including genetically modified mice, hamsters, ferrets and various nonhuman primates.
Dipeptidyl peptidase 4 (DPP4) is an essential protein for Middle East respiratory syndrome coronavirus (MERS-CoV) entry and cellular infection. Given interspecies differences in the amino acid sequence of this protein,25 providing selective animal models for evaluating MERS infection appears reasonable. Given that their DPP4 is ∼96% similar to its human ortholog, marmosets have been used to model the pathogenesis of, and therapeutic options for, MERS-CoV infections (Fig. 2 a).26 This demonstrated the beneficial effects of lopinavir/ritonavir treatment on clinical disease manifestations in the treated groups.27 Likewise, clinical benefits of remdesivir (GS-5734) were observed in a Rhesus macaque model of COVID-19.28
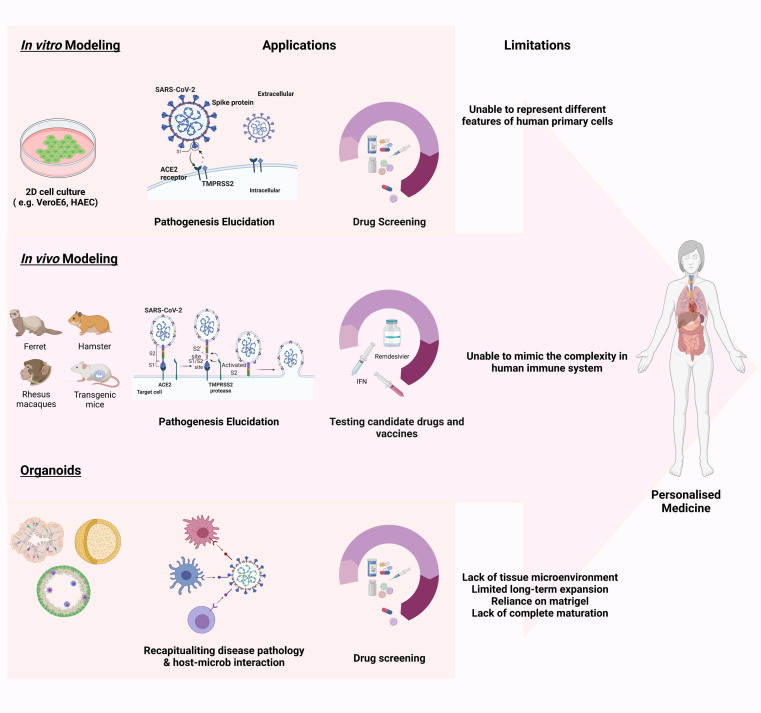
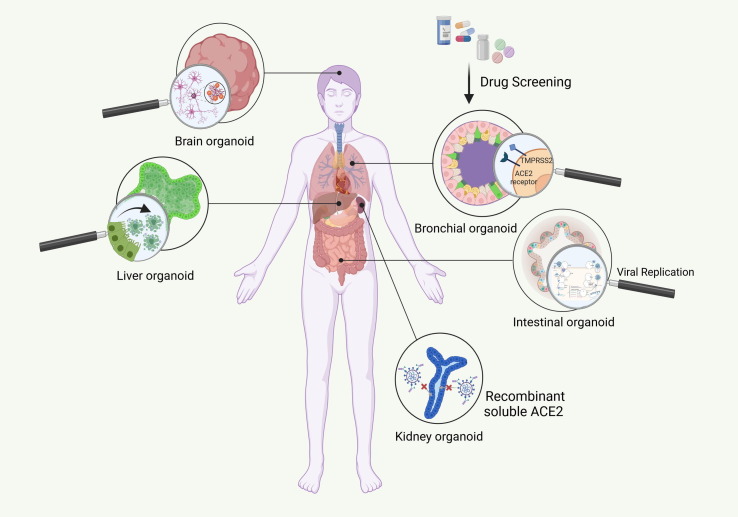
Figure 2.
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) disease modeling. (a) Platforms for disease modeling. In vitro (2D adherent cultures and 3D organoids), and in vivo modeling have thus far been widely used to assess pathogenesis and test candidate drugs. In summary, Vero E6 cells and human airway epithelial cells (HAECs) are two most common cell lines used in SARS-CoV-2 studies. Genetically modified mice, hamsters, ferrets, and various nonhuman primates are among the animals used for assessing the pathogenesis and testing candidate drugs. For instance, mRNA vaccine candidates for COVID-19 have been evaluated in mice. In addition, clinical benefits of remdesivir have been shown in the Rhesus macaque model of coronavirus 2019 (COVID-19). Considering the limitations of these models, including failure to represent the multiple aspects of the infection and the inability to mimic the complex nature of human immunity, organoids are attracting attention because of their ability to show host–microbe interactions and their potential for drug screening. Organoids also have limitations such as in mimicking native tissue microenvironment, long-term culture and expansion, reliance on Matrigel, and lack of full maturity. These need to be addressed in future. (b) Organoids for mechanism elucidation in COVID-19. Given the notable manifestation ov COVID-19 in lungs, kidneys, gastrointestinal system, liver, and brain, among others, scientists have focused on creating the corresponding organoids. Thus far, human bronchial, kidney, intestinal, liver ductal, and brain organoids contain various types of cell, have been developed to mimic virus entry and recapitulate the viral life cycle, along with the possible mechanism involved in pathogenesis, including the direct cytopathic effects. Abbreviation: ACE, angiotensin-converting enzyme.
Although mice have various advantages for disease modeling, including their small size, rapid breeding, and the potential to be easily genetically manipulated, there are barriers regarding their use for COVID-19 modeling. Mice are poorly susceptible to SARS-CoV because of amino acid differences in ACE2 between mice and humans.29 By taking advantage of reverse genetics, Dinnon et al. remodeled both the S protein of SARS-CoV-2 and the murine ACE2 protein. This then enabled mice to be infected with the recombinant virus. This mouse-adapted SARS-CoV-2 model reflects the age-related pathogenesis in terms of severity and has been used to test the prophylactic and therapeutic efficiency of IFN λ1a.30 In addition, BtCoV-HKU5 recombinant virus, used to infect Vero E6 cells, also demonstrated enhanced virulence and airway epithelial cell damage in mice (Fig. 2a).22 Humanized mice are another promising animal model to recapitulate complex biological responses and improving our understanding of various dimensions of pathological disorders.31 The Ad5-hACE2 humanized mouse can be used to investigate COVID-19 pathogenesis and therapeutic modalities as well as vaccine candidates.32 In addition, the Syrian hamster can also mirror various dimensions of COVID-19 infection in humans.33 Using in silico analysis, Lee et al. found that hamster ACE2 can interact with the receptor-binding domain of the surface S protein of SARS-CoV-2. Interestingly, clinical and histopathological findings in this animal model highly mimic symptoms in patients with COVID-19. Finally, vaccine development, as an urgent need to control the speed of spread of the SARS-CoV-2 pandemic, has benefited enormously from animal models, as recently summarized.34
Advantages and limitations of current models
Animal models and human cell lines have thus far contributed extensively to the study of respiratory viral infections and the genetic diversity that affects the clinical manifestations of an infection. However, both strategies have limitations, such as the translation of murine model results to human diseases is not always possible given that human immunity is more redundant compared with that of inbred mouse lines. By contrast, human cell lines might also not fully recapitulate all characteristics of primary human cells.35 Therefore, newer models are vital for development of anti-COVID-19 drugs.
An effective platform should not only recapitulate the pathogenesis, but also unravel various aspects of the toxicology, PK, PD, and immunology of prophylactic and therapeutic approaches. However, in terms of COVID-19, one major challenge is that a single platform might not meet all the facets of the infection in humans because of complicated crosstalk between the host and virus. Therefore, using a wide range of platforms is required. In addition, the selection of the final platform depends on the initial question and, as a result, different classes of drug and vaccine must be tested in specific models. The model used in preclinical studies should also precisely reflect the PK of the drugs in humans.29
Here, we discuss several approaches that could be used to improve pathogenesis studies and drug development, including genome-based and computational platforms suitable for drug discovery and the use of more complex human cell culture models, also termed organoid/spheroid models. Finally, we address the possibility of using the latter in an ‘organ on a chip’ (OoC) or even ‘human on chip’ (HoC) culture system to fully understand COVID-19 pathogenesis in a human context as well as to develop effective drugs.
Genome-based and computational platforms for drug screening
The lack of reliable drug-screening platforms highlights the crucial need to evaluate possible tools in the context of COVID-19. Here, we describe two main drug-screening approaches that have already been used for COVID-19 drug discovery. The use of cellular modelling platforms and especially advanced 3D models, (e.g., organoids and microfluidic devices), in combination with the following strategies, could pave the way to uncover more efficient drugs.
