Abstract
An accurate identification of poisonous mushrooms and the confirmation of the toxins involved are both of great importance in the treatment of mushroom poisoning incidents. In recent years, cases of mushroom poisoning by Inosperma spp. have been repeatedly reported from tropical Asia. It is urgent to know the real species diversity of Inosperma in this region. In the present study, we proposed two new Inosperma species from tropical Asia, namely I.muscarium and I.hainanense. They were described based on morphology and multilocus phylogeny. Detailed descriptions, color photographs and the discussion with other closely related species of the two new taxa were provided. In addition, a comprehensive muscarine determination of these two new species using ultrahigh performance liquid chromatography tandem mass spectrometry (UPLC-MS/MS) approach has been performed. Results showed that these two species were muscarine positive, with a content of 16.03 ± 1.23 g/kg in I.muscarium and a content of 11.87 ± 3.02 g/kg in I.hainanense, much higher than the known species I.virosum. Recovery of muscarine ranged from 93.45% to 97.25%, and the average recovery is 95.56%.
Keywords: Agaricales, muscarine, new species, phylogeny, taxonomy
Introduction
Muscarine C9H20NO2+, CAS number: 300–54–9, is a toxic alkaloid found in Inocybaceae, Clitocybe and several other mushroom genera (Patocka et al. 2021). The ingestion of muscarine-containing mushrooms would cause diaphoresis, salivation, urination, nausea, vomiting, gastrointestinal effects and muscular cramp, and fatal muscarinic syndromes like miosis, bronchoconstriction, and bradycardias in humans (Wilson 1947; Lurie et al. 2009; Chandrasekharan et al. 2020; Latha et al. 2020; Patocka et al. 2021), or even death (Pauli et al. 2005; Işıloğlu et al. 2009; Zosel et al. 2015). Many species of Inocybaceae are known to contain muscarine (Malone et al. 1962), especially in Inocybesensu stricto, and Pseudosperma (Kosentka et al. 2013; Matheny et al. 2020). Inosperma, a genus in Inocybaceae, is supposed to contain only a small number of muscarine positive species (Kosentka et al. 2013). However, mushroom poisoning events caused by Inosperma species were repeatedly reported from tropical Asia in recent years (Chandrasekharan et al. 2020; Li et al. 2021; Parnmen et al. 2021). Accordingly, it is urgent to enrich the knowledge of species diversity of the genus and to detect their muscarine toxin contents in tropical Asia.
Inosperma was erected as a subgenus of Inocybe with Inocybecalamistrata (Fr.) Gillet as type (Kühner 1980), and is now treated as genus rank (Matheny et al. 2020). Members in this genus are characterized by small to medium-sized basidiomata, rimose to scaly pileus, often rubescent context, phaseoliform to subglobose basidiospores, thin-walled cheilocystidia, lack of pleurocystidia, and often with distinctive odors. Inosperma species are widespread and there are seventy-one taxa documented globally (http://www.indexfungorum.org, retrieved 7 Oct. 2021). The tropical elements of Inosperma comprise several recently described, and still a few undescribed taxa, which were divided into two separate Old World tropical clades (Kropp et al. 2013; Matheny et al. 2020; Aïgnon et al. 2021; Deng et al. 2021). Interestingly, most of the taxa from Old World tropical clade 1 were mainly distributed in western Africa (Matheny et al. 2020; Aïgnon et al. 2021), and species in Old World tropical clade 2 were mainly from tropical Asia (Deng et al. 2021).
During our field works around the tropical China, two new Inosperma species were discovered. The present study aims to describe these two new tropical species using a combined data of morphology and phylogeny, and to determine their muscarine contents, in order to provide an accurate data for the prevention and clinical treatment of potential Inosperma poisoning accidents.
Materials and methods
Research area and specimens sampling
Our collections were made from Castanopsis dominated forests in Hainan, Guangdong Provinces, and Guangxi Zhuang Autonomous Region of China, with a tropical or subtropical climate. Specimens were photographed in the field using a digital camera and then described soon after collection. The specimens were dried through an electronic drier at 45 °C overnight, and were then preserved in plastic bags and sealed. After study, dried specimens were deposited in the Fungal Herbarium of Hainan Medical University (FHMU), Haikou City, Hainan Province of China, or in the Fungarium of Guangdong Institute of Microbiology (GDGM), Guangzhou, China.
Morphological study
Marcoscopic features were made from field notes and photographs. Color notations follow Kornerup and Wanscher (1978). Microscopic characters from dried materials mounted in KOH (5%) or mixed with Congo Red (1%) solution were observed with a microscope and photographed using a digital camera. Randomly selected twenty basidiospores and ten basidia for each specimen, the length and width of each basidiospore and basidium were measured, excluding the apiculus and sterigmata respectively (Kobayashi 2009). Numbers in square brackets [n/m/p] represent “n” basidiospores measured from “m” basidiomata of “p” specimens (Zhang et al. 2019). The dimensions of basidiospores and Q values are expressed as (a) b–c (d), “a” and “d” denote extreme values (“a” < 5th percentile; “d” > 95th percentile), while the ranges “b–c” means 5th to 95th percentile values. The quotient Q = length/width ratio for individual basidiospore, and Qm means the average of Q values (Dramani et al. 2020).
DNA extraction, PCR and sequencing
Genomic DNA was extracted from dried specimens using the NuClean Plant Genomic DNA kit (ComWin Biotech, Beijing). The following primers were used: ITS1F/ITS4 for ITS (Gardes and Bruns 1993), LR0R/LR7 for LSU (Vilgalys and Herster 1990), bRPB2-6F/bRPB2-7.1R for rpb2 (Matheny 2005). The volume of polymerase chain reaction (PCR) mixture solution was 25 μL, containing 9.5 μL dd H2O, 12.5 μL 2×Taq Plus MasterMix (Dye), 1 μL of each primer, and 1 μL of template DNA. PCR conditions for ITS, LSU and rpb2 followed Wang et al. (2021), that the conditions of PCR for three different gene regions are all the same as denaturation at 95 °C for 1 min at first, then followed by 35 cycles of denaturation at 95 °C for 30 s, annealing at 52 °C for 1 min, extension at 72 °C for 1 min, and a final extension at 72 °C for 8 min. Afterwards, the products of amplifications were sent to the Beijing Genomics Institute for purification and sequenced as soon as possible.
Analysis of sequence data
Sequences in this study were prepared and compared with closely related Inosperma sequences that were retrieved from GenBank (https://www.ncbi.nlm.nih.gov/) through BLAST tool (https://blast.ncbi.nlm.nih.gov/Blast.cgi) or literature survey (Larsson et al. 2009; Kropp et al. 2013; Horak et al. 2015; Nasser et al. 2017; Bau and Fan 2018; Matheny and Kudzma 2019; Matheny et al. 2020; Deng et al. 2021; Aïgnon et al. 2021; Cervini et al. 2021; Bandini et al. 2021). Then sequences from three genes were aligned respectively using MAFFT online service (https://mafft.cbrc.jp/alignment/server/) (Katoh et al. 2019) and were edited by BioEdit version 7.0.9.0 (Hall 1999). Two taxa in Auritella (A.hispida and A.spiculosa) were served as outgroups (Matheny et al. 2020). MrModeltest v2.3 was used to select the best-fit model for each gene partition for Bayes analysis (Nylander 2004). The datasets of each locus were combined in MEGA 5.02 (Tamura 2011). Maximum likelihood (ML) was inferred under partitioned models using W-IQ-TREE Web Service (http://iqtree.cibiv.univie.ac.at/), and the ultrafast bootstrapping was done with 1000 replicates (Trifinopoulos et al. 2016). Bayesian analysis was performed in MrBayes v.3.2.7a (Ronquist et al. 2012).
