Abstract
Hepatocellular carcinoma (HCC) has a poor prognosis due to its high malignancy, rapid disease progression, and the presence of chemotherapy resistance. Long-stranded non-coding RNAs (lncRNAs) affect many malignant tumors, including HCC. However, their mechanism of action in HCC remains unclear. This study aimed to clarify the role of DUXAP8 in regulating the malignant phenotype and chemotherapy resistance in HCC. Using an in vivo xenograft tumor model, the regulatory functions and mechanisms of lncRNA DUXAP8 in the progression and response of HCC to chemotherapy were explored. It was found that DUXAP8 was significantly upregulated in a patient-derived xenograft tumor model based on sorafenib treatment, which is usually associated with a relatively poor prognosis in patients. In HCC, DUXAP8 maintained its upregulation in the expression by increasing the stability of m6A methylation-mediated RNA. DUXAP8 levels were positively correlated with the proliferation, migration, invasion, and chemotherapy resistance of HCC in vivo and in vitro. In the mechanistic study, it was found that DUXAP8 competitively binds to miR-584-5p through a competing endogenous RNA (ceRNA) mechanism, thus acting as a molecular sponge for miR-584-5p to regulate MAPK1 expression, which in turn activates the MAPK/ERK pathway. These findings can provide ideas for finding new prognostic indicators and therapeutic targets for patients with HCC.
Keywords: DUXAP8, hepatocellular carcinoma, m6A methylation modification, chemotherapy resistance, malignant phenotype, miR-584-5p, MAPK1
Introduction
Hepatocellular carcinoma (HCC) is one of the five most common malignant tumors globally (Zhu et al., 2019). Worldwide, nearly 700,000 people die each year from HCC (Fang et al., 2018; Wu et al., 2019), and its death toll ranks second among all malignant tumor-associated deaths. HCC poses a serious health burden globally (Petrick et al., 2020; Reinders et al., 2020; Yang et al., 2020). In China, the number of deaths due to HCC is the second-highest among all malignant tumors, which come right after lung cancer (Raoul and Edeline, 2020). Despite recent improvements in surgical techniques and chemotherapy regimens, the prognosis of HCC remains poor, and its prevalence is approaching its mortality (Jemal et al., 2011). Particularly, the occurrence of chemotherapy resistance has severely hampered the therapeutic outcome of HCC. Sorafenib, a tyrosine kinase inhibitor, is currently the first-line medication for treating unresectable HCC (Bruix et al., 2019) and can extend the survival of HCC patients by an average of 12.3 months (Kudo et al., 2018; Rimassa and Worns, 2020), but this benefit of increased survival may be lost due to the emergence of drug resistance. The etiology of HCC and the mechanisms of chemotherapy resistance are still unclear; thus, a deeper understanding of genetics and epigenetic mechanisms behind the development and progression of HCC and chemotherapy resistance is required to develop more effective therapeutic intervention strategies.
The development of whole-genome and transcriptome sequencing technologies has revealed the composition of the human genome, where less than 2% of genes are protein-coding genes and the rest are all non-coding genes (Harrow et al., 2012; Shi et al., 2016). There is increasing evidence that complex functions in living organisms may be regulated by a range of RNAs from non-coding regions of the genome. Long non-coding RNAs (LncRNAs) represent a group of RNA molecules with a transcript length exceeding 200 nucleotides and encode only short polypeptides or do not encode proteins (Zheng et al., 2015). Recent studies have indicated that lncRNAs are involved in many critical biological processes, such as X-chromosome inactivation, stem cell maintenance, transcriptional regulation, and epigenetic regulation (Zhang et al., 2018). Additionally, lncRNAs are involved in the regulation of many diseases, especially playing an essential role in the occurrence and development of malignant tumors. Some lncRNAs act as protein co-regulators directly binding to their helper proteins and regulating the expression of downstream tumor-associated genes (Chen et al., 2016; Yuan et al., 2016; Zhu et al., 2016). Another part of lncRNAs act as competitive endogenous RNAs (ceRNAs) (Yuan et al., 2014; Li et al., 2015b), and such lncRNAs achieve microRNA detachment and regulate the expression of microRNA-targeted oncogenes or tumor suppressor genes through molecular sponge effects. These studies suggested that the abnormal expression of lncRNAs plays an essential role in tumor development.
The pseudogene-derived lncRNA, Double Homeobox A Pseudogene 8 (DUXAP8), is located on chromosome 22q11.1 with a full length of 2,107 bp (Sun et al., 2017). Recently, DUXAP8 has been shown to be highly expressed in various malignant tumors. It has been shown that DUXAP8 can significantly inhibit the expression of PLEKHO1 in gastric cancer, enhancing the proliferation and migration of tumor cells (Ma et al., 2017a). In glioma, downregulation of DUXAP8 inhibits the proliferation of tumor cells (Zhao et al., 2019). In non-small cell lung cancer (NSCLC), DUXAP8 promotes tumor cell proliferation and invasion through epigenetic silencing of Egr1 and RHOB (Sun et al., 2017). In pancreatic cancer, DUXAP8 promotes tumor growth through epigenetic silencing of CDKN1A and KLF2 (Lian et al., 2018). While the mechanism of action of DUXAP8 in HCC is unclear, Wang et al. (2020) reported that DUXAP8 can be used for the diagnosis of HCC and can predict the prognosis of HCC. In this study, it was found that DUXAP8 was highly expressed in liver cancer tissues and cells, where high expression of DUXAP8 often predicts poor prognosis in patients. In in vivo and in vitro experiments, it was found that overexpression of DUXAP8 promoted the malignant phenotype and chemotherapy resistance in HCC. Mechanistically, it was found that DUXAP8 upregulated MAPK1 through competitive binding to miR-584-5p, which activated the MAPK/ERK pathway and promoted the proliferation, invasive migration, stemness maintenance, and chemotherapy resistance of HCC. These results provided a deeper understanding of the driving mechanisms of lncRNAs in HCC development, progression, and resistance to chemotherapy. They revealed the importance of DUXAP8 in HCC progression, which can help identify new prognostic indicators and therapeutic targets for HCC patients.
Materials and Methods
Cell Lines and Cell Culture
Standard human hepatocytes THLE-3 and HCC cell lines MHCC-97H, Huh7, HCC-LM3, Bel-7405, SNU-449, SK-Hep-1, and SNU-182 were purchased from the American Type Culture Collection or the Cell Bank of the National Collection of Authenticated Cell Cultures (Shanghai, China). The conventional culture of HCC cell lines was performed (Li et al., 2015a). Cells were cultured in appropriate culture media containing 10% fetal bovine serum (FBS; Gibco) supplemented with penicillin (100 U/ml) and streptomycin (100 μg/ml). All cell lines were cultured at 37°C in an incubator using 5% CO2/95% air and 100% humidity to minimize the chances of bacterial contamination. Cells were passaged every 1–2 days to maintain logarithmic growth.
Patients and Specimens
All studies involving human samples were reviewed and approved by the Ethics Committee of the Second Hospital of Jilin University. The study protocol conformed to the ethical standards of the Declaration of Helsinki. Written informed consent was also obtained from all patients according to the Helsinki Declaration. Liver cancer and pericarcinomatous tissues were obtained from HCC patients at the Second Hospital of Jilin University (n = 79). The examined clinicopathological features included age, sex, number of tumor nodules, etiology, serum alpha fetoprotein (AFP) level, cancer staging, tumor size, and the presence of vascular invasion. Cancer staging was based on the 6th edition of the International Union Against Cancer tumor–node–metastasis (TNM) Classification.
Chemical Reagents and Antibodies
Lipofectamine 2000 transfection reagent and total RNA extractant (TRIzol) were purchased from Invitrogen (Grand Island, NY, United States); P-CREB antibodies were purchased from Cell Signaling Technology (Danvers, MA, United States); P38 and P-P38 antibodies were purchased from Abcam Corporation (Cambridge, MA, United States); and all other antibodies were purchased from Proteintech (Rosemont, IL, United States). Unless otherwise stated, all other chemical reagents were purchased from Sigma-Aldrich (St. Louis, MO, United States).
Reverse Transcription Quantitative Polymerase Chain Reaction (RT–qPCR)
Total RNA was extracted using the RNA Simple Total RNA Kit (TIANGEN, DP419), and RNA in the cytoplasm and nucleus was isolated using the Nuclear/Cytoplasmic Isolation Kit (BioVision, San Francisco, CA, United States) according to the manufacturer’s instructions. Complementary DNA (cDNA) was synthesized using the RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific, #K1622) and Poly(A) Polymerase Reaction Buffer (NEB, M0276s) according to the manufacturer’s instructions. According to the manufacturer’s instructions, real-time qRT–PCR analysis was performed using the Platinum SYBR Green qPCR SuperMix-UDG kit (Life Technologies, Gaithersburg, MD, United States). Fold changes in RNA expression were quantified using the 2−ΔΔCt method. The primer sequences used in this study are listed in Supplementary Table S1.
