Abstract
Infections caused by the opportunistic yeast Candida albicans are one of the major life threats for hospitalized and immunocompromised patients, as a result of antibiotic and long-term antifungal treatment abuse. Odorant binding proteins can be considered interesting candidates to develop systems able to reduce the proliferation and virulence of this yeast, because of their intrinsic antimicrobial properties and complexation capabilities toward farnesol, the major quorum sensing molecule of Candida albicans. In the present study, a hybrid system characterized by a superparamagnetic iron oxide core functionalized with bovine odorant binding protein (bOBP) was successfully developed. The nanoparticles were designed to be suitable for magnetic protein delivery to inflamed areas of the body. The inorganic superparamagnetic core was characterized by an average diameter of 6.5 ± 1.1 nm and a spherical shape. Nanoparticles were functionalized by using 11-phosphonoundecanoic acid as spacer and linked to bOBP via amide bonds, resulting in a concentration level of 26.0 ± 1.2 mg bOBP/g SPIONs. Finally, both the biocompatibility of the developed hybrid system and the fungistatic activity against Candida albicans by submicromolar OBP levels were demonstrated by in vitro experiments.
Hybrid superparamagnetic nanoparticles functionalized with odorant binding proteins for Candida albicans treatment.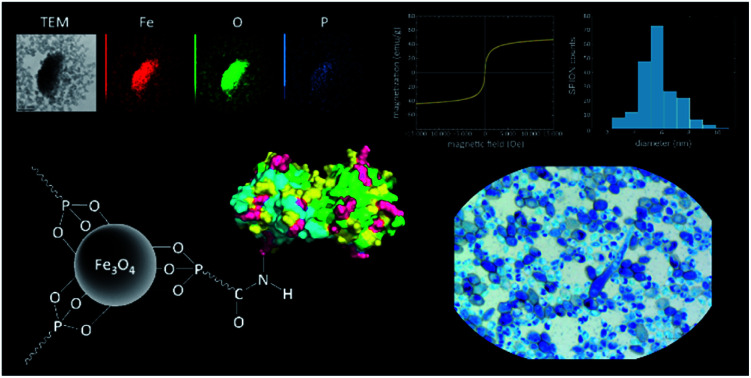
Introduction
Infections caused by opportunistic pathogens such as Pseudomonas aeruginosa, Candida albicans and Staphylococcus aureus are responsible for most of the morbidity and mortality in immunocompromised patients. In recent years, the antibiotic abuse and the release of pharmaceuticals in the environment resulted in the change of the microbial community affecting humans and the emergence of multidrug-resistant bacteria and fungi.1–3 The fight against these microorganisms is challenging since it has been reported that more than 70% of the human pathogenic bacteria exhibit resistance to at least one commercial antibiotic drug.4 The increasing rates of invasive candidiasis and sepsis caused by fungal microorganisms are emerging problems for hospital treatments5 and Candida species are considered one of the main causes of nosocomial bloodstream and dermal invasive fungal infections.
C. albicans in healthy subjects is a harmless commensal opportunistic fungus of the human microbiota, asymptomatically colonizing different tissues and organs, such as gastrointestinal tract, oral cavity and skin. Being able to rapidly proliferate, switching from benign to pathogenic forms especially in diseased subjects as well as in case of broad-spectrum antibiotics abuse,6–8 a strong increase of the mortality rates of immunocompromised patients in case of C. albicans fungal infections has been reported.5,7,9
The symptoms of infections are protean, including oral and genital infections, fungemia and invasive systemic diseases, affecting different organs:5C. albicans has been related to oropharyngeal, esophageal and vulvovaginal candidiasis5 and acute lung injuries.9 Antifungal treatment of this yeast infections accounts for tens of drugs belonging to the following four main classes: polyenes, azoles, echinocandins, and antimetabolites.5 However, in recent years, multi-drug resistant strains have been reported in response to the long-term antifungal treatments resulting in recurrent infections:10,11 in this context, the development of new fungistatic treatments against C. albicans is an open challenge. Taking into account that C. albicans is reported to produce and secrete extracellular molecules that: (i) increase the antibiotic sensitivity of other bacteria, such as Staphylococcus aureus; (ii) affect the growth of other Candida and Aspergillus species and (iii) induce apoptosis of cancerous cells as well as of S. cerevisiae and A. nidulans,12 its complete removal from the microbiota could be also harmful, causing the proliferation of other pathogenic microorganisms. An interesting challenge is related to the reduction of both the proliferation and the virulence of the yeast in inflamed areas, while maintaining the overall commensal form. It has been hypothesized that this could be achieved by using antimicrobial agents able to bind or react with signalling molecules typical of C. albicans, such as farnesol.12–17
Odorant Binding Proteins (OBPs), belonging to the superfamily of carrier proteins called lipocalins, are multifunctional scavengers for small hydrophobic molecules dissolved in the mucus lining the nasal epithelia of vertebrates. They are able to bind a wide range of natural and synthetic compounds belonging to different chemical classes.18–23 In this context, OBPs scavenging capabilities have been recently demonstrated in vitro toward several signalling molecules secreted by different pathogenic microorganisms, known as quorum sensing molecules (QSMs), and virulence factors, namely acyl homoserine lactones (AHLs), oxo-AHLs, farnesol, and pyocyanin. In addition, both fungistatic and bacteriostatic activities of OBPs have been demonstrated by performing time kill assays (TKA) against different bacteria and fungi19 using proteins at a concentration of 50 μM. Therefore, it has been hypothesized that this protein might physiologically contribute to the preservation of the microflora colonizing the respiratory tract, behaving as a component of innate immunity. The reported data suggest that OBPs might have a potential as natural antimicrobial drugs, interfering with both communication and toxicity of pathogenic microorganisms, thus decreasing their activity, proliferation and virulence. Taking into account the scavenging capabilities toward farnesol, its major QSM and the demonstrated fungistatic activity,19 OBPs could be considered as good candidates to develop antimicrobial agents against C. albicans strains. However, these proteins have to be engineered in order to be targeted to infected and inflamed areas, thus increasing their local concentrations.
In this context, the development of nanodrugs and nanocarriers has opened new perspective in biomedical science. A wide range of nanoparticles presenting an inorganic core has been proposed, including noble metals, magnetic materials, metal oxide nanoparticles, quantum dots and semiconductor nanocrystals.24–28 Metallic nanoparticles are characterized by high chemical and thermal stabilities, low-cost, very high area-to-volume and area-to-weight ratio, and by the possibility of engineering the nanoparticle surface with both natural and synthetic antimicrobial agents, obtaining core–shell nanoparticles. In addition, the presence of a shell also reduces agglomeration phenomena and increases nanoparticle biocompatibility.24,29 In particular, iron oxide superparamagnetic nanoparticles (SPIONs) can be properly functionalized by specific receptor and/or active components and targeted to localized area of the body by applying an external magnetic field.26,27,30 In fact, the main advantage of SPIONs is the possibility to increase the local concentration of the drug in the infection site with minimum side effects to the whole organism.26,27,31 In addition, iron oxide nanoparticles have been shown to be an effective tool for the disruption or inactivation of biofilms, increasing drug permeability and causing pathogens detachment.32–34 Magnetic nanoparticles have been proposed as antimicrobial agents against several pathogenic microorganisms, such as bacteria, viruses and fungi.24,27,35,36 A promising approach relies on the use of aerosol formulations containing SPIONs (nanomagnetosols) in order to improve the targeting of the aerosol toward a specific region of lungs.37–39 The use of functionalized nanoparticles able to interfere with pathogen communication, proliferation and virulence systems is a new frontier in antimicrobial design.40
In this context, the aim of this research study is the development of OBP-functionalized SPIONs for the treatment of C. albicans, chosen as target pathogenic fungus because of the increased occurrence of candidiasis, sepsis and repeated infections in hospitalized and immunocompromised patients, and for its multi-drug resistance. This innovative approach combines the local targeting potential of SPIONs with the natural antimicrobial capabilities of the OBPs.