Genome-based strategies for drug screening
The virus genome comprises two ORFs, ORF 1a and ORF1b, encoding nonstructural proteins and replication-related proteins, respectively. After translation of ORF1a, ribosomes bypass the stop codon to act on ORF1b through programmed ribosomal frameshifting (PRF). Taking advantage of PRF, several studies have evaluated the ability of a variety of drugs to reduce PRF efficiency and, hence, inhibit viral infection. One example is anti-frameshift compounds with the potential to be used as antiviral agents against SARS-CoV.36 An in vitro drug-screening tool has been developed based on reporter system activity, which works under the control of the PRF element in SARS-CoV-2. Using this novel system, high-throughput screening of a series of US Food and Drug Administration (FDA)-approved antiviral drugs against COVID-19 was carried out. These studies demonstrated that ivacaftor and huperzine A could change effectively the 1-ribosomal frame shift of SARS-CoV-2 in vitro.37
Xie et al. designed a screening platform in which a nanoluciferase (Nluc) cassette was inserted in the SARS-CoV-2 to create a high-throughput platform for both diagnostic and drug-screening applications. Bioluminescence was then used to determine inhibitory effects of candidate drugs.38 A similar platform, termed amplified luminescent proximity homogenous assay (AlphaLISA) technology, also offers diagnostic and drug-screening applications for SARS-CoV-2. In this system, excitation of the donor bead at a certain wavelength produces a radical oxygen species that, in turn, activates the acceptor bead to produce light. Gorshkov et al. developed a N-based AlphaLISA assay for detecting COVID-19 infection and screening for viral inhibitors. This platform is based on the vicinity of two labeled antibodies taht bind to NP in the samples to generate luminescent signals. The authors suggest that such a system has the potential to identify inhibitors of N in SARS-CoV-2 (Fig. 3 ).39
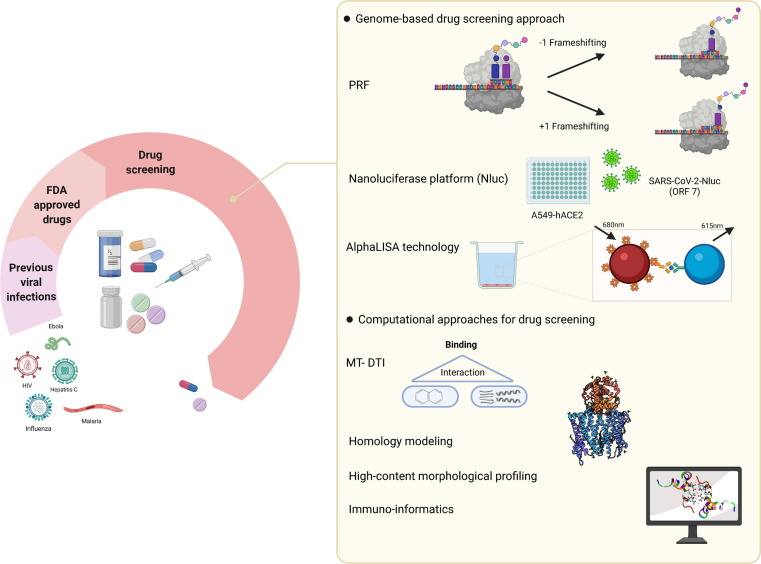
Figure 3.
Drug-screening platforms. Genome-based strategies and computational approach are two main measures for drug-screening against coronavirus 2019 (COVID-19). Using programmed ribosomal frameshifting (PRF), a strategy for the translation of nonstructural viral proteins, the efficacy of many US Food and Drug Administration (FDA)-approved drugs have been evaluated in the context of COVID-19. Another genome-based strategy, Nluc, is taking advantage of nanoluciferase gene insertion in viral genome and provides both diagnostic and therapeutic applications for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). In AlphaLISA technology, excitation of the donor bead at a certain wavelength leads to generation of radical oxygen species, which activates the acceptor bead to produce light. Computational approaches comprise MT-DTI, homology modeling, high-content morphological profiling, and immunoinformatics. The first three strategies have been used so far to evaluate a range of FDA-approved drugs against SARS-CoV-2, whereas immunoinformatics is applied for the detection of the most reliable antigens able to confer the ability to produce strong immune responses and can be used in vaccine development. Abbreviation: AlphaLISA, amplified luminescent proximity homogenous assay.
Computational platforms for drug screening
Artificial intelligence (AI) has recently gained attention for accelerating drug repurposing against a variety of challenging diseases, including COVID-19. AI can be used to mimic a complex human biological system in terms of drug–target interactions in an integrating platform in which different aspects of possible effects can be evaluated.40 Using AI technology, the inhibitory ability of a range of antiviral drugs has been tested against COVID-19.41, 42 Molecule Transformer-Drug Target Interaction (MT-DTI) is a deep learning drug target interaction-based model through which inhibitory effects of a range of commercially available drugs were assessed. This in silico approach identified the antiretroviral compound, atazanavir, as a powerful candidate agent against SARS-CoV-2.43 In addition, taking advantage of homology modeling, Wu et al. conducted docking analysis of the ZINC drug database and a Chinese natural products database for 21 known targets of SARs-CoV-2. As a result of such computational modeling, several new compounds with possible antiviral effects were proposed.44 Recently, a quantitative high-throughput platform was developed to detect antiviral activity of FDA-approved compounds. Quantitative high-content morphological profiling, an image-based technique that enables identification of the underlying pathways during infection, was added to an AI-based machine learning strategy to determine the type of viral mechanism of action that would be affected, including viral entry, propagation, and modulation of host cellular responses. Using this technology, lactoferrin has been suggested to be a powerful inhibitor of the infection, which might enhance the efficacy of, for example, remdesivir (Fig. 3).45
Finally, immuno-informatics offers opportunities to screen antigens to confer strong immune responses against viral infections with the aim of efficient vaccine development. Several studies have focused on the identification of vaccine candidates for COVID-19 through this approach, which was recently summarized elsewhere.46
The use of the above-described computational platforms for drug screening allows fast screens of candidate drugs for their efficacy against COVID-19. However, additional downstream studies are required to demonstrate the effectiveness and/or lack of toxicity of these drugs in human models. As discussed earlier, although 2D cell line models are easy to use, they do not recreate several aspects and the complexity of human disease. Therefore, we go on to discuss human organoid models that have been developed during the past 5–10 years, that better mimic different aspects of human tissues, and are/will be an important asset in COVID-19 disease modeling and drug development strategies.
Organoids
Organoids can be generated from pluripotent stem cells (PSCs) [i.e., embryonic stem cells (ESCs) or induced pluripotent stem cells (iPSCs)] embryonic progenitors, as well as organ-specific adult stem cells (ASCs).47 Spontaneous cell sorting and spatially restricted lineage commitment, mimicking processes occurring during in vivo tissue development, are the main underlying mechanisms through which these structures self-organize in vitro. Generally, organoids are defined by the fact that they comprise multiple tissue-specific cell types and display similar organization of cells as well as key tissue-specific functions compared with their in vivo correspondents (Fig. 2a,b).48
In addition, organoid platforms can be further engineered to incorporate an appropriate microenvironment that biomimics the in vivo environment of the related tissue. This can be achieved by, for instance, the incorporation of scaffolds [such as using microcontact printing to recapitulate the natural extracellular matrix (ECM)] with high specificity and flexibility. The spatiotemporal control of signals to cells should be taken into account in such bioengineering processes.49
PSC-derived organoids
When factors that maintain the pluripotency of PSCs are removed from cultures, spontaneous differentiation of PSCs in 3D embryoid bodies (EBs) containing cells from the three germ layers ensues. When such EBs are exposed to specific growth factors, cytokines, and media, directed differentiation of the cells in an EB to cells of a given germ layer, or tissue can be achieved, to form so-called ‘organoids’.47
ASC-derived organoids
Organoids can also be generated from adult tissue (stem) cells, including endoderm-derived organoids (e.g., intestinal, gastric, liver, pancreatic, and lung), mesoderm-derived organoids (e.g., bone spheroids), and ectoderm-derived organoids (e.g., mammary organoids organoids).47 In contrast to organoids derived from PSCs, the stem cell giving rise to the adult tissue-derived organoids has a more limited differentiation potential.