Muscarine toxin detection
Methods for sample preparation and analysis through UPLC-MS/MS were followed by Xu et al. (2020) with some modifications. Dried samples were ground to a fine power respectively, to 20 mg of each homogenised portion, 2 mL methanol-water solution (5:95 v/v) was added. The extraction was vortexed in a vortex mixer for 30 min, the mixture was further extracted by using an ultrasonic bath for another 30 min, and centrifuged for 5 min with 10000 rpm speed. Total supernatant was collected, using 0.22 μm organic filter membrane to filtrate for UPLC-MS/MS analysis and diluted with methanol-water (5:95, v/v) when necessary. The blank sample used here was Lentinulaedodes. The optimal MS parameters and product ion confirmation settings followed Xu et al. (2020), while the chromatographic column we used was ACQUITY UPLC BEH Amide (2.1 mm × 100 mm, 1.7 µm). The muscarine content was estimated in the mushroom extract by using standard muscarine (Sigma-Aldrich, Chemical purity ≥ 98%). The analytical results are reported as Mean ± SD g/kg, where Mean is the average content of muscarine in the mushroom from each experimental species, and SD represents its standard deviation.
Results
Phylogenetic inference
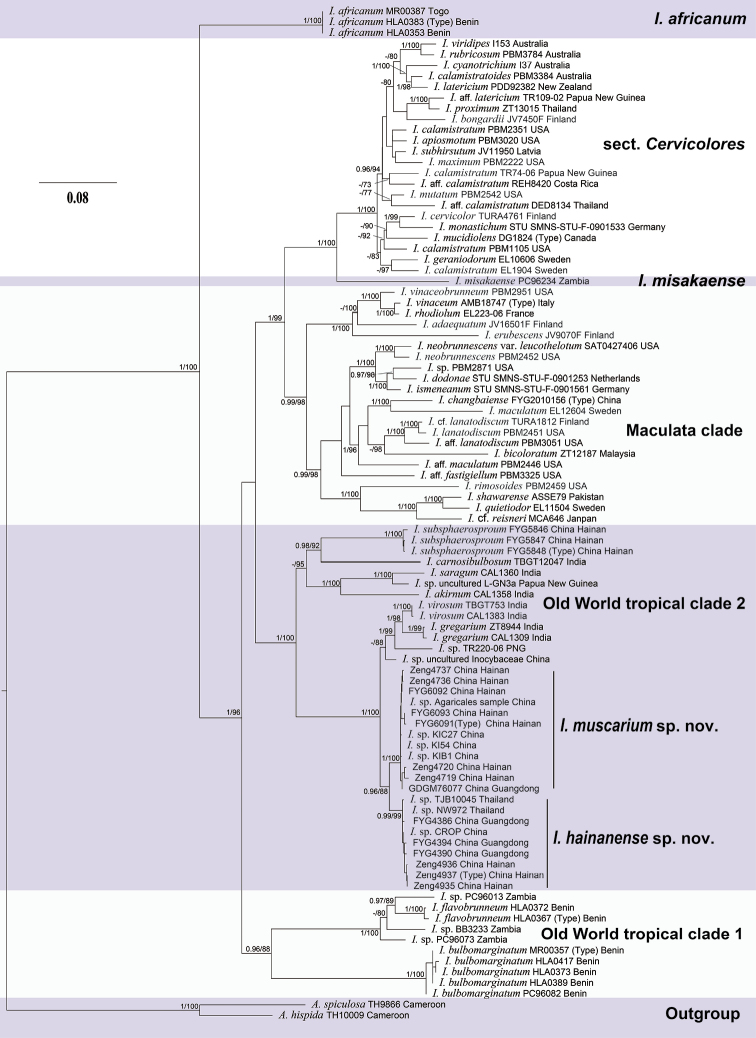
The final multilocus dataset (Table 1) includes 94 taxa and 3130 characters, and 37 new sequences (14 ITS, 12 LSU and 11 rpb2) were generated in this study and then submitted to GenBank. The alignment was deposited in TreeBase (28515). The best-fit models for each gene selected by MrModelGUI are GTR+I+G equally. The Maximum likelihood (ML) and Bayesian analyses for the combined dataset provide a best scoring tree is shown in Fig. 1. Three ectomycorrhizal samples (KIC27, KI54, and KIB1) and an environmental sample grouped together with eight specimens of I.muscarium with significant support (BP = 100%, PP = 1). In addition, two specimens (TJB10045 and NW972) from Thailand and an environmental sample (CROP denovo 1461) from China grouped together with six specimens of I.hainanense with high support (BP = 99%, PP = 0.99). The two new Inosperma species formed separate lineages and were sister with significant support (BP = 88%, PP = 0.96) to each other. These two new species formed a subclade in the Old World tropical clade 2. The subclade was sister to I.virosum (K.B. Vrinda, C.K. Pradeep, A.V. Joseph & T.K. Abraham ex C.K. Pradeep, K.B. Vrinda & Matheny) Matheny & Esteve-Rav., I.gregarium (K.P.D. Latha & Manimohan) Matheny & Esteve-Rav., and an undescribed specimen I. sp. (TR220-06) from Papua New Guinea with full support (BP = 100%, PP = 1).
Table 1.
Taxon sampling information and DNA sequences used for phylogenetic analyses
| Taxa | Collection number/Herbaium | Locality | GenBank accession number | Reference | ||
|---|---|---|---|---|---|---|
| ITS | LSU | rpb2 | ||||
| Auritellahispida | TH10009 | Cameroon | KT378203 | KT378207 | KT378215 | Matheny et al. (2020) |
| Auritellaspiculosa | TH9866 | Cameroon | KT378204 | KT378206 | KT378214 | Matheny et al. (2020) |
| Inospermaadaequatum | JV16501F | Finland | – | AY380364 | AY333771 | Matheny et al. (2020) |
| Inospermaaff.lanatodiscum | PBM3051 | USA | JQ801401 | JN975026 | JQ846485 | Pradeep et al. (2016) |
| Inospermaaff.calamistratum | DED8134 | Thailand | GQ892983 | GQ892937 | – | Pradeep et al. (2016) |
| Inospermaaff.calamistratum | REH8420 | Costa Rica | JQ801390 | JN975018 | JQ846471 | Pradeep et al. (2016) |
| Inospermaaff.fastigiellum | PBM3325 | USA | JQ801399 | JQ815419 | JQ846477 | Pradeep et al. (2016) |
| Inospermaaff.latericium | TR109-02 | Papua New Guinea | JQ801405 | JN975023 | JQ846487 | Pradeep et al. (2016) |
| Inospermaaff.maculatum | PBM2446 | USA | DQ241778 | AY745700 | EU569863 | Pradeep et al. (2016) |
| Inospermaafricanum | MR00387 | Togo | MN096189 | MN097881 | MT770739 | Aïgnon et al. (2021) |
| Inospermaafricanum | HLA0383 (Type) | Benin | MT534298 | MT560733 | – | Aïgnon et al. (2021) |
| Inospermaafricanum | HLA0353 | Benin | MT534299 | – | – | Aïgnon et al. (2021) |
| Inospermaakirnum | CAL1358 | India | KY440085 | KY549115 | KY553236 | Matheny et al. (2020) |
| Inospermaapiosmotum | PBM3020 | USA | JQ801385 | JN975021 | JQ846463 | Matheny et al. (2020) |
| Inospermabicoloratum | ZT12187 | Malaysia | GQ892984 | GQ892938 | JQ846464 | Pradeep et al. (2016) |
| Inospermabongardii | JV7450F | Finland | – | EU555448 | – | Pradeep et al. (2016) |
| Inospermabulbomarginatum | MR00357 (Type) | Benin | MN096190 | MN097882 | MN200775 | Aïgnon et al. (2021) |
| Inospermabulbomarginatum | HLA0417 | Benin | MT534300 | MT560734 | – | Aïgnon et al. (2021) |
| Inospermabulbomarginatum | HLA0373 | Benin | MT534301 | – | – | Aïgnon et al. (2021) |
| Inospermabulbomarginatum | HLA0389 | Benin | MT534302 | – | – | Aïgnon et al. (2021) |
| Inospermabulbomarginatum | PC96082 | Benin | JQ801412 | JN975027 | – | Aïgnon et al. (2021) |
| Inospermacalamistratoides | PBM3384 | Australia | JQ801393 | JQ815415 | KJ729949 | Pradeep et al. (2016) |
| Inospermacalamistratum | PBM1105 | USA | JQ801386 | JQ815409 | JQ846466 | Pradeep et al. (2016) |
| Inospermacalamistratum | EL1904 | Sweden | AM882938 | AM882938 | – | Pradeep et al. (2016) |
| Inospermacalamistratum | PBM2351 | USA | – | AY380368 | AY333764 | Pradeep et al. (2016) |
| Inospermacalamistratum | TR74-06 | Papua New Guinea | JQ801391 | JN975020 | JQ846472 | Pradeep et al. (2016) |
| Inospermacarnosibulbosum | TBGT12047 | India | KT329448 | KT329454 | KT329443 | Pradeep et al. (2016) |
| Inospermacervicolor | TURA4761 | Finland | JQ801395 | JQ815417 | JQ846474 | Pradeep et al. (2016) |
| Inospermacf.lanatodiscum | TURA1812 | Finland | JQ408763 | JQ319694 | JQ846484 | Pradeep et al. (2016) |
| Inospermacf.reisneri | MCA646 | Japan | – | EU555463 | – | Pradeep et al. (2016) |
