Abstract
Hibiscus cannabinus L. leaves (HCLLs) are considered a favorable source of natural antiobesity substances. However, actual bioactive compound(s) in it and their mechanism(s) against obesity have not been confirmed. Hence, network pharmacology was conducted to identify its key compounds and mechanism(s) against obesity. Compounds in HCLLs were identified through GC-MS analysis and screened by Lipinski's rule. Genes related to the selected compounds and obesity were obtained from public databases, and overlapping genes between HCLL compound-related genes and obesity target genes were selected using a Venn diagram. The networking between selected compounds and overlapping genes was then constructed, visualized, and analyzed by RStudio. Finally, the binding affinity between compounds and genes was evaluated via molecular docking (MD). A total of 30 compounds in HCLLs were detected via GC-MS, and Lipinski's rule accepted all compounds. The compound-related genes (570 genes) and obesity targeted genes (3028 genes) were identified, and between them, 64 overlapping genes were selected. Gene Set Enrichment Analysis (GSEA) displayed that the mechanisms of HCLLs against obesity were associated with 13 signaling pathways on 22 compounds in HCLLs. Superficially, AKT1, vitamin E, and RAS signaling pathways were noted as a hub gene, an uppermost bioactive compound, and a hub signaling pathway, respectively. However, the binding affinity of ligands and proteins on the RAS signaling pathway was very low; instead, the PPAR signalling pathway was evaluated with potent efficacy against obesity through MD. On the PPAR signaling pathway, α-amyrin was found as the most significant compound for the amelioration of obesity. α-Amyrin manifested the strongest binding affinity on six target proteins associated with the PPAR signaling pathway. Our study suggests that an auxiliary (PPAR) signaling pathway of HCLLs might intervene efficiently against obesity over the hub (RAS) signaling pathway.
Hibiscus cannabinus L. leaves (HCLLs) are considered a favorable source of natural antiobesity substances.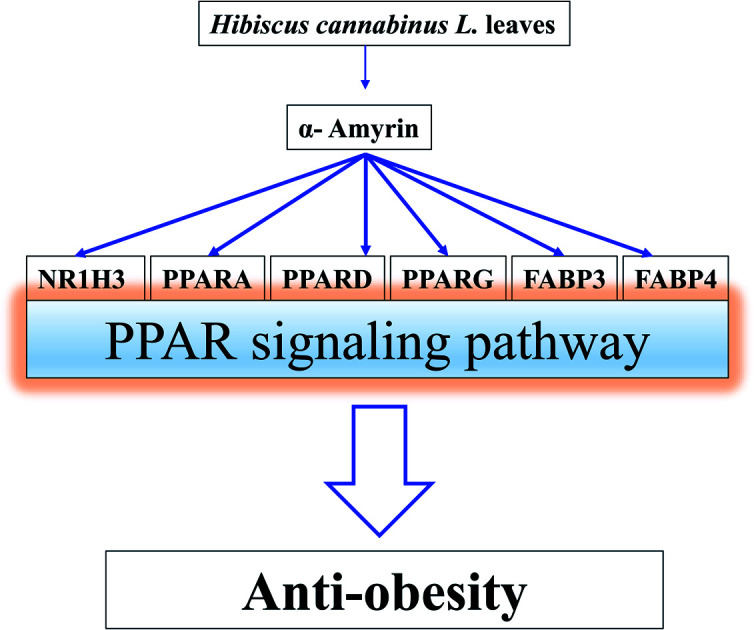
1. Introduction
Obesity is a perplexing health issue, it can cause diabetes, heart failure, stroke, and some types of cancer, and is linked to deaths worldwide.1 Obesity is characterized by excessive body fat, wherein the body mass index (BMI) reaching more than thirty is considered obesity.2 Obesity can appear at any age; about 13% (11% of men and 15% of women) of adults and 7% (6% of girls and 8% of boys) of children and adolescents were obese in 2016.3 Obesity also gives to rise an enormous burden on individual, family members, and even nations, resulting in economic disadvantages.4 The main driving factor of obesity is the breaking down oily compounds (from food sources) into small glycerol molecules and fatty acids that are absorbed quickly in human cells and accumulate in the body. In this regard, the inhibition of metabolism can be a critical therapeutic strategy.5 Currently, antiobesity drugs are being used to inhibit intestinal fat absorption (Orlistat). Also, suppressing appetite (diethylpropion, fenfluramine, sibutramine, rimonabant) is another strategy followed by most countries to control obesity.6 However, recent researches revealed that herbal plants might play a significant role in treating obesity, and among various plants, Kenaf (Hibiscus cannabinus L.) leaves are reported to be effective for antiobesity.7,8 Moreover, our previous studies proved that various kenaf parts have promising biological activities.9–12 To date, natural plant products are used as dietary supplements and further therapeutic agents for health promotion to prevent obesity.13
A study showed that administration of H. cannabinus extract to female Wistar albino rats fed with a high cholesterol diet significantly reduced the level of serum cholesterol, triglycerides, and low-density lipoprotein cholesterol (LDL-C).14 In addition, obesity-induced heart failure and hypertension can be ameliorated by the presence of abundant polyphenols and flavonoids in HCLLs.15 At present, uppermost bioactive compounds and mechanisms of HCLLs against obesity have remained unknown, and should be strengthened to foster pharmacological evidence to provide its therapeutic application in ameliorating obesity.
By utilizing a suitable analytical method, “network pharmacology”, it is possible to systematically evaluate the complex compound-gene and compound–therapeutics interactions, thus, determining a prototype for efficient treatment.16,17 Network pharmacology can be used to discover novel mechanisms of drugs, holistically, which is directed to a paradigm instead of “multiple targets, multiple diseases” of “one target, one disease”.18 Therefore, it is an efficient approach to select potential lead compounds (from natural sources) with a specific mechanism of action for the amelioration of various diseases, and mostly to elucidate the synergistic efficacy of bioactive compounds.19
A report indicated that progressive development of bioinformatics, systems biology, and poly-pharmacology facilitates a network-based drug discovery, which is considered as an efficient new drug development method.20 Another report explicated that network pharmacology is utilized as a powerful tool to understand the pharmacological mechanism(s) between the traditional Ge-Gen-Qin-Lian Decoction (GGQLD) formula and target genes.21 Furthermore, the development of bioinformatics contributes significantly to the methodology of network pharmacology. For instance, Dr Shoichet's group developed the ‘Similarity Ensemble Approach’ (SEA) to search the ligand–target interaction and ultimately understand its association.22 Also, SwissTargetPrediction (STP) is an online tool developed by SIB (Swiss Institute of Bioinformatics), which uploaded 376 342 experimentally approved molecules and 3068 protein targets since 2014.23 In this work, we utilized the two-bioinformatics tools to construct the ligand–protein interaction. To sum things up, network pharmacology via the bioinformatics tool was utilized to investigate the chemical constituents and mechanism(s) of HCLLs against obesity. The analysis processing is shown in Fig. 1.
Fig. 1. Analytical processing steps of HCLLs against obesity.
2. Materials and methods
2.1. Plant material collection and identification
Hibiscus cannabinus L. leaves (HCLLs) were collected from Pyeonghwa-ro of Chuncheon-si (latitude: 37.885596, longitude: 127.730032), Gangwon-do, Republic of Korea, in September 2020, and the plant was identified by Dr Dong Ha Cho, Plant biologist and Professor, Department of Bio-Health Convergence, College of Biomedical Science, Kangwon National University. A voucher number (KEN 305) has been deposited at Kenaf Corporation in the Department of Bio-Health Convergence, and the material can be only used for research.
2.2. Plant preparation, extraction
The collected HCLLs were dried at a shady area at room temperature (20–22 °C), and dried leaves were powdered using an electric blender. Approximately 200 g of HCLLs powder was soaked in 500 mL of 100% methanol (Daejung, Korea) for 3 days and repeated 3 times to collect the extraction. The solvent extract was collected, filtered, and evaporated using a vacuum evaporator (IKA-RV8, Japan). The evaporated sample was dried under a boiling water bath (IKA-HB10, Japan) at 40 °C to obtain the yield.
2.3. GC-MS analysis condition
Agilent 7890A was used to perform GC-MS analysis. The GC was equipped with a DB-5 (30 m × 0.25 mm × 0.25 μm) capillary column. Initially, the instrument was maintained at a temperature of 100 °C for 2.1 minutes. The temperature rose to 300 °C at a rate of 25°C min−1 and was maintained for 20 minutes. Injection port temperature and helium flow rate were ensured as 250 °C and 1.5 mL min−1, respectively. The ionization voltage was 70 eV. The samples were injected in a split mode at 10 : 1. The MS scan range was set at 35–900 (m/z). The fragmentation patterns of mass spectra were compared with those stored in the W8N05ST library MS database. The percentage of each compound was calculated from the relative peak area of each compound in the chromatogram. The concept of integration was used the ChemStation integrater algorithms.24
2.4. Chemical compounds database construction and drug-likeness filtering
The information about chemical compounds from HCLLs was identified through GC-MS analysis. GC-MS detected 30 compounds filtered according to Lipinski's rule through SwissADME (http://www.swissadme.ch/) to identify the “Drug-likeness” property. The PubChem (https://pubchem.ncbi.nlm.nih.gov/) was utilized to identify the SMILES (Simplified Molecular Input Line Entry System) of compounds.
2.5. Target genes related to selected compounds or obesity
Based on the SMILES, target genes linked to the compounds were selected through both Similarity Ensemble Approach (SEA) (http://sea.bkslab.org/) and SwissTargetPrediction (STP) (http://www.swisstargetprediction.ch/) with “Homo Sapiens” setting. Obesityrelated genes were identified by DisGeNET (https://www.disgenet.org/search) and OMIM (https://www.ncbi.nlm.nih.gov/omim) databases. The overlapping genes between compounds of HCLLs and obesity target genes were identified and visualized by VENNY 2.1 (https://bioinfogp.cnb.csic.es/tools/venny/).
2.6. Network construction of interacted overlapping genes
The final overlapping genes input in the STRING database with Homo sapience mode identified signaling pathways connected to the final overlapping genes. A bubble chart analyzed the identified signaling pathways through RStudio. The bubble chart shows a hub signaling pathway (the lowest rich factor) and an auxiliary signaling pathway (the highest rich factor) between bioactive compounds and obesity targeted genes of HCLLs.
2.7. Preparation for MD of target proteins
Four target proteins of a hub signaling pathway i.e., AKT1 (PDB ID: 1UNQ), PRKCA (PDB ID: 3IW4), PLA2G4A (PDB ID: 1KVO), and PLA2G2A (PDB ID: IBCI) were selected on STRING via RCSB PDB (https://www.rcsb.org/). Six target proteins of an auxiliary signaling pathway, i.e., NR1H3 (PDB ID: 2ACL), PPARA (PDB ID: 3SP6), PPARD (PDB ID: 5U3Q), PPARG (PDB ID: 3E00), FABP3 (PDB ID: 5HZ9), and FABP4 (PDB ID: 3P6D) were selected on STRING via RCSB PDB (https://www.rcsb.org/). The proteins selected in the .pdb format were converted into the .pdbqt format via Autodock (http://autodock.scripps.edu/).
2.8. Preparation for MD of positive control compounds
Five positive control compounds on PPARA agonists, i.e., clofibrate (PubChem ID: 2196), gemfibrozil (PubChem ID: 3463), cprofibrate (PubChem ID: 2763), bezafibrate (PubChem ID: 39042), fenofibrate (PubChem ID: 3339) were selected to identify the docking score. One positive control compound on PPARD agonist, i.e., cardarine (PubChem ID: 9803963), was selected to identify the docking score. Three positive control compounds on PPARG agonists, i.e., pioglitazone (PubChem ID: 4829), Rosiglitazone (PubChem ID: 77999), Lobeglitazone (PubChem ID: 9826451), were selected to identify the docking score.
2.9. Preparation for MD of ligand molecules
The ligand molecules were converted from .sdf from PubChem into .pdb format using Pymol, and the ligand molecules were converted into .pdbqt format through Autodock.