Immunohistochemistry (IHC)
The paraffin sections of tissues were dewaxed in xylene and hydrated using graded ethanol. The heat-mediated and antigen repair citrate (0.01 M, pH 6.0) was used in performing the IHC experiment. Endogenous peroxidase activity was blocked using 3% H2O2 at room temperature for 15 min. Thereafter, the sections were incubated with goat serum for 1 h to block the nonspecific binding site and then incubated overnight at 4°C using the primary antibody. The sections were then re-warmed at room temperature for 30 min, rinsed three times with phosphate-buffered saline (PBS) for 5 min each, and incubated with horseradish peroxidase (HRP)-labeled secondary antibody at room temperature for 1 h. Each section was rinsed thrice again with PBS for 5 min each. Color development was conducted using the HRP DAB kit (Thermo Science, Shanghai, China). Nuclei were re-stained with hematoxylin, and sections were sealed with neutral glue. Images were obtained using an Olympus X71 inverted microscope (Olympus Corp., Tokyo, Japan).
Western Blot
Tissues or cells were homogenized and lysed using a lysis buffer. After the protein concentration was determined using the bicinchoninic acid method, β-mercaptoethanol, and bromophenol blue were added to the sample buffer for electrophoresis. Proteins were separated through 10% polyacrylamide gel electrophoresis and then transferred to polyvinylidene difluoride membranes (Bio-Rad, Shanghai, China). The membranes were incubated overnight at 4°C with primary antibodies. After incubation with secondary antibodies for 2 h at room temperature, the reaction bands were visualized using an enhanced chemiluminescence system, and the intensity of the bands was quantified using an image analysis system.
Methylated RNA Immunoprecipitation qPCR (MeRIP–qPCR)
The level of m6A in DUXAP8 was determined using MeRIP–qPCR. The intracellular RNA was first extracted using the TRIzol reagent and then bound to protein A/G magnetic beads with anti-m6A antibody or immunoglobulins (IgG; Cell Signaling Technology) (3 μg), and mixed with 100-μg total RNA in an immunoprecipitation buffer containing RNase/protease. The m6A-modified RNA was eluted twice with 6.7-mM N6-methyladenosine 5′-monophosphate disodium salt at 4°C for 1 h. RT–qPCR analysis was subsequently performed to determine the extent of m6A enrichment on DUXAP8.
Luciferase Assay
The complementary DNA fragment containing wild-type or mutant DUXAP8 fragment/MAPK1 3′ untranslated region (UTR) was subcloned to the downstream of luciferase in the pGL3-basic luciferase reporter gene (Promega, Beijing, China). The firefly luciferase gene containing wild-type or mutant DUXAP8 fragment/MAPK1 was cotransfected with an empty vector or miR-584-5p mimics, and luciferase activity was measured using the Dual-Luciferase Kit (Promega, Beijing, China) 48 h after transfection.
RNA Immunoprecipitation (RIP) Analysis
According to the manufacturer’s instructions, the Magna RIPTM RNA Binding Protein Immunoprecipitation Kit (Millipore, United States) was used. Briefly, cell extracts were immunoprecipitated with antibodies against AGO2 or IgG and magnetic beads at 4°C for 6 h. The proteins in the complex were removed using 0.1% SDS/proteinase K (0.5-mg/ml, 55°C for 30 min), and immunoprecipitated proteins and RNA were detected using Western blot and RT–qPCR, respectively.
RNA Pull-Down Experiments
The Pierce Magnetic RNA-Protein Pull-Down Kit (Thermo Fisher Scientific, 20,164) was used according to the manufacturer’s instructions. Briefly, cell lysates were treated with RNAase-free DNAase I and then incubated with the treated cell lysate, streptavidin-labeled magnetic beads, and a biotin-labeled DUXAP8 probe. The magnetic beads could capture proteins/miRNAs that interact with DUXAP8. A Pierce™ RNA 3′ End Desthiobiotinylation Kit (Thermo, 20,163) was used for DUXAP8 biotinylation labeling. The proteins and RNA in the captured protein-RNA complexes were analyzed using Western blot and RT–qPCR.
Establishment of a Subcutaneous Xenotransplanted Tumor Model in Mice
Four-week-old BALB/c nude mice were purchased from the Experimental Animal Center of Jilin University (Changchun, China). All experimental animal protocols were performed according to the “Guide for the Care and Use of Laboratory Animals” issued by the National Institutes of Health. Experimental animal protocols for this study were reviewed and approved by the Animal Experiment Ethics Committee of the First Hospital of Jilin University. During modeling, 2 × 106 Huh7 cells with silenced DUXAP8 expression or negative control Huh7 cells were injected subcutaneously into the lateral abdominal region of each mouse. The tumor volume was measured weekly and calculated as V = (length × width2)/2. The tumors were then excised and weighed 4 weeks later.
Statistical Analysis
All values were expressed as mean ± standard deviation. Comparisons between groups were made using a t-test or one-way analysis of variance. Qualitative data were analyzed using the chi-square test. Linear regression analysis was conducted for correlations between gene expression levels. Statistical analyses were performed using GraphPad Prism v8.0 (GraphPad, Inc., United States) and Statistical Software Package for Social Sciences (v 22.0; SPSS, Inc., Chicago, IL, United States). Differences were statistically significant when p < 0.05.
Results
High Expression of DUXAP8 in an Animal Model of Patient-Derived Xenograft (PDX) HCC Treated With Sorafenib
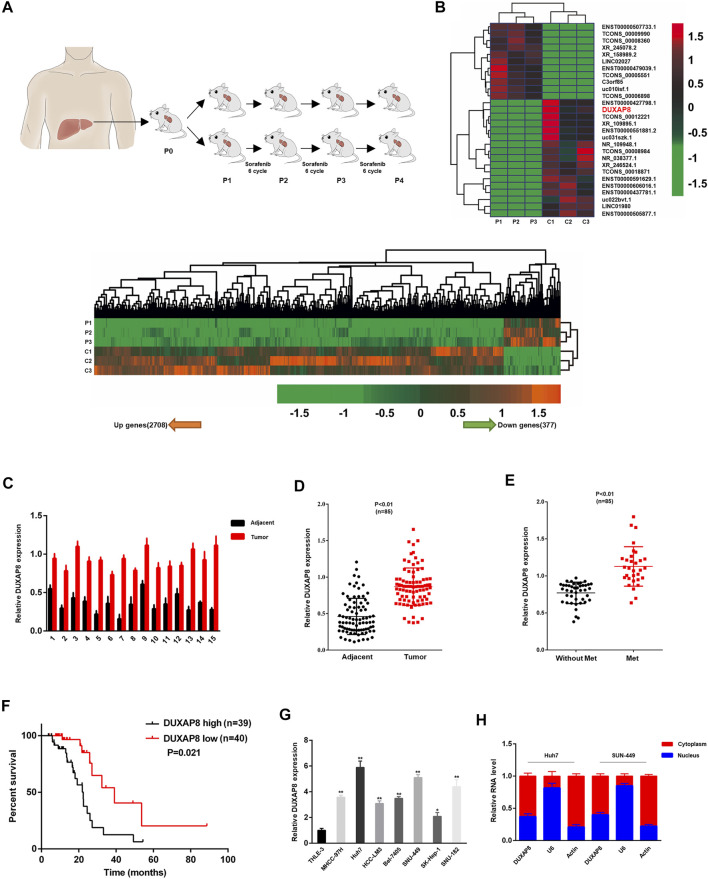
Sorafenib is an oral multi-kinase inhibitor. As the standard FDA-approved targeted therapy for HCC, sorafenib exhibits a survival benefit and has dramatically improved the prognosis of HCC patients, especially in patients with advanced HCC. However, acquired or intrinsic chemotherapy resistance severely affects its overall efficacy in treating HCC (Liu et al., 2019; Wei et al., 2019; Xiang et al., 2019). To clarify the molecular mechanism of drug resistance to sorafenib in HCC, a PDX model for sorafenib treatment was first established, in which surgically resected primary HCC tissue was finely trimmed and directly transplanted into mice with immunodeficiency. Tumor-bearing mice were treated with saline (vector) or sorafenib for several generations (Figure 1A). Differentially expressed lncRNAs between P4-PDX treated with vector or sorafenib were identified using lncRNA sequencing analysis (Figure 1B). Among identified lncRNAs, the significantly upregulated DUXAP8 (ENST00000607933.1) was selected to investigate its role in HCC and chemosensitivity further (Supplementary Figure S1A).
FIGURE 1.