| Inospermachangbaiense | FYG2010156 (Type) | China | MH047251 | MG844976 | MT086755 | Bau and Fan (2018) |
| Inospermacyanotrichium | I37 | Australia | JQ801396 | JN975033 | JQ846476 | Pradeep et al. (2016) |
| Inospermadodonae | SMNS-STU-F-0901253 | Netherlands | MW647615 | – | – | Bandini et al. (2021) |
| Inospermaerubescens | JV9070F | Finland | EU569846 | – | Pradeep et al. (2016) | |
| Inospermaflavobrunneum | HLA0372 | Benin | MT534290 | MT536756 | – | Aïgnon et al. (2021) |
| Inospermaflavobrunneum | HLA0367 (Type) | Benin | MN096199 | MT536754 | – | Aïgnon et al. (2021) |
| Inospermageraniodorum | EL10606 | Sweden | FN550945 | FN550945 | – | Pradeep et al. (2016) |
| Inospermagregarium | ZT8944 | India | – | EU600903 | EU600902 | Pradeep et al. (2016) |
| Inospermagregarium | CAL1309 | India | KX852305 | KX852306 | KX852307 | Latha and Manimohan. (2016) |
| Inospermahainanense | Zeng4936 | China | MZ374069 | MZ374760 | MZ388103 | The present study |
| Inospermahainanense | Zeng4937 (Type) | China | MZ374070 | MZ374761 | MZ388104 | The present study |
| Inospermahainanense | Zeng4935 | China | MZ374071 | MZ374762 | MZ388105 | The present study |
| Inospermahainanense | FYG4386 | China | MZ374072 | – | – | The present study |
| Inospermahainanense | FYG4390 | China | MZ374073 | MZ374763 | – | The present study |
| Inospermahainanense | FYG4394 | China | MZ374068 | – | – | The present study |
| Inospermaismeneanum | STU:SMNS-STU-F-0901561 | Germany | MW647625 | – | – | Bandini et al. (2021) |
| Inospermalanatodiscum | PBM2451 | USA | JQ408759 | JQ319690 | JQ846483 | Pradeep et al. (2016) |
| Inospermalatericium | PDD92382 | New Zealand | GU233367 | GU233413 | – | Pradeep et al. (2016) |
| Inospermamaculatum | EL12604 | Sweden | AM882964 | AM882964 | – | Pradeep et al. (2016) |
| Inospermamaximum | PBM2222 | USA | EU569854 | – | Pradeep et al. (2016) | |
| Inospermamisakaense | PC96234 | Zambia | JQ801409 | EU569875 | AY333767 | Pradeep et al. (2016) |
| Inospermamonastichum | STU:SMNS-STU-F-0901533 | Germany | MW647631 | – | – | Bandini et al. (2021) |
| Inospermamucidiolens | DG1824 (Type) | Canada | HQ201339 | HQ201340 | – | Pradeep et al. (2016) |
| Inospermamuscarium | Zeng4720 | China | MZ373978 | MZ373988 | MZ388089 | The present study |
| Inospermamuscarium | Zeng4736 | China | MZ373979 | MZ373989 | MZ388090 | The present study |
| Inospermamuscarium | Zeng4737 | China | MZ373980 | – | MZ388091 | The present study |
| Inospermamuscarium | Zeng4719 | China | MZ373981 | MZ373990 | MZ388092 | The present study |
| Inospermamuscarium | FYG6091 (Type) | China | MZ373982 | MZ373991 | MZ388093 | The present study |
| Inospermamuscarium | FYG6092 | China | MZ373983 | MZ373992 | MZ388094 | The present study |
| Inospermamuscarium | FYG6093 | China | MZ373984 | MZ373993 | MZ388095 | The present study |
| Inospermamuscarium | GDGM76077 | China | MZ520549 | MZ520550 | MZ542730 | The present study |
| Inospermaneobrunnescens | PBM2452 | USA | – | EU569868 | EU569867 | Pradeep et al. (2016) |
| Inospermaneobrunnescensvar.leucothelotum | SAT0427406 | USA | JQ801411 | JN975025 | JQ846489 | Pradeep et al. (2016) |
| Inospermaproximum | ZT13015 | Thailand | EU600839 | EU600840 | Matheny et al. (2020) | |
| Inospermaquietiodor | EL11504 | Sweden | AM882960 | AM882960 | Pradeep et al. (2016) | |
| Inospermarhodiolum | EL223-06 | France | FJ904175 | FJ904175 | Pradeep et al. (2016) | |
| Inospermarimosoides | PBM2459 | USA | DQ404391 | AY702014 | DQ385884 | Pradeep et al. (2016) |
| Inospermarubricosum | PBM3784 | Australia | KP308817 | KP170990 | KM406230 | Pradeep et al. (2016) |
| Inospermasaragum | CAL1360 | India | KY440103 | KY549133 | KY553249 | Latha and Manimohan (2017) |
| Inospermashawarense | ASSE79 | Pakistan | KY616964 | KY616966 | Naseer et al. (2018) | |
| Inosperma sp. | PBM2871 | USA | HQ201348 | HQ201348 | JQ846475 | Pradeep et al. (2016) |
| Inosperma sp. | BB3233 | Zambia | JQ801415 | EU600885 | Pradeep et al. (2016) | |
| Inosperma sp. | L-GN3a | Papua New Guinea | JX316732 | JX316732 | Pradeep et al. (2016) | |
| Inosperma sp. | TJB10045 | Thailand | KT600658 | KT600659 | KT600660 | Pradeep et al. (2016) |
| Inosperma sp. | TR22006 | Papua New Guinea | JQ801416 | JN975017 | JQ846496 | Pradeep et al. (2016) |
| Inosperma sp. | China | LS983441 | Unpublished | |||
| Inosperma sp. | CROP | China | MF532817 | Unpublished | ||
| Inosperma sp. | China | LS975930 | Unpublished | |||
| Inosperma sp. | NW972 | Thailand | MN492637 | Unpublished | ||
| Inosperma sp. | KIB1 | China | JX456867 | Unpublished | ||
| Inosperma sp. | KIC27 | China | JX456949 | Unpublished | ||
| Inosperma sp. | KI54 | China | JX456860 | Unpublished | ||
| Inosperma sp. | PC96013 | Zambia | JQ801383 | EU600883 | EU600882 | Pradeep et al. (2016) |
| Inosperma sp. | PC96073 | Zambia | JQ801417 | EU600870 | EU600869 | Pradeep et al. (2016) |
| Inospermasubhirsutum | JV11950 | Latvia | EU555452 | AY333763 | Pradeep et al. (2016) | |
| Inospermasubsphaerosproum | FYG5848 (Type) | China | MW403825 | MW397171 | MW404237 | Deng et al. (2021) |
| Inospermasubsphaerosproum | FYG5847 | China | MW403826 | MW397172 | MW404238 | Deng et al. (2021) |
| Inospermasubsphaerosproum | FYG5846 | China | MW403827 | MW397173 | MW404239 | Deng et al. (2021) |
| Inospermavinaceobrunneum | PBM2951 | USA | HQ201353 | JQ846478 | Pradeep et al. (2016) | |
| Inospermavinaceum | AMB18747 | Italy | MW561108 | MW561120 | Cervini et al. (2021) | |
| Inospermaviridipes | I153 | Australia | KP641646 | KP171095 | KM656139 | Pradeep et al. (2016) |
| Inospermavirosum | TBGT753 | India | KT329452 | KT329458 | KT329446 | Pradeep et al. (2016) |
| Inospermavirosum | CAL1383 | India | KY440108 | KY549138 | KY553253 | Latha and Manimohan (2017) |
Figure 1.