2.10. Ligand–protein docking
The ligand molecules were docked with target proteins utilizing autodock4 by setting-up 4 energy ranges and 8 exhaustiveness as default to obtain 10 different poses of ligand molecules.25 The active site's grid box size was x = 20.973 Å, y = 25.96 Å and z = 41.239 Å. The 2D-binding interactions were identified through LigPlot+ v.2.2 (https://www.ebi.ac.uk/thornton-srv/software/LigPlus/). After docking, ligands of the lowest binding energy (highest affinity) were selected to visualize the ligand–protein interaction in pymol.
2.11. Chemicals and reagents for HPLC analysis
Standard α-amyrin was purchased from Sigma Aldrich (USA). HPLC grade acetonitrile was obtained from Burdick & Jackson. Ultrapure water obtained using a Milli-Q UF-Plus instrumentation (Millipore, MA, USA) was utilized to prepare all solutions for the method.
2.12. Instrumentation and chromatographic conditions
HPLC Agilent 1260 series chromatographic instrumentation was used in this research. Data were collected and processed with the Agilent 1260 chemstation. The HPLC system was equipped with an injection valve, quaternary gradient pump system, and UV dual λ absorbance detector. Chromatographic separation was performed on a C18 column 4.6 × 150 mm, 3.5 μm. The mobile phase was isocratic acetonitrile 95% (95 : 5, v/v, acetonitrile : water) at a flow rate of 10 μL min−1. Its analysis was performed at ambient temperature, and detection was made at 200 nm. The injected volume was 20 μL.
2.13. Preparation of standard solution
A stock solution of standard (α-amyrin) was prepared in MeOH. The prepared stock solutions of concentrations 7.8125, 16.625, 31.25, 62.5, 125, and 250 ppm were made to plot the standard curve.
2.14. Preparation of plant extraction for HPLC analysis
The 800 mg of HCLLs MeOH extraction was taken in a flask, 40 mL of MeOH was added and kept for 3 hours. After shaking several times, the extraction was left for 2 days at room temperature. The solution from the flask was filtered through a Whatman No. 1 filter paper. The filtered solution was passed through a 0.2 μm syringe filter and HPLC analysis was performed.
3. Results and discussion
3.1. Composition of potentially bioactive compounds from HCLLs
A total of 30 compounds in HCLLs were detected using the GC-MS analysis (Fig. 2), and the name of compounds, PubChem ID, retention time, peak area (%) are enlisted in Table 1. All 30 compounds were checked and accepted by Lipinski's rule (molecular weight ≤500 g mol−1; moriguchi octanol–water partition coefficient ≤4.15; the number of nitrogen or oxygen ≤10; the number of NH or OH ≤5), and all compounds passed the standard of “Abbott Bioavailability Score (>0.1)” verified through SwissADME (Table 2).
Fig. 2. GC-MS chromatogram of methanolic extract of HCLLs and indication of α-amyrin.
A list of the identified 30 chemical compounds from HCLLs through GC-MS.
| No. | Compounds | PubChem ID | RT (mins) | Area (%) |
|---|---|---|---|---|
| 1 | 1-Methoxycyclohexa-1,3-diene | 75098 | 3.414 | 5.23 |
| 2 | 1-Methyl-3-piperidinol | 98016 | 4.01 | 2.74 |
| 3 | 2,4-Diamino-6-pyrimidinone | 135408763 | 4.241 | 1.79 |
| 4 | 3,4-Pentadienal | 534089 | 4.549 | 4.19 |
| 5 | 3-Hydroxy-2,3-dihydromaltol | 119838 | 4.77 | 4.99 |
| 6 | 5-(Hydroxymethyl)furfural | 237332 | 5.347 | 9.54 |
| 7 | 1-Octanamine | 8143 | 6.847 | 2.58 |
| 8 | Propanoic acid, 3-hydroxy- | 68152 | 7.077 | 2.83 |
| 9 | 2-Thio-6-azauracil | 1275976 | 7.174 | 0.58 |
| 10 | 4-Ethyl-2,5-dimethylisoxazolidine, (E)- | 22212544 | 7.202 | 0.52 |
| 11 | 2,5-Dimethoxy-4-methylbenzaldehyde | 602019 | 7.25 | 0.78 |
| 12 | N-Hydroxymethylacetamide | 69365 | 7.318 | 0.32 |
| 13 | 2-Furanone | 140765 | 7.741 | 2.57 |
| 14 | 2,3-Dihydro-5H-1,4-dioxepine | 536111 | 8.202 | 1.35 |
| 15 | Oleic acid | 445639 | 8.279 | 0.54 |
| 16 | Loliolide | 100332 | 8.327 | 1.77 |
| 17 | Citronellylacetone | 102604 | 8.395 | 1.23 |
| 18 | Cyclopentaneundecanoic acid, methyl ester | 535041 | 8.75 | 0.88 |
| 19 | Palimitic acid | 985 | 9.039 | 0.85 |
| 20 | Methyl elaidolinolenate | 5367462 | 9.443 | 1.65 |
| 21 | Phytol | 5366244 | 9.481 | 0.93 |
| 22 | Linolenyl alcohol | 6436081 | 9.616 | 4.26 |
| 23 | Stearic acid | 5281 | 9.674 | 3.31 |
| 24 | Octyl adipate | 7641 | 10.424 | 0.78 |
| 25 | Monopalmitin | 14900 | 10.933 | 3.66 |
| 26 | 9,12,15-Octadecatrienol | 68169 | 11.75 | 7.31 |
| 27 | Squalene | 638072 | 12.25 | 1.72 |
| 28 | Vitamin E | 14985 | 14.664 | 5.19 |
| 29 | Clionasterol | 457801 | 17.058 | 4.70 |
| 30 | α-Amyrin | 73170 | 18.558 | 4.59 |
Physicochemical properties of the 20 compounds for good oral bioavailabilitya.
| No. | Compounds | Lipinski's rules | Lipinski's violations | Bioavailability score | |||
|---|---|---|---|---|---|---|---|
| MW | HBA | HBD | Mlog P | ||||
| <500 | <10 | ≤5 | ≤4.15 | ≤1 | >0.1 | ||
| 1 | 1-Methoxycyclohexa-1,3-diene | 110.15 | 1 | 0 | 1.24 | 0 | 0.55 |
| 2 | 1-Methyl-3-piperidinol | 115.17 | 2 | 1 | 0.21 | 0 | 0.55 |
| 3 | 2,4-Diamino-6-pyrimidinone | 126.12 | 2 | 3 | −1.82 | 0 | 0.55 |
| 4 | 3,4-Pentadienal | 82.10 | 2 | 1 | 0.81 | 0 | 0.55 |
| 5 | 3-Hydroxy-2,3-dihydromaltol | 144.13 | 4 | 2 | −1.77 | 0 | 0.85 |
| 6 | 5-(Hydroxymethyl)furfural | 126.11 | 3 | 1 | −1.06 | 0 | 0.55 |
| 7 | 1-Octanamine | 129.24 | 1 | 1 | 2.22 | 0 | 0.55 |
| 8 | Propanoic acid, 3-hydroxy- | 90.08 | 3 | 2 | −0.85 | 0 | 0.85 |
| 9 | 2-Thio-6-azauracil | 129.14 | 2 | 2 | −1.25 | 0 | 0.55 |
| 10 | 4-Ethyl-2,5-dimethylisoxazolidine, (E)- | 129.20 | 2 | 0 | 1.38 | 0 | 0.55 |
| 11 | 2,5-Dimethoxy-4-methylbenzaldehyde | 180.20 | 3 | 0 | 1.13 | 0 | 0.55 |
| 12 | N-Hydroxymethylacetamide | 89.09 | 2 | 2 | −0.85 | 0 | 0.55 |
| 13 | 2-Furanone | 84.07 | 2 | 0 | −0.01 | 0 | 0.55 |
| 14 | 2,3-Dihydro-5H-1,4-dioxepine | 100.01 | 2 | 0 | −0.31 | 0 | 0.55 |
| 15 | Oleic acid | 282.46 | 2 | 1 | 4.57 | 1 | 0.85 |
| 16 | Loliolide | 196.24 | 3 | 1 | 1.49 | 0 | 0.55 |
| 17 | Citronellylacetone | 196.33 | 1 | 0 | 3.43 | 0 | 0.55 |
| 18 | Cyclopentaneundecanoic acid, methyl ester | 268.43 | 2 | 0 | 4.04 | 0 | 0.55 |
| 19 | Palmitic acid | 256.42 | 2 | 1 | 4.19 | 1 | 0.85 |
| 20 | Methyl elaidolinolenate | 292.46 | 2 | 0 | 4.61 | 1 | 0.55 |
| 21 | Phytol | 296.53 | 1 | 1 | 5.25 | 1 | 0.55 |
| 22 | Linolenyl alcohol | 264.45 | 1 | 1 | 4.59 | 1 | 0.55 |
| 23 | Stearic acid | 284.48 | 2 | 1 | 4.67 | 1 | 0.85 |
| 24 | Octyl adipate | 370.57 | 4 | 0 | 4.55 | 1 | 0.55 |
| 25 | Monopalmitin | 330.50 | 4 | 2 | 3.18 | 0 | 0.55 |
| 26 | 9,12,15-Octadecatrienol | 264.45 | 1 | 1 | 4.59 | 1 | 0.55 |
| 27 | Squalene | 410.72 | 0 | 0 | 7.93 | 1 | 0.55 |
| 28 | Vitamin E | 430.71 | 2 | 1 | 6.14 | 1 | 0.55 |
| 29 | Clionasterol | 414.71 | 1 | 1 | 6.73 | 1 | 0.55 |
| 30 | α-Amyrin | 426.72 | 1 | 1 | 6.92 | 1 | 0.55 |
MW, molecular weight (g mol−1); HBA, hydrogen bond acceptor; HBD, hydrogen bond donor; log P, lipophilicity; bioavailability score, the ability of a drug or other substance to be absorbed and used by the body.
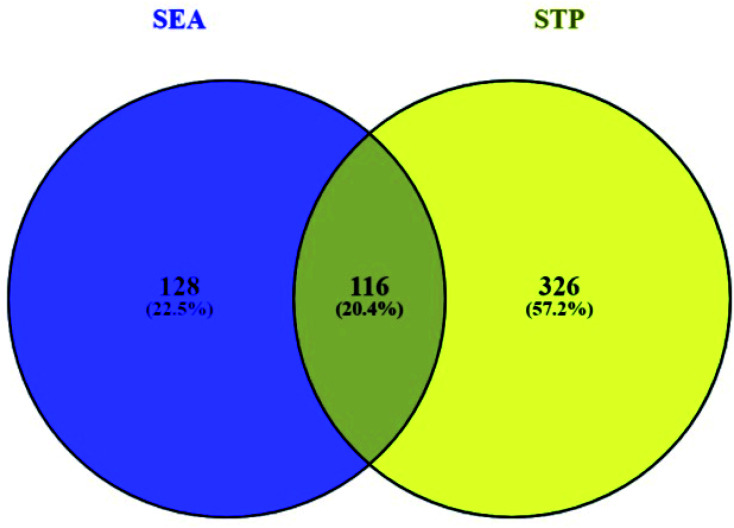
3.2. Overlapping genes between SEA and STP networked with 30 compounds
Based on the SMILES, a total of 244 genes from SEA and 442 genes from STP connected to 30 compounds were extracted (ESI Table S1†). A Venn diagram showed that 116 genes were overlapped between the two public databases (Fig. 3).
Fig. 3. Overlapping genes (116 genes) between SEA (244 genes) and STP (442 genes).