DUXAP8 is highly expressed in sorafenib-treated patient-derived xenograft (PDX) hepatocellular carcinoma model, and its upregulation predicts poor prognosis of HCC. (A) Experimental procedure of the sorafenib-treated PDX HCC model. (B) The heat map indicates the differential expression of lncRNAs in sorafenib-treated and control P4-PDX mice. (C) Comparison of DUXAP8 expression in liver cancer tissues of 15 HCC patients with paired pericarcinomatous normal tissues. (D) Comparison of DUXAP8 expression in HCC tissues of 85 HCC patients with paired pericarcinomatous normal tissues. (E) Relationship between DUXAP8 expression in liver cancer tissues of 85 patients and the presence of distant metastasis. (F) Kaplan-Meier analysis of the correlation between DUXAP8 expression levels and HCC prognosis in 79 patients. (G) Comparison of DUXAP8 expression in HCC cell lines and THLE-3 cells. (H) RT–qPCR assay of the distribution of DUXAP8 in the nucleus and cytoplasm of HCC cells. Data are expressed as mean, *p < 0.05, **p < 0.01.
It was confirmed by RT–qPCR that DUXAP8 expression in HCC tissues was significantly higher than that of normal tissues adjacent to cancer, and the same results were obtained after expanding the sample size (Figures 1C,D). Correlation analysis with clinicopathological parameters showed that high levels of DUXAP8 correlated with the TNM stage of cancer, tumor size, microvascular invasion, and distant metastasis (Figure 1E; Supplementary Table S2), and that HCC patients with high DUXAP8 expression tended to have a poorer prognosis (Figure 1F). which is consistent with the results of the GEPIA database (Supplementary Figures S1B,C). We found that the expression of DUXAP8 in seven HCC cell lines (MHCC-97H, Huh7, HCC-LM3, Bel-7405, SNU-449, SK-Hep-1, SNU-182) was significantly higher than its expression in the normal hepatocyte cell line THLE-3 (Figure 1G), and further confirmed that DUXAP8 was mainly localized in the cytoplasm (Figure 1H). In conclusion, these findings suggested that DUXAP8 is highly expressed in HCC tissues and positively correlates with the malignancy of HCC, and may function as a chemotherapy-resistant molecule in HCC.
Overexpression of DUXAP8 Promoted the Characteristics of Migration, Invasion, and Stemness of HCC Cells
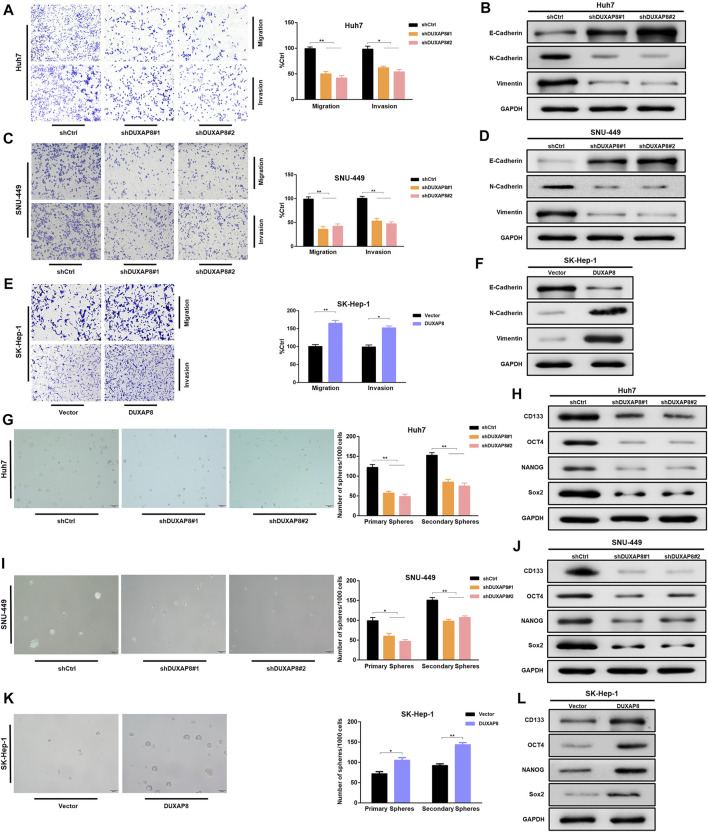
To investigate the effect of DUXAP8 on the biological behavior of HCC cells, shRNAs, and lentiviral vectors of DUXAP8 mimics were transfected into DUXAP8 high-expressing HCC cell lines (Huh7, SNU-449) and DUXAP8 low-expressing HCC cell lines (SK-Hep-1, HCC-LM3), respectively, and the transfection efficiency was verified by RT-qPCR (Supplementary Figures S2A,B). To clarify whether DUXAP8 affects the features of migration and invasion of HCC cells, Transwell assays were performed, which showed that the knockdown of DUXAP8 significantly decreased the migratory and invasive ability of Huh7 and SNU-449 cells. In contrast, overexpression promoted the migration and invasion of SK-Hep-1 and HCC-LM3 cells (Figures 1A,C,E; Supplementary Figure S2C). Epithelial–mesenchymal transition (EMT) is closely related to the migration and invasion of tumor cells, and it was found through Western blot that when DUXAP8 was knocked down, the epithelial marker E-cadherin increased and the mesenchymal marker N-cadherin and vimentin decreased in HCC cell lines (Figures 2B,D); when DUXAP8 was overexpressed, it produced the opposite effect (Figure 2F; Supplementary Figure S2D), indicating that DUXAP8 positively regulated EMT process in HCC cells. Altogether, DUXAP8 overexpression promoted the migration and invasion of HCC cells.
FIGURE 2.
DUXAP8 can regulate the characteristics of migration, invasion, and stemness of HCC cells. (A,C,E) DUXAP8 or shDUXAP8 and the control cells were subjected to Transwell migration and Matrigel invasion assays. (B,D,F) Western blot detects the expression of epithelial–mesenchymal transformation markers in DUXAP8-downregulated Huh7 and SNU-449 cells and DUXAP8-overxpressed SK-Hep-1 cells. (G,I,K) Statistical analysis of the primary and secondary spheroid-forming ability of DUXAP8-downregulated Huh7 and SNU-449 cells and DUXAP8-overexpressed SK-Hep-1 cells, with representative images showing secondary spheroid formation in these cells. (H,J,L) Western blot detects the expression levels of stemness-related genes in DUXAP8-downregulated Huh7 and SNU-449 cells and DUXAP8-overexpressed SK-Hep-1 cells. Data are expressed as mean, *p < 0.05, **p < 0.01.
Next, the effect of DUXAP8 on the stem cell characteristics of HCC cells was investigated. The spheroid-forming ability was reduced when DUXAP8 was knocked down in Huh7 and SNU-449 cells (Figures 2G,I). In contrast, DUXAP8 overexpression significantly improved the spheroid-forming ability of SK-Hep-1 and HCC-LM3 cells (Figure 2K; Supplementary Figure S2E). The potential regulatory effects of DUXAP8 on the expression of the cancer stem cell marker, CD133, and stem cell-related genes, including OCT4, NANOG, and Sox2 were then investigated. It can be seen that DUXAP8 knockdown significantly suppressed the expression of CD133, OCT4, NANOG, and Sox2 (Figures 2H,J, Supplementary Figure S2G), while the expression of CD133, OCT4, NANOG, and Sox2 was promoted when DUXAP8 was overexpressed (Figure 2L, Supplementary Figures S2F,H). Thus, DUXAP8 overexpression promoted the stem cell features of HCC cells.