Phylogram generated by Bayesian Inference (BI) analyses based on sequences of a combined data set from nuclear genes (rDNA-ITS, nrLSU, and rpb2), rooted with Auritellahispida and A.spiculosa. Bayesian Inference posterior probabilities (BI-PP) ≥0.95 and ML bootstrap proportions (ML-BP) ≥70 are represented as BI-PP/ML-BP. I.muscarium sp. nov. and I.hainanense sp. nov. are two newly described taxa.
Taxonomy
. Inosperma muscarium
Y.G. Fan, L.S. Deng, W.J. Yu & N.K. Zeng sp. nov.
0B930071-9800-5B82-8872-66B1A6353BA0
MB840527
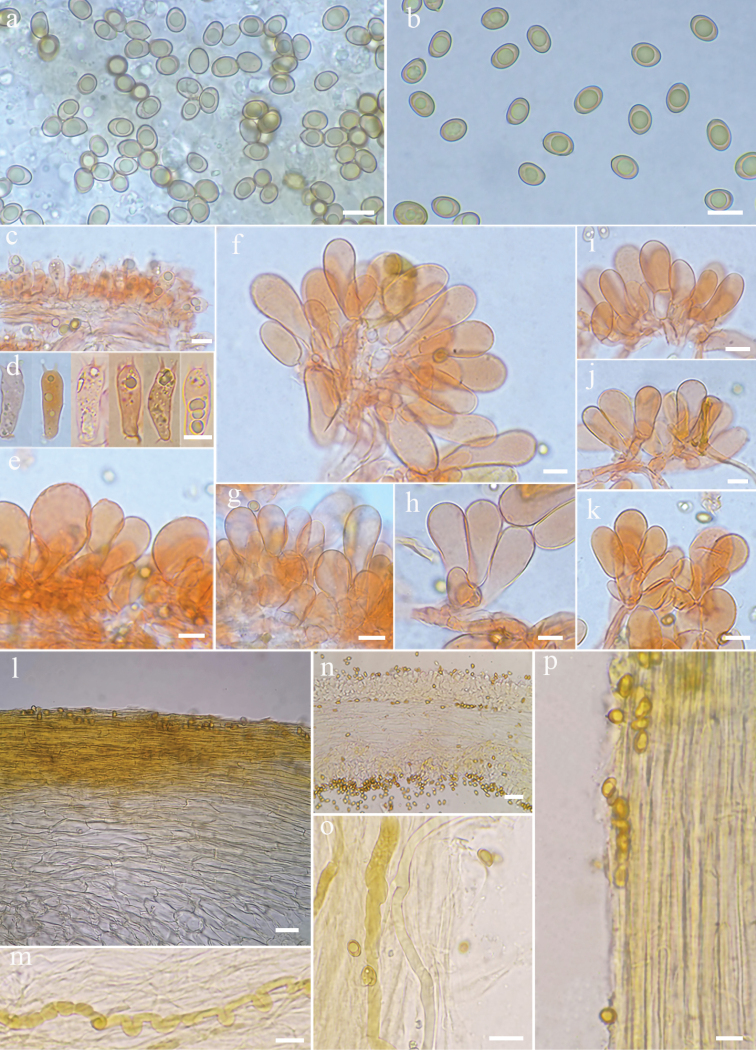
Figure 2.
Basidiomata of Inospermamuscariuma–e basidiomata f–h rimose to rimulose pileus i lamellae j–k lamellae edge l–m stipe surface. a–b, d, f–g, i–m FHMU3162 (holotype) c, e FYG6092 (FHMU3163) h FYG6093 (FHMU3164). Scale bars: 10 mm (a–m). Photos by Y.-G. Fan.
Figure 3.
Microscopic features of Inospermamuscarium (FHMU3162, holotype) a–b basidiospores c–d basidia e–h cheilocystidia in clusters i oleiferous hyphae j pileipellis and pileal trama k terminal hyphae at the stipe apex l hymenophoral trama m stipitipellis and stipe trama. Scale bars: 10 μm (a–m). Photos by L.-S. Deng
Etymology.
“muscarium” refers to its high content of muscarine.
Holotype. China, Hainan Province, Ledong Li Autonomous County, Yinggeling substation of Hainan Tropical Rainforest National Park, under Castanopsis forest, at 19°1'20"N, 109°23'33"E, alt. 550 m, 26 April 2021, FYG6091 (FHMU3162), GenBank accession number: ITS (MZ373982); LSU (MZ373991) and rpb2 (MZ388093).
Diagnosis.
Basidiomata small to medium-sized. Pileus rimulose to rimose with an indistinct umbo, lamellae rather crowded. Basidiospores smooth, enlongate ellipsoid to ellipsoid. Cheilocystidia clavate. Under Castanopsis forest. Differs from I.hainanense by its more robust habit, elongate basidiospores, and narrower cheilocystidia.
Basidiomata.
small to medium-sized. Pileus 25–60 mm diam., conical convex to convex when young, becoming broadly convex to plano-convex with a small indistinct umbo when mature, margin slightly incurved when young, becoming somewhat reflexed with age. Surface dry, smooth with distinct ivory white (5A1) veil layer around the disc when young, then appressed with indistinct veil remnants, fibrillose-rimulose elsewhere, margin usually strongly rimose with age; yellowish brown (5D8) to chocolate brown (5E8) around the center and on the fibrils, yellowish brown (5C6) elsewhere, yellowish brown (6C6) to slightly dark brown (6E7) all over the basidiomata when overmatured. Lamellae rather crowded, adnexed, initially pure white to pale off-white (4B1), becoming grayish white (5B1) to yellowish white (4A2), dirty yellow (4A3) to yellowish brown (5B4) when overmatured, 1.5–3 mm wide, edge fimbriate, faint serrate to somewhat wavy. Stipe 35–72 × 3–8 mm, central, solid, terete, equal with a slightly swollen apex and base; with sparse fibrils at apex, longitudinally fibrillose downwards the stipe, with white tomentose hyphae at the base; initially white (5A1) to cream white(3A2), yellowish (4A3) or brownish (5A3) with age, brown (5B6) to dark brown (5C5) when old. Context solid, fleshy in pileus, 0.5–1 mm thick at mid-radius, 1.5–4.5 mm under the umbo, white to ivory white (5A1) at first, becoming brownish white (5B2); fibrillose and striate in the stipe, white to yellowish (4A2) or flesh color (4B3). Odor fungoid, slightly grassy or mild.
Basidiospores.
[180/9/9] 8–10(11) × 5–6 (6.5) μm, Q = (1.15)1.42–1.86(2.00), Qm=1.63, mostly ellipsoid to enlongate ellipsoid, occasionally sub-phaseoliform, smooth, thick-walled, yellowish, apiculus small, indistinct, with a spherical to ellipsoid yellowish brown oil-droplet inside. Basidia 17–24 × 7–9 μm, clavate to broadly clavate, obtuse at apex, slightly tapering towards the base, 4-spored, sterigmata 2–4 μm in length, thin-walled, hyaline or pale yellow, with oily drops in various sizes with age. Pleurocystidia none. Lamella edge sterile. Cheilocystidia 36–50 × 9–14 μm, abundant and crowded, mostly clavate, broadly clavate to enlongate-clavate, rarely balloon-shaped, apices rounded to obtuse, or occasionally subcapitate, thin- to slightly thick-walled, septate, often constricted at septa, colorless to yellowish, sometimes with golden yellow inclusions. Hymenophoral trama 75–108 μm thick, sub-regular, colorless to yellowish, composed of thin-walled, smooth, cylindric to mostly inflated, hyphae 12–25 μm wide, somewhat constricted at the both ends of per hyphae. Pileipellis a cutis, sub-regular, composed of thin-walled, brown to yellowish brown, cylindrical, slightly encrusted hyphae 4–10 μm wide. Pileal trama colorless, regular to subregular, hyphae 12–25 μm wide. Stipitipellis a cutis, regularly arranged, occasionally with small clusters of terminal cheilocystidoid cells at the stipe apex, cheilocystidoid cells 31–47 × 9–10 μm, rare, clavate to enlongate clavate, hyaline or pale yellow, thin- to slightly thick-walled, some with golden yellow inclusions. Caulocystidia not observed. Oleiferous hyphae 4–13 μm wide, scattered in pileus and stipe tramal tissue, yellow or bright golden yellow, smooth, often bent, sometimes diverticulate. Clamp connections present, common in all tissues.