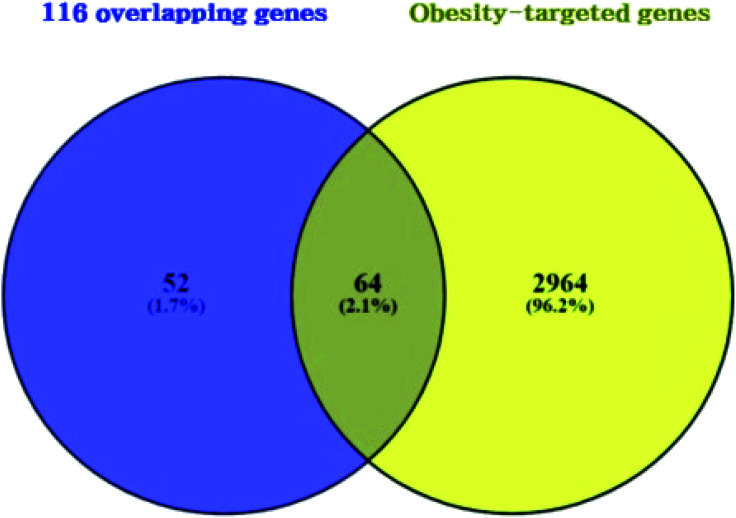
3.3. Overlapping genes between obesity related genes and 116 overlapping genes
A total of 3028 genes related to T2DM were sorted by browsing DisGeNET and OMIM databases (ESI Table S2†). Venn diagram's result unveiled 64 overlapping genes that were identified between 3028 genes related to obesity and the 116 overlapping genes (Fig. 4) (ESI Table S3†). As shown in ESI Table S4.† A total of 64 overlapping genes linked to 22 compounds from the 30 compounds mentioned above were identified, retrieving from both SEA and STP public databases, and no genes were found associated with the other 8 compounds (1-methoxycyclohexa-1,3-diene; 2,4-diamino-6-pyrimidinone; 3,4-pentadienal; 3-hydroxy-2,3-dihydromaltol; 5-(hydroxymethyl)furfural; 2-thio-6-azauracil; 2-furanone; 2,3-dihydro-5H-1,4-dioxepine) in the two databases.
Fig. 4. Overlapping genes between 116 overlapping genes (figure as mentioned earlier 3) from two databases (A) and obesity targeted genes (3028 genes).
3.4. Protein–protein network analysis of 22 compounds of HCLLs against obesity
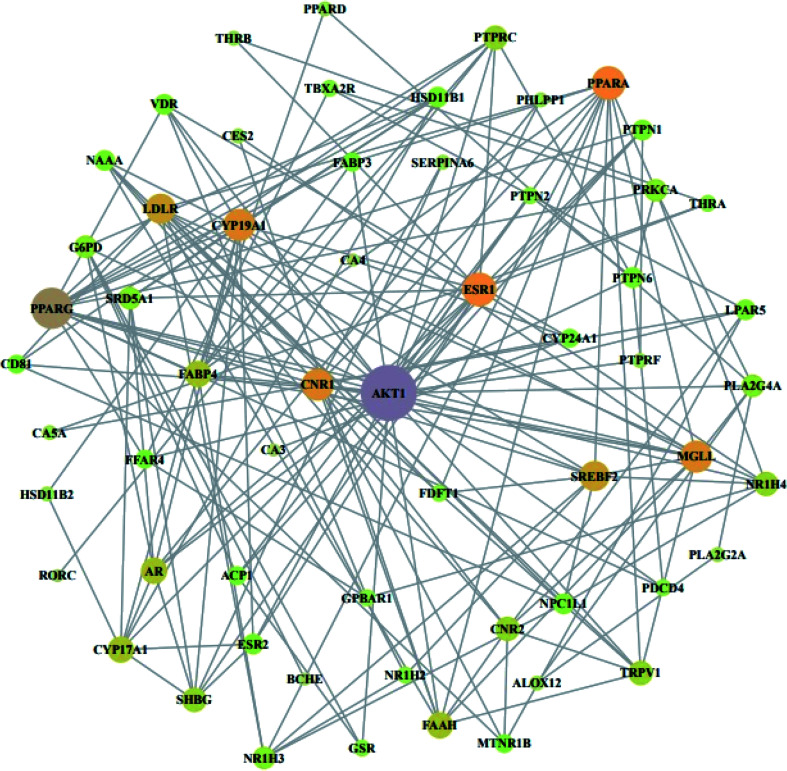
From STRING analysis, 59 out of 64 overlapping genes were closely associated, indicating 59 nodes and 163 edges (Fig. 5). The other (removed) 5 genes (ERN1, SLC22A6, PAM, ADH1B, and GSTK) had no association with the overlapping 64 genes. In the protein–protein interaction (PPI), the AKT1 gene with the highest degree (24) was considered as a hub gene (Table 3).
Fig. 5. PPI network against obesity. Nodes: the number of networks of compounds; edges: interactions between compounds and genes.
Degree of values of target proteins.
| No. | Gene symbol | Degree | No. | Gene symbol | Degree |
|---|---|---|---|---|---|
| 1 | AKT1 | 24 | 31 | NAAA | 5 |
| 2 | PPARG | 16 | 32 | ACP1 | 4 |
| 3 | ESR1 | 12 | 33 | CD81 | 4 |
| 4 | PPARA | 12 | 34 | CYP24A1 | 4 |
| 5 | CNR1 | 11 | 35 | FABP3 | 4 |
| 6 | CYP19A1 | 11 | 36 | FFAR4 | 4 |
| 7 | MGLL | 11 | 37 | MTNR1B | 4 |
| 8 | LDLR | 10 | 38 | FDFT1 | 3 |
| 9 | SREBF2 | 10 | 39 | GPBAR1 | 3 |
| 10 | AR | 8 | 40 | NR1H2 | 3 |
| 11 | CYP17A1 | 8 | 41 | PDCD4 | 3 |
| 12 | FAAH | 8 | 42 | PTPN2 | 3 |
| 13 | FABP4 | 8 | 43 | THRA | 3 |
| 14 | CNR2 | 7 | 44 | GSR | 3 |
| 15 | TRPV1 | 7 | 45 | TBXA2R | 3 |
| 16 | NR1H4 | 7 | 46 | ALOX12 | 2 |
| 17 | PTPRC | 7 | 47 | CA5A | 2 |
| 18 | SHBG | 7 | 48 | CES2 | 2 |
| 19 | G6PD | 6 | 49 | PLA2G2A | 2 |
| 20 | NR1H3 | 6 | 50 | SERPINA6 | 2 |
| 21 | PLA2G4A | 6 | 51 | PHLPP1 | 2 |
| 22 | PRKCA | 6 | 52 | PTPRF | 2 |
| 23 | SRD5A1 | 6 | 53 | HSD11B2 | 2 |
| 24 | ESR2 | 5 | 54 | THRB | 2 |
| 25 | HSD11B1 | 5 | 55 | PPARD | 2 |
| 26 | LPAR5 | 5 | 56 | BCHE | 1 |
| 27 | NPC1L1 | 5 | 57 | CA3 | 1 |
| 28 | PTPN1 | 5 | 58 | CA4 | 1 |
| 29 | PTPN6 | 5 | 59 | RORC | 1 |
| 30 | VDR | 5 |
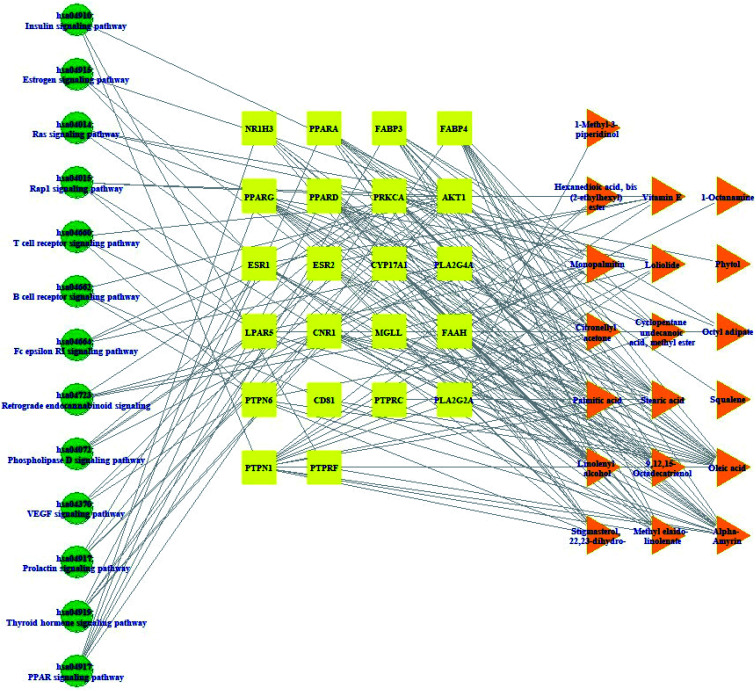
3.5. Pathway(s)–target protein(s)–compound(s) network analysis of HCLLs against obesity
Pathway(s)–target protein(s)–compound(s) of HCLLs are exhibited in Fig. 6. There were 19 bioactives, 22 target proteins, and 13 pathways (54 nodes, 200 edges). The nodes represented a total number of bioactives, target proteins, and pathways. The edges represented relationships of the three components. The pathway(s)–target protein(s)–compound(s) relationship suggested that the network might interact with therapeutic efficacy against obesity.
Fig. 6. Pathway(s)–target protein(s)–compound(s) network.
3.6. Finding of a hub signaling and hub gene of HCLLs against obesity
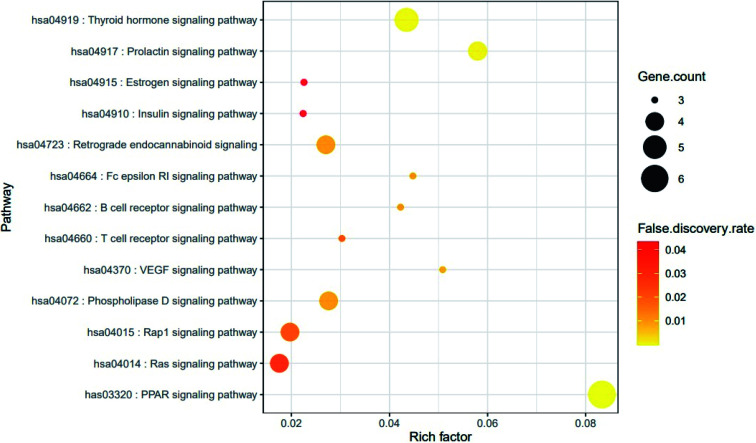
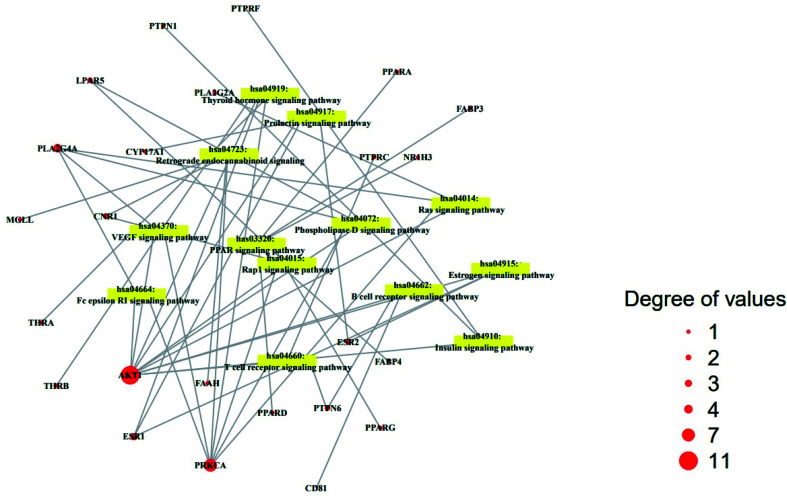
The result of KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway enrichment analysis demonstrated that 64 genes were related to 13 signaling pathways (false discovery rate < 0.05). The 13 signaling pathways were directly associated with obesity and indicated that these 13 signaling pathways might be the significant pathways of HCLLs against obesity. The description of 13 signaling pathways is shown in Table 4. Additionally, a bubble chart suggested that the RAS signaling pathway might be a hub signaling pathway of HCLLs against T2DM (Fig. 7). Among 13 signaling pathways, the AKT1 gene was associated with 11 signaling pathways, representing the highest degree of value (Fig. 8). It suggested that AKT1 might be a1 hub gene of HCLLs against obesity.