Experimental
Chemicals and materials
Benzyl alcohol anhydrous (99.8%), iron(iii) acetylacetonate (Fe(acac)3), dichloromethane anhydrous (≥99.8%), ethanol anhydrous (>99.8%), hexane anhydrous (95%), 2-propanol anhydrous (99.5%), potassium phosphate dibasic trihydrate (≥99.0%) and potassium dihydrogen phosphate (>99%) were purchased from Merck (Milan, Italy). 11-Phosphonoundecanoic acid (>98%), 3-phosphonopropionic acid (>94%) and 1-(3-dimethyl-aminopropyl)-3-ethylcarbodiimide (EDC) (97%) were from abcr (76187 Karlsruhe, Germany). HisPur™ Ni-NTA Resin was from Thermo Fisher (Waltham, MA, USA). Commercial fibers 1 cm to 50/30 μm DVB–CAR–PDMS were purchased from Supelco (Bellefonte, PA, USA).
bOBP biosynthesis
The recombinant form of bovine OBP tagged with six histidine residues at the amino terminal (6-His-bOBP) was obtained from a BL21-DE3 E. coli strain transformed with the expression vector pT7-7 containing the appropriate OBP cDNA, as previously reported.41 Purification of the protein was carried out by affinity chromatography using Ni-NTA agarose according to the instructions of the manufacturer, followed by dialysis in 50 mM sodium phosphate pH = 7.3. Protein purity was assessed by SDS-PAGE and its concentration was determined by measuring the absorbance value at 280 nm.
Functionality of the protein in solution was determined by direct titrations using the fluorescent ligand 1-aminoanthracene (AMA), as previously reported.18 Briefly, samples of 0.5 μM 6-His-bOBP in 20 mM Tris–HCl buffer pH = 7.3 were incubated overnight at 4 °C with increasing concentrations of AMA (0.15–20 μM). Fluorescence emission spectra were recorded between 450 and 550 nm with a PerkinElmer LS 50 luminescence spectrometer (PerkinElmer, Waltham, MA, USA) (excitation and emission slits at 5 nm) at a fixed excitation wavelength of 380 nm. The formation of the binding complex between 6-His-bOBP and AMA was monitored as an increase in the fluorescence emission intensity at 480 nm.
Sample characterization
Magnetization was determined by using a Vibrating Sample Magnetometer (VSM) model 7400 from Lake Shore (Westerville, USA). The maximum applied field was 1.8 T.
X-ray powder diffraction (XRPD) data were collected on a Thermo ARL X'TRA X-ray diffractometer CuKα X-radiation at 40 kV and 30 mA. X-ray photoelectron spectroscopy (XPS) analyses were performed by using a high intensity twin anode source, XR 50 and a hemispherical energy analyzer with wide-angle lens – Phoibos 150 Wal (SPECS GmbH, Berlin, Germany). The measurements were performed by using the Kα emission of Al (1486.6 eV).
Conventional and high-resolution transmission electron microscopy (TEM) and energy dispersive X-ray spectroscopy (EDX) in scanning mode were carried out on the solution of the nanoparticles using a JEOL 2200FS microscope working at 200 keV. Samples were prepared by depositing a drop of a suspension of the particles in hexane on carbon-coated copper grids.
UV-vis analyses were performed by using a Thermo Evolution 260 Bio + SPE 8w Peltier Water Cooled Cell-Changer, Thermo Fisher (Waltham, MA, USA).
Infrared spectra were recorded by means of attenuated total reflectance-Fourier transform infrared spectroscopy (ATR/FT-IR) using a FT-IR Nicolet 5700 spectrometer (Thermo Fisher Scientific, Inc., Waltham, MA, USA), equipped with an ATR Smart Orbit™, operating between 400 and 4000 cm−1.
Synthesis of superparamagnetic iron oxide nanoparticles
SPIONs were obtained by thermal decomposition and reduction of iron(iii) acetylacetone under nitrogen atmosphere: 1.00 g of Fe(acac)3 and 20 ml of benzyl alcohol were added in a three-neck round-bottom flask.42,43 The solution was vigorously stirred at ambient temperature under nitrogen flow for 1 h, then it was heated at 175 °C for 48 h. Then the mixture was cooled at ambient temperature by keeping the magnetic stirring and the inert atmosphere. The black precipitate was sonicated, centrifuged and magnetically decanted; finally, the precipitate was washed with dichloromethane (1 time) and absolute ethanol (3 times). The obtained nanoparticles were stored in ethanol and were stable up to 1 month.
SPION functionalization with carboxyalkylphosphonic acids
A solution of 11-phosphonoundecanoic acid or 3-phosphonopropionic acid in 2-propanol anhydrous (1 mM) was prepared under nitrogen. Then the dried SPIONs were added to the solution under vigorous stirring and sonicated for 1 h. A 2 : 1 carboxyalkylphosphonic acid/SPIONs molar ratio was found to be optimal. The nanoparticles were stirred at room temperature under nitrogen for 16 h, then they were magnetically decanted and washed with water 10 times. The nanoparticles were characterized by VSM, TEM, EDX, XPS and ATR FT-IR analyses.
Analysis of solvent residues by SPME-GC-MS analysis
The presence of solvent residues, namely benzyl alcohol, hexane and isopropanol, on the nanoparticles was investigated via solid phase microextraction-gas chromatography-mass spectrometry (SPME-GC-MS). Briefly, headspace analyses of 20 mg nanoparticles in 10 ml amber glass vials were carried out: samples were incubated for 10 minutes at 50 °C under an agitation speed of 250 rpm. SPME extraction was performed using a 1 cm to 50/30 μm DVB–CAR–PDMS fiber at 50 °C for 15 min under stirring. Finally, the fiber was desorbed in the GC injection port at 250 °C for 2 min. Five replicate measurements were performed. Detection (yD) and quantitation (yQ) limits were expressed as signals based on the mean blank (xb) and the standard deviation of blank responses (sb) as follows: yD = xb + 3sb and yQ = xb + 10sb. The value of xb and sb were calculated performing ten blank measurements. Instrumental conditions are reported in ESI.†
SPION–bOBP functionalization
The nanoparticles functionalized with the carboxyalkylphosphonic acids were conjugated with bOBP by using EDC as coupling agent. The w/w ratio between the SPIONs and the protein is 8.9 : 0.15, whereas the molar ratio SPIONs : EDC is 1 : 2.44 The reaction was performed in phosphonate buffer (pH = 7.3) at 4 °C for 24 h under mechanical agitation. The nanoparticles were magnetically decanted, washed with phosphonate buffer (3 times 5 ml) and stored at 4 °C for up to one month. The supernatant solutions were analyzed by Pierce-BCA protein assay kit (Thermo Scientific, USA) in order to obtain by difference the concentration of nanoparticles loaded on the SPIONs. The analysis was performed by UV-vis spectroscopy at 562 nm.
Calibration curves (eight concentration levels, three replicated measurements for each level) were established by analyzing blank and standard solutions of bOBP in phosphonate buffer in the 0.02–0.64 μg l−1 range, corresponding to a 0–45.40 μg OBP/g SPION.
Shapiro–Wilk and Bartlett tests were performed to assess the normal distribution of data and the homoscedasticity condition, respectively. Mandel's fitting test was also performed to check the goodness of fit and linearity. The significance of the intercept (significance level 5%) was established by running a Student's t-test. Repeatability was calculated in terms of RSD% on two concentration levels, i.e. 2.84 and 11.35 μg l−1, by performing six replicated measurements for each level. The concentration of bOBP grafted onto the nanoparticles (6-His-bOBP–SPIONs) was considered by three different samples from two protein batches. Finally, the magnetization and hysteresis properties of the functionalized 6-His-bOBP–SPIONs were evaluated from VSM measurements.