Habitat.
Gregarious in clusters, usually scattered with numerous clusters under Castanopsis forest, late March to August in tropical China.
Known distribution.
China (Hainan, Guangdong, Guangxi), Thailand.
Additional materials examined.
China. Hainan Province, Ledong Li Autonomous County, Yinggeling substation of Hainan Tropical Rainforest National Forest Park, under Castanopsis forest, 13 August 2020, N.K. Zeng, Zeng4720 (FHMU3158); Same location, under Castanopsis forest, 14 August 2020, N.K. Zeng Zeng4736 (FHMU3159); Zeng4737 (FHMU3160), Same location, 26 April 2021, Y.G. Fan, L.S. Deng & Q.Q. Chen, FYG6092 (FHMU3163); FYG6093 (FHMU3164); FYG6094 (FHMU3173); Guangdong Province, Yangchun City, Gangmei Town, Lunshui Village, under Castanopsis forest, 29 March 2019, W.Y. Huang, GDGM76077; Guangxi Zhuang Autonomous Region: Wuzhou City, Cangwu Country, Wangfu Town, 23°40'28"N, 111°29'6"E, alt. 30 m, Under Castanopsis dominated forest, 29 May 2021, L.L. Qi, WSW10286, (FHMU3174).
. Inosperma hainanense
Y.G. Fan, L.S. Deng, W.J. Yu & N.K. Zeng sp. nov.
BAF0240A-9583-5E18-B038-28AD3D315F33
MB840528
Figure 4.
Basidiomata of Inospermahainanensea–e basidiomata f–g rimose to rimulose pileus h lamellae i lamellae edge j–k stipe surface. c FHMU3166 (holotype) a–b, d–g, i–k FHMU6511 h FHMU3168. Scale bars: 10 mm (a–k). a–b, d–k: photos by L.-S. Deng; c: photos by N.-K. Zeng
Figure 5.
Microscopic features of Inospermahainanense (FHMU3166, holotype) a–b basidiospores c–d basidia e–k cheilocystidia in clusters l pileipellis and pileal trama n hymenophoral trama m, o oleiferous hyphae p stipitipellis and stipe trama. Scale bars: 10 μm (a–k). Photos by L.-S. Deng
Etymology.
“hainanense” refers to the its type locality.
Holotype. China, Hainan Province, Changjiang Li Autonomous County, Bawangling substation of Hainan Tropical Rainforest National Park, under Castanopsis dominated forest, at 19°7'12.43"N, 109°7'6.29"E, alt. 630 m, 2 September, 2020, N.K. Zeng, Zeng4937 (FHMU3166), GenBank accession number: ITS (MZ374070); LSU (MZ374761) and rpb2 (MZ388104).
Diagnosis.
Distinguishes from I.muscarium by its slender basidiomata, ellipsoid to ovoid basidiospores, and mostly vesiculose cheilocystidia.
Basidiomata.
small to medium-sized. Pileus 25–53 mm diam., conical to convex at young age, becoming applanate to uplifted with age, with a broad to subacute umbo, margin initially decurved, straight to somewhat wavy when mature; surface dry, smooth when young, fibrillose-rimulose elsewhere, strongly rimose towards the margin with age; chocolate brown (5D8) to somewhat dark brown (5F7) around the disc, straw yellow (4A6) to yellowish brown (4B5) elsewhere, background pallid to cream white (4B1), becoming brown (5B4) to dark brown (5C6) with age; Lamellae rather crowded, adnexed, initially ivory white (5A1) to grayish white (5B2), becoming dirty yellowish (5B5) to brownish (5C7) when matured, completely brown (5D6) after drying, 2–3 mm in width, edge fimbriate, slightly serrate. Stipe 40–72 × 3–5 mm, central, nearly terete, equal with a slightly swollen apex, base somewhat swollen; nearly smooth and longitudinally striate all over the stipe; initially ivory (5A1) to yellowish white (5A2) at the upper half, yellowish to brownish (4B5) downwards, becoming uniformly yellowish brown (4B7) to brown (4C7) with age. Context solid, fleshy in pileus, white to grayish white (4B1), pale brown under the umbo (4B2), 1–2 mm thick at mid-radius, 4–5 mm thick under the umbo, fibrillose in stipe, pallid to yellowish (4A2) or brownish (4B2), striate, shiny. Odor indistinct or slightly acid.
Basidiospores.
[180/9/9] 8–9(10.5) × 5–7 μm, Q = (1.18)1.28–1.64 (1.78), Qm = 1.43, mostly ellipsoid to ovoid, occasionally subphaseoliform, smooth, slightly thick-walled, brown to yellowish brown, apiculus small, indistinct, with a spherical to ellipsoid yellowish brown oil-droplet. Basidia 21–28 × 6–9 μm, clavate, often obtuse at apex, slightly tapered towards the base, thin-walled, 4-spored, sometimes 2-spored, sterigmata 4–6 μm in length, with spherical yellowish brown to golden yellow brown oily inclusions. Pleurocystidia absent. Lamella edge sterile. Cheilocystidia 34–55 × 15–25 μm, abundant and crowded, mostly obovoid to balloon-shaped, occasionally broadly clavate, rarely enlongate-clavate, thin- to slightly thick-walled (up to 1 μm thick); often rounded or slightly obtuse at apex, colorless to pale yellow, sometimes with golden yellow pigments. Hymenophoral trama 75–138 μm thick, sub-regular, hyaline to slightly yellow, composed of cylindric to inflated hyphae 20–33 μm wide, slightly constricted at septa. Pileipellis a cutis, hyphae 2.5–10 μm wide, thin-walled, pale yellow to yellowish brown, cylindrical, sometimes slightly encrusted. Pileal trama regular to subregular, hyphae 12–30 μm wide, thin-walled, colorless. Stipitipellis a cutis, regularly arranged, walls yellowish to bright yellow. Oleiferous hyphae 2.5–10 μm wide, commonly scattered in pileus and stipe tramal tissues, straw yellow or bright golden yellow, smooth, often bent or diverticulate. Clamp connections observed in all tissues.
Habitat.
Scattered or gregarious in small clusters under Castanopsis dominated forest, June to September in tropical China.
Known distribution.
China (Hainan, Guangdong).
Additional materials examined.
China. Hainan Province, Wuzhishan City, Maoyang Town, Maoyang Village, 11 August 2021, Y.G. Fan & L.S. Deng, FYG6440 (FHMU6513); Ganshiling Provincial Nature Reserve, L.S. Deng & Y.G. Fan, DLS0043 (FHMU6512); Changjiang Li Autonomous County, Bawangling substation of Hainan Tropical Rainforest National Park, under Castanopsis dominated forest, 2 September 2020, N.K. Zeng, Zeng4936 (FHMU3165); Zeng4935 (FHMU3167); Guangdong Province, Guangzhou City, Tianluhu Forest Park, 2 June 2019, Y.G. Fan & W.J. Yu, FYG4386 (FHMU3168); Shaoguan City, Danxiashan Nature Reserve, 4 June 2019, Y.G. Fan & W.J. Yu, FYG4388 (FHMU3175); 4390 (FHMU3169); FYG4394 (FHMU3170).