Target genes in 13 signaling pathways enrichment related to obesity.
| KEGG ID & description | Target genes | False discovery rate |
|---|---|---|
| has03320: PPAR signaling pathway | NR1H3, PPARA, FABP3, FABP4, PPARG, PPARD | 0.0000333 |
| hsa04919: thyroid hormone signaling pathway | PRKCA, AKT1, ESR1, THRA, THRB | 0.0012 |
| hsa04917: prolactin signaling pathway | AKT1, ESR1, ESR2, CYP17A1 | 0.0019 |
| hsa04370: VEGF signaling pathway | AKT1, PRKCA, PLA2G4A | 0.012 |
| hsa04072: phospholipase D signaling pathway | AKT1, PRKCA, PLA2G4A, LPAR5 | 0.0138 |
| hsa04723: retrograde endocannabinoid signaling | PRKCA, CNR1, MGLL, FAAH | 0.014 |
| hsa04664: Fc epsilon RI signaling pathway | AKT1, PRKCA, PLA2G4A | 0.014 |
| hsa04662: B cell receptor signaling pathway | AKT1, PTPN6, CD81 | 0.0143 |
| hsa04660: T cell receptor signaling pathway | AKT1, PTPRC, PTPN6 | 0.0259 |
| hsa04015: Rap1 signaling pathway | AKT1, PRKCA, CNR1, LPAR5 | 0.0259 |
| hsa04014: ras signaling pathway | AKT1, PRKCA, PLA2G4A, PLA2G2A | 0.0347 |
| hsa04915: estrogen signaling pathway | AKT1, ESR1, ESR2 | 0.0433 |
| hsa04910: insulin signaling pathway | AKT1, PTPN1, PTPRF | 0.0433 |
Fig. 7. Bubble chart of 13 signaling pathways connected to the occurrence and development of obesity.
Fig. 8. Degree values of 22 compounds connected to 13 signaling pathways.
3.7. MD of 4 genes and 11 compounds related to the RAS signaling pathway
From the SEA and STP databases, it was revealed that the AKT1 gene was associated with three compounds (vitamin E, octyl adipate, and monopalmitin), the PRKCA gene with eight compounds (octyl adipate, monopalmitin, methyl elaidolinolenate, oleic acid, stearic acid, palmitic acid, phytol, and loliolid), the PLA2G2A gene with six compounds (octyl adipate, oleic acid, stearic acid, palmitic acid, 1-methyl-3-piperidinol, and loliolid), and the PLA2G4A gene with three compounds (oleic acid, stearic acid, and palmitic acid).
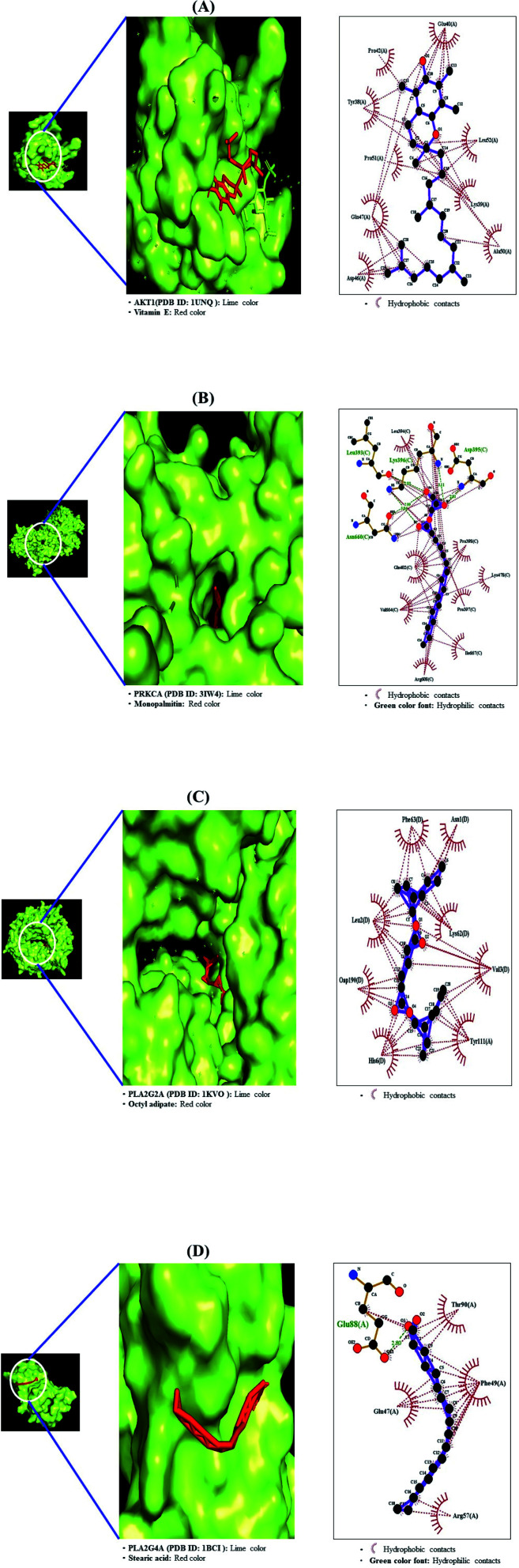
The MD was performed to evaluate the ligand–protein affinity of these four genes against their association with each gene and the highest ligand–protein docking figures are depicted in Fig. 9. AKT1 protein (PDB ID: 1UNQ) connected to the three compounds on the RAS signaling pathway was subjected to MD. It was observed that vitamin E (−5.5 kcal mol−1) docked on AKT1 protein (PDB ID: 1UNQ) had the greatest binding energy, followed by monopalmitin (−5.3 kcal mol−1) and octyl adipate (−4.8 kcal mol−1). The MD score of eight compounds on PRKCA protein (PDB ID: 3IW4) was analyzed in the “Homo Sapiens” setting. It was exposed that monopalmitin (−6.7 kcal mol−1) docked on PRKCA protein (PDB ID: 3IW4) had the highest binding energy, followed by octyl adipate (−6.2 kcal mol−1), loliolid (−5.8 kcal mol−1), phytol (−5.6 kcal mol−1), methyl elaidolinolenate (−5.3 kcal mol−1), oleic acid (−4.9 kcal mol−1), palmitic acid (−4.7 kcal mol−1), and stearic acid (−4.2 kcal mol−1). The MD score of six compounds on PLA2G2A protein (PDB ID: 1KVO) was analyzed in the “Homo Sapiens” setting. It was revealed that octyl adipate (−6.2 kcal mol−1) docked on PLA2G2A protein (PDB ID: 1KVO) had the highest binding energy, followed by stearic acid (−6.0 kcal mol−1), loliolid (−5.8 kcal mol−1), oleic acid (−5.2 kcal mol−1), palmitic acid (−5.2 kcal mol−1), and 1-methyl-3-piperidinol (−4.3 kcal mol−1). The MD score of the three compounds on PLA2G4A protein (PDB ID: 1BCI) was analyzed in the “Homo Sapiens” setting. It was observed that stearic acid (−4.4 kcal mol−1) docked on PLA2G4A protein (PDB ID: 1BCI) had the highest binding energy, followed by oleic acid (−3.4 kcal mol−1) and palmitic acid (−3.1 kcal mol−1). Collectively, these results showed that the affinity of each compound was not striking a binding score. The docking results are shown in Table 5. It implies that 11 compounds connected to the RAS signaling pathway (a hub signaling pathway) might have a low potency level on target proteins (AKT1, PRKCA, PLA2G2A, PLA2G4A).
Fig. 9. Molecular docking interaction on RAS signaling pathway between best-docked compounds from HCLLs and target proteins. (A) Vitamin E on AKT1 (PDB ID: 1UNQ) (B) monopalmitin on PRKCA (PDB ID: 3IW4) (C) 1-methyl-3-piperidinol on PLA2G2A (PDB ID: 1KVO) (D) stearic acid on PLA2G4A (PDB ID: 1BCI).
Binding energy and interactions of potential active compounds on RAS signaling pathway.
| Protein | Ligand | PubChem ID | Binding energy (kcal mol−1) | Hydrogen bond interactions | Hydrophobic interactions |
|---|---|---|---|---|---|
| Amino acid residue | Amino acid residue | ||||
| AKT1 (PDB ID: 1UNQ) | Vitamin E | 14985 | −5.5 | N/A | Glu40, Leu52, Lys39 |
| Ala50, Asp46, Gln47 | |||||
| Pro51, Tyr38, Pro42 | |||||
| Monopalmitin | 14900 | −5.3 | Glu95, Glu91 | Val90, Leu12, Trp11 | |
| Glu40, Lys39, Pro24 | |||||
| His13 | |||||
| Octyl adipate | 7641 | −4.8 | Leu52, Lys39 | Ala50, Glu40, Pro42 | |
| Tyr38, Gln47 | |||||
| PRKCA (PDB ID: 3IW4) | Monopalmitin | 14900 | −6.7 | Leu393, Lys396, Asp395 | Leu394, Pro398, Lys478 |
| Asn660 | Pro397, Ile667, Arg608 | ||||
| Val664, Gln402 | |||||
| Octyl adipate | 7641 | −6.2 | N/A | Pro397, Pro398, Gln402 | |
| Asn660, Lys396, Leu394 | |||||
| Gln662, Asp395, Glu552 | |||||
| Val664 | |||||
| Loliolid | 100332 | −5.8 | Arg608, His476, Asp472 | Glu474, Glu609, Gln548 | |
| Glu552 | Asn607, Met551, Ile667 | ||||
| Phytol | 5366244 | −5.6 | Asp395, Leu393, Lys396 | Asn660, Glu552, Gln662 | |
| His553, Ser549, Gln548 | |||||
| Val664, Gln402, Pro398 | |||||
| Pro397 | |||||
| Methyl elaidolinoleate | 5367462 | −5.3 | Lys396, Asn660 | Gln402, Val664, Pro397 | |
| Pro398, Glu552, Gln662 | |||||
| His553, Glu545, Ser549 | |||||
| Asp539, Leu393, Asp395 | |||||
| Oleic acid | 445639 | −4.9 | Asp395, Asn660 | Gln402, Leu394, Lys396 | |
| Gln662, Glu552, Pro398 | |||||
| Pro397, Val664 | |||||
| Palmitic acid | 985 | −4.7 | Asn660, Lys396, Leu393 | Leu394, Gln662, Asp395 | |
| Gln402, Pro398, Glu552 | |||||
| Val664, Gln548 | |||||
| Stearic acid | 5281 | −4.2 | Asn660, Leu393, Lys396 | Gln402, Arg608, Pro398 | |
| Glu418, Lys478, Pro666 | |||||
| His665, Val664 | |||||
| PLA2G2A (PDB ID: 1KVO) | Octyl adipate | 7641 | −6.2 | N/A | Phe63, Asn1, Lys62 |
| Val3, Tyr111, His6 | |||||
| Leu2 | |||||
| Stearic acid | 5281 | −6.0 | Val30 | Cys28, Phe23, Gly29 | |
| Tyr111, Val3, Tyr112 | |||||
| Asn114 | |||||
| Loliolid | 100332 | −5.8 | Lys115 | Thr121, Gly32, Arg33 | |
| Lys52, Asp48, Cys49 | |||||
| Oleic acid | 445639 | −5.2 | Val3 | Phe63, Lys62, Phe23 | |
| Tyr111, His6, Leu2 | |||||
| Asn1 | |||||
| Palmitic acid | 985 | −4.2 | Tyr112, Gly25, Asn114, Phe23 | Tyr111, His6, Val3 | |
| Leu2, Ser113 | |||||
| 1-Methyl-3-piperidinol | 98016 | −4.3 | Cys28, Phe23, Gly25 | Val30 | |
| Tyr112, Asn114 | |||||
| PLA2G4A (PDB ID: 1BCI) | Stearic acid | 5281 | −4.4 | Glu88 | Thr90, Phe49, Arg57 |
| Glu47 | |||||
| Oleic acid | 445639 | −3.4 | Ile78 | Glu76, Phe77, Pro54 | |
| Thr53, Leu79, Tyr16 | |||||
| Palmitic acid | 985 | −3.1 | Ala94 | Asn64, Asn95, Arg61 | |
| His62, Phe63, Asp43 |
3.8. Finding auxiliary signaling over a hub-signaling pathway against obesity
Based on the RichFactor value on each signalling pathway's bubble chart, another distinguished signaling pathway was the PPAR signaling pathway with the highest RichFactor. Noticeably, the AKT1 gene was not associated with the PPAR signaling pathway on the lowest false discovery rate (3.33 × 10−7). It implies that the PPAR signaling pathway not related to a hub gene might function as auxiliary signaling.