Advantages
Given the similarities in the developmental process of organoids and their original correspondent tissues, they yield models in which the structure and function of the tissue are recapitulated, at least to some extent. As a result, organoids might enable the culture of viruses that are only poorly maintained in commonly used 2D-cultured models, because organoids provide a more natural virus host environment. Such virus-infected organoids are believed to be valuable for disease modeling because they can better mimic the clinical observations in vivo and virus–host tissue interactions.50 Numerous infectious conditions have been modeled in organoid cultures.47, 51, 52 In addition, organoids, to some extent, also enable assessment of drug diffusion and delivery, enhancing the predictability of drug screens in 3D compared with 2D cultures.53 Hence, organoids could overcome challenges in preclinical drug screening in terms of safety, efficiency, and cost-effectiveness, as well as genotype–phenotype testing, with the aim of personalized and regenerative medicine.51, 54
Limitations
Despite these advantages, several shortcomings exist. These include absence of a complete tissue microenvironment (such as incorporation of the immune system, innervation, among others; and in ASC organoids, the limitation of a single germ layer, hence precluding, for instance, the stromal niche present in epithelial organs). Given the importance of the immune system (not present in organoids) in antiviral defense, there is a need to develop even more complete modeling systems such as microfluidic platforms. Moreover, incomplete maturation and presence of the often-used Matrigel, which is a variable source of ECM, and the often limited long-term maintenance of some organoids, constitute other limitations of organoid models. Finally, crosstalk between different organs cannot be modeled via organoids, unless they are incorporated in microfluidic systems.55
Organoids in modeling SARS-CoV-2 infection
The use of organoids in COVID-19 could aid the following three aims: (i) elucidating the pathogenesis of organ disease caused by COVID-19; (ii) performing drug screens; and (iii) validating potential therapeutic strategies. When combined with CRISPR technology, organoids may answer many unsolved questions related to SARS-CoV-2 (Fig. 2b). Here, we describe how organoids have been used to elucidate disease mechanisms and as tools for testing therapeutic interventions specifically for COVID-19.
Use of organoids to elucidate COVID-19 pathophysiology
Organoids have already been used to model viral infections (Table S2 in the supplemental information online). Several mechanisms by which SARS-CoV-2 affects various organs have been proposed, even though our understanding of COVID-19 pathogenesis and susceptibility-determining factor(s), among others, is far from complete. This is caused, at least in part, by a lack of appropriate disease models. Models successfully used so far include monkey kidney-derived cell lines that are susceptible to SARS-CoV infection and support its replication because of the high expression of ACE-2 in kidney tissue.56 In addition, the human Huh7 cell line and human airway epithelial cells have also been useful for isolating the virus.57 However, more complete models, such as organoids, will likely increase our understanding of viral tissue tropism, susceptibility of different tissues to infection, ability of different tissues to support viral replication, as well as differences in response by different tissues to SARS-CoV-2. When co-cultured with immune cells, in vitro imitation of immune response to viral particles might also be possible.58 Here, we provide an overview of how organoids have been used in COVID-19 pathogenesis studies.
Human lung organoids
Given that COVID-19 is a respiratory-based disease, human lung organoids generated from PSCs containing both bronchial and alveolar epithelial cells have been used. The presence of one or the other of these two cell types can affect viral entry, because single cell RNA-sequencing of human lung demonstrated that, although both cell types have strong expression of TMPRSS2, ACE2 expression is predominant in bronchial cells.59 Infecting ACE2-expressing human alveolar organoids with SARS-CoV-2 was shown to cause a robust increase in cytokine and chemokine response. Moreover, screening of FDA-approved drugs demonstrated that two drugs, namely imatinib and mycophenolic acid, could inhibit viral entry.60 In another study, Suzuki et al. used human bronchial organoids (hBOs), containing basal, club, ciliated, and goblet cells, to recapitulate the viral life cycle and assess the effect of the virus on the survival of the cells. Five days after infection, significant cytotoxicity was observed, which could be prevented by the TMPRSS2 inhibitor, camostat. Hence, this models might be useful to screen for antiviral agents.61
Intestinal organoids
Gastrointestinal (GI) manifestations are present in a subset of patients with COVID-19. Moreover, the highest levels of ACE2 expression are seen in intestinal enterocytes.62, 63 Lamers et al. established human small intestinal organoids (hSIOs) under four different culture conditions. They demonstrated that the organoids can be infected by SARS-CoV-2. The infected cells were proliferating progenitors and enterocytes, and their number increased over time with concomitant production of new virus. In addition, transcriptome studies demonstrated the induction of generic viral response pathways.62 Of note, intestinal organoids can also be established from horseshoe bats, the animal believed to be the original host of SARS-CoV-2. Following the infection of bat intestinal organoids with SARS-CoV-2, a robust viral replication was observed along with an increasing cytopathogenic effect,64 suggesting that viral infection is possible also in bat intestine, and that this too could be used to model COVID-19 infection.
Kidney organoids
Multiple indirect mechanisms have been proposed to underlay kidney damage and failure in patients with COVID-19 and include damage caused by the cytokine storm, endothelial damage, hemodynamic instability, among others.65 However, because ACE2 is highly expressed in kidney tubules, direct infection of kidney tissue might be a possibility. Therefore, Monteil et al. developed kidney organoids that have tubular networks to assess the direct infectivity of SARS-CoV-2. They demonstrated that tubular- and podocyte-like cells expressed ACE2. Furthermore, they found that virus entry and production of infectious viral progeny were possible in these organoids, and that these could be blocked by human recombinant soluble ACE2.63 However, a direct cytopathic effect (CPE) of the virus was not reported.
Liver organoids
The exact cause of liver damage in patients with COVID-19 is not yet clear. As seen for renal damage, this could be a secondary phenomenon; however, given that ACE and TMPRSS2 are highly expressed on biliary cells, direct cytopathotoxicity might also be possible. However, mature hepatocytes do not express ACE2.66 Zhao et al. investigated long-term expandable liver ductal organoids that express both ACE2 and TMPRSS2. These organoids were infectable and supported SARS-COV-2 viral replication. Moreover, CPE by SARS-COV-2 was observed, suggesting that cholangiocytes might explain, at least in part, observed liver dysfunction in patients with COVID-19.67
Brain organoids
Numerous neurological symptoms have been reported among patients with COVID-19, suggesting that the central nervous system (CNS) is affected by the infection process.68 Again, whether these are directly caused by viral infections of cells within the CNS or are secondary effects is unclear. Ramani et al. recently reported that neurons in brain organoids, which contain cells with cortical neuronal identity as well as glial cells, can be infected with SARS-CoV-2. Interestingly, the phenotypic changes observed were reminiscent of tauopathies, with altered Tau localization and hyperphosphorylation along with neuronal death,69 but no viral replication was seen. Zhang et al. demonstrated that SARS-COV-2 also infects neuronal progenitors in typical brain organoids, and that the virus also replicates in these organoids.70
Vascular organoids
Coagulopathy is one of the hallmarks observed in patients with COVID-19.40 Endothelial damage is a well-known cause of coagulopathies, and ACE2 is expressed on endothelium throughout the body.71 Endothelium also expresses the TMPRSS2 receptor.71 To assess whether SARS-COV-2 can infect endothelial cells and cause endothelial damage, Monteil et al. created vascular organoids and exposed these to SARS-CoV-2. They demonstrated successful infection, which, as seen for in kidney organoids, could be prevented by human recombinant soluble ACE2.63 Whether the infection causes CPE was not described. Nevertheless, studies in 2D cultures suggest that endothelial cells are not productively infected by SARS-COV-2, but that the virus causes an inflammatory response, which might then lay at the basis of the coagulopathy.71
Therefore, organoids are promising tools for investigating SARS-COV-2 infections and specific host–virus interactions. The initial studies described above confirm this promise. In addition, because organoids can be created in a patient-specific way, they might ultimately also aid in understanding different individual susceptibilities. They could also form an interesting platform for drug screening and developing personalized medicine.
Mechanism elucidation
Further assessment of organoids
The field of SARS-COV-2 disease modeling is young, given that the virus was only detected 2 years ago. Therefore, many of the studies listed above, although promising, have not yet been able to fully define the usefulness of organoids of different tissues for modeling disease pathogenesis. Therefore, additional studies will shed ever-increasing light on which cells/tissues are infected robustly be the virus, whether the infection can be propagated in these tissues, and how infection affects the function of a given cell, in a tissue-like setting. In addition, by using progressively advancing single cell ‘omics technologies, including spatial ‘omics, it will become possible to understand how infection of a given cell in an organoid affects the function of not only the infected cell, but also its nearest neighbors, to fully comprehend how tissue damage occurs in patients with SARS-COV-2. As discussed below, once these tissues can be co-cultured on ‘body-on-a-chip’ systems, the effects of infection of one tissue on distant tissues will also be testable.