Fungal inoculum preparation
Fungal colonies of C. albicans ATCC 11006 and of the human field strain FUN RE RB4 from fresh cultures were inoculated in RPMI broth and incubated 48 hours at 37 °C in aerobic atmosphere. After incubation, suspension was centrifuged 20 min at 2000 rpm and 4 °C and the pellet resuspended in phosphate buffer (PB) 10 mM pH 7. Fungal suspension was diluted to a concentration of 108 CFU ml−1 corresponding to the 5 McFarland turbidity scale,45 then further diluted 1 : 10 in RPMI broth to obtain a final concentration of 107 CFU ml−1. Within 30 minutes after preparation, fungal suspensions were used for the microdilution assay.45
6-His-bOBP–SPIONs time-kill assay
The antimicrobial activity of 6-His-bOBP–SPIONs was evaluated through time-kill assay.46 In a 96 U bottomed wells microtiter plate, 95 μl of OBP-SPION solution in PB were inoculated with 5 μl of fungal suspension prepared as previously described. Final protein concentrations were 0.1 and 0.5 μM, respectively. The plates were incubated at 37 °C in aerobic atmosphere for the whole duration of the test. After 8 hours of contact, 20 μl of the suspension were plated on solid agar medium (Sabouraud agar). For CFU count, agar plates were incubated for 48 hours at 37 °C in aerobic atmosphere. For each assay three replicates in three independent experiments were set up. Kruskal–Wallis test followed by pairwise analysis were carried out to check for the presence of significant differences among the obtained results.
Cytotoxicity assay
A concentration of 0.1 μM bOBP was added in a flat bottomed 96 wells sterile plate treated for cells containing a number of 8000 cells per wells of adenocarcinomic human alveolar basal epithelial cells A549 (ATCC CCL-185) in EMEM medium. The same assay was performed with SPIONs and 6-His-bOBP–SPIONs. Plates were incubated for 24 hours at 37 °C in presence of 5% CO2. After incubation, 10 μl of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) were added in each well of the assay and reincubated at 37 °C for 6 hours.47 At the end of the incubation, 100 μl of a 10% SDS in HCl 0.01 M solution were added to each well and then reincubated overnight. The yellow tetrazolium MTT salt is reduced in metabolically active cells to form insoluble purple formazan crystals, which are solubilized by the addition of a detergent. After incubation the plates were read with a spectrophotometer at 620 nm. A negative control containing 50 μl PB (equivalent to the volume in which the proteins and the nanoparticles are suspended) was performed for each plate. Two independent experiments and three replicate measurements were performed for each assay. The results were investigated in terms of survival rate of the cells treated with bOBPs and carboxyalkylphosphonic acid functionalized nanoparticles, respectively. For each assay, a comparison with the data obtained from the control experiments was performed. In case of data homoscedasticity, an independent samples homoscedastic Student's t-test was carried out, whereas a heteroscedastic t-test was performed in presence of unequal variances.
Results and discussion
In this study, hybrid nanoparticles were developed and tested as fungistatic agents against C. albicans. Differently from Miller and coworkers,40 who developed nanoparticles capable of binding AHLs using synthetic receptors, OBPs have been proposed as natural receptor due to their affinity towards several QSMs, among which farnesol,18,19,41 a QSM central in C. albicans metabolism and virulence. Since OBPs are proteins naturally present in respiratory apparatus of mammals, no problem related to their biocompatibility is expected.
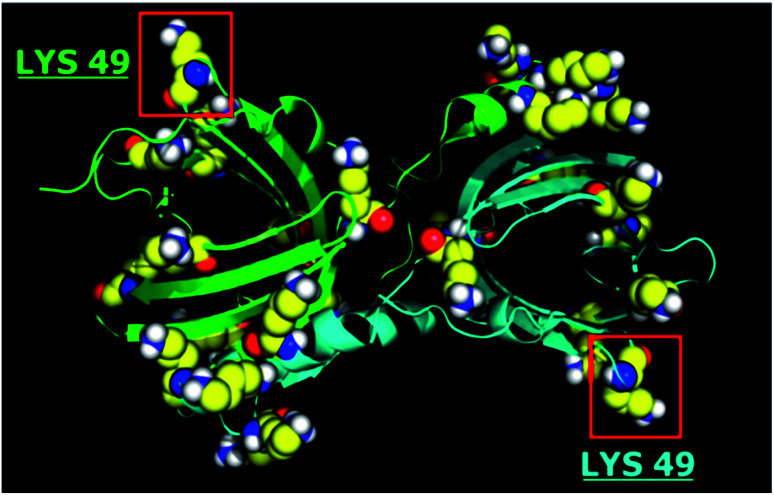
Among OBPs, the bovine form was characterized by enhanced binding capabilities over QSMs,19 therefore in the present study it was used as antimicrobial agent. The bOBP is a homodimer characterized by several lysine residues on the surface suitable for functionalization, such as two Lys49 groups.41 The recombinant six-tagged histidine form was proposed instead of the native protein since it can be purified at higher yields from Escherichia coli soluble extracts.48 In addition, increased antimicrobial and antifungal activities have been previously demonstrated for the mutant protein, probably due to the chelation of metals that are essential for the metabolism of several microorganisms.19 This mechanism could be similar to that reported for the copper scavenging activity of the histatins.19,49,50 The overall dimension of the dimeric protein is 40 × 40 × 70 Å.51 A 3D representation of the protein is depicted in Fig. 1.
Fig. 1. 3D structure of bOBP: lysine amino acid residues highlighted.
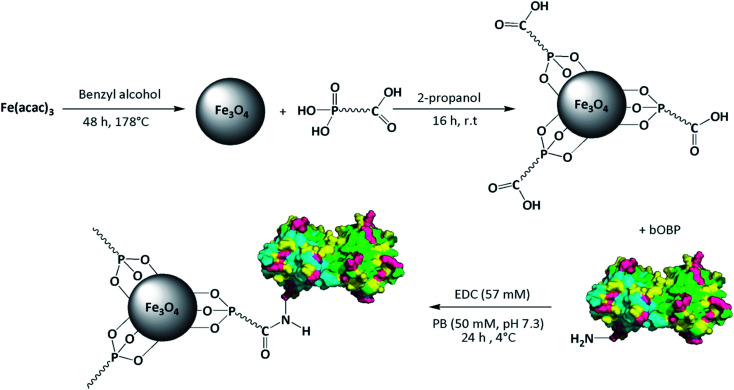
The hybrid system was obtained by functionalization of the superparamagnetic magnetite (Fe3O4) core with carboxyalkylphosphonic acid and bOBP, with the final aim of driving the antimicrobial agent directly to the infected area, thus increasing its local concentration. The SPIONs were functionalized using carboxyalkylphosphonic acids as spacers, being the phosphonate group able to effectively bind the iron oxide core,52,53 while the protein was linked to the carboxyl group by amide bond. The synthetic path of the functionalization is summarized in Fig. 2.
Fig. 2. Synthesis of bOBP-functionalized SPIONs.