3.9. MD of 6 genes and 14 compounds related to the PPAR signaling pathway
From SEA and STP databases, it was observed that the NR1H3 gene was connected to six compounds (clionasterol, methyl elaidolinolenate, α-amyrin, linolenyl alcohol, 9,12,15-octadecatrienol, and oleic acid), the PPARA gene was related to eleven compounds (oleic acid, palmitic acid, stearic acid, squalene, linolenyl alcohol, 9,12,15-octadecatrienol, methyl elaidolinolenate, citronellylacetone, α-amyrin, clionasterol, and cyclopentaneundecanoic acid, methyl ester), the PPARD gene was associated with ten compounds (oleic acid, palmitic acid, stearic acid, linolenyl alcohol, 9,12,15-octadecatrienol, methyl elaidolinolenate, citronellylacetone, clionasterol, α-amyrin, and cyclopentaneundecanoic acid, methyl ester), the PPARG gene was involved with ten compounds (oleic acid, palmitic acid, stearic acid, methyl elaidolinolenate, loliolide, linolenyl alcohol, 9,12,15-octadecatrienol, citronellylacetone, α-amyrin, and clionasterol), the FABP3 gene was linked to eleven compounds (oleic acid, linolenyl alcohol, 9,12,15-octadecatrienol, methyl elaidolinolenate, octyl adipate, monopalmitin, palmitic acid, stearic acid, cyclopentaneundecanoic acid, methyl ester, citronellylacetone, and α-amyrin), and the FABP4 gene was connected to nine compounds (oleic acid, palmitic acid, stearic acid, methyl elaidolinolenate, linolenyl alcohol, 9,12,15-octadecatrienol, cyclopentaneundecanoic acid, methyl ester, citronellylacetone, and α-amyrin). In addition, the STRING analysis exhibited an inter-correlation between six genes (PPI enrichment p-value: 2.96 × 10−9) (Fig. 10). MD was performed to evaluate the ligand-protein affinity of these six genes against their associated ones with each gene, respectively, and the highest ligand–protein docking affinity figures are depicted in Fig. 11. The MD score of six compounds on NR1H3 protein (PDB ID: 2ACL) was analyzed in the “Homo Sapiens” setting. It was revealed that α-amyrin (−9.7 kcal mol−1) docked on NR1H3 protein (PDB ID: 2ACL) showed the highest binding energy, followed by clionasterol (−8.1 kcal mol−1), methyl elaidolinolenate (−5.3 kcal mol−1), oleic acid (−4.9 kcal mol−1), 9,12,15-octadecatrienol (−4.8 kcal mol−1) and linolenyl alcohol (−4.8 kcal mol−1). It was observed that α-amyrin (−7.4 kcal mol−1) docked on PPARA protein (PDB ID: 3SP6) manifested the highest binding energy, followed by clionasterol (−6.7 kcal mol−1), squalene (−6.0 kcal mol−1), stearic acid (−5.7 kcal mol−1), oleic acid (−5.3 kcal mol−1), cyclopentaneundecanoic acid methyl ester (−5.2 kcal mol−1), methyl elaidolinolenate (−4.7 kcal mol−1), palmitic acid (−4.5 kcal mol−1), citronellylacetone (−4.4 kcal mol−1), 9,12,15-octadecatrienol (−4.4 kcal mol−1), and linolenyl alcohol (−3.6 kcal mol−1). It was found that α-amyrin (−8.5 kcal mol−1) docked on PPARD protein (PDB ID: 5U3Q) exhibited the highest binding energy, followed by clionasterol (−6.8 kcal mol−1), stearic acid (−5.7 kcal mol−1), 9,12,15-octadecatrienol (−5.2 kcal mol−1), palmitic acid (−5.0 kcal mol−1), oleic acid (−4.9 kcal mol−1), citronellylacetone (−4.8 kcal mol−1), methyl elaidolinolenate (−4.7 kcal mol−1), cyclopentaneundecanoic acid methyl ester (−4.7 kcal mol−1), and linolenyl alcohol (−4.6 kcal mol−1). It was revealed that α-amyrin (−8.4 kcal mol−1) docked on PPARG protein (PDB ID: 3E00) showed the highest binding energy, followed by clionasterol (−7.3 kcal mol−1), loliolide (−7.1 kcal mol−1), stearic acid (−5.6 kcal mol−1), citronellylacetone (−5.0 kcal mol−1), oleic acid (−4.9 kcal mol−1), 9,12,15-octadecatrienol (−4.8 kcal mol−1), linolenyl alcohol (−4.7 kcal mol−1), palmitic acid (−4.6 kcal mol−1), and methyl elaidolinolenate (−4.3 kcal mol−1). It was observed that α-amyrin (−10.0 kcal mol−1) docked on FABP3 protein (PDB ID: 5HZ9) had the highest binding energy, followed by stearic acid (−9.0 kcal mol−1), monopalmitin (−8.6 kcal mol−1), octyl adipate (−8.5 kcal mol−1), 9,12,15-octadecatrienol (−7.6 kcal mol−1), cyclopentaneundecanoic acid methyl ester (−7.4 kcal mol−1), citronellylacetone (−7.2 kcal mol−1), oleic acid (−7.1 kcal mol−1), methyl elaidolinolenate (−7.0 kcal mol−1), and palmitic acid (−6.9 kcal mol−1). Finally, it was also exposed that α-amyrin (−8.5 kcal mol−1) docked on FABP4 protein (PDB ID: 3P6D) demonstrated the highest binding energy, followed by stearic acid (−6.5 kcal mol−1), methyl elaidolinolenate (−5.3 kcal mol−1), cyclopentaneundecanoic acid methyl ester (−5.0 kcal mol−1), citronellylacetone (−4.5 kcal mol−1), 9,12,15-octadecatrienol (−4.4 kcal mol−1), oleic acid (−4.4 kcal mol−1), linolenyl alcohol (−4.4 kcal mol−1), and palmitic acid (−4.0 kcal mol−1). Noticeably, α-amyrin had the greatest docking score on all six proteins. The docking results are displayed in Table 6.
Fig. 10. Molecular docking interaction on PPAR signaling pathway between best-docked compounds from HCLLs and target proteins. (A) α-Amyrin on NR1H3 (PDB ID: 2ACL) (B) α-amyrin on PPARA (PDB ID: 3SP6) (C) α-amyrin on PPARB (PDB ID: 5U3Q) (D) α-amyrin on PPARG (PDB ID: 3E00) (E) α-amyrin on FABP3 (PDB ID: 5HZ9) (F) α-amyrin on FABP4 (PDB ID: 3P6D).
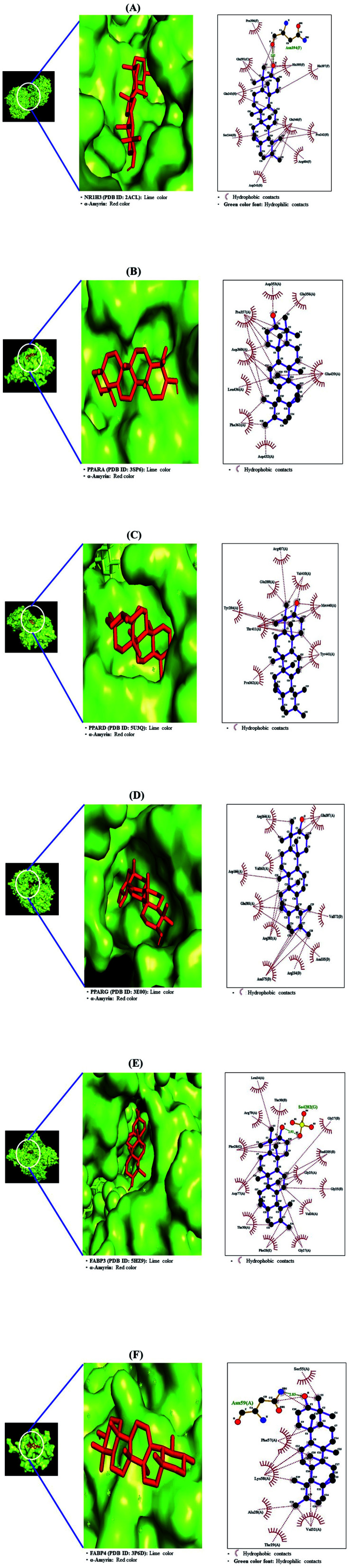
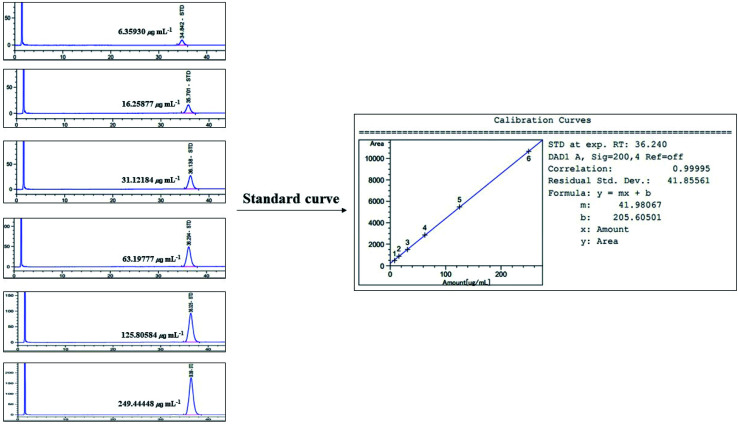
Fig. 11. Standard curve for HPLC/UV analysis of α-amyrin (200 nm).
Binding energy and interactions of potential active compounds on PPAR signaling pathway.