Creating genetically diverse organoid/iPSC banks
Using organoids, whether from different ASCs or PSCs, offers advantages for studying the host immune responses to viral infections. Although we do not yet understand why some patients develop more severe disease than others, it should theoretically be possible to start elucidating an individual’s response to SARS-COV-2 infections, and to determine the contribution of underlying genetic traits of an individual to the severity of disease development. Having a bank of organoids or initial PSCs available that cover a range of racially and genetically different individuals will be vital for studying the genetic predisposition of COVID-19 and subsequently its pharmacogenomics.35
An alternative way to define susceptibility risks for SARS-COV-2 is to use precise gene editing to mimic allelic variation via CRISPR technology in organoids, either in the original stem cells or in single cells dissociated from organoids.72 Moreover, CRSIPR technology should also enable identification of the role of host proteins, or signaling pathways in SARS-COV-2-mediated pathophysiology, via gene knockout studies, or by exploiting CRISPRa/i technology, which allows gene activation or inhibition.73
Therapeutics validation and drug screening
The first studies describing the use of organoids to study SARS-COV-2 have already yielded novel information regarding how the infection can be blocked or mitigated. However, the merger of SARS-COV-2 infection and organoid modeling research is very young. Nevertheless, further development of the combination of organoid modeling, identifying robust phenotypic alterations, followed by phenotypic drug-screening approaches, will be vital for identifying novel therapeutics that can interfere in many of the steps during SARS-COV-2 disease progression, and could lead to the development of new and powerful antiviral drug entities.74
Prospects for COVID-19 modeling: toward novel platforms for disease modelling and drug screening
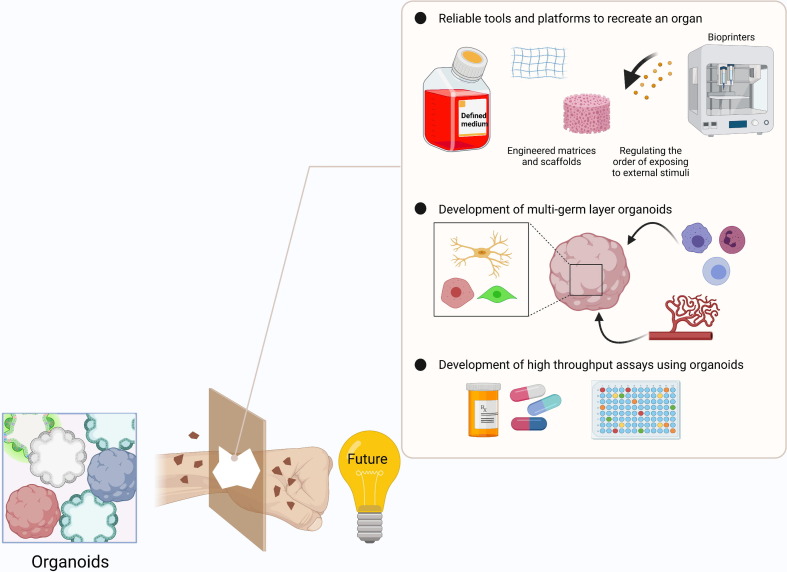
Currently, the heterogeneity of organoids in terms of their size and/or cellular composition remains a major challenge, and prevents their wide and robust use for disease modeling.75 Moreover, most organoids contain cells from only one germ layer (e.g., epithelium), but not the surrounding mesodermal niche cells, which also hampers full disease modeling (Fig. 4 ). These shortcomings are addressed herein.
Figure 4.
Future perspectives for coronavirus 2019 (COVID-19) modeling. Variability, multigerm-layer composition, and high-throughput analysis are three main subjects that need further investigation in organoid platforms. Reliable recreation of an organoid is an important requirement for achieving comparable results. Thus far, different strategies have been used to address this challenge. These contain optimization of the culture media and using certain matrices, among others. Using different components of germ layers is another promising area to create fully functional organoids with the most similarity to the human body environment. High-throughput screening of candidate drugs is another promised field for the future applications of organoids.
Reliable recreation of an organ
The consistent differentiation of organoids is one of the main challenges that needs to be addressed so that organoids can be robustly used for disease modeling and drug-screening approaches.76 For instance, the development of methods for unidirectional lineage commitment and further differentiation and maturation has already demonstrated that more consistent differentiation might be possible. Variability mitigation has gained attention especially in the field of brain organoids. For instance, Nickels et al. generated human midbrains using a defined patterning strategy, and starting from neural committed progenitors, rather than PSCs, which reduced the variability of the midbrain organoids significantly.75 The use of not only cocktails of small molecules to drive lineage differentiation, but also PSC culture conditions that are feeder and xeno free enhanced the consistency of human cortical organoids generated from 15 different hiPSC lines.76 Similarly, human T cell differentiation also appeared to be more consistent when using a serum-free system to culture thymic organoids.77 Several tissue-engineering approaches might also improve consistency. For instance, replacement of less well-defined natural hydrogels, such as Matrigel, with synthetic hydrogels also decreases batch-to-batch variability. Alternatively, the use of chemically defined scaffolds that enable the spatial and temporal controlled presentation of external stimuli could improve differentiation, which can be created, for instance, by bioprinting.78
Multigerm layer composition
Although the reductionist approach of having only a single germ-layer population in an organoid has the merit of being able to better control the make-up of that organoid, the lack of cells of other germ layers, such as stromal niche cells, vasculature, or immune cells, makes disease modeling incomplete. Incorporating the stromal compartment into organoids, such as vascularization of brain organoids, has demonstrated how it might be possible to improve organoid culture models.79 Not only will incorporation of vascular cells in organoids enable assessment of the crucial role of vascular tissue in COVID-19, but also the perfusion of immune cells into a range of organoids.80 Indeed, as illustrated by Nozaki et al. for gut organoids,81 incorporation of immune system components into organoids might provide insights into the role of inflammation in COVID-19.
Medium/high-throughput assays
Once robust and consistent organoid differentiation becomes possible, organoids could be deployed for medium/high-throughput screens to identify therapeutic drugs against COVID-19. As alluded to above, this will require biobanks of stem cells or derived organoids with different genetic backgrounds.82 Initial studies on lung organoids demonstrated that this will become feasible. For instance, hPSC-derived lung organoids infected with SARS-CoV-2 were used to identify drugs in the list of FDA-approved drugs that might block SARS-CoV-2 viral entry.60 In another study, Mulay et al. demonstrated significant IFN signaling activation in lung organoids infected with SARS-CoV-2. The viral infection/replication was shown to be strongly inhibited by remdesivir.83 Given the significance of drug-induced liver injury (DILI) in the context of liver pathophysiology, Shinozawa et al. developed a hiPSC-derived liver organoid (HLO) suitable for mid/high-throughput DILI compound screening.84
What tissue engineering holds for COVID-19 investigations
Engineered tissues
Tissue engineering holds great promise for disease detection and drug screening. The possible use of tissue-engineered products, given the widespread organ complications in COVID-19 (lung, heart, liver, gastrointestinal system, kidney, etc.), has been described in detail elsewhere.85 However, there are still limited numbers of studies that used engineered tissues in COVID-19, although a recent study demonstrated cardiomyocyte tropism of SARS-CoV-2 using a heart tissue model.86
From organ-on-chip to human-on-chip systems
Given the recent development of robust organoid differentiation protocols for a variety of human organs, it now also becomes possible to create so-called single-organoid OoC and complex multiorganoid HoC systems. OoC/HoC represent an unprecedented platform that exploits advances made in organoid culture and complex tissue engineering, microfluidic chip biofabrication, and micro-electromechanical system integration. These microphysiological systems house either single organoids (OoC) or multiple organoids each situated within specific compartments that are interconnected via microchannels and mimic the systemic blood circulation, organ proximity, connectivity, and functionality of a human body (HoC). Multiorganoid systems are necessary because many physiological functions and pathological conditions are not attributable to one specific organ but rather act at higher levels of tissue–tissue or multiorgan interactions. Full exploitation of OoC and HoC models will require further optimization of medium, standardization, and automation of injected media, compounds, and samples, fabrication as well as scale-up, and integration of online analytics/sensors.87
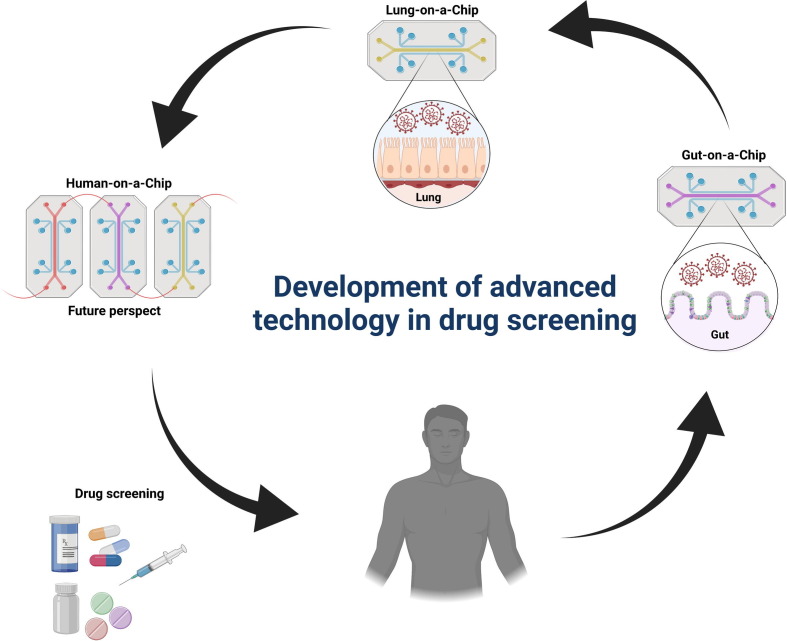
Given that COVID-19 is a prime example of how disease starting in one organ can affect many organs, development of HoC models will significantly enhance our ability to study this interorgan communication (Fig. 5 ). Whereas entry of SARS-CoV-2 in cells via the ACE2 receptor (and TMPRSS2) might cause CPE in infected cells of patients with COVID-19, whether this occurs in all organs affected is not clear; and organ failure might be secondary to factors produced by one infected and dying cell affecting a neighboring cell or cells at a distance. Therefore, OoC/HoC models are clinically relevant preclinical models to study virus entry and spreading routes, and the secondary effects of infection of one tissue on another tissue, as well as for assessing prophylactic or therapeutic candidate drugs.88 Given the prevalence of lung involvement in the pathogenesis of COVID-19, lung-on-chips should enable evaluation of immune cell responses to SARS-CoV-2, as well as drug screening. These chips enable more accurate modeling of lung organs, compared with lung organoids, by, for example, culturing of bronchial-airway epithelium at an air–liquid interface (ALI) and mimicking some of the relevant features of the organ. For instance, a human lung airway-on-chip model comprising ALI of human lung airway epithelium and a perfusion system has been developed to model SARS-CoV-2 virus entry, replication, strain-dependent virulence, host cytokine production, and recruitment of circulating immune cells, as well as to assess clinically approved ACE2 inhibitors.88 Similarly, Zhang et al. created a biomimetic alveolar–capillary barrier-on-chip and revealed innate immune responses upon alveolar barrier injury resulting from SARS-CoV-2 infection, which could be alleviated by inhibiting viral replication via remdesivir treatment.89 As another example, a human gut-on-chip was created to model the intestinal epithelium–vascular endothelium barrier to study the permissiveness of SARS-CoV-2 infection and intestinal barrier injury and dysfunction resulting from morphological and genetic abnormalities induced by the virus.90
Figure 5.