Overexpression of DUXAP8 Promoted the Proliferation of HCC Cells and Reduced the Chemosensitivity of HCC Cells to Sorafenib
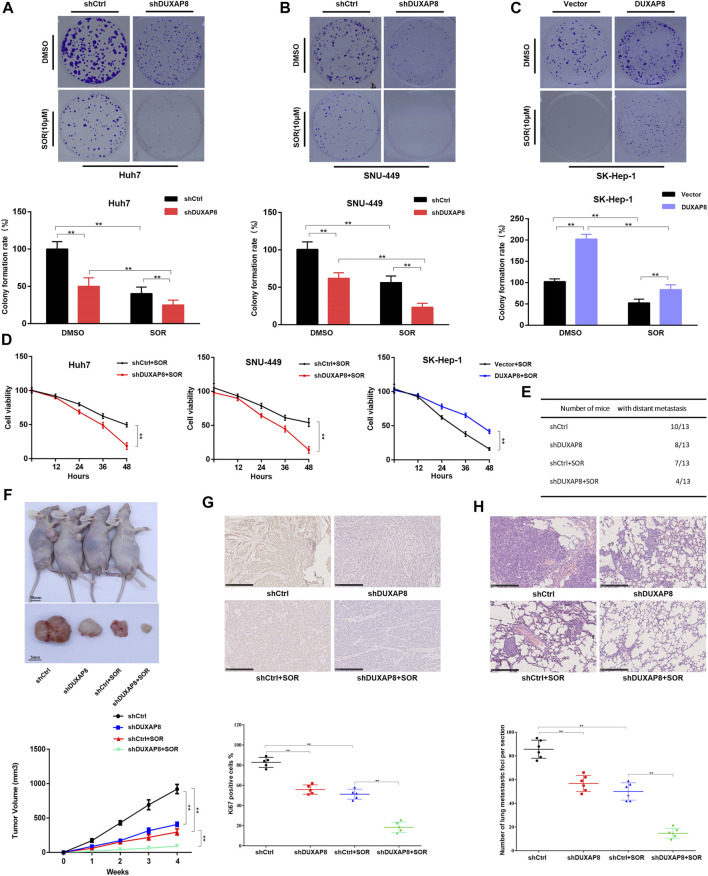
It was further investigated whether DUXAP8 affected the chemosensitivity of HCC cells to sorafenib. It was observed that the knockdown of DUXAP8 inhibited the proliferation ability of HCC cells and enhanced the chemosensitivity of HCC cells to sorafenib, while its overexpression promoted the proliferation of HCC cells and decreased the chemosensitivity of HCC cells to sorafenib (Figures 3A–D, Supplementary Figure S3A–C), indicating that DUXAP8 mediated the development of chemotherapy resistance to sorafenib in HCC cells. Furthermore, consistent with the above in vitro findings, in in vivo experiments, knockdown of DUXAP8 decreased the size and Ki67-positive rate of transplanted tumors in saline (NS) and sorafenib-treated nude mice (Figures 3F,G). Additionally, in the pulmonary metastasis model, it was found that knockdown of DUXAP8 significantly inhibited the ability of pulmonary metastasis of HCC cells, as evidenced by the reduction in the number of mice with metastatic lung tumors and the decrease in the number of lung tumors in mice treated with NS or sorafenib (Figures 3E,H, Supplementary Figure S3D). In summary, these results suggested that overexpression of DUXAP8 promoted the proliferation of HCC cells and reduced the chemosensitivity of HCC cells against sorafenib.
FIGURE 3.
DUXAP8 promotes the chemotherapy resistance of HCC to sorafenib in vivo and in vitro. (A,B) Colony formation of cells transfected with shDUXAP8 after sorafenib treatment. (C) Colony formation of cells transfected with DUXAP8-overexpressing plasmid after sorafenib treatment. (D) Cell viability of Huh7/SNU-449 cells transfected with shDUXAP8 and SK-Hep-1 cells transfected with DUXAP8-overexpressed plasmid under the effect of sorafenib wasanalyzedusingCCK8. (E) Summary table of pulmonary metastases. (F) Typical images of xenograft tumors formed by liver cancer cells transfected with shCtrl or shDUXAP8 in sorafenib-treated nude mice. Tumor growth curves of HCC cells transfected with shCtrl or shDUXAP8 in sorafenib-treated nude mice. (G) Ki67 expression in xenograft tumors formed by shCtrl- or shDUXAP8-transfected HCC cells in sorafenib-treated nude mice (H) Number of metastatic foci per section in the lungs of nude mice treated with sorafenib after caudal vein injection of HCC cells transfected with shCtrl or shDUXAP8. Data are expressed as mean, **p < 0.01.
Mettl3-Mediated m6A Modifications Involved in the Upregulation of DUXAP8
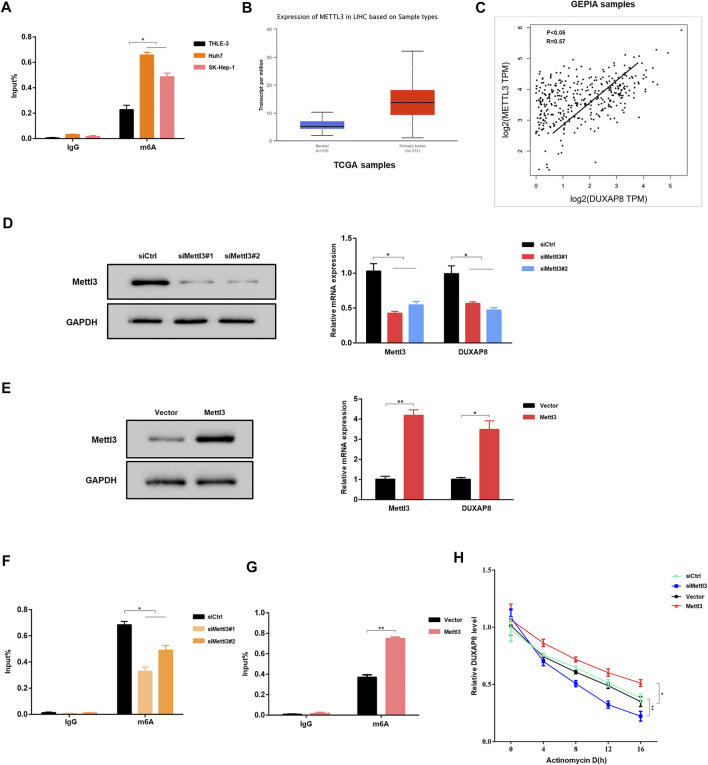
Subsequently, the mechanism of DUXAP8 upregulation in HCC cells was investigated. N6-methyladenosine (m6A) is the most common internal post-transcriptional modification in eukaryotic RNA (Wei et al., 1975; Schwartz et al., 2013), affecting RNA transcription, processing, translation, and metabolism. As a novel RNA epigenetic modification, the m6A modification plays an essential role in gene expression, including mRNA metabolism and other fundamental life processes and in the development of malignant tumors. RMBase (http://rna.sysu.edu.cn/rmbase/index.php) was used to find that DUXAP8 has many m6A-binding sites. Therefore, it was hypothesized that the upregulation of DUXAP8 in HCC might be associated with its m6A modification. The experimental results of MeRIP–qPCR verified the hypothesis that HCC cell lines (Huh7, SK-Hep-1) had a significantly higher level of m6A enrichment of DUXAP8 than that of the normal hepatocyte line (THLE-3) (Figure 4A). Methyltransferase 3 (Mettl3) is a methyltransferase in m6A modification, which plays a vital role in mediating the m6A modification process in mammalian cells. By searching the Cancer Genome Atlas (TCGA) database, it was found that the expression level of Mettl3 was significantly higher in liver cancer tissues than in the corresponding pericarcinomatous tissues (Figure 4B), and there was a significant positive correlation between the expression levels of Mettl3 and DUXAP8 (Figure 4C). When Mettl3 was silenced, it was observed that the expression level of DUXAP8 also showed a significant decrease (Figure 4D); when Mettl3was overexpressed, the expression level of DUXAP8 also indicated a corresponding increase (Figure 4E). It was also found that the m6A enrichment of DUXAP8 in HCC cells decreased also when Mettl3 was silenced (Figure 4F), while the m6A enrichment of DUXAP8 in HCC cells was correspondingly increased when Mettl3 was overexpressed (Figure 4G). The silencing of Mettl3 in the presence of dactinomycin, a drug that inhibits RNA synthesis, decreased the stability of DUXAP8, while Mettl3 overexpression produced the opposite result (Figure 4H). Altogether, as a methyltransferase in m6A modification, Mettl3 is essential to increase DUXAP8 expression in HCC cells.
FIGURE 4.
Mettl3-mediated m6A modifications are involved in the upregulation of DUXAP8. (A) MeRIP–qPCR results showed that the m6A enrichment of DUXAP8 was higher in Huh7 and SK-Hep-1 cells than in THLE-3 cells. (B) The expression of methyltransferase 3 (Mettl3) in HCC was analyzed using the TCGA database. (C) The results of TCGA database showed that the expression level of Mettl3 was positively correlated with DUXAP8. (D,E) Effect of Mettl3 knockdown and overexpression on DUXAP8 expression. (F,G) Effect of Mettl3 knockdown and overexpression on the degree of m6A enrichment of DUXAP8. (H) Effect of Mettl3 knockdown and overexpression on DUXAP8 stability in the presence of dactinomycin. Data are expressed as mean, *p < 0.05, **p < 0.01.