| Protein | Ligand | PubChem ID | Binding energy (kcal mol−1) | Hydrogen bond interactions | Hydrophobic interactions |
|---|---|---|---|---|---|
| amino acid residue | Amino acid residue | ||||
| NR1H3 (PDB ID: 2ACL) | α-Amyrin | 73170 | −9.7 | Asn394 | His395, His397, Glu346 |
| Pro242, Arg404, Asp241 | |||||
| Ser244, Gln243, Glu291 | |||||
| Clionasterol | 457801 | −8.1 | Asn385 | Arg251, Trp236, Lys326 | |
| Ile238, Glu394, Pro240 | |||||
| Ala391, Leu400, Ala398 | |||||
| Asp229, Lys395, Glu322 | |||||
| Glu388, Pro237 | |||||
| Methyl elaidolinoleate | 5367462 | −5.3 | Lys408 | Glu390, Arg404, Phe340 | |
| Pro336, Glu339, Ala343 | |||||
| Asp379, Pro386 | |||||
| Oleic acid | 445639 | −4.9 | N/A | Glu390, Lys408, Arg342 | |
| Glu339, Pro386, Pro240 | |||||
| Glu346, Tyr397, Met407 | |||||
| Arg404 | |||||
| 9,12,15-Octadecatrienol | 5367327 | −4.8 | Glu339 | Ala343, Asp379, Glu390 | |
| Lys408, Glu346, Ala387 | |||||
| Pro240, Pro386, Arg342 | |||||
| Linolenyl alcohol | 6436081 | −4.8 | N/A | Arg248, Leu294, Gln429 | |
| Ile299, Val331, Leu329 | |||||
| Ala325, Gln33, Arg302 | |||||
| Asp295, Val298, Lys431 | |||||
| PPARA (PDB ID: 3SP6) | α-Amyrin | 73170 | −7.4 | N/A | Asp353, Glu356, Glu439 |
| Asp432, Phe361, Leu436 | |||||
| Asp360, Pro357 | |||||
| Clionasterol | 457801 | −6.7 | Lys345 | Asp360, Pro357, Glu439 | |
| His440, Leu443, Asp353 | |||||
| Glu356 | |||||
| Squalene | 638072 | −6.0 | N/A | Tyr334, Asn336, Ala333 | |
| Thr279, Leu254, Il2901 | |||||
| Val332, Ile241, Glu251 | |||||
| Ala250, Cys275, Cys278 | |||||
| Val255 | |||||
| Stearic acid | 5281 | −5.7 | N/A | Phe361, Asp432, Leu436 | |
| Glu439, His440, Leu443 | |||||
| Asp353, Gln442, Ile446 | |||||
| Pro357, Lys358 | |||||
| Oleic acid | 445639 | −5.3 | N/A | Leu254, Val255, Il2901 | |
| Ala250, Ala333, Asn219 | |||||
| Thr283, Met320, Leu321 | |||||
| Val324, Ile317, Thr279 | |||||
| Tyr334, Cys275, Glu251 | |||||
| Cyclopentaneundecanoic acid methyl ester | 535041 | −5.2 | Ala333 | Asn219, Gly335, Leu331 | |
| Tyr334, Thr283, Met220 | |||||
| Glu286, Phe218, Met320 | |||||
| Leu321, Ile317, Thr279 | |||||
| Val324, Val332 | |||||
| Methyl elaidolinoleate | 5367462 | −4.7 | N/A | Asp432, Leu436, Phe361 | |
| Asp360, Leu443, Gln442 | |||||
| Glu439, Pro357, Gln435 | |||||
| Palmitic acid | 985 | −4.5 | N/A | Val255, Lys257, Leu258 | |
| Leu254, Ile241, Ala333 | |||||
| Il2901, Cys275, Thr279 | |||||
| Tyr334, Cys278 | |||||
| Citronellylacetone | 102604 | −4.4 | N/A | Lys364, Asp360, Pro357 | |
| Phe361, Glu439, Gln435 | |||||
| Asp432, Leu436 | |||||
| 9,12,15-Octadecatrienol | 5367327 | −4.4 | Thr307, Asn303 | Gly390, Leu690, Ser688 | |
| Gln691, Val306, Glu462 | |||||
| Arg465, Asp466, Lys310 | |||||
| Pro389 | |||||
| Linolenyl alcohol | 6436081 | −3.6 | Asn393, Asp304 | Ile397, Leu391, Leu300 | |
| Leu302, Asn303, Val394 | |||||
| PPARB (PDB ID: 5U3Q) | α-Amyrin | 73170 | −8.5 | N/A | Arg407, Val410, Met440 |
| Tyr441, Pro362, Thr411 | |||||
| Tyr284, Glu288 | |||||
| Clionasterol | 457801 | −6.8 | N/A | Pro268, His181, Lys265 | |
| Ser266, Ser271, Glu262 | |||||
| Lys265, Glu262, Ser271 | |||||
| Stearic acid | 5281 | −5.7 | N/A | Ser271, Glu262, Lys265 | |
| Ser266, Pro268, Ser271 | |||||
| 9,12,15-Octadecatrienol | 5367327 | −5.2 | Thr411, Tyr441 | Met440, Pro362, Glu288 | |
| Tyr284, Arg361 | |||||
| Palmitic acid | 985 | −5.0 | Thr411, Arg407 | Val410, Arg361, Tyr284 | |
| Asp360, Pro362, Tyr441 | |||||
| Met440 | |||||
| Oleic acid | 445639 | −4.9 | N/A | Asp360, Pro362, Tyr284 | |
| Val410, Met440, Tyr441 | |||||
| Thr411 | |||||
| Citronellylacetone | 102604 | −4.8 | Tyr441 | Met440, Thr411, Tyr284 | |
| Asp360, Pro362 | |||||
| Methyl elaidolinoleate | 5367462 | −4.7 | N/A | Tyr441, Met440, Thr411 | |
| Arg407, Val410, Glu288 | |||||
| Tyr284, Pro362, Arg361 | |||||
| Cyclopentaneundecanoic acid methyl ester | 535041 | −4.7 | N/A | Pro362, Tyr441, Thr411 | |
| Met440, Val410, Arg407 | |||||
| Arg361, Glu288, Tyr284 | |||||
| Linolenyl alcohol | 6436081 | −4.6 | Tyr441 | Arg361, Tyr284, Met440 | |
| Glu288, Thr411, Pro362 | |||||
| PPARG (PDB ID: 3E00) | α-Amyrin | 73170 | −8.4 | N/A | Glu207, Val372, Asn335 |
| Arg234, Asn375, Arg202 | |||||
| Glu203, Asp166, Val163 | |||||
| Arg164 | |||||
| Clionasterol | 457801 | −7.3 | Asn375 | Asp166, Val372, Val163 | |
| Arg164, Lys165, Glu208 | |||||
| Glu207, Arg202, Glu203 | |||||
| Lys336, Asn335, Ala371 | |||||
| Loliolide | 100332 | −7.1 | Arg202, Glu351 | Thr162, Leu167, Asp337 | |
| Lys336, Gln193, Tyr189 | |||||
| Thr168, Tyr169, Tyr192 | |||||
| Stearic acid | 5281 | −5.6 | N/A | Val163, Arg164, Val205 | |
| Glu203, Arg202, Lys336 | |||||
| Val372, Ala376, Asn375 | |||||
| Asp166, Glu208, Glu207 | |||||
| Gln206 | |||||
| Citronellylacetone | 102604 | −5.0 | N/A | Lys354, Lys336, Arg350 | |
| Leu167, Tyr192, Gln193 | |||||
| Tyr169, Tyr189, Asp337 | |||||
| Thr168 | |||||
| Oleic acid | 445639 | −4.9 | Lys354, Glu351 | Thr168, Lys336, Arg202 | |
| Tyr192, Leu167, Tyr169 | |||||
| Asp337, Arg350, Gln193 | |||||
| 9,12,15-Octadecatrienol | 5367327 | −4.8 | Asp337 | Tyr192, Tyr169, Tyr189 | |
| Thr168, Glu351, Glu369 | |||||
| Val372, Lys336, Arg350 | |||||
| Leu167, Gln193 | |||||
| Linolenyl alcohol | 6436081 | −4.7 | Glu448 | Ser380, Lys381, Pro366 | |
| Asp441, Phe370, Glu369 | |||||
| Lys373, Gln444, Asp379 | |||||
| Asn377 | |||||
| Palmitic acid | 985 | −4.6 | N/A | Tyr189, Tyr169, Glu351 | |
| Gln193, Lys354, Lys336 | |||||
| Arg350, Thr168, Leu167 | |||||
| Asp337, Tyr192 | |||||
| Methyl elaidolinoleate | 5367462 | −4.3 | N/A | Lys336, Arg202, Asn335 | |
| Glu203, Arg234, Asn375 | |||||
| Glu207, Lys157, Val205 | |||||
| Val372, Glu378, Gln206 | |||||
| Val163 | |||||
| FABP3 (PDB ID: 5HZ9) | α-Amyrin | 73170 | −10.0 | N/A | Leu24, Thr30, Gly27 |
| Gly25, Val26, Phe28 | |||||
| Thr30, Asp77, Phe28 | |||||
| Arg79 | |||||
| Stearic acid | 5281 | −9.0 | N/A | Phe28, Gln32, Phe58 | |
| Val26, Thr122, Asp77 | |||||
| Thr30, Lys59, Val33 | |||||
| Thr57, Lys22, Ala29 | |||||
| Monopalmitin | 14900 | −8.6 | Phe58, Thr57 | Phe28, Gln32, Val33 | |
| Octyl adipate | 7641 | −8.5 | N/A | Phe28, Val33, Gln32 | |
| Ala29, Phe58, Met36 | |||||
| Val33, Thr57 | |||||
| 9,12,15-Octadecatrienol | 5367327 | −7.6 | Thr57, Phe58 | Gly27, Gly25, Phe28 | |
| Gln32, Ala29 | |||||
| Cyclopentaneundecanoic acid methyl ester | 535041 | −7.4 | N/A | Gly27, Phe28, Phe58 | |
| Lys22, Thr57, Gln32 | |||||
| Citronellylacetone | 102604 | −7.2 | Lys22 | Thr57, Gln32, Ala29 | |
| Oleic acid | 445639 | −7.1 | N/A | Gly27, Gly25, Gln32 | |
| Ala29, Phe28 | |||||
| Methyl elaidolinoleate | 5367462 | −7.0 | N/A | Gly25, Gly27, Ala29 | |
| Phe58, Lys22, Phe28 | |||||
| Palmitic acid | 985 | −6.9 | Gly25 | Phe28, Gly27, Gln32 | |
| Ala29, Val33, Met36 | |||||
| Thr57 | |||||
| FABP4 (PDB ID: 3P6D) | α-Amyrin | 73170 | −8.5 | Asn59 | Ser55, Val32, Thr29 |
| Ala28, Lys58, Phe57 | |||||
| Stearic acid | 5281 | −6.5 | N/A | Ser1, Ile49, Asp47 | |
| Leu66, Leu86, Gly88 | |||||
| Met0 | |||||
| Methyl elaidolinoleate | 5367462 | −5.3 | Gly88 | Asp87, Leu86, Ser1 | |
| Asp47, Ile65, Leu66 | |||||
| Met0 | |||||
| Cyclopentaneundecanoic acid methyl ester | 535041 | −5.0 | N/A | Ser1, Met0, Leu86 | |
| Leu66, Asp47, Ile49 | |||||
| Citronellylacetone | 102604 | −4.5 | Lys120 | Lys100, Met119, Ser101 | |
| 9,12,15-Octadecatrienol | 5367327 | −4.4 | N/A | Leu86, Leu66, Asp47 | |
| Ile49, Ser1 | |||||
| Oleic acid | 445639 | −4.4 | Leu86, Gly88 | Asp87, Leu66, Asp47 | |
| Ser1 | |||||
| Linolenyl alcohol | 6436081 | −4.4 | Gly88, Leu86 | Asp87, Cys1, Leu66 | |
| Ser1, Met0 | |||||
| Palmitic acid | 985 | −4.0 | Leu86 | Thr85, Gly88, Met0 | |
| Leu66 |
3.10. Comparative investigation of the docking score against positive controls on target proteins
A comparative MD was performed to evaluate the affinity strength of the highest docking score ligand (α-amyrin) – target proteins. Each positive control of the target protein is as follows. The MD score of GW3965 (PubChem ID: 16078973) on NR1H3 protein (PDB ID: 2ACL) was −11.9 kcal mol−1, where α-amyrin had −9.7 kcal mol−1. The GW3965 (PubChem ID: 16078973) showed a stronger binding affinity than α-amyrin. The MD scores of clofibrate (PubChem ID: 2796), gemfibrozil (PubChem ID: 3463), ciprofibrate (PubChem ID: 2763), bezafibrate (PubChem ID: 39042), and fenofibrate (PubChem ID: 3339) on PPARA protein (PDB ID: 3SP6) were −6.4 kcal mol−1, −6.3 kcal mol−1, −5.4 kcal mol−1, −5.8 kcal mol−1, and −5.4 kcal mol−1, respectively. Interestingly, the MD score of α-amyrin on PPARA protein was more substantial (−7.4 kcal mol−1) than that of five standard drugs. The MD score of cardarine (PubChem ID: 9803963) on PPARD (PDB ID: 5U3Q) was −8.5 kcal mol−1, while α-amyrin docking score was the same as cardarine (PubChem ID: 9803963). The MD scores of pioglitazone (PubChem ID: 4829), rosiglitazone (PubChem ID: 77999), and lobeglitazone (PubChem ID: 9826451) on PPARG (PDB ID: 3E00) were −7.7 kcal mol−1, −7.4 kcal mol−1, and −7.3 kcal mol−1, respectively. Noticeably, the MD score of α-amyrin exposed a stronger binding affinity (−8.4 kcal mol−1) than the three standard drugs. The detailed information is enlisted in Table 7.