Synthesis of superparamagnetic iron oxide nanoparticles
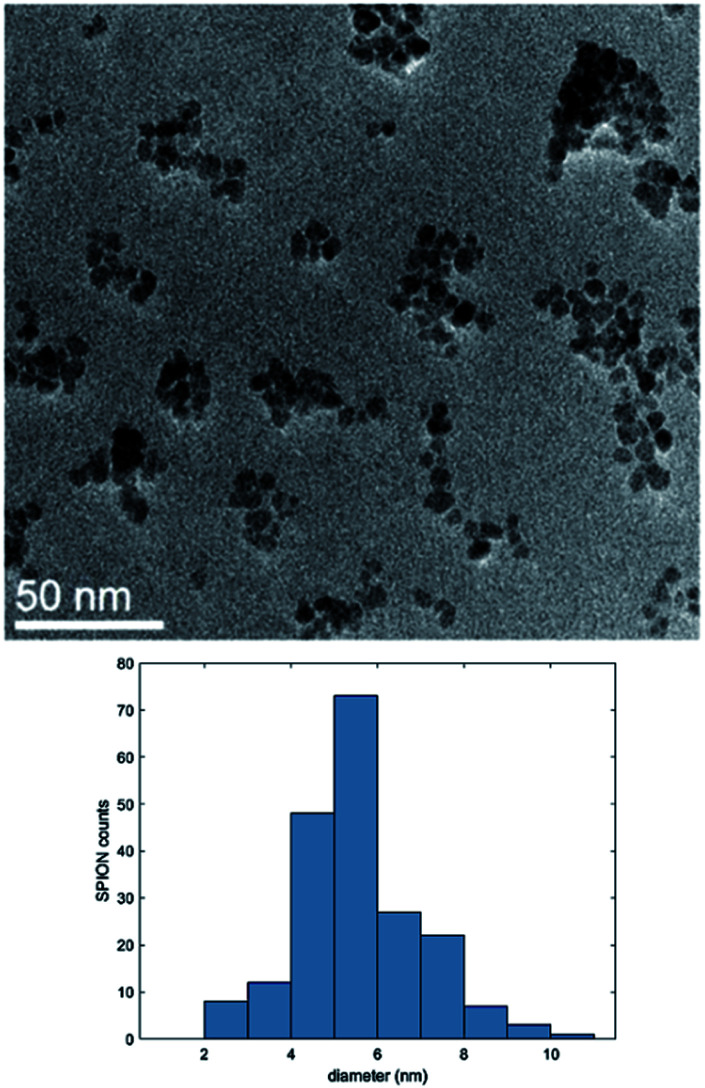
Magnetic nanoparticles were synthetized by thermal decomposition and reduction of Fe(acac)3 in benzyl alcohol.42,43 TEM analysis of the bare nanoparticles revealed a spherical shape with a very narrow size distribution, characterized by an average diameter of 6.5 ± 1.1 nm (Fig. 3). The dimensions and geometry of the obtained beads have been designed to obtain drug nanocarriers able to reach different areas of the body, thus comprising the alveoli region of the lungs,37,54 and to be removed by kidneys.29,55
Fig. 3. TEM image and size distribution of the synthetized SPIONs.
The synthesis of Fe3O4 nanoparticles was assessed both by means of XRPD and XPS measurements. XRPD analysis (Fig. S1†) resulted in peaks corresponding to (2 2 0), (3 1 1), (4 0 0), (4 2 2), (5 1 1) and (4 4 0) reflections of Fe3O4 (JCPDS 82-1533). XPS spectrum of Fe 2p (Fig. S2†) showed two peaks attributable to magnetite at 711 and 725 eV related to Fe 2p3/2 and Fe 2p1/2, respectively.
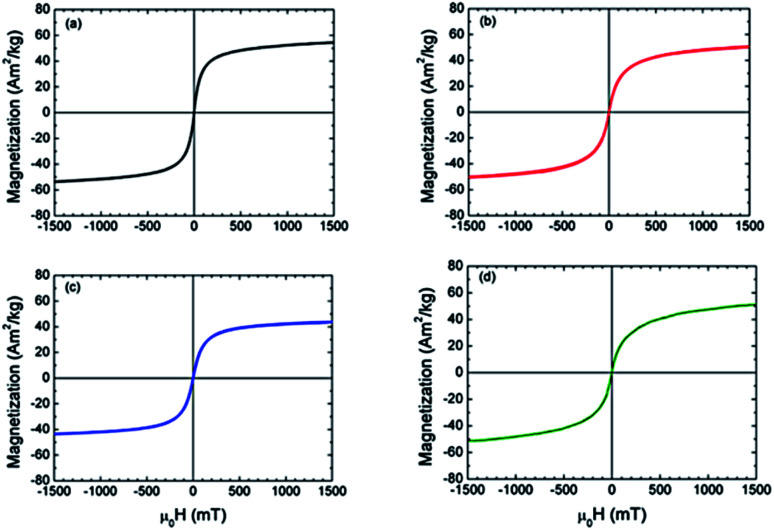
The nanoparticles were also characterized by ATR/FT-IR analysis (Fig. S3†), showing a band at ν = 555 cm−1 that can be ascribed to the stretching of Fe–O groups. Finally, magnetization studies were performed by using a VSM. At room temperature the nanoparticles show a superparamagnetic behavior, evidenced by the absence of open hysteresis in the magnetization curves, and a specific magnetization value of 55 A m2 kg−1 at 1.5 T (Fig. 4a). The superparamagnetic behavior guarantees the stability of the ferrofluid suspension and prevents nanoparticle aggregation phenomena. At the same time, the high magnetization value at room temperature is suitable for effectively isolating the nanoparticles from the matrix solution by means of an external magnetic field.
Fig. 4. Magnetization curve of the: (a) bare nanoparticles; (b) 11-phosphonoundecanoic-; (c) 3-phosphonopropionic-functionalized nanoparticles; (d) bOBP-functionalized nanoparticles.
Core–shell SPIONs
The first step for SPIONs functionalization was performed by using carboxyalkylphosphonic acids with the aim to: (i) protect the core of the nanoparticles from oxidation and aggregation phenomena; (ii) increase the stability in water and phosphate buffer solutions (PB, pH = 7.3); (iii) provide free carboxylic groups for further protein conjugation. The use of phosphonate compounds as spacers to bind to iron oxide nanoparticles has been already reported in literature both to obtain a strong coordination bond between the –PO3 group of the spacer and the core of the material, and to functionalize the terminal groups with a proper organic molecule.52,53
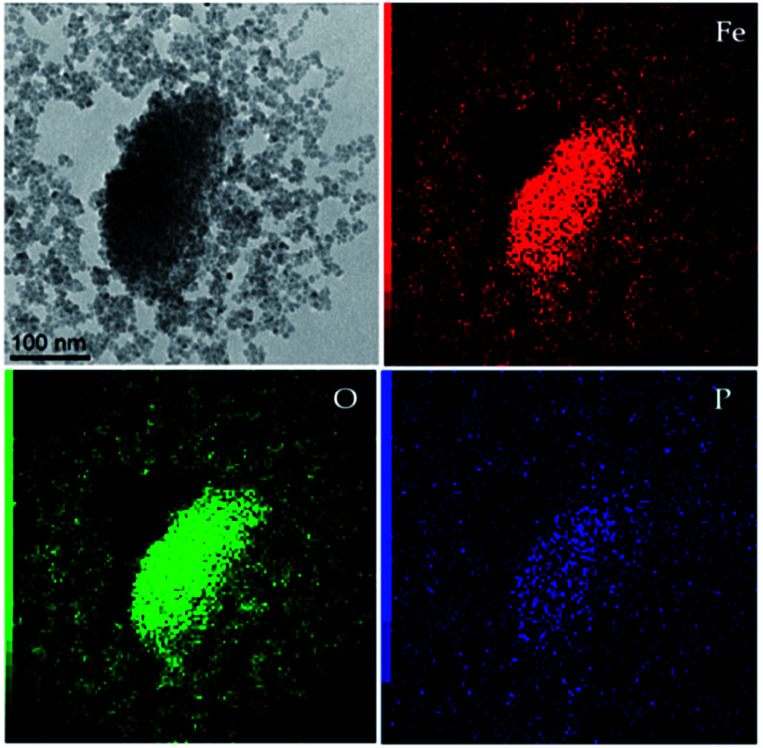
The reaction was performed at room temperature in 2-propanol using two different carboxyalkylphosphonic acids, namely 11-phosphonoundecanoic and 3-phosphonopropionic acids, in order to evaluate the effect of the spacer chain length at the concentration of 1 mM. The reaction conditions were compatible with the potential pharmaceutical application of the drug, avoiding the use of toxic and low-volatile solvents, such as dimethylformamide. Different carboxyalkylphosphonic acids/SPIONs ratios were tested and the best results were achieved using 1 : 2 acid/SPION. The functionalized nanoparticles were investigated by TEM-EDX: the EDX spectrum (Fig. S4†) showed a peak at 2.00 keV attributable to phosphorus. Elemental mapping of Fe, O and P (Fig. 5) demonstrated a strong correlation in the intensities and distributions of the investigated elements, thus supporting the binding of the spacer onto the magnetite core.