Muscarine detection
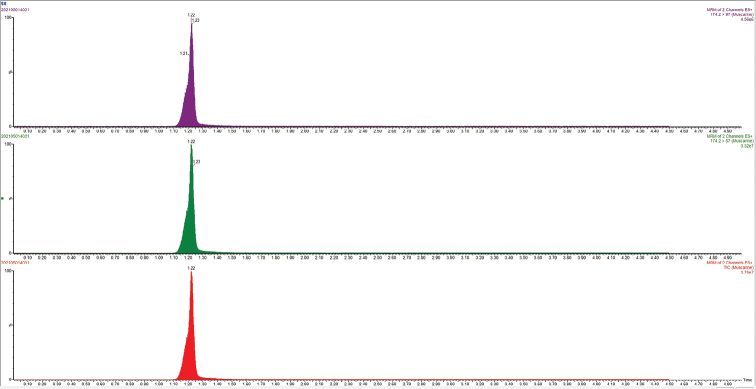
Representative chromatograms of muscarine were shown in Fig. 8. The muscarine toxin content was confirmed by linear equation according to the analysis of UPLC-MS/MS, it was found that both of the two new species contained muscarine toxin, and the content of Inospermamuscarium was 16.03 ± 1.23 g/kg while I.hainanense was 11.87 ± 3.02 g/kg. Muscarine was identified by comparing retention time (1.22 min) and relative deviation (0.82%) in the allowable relative range of 25 % base on the qualitative analysis. The calibration curve for muscarine generated during the validation was y = 2083.17 x–209.297 (r = 0.9988) for muscarine concentration in the range of 2–200 ng/mL (y represents the peak area, and x is muscarine concentration, r is correlation coefficient). Recovery of muscarine ranged from 93.45% to 97.25%, and the average recovery was 95.56%.
Figure 8.
Representative chromatograms of muscarine.
Discussion
New species delimitation
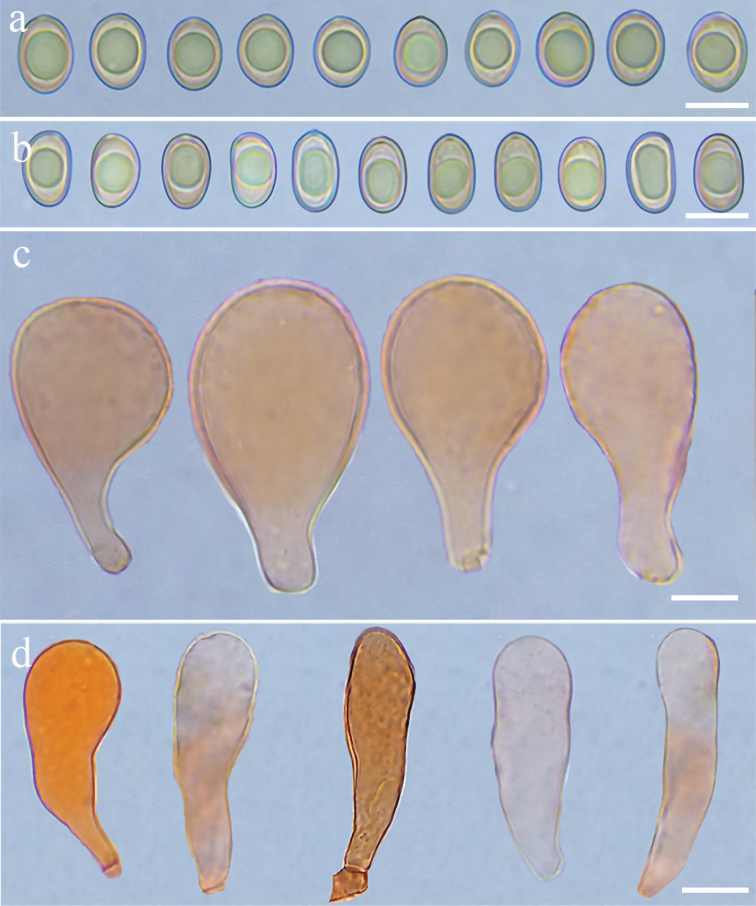
The phylogenetic results place both the two new species in the Old World tropical clade 2 in genus Inosperma (Kropp et al. 2013; Pradeep et al. 2016; Deng et al. 2021), and they are sister to each other with significant support (BP = 88%, PP = 0.96). Morphologically, they share yellowish brown pileus, longitudinally striate stipe, crowded lamellae, and elliptic basidiospores. It is really difficult to distinguish the two new species by their macromorphology, in spite of the fact that I.hainanense has a relatively more slender habit, more finely rimulose in pileus, and a smoother stipe surface. However, they could be easily distinguished by their outlines of basidiospores and cheilocystidia. As is shown in Figs 6–7, I.muscarium has more elongated basidiospores in outline, as well as narrower cheilocystidia (I.muscarium: 36–50 × 9–14 μm; I.hainanense: 34–55 × 15–25 μm).
Figure 6.
The comparisons of the two new species in their outline of basidiospores and cheilocystidia shape a, c basidiospores and cheilocystidia of I.hainanense (FHMU3162, holotype); b, d Basidiospores and cheilocystidia of I.muscarium (FHMU3166, holotype). Scale bars: 10 μm (a–d). Photos by L.-S. Deng
Figure 7.
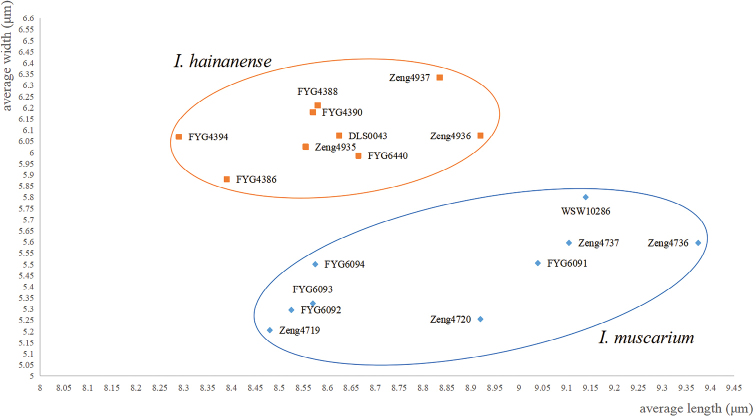
The comparisons of the two new species in their dimensions of basidiospores.
In Old World tropical clade 2, I.gregarium and I.virosum, both of which described from India, formed a sister lineage with the two new species. They also share fibrillose-rimose pileus, longitudinally striate stipe, crowded lamellae, and elliptic basidiospores (Vrinda et al. 1996; Latha and Manimohan 2016). However, I.gregarium differs from the two new species by its smaller basidiospores (7–8.5 × 5–5.5 μm, Q = 1.3–1.8, Qm = 1.6), versiform and longer cheilocystidia (24–60 × 16–24 µm), the presence of caulocystidia, and an association with Dipterocarpaceae trees (Latha and Manimohan 2016). Inospermavirosum differs in having smaller basidiospores (6.5–8.5 × 5–6 µm, Q = 1.3–1.6, Qm = 1.4), and an association also with Dipterocarpaceae trees (Vrinda et al. 1996; Latha and Manimohan 2017). The remaining species in this subgrouping resemble the two new species to some extent; however, they have appressed-scaly or appressed-fibrillose pileus and different phylogenetic positions (Latha and Manimohan 2017).
There are eight described species in Old World tropical clade 2 so far, three of which were described from China in Fagaceae forest (Deng et al. 2021), and the rest five species were all described from India under Dipterocarpaceae forest or among ginger plants (Pradeep et al. 2016; Latha and Manimohan 2017). By our current knowledge, members in this subgrouping usually have medium-sized basidiomata, gregarious habit, appressed-scaly or fibrillose-rimose pileus, rather crowded lamellae, longitudinally striate stipe, non-changing context, subglobose to elliptic basidiospores, and the lack of distinctive odors (Pradeep et al. 2016; Latha and Manimohan 2017; Deng et al. 2021).