The Effect of DUXAP8 on the Malignant Phenotype and Chemosensitivity of HCC Through Competitive Binding to miR-584-5p
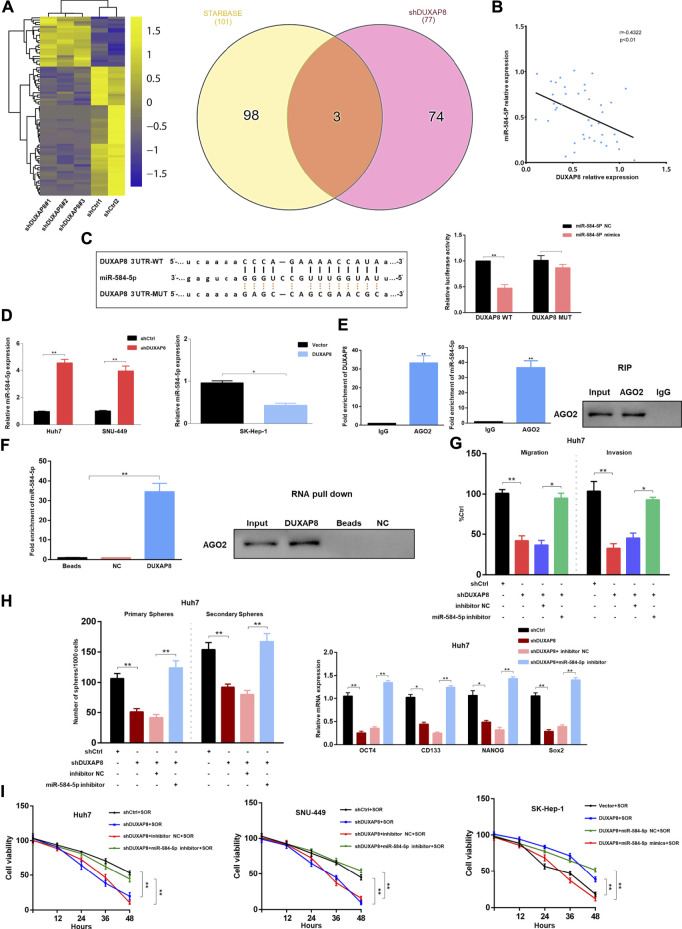
Cytoplasmic lncRNAs are often assumed to be competitive endogenous RNAs that can competitively bind to microRNAs, thereby producing an “isolation” effect on these microRNAs, reducing their regulatory effect on target genes (Tay et al., 2014). Considering the localization of DUXAP8 in the nucleus and cytoplasm (Figure 1H), it was hypothesized that DUXAP8 also exerts oncogenic effects in HCC through the ceRNA mechanism. To test this hypothesis, RNA sequencing analysis of DUXAP8-knockdown HCC cells was conducted to screen out significantly upregulated microRNAs, and the bioinformatics database, Encyclopedia of RNA Interactomes (previously known as starBase v2.0) was used, to predict the microRNAs that can target and bind to DUXAP8. In this study, two microRNAs were intersected, and three microRNAs (miR-584-5p, miR-409-3p, miR-374b-5p) were screened. miR-584-5p was selected, which exhibited the largest fold change, for subsequent experimental validation (Figure 5A, Supplementary Figure S4A). MiR-584-5p has been shown to be involved in regulating multiple malignant tumors (Xiang et al., 2015; Zhang et al., 2020), and it was found that in HCC tissues, DUXAP8 showed a significant negative correlation with miR-584-5p expression (Figure 5B). Additionally, the overexpression of miR-584-5p could significantly reduce the luciferase activity of DUXAP8 WT, but it had no effect on the luciferase activity of DUXAP8 MUT (Figure 5C). The expression of miR-584-5p was significantly increased in HCC cells after the knockdown of DUXAP8, but miR-584-5p expression was significantly reduced when DUXAP8 was overexpressed (Figure 5D). In the RIP, it was found that DUXAP8 and miR-584-5p were highly enriched in AGO2 precipitation (Figure 5E). In the results of RNA pull-down assay, it was found that miR-584-5p and AGO2 were highly enriched in the biotin-labeled DUXAP8 group (Figure 5F). The above experimental results suggested that DUXAP8 acted as a molecular sponge for miR-584-5p in HCC through the ceRNA mechanism.
FIGURE 5.
In HCC, DUXAP8 acts as a molecular sponge for miR-584-5p, which downregulates miR-584-5p and affects the malignant phenotype and chemosensitivity of HCC. (A) The heat map demonstrating the differentially expressed microRNAs in HCC after DUXAP8 downregulation, and Wayne plot showing the differentially expressed microRNAs after the intersection of RNA sequencing results with the bioinformatics database. (B) Correlation analysis between the expression of DUXAP8 and miR-584-5p. (C) Dual-luciferase reporter assay detecting the relative activity of luciferase in transfected HCC cells. (D) Effect of DUXAP8 downregulation and overexpression on miR-584-5p expression in HCC cells. (E) RIP assay detecting the relative enrichment of DUXAP8 and miR-584-5p in anti-IgG or anti-AGO2 specific immunoprecipitates. (F) RNA pull-down assay was used to detect the interrelationship between DUXAP8, miR-584-5p, and AGO2. (G,H) Effects of DUXAP8 and miR-584-5p downregulation on the migration, invasion, primary and secondary spheroid-forming ability, and stemness-related gene expression levels in Huh7 cells. (I) The viability of HCC cells with downregulated DUXAP8 and miR-584-5p under the effect of sorafenib. Data are expressed as mean, *p < 0.05, **p < 0.01.
Next, it was investigated whether DUXAP8 affects the malignant phenotype of HCC by competitively binding to miR-584-5p. When DUXAP8 was knocked down, it significantly reduced the migration and invasion ability of Huh7 and SNU-449 cells while decreasing their spheroid-forming ability, and inhibited the expression of CD133, OCT4, NANOG, and Sox2, although such inhibitory effects of DUXAP8 could be reversed using the knockdown of miR-584-5p (Figures 5G,H, Supplementary Figure S4B–D). When DUXAP8 was overexpressed, it promoted the migration, invasion, and spheroid-forming ability of SK-Hep-1 cells and the expression of CD133, OCT4, NANOG, and Sox2. And this promotion effect could also be reversed through the overexpression of miR-584-5p (Supplementary Figure S4E–G).
As shown in Figure 5I, the knockdown of DUXAP8 enhanced the chemosensitivity of HCC cells to sorafenib, while DUXAP8 overexpression decreased the chemosensitivity of HCC cells to sorafenib, both results could be reversed by the knockdown and overexpression of miR-584-5p, respectively. In conclusion, DUXAP8 could competitively bind to miR-584-5p through the ceRNA mechanism, thus affecting the malignant phenotype and chemosensitivity of HCC.
miR-584-5p Affects the MAPK/ERK Pathway by Targeting MAPK1, a Protein Involved in DUXAP8 Regulation of the Malignant Phenotype and Chemosensitivity in HCC
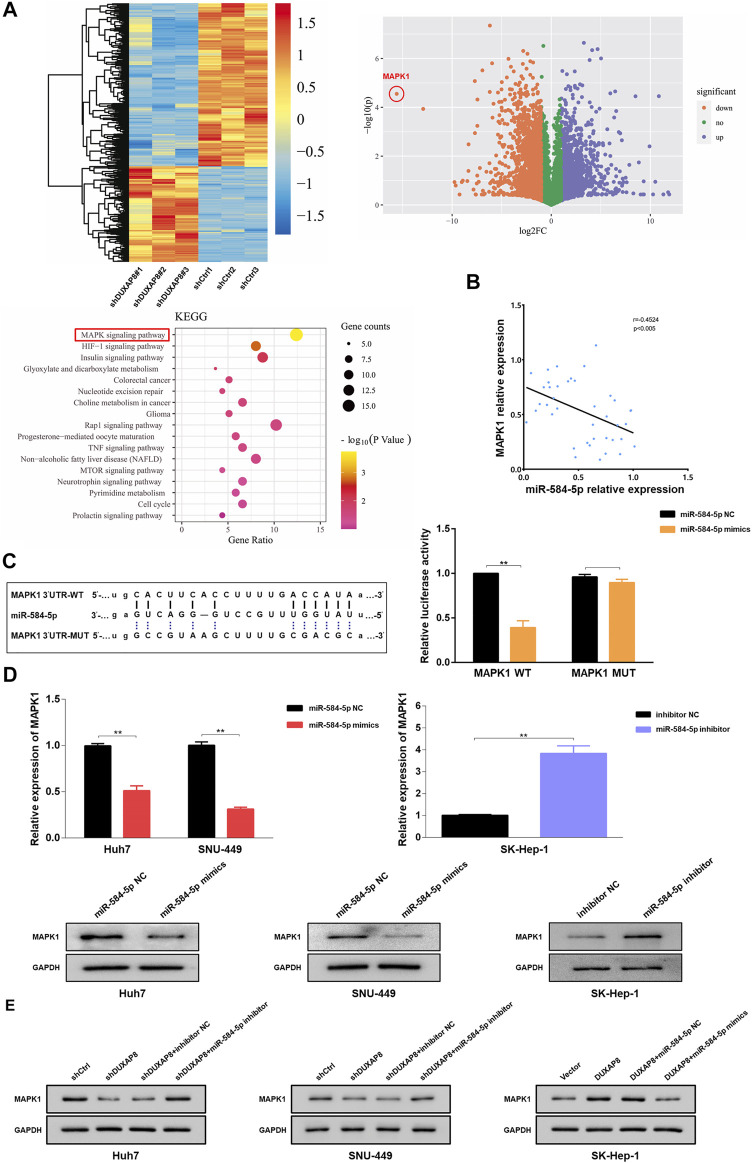
To investigate the mechanism by which DUXAP8 affects the malignant phenotype and chemotherapy resistance of HCC, RNA sequencing analysis of DUXAP8-knockdown HCC cells was performed. The results showed significant differences in the number of genes regulated by DUXAP8 expression, and the MAPK/ERK pathway was the predominantly relevant signaling pathway (Figure 6A). MAPK1 (mitogen-activated protein kinase 1, ERK2) is an essential biomarker of the MAPK/ERK pathway and can serve as a binding site for numerous biochemical signals (Jung et al., 2016; Wu et al., 2016). It has been shown that the activation of the MAPK/ERK pathway is closely associated with the invasion and migration, proliferation, drug resistance, and glycolysis in HCC (Liet al., 2020a; Li et al., 2020b; He et al., 2020). Therefore, it was hypothesized that miR-584-5p regulates the malignant phenotype and chemosensitivity of HCC by targeting MAPK1 and affecting the MAPK/ERK pathway. it was that MAPK1 negatively correlated with miR-584-5p expression in HCC tissues (Figure 6B), and the overexpression of miR-584-5p could significantly reduce the luciferase activity of MAPK1 WT, but had no effect on the luciferase activity of MAPK1 MUT (Figure 6C). Overexpression of miR-584-5p significantly reduced the expression of MAPK1 of Huh7 and SNU- 449, whereas its knockdown produced the opposite result in SK-Hep-1 (Figure 6D). We also found that the knockdown of DUXAP8 significantly reduced MAPK1 expression in Huh7 and SNU-449 cells, however, this effect could be reversed by the downregulation of miR-584-5p. In contrast, overexpression of DUXAP8 increased the expression of MAPK1 in SK-Hep-1 cells, while overexpression of miR-584-5p reversed this effect (Figure 6E). The above experimental results showed that miR-584-5p could directly target MAPK1 in HCC cells.