Fig. 5. TEM image (top left) and corresponding EDX mapping for different elements (Fe, O and P).
Subsequent ATR/FT-IR analyses (Fig. S5†) showed the presence of two sharp bands at 2920 and 2855 cm−1 attributable to the νa(CH2) asymmetric and νs(CH2) symmetric stretches, respectively. The peak at 1710 cm−1 can be assigned to the C O stretching, whereas the single and broad band at 1035 cm−1 can be ascribed to the phosphonic acid anchoring onto the SPIONs via multidentate bonding, involving both P O and P–O groups.
The absence of peaks at 1250 and 920 cm−1 related to P O and P–O–H stretches respectively, confirmed the interaction between the phosphonate group and the iron oxide surface,56 thus preserving the carboxylic acid unbounded for protein conjugation. Finally, the magnetization curves for both 11-phosphonoundecanoic- and 3-phosphonopropionic-functionalized nanoparticles were obtained by VSM. After functionalization, the superparamagnetic behavior is maintained, confirming the absence of nanoparticles aggregation and collective magnetic phenomena driven by strong interparticle interactions (Fig. 4b and c).57 The specific magnetization value at 1.5 T is slightly reduced to 45 and 50 A m2 kg−1 for the long- and short-chain acids respectively, because of the mass of the coating.
SPION–bOBP functionalization
Prior to protein functionalization, the absence of residual solvents, namely hexane, isopropanol and benzyl alcohol used for the synthesis of the core–shell nanoparticles, was assessed via SPME-GC-MS analysis. Detection limits in the 0.06–0.19 μg l−1 range were obtained: since no signal related to the investigated solvents was detected, the efficacy of the washing cycles was demonstrated.
Finally, the carboxyalkylphosphonic-functionalized nanoparticles were conjugated with the bOBP by using EDC as coupling agent, performing the reaction under the conditions proposed by Wang et al.44
The conjugation was achieved via amide bond formation between the carboxylic group of the spacer and the lysine residues of the bOBP. The use of EDC was required to obtain the protein functionalization with good yields under very mild conditions, thus preventing extended unfolding of the bOBP. This aspect is of paramount importance in order to maintain the affinity of the ligand binding site towards QSMs. The use of N-hydroxysuccinimide was avoided since it was reported to reduce the Cu2+ ions used for protein quantitation in the BCA protein assay kit.58
The bOBP-functionalized nanoparticles were magnetically decanted and washed three times with PB to remove unreacted reagents and unbounded protein. Then the concentration of the protein functionalizing the nanoparticles was determined using the BCA protein assay kit.59 However, the high absorbance values of the iron oxide core did not allow the direct assessment of the bound bOBP, thus the concentration of the not-conjugated protein in both the reaction and washing solutions was evaluated. The obtained regression curve, i.e. y = 0.00238 (±0.00007)x + 0.0764 (±0.0015), was used to quantify the concentration of the loaded protein, by calculating the difference of the OBP present in the mixture before the reaction and that present after the removal of the functionalized SPIONs and in the washing solutions. As for precision, a very good repeatability was obtained with RSD always lower than 5% (n = 3). The calculated concentration of bOBP on the functionalized nanoparticles was 26.0 ± 1.2 mg bOBP/g SPIONs: this result is confident with outcomes published in literature for similar proteins.44 As for the use of the 3-phosphonopropionic acid, it was demonstrated that by using this spacer, SPIONs could not be effectively conjugated, probably due to the high steric hindrance of the bOBP and the reduced length of the spacer.
The magnetization measurements performed on the bOBP-functionalized nanoparticles confirmed the superparamagnetic behavior with a specific magnetization value of 51 A m2 kg−1 at 1.5 T (Fig. 3d), coherently with the magnetic features evidenced in the previous steps.
Antimicrobial activity
The antimicrobial activity of both free bOBP and 6-His-bOBP–SPIONs was evaluated over a period of 8 hours against C. albicans reference strain ATCC 11006 and C. albicans human field strain FUN RE RB4, in order to assess the inhibitory efficacy of the developed systems.
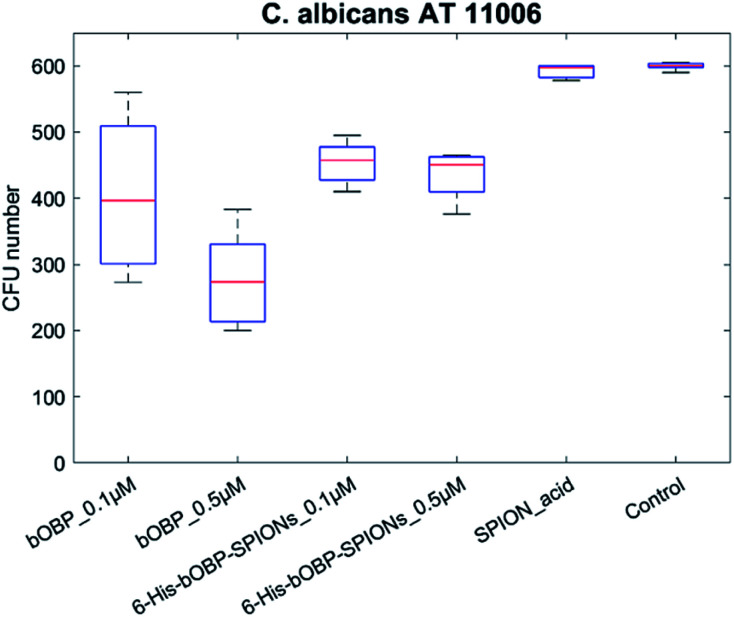
As for the reference strain AT11006, two different concentrations of bOBP were tested, namely 0.1 and 0.5 μM (Fig. 6). The presence of significant differences in the responses was verified by applying the non-parametrical Kruskal–Wallis test, followed by pairwise analysis.
Fig. 6. Antimicrobial activity of unbound bOBP (0.1 and 0.5 μM), 6-His-bOBP–SPIONs (0.1 and 0.5 μM) and carboxyalkylphosphonic-functionalized nanoparticles (SPION_acid) against C. albicans reference strain ATCC 11006.
The obtained results showed the absence of significant difference between the carboxyalkylphosphonic-functionalized nanoparticles and the C. albicans growth control (p > 0.05), thus highlighting that SPION_acid nanoparticles were not able to exert any antimicrobial activity. In contrast, both the unbounded bOBP and the 6-His-bOBP–SPIONs were able to inhibit C. albicans strain AT11006 already at 0.1 μM, with inhibitions of 34 and 24%, respectively (p < 0.05).