Comparative binding energy between positive controls and α-Amyrin on PPAR signaling pathway.
| Compounds | PubChem ID | Docking score (kcal mol−1) | |||||
|---|---|---|---|---|---|---|---|
| NR1H3 (PDB ID: 2ACL) | PPARA (PDB ID: 3SP6) | PPARB (PDB ID: 5U3Q) | PPARG (PDB ID: 3E00) | FABP3 (PDB ID: 5HZ9) | FABP4 (PDB ID: 3P6D) | ||
| α-Amyrin | 225688 | −9.7 | — | — | — | — | — |
| aGW3965 | 16078973 | −11.9 | — | — | — | — | — |
| α-Amyrin | 225688 | — | −7.4 | — | — | — | — |
| bClofibrate | 2796 | — | −6.4 | — | — | — | — |
| bGemfibrozil | 3463 | — | −6.3 | — | — | — | — |
| bCiprofibrate | 2763 | — | −5.4 | — | — | — | — |
| bBezafibrate | 39042 | — | −5.8 | — | — | — | — |
| bFenofibrate | 3339 | — | −5.4 | — | — | — | — |
| α-Amyrin | 225688 | — | — | −8.5 | — | — | — |
| cCardarine | 9803963 | — | — | −8.5 | — | — | — |
| α-Amyrin | 225688 | — | — | — | −8.4 | — | — |
| dPioglitazone | 4829 | — | — | — | −7.7 | — | — |
| dRosiglitazone | 77999 | — | — | — | −7.4 | — | — |
| dLobeglitazone | 9826451 | — | — | — | −7.3 | — | — |
NR1H3 agonist.
PPARA agonist.
PPARD agonist.
PPARG agonist.
3.11. Potential bioactive and signaling pathways of HCLLs against obesity
The MD results of α-amyrin on the PPAR signaling pathway revealed a high binding affinity score. In addition, α-amyrin on 4 proteins (NR1H3, PPARA, PPARD, and PPARG) out of 6 proteins on the PPAR signaling pathway indicated a higher docking score than that of the positive controls (standard drugs). The other 2 proteins (FABP3, FABP4) were not the standard drugs to compare with α-amyrin. Coincidently, α-amyrin might play an essential role in PPAR signaling pathways, suggesting that the PPAR signaling pathway might be an auxiliary signaling pathway of HCLLs against obesity, instead of the RAS signaling pathway.
3.12. Linearity of standard α-amyrin
Linearity was evaluated by the standard curve, determined using 6 different concentrations of α-amyrin dissolved in MeOH. The peak area was acquired to calculate the correlation coefficient of square linear regression analysis. The linearity of peak area responses versus concentrations was identified in the range of 6.359 μg mL−1 to 249.444 μg mL−1 (r = 0.99995, n = 6) (Fig. 11).
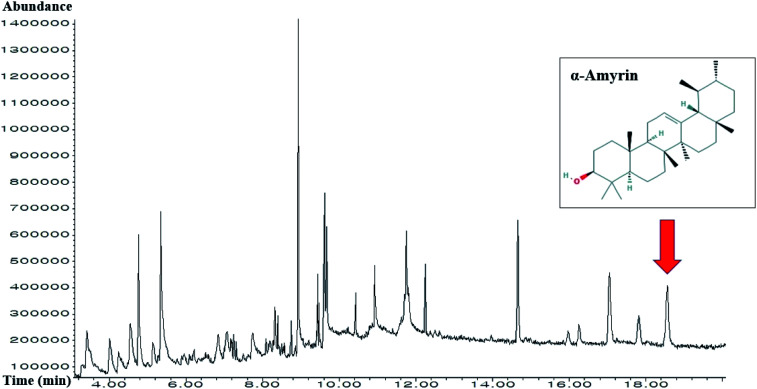
3.13. The identification of α-amyrin from HCLLs
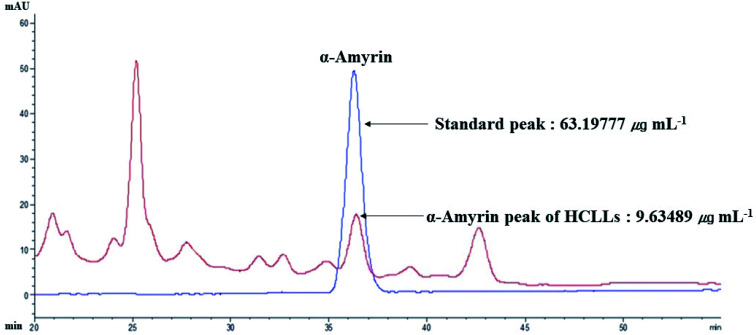
The retention time of α-amyrin was 36.294 min during the HPLC analysis, which exactly overlapped with that of the standard solution. The α-amyrin amount was 9.63489 μg mL−1 in HCLLs MeOH extraction (20 μg mL−1) (Fig. 12). The amount of α-amyrin was about 0.05% in HCLLs MeOH extract.
Fig. 12. Overlapping HPLC chromatograms obtained by standard α-amyrin (blue curve) and α-amyrin (red curve) in HCLLs MeOH extraction, wavelength = 200 nm.
3.14. A hub and an auxiliary signaling pathway among 13 signaling pathways
The selection of the optimal organic solvent is crucial to obtain various bioactive compounds from medicinal plants. Several essential metabolites (alkaloids, flavonoids, tannins and terpenoids) and some non-polar natural compounds are highly soluble in methanol. The high polarity index value of methanol stimulates the extraction rate. Hence, methanol is substantially utilized in medicinal plant extraction to prove medicinal plants' potentiality, and we used MeOH for the extraction of HCLLs.26 Then, we performed GC-MS analysis, PPI network, signaling pathway analysis, and MD was finally performed through HPLC analysis for an uppermost bioactive compound (α-amyrin).
Compound-genes network indicated that the therapeutic effect of HCLLs on obesity was directly associated with 22 compounds, 64 genes, and 13 signaling pathways against obesity. Based on each gene's degree of value in 13 signaling pathways, AKT1 on the RAS signaling pathway was considered a hub gene of HCLLs against obesity. However, each compound's binding affinity (vitamin E, octyl adipate, and monopalmitin) connected to AKT1 on the RAS signaling pathway revealed a low score through MD. It might be to inhibit the RAS signaling pathway. Meanwhile, the PPAR signaling pathway might function as an auxiliary mode over the RAS signaling pathway. The measurement of ligand–protein binding affinity demonstrated that α-amyrin had a stronger affinity than PPAR agonists. A study suggested that mice (fed a high-fat diet) treated with α,β-amyrin exhibited a significant reduction in body weights, visceral fat, levels of blood glucose, plasma lipids, amylase and lipase activation relative to their respective controls (no treatment of α,β-amyrin).27 Another animal study indicated that mice treated with α,β-amyrin had a significant drop in blood glucose, total cholesterol, and serum triglyceride levels. Hence, the authors concluded that it could be a lead compound for drug development in metabolic disorders.28 Additionally, it was reported that treatment of α,β-amyrin (6.25–50 μg mL−1) exerted an anti-adipogenic efficacy via the control of lipid and carbohydrate metabolism in 3T3-L1 pre-adipocytes.29 A study suggested that α,β-amyrin showed a pronounced antiobesity effect on high-fat diet mice,30 suggesting that α,β-amyrin exposed high enough pharmacological relevance.
Compounds-gene network showed that the therapeutic effect of HCLLs on obesity was directly connected to 64 genes. The results of the KEGG pathway enrichment analysis of 64 genes demonstrated that 13 signaling pathways were directly associated with the occurrence and development of obesity, suggesting that these signaling pathways might be the mechanisms of HCLLs against obesity. The relationships of the 13 signaling pathways with obesity are succinctly discussed as follows. The thyroid hormone signaling pathway: the fast weight loss is related to reducing TSH (Thyroid Stimulating Hormone) and T3 (Triiodothyronine). Moreover, due to the Resting Energy Expenditure (REE) decrease, weight loss may be difficult to maintain.31 Prolactin signaling pathway: prolactin treatment to diet-induced obese rats enhanced the insulin sensitivity that promoted the antiobesity.32 Estrogen signaling pathway: estrogen receptor α (ER α) mRNA expression levels in non-obese women were lower in obese women in subcutaneous adipose tissue and adipocytes.33 A study showed that liver ER α–knockout (LKO) male mice had weak insulin sensitivity compared to the wild-type, where estrogens played an important biological role in the modulation of metabolism and obesity.34 The insulin signaling pathway: in chronic obesity, the expression level of diverse insulin signaling molecules decreased in skeletal muscle that dysregulated the insulin receptor.35 Retrograde endocannabinoid signaling: cannabinoid type 1 receptor (CB1R) -induced signaling had been exhibited to lead to metabolic inactivation. In contrast, the interruption of the CB1R function could alleviate several obesity-induced causal factors in metabolism.36 Fc epsilon RI (FcεRI) signaling pathway: FcεRI suppressed body weight gain and enhanced glucose tolerance in diet-induced obese mice. It implies that FcεRI is an efficient mediator to mitigate obesity.37 B cell receptor signaling pathway: obese individuals have lower antibody production to influenza vaccination than non-obese individuals. Besides, mice experiments demonstrated that obese mice had lower productivity of haemagglutination inhibition (HAI) antibodies.38 T cell receptor signaling pathway: T cell dysfunction due to obesity induces proinflammatory cytokines, which correlate with abnormal immunity.39 VEGF signaling pathway: VEGF release rate from obese subjects of visceral adipose tissue (VAT) was found higher in comparison to subcutaneous adipose tissue (SAT) and VAT of non-obese controls.40 Phospholipase D signaling pathway: PLD1 (phospholipase D1) or PLD2 (phospholipase D2) deleted mice consumed more food than control groups, specifically, PLD1 and PLD2 repressed appetite and protected against obesity.41 Rap1 signaling pathway: Rap1-lacking mice accumulated fat in the abdomen, hepatic steatosis, and rapid-fasting plasma levels of insulin, glucose, cholesterol, and alanine aminotransferase.42 RAS signaling pathway: Obesity-associated disorders over-activate RAS, suggesting a significant RAS role in body weight control and enhancing insulin sensitivity.43 PPAR signaling pathway: PPARs modulate the inflammatory responses, which mitigate obesity-induced inflammation.44 PPARA or PPARD agonist treatment induced a decrease in fat mass (FM) and PPARB agonist-improved insulin resistance.45,46 Reports indicated that all the 13 signaling pathways with 22 compounds in HCLLs were connected to the suppression of obesity. Outwardly, the AKT1 gene and vitamin E are related to inactivating the RAS signaling pathway and might be a key mechanism against obesity. However, RAS signaling pathway related compounds and proteins did not result in viable predicted binding energies (>−7.0 kcal mol−1) during MD.
The threshold value of docking binding energy was set up <−7.0 kcal mol−1, which was significantly favorable binding energy.47 As an alternative approach, 14 compounds and 6 genes associated with the PPAR signaling pathway were selected to generate binding energy. Among 14 compounds, α-amyrin with all 6 proteins (NR1H3, PPARA, PPARD, PPARG, FABP3, and FABP4) showed significant binding energy. From comparative MD, α-amyrin had a higher binding affinity than that of the standard drugs (NR1H3 agonist, PPARA agonists, and PPARG agonists). Only PPARD agonist binding energy was the same as α-amyrin. Although this study identified 14 molecules, α-amyrin exhibited the highest binding affinity mutually in 6 proteins on PPAR signaling pathways. Furthermore, we identified the amount of α-amyrin (9.63489 μg mL−1) from HCLLs (20 mg) MeOH extraction, which was confirmed via standard curve validation. Altogether, our study demonstrated the potential effectiveness of the auxiliary signaling pathway of HCLLS in suppressing obesity through network pharmacology analysis.
4. Conclusion
The active molecules and mechanisms of HCLLs against obesity were first confirmed by utilizing network pharmacology. Furthermore, this research suggested that activation of the PPAR signaling pathway is a comparatively promising mechanism than the inactivation of the RAS signaling pathway of HCLLs for antiobesity. A prominent molecule, α-amyrin, might be considered a triple agonist (PPARA, PPARD, PPARG) against obesity. Finally, HPLC analysis identified the amount (0.05%) of α-amyrin in HCLLs, which confirmed the presence of this compound in HCLLs. Therefore, this analysis provides pharmacological evidence to support the efficacy of HCLLs on obesity and expounds an uppermost bioactive compound and mechanism(s) of HCLLs against obesity.