Muscarine toxin in Inosperma
The compound muscarine was initially isolated and identified from Amanitamuscaria with the content at about 0.0003% of the fresh weight (Spoerke and Rumack 1994). However, muscarine was more commonly found in Inocybaceae and Clitocybe spp. with significant concentrations reached the highest record of 1.6%. (Lurie et al. 2009). Many Inocybaceae species were well known to contain muscarine (Peredy et al. 2014; Patocka et al. 2021), and various methods have been used to detect this toxin in the past years (Fahrig 1920; Eugster 1957; Brown et al. 1962; Robbers 1964; Kosentka et al. 2013; Latha et al. 2020). Five Inosperma species were reported as muscarine positive, including I.cervicolor (Pers.) Matheny & Esteve-Rav., I.erubescens (A. Blytt) Matheny & Esteve-Rav., I.maculatum (Boud.) Matheny & Esteve-Rav., I.vinaceobrunneum (Matheny, Ovrebo & Kudzma) Haelew. and I.virosum (K.B. Vrinda, C.K. Pradeep, A.V. Joseph & T.K. Abraham ex C.K. Pradeep, K.B. Vrinda & Matheny) Matheny & Esteve-Rav. (Kosentka et al. 2013; Latha et al. 2020). In addition, I.carnosibulbosum (C.K. Pradeep & Matheny) Matheny & Esteve-Rav., a species described from India, is probably a muscarine positive species due to a recent report of poisonous case (Chandrasekharan et al. 2020). Among these muscarine positive species in Inosperma, I.virosum described from India, is more extensively studied in toxin detection, toxicity in vitro using NCM460 colon epithelial cell line, toxic effects in vivo and pharmacokinetics of muscarine (Latha et al. 2020). The muscarine content of I.virosum is 270 or 300 mg/kg reported by separate studies (Sailatha et al. 2014; Latha et al. 2020).
Surprisingly, of the two new species we assayed, both of them have a high content of muscarine that is about 30 to 50 times higher than I.virosum (Sailatha et al. 2014; Latha et al. 2020). For humans, a lethal dose of muscarine is estimated from 40 mg to 495 mg (Pauli et al. 2005). Based on the muscarine concentrations of between 0.1% to 0.33% (dry weight) in Inocybaceae spp., a single mushroom can be lethal (Puschner 2018; Patocka et al. 2021). Consequently, the two new species proposed by the present study were considered to be more dangerous when mistakenly ingested by humans. In particular, for I.muscarium, a species often with a medium-sized basidiomata, a gregarious, large, discrete clusters habitat, and the lack of aposematic coloration make it extremely easily collected by local people as an edible mushroom. The publicity and education of the two new species were essential to prevent mushroom poisoning from tropical areas where they distributed.
The accurate identification of poisonous mushrooms and the knowledge of toxin type and contents are crucial for the treatment of mushroom poisoning patients (Li et al. 2021). However, species identification can usually be difficult for doctors when faced with mushroom-poisoned patients, mainly because of the insufficient identification data of wild poisoning mushrooms (Hall et al. 1987). Our present study provides detailed knowledge for a better prevention of potential Inosperma poisoning from tropical Asia.
Supplementary Material
Acknowledgements
The authors thank Dr. Shuai Jiang, Mr. Yongqing Fu (Hainan tropical rainforest National Park, China) and Mr. Weiyong Huang (Yangchun Center for Disease Control and Prevention, China) for their kind help in field work, and to Dr. Junqing Yan (Jiangxi Agricultural University, China) and Dr. Yupeng Ge (Ludong University, China) for their kind help in the phylogenetic analysis. This work was supported by the National Natural Science Foundation of China (Grant Nos. 31860009 & 31400024), Hainan Basic and applied research project for cultivating high level talents (2019RC230), The Innovative Research Projects for Graduate Students in Hainan Medical University, Hainan China (HYYS2020-42), and Jilin Provincial Foundation for Excellent Scholars (20180520035JH). We also thank the anonymous reviewers for their corrections and suggestions to improve our work.
Citation
Deng L-S, Kang R, Zeng N-K, Yu W-J, Chang C, Xu F, Deng W-Q, Qi L-L, Zhou Y-L, Fan Y-G (2021) Two new Inosperma (Inocybaceae) species with unexpected muscarine contents from tropical China. MycoKeys 85: 87–108. https://doi.org/10.3897/mycokeys.85.71957
References
- Aïgnon HL, Jabeen S, Naseer A, Yorou NS, Ryberg M. (2021) Three new species of Inosperma (Agaricales, Inocybaceae) from Tropical Africa. MycoKeys 77(1): 97–116. 10.3897/mycokeys.77.60084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandini D, Oertel B, Eberhardt U. (2021) Even more fibre-caps (2): Thirteen new species of the family Inocybaceae. Mycologia Bavarica 21: 27–98. [Google Scholar]
- Bau T, Fan YG. (2018) Three new species of Inocybesect.Rimosae from China. Mycosystema 37(6): 693–702. 10.13346/j.mycosystema.180033. [DOI] [Google Scholar]
- Brown J, Malone M, Stuntz D, Tyler V. (1962) Paper chromatographic determination of muscarine in Inocybe species. Journal of Pharmaceutical Sciences 51(9): 853–856. 10.1002/jps.2600510908. [DOI] [PubMed] [Google Scholar]
- Cervini M, Carbone M, Bizio E. (2021) Inospermavinaceum, una nuova specie distinta da I.rhodiolum e I.adaequatum. Rivista di Micologia 63(3): 215–241. [Google Scholar]
- Chandrasekharan B, Pradeep C, Vrinda B. (2020) Inocybe poisoning from Kerala– a case study. Journal of Mycopathological Research 57(4): 255–258. [Google Scholar]
- Deng LS, Yu WJ, Zeng NK, Liu LJ, Liu LY, Fan YG. (2021) Inospermasubsphaerosporum (Inocybaceae), a new species from Hainan, tropical China. Phytotaxa 502(2): 169–178. 10.11646/phytotaxa.502.2.5. [DOI] [Google Scholar]
- Dramani R, Hegbe ADMT, Tabe A, Badou AS, Furneaux B, Ryberg M, Yorou NS. (2020) How are basidiospore size measurements affected by drying? Current Research in Environmental and Applied Mycology 10(1): 63–70. 10.5943/cream/10/1/7 [DOI]
- Eugster CH. (1957) Isolierung von muscarin aus Inocybe Patouillardi (Bres.) 4. Mitteilung ber Muscarin. Helvetica Chimica Acta 40(4): 886–887. 10.1002/hlca.19570400403. [DOI] [Google Scholar]
- Fahrig C. (1920) Über die Vergiftung durch pilze aus der gattung Inocybe (Rißpilze und Faserköpfe). Archiv Für Experimentelle Pathologie Und Pharmakologie 88(5): 227–246. 10.1007/BF01864886. [DOI] [Google Scholar]
- Gardes M, Bruns TD. (1993) ITS primers with enhanced specificity for basidiomycetes – application to the identification of mycorrhizae and rusts. Molecular Ecology 2(2): 113–118. 10.1111/j.1365-294X.1993.tb00005.x [DOI] [PubMed] [Google Scholar]
- Hall A, Spoerke D, Rumack B. (1987) Mushroom poisoning: identification, diagnosis, and treatment. Pediatrics in review / American Academy of Pediatrics 8(10): 291–298. 10.1542/pir.8-10-291. [DOI] [PubMed] [Google Scholar]
- Hall TA. (1999) BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 41(41): 95–98. [Google Scholar]
- Horak E, Matheny PB, Desjardin DE, Soytong K. (2015) The genus Inocybe (Inocybaceae, Agaricales, Basidiomycota) in Thailand and Malaysia. Phytotaxa 230(3): 201–238. 10.11646/phytotaxa.230.3.1. [DOI] [Google Scholar]
- Işiloğlu M, Helfer S, Alli H, Yilmaz F. (2009) A fatal Inocybe (Fr.) Fr. poisoning in mediterranean Turkey. Turkish Journal of Botany 33(1): 71–73. 10.3906/bot-0605-2. [DOI] [Google Scholar]
- Katoh K, Rozewicki J, Yamada KD. (2019) MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Briefings in Bioinformatics 20(4): 1160–1166. 10.1093/bib/bbx108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T. (2009) Notes on the genus Inocybe of Japan. IV. Species having metuloids collected from Hokkaido, Honshu, and Kyushu. Mycoscience 50(3): 203–211. 10.1007/s10267-008-0472-y [DOI] [Google Scholar]