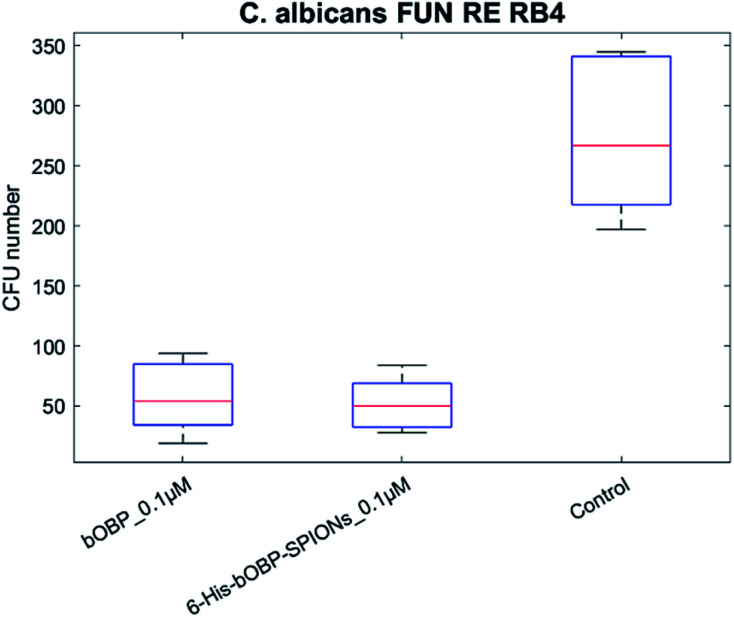
Considering the results obtained with the reference strain, the field strain FUN RE RB4 was tested using only a bOBP concentration of 0.1 μM (Fig. 7).
Fig. 7. Antimicrobial activity of unbound bOBP (0.1 μM), 6-His-bOBP–SPIONs (0.1 μM) against C. albicans field strain FUN RE RB4.
Although human field strains such as FUN RE RB4 are known to be hard to grow in vitro, thus explaining a control having a median CFU number of 267, the achieved results highlighted the antimicrobial activity for both the bOBP and the 6-His-bOBP–SPIONs.
These findings suggest that the protein might be effective in vivo against C. albicans reference strain ATCC 11006 and human field strain FUN RE RB4 already at concentrations lower than those present in the mucus layering the epithelium of the respiratory apparatus of vertebrates.19,20 In addition, the possibility to drive the nanoparticles to a specific area of the body by the application of an external magnetic field plays an important role to strongly increase the local concentration of the OBP, thus enhancing the on-site efficacy of the antimicrobial activity.
Cytotoxicity array
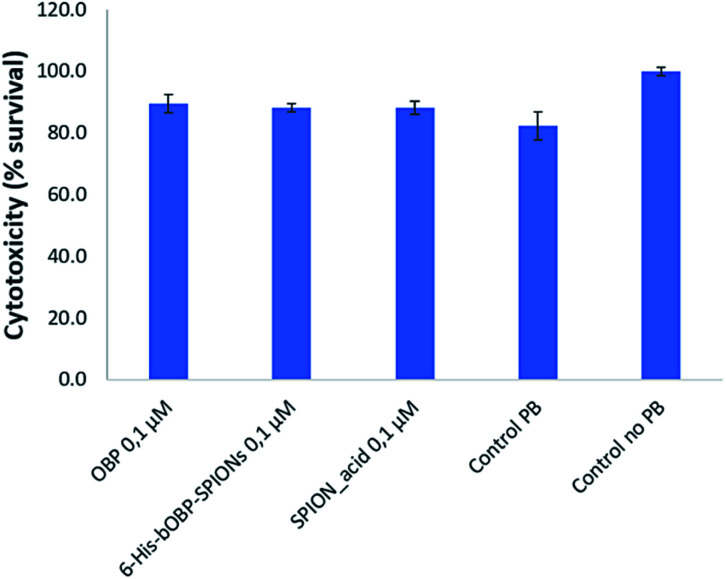
Since the developed hybrid system could be used to exert antimicrobial activity in humans by combining the use of bOBP with the capabilities of magnetic nanoparticles, cytotoxicity evaluation was performed. The human pulmonary adenocarcinoma A549 human cell line was selected for this study in order to test the cytotoxicity in a cellular substrate similar to the natural site of production of bOBP, i.e. respiratory tract of vertebrates.
To test the presence of inherent cytotoxicity, unbounded bOBP, carboxyalkylphosphonic-functionalized nanoparticles 6-His-bOBP–SPIONs were tested at the concentration of 0.1 μM. Controls without and with 50 μl PB, i.e. the amount of buffer used to suspend both the protein and the nanoparticles, were used as references. As shown in Fig. 8, no significant differences were observed for unbounded bOBP, carboxyalkylphosphonic-functionalized nanoparticles and for the 6-His-bOBP–SPIONs compared to the 50 μl PB control (p > 0.05). However, a decrease in cell viability of about 15% was observed in comparison to the EMEM medium without PB. The achieved results demonstrate that the hybrid nanoparticles show good biocompatibility.
Fig. 8. Cytotoxicity of bOBP, 6-His-bOBP–SPIONs and SPION_acid at the concentration of 0.1 μM vs. control with 50 μl PB and without PB. The results are expressed as cell survival rate.
The obtained results demonstrate that the functionalization of the superparamagnetic nanoparticles with a natural binding protein can be an interesting tool to develop antimicrobial biocompatible systems capable of driving the protein by the application of an external magnetic field and removing localized candidiasis, by controlling the proliferation of the yeasts.
Conclusions
A new hybrid system composed by a superparamagnetic iron oxide core, a shell of phosphonoundecanoic acid and bovine odorant binding protein as active component was developed and tested as antimicrobial agent against a reference and a field strain of C. albicans, a commensal yeast responsible of detrimental candidiasis and inflammations. Histidine tagged bOBP was selected because of its enhanced antimicrobial activity toward a large panel of microorganisms.
The synthesis of superparamagnetic iron oxide core resulted in spherical nanoparticles, potentially able to reach targeted localized inflammations under the effect of an external magnetic field. The use of a long spacer i.e. the phosphonoundecanoic acid allowed an efficient conjugation of the SPIONs with the bOBP.
The antimicrobial activity of the developed hybrid system was tested against both reference strain and field strain of C. albicans, demonstrating a good inhibition rate at 0.1 μM, while no cytotoxic effect was obtained for pulmonary adenocarcinoma cells. The main features of 6-His-bOBP–SPIONs hybrid nanoparticles are related both to the possibility of targeting localized infected areas of the body thanks to the superparamagnetic core and to exploit the natural complexation capabilities of OBPs toward QSMs to decrease the activity and proliferation of pathogenic yeasts, inhibiting the growth of C. albicans localized infection foci. In addition, 6-His-bOBP–SPIONs has proven to be not cytotoxic.
The development of bOBP-functionalized SPIONs can be considered a first step to obtain hybrid systems able to bind different inducers or metabolites produced by antibiotic resistant microorganisms. The obtained results suggest that the developed hybrid system could be used in combination with conventional antimicrobial drugs, aiming at enhancing their efficacy for the treatment against pathogenic microorganisms.
Future efforts will be devoted to test the developed hybrid nanoparticles against other multi-drug resistant microorganisms both in vitro and in animal models. Finally, further binding tests of antibiotics, autoinflammatory drugs, as well as drugs used for cancer treatment, will be performed to evaluate the potentialities of the OBP-SPIONs as drug delivery system for compounds having relevant side effects when administered systemically.