FIGURE 6.
miR-584-5p targets MAPK1 in HCC. (A) The heat map and volcano plot demonstrating the differentially expressed genes in HCC after DUXAP8 downregulation, and Kyoto Encyclopedia of Genes and Genomes analysis of the main associated pathways. (B) The result of the correlation analysis showing that miR-584-5p was negatively correlated with MAPK1 expression. (C) Dual-luciferase reporter assay detecting the relative activity of luciferase in transfected HCC cells. (D) Effect of miR-584-5p downregulation and overexpression on MAPK1 expression in HCC cells. (E) Effect of down-regulation or overexpression of DUXAP8 and miR-584-5p on MAPK1 expression in HCC during co-transfection. Data are expressed as mean, **p < 0.01.
When miR-584-5p was overexpressed, it significantly reduced the migration and invasion ability of Huh7 and SNU-449 cells while reducing their spheroid-forming ability and also inhibited the expression of CD133, OCT4, NANOG, and Sox2, although this inhibitory effect could be reversed by MAPK1 overexpression (Supplementary Figure S5). When miR-584-5p was knocked down, it promoted the migration, invasion, and spheroid-forming ability of SK-Hep-1 cells and the expression of CD133, OCT4, NANOG, and Sox2, and this effect could also be reversed by the knockdown of MAPK1. The overexpression of miR-584-5p enhanced the chemosensitivity of HCC cells to sorafenib, while its knockdown decreased the chemosensitivity of HCC cells to sorafenib, and both results could also be reversed through the overexpression and knockdown of MAPK1, respectively (Supplementary Figure S5).
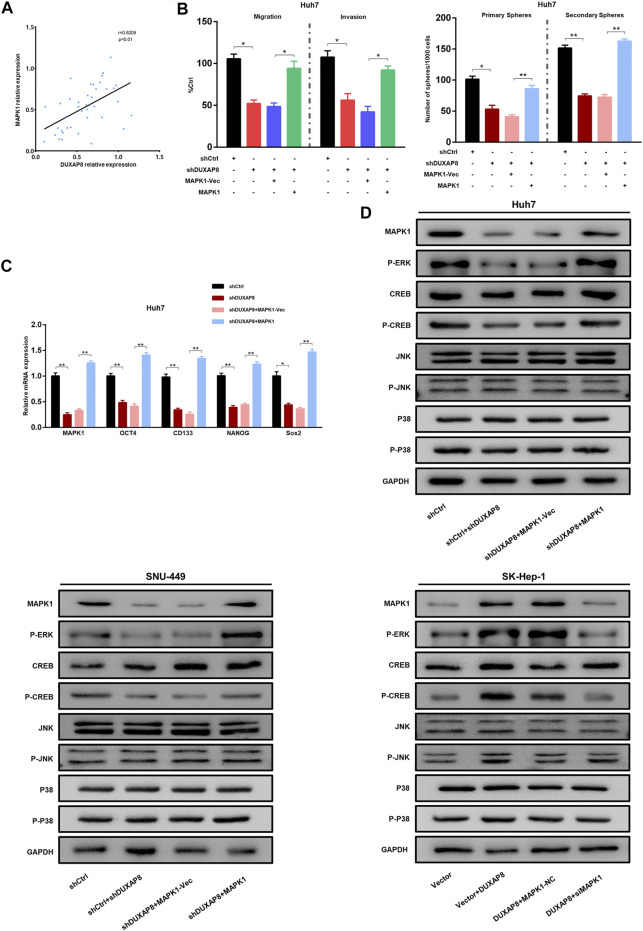
Correlation analyses show that the expression of DUXAP8 was positively correlated with MAPK1 in HCC tissues (Figure 7A). Western blot results showed that the knockdown of DUXAP8 downregulated the phosphorylation level of ERK/CREB, while the overexpression of MAPK1 abolished the inhibitory effect of DUXAP8 knockdown on the phosphorylation level of above proteins. However, there was no effect on the phosphorylation level of JNK/p-38 (Figure 7D). This suggested that DUXAP8 exerts its biological function by activating the MAPK/ERK pathway. To verify this conclusion, a rescue experiment was performed. When DUXAP8 was knocked down, it significantly reduced the migration, invasion, and spheroid-forming ability of Huh7 and SNU-449 cells and the expression of CD133, OCT4, NANOG, and Sox2, but this inhibitory effect could be reversed by the overexpression of MAPK1 (Figures 7B,C, Supplementary Figure S6). When DUXAP8 was overexpressed, it promoted the migration, invasion and spheroid-forming ability of SK-Hep-1 cells and the expression of CD133, OCT4, NANOG, and Sox2, and this effect could also be reversed by the knockdown of MAPK1 (Supplementary Figure S6). The knockdown of DUXAP8 enhanced the chemosensitivity of HCC cells to sorafenib, while its overexpression reduced the chemosensitivity of HCC cells to sorafenib, and both results could be reversed by the overexpression and knockdown of MAPK1, respectively (Supplementary Figure S6).
FIGURE 7.
The DUXAP8/miR-584-5p/MAPK1 axis regulates the malignant phenotype of HCC. (A) The result of the correlation analysis showing that DUXAP8 was positively correlated with the expression of MAPK1. (B) Effects of DUXAP8 downregulation and MAPK1 overexpression on the migration, invasion, and primary and secondary spheroid-forming ability of Huh7 cells. (C) Effects of DUXAP8 downregulation and MAPK1 overexpression on the expression levels of stemness-related genes in Huh7 cells. (D) Effects of DUXAP8 downregulation and MAPK1 overexpression on ERK/CREB/JNK/P38 and their phosphorylation levels in HCC cell lines. Data are expressed as mean, *p < 0.05, **p < 0.01.
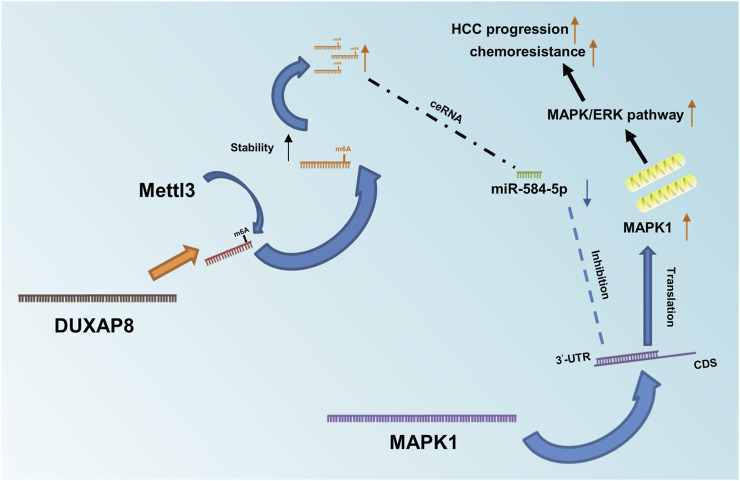
In summary, DUXAP8 competitively bound to miR-584-5P through the ceRNA mechanism (Figure 8), thus targeting MAPK1 and activating the MAPK/ERK pathway to regulate the malignant phenotype and chemotherapy resistance of HCC.