- Kornerup A, Wanscher JH. (1978) The methuen handbook of colour 3rd edn. Eyre Methuen Ltd. Reprint, London, 252 pp. [Google Scholar]
- Kosentka P, Sprague S, Ryberg M, Gartz J, May A, Campagna S, Matheny P. (2013) Evolution of the toxins muscarine and psilocybin in a family of mushroom-forming fungi. PLoS ONE 8(5): e64646. 10.1371/journal.pone.0064646. [DOI] [PMC free article] [PubMed]
- Kropp BR, Matheny PB, Hutchison LJ. (2013) InocybesectionRimosae in Utah: phylogenetic affinities and new species. Mycologia 105(3): 728–747. 10.3852/12-185. [DOI] [PubMed] [Google Scholar]
- Kühner R. (1980) Les Hyménomycètesagaricoïdes (Agaricales, Tricholomatales, Plutéales, Russulales). Etude générale et classification. Bulletin de la Société Linnéenne de Lyon 50(4): 1–1927. [Google Scholar]
- Larsson E, Ryberg M, Moreau PA, Mathiesen AD, Jacobsson S. (2009) Taxonomy and evolutionary relationships within species of section Rimosae (Inocybe) based on ITS, LSU and mtSSU sequence data. Persoonia 23(1): 86–98. 10.3767/003158509X475913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latha KPD, Manimohan P. (2017) Inocybes of Kerala. SporePrint Books, Calicut, 181 pp. [Google Scholar]
- Latha KPD, Manimohan P. (2016) Inocybegregaria, a new species of the Inosperma clade from tropical India. Phytotaxa 286(2): 107–115. 10.11646/phytotaxa.286.2.5. [DOI] [Google Scholar]
- Latha S, Shivanna N, Naika M, Anilakumar K, Kaul A, Mittal G. (2020) Toxic metabolite profiling of Inocybevirosa. Scientific Reports 10(1): 13669. 10.1038/s41598-020-70196-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HJ, Zhang HS, Zhang YZ, Zhou J, Yin Y, He Q, Jiang SF, Ma PB, Zhang YT, Wen K, Yuan Y, Lang N, Cheng BW, Lu JJ, Sun CY. (2021) Mushroom poisoning outbreaks – China, 2020. China CDC Weekly 3(3): 41–45. 10.46234/ccdcw2021.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lurie Y, Wasser S, Taha M, Shehade H, Nijim J, Hoffmann Y, Basis F, Vardi M, Lavon O, Suaed S, Bisharat B, Bentur Y. (2009) Mushroom poisoning from species of genus Inocybe (fiber head mushroom): A case series with exact species identification Inocybe mushroom poisoning. Clinical Toxicology 47(6): 562–565. 10.1080/15563650903008448. [DOI] [PubMed] [Google Scholar]
- Malone MH, Robichaud RC, Tyler Jr V, Brady LR. (1962) Relative muscarinic content of thirty Inocybe species. Lloydia 25: 231–237. [Google Scholar]
- Matheny PB. (2005) Improving phylogenetic inference of mushrooms with RPB1 and RPB2 nucleotide sequences (Inocybe, Agaricales). Molecular Phylogenetics and Evolution 35(1): 1–20. 10.1016/j.ympev.2004.11.014 [DOI] [PubMed] [Google Scholar]
- Matheny PB, Hobbs AM, Esteve-Raventós F. (2020) Genera of Inocybaceae: New skin for the old ceremony. Mycologia 112(1): 83–120. 10.1080/00275514.2019.1668906 [DOI] [PubMed] [Google Scholar]
- Matheny PB, Kudzma LV. (2019) New species of Inocybe (Inocybaceae) from eastern North America. The Journal of the Torrey Botanical Society 146(3): 213–235. 10.3159/TORREY-D-18-00060.1 [DOI] [Google Scholar]
- Naseer A, Khalid AN, Smith ME. (2017) Inocybeshawarensis sp. nov. in the Inosperma clade from Pakistan. Mycotaxon 132(4): 909–918. 10.5248/132.909 [DOI] [Google Scholar]
- Nylander J. (2004) MrModeltest V2. program distributed by the author. Bioinformatics 24: 581–583. 10.1093/bioinformatics/btm388 [DOI] [Google Scholar]
- Parnmen S, Nooron N, Leudang S, Sikaphan S, Polputpisatkul D, Pringsulaka O, Binchai S, Rangsiruji A. (2021) Foodborne illness caused by muscarine-containing mushrooms and identification of mushroom remnants using phylogenetics and LC-MS/MS. Food Control 128(4): 108182. 10.1016/j.foodcont.2021.108182. [DOI] [Google Scholar]
- Patocka J, Wu R, Nepovimova E, Valis M, Wu W, Kuca K. (2021) Chemistry and toxicology of major bioactive substances in Inocybe mushrooms. International Journal of Molecular Sciences 22(4): 2218. 10.3390/ijms22042218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli J, Foot C. (2005) Fatal muscarinic syndrome after eating wild mushrooms. The Medical Journal of Australia 182(6): 294–295. 10.5694/j.1326-5377.2005.tb06705.x. [DOI] [PubMed] [Google Scholar]
- Peredy T, Bradford H. (2014) Mushrooms, Muscarine. Encyclopedia of Toxicology 3: 416–417. 10.1016/B978-0-12-386454-3.00758-2. [DOI] [Google Scholar]
- Pradeep CK, Vrinda KB, Varghese SP, Korotkin HB, Matheny PB. (2016) New and noteworthy species of Inocybe (Agaricales) from tropical India. Mycological Progress 15(3): 1–25. 10.1007/s11557-016-1174-z. [DOI] [Google Scholar]
- Puschner B. (2018) Mushroom Toxins. In: Gupta RC. (Ed.) Veterinary toxicology: basic and clinical principles: 3rd edn.Acdemic Press, Hopkinsville, KY, 955–966. 10.1016/B978-0-12-811410-0.00067-2. [DOI]
- Robbers JE, Brady L, Tyler Jr V. (1964) A chemical and chemotaxonomic evaluation of Inocybe species. Lloydia 27: 192–202. [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. (2012) MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice across a Large Model Space. Systematic Biology 61: 539–542. 10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sailatha S, Naveen S, Naika M, Anilakumar KR, Singh M. (2014) Toxicological evaluation of Inocybevirosa. In: Manjit S, Ramesh U, Parkash SV, Ahlawat OP, Satish K, Shwet K, Bindvi A, Mamta G. (Eds) Proceedings of the 8th international conference on mushroom biology and mushroom products (icmbmp8) volume ii.Papers presented in 8th ICMBMP, NASC, New Delhi (India), November 2014, Yugantar Prakashan Pvt. Ltd., 467–472.
- Spoerke DG, Rumack BH. (1994) Handbook of mushroom poisoning: diagnosis and treatment. CRC Press, London, 456 pp. [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. (2011) Mega5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution 28(10): 2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trifinopoulos J, Nguyen LT, von Haeseler A, Minh BQ. (2016) W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Research 44(W1): 1–4. 10.1093/nar/gkw256. [DOI] [PMC free article] [PubMed]
- Vilgalys R, Hester M. (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172(8): 4238–4246. 10.1128/jb.172.8.4238-4246.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrinda KB, Pradeep CK, Joseph AV, Abraham TK. (1996) A new Inocybe (Cortinariaceae) from Kerala state, India. Mycotaxon 57: 171–174. [Google Scholar]
- Wang SN, Hu YP, Chen JL, Qi LL, Zeng H, Ding H, Huo GH, Zhang LP, Chen FS, Yan JQ. (2021) First record of the rare genus Typhrasa (Psathyrellaceae, Agaricales) from China with description of two new species. MycoKeys 79: 119–128. 10.3897/mycokeys.79.63700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson D. (1947) Poisoning by Inocybefastigiata. British Medical Journal 2(4520): 297. 10.1136/bmj.2.4520.297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F, Zhang YZ, Zhang YH, Guan GY, Zhang KP, Li HJ, Wang JJ. (2020) Mushroom poisoning from Inocybeserotina: A case report from Ningxia, northwest China with exact species identification and muscarine detection. Toxion 179: 72–75. 10.1016/j.toxicon.2020.03.003. [DOI] [PubMed] [Google Scholar]
- Zhang M, Li TH, Wang CQ, Zeng NK, Deng WQ. (2019) Phylogenetic overview of Aureoboletus (Boletaceae, Boletales), with descriptions of six new species from China. MycoKeys 61(1): 111–145. 10.3897/mycokeys.61.47520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zosel A, Stanton M, Klager A, Gummin D. (2015) Death following ingestion of Clitocybe species mushroom. Clinical Toxicology 53(7): 735–736. 10.4172/2161-0495.1000308. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.