Author contributions
K. K. O.: conceptualization, methodology, formal analysis, investigation, data curation, writing – original draft. K. K. O. and M. A. and I. S. J.: software, investigation, data curation. M. A.: validation, writing – review & editing. D. H. C.: supervision, project administration.
Conflicts of interest
The authors have declared no conflict of interest. They have no known competing financial interests or personal relationships that could have appeared to influence the research reported in this publication.
Abbreviations
- BMI
Body mass index
- CB1R
Cannabinoid type 1 receptor
- GC-MS
Gas chromatography-mass spectrometer
- ER-α
Estrogen receptor α
- FM
Fat mass
- GGQLD
Ge-Gen-Qin-Lian decoction
- GSEA
Gene set enrichment analysis
- HAI
Haem agglutination inhibition
- HCLLs
Hibiscus cannabinus L. leaves
- KEGG
Kyoto encyclopedia of genes and genomes
- LDL-C
Low density lipoprotein – cholesterol
- LKO
Liver ER α-knockout
- MD
Molecular docking
- PDB
Protein data bank
- PLD1
Phospholipase D1
- PLD2
Phospholipase D2
- PPAR
Peroxisome proliferator activated receptor
- PPARA
Peroxisome proliferator activated receptor alpha
- PPARD
Peroxisome proliferator activated receptor delta
- PPARG
Peroxisome proliferator activated receptor gamma
- PPI
Protein–protein interaction
- RAP1
Repressor activator protein 1
- RAS
Renin angiotensin system
- REE
Resting energy expenditure
- SAT
Subcutaneous adipose tissue
- SEA
Similarity ensemble approach
- SMILES
Simplified molecular input line entry system
- STP
SwissTargetPrediction
- TSH
Thyroid stimulating hormone
- VEGF
Vascular endothelial growth factor
Supplementary Material
Acknowledgments
This research was acknowledged by the Department of Bio-Health Convergence, Kangwon National University, Chuncheon 24341, Republic of Korea.
Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra10932k
Notes and references
- Bhaskaran K. Douglas I. Forbes H. Dos-Santos-Silva I. Leon D. A. Smeeth L. Lancet. 2014;384:755–765. doi: 10.1016/S0140-6736(14)60892-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K. B. Smith M. S. Primary Care: Clinics in Office Practice. 2016;43:121–135. doi: 10.1016/j.pop.2015.10.001. [DOI] [PubMed] [Google Scholar]
- Obesity and overweight
- Tremmel M. Gerdtham U. G. Nilsson P. M. Saha S. Int. J. Environ. Res. Public Health. 2017;14:435. doi: 10.3390/ijerph14040435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T. T. Liu X. T. Chen Q. X. Shi Y. Biomed. Pharmacother. 2020;128:110314. doi: 10.1016/j.biopha.2020.110314. [DOI] [PubMed] [Google Scholar]
- Antiobesity Drugs – an overview, ScienceDirect Topics
- Liu Y. Sun M. Yao H. Liu Y. Gao R. J. Evidence-Based Complementary Altern. Med. 2017;2017:1–17. [Google Scholar]
- Kai N. S. Nee T. A. Ling E. L. C. Ping T. C. Kamariah L. Lin N. K. Asian Pac. J. Trop. Med. 2015;8:6–13. doi: 10.1016/S1995-7645(14)60179-6. [DOI] [PubMed] [Google Scholar]
- Son J. M. Ju H. S. Gung H. J. N. Azad M. O. K. Adnan M. D. Cho D. H. Korean Journal of Medicinal Crop Science. 2019;27:89. [Google Scholar]
- Adnan M. Azad M. O. K. Ju H. S. Son J. M. Park C. H. Shin M. H. Alle M. Cho D. H. Appl. Nanosci. 2019:1–13. [Google Scholar]
- Adnan M. C. D. Azad M. O. K. Madhusudhan A. Saravanakumar K. Hu X. Wang M. H. Cho D. H. Nanotechnology. 2020;31:265101. doi: 10.1088/1361-6528/ab7d72. [DOI] [PubMed] [Google Scholar]
- Adnan M. Oh K. K. Azad M. O. K. Shin M. H. Wang M.-H. Cho D. H. Life. 2020;10:223. doi: 10.3390/life10100223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P. Bhandari U. Ancient Science of Life. 2015;35:58. doi: 10.4103/0257-7941.165629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Effect of ethanolic extract of Hibiscus cannabinus leaf on high cholesterol diet induced obesity in female albino rats
- Giwa Ibrahim S. Karim R. Saari N. Wan Abdullah W. Z. Zawawi N. Ab Razak A. F. Hamim N. A. Umar R. A. J. Food Sci. 2019;84(8):2015–2023. doi: 10.1111/1750-3841.14714. [DOI] [PubMed] [Google Scholar]
- Zhang R. Zhu X. Bai H. Ning K. Front. Pharmacol. 2019;10:1–15. doi: 10.3389/fphar.2019.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh K. K. Adnan M. Cho D. H. Gene Reports. 2020:100851. doi: 10.1016/j.genrep.2020.100851. [DOI] [Google Scholar]
- Hopkins A. L. Nat. Chem. Biol. 2008;4(11):682–690. doi: 10.1038/nchembio.118. [DOI] [PubMed] [Google Scholar]
- Oh K. K. Adnan M. Cho D. H. PLoS One. 2020;15:e0240873. doi: 10.1371/journal.pone.0240873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S. Zhang B. Chin. J. Nat. Med. 2013;11:110–120. doi: 10.3724/SP.J.1009.2013.00110. [DOI] [PubMed] [Google Scholar]
- Li H. Zhao L. Zhang B. Jiang Y. Wang X. Guo Y. Liu H. Li S. Tong X. J. Evidence-Based Complementary Altern. Med. 2014;2014:495840. doi: 10.1155/2014/495840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiser M. J. Setola V. Irwin J. J. Laggner C. Abbas A. I. Hufeisen S. J. Jensen N. H. Kuijer M. B. Matos R. C. Tran T. B. Whaley R. Glennon R. A. Hert J. Thomas K. L. H. Edwards D. D. Shoichet B. K. Roth B. L. Nature. 2009;462:175–181. doi: 10.1038/nature08506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daina A. Michielin O. Zoete V. Nucleic Acids Res. 2019;47:W357–W3664. doi: 10.1093/nar/gkz382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh K. K. Adnan M. Cho D. H. Gene. 2020:145320. doi: 10.1016/j.gene.2020.145320. [DOI] [PubMed] [Google Scholar]
- Khanal P. Patil B. M. Chand J. Naaz Y. Nat. Prod. Bioprospect. 2020;10:325–335. doi: 10.1007/s13659-020-00260-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adnan M. Chy M. N. U. Kamal A. T. M. M. Barlow J. W. Faruque M. O. Yang X. Uddin S. B. J. Ethnopharmacol. 2019;236:401–411. doi: 10.1016/j.jep.2019.01.025. [DOI] [PubMed] [Google Scholar]
- Santos F. Rao V. Carvalho K. Morais T. da Silva A. Chaves M. Planta Med. 2013;79:PE14. [Google Scholar]
- Santos F. A. Frota J. T. Arruda B. R. De Melo T. S. Da Silva A. A. D. C. A. Brito G. A. D. C. Chaves M. H. Rao V. S. Lipids Health Dis. 2012;11(98):1–8. doi: 10.1186/1476-511X-11-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Melo K. M. de Oliveira F. T. B. Costa Silva R. A. Gomes Quinderé A. L. Marinho Filho J. D. B. Araújo A. J. Barros Pereira E. D. Carvalho A. A. Chaves M. H. Rao V. S. Santos F. A. Biomed. Pharmacother. 2019;109:1860–1866. doi: 10.1016/j.biopha.2018.11.027. [DOI] [PubMed] [Google Scholar]
- Karine Maria Martins Bezerra Carvalho A. Sousa de Melo T. Moura de Melo K. Luiza Gomes Quinderé A. Tuelly Bandeira de Oliveira F. Flávia Seraine Custódio Viana A. Iury Gomes Nunes P. da Silva Quetz J. de Araújo Viana D. André de Carvalho Almeida da Silva A. Havt A. Gonçalves da Cruz Fonseca S. Helena Chaves M. Satyanarayana Rao V. Almeida Santos F. Planta Med. 2017;83:285–291. doi: 10.1055/s-0042-114222. [DOI] [PubMed] [Google Scholar]
- Reinehr T. Mol. Cell. Endocrinol. 2010;316:165–171. doi: 10.1016/j.mce.2009.06.005. [DOI] [PubMed] [Google Scholar]
- Ruiz-Herrera X. de los Ríos E. A. Díaz J. M. Lerma-Alvarado R. M. de la Escalera L. M. López-Barrera F. Lemini M. Arnold E. de la Escalera G. M. Clapp C. Macotela Y. Endocrinology. 2016;158:1444. doi: 10.1210/en.2016-1444. [DOI] [PubMed] [Google Scholar]
- Nilsson M. Dahlman I. Rydén M. Nordström E. A. Gustafsson J. Å. Arner P. Dahlman-Wright K. Int. J. Obes. 2007;31:900–907. doi: 10.1038/sj.ijo.0803528. [DOI] [PubMed] [Google Scholar]
- Zhu L. Martinez M. N. Emfinger C. H. Palmisano B. T. Stafford J. M. Am. J. Physiol.: Endocrinol. Metab. 2014;306:E1188. doi: 10.1152/ajpendo.00579.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn B. B. Flier J. S. J. Clin. Invest. 2000;106:473–481. doi: 10.1172/JCI10842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipina C. Rastedt W. Irving A. J. Hundal H. S. Wiley Interdiscip. Rev.: Membr. Transp. Signaling. 2013;2:49–63. [Google Scholar]
- Lee Y. J. Liu C. Liao M. Sukhova G. K. Shirakawa J. Abdennour M. Iamarene K. Andre S. Inouye K. Clement K. Kulkarni R. N. Banks A. S. Libby P. Shi G. P. Endocrinology. 2015;156:4047–4058. doi: 10.1210/en.2015-1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaikh S. R. Haas K. M. Beck M. A. Teague H. Clin. Exp. Immunol. 2015;179:90–99. doi: 10.1111/cei.12444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilar E. G. Murphy W. J. Curr. Opin. Immunol. 2018;51:181–186. doi: 10.1016/j.coi.2018.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlich R. Willems M. Greulich S. Ruppe F. Knoefel W. T. Ouwens D. M. Maxhera B. Lichtenberg A. Eckel J. Sell H. Mediators Inflammation. 2013;2:1–11. doi: 10.1155/2013/982458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viera J. T. El-Merahbi R. Nieswandt B. Stegner D. Sumara G. PLoS One. 2016;11(6):1–13. doi: 10.1371/journal.pone.0157607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez P. Gómez-López G. García F. Mercken E. Mitchell S. Flores J. M. deCabo R. Blasco M. A. Cell Rep. 2013;3:2059–2074. doi: 10.1016/j.celrep.2013.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gul R. Dekhil H. Alfadda A. Saudi Journal of Obesity. 2018;6:5. doi: 10.4103/sjo.sjo_15_18. [DOI] [Google Scholar]
- Stienstra R. Duval C. Kersten S. PPAR Res. 2007;2007:95974. doi: 10.1155/2007/95974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stienstra R. Duval C. Müller M. Kersten S. PPAR Res. 2007;2007:95974. doi: 10.1155/2007/95974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva-Veiga F. M. Rachid T. L. de Oliveira L. Graus-Nunes F. Mandarim-de-Lacerda C. A. Souza-Mello V. Mol. Cell. Endocrinol. 2018;474:227–237. doi: 10.1016/j.mce.2018.03.013. [DOI] [PubMed] [Google Scholar]
- Mohan R. R. Wilson M. Gorham R. D. Harrison R. E. S. Morikis V. A. Kieslich C. A. Orr A. A. Coley A. V. Tamamis P. Morikis D. Mcferrin A. ACS Omega. 2018;3:6427–6438. doi: 10.1021/acsomega.8b00606. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.