Development of advanced technology in drug screening. Given limitations of animal research, using novel and more practical strategies appears imperative. To this end, chip platforms [from organ-on-chip (OoC) to human-on-chip (HoC)] have been progressed extensively to emulate the complicated characteristics of human body and provide a more reliable platform in the translational clinical setting. Human lung-on-chip and gut-on-chip are two examples of such advanced technology that have been used in COVID-19 studies. OoC/HoC technology offers great opportunities for high-throughput screening of candidate drugs.
As discussed above, the coagulopathy caused by SARS-CoV-2 might be caused by direct viral entry in endothelium or an indirect consequence of the cytokine storm. To start to tackle this question, Thacker et al. created vascularized lung-on-chip to simulate SARS-CoV-2 infection and found a transient inflammatory response in epithelium but a persistent inflammatory response in endothelium because of IL-6 secretion even in the absence of immune cells.91 Interestingly, IL-6 signaling could be reduced with tocilizumab treatment, although this did not prevent loss of barrier integrity. This might suggest that direct viral infection in endothelial cells is also responsible for endothelial death. OoC models might also provide novel insights into mechanisms underlying the major cardiac and arrhythmic complications often seen in patients with COVID-19, of which the cause is currently unknown. By exposing heart organoid on-chip to the virus should both provide important pathophysiological information and enable the identification of drugs that counteract these phenotypes.
Multiorgan systems or the HoC could be designed in such a way to enable testing of biotransformed drugs, whereby the drugs are subjected initially to intestinal and liver organoid on-chip for bioabsorption and biotransformation into active compounds before entering the downstream targeted organoids, thus emulating drug biodistribution in the heart, brain, kidney, retina, and lung, including excretion of metabolized compound waste. Such studies can currently only be performed in animal models. However, because, for example, biotransformation enzyme activities differ significantly between species, animal models do not fully allow such evaluation. Similarly, such a multiorgan system could be useful to test the organ toxicity of candidate drugs and vaccines (in terms of dosing), potentially allowing scientists to reduce the dependency on cost-inefficient nonhuman primate toxicity studies before conducting clinical trials.
There is no doubt that OoC/HoC could have important roles in the pharmaceutical industry. They could serve as exploratory humanized assays to identify potential human-specific hazards (e.g., cyto- or mitotoxicity screening, or cytokine release assays) of antiviral drugs by filtering early compounds and optimizing molecular structure. This will help to replace pivotal animal studies and justify the subsequent test animal species both cost-wise and ethically, which coincides with the concept of animal-free safety testing.92 At a regulatory level, OoC/HoC could be useful to complement, to certain extent, the mandatory requirements for safety risk assessment and regulatory decision-making, in particular on the PK and PD, genotoxicity, phototoxicity, as well as immunotoxicity. OoC/HoC also has the advantages of high-throughput assays, whereby multiplexing and parallelization of the microfluidic systems could be implemented to achieve upscaling and/or outscaling of the screening platform.93 They also represent enclosed and clean testing platforms, which are vital to meet the standards for good laboratory practice (GLP). Furthermore, operation at a minute volume within the microfluidic system offers versatility to screen more candidate drugs at lower cost and also allows drug gradient and dose studies, rendering them an attractive one-stop in vitro system to test drug/vaccine efficacy and organ toxicity in place of preclinical animal models.
Nonetheless, as also remains true for simple organoid models, OoC/HoC technology is still in its infancy and industrial stakeholders are adopting it slowly for obvious reasons: (i) the technology is seen more as a tool for research and development (R&D); (ii) knowledge gaps between biologists and engineers remained to be bridged to avoid mismatch between biology and engineering aspects of the chip design; (iii) concerns over the reproducibility, stability, logistic issues on cell viability, and user-friendly aspects because of the complexity of the technology; and (iv) unidentified translational demands and regulatory requirements. Several scientific and technical challenges are foreseen94: (i) implementation of a universal medium for simultaneous co-culturing of different organoid types on-chip; otherwise, a relative more complex nutrient-waste management system for different media on-chip, including miniature automatization, will be required; (ii) upscaling of microfluidic to millifluidic models to accommodate large or ever-growing organoids, an intrinsic technical challenge for fabricating upscaled microfluidic with micro-precision; alternatively, 3D printing technology can be used at the expense of lower resolution95; (iii) integration of an on-chip real-time monitoring system, including built-in microsensors within the perfusion system or underneath the chip to record cellular activity (e.g., cell vitality, metabolic, inflammatory, and electrophysiology) of each organoids as well as for assessing potential biomarker profiling as diagnosis or prognosis of diseases96, 97; (iv) implementation of interconnected and perfusable endothelial vasculature between the different organoids on-chip within the microfluidic microchannels, whereby the hematopoietic and immune cells could be integrated into the flow lines to simulate systemic circulation system of human body.98 Herein, the flow lines could also serve as entry point for virus infection, therapeutics, or toxic compound infusion; (iv) the need to develop culture medium with physicochemical properties (e.g. liquid viscosity, osmolarity, pH) close to human blood and the optimization of the flow rate to mimic interorgan and interstitial hemodynamics of the blood circulation.
Concluding remarks
Despite extensive efforts to find efficient, safe, and reliable therapies for COVID-19, no decisive treatment has thus far been found. Using standard platforms with the closest resemblance to human organs to recapitulate the complex in vivo conditions, will be vital for identifying pathophysiological phenotypes of SARS-CoV-2-infected tissues and drugs that efficiently interfere with these events. Here, we discussed some novel platforms, both genome based and computational platforms, to predict candidate drugs that, when validated with advanced multicellular models of tissues, or even combined tissue models, such as in HoC, could hasten the search to finding effective therapies and prophylaxis for COVID-19, as well as many other diseases.
Biographies

Catherine Verfaillie is professor in the Faculty of Medicine and head of Stem Cell and Developmental Biology at the Katholieke Universiteit Leuven (KU Leuven), Belgium. She is also a member of the LKI - KU Leuven Cancer Institute. Current research interests in Professor Verfaillie’s group include: hematopoiesis, with the ultimate aim of applying this knowledge to innovative cord blood expansion schemes; pluripotent stem cell (PSC)-derived liver (hepatocytes and nonparenchymal liver cells) models; and PSC-derived neurodegeneration models.