Author contributions
Conceptualization: F. Bianchi, N. Riboni, S. Grolli., C. S. Cabassi, R. Ramoni; data analysis: F. Bianchi, M. Careri, N. Riboni, S. Grolli., C. S. Cabassi, R. Ramoni, F. Casoli, L. Nasi, C. de Julián Fernández, P. Luches. Investigation: F. Bianchi, N. Riboni, C. Spadini, V. Conti, F. Casoli, L. Nasi, C. de Julián Fernández, P. Luches. Methodology: F. Bianchi, R. Ramoni, C. S. Cabassi, F. Casoli, L. Nasi. Writing – original draft: F. Bianchi, N. Riboni, C. S. Cabassi, R. Ramoni, F. Casoli, L. Nasi. All authors edited and reviewed the manuscript.
Conflicts of interest
Giuseppina Basini, Federica Bianchi, Maria Careri, Virna Conti, Stefano Grolli, and Roberto Ramoni are inventors of a Patent (“Agenti antibatterici e antimicotici ed usi derivati” no. 102015000038896, date of publication: 28.01.2017) concerning the use of the OBP and/or its variants, or a composition containing these OBP forms, as inactivator or inhibitor of bacterial quorum sensing (QS) communication, by scavenging at least one Quorum Sensing Molecule (QSM).
Supplementary Material
Acknowledgments
This work has benefited from the equipment and framework of the COMP-HUB Initiative, funded by the ‘Departments of Excellence’ program of the Italian Ministry for Education, University and Research (MIUR, 2018–2022).
Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ra01112j
Notes and references
- Bittar F. Richet H. Dubus J. C. Reynaud-Gaubert M. Stremler N. Sarles J. Raoult D. Rolain J. M. PLoS One. 2008;3:e2908. doi: 10.1371/journal.pone.0002908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escolà-Vergé L. Los-Arcos I. Almirante B. Med. Clínica. 2020;154:351–357. doi: 10.1016/j.medcli.2019.11.002. [DOI] [PubMed] [Google Scholar]
- Grossi P. A. Tebini A. Dalla Gasperina D. Minerva Anestesiol. 2015;81:52–64. [PubMed] [Google Scholar]
- Haque S. Ahmad F. Dar S. A. Jawed A. Mandal R. K. Wahid M. Lohani M. Khan S. Singh V. Akhter N. Microb. Pathog. 2018;121:293–302. doi: 10.1016/j.micpath.2018.05.046. [DOI] [PubMed] [Google Scholar]
- Moudgal V. Sobel J. Expert Opin. Pharmacother. 2010;11:2037–2048. doi: 10.1517/14656566.2010.493875. [DOI] [PubMed] [Google Scholar]
- Gulati M. Nobile C. J. Microbes Infect. 2016;18:310–321. doi: 10.1016/j.micinf.2016.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han T. L. Cannon R. D. Villas-Bôas S. G. Fungal Genet. Biol. 2011;48:747–763. doi: 10.1016/j.fgb.2011.04.002. [DOI] [PubMed] [Google Scholar]
- Wall G. Montelongo-Jauregui D. Vidal Bonifacio B. Lopez-Ribot J. L. Uppuluri P. Curr. Opin. Microbiol. 2019;52:1–6. doi: 10.1016/j.mib.2019.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z. L. Li S. R. Fu L. Zheng L. Ye J. Bin Li J. Int. Immunopharmacol. 2019;72:275–283. doi: 10.1016/j.intimp.2019.04.026. [DOI] [PubMed] [Google Scholar]
- Mishra N. N. Prasad T. Sharma N. Payasi A. Prasad R. Gupta D. K. Singh R. Acta Microbiol. Immunol. Hung. 2007;54:201–235. doi: 10.1556/AMicr.54.2007.3.1. [DOI] [PubMed] [Google Scholar]
- Arendrup M. C. Patterson T. F. J. Infect. Dis. 2017;216:S445–S451. doi: 10.1093/infdis/jix131. [DOI] [PubMed] [Google Scholar]
- Padder S. A. Prasad R. Shah A. H. Microbiol. Res. 2018;210:51–58. doi: 10.1016/j.micres.2018.03.007. [DOI] [PubMed] [Google Scholar]
- Albuquerque P. Casadevall A. Med. Mycol. 2012;50:337–345. doi: 10.3109/13693786.2011.652201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornby J. M. Jensen E. C. Lisec A. D. Tasto J. J. Shoemaker R. Dussault P. Nickerson K. W. Appl. Environ. Microbiol. 2001;67:2982–2992. doi: 10.1128/AEM.67.7.2982-2992.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wongsuk T. Pumeesat P. Luplertlop N. J. Basic Microbiol. 2016;56:440–447. doi: 10.1002/jobm.201500759. [DOI] [PubMed] [Google Scholar]
- Cho T. Nagao J. Imayoshi R. Kaminishi H. Aoyama T. Nakayama H. J. Oral Biosci. 2010;52:233–239. doi: 10.1016/S1349-0079(10)80026-X. [DOI] [Google Scholar]
- Nickerson K. W. Atkin A. L. Mol. Microbiol. 2017;103:567–575. doi: 10.1111/mmi.13601. [DOI] [PubMed] [Google Scholar]
- Bianchi F. Basini G. Grolli S. Conti V. Bianchi F. Grasselli F. Careri M. Ramoni R. Anal. Bioanal. Chem. 2013;405:1067–1075. doi: 10.1007/s00216-012-6499-0. [DOI] [PubMed] [Google Scholar]
- Bianchi F. Flisi S. Careri M. Riboni N. Resimini S. Sala A. Conti V. Mattarozzi M. Taddei S. Spadini C. Basini G. Grolli S. Cabassi C. S. Ramoni R. PLoS One. 2019;14:e0213545. doi: 10.1371/journal.pone.0213545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegoni M. Pelosi P. Vincent F. Spinelli S. Campanacci V. Grolli S. Ramoni R. Cambillau C. Biochim. Biophys. Acta, Protein Struct. Mol. Enzymol. 2000;1482:229–240. doi: 10.1016/S0167-4838(00)00167-9. [DOI] [PubMed] [Google Scholar]
- Kotlowski C. Larisika M. Guerin P. M. Kleber C. Kröber T. Mastrogiacomo R. Nowak C. Pelosi P. Schütz S. Schwaighofer A. Knoll W. Sens. Actuators, B. 2018;256:564–572. doi: 10.1016/j.snb.2017.10.093. [DOI] [Google Scholar]
- Pelosi P. Zhu J. Knoll W. Appl. Microbiol. Biotechnol. 2018;102:8213–8227. doi: 10.1007/s00253-018-9253-5. [DOI] [PubMed] [Google Scholar]
- Pelosi P. Zhu J. Knoll W. Sensors. 2018;18:3248. doi: 10.3390/s18103248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatti M. McHugh T. D. Milanesi L. Tomas S. Chem. Commun. 2014;50:7649–7651. doi: 10.1039/C4CC00349G. [DOI] [PubMed] [Google Scholar]
- Rodrigues G. R. López-Abarrategui C. de la Serna Gómez I. Dias S. C. Otero-González A. J. Franco O. L. Int. J. Pharm. 2019;555:356–367. doi: 10.1016/j.ijpharm.2018.11.043. [DOI] [PubMed] [Google Scholar]
- Cotin G., Piant S., Mertz D., Felder-Flesch D. and Begin-Colin S., in Iron Oxide Nanoparticles for Biomedical Applications, Elsevier, 2018, pp. 43–88 [Google Scholar]