FIGURE 8.
Diagram of DUXAP8’s role in HCC and its mechanism. Mettl3-mediated m6A methylation modification stabilizes and upregulates DUXAP8, which competitively binds to miR-584-5p through a ceRNA mechanism, thereby targeting MAPK1 and activating the MAPK/ERK pathway to regulate the malignant phenotype and chemotherapy resistance of HCC.
Discussion
The mechanisms of drug resistance in HCC are quite complex, involving the increased expression of drug efflux transporters (a type of protein that recognizes and pumps anticancer drugs out of tumor cells), intracellular redistribution of drugs, inactivation of the apoptosis signaling pathway, and activation of tumor stem cells (Butler et al., 2013; Salehan and Morse, 2013; Llovet et al., 2015). However, to date, the exact mechanism of drug resistance in HCC remains to be investigated.
In this study, the role of DUXAP8 in promoting malignant phenotype and chemotherapy resistance in HCC was revealed, and it was confirmed for the first time that Mettl3-mediated m6A modifications were involved in the upregulation of DUXAP8. In summary, it was found that DUXAP8 exhibited high expression in sorafenib-treated PDX models and was positively correlated with the TNM cancer stage, tumor size, microvascular invasion, and distant metastasis. The results of in vivo and in vitro experiments showed that DUXAP8 is positively correlated with the proliferation, migration, invasion, and sorafenib resistance of HCC. DUXAP8 upregulates MAPK1 through competitively binding to miR-584-5p, which activates the MAPK/ERK pathway to perform its biological functions.
There is increasing evidence supporting that lncRNAs play an essential role in the occurrence and development of malignant tumors (Yuan et al., 2014; Li et al., 2015b; Chen et al., 2016; Yuan et al., 2016; Zhu et al., 2016). Until now, DUXAP8 is highly expressed in various malignant tumors. DUXAP8 can significantly inhibit the expression of PLEKHO1 in gastric cancer, which enhances the proliferation and migration of tumor cells (Ma et al., 2017a). In glioma, the downregulation of DUXAP8 inhibits the proliferation of tumor cells (Zhao et al., 2019). In non-small cell lung cancer (NSCLC), DUXAP8 promotes tumor cell proliferation and invasion through epigenetically silencing Egr1 and RHOB (Sun et al., 2017). In pancreatic carcinoma, DUXAP8 promotes tumor growth through epigenetically silencing CDKN1A and KLF2 (Lian et al., 2018). This study found significantly elevated expression of DUXAP8 in HCC compared to that of the adjacent normal liver tissue, and this result is consistent with that in previous reports.
As a novel epigenetic RNA modification, m6A is closely related to the phenotype and mechanism of malignant tumors and plays an essential role in the self-renewal of tumor stem cells and the metabolism, recurrence and metastasis of various malignant tumors. The dynamic regulatory proteins modified by m6A include methyltransferases (Writers), demethylases (Erasers), and reading genes (Readers) (Zhao et al., 2017). The methyltransferases are multicomponent compounds consisting of METTL3, METTL14, WTAP, KIAA1429, RBM15, and ZC3H13 (Knuckles et al., 2018). Recent studies (Jo et al., 2013; Bansal et al., 2014; Ma et al., 2017b; Cui et al., 2017; Visvanathan et al., 2018) have shown that m6A-related proteins are involved in the development and progression of different types of malignant tumors, such as acute myeloid leukemia, cholangiocarcinoma, glioblastoma, and hepatocellular carcinoma (HCC). In this study, it was demonstrated for the first time that DUXAP8 is overexpressed in HCC because Mettl3-mediated m6A modification confers its stability.
Existing studies have demonstrated that lncRNAs are involved in the regulation of many diseases and play an essential role in the occurrence and development of malignant tumors. Some lncRNAs act as protein co-regulators by directly binding to helper proteins and regulating the expression of downstream tumor-associated genes (Chen et al., 2016; Yuan et al., 2016; Zhu et al., 2016). Another part of lncRNAs functions as competitive endogenous RNAs (ceRNAs) (Yuan et al., 2014; Li et al., 2015b). Such lncRNAs isolate microRNAs and regulate the expression of microRNA-targeted oncogenes or tumor suppressor genes through molecular sponge effects. Argonaute 2 (AGO2) is a core component of the microRNA (miRNA)-induced silencing complex (RISC), linking miRNAs and their mRNA target sites(Meister et al., 2004). And the mechanisms through which lncRNAs regulate the biological functions of malignant tumors largely depend on their subcellular localization (Statello et al., 2021). Considering that DUXAP8 is mainly localized in the cytoplasm, while RNA sequencing analysis and bioinformatics analysis also indicate that DUXAP8 has a putative binding site for miR-584-5p, it was therefore suspected that DUXAP8 also affects HCC progression through a ceRNA mechanism. The luciferase reporter assay, RIP and RNA pull-down experiments confirmed this speculation. Meanwhile, the expression of miR-584-5p was correspondingly reversed when the expression of DUXAP8 was changed. These data suggested that DUXAP8 acts as a molecular sponge, which binds to miR-584-5p to perform its biological function. miR-584-5p was further investigated and it was found that miR-584-5p significantly inhibited the features of migration, invasion, and stemness of HCC and correspondingly enhanced its chemosensitivity to sorafenib. RNA sequencing analysis revealed that the downregulation of DUXAP8 had a significant effect on the MAPK/ERK pathway, accompanied by the downregulation of MAPK1. Previous studies have reported that MAPK/ERK pathway is closely related to the invasion and migration, proliferation, drug resistance, and glycolysis in HCC (Li et al., 2020a; Li et al., 2020b; He et al., 2020). Additionally, MAPK1 is an essential biomarker of the MAPK/ERK pathway and can serve as a binding site for numerous biochemical signals (Jung et al., 2016; Wu et al., 2016). Therefore, it was suggested that miR-584-5p might play an oncogenic role in HCC by targeting the downregulation of MAPK1, and subsequent findings supported this hypothesis. Through luciferase reporter assay, expression correlation analysis and cell function up-and downregulation assays in HCC, it was demonstrated that miR-584-5p could indeed directly affect the expression level of MAPK1 in HCC cells. Meanwhile, the knockdown and overexpression of MAPK1 could effectively reverse the effects of miR-584-5p knockdown and overexpression on the malignant phenotype and chemosensitivity of HCC cells to sorafenib. Then, it was further shown that the knockdown of DUXAP8 inhibited the phosphorylation level of ERK/CREB, while the overexpression of MAPK1 reduced the inhibitory effect of DUXAP8 knockdown on the phosphorylation level of ERK/CREB. It was also suggested that DUXAP8 exerts its biological function by activating the MAPK/ERK pathway.
In conclusion, this study revealed the role and potential mechanism of DUXAP8 in promoting the malignant phenotype of HCC and chemoresistance. Mechanistically, DUXAP8 endogenously competes for the binding of miR-584-5p through the ceRNA mechanism, reducing its inhibitory effect -on MAPK1 and thus activating the MAPK/ERK pathway to promote the proliferation, invasive migration, stemness maintenance, and chemoresistance in HCC. These results have deepened the understanding of the driving mechanisms of lncRNAs in the development, progression, and chemotherapy resistance of HCC and provided greater insight into the importance of DUXAP8 in HCC progression, which may help provide ideas for finding new prognostic indicators and therapeutic targets for HCC patients.
Acknowledgments
We thank Enago—https://www.enago.cn/fanyi/tougaoji-fanyi.htm for their assistance in manuscript translation and editing.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://doi.org/10.6084/m9.figshare.17049722.v1, https://doi.org/10.6084/m9.figshare.17049716.v1, https://doi.org/10.6084/m9.figshare.17049686.v1.
Ethics Statement
The studies involving human participants were reviewed and approved by Ethics Committee of the Second Hospital of Jilin University. The patients/participants provided their written informed consent to participate in this study. The animal study was reviewed and approved by Animal Experiment Ethics Committee of the First Hospital of Jilin University.
Author Contributions
ZL, YY, BT, and XZ developed the original hypothesis and supervised the experimental design. ZL, JL, and HF performed in vitro and in vivo experiments. ZL, JS, and MC analyzed the data. ZL, BT, and XZ wrote and revise the manuscript. All authors read and approved the final.