Massoud Vosough is an assistant professor in the Stem Cells Biology and Technology Institute, Royan Institute and head of the Regenerative Medicine Department, Tehran, Iran. Current research interests in his group include: interdisciplinary research in stem cell biology and regenerative medicine for the efficient large-scale differentiation of human PSCs toward hepatocyte-like cells, and using human PSC-derived hepatocytes for regenerative medicine and drug assays, to achieve engineered liver structures using hydrogels derived from liver specific-extracellular matrix and to construct a microfluidic device for medical applications.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.drudis.2021.12.014.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Gorbalenya A.E., Baker S.C., Baric R.S., de Groot R.J., Drosten C., Gulyaeva A.A., et al. The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zumla A., Chan J.F.W., Azhar E.I., Hui D.S.C., Yuen K.-Y. Coronaviruses — drug discovery and therapeutic options. Nat Rev Drug Discov. 2016;15:327–347. doi: 10.1038/nrd.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shahriari Felordi M., Memarnejadian A., Najimi M., Vosough M. Is there any alternative receptor for SARS-CoV-2? Cell J. 2021;23:247–250. doi: 10.22074/cellj.2021.7977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tay M.Z., Poh C.M., Rénia L., MacAry P.A., Ng L.F.P. The trinity of COVID-19: immunity, inflammation and intervention. Nat Rev Immunol. 2020;20:363–374. doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Astashkina A., Mann B., Grainger D.W. A critical evaluation of in vitro cell culture models for high-throughput drug screening and toxicity. Pharmacol Ther. 2012;134:82–106. doi: 10.1016/j.pharmthera.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 6.Ikitimur H., Borku Uysal B., Cengiz M., Ikitimur B., Uysal H., Ozcan E., et al. Determining host factors contributing to disease severity in a family cluster of 29 hospitalized SARS-CoV-2 patients: could genetic factors be relevant in the clinical course of COVID-19? J Med Virol. 2021;93:357–365. doi: 10.1002/jmv.26106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.LoPresti M., Beck D.B., Duggal P., Cummings D.A.T., Solomon B.D. The role of host genetic factors in coronavirus susceptibility: review of animal and systematic review of human literature. Am J Hum Genet. 2020;107:381–402. doi: 10.1016/j.ajhg.2020.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benetti E., Tita R., Spiga O., Ciolfi A., Birolo G., Bruselles A., et al. ACE2 gene variants may underlie interindividual variability and susceptibility to COVID-19 in the Italian population. Eur J Hum Genet. 2020;28:1602–1614. doi: 10.1038/s41431-020-0691-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hussain M., Jabeen N., Raza F., Shabbir S., Baig A.A., Amanullah A., et al. Structural variations in human ACE2 may influence its binding with SARS-CoV-2 spike protein. J Med Virol. 2020;92:1580–1586. doi: 10.1002/jmv.25832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Devaux C.A., Rolain J.-M., Raoult D. ACE2 receptor polymorphism: Susceptibility to SARS-CoV-2, hypertension, multi-organ failure, and COVID-19 disease outcome. J Microbiol Immunol Infect. 2020;53:425–435. doi: 10.1016/j.jmii.2020.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao Y., Zhao Z., Wang Y., Zhou Y., Ma Y., Zuo W. Single-cell RNA expression profiling of ACE2 the receptor of SARS-CoV-2. Am J Respir Crit Care Med. 2020;202:756–759. doi: 10.1164/rccm.202001-0179LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang F., Huang S., Gao R., Zhou Y., Lai C., Li Z., et al. Initial whole-genome sequencing and analysis of the host genetic contribution to COVID-19 severity and susceptibility. Cell Discovery. 2020;6:83. doi: 10.1038/s41421-020-00231-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Senapati S., Kumar S., Singh A.K., Banerjee P., Bhagavatula S. Assessment of risk conferred by coding and regulatory variations of TMPRSS2 and CD26 in susceptibility to SARS-CoV-2 infection in human. J Genet. 2020;99:53. doi: 10.1007/s12041-020-01217-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amoroso A., Magistroni P., Vespasiano F., Bella A., Bellino S., Puoti F., et al. HLA and AB0 polymorphisms may influence SARS-CoV-2 infection and COVID-19 severity. Transplantation. 2021;105:193–200. doi: 10.1097/TP.0000000000003507. [DOI] [PubMed] [Google Scholar]
- 15.Iturrieta-Zuazo I., Rita C.G., García-Soidán A., de Malet P.-F., Alonso-Alarcón N., Pariente-Rodríguez R., et al. Possible role of HLA class-I genotype in SARS-CoV-2 infection and progression: a pilot study in a cohort of Covid-19 Spanish patients. Clin Immunol (Orlando, Fla) 2020;219 doi: 10.1016/j.clim.2020.108572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gómez J., Albaiceta G.M., Cuesta-Llavona E., García-Clemente M., López-Larrea C., Amado-Rodríguez L., et al. The Interferon-induced transmembrane protein 3 gene (IFITM3) rs12252 C variant is associated with COVID-19. Cytokine. 2021;137 doi: 10.1016/j.cyto.2020.155354. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Q., Bastard P., Liu Z., Le Pen J., Moncada-Velez M., Chen J., et al. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science. 2020;370:eabd4570. doi: 10.1126/science.abd4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wake D.T., Ilbawi N., Dunnenberger H.M., Hulick P.J. Pharmacogenomics: prescribing precisely. Med Clinics North Am. 2019;103:977–990. doi: 10.1016/j.mcna.2019.07.002. [DOI] [PubMed] [Google Scholar]
- 19.Mallal S., Nolan D., Witt C., Masel G., Martin A.M., Moore C., et al. Association between presence of HLA-B*5701, HLA-DR7, and HLA-DQ3 and hypersensitivity to HIV-1 reverse-transcriptase inhibitor abacavir. Lancet (London, England) 2002;359:727–732. doi: 10.1016/s0140-6736(02)07873-x. [DOI] [PubMed] [Google Scholar]
- 20.Cafiero C., Re A., Micera A., Palmirotta R., Monaco D., Romano F., et al. Pharmacogenomics and pharmacogenetics: in silico prediction of drug effects in treatments for novel coronavirus SARS-CoV2 disease. Pharmacogenomics Pers Med. 2020;13:463–484. doi: 10.2147/PGPM.S270069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pizzorno A., Padey B., Julien T., Trouillet-Assant S., Traversier A., Errazuriz-Cerda E., et al. Characterization and treatment of SARS-CoV-2 in nasal and bronchial human airway epithelia. Cell Rep Med. 2020;1 doi: 10.1016/j.xcrm.2020.100059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Agnihothram S., Yount B.L., Donaldson E.F., Huynh J., Menachery V.D., Gralinski L.E., et al. A mouse model for betacoronavirus subgroup 2c using a bat coronavirus strain HKU5 variant. mBio. 2014;5:e00047–e114. doi: 10.1128/mBio.00047-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Riva L., Yuan S., Yin X., Martin-Sancho L., Matsunaga N., Pache L., et al. Discovery of SARS-CoV-2 antiviral drugs through large-scale compound repurposing. Nature. 2020;586:113–119. doi: 10.1038/s41586-020-2577-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garcia G., Jr., Sharma A., Ramaiah A., Sen C., Purkayastha A., Kohn D.B., et al. Antiviral drug screen identifies DNA-damage response inhibitor as potent blocker of SARS-CoV-2 replication. Cell Rep. 2021;35 doi: 10.1016/j.celrep.2021.108940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barlan A., Zhao J., Sarkar M.K., Li K., McCray P.B., Jr., Perlman S., et al. Receptor variation and susceptibility to Middle East respiratory syndrome coronavirus infection. J Virol. 2014;88:4953–4961. doi: 10.1128/JVI.00161-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Falzarano D., de Wit E., Feldmann F., Rasmussen A.L., Okumura A., Peng X., et al. Infection with MERS–CoV causes lethal pneumonia in the common marmoset. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1004250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chan J.F., Yao Y., Yeung M.L., Deng W., Bao L., Jia L., et al. Treatment With lopinavir/ritonavir or interferon-β1b improves outcome of MERS-CoV infection in a nonhuman primate model of common marmoset. J Infect Dis. 2015;212:1904–1913. doi: 10.1093/infdis/jiv392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Williamson B.N., Feldmann F., Schwarz B., Meade-White K., Porter D.P., Schulz J., et al. Clinical benefit of remdesivir in rhesus macaques infected with SARS-CoV-2. Nature. 2020;585:273–276. doi: 10.1038/s41586-020-2423-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hewitt J.A., Lutz C., Florence W.C., Pitt M.L.M., Rao S., Rappaport J., et al. ACTIVating resources for the COVID-19 pandemic: in vivo models for vaccines and therapeutics. Cell Host Microbe. 2020;28:646–659. doi: 10.1016/j.chom.2020.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dinnon K.H., Leist S.R., Schäfer A., Edwards C.E., Martinez D.R., Montgomery S.A., et al. A mouse-adapted SARS-CoV-2 model for the evaluation of COVID-19 medical countermeasures. Nature. 2020;586:560–566. doi: 10.1038/s41586-020-2708-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shultz L.D., Ishikawa F., Greiner D.L. Humanized mice in translational biomedical research. Nat Rev Immunol. 2007;7:118–130. doi: 10.1038/nri2017. [DOI] [PubMed] [Google Scholar]