- Seabra A. B., Pelegrino M. T. and Haddad P. S., Antimicrobial Applications of Superparamagnetic Iron Oxide Nanoparticles: Perspectives and Challenges, Elsevier Inc., 2017 [Google Scholar]
- Gao W. Chen Y. Zhang Y. Zhang Q. Zhang L. Adv. Drug Delivery Rev. 2018;127:46–57. doi: 10.1016/j.addr.2017.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues G. R. López-Abarrategui C. de la Serna Gómez I. Dias S. C. Otero-González A. J. Franco O. L. Int. J. Pharm. 2019;555:356–367. doi: 10.1016/j.ijpharm.2018.11.043. [DOI] [PubMed] [Google Scholar]
- Rühle B. Datz S. Argyo C. Bein T. Zink J. I. Chem. Commun. 2016;52:1843–1846. doi: 10.1039/C5CC08636A. [DOI] [PubMed] [Google Scholar]
- Ivashchenko O. Lewandowski M. Peplińska B. Jarek M. Nowaczyk G. Wiesner M. Załęski K. Babutina T. Warowicka A. Jurga S. Mater. Sci. Eng., C. 2015;55:343–359. doi: 10.1016/j.msec.2015.05.023. [DOI] [PubMed] [Google Scholar]
- Quan K. Zhang Z. Chen H. Ren X. Ren Y. Peterson B. W. van der Mei H. C. Busscher H. J. Small. 2019;15:1–6. doi: 10.1002/smll.201902313. [DOI] [PubMed] [Google Scholar]
- Li J. Nickel R. Wu J. Lin F. Van Lierop J. Liu S. Nanoscale. 2019;11:6905–6915. doi: 10.1039/C8NR09802F. [DOI] [PubMed] [Google Scholar]
- Thukkaram M. Sitaram S. Kannaiyan S. k. Subbiahdoss G. Int. J. Biomater. 2014;2014:1–6. doi: 10.1155/2014/716080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khezerlou A. Alizadeh-Sani M. Azizi-Lalabadi M. Ehsani A. Microb. Pathog. 2018;123:505–526. doi: 10.1016/j.micpath.2018.08.008. [DOI] [PubMed] [Google Scholar]
- Rispail N. De Matteis L. Santos R. Miguel A. S. Custardoy L. Testillano P. S. Risueño M. C. Pérez-De-Luque A. Maycock C. Fevereiro P. Oliva A. Fernández-Pacheco R. Ibarra M. R. De La Fuente J. M. Marquina C. Rubiales D. Prats E. ACS Appl. Mater. Interfaces. 2014;6:9100–9110. doi: 10.1021/am501029g. [DOI] [PubMed] [Google Scholar]
- Abdelaziz H. M. Gaber M. Abd-Elwakil M. M. Mabrouk M. T. Elgohary M. M. Kamel N. M. Kabary D. M. Freag M. S. Samaha M. W. Mortada S. M. Elkhodairy K. A. Fang J. Y. Elzoghby A. O. J. Controlled Release. 2018;269:374–392. doi: 10.1016/j.jconrel.2017.11.036. [DOI] [PubMed] [Google Scholar]
- Dames P. Gleich B. Flemmer A. Hajek K. Seidl N. Wiekhorst F. Eberbeck D. Bittmann I. Bergemann C. Weyh T. Trahms L. Rosenecker J. Rudolph C. Nat. Nanotechnol. 2007;2:495–499. doi: 10.1038/nnano.2007.217. [DOI] [PubMed] [Google Scholar]
- Tewes F. Ehrhardt C. Healy A. M. Eur. J. Pharm. Biopharm. 2014;86:98–104. doi: 10.1016/j.ejpb.2013.09.004. [DOI] [PubMed] [Google Scholar]
- Miller K. P. Wang L. Chen Y. P. Pellechia P. J. Benicewicz B. C. Decho A. W. Front. Microbiol. 2015;6:1–7. doi: 10.3389/fmicb.2015.00189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grolli S. Merli E. Conti V. Scaltriti E. Ramoni R. FEBS J. 2006;273:5131–5142. doi: 10.1111/j.1742-4658.2006.05510.x. [DOI] [PubMed] [Google Scholar]
- Bianchi F. Chiesi V. Casoli F. Luches P. Nasi L. Careri M. Mangia A. J. Chromatogr. A. 2012;1231:8–15. doi: 10.1016/j.chroma.2012.02.015. [DOI] [PubMed] [Google Scholar]
- Pinna N. Grancharov S. Beato P. Bonville P. Antonietti M. Niederberger M. Chem. Mater. 2005;17:3044–3049. doi: 10.1021/cm050060+. [DOI] [Google Scholar]
- Wang T. H. Lee W. C. Biotechnol. Bioprocess Eng. 2003;8:263–267. doi: 10.1007/BF02942276. [DOI] [Google Scholar]
- Clinical and Laboratory Standards Institute (CLSI), Reference method for broth dilution, Clinical and Laboratory Standards Institute, 3rd edn, 2008, vol. 28 [Google Scholar]
- Balouiri M. Sadiki M. Ibnsouda S. K. J. Pharm. Anal. 2016;6:71–79. doi: 10.1016/j.jpha.2015.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donofrio G. Franceschi V. Capocefalo A. Cavirani S. Sheldon I. M. Reprod. Biol. Endocrinol. 2008;6:1–9. doi: 10.1186/1477-7827-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegoni M. Cambillau C. Vincent F. Grolli S. Accornero P. Ashcroft A. E. Ramoni R. Valencia C. Biochem. J. 2002;365:739–748. doi: 10.1042/bj20011631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valensin D. Kamysz W. Gaggelli E. Kozłowski H. Kulon K. Nadolny R. Valensin G. Dalton Trans. 2008:5323. doi: 10.1039/b806851h. [DOI] [PubMed] [Google Scholar]
- Su Q. Riggs-Gelasco P. Conklin S. E. Haas K. L. Franz K. J. Bridgman E. C. Su Q. Riggs-Gelasco P. Haas K. L. Franz K. J. Biochemistry. 2017;56:4244–4255. doi: 10.1021/acs.biochem.7b00958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegoni M. Ramoni R. Bignetti E. Spinelli S. Cambillau C. Nat. Struct. Biol. 1996;3:863–867. doi: 10.1038/nsb1096-863. [DOI] [PubMed] [Google Scholar]
- Tudisco C. Cambria M. T. Sinatra F. Bertani F. Alba A. Giuffrida A. E. Saccone S. Fantechi E. Innocenti C. Sangregorio C. Dalcanale E. Condorelli G. G. J. Mater. Chem. B. 2015;3:4134–4145. doi: 10.1039/C5TB00547G. [DOI] [PubMed] [Google Scholar]
- Gharbi K. Salles F. Mathieu P. Amiens C. Collière V. Coppel Y. Philippot K. Fontaine L. Montembault V. Smiri L. S. Ciuculescu-Pradines D. New J. Chem. 2017;41:11898–11905. doi: 10.1039/C7NJ02482G. [DOI] [Google Scholar]
- Plank C. Trends Biotechnol. 2008;26:59–63. doi: 10.1016/j.tibtech.2007.11.001. [DOI] [PubMed] [Google Scholar]
- Mohammed L. Gomaa H. G. Ragab D. Zhu J. Particuology. 2017;30:1–14. doi: 10.1016/j.partic.2016.06.001. [DOI] [Google Scholar]
- Smecca E. Motta A. Fragalaì M. E. Aleeva Y. Condorelli G. G. J. Phys. Chem. C. 2013;117:5364–5372. doi: 10.1021/jp308983p. [DOI] [Google Scholar]
- Bedanta S. Kleemann W. J. Phys. D: Appl. Phys. 2009;42:013001. doi: 10.1088/0022-3727/42/1/013001. [DOI] [Google Scholar]
- Vashist S. K. Dixit C. K. Biochem. Biophys. Res. Commun. 2011;411:455–457. doi: 10.1016/j.bbrc.2011.06.187. [DOI] [PubMed] [Google Scholar]
- Wiechelman K. J. Braun R. D. Fitzpatrick J. D. Anal. Biochem. 1988;175:231–237. doi: 10.1016/0003-2697(88)90383-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.