- 32.Sun J., Zhuang Z., Zheng J., Li K., Wong R.-L.-Y., Liu D., et al. Generation of a broadly useful model for COVID-19 pathogenesis, vaccination, and treatment. Cell. 2020;182:734–743. doi: 10.1016/j.cell.2020.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chan J.F., Zhang A.J., Yuan S., Poon V.K., Chan C.C., Lee A.C., et al. Simulation of the clinical and pathological manifestations of coronavirus disease 2019 (COVID-19) in a golden Syrian hamster model: implications for disease pathogenesis and transmissibility. Clin Infect Dis. 2020;71:2428–2446. doi: 10.1093/cid/ciaa325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muñoz-Fontela C., Dowling W.E., Funnell S.G.P., Gsell P.-S., Riveros-Balta A.X., Albrecht R.A., et al. Animal models for COVID-19. Nature. 2020;586:509–515. doi: 10.1038/s41586-020-2787-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Forbester J.L., Humphreys I.R. Genetic influences on viral-induced cytokine responses in the lung. Mucosal Immunol. 2021;14:14–25. doi: 10.1038/s41385-020-00355-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park S.-J., Kim Y.-G., Park H.-J. Identification of RNA Pseudoknot-binding ligand that inhibits the -1 ribosomal frameshifting of SARS-coronavirus by structure-based virtual screening. J Am Chem Soc. 2011;133:10094–10100. doi: 10.1021/ja1098325. [DOI] [PubMed] [Google Scholar]
- 37.Chen Y., Tao H., Shen S., Miao Z., Li L., Jia Y., et al. A drug screening toolkit based on the -1 ribosomal frameshifting of SARS-CoV-2. Heliyon. 2020;6 doi: 10.1016/j.heliyon.2020.e04793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xie X., Muruato A.E., Zhang X., Lokugamage K.G., Fontes-Garfias C.R., Zou J., et al. A nanoluciferase SARS-CoV-2 for rapid neutralization testing and screening of anti-infective drugs for COVID–19. Nat Commun. 2020;11:5214. doi: 10.1038/s41467-020-19055-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gorshkov K., Chen C.Z., de la Torre J.C., Martinez-Sobrido L., Moran T., Zheng W. Development of a high-throughput homogeneous AlphaLISA drug screening assay for the detection of SARS-CoV-2 nucleocapsid. ACS Pharmacol Transl Sci. 2020;3:1233–1241. doi: 10.1021/acsptsci.0c00122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Song J.-C., Wang G., Zhang W., Zhang Y., Li W.-Q., Zhou Z., et al. Chinese expert consensus on diagnosis and treatment of coagulation dysfunction in COVID-19. Mil Med Res. 2020;7:19. doi: 10.1186/s40779-020-00247-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gysi D.M., Do Valle Í., Zitnik M., Ameli A., Gan X., Varol O., et al. Network medicine framework for identifying drug repurposing opportunities for COVID-19. Proc Natl Acad Sci USA. 2021;118 doi: 10.1073/pnas.2025581118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou Y., Hou Y., Shen J., Huang Y., Martin W., Cheng F. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discovery. 2020;6:14. doi: 10.1038/s41421-020-0153-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beck B.R., Shin B., Choi Y., Park S., Kang K. Predicting commercially available antiviral drugs that may act on the novel coronavirus (SARS-CoV-2) through a drug-target interaction deep learning model. Comput Struct Biotechnol J. 2020;18:784–790. doi: 10.1016/j.csbj.2020.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu C., Liu Y., Yang Y., Zhang P., Zhong W., Wang Y., et al. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharmaceutica Sinica B. 2020;10:766–788. doi: 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mirabelli C., Wotring J.W., Zhang C.J., McCarty S.M., Fursmidt R., Frum T., et al. Morphological cell profiling of SARS-CoV-2 infection identifies drug repurposing candidates for COVID-19. Proc Natl Acad Sci USA. 2021;118 doi: 10.1073/pnas.2105815118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rezaei S., Sefidbakht Y., Uskoković V. Tracking the pipeline: immunoinformatics and the COVID-19 vaccine design. Briefings Bioinf. 2021;22:bbab241. doi: 10.1093/bib/bbab241. [DOI] [PubMed] [Google Scholar]
- 47.Lancaster M.A., Huch M. Disease modelling in human organoids. Dis Models Mech. 2019;12:dmm039347. doi: 10.1242/dmm.039347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lancaster M.A., Knoblich J.A. Organogenesis in a dish: modeling development and disease using organoid technologies. Science. 2014;345:1247125. doi: 10.1126/science.1247125. [DOI] [PubMed] [Google Scholar]
- 49.Yin X., Mead B.E., Safaee H., Langer R., Karp J.M., Levy O. engineering stem cell organoids. Cell Stem Cell. 2016;18:25–38. doi: 10.1016/j.stem.2015.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sridhar A., Simmini S., Ribeiro C.M.S., Tapparel C., Evers M.M., Pajkrt D., et al. A perspective on organoids for virology research. Viruses. 2020;12:1341. doi: 10.3390/v12111341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dutta D., Heo I., Clevers H. Disease modeling in stem cell-derived 3D organoid systems. Trends Mol Med. 2017;23:393–410. doi: 10.1016/j.molmed.2017.02.007. [DOI] [PubMed] [Google Scholar]
- 52.Fatehullah A., Tan S.H., Barker N. Organoids as an in vitro model of human development and disease. Nat Cell Biol. 2016;18:246–254. doi: 10.1038/ncb3312. [DOI] [PubMed] [Google Scholar]
- 53.Yu J. Organoids: a new model for SARS-CoV-2 translational research. Int J Stem Cells. 2021;14:138–149. doi: 10.15283/ijsc20169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lou Y.-R., Leung A.W. Next generation organoids for biomedical research and applications. Biotechnol Adv. 2018;36:132–149. doi: 10.1016/j.biotechadv.2017.10.005. [DOI] [PubMed] [Google Scholar]
- 55.Xu H., Jiao Y., Qin S., Zhao W., Chu Q., Wu K. Organoid technology in disease modelling, drug development, personalized treatment and regeneration medicine. Exp Hematol Oncol. 2018;7 doi: 10.1186/s40164-018-0122-9. 30– 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kaye M. SARS-associated coronavirus replication in cell lines. Emerg Infect Dis. 2006;12:128–133. doi: 10.3201/eid1201.050496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Matsuyama S., Nao N., Shirato K., Kawase M., Saito S., Takayama I., et al. Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc Natl Acad Sci USA. 2020;117:7001. doi: 10.1073/pnas.2002589117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Elbadawi M., Efferth T. Organoids of human airways to study infectivity and cytopathy of SARS-CoV-2. Lancet Respiratory Med. 2020;8:e55–e56. doi: 10.1016/S2213-2600(20)30238-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lukassen S., Chua R.L., Trefzer T., Kahn N.C., Schneider M.A., Muley T., et al. SARS-CoV-2 receptor ACE2 and TMPRSS2 are primarily expressed in bronchial transient secretory cells. EMBO J. 2020;39 doi: 10.15252/embj.20105114. e105114–e105114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Han Y., Yang L., Duan X., Duan F., Nilsson-Payant B.E., Yaron T.M., et al. Identification of-candidate COVID-19 therapeutics using hPSC-derived lung organoids. bioRxiv. 2020 doi: 10.1101/2020.05.05.079095. (Published online May 5, 2020) [DOI] [Google Scholar]
- 61.Suzuki T., Itoh Y., Sakai Y., Saito A., Okuzaki D., Motooka D. et al.Generation of human bronchial organoids for SARS-CoV-2 research. bioRxiv. 2020 doi: 10.1101/2020.05.25.115600. Published online June 1, 2020. [DOI] [Google Scholar]
- 62.Lamers M.M., Beumer J., van der Vaart J., Knoops K., Puschhof J., Breugem T.I., et al. SARS-CoV-2 productively infects human gut enterocytes. Science. 2020::eabc1669. doi: 10.1126/science.abc1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Monteil V., Kwon H., Prado P., Hagelkrüys A., Wimmer R.A., Stahl M., et al. Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2. Cell. 2020;181:905–913. doi: 10.1016/j.cell.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhou J., Li C., Liu X., Chiu M.C., Zhao X., Wang D., et al. Infection of bat and human intestinal organoids by SARS-CoV-2. Nat Med. 2020;26:1077–1083. doi: 10.1038/s41591-020-0912-6. [DOI] [PubMed] [Google Scholar]
- 65.Ronco C., Reis T. Kidney involvement in COVID-19 and rationale for extracorporeal therapies. Nat Rev Nephrol. 2020;16:308–310. doi: 10.1038/s41581-020-0284-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wen Seow J.J., Pai R., Mishra A., Shepherdson E., Hon Lim T.K., Goh B.K.P., et al. scRNA-seq reveals ACE2 and TMPRSS2 expression in TROP2+ liver progenitor cells: implications in COVID-19 associated liver dysfunction. Front Med. 2021;8 doi: 10.3389/fmed.2021.603374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhao B., Ni C., Gao R., Wang Y., Yang L., Wei J., et al. Recapitulation of SARS-CoV-2 infection and cholangiocyte damage with human liver ductal organoids. Protein Cell. 2020;11:771–775. doi: 10.1007/s13238-020-00718-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liu K., Pan M., Xiao Z., Xu X. Neurological manifestations of the coronavirus (SARS-CoV–2) pandemic 2019–2020. J Neurol Neurosurg Psychiatry. 2020;91:669. doi: 10.1136/jnnp-2020-323177. [DOI] [PubMed] [Google Scholar]
- 69.Ramani A., Müller L., Ostermann P.N., Gabriel E., Abida-Islam P., Müller-Schiffmann A., et al. SARS-CoV-2 targets cortical neurons of 3D human brain organoids and shows neurodegeneration-like effects. EMBO J. 2020;39 doi: 10.15252/embj.2020106230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang B.-Z., Chu H., Han S., Shuai H., Deng J., Hu Y.-F., et al. SARS-CoV-2 infects human neural progenitor cells and brain organoids. Cell Res. 2020;30:928–931. doi: 10.1038/s41422-020-0390-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schimmel L., Chew K.Y., Stocks C., Yordanov T., Essebier T., Kulasinghe A. Endothelial cells elicit a pro-inflammatory response to SARS-CoV-2 without productive viral infection. bioRxiv. 2021 doi: 10.1101/2021.02.14.431177. Published online May 6, 2021. [DOI] [Google Scholar]
- 72.Matano M., Date S., Shimokawa M., Takano A., Fujii M., Ohta Y., et al. Modeling colorectal cancer using CRISPR-Cas9-mediated engineering of human intestinal organoids. Nat Med. 2015;21:256–262. doi: 10.1038/nm.3802. [DOI] [PubMed] [Google Scholar]
- 73.Cámara E., Lenitz I., Nygård Y. A CRISPR activation and interference toolkit for industrial Saccharomyces cerevisiae strain KE6-12. Sci Rep. 2020;10:14605. doi: 10.1038/s41598-020-71648-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Abbott T.R., Dhamdhere G., Liu Y., Lin X., Goudy L., Zeng L., et al. Development of CRISPR as an antiviral strategy to combat SARS-CoV-2 and influenza. Cell. 2020;181:865–876. doi: 10.1016/j.cell.2020.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nickels S.L., Modamio J., Mendes-Pinheiro B., Monzel A.S., Betsou F., Schwamborn J.C. Reproducible generation of human midbrain organoids for in vitro modeling of Parkinson’s disease. Stem Cell Res. 2020;46 doi: 10.1016/j.scr.2020.101870. [DOI] [PubMed] [Google Scholar]
- 76.Yoon S.-J., Elahi L.S., Pașca A.M., Marton R.M., Gordon A., Revah O., et al. Reliability of human cortical organoid generation. Nat Methods. 2019;16:75–78. doi: 10.1038/s41592-018-0255-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bosticardo M., Pala F., Calzoni E., Delmonte O.M., Dobbs K., Gardner C.L., et al. Artificial thymic organoids represent a reliable tool to study T-cell differentiation in patients with severe T-cell lymphopenia. Blood Adv. 2020;4:2611–2616. doi: 10.1182/bloodadvances.2020001730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hofer M., Lutolf M.P. Engineering organoids. Nat Rev Mater. 2021;6:402–420. doi: 10.1038/s41578-021-00279-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wörsdörfer P., Asahina I.T., Sumita Y., Ergün S. Do not keep it simple: recent advances in the generation of complex organoids. J Neural Transmis. 2020;127:1569–1577. doi: 10.1007/s00702-020-02198-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rossi G., Manfrin A., Lutolf M.P. Progress and potential in organoid research. Nat Rev Genet. 2018;19:671–687. doi: 10.1038/s41576-018-0051-9. [DOI] [PubMed] [Google Scholar]
- 81.Nozaki K., Mochizuki W., Matsumoto Y., Matsumoto T., Fukuda M., Mizutani T., et al. Co-culture with intestinal epithelial organoids allows efficient expansion and motility analysis of intraepithelial lymphocytes. J Gastroenterol. 2016;51:206–213. doi: 10.1007/s00535-016-1170-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Takahashi T. Organoids for drug discovery and personalized medicine. Annu Rev Pharmacol Toxicol. 2019;59:447–462. doi: 10.1146/annurev-pharmtox-010818-021108. [DOI] [PubMed] [Google Scholar]
- 83.Mulay A., Konda B., Garcia G., Yao C., Beil S., Villalba J.M., et al. SARS-CoV-2 infection of primary human lung epithelium for COVID-19 modeling and drug discovery. Cell Rep. 2021;35 doi: 10.1016/j.celrep.2021.109055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Khetani S.R. Pluripotent stem cell-derived human liver organoids enter the realm of high-throughput drug screening. Gastroenterology. 2021;160:653–655. doi: 10.1053/j.gastro.2020.12.005. [DOI] [PubMed] [Google Scholar]
- 85.Aydin A., Cebi G., Demirtas Z.E., Erkus H., Kucukay A., Ok M., et al. Combating COVID-19 with tissue engineering: a review. Emerg Mater. 2021;4:329–349. doi: 10.1007/s42247-020-00138-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bailey A.L., Dmytrenko O., Greenberg L., Bredemeyer A.L., Ma P., Liu J., et al. SARS-CoV-2 infects human engineered heart tissues and models COVID-19 myocarditis. JACC: Basic Translati Sci. 2021;6:331–345. doi: 10.1016/j.jacbts.2021.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Probst C., Schneider S., Loskill P. High-throughput organ-on-a-chip systems: current status and remaining challenges. Curr Opin Biomed Eng. 2018;6:33–41. [Google Scholar]
- 88.Si L., Bai H., Rodas M., Cao W., Oh C.Y., Jiang A., et al. A human-airway-on-a-chip for the rapid identification of candidate antiviral therapeutics and prophylactics. Nat Biomed Eng. 2021;5:815–829. doi: 10.1038/s41551-021-00718-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhang M., Wang P., Luo R., Wang Y., Li Z., Guo Y., et al. Biomimetic human disease model of SARS-CoV-2-induced lung injury and immune responses on organ chip system. Adv Sci. 2021;8:2002928. doi: 10.1002/advs.202002928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Guo Y., Luo R., Wang Y., Deng P., Song T., Zhang M., et al. SARS-CoV-2 induced intestinal responses with a biomimetic human gut-on-chip. Sci Bull (Beijing) 2021;66:783–793. doi: 10.1016/j.scib.2020.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Thacker V.V., Sharma K., Dhar N., Mancini G.-F., Sordet-Dessimoz J., McKinney J.D. Rapid endotheliitis and vascular damage characterize SARS-CoV-2 infection in a human lung-on-chip model. EMBO Rep. 2021;22 doi: 10.15252/embr.202152744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Beilmann M., Boonen H., Czich A., Dear G., Hewitt P., Mow T., et al. Optimizing drug discovery by investigative toxicology: current and future trends. Altex. 2019;36:289–313. doi: 10.14573/altex.1808181. [DOI] [PubMed] [Google Scholar]
- 93.Migliozzi D., Pelz B., Dupouy D.G., Leblond A.-L., Soltermann A., Gijs M.A.M. Microfluidics–assisted multiplexed biomarker detection for in situ mapping of immune cells in tumor sections. Microsyst Nanoeng. 2019;5:59. doi: 10.1038/s41378-019-0104-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Halldorsson S., Lucumi E., Gómez-Sjöberg R., Fleming R.M.T. Advantages and challenges of microfluidic cell culture in polydimethylsiloxane devices. Biosens Bioelectron. 2015;63:218–231. doi: 10.1016/j.bios.2014.07.029. [DOI] [PubMed] [Google Scholar]
- 95.Berger E., Magliaro C., Paczia N., Monzel A.S., Antony P., Linster C.L., et al. Millifluidic culture improves human midbrain organoid vitality and differentiation. Lab Chip. 2018;18:3172–3183. doi: 10.1039/c8lc00206a. [DOI] [PubMed] [Google Scholar]
- 96.Clarke G.A., Hartse B.X., Niaraki Asli A.E., Taghavimehr M., Hashemi N., Abbasi Shirsavar M., et al. Advancement of sensor integrated organ-on-chip devices. Sensors (Basel, Switzerland) 2021;21:1367. doi: 10.3390/s21041367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhang Y.S., Aleman J., Shin S.R., Kilic T., Kim D., Mousavi Shaegh S.A., et al. Multisensor-integrated organs-on–chips platform for automated and continual in situ monitoring of organoid behaviors. Proc Natl Acad Sci USA. 2017;114:E2293. doi: 10.1073/pnas.1612906114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hasan A., Paul A., Memic A., Khademhosseini A. A multilayered microfluidic blood vessel-like structure. Biomed Microdevices. 2015;17:88. doi: 10.1007/s10544-015-9993-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.