Funding
The work was funded by the project of Hepatobiliary and Pancreatic Disease Translational Medicine Platform Construction (2017F009), This work was supported by grants from Medical and Health Talents Project of Jilin Province (2019SCZT003), Health special project of Jilin Province (2020sczt017), Sponsored by National Science Foundation of The First Hospital of Jilin University (JDYY92018041).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2021.783385/full#supplementary-material
References
- Bansal H., Yihua Q., Iyer S. P., Ganapathy S., Proia D., Penalva L. O., et al. (2014). WTAP Is a Novel Oncogenic Protein in Acute Myeloid Leukemia. Leukemia 28, 1171–1174. 10.1038/leu.2014.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruix J., da Fonseca L. G., Reig M. (2019). Insights into the success and Failure of Systemic Therapy for Hepatocellular Carcinoma. Nat. Rev. Gastroenterol. Hepatol. 16, 617–630. 10.1038/s41575-019-0179-x [DOI] [PubMed] [Google Scholar]
- Butler E. B., Zhao Y., Muñoz-Pinedo C., Lu J., Tan M. (2013). Stalling the Engine of Resistance: Targeting Cancer Metabolism to Overcome Therapeutic Resistance. Cancer Res. 73, 2709–2717. 10.1158/0008-5472.CAN-12-3009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z.-z., Huang L., Wu Y.-h., Zhai W.-j., Zhu P.-p., Gao Y.-f. (2016). LncSox4 Promotes the Self-Renewal of Liver Tumour-Initiating Cells through Stat3-Mediated Sox4 Expression. Nat. Commun. 7, 12598. 10.1038/ncomms12598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Q., Shi H., Ye P., Li L., Qu Q., Sun G., et al. (2017). m 6 A RNA Methylation Regulates the Self-Renewal and Tumorigenesis of Glioblastoma Stem Cells. Cel Rep. 18, 2622–2634. 10.1016/j.celrep.2017.02.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang K.-C., Kao W.-Y., Su C.-W., Chen P.-C., Lee P.-C., Huang Y.-H., et al. (2018). The Prognosis of Single Large Hepatocellular Carcinoma Was Distinct from Barcelona Clinic Liver Cancer Stage A or B: The Role of Albumin-Bilirubin Grade. Liver Cancer 7, 335–358. 10.1159/000487407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrow J., Frankish A., Gonzalez J. M., Tapanari E., Diekhans M., Kokocinski F., et al. (2012). GENCODE: the Reference Human Genome Annotation for the ENCODE Project. Genome Res. 22, 1760–1774. 10.1101/gr.135350.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H., Chen T., Mo H., Chen S., Liu Q., Guo C. (2020). Hypoxia-inducible Long Noncoding RNA NPSR1-AS1 Promotes the Proliferation and Glycolysis of Hepatocellular Carcinoma Cells by Regulating the MAPK/ERK Pathway. Biochem. Biophysical Res. Commun. 533, 886–892. 10.1016/j.bbrc.2020.09.076 [DOI] [PubMed] [Google Scholar]
- Jemal A., Bray F., Center M. M., Ferlay J., Ward E., Forman D. (2011). Global Cancer Statistics. CA: A Cancer J. Clinicians 61, 69–90. 10.3322/caac.20107 [DOI] [PubMed] [Google Scholar]
- Jo H.-J., Shim H.-E., Han M.-E., Kim H.-J., Kim K.-S., Baek S., et al. (2013). WTAP Regulates Migration and Invasion of Cholangiocarcinoma Cells. J. Gastroenterol. 48, 1271–1282. 10.1007/s00535-013-0748-7 [DOI] [PubMed] [Google Scholar]
- Jung Y.-C., Han S., Hua L., Ahn Y.-H., Cho H., Lee C.-J., et al. (2016). Kazinol-E Is a Specific Inhibitor of ERK that Suppresses the Enrichment of a Breast Cancer Stem-like Cell Population. Biochem. Biophysical Res. Commun. 470, 294–299. 10.1016/j.bbrc.2016.01.066 [DOI] [PubMed] [Google Scholar]
- Knuckles P., Lence T., Haussmann I. U., Jacob D., Kreim N., Carl S. H., et al. (2018). Zc3h13/Flacc Is Required for Adenosine Methylation by Bridging the mRNA-Binding Factor Rbm15/Spenito to the m6A Machinery Component Wtap/Fl(2)d. Genes Dev. 32, 415–429. 10.1101/gad.309146.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudo M., Finn R. S., Qin S., Han K.-H., Ikeda K., Piscaglia F., et al. (2018). Lenvatinib versus Sorafenib in First-Line Treatment of Patients with Unresectable Hepatocellular Carcinoma: a Randomised Phase 3 Non-inferiority Trial. The Lancet 391, 1163–1173. 10.1016/S0140-6736(18)30207-1 [DOI] [PubMed] [Google Scholar]
- Li L., Tang J., Zhang B., Yang W., LiuGao M., Wang R., et al. (2015a). Epigenetic Modification of MiR-429 Promotes Liver Tumour-Initiating Cell Properties by Targeting Rb Binding Protein 4. Gut 64, 156–167. 10.1136/gutjnl-2013-305715 [DOI] [PubMed] [Google Scholar]
- Li Q.-T., Feng Y.-M., Ke Z.-H., Qiu M.-J., He X.-X., Wang M.-M., et al. (2020b). KCNN4 Promotes Invasion and Metastasis through the MAPK/ERK Pathway in Hepatocellular Carcinoma. J. Investig. Med. 68, 68–74. 10.1136/jim-2019-001073 [DOI] [PubMed] [Google Scholar]
- Li Q., Ren B., Gui Q., Zhao J., Wu M., Shen M., et al. (2020a). Blocking MAPK/ERK Pathway Sensitizes Hepatocellular Carcinoma Cells to Temozolomide via Downregulating MGMT Expression. Ann. Transl Med. 8, 1305. 10.21037/atm-20-5478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Xie J., Shen C., Cheng D., Shi Y., Wu Z., et al. (2015b). Amplification of Long Noncoding RNA ZFAS1 Promotes Metastasis in Hepatocellular Carcinoma. Cancer Res. 75, 3181–3191. 10.1158/0008-5472.CAN-14-3721 [DOI] [PubMed] [Google Scholar]
- Lian Y., Yang J., Lian Y., Xiao C., Hu X., Xu H. (2018). DUXAP8, a Pseudogene Derived lncRNA, Promotes Growth of Pancreatic Carcinoma Cells by Epigenetically Silencing CDKN1A and KLF2. Cancer Commun. 38, 64. 10.1186/s40880-018-0333-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Wang M., Liang N., Guan L. (2019). PDCD2 Sensitizes HepG2 Cells to Sorafenib by Suppressing Epithelial-mesenchymal T-ransition. Mol. Med. Rep. 19, 2173–2179. 10.3892/mmr.2019.9860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llovet J. M., Villanueva A., Lachenmayer A., Finn R. S. (2015). Advances in Targeted Therapies for Hepatocellular Carcinoma in the Genomic Era. Nat. Rev. Clin. Oncol. 12, 408–424. 10.1038/nrclinonc.2015.103 [DOI] [PubMed] [Google Scholar]
- Ma H.-w., Xie M., Sun M., Chen T.-y., Jin R.-r., Ma T.-s., et al. (2017a). The Pseudogene Derived Long Noncoding RNA DUXAP8 Promotes Gastric Cancer Cell Proliferation and Migration via Epigenetically Silencing PLEKHO1 Expression. Oncotarget 8, 52211–52224. 10.18632/oncotarget.11075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J. z., Yang F., Zhou C. c., Liu F., Yuan J. h., Wang F., et al. (2017b). METTL14 Suppresses the Metastatic Potential of Hepatocellular Carcinoma by Modulating N 6 ‐methyladenosine‐dependent Primary MicroRNA Processing. Hepatology 65, 529–543. 10.1002/hep.28885 [DOI] [PubMed] [Google Scholar]
- Meister G., Landthaler M., Patkaniowska A., Dorsett Y., Teng G., Tuschl T. (2004). Human Argonaute2 Mediates RNA Cleavage Targeted by miRNAs and siRNAs. Mol. Cel 15, 185–197. 10.1016/j.molcel.2004.07.007 [DOI] [PubMed] [Google Scholar]
- Petrick J. L., Florio A. A., Znaor A., Ruggieri D., Laversanne M., Alvarez C. S., et al. (2020). International Trends in Hepatocellular Carcinoma Incidence, 1978-2012. Int. J. Cancer 147, 317–330. 10.1002/ijc.32723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raoul J.-L., Edeline J. (2020). Systemic Treatment of Hepatocellular Carcinoma: Standard of Care in China and Elsewhere. Lancet Oncol. 21, 479–481. 10.1016/S1470-2045(20)30082-6 [DOI] [PubMed] [Google Scholar]
- Reinders M. T. M., van Meer S., Burgmans M. C., de Jong K. P., Klümpen H.-J., de Man R. A., et al. (2020). Trends in Incidence, Diagnosis, Treatment and Survival of Hepatocellular Carcinoma in a Low-Incidence Country: Data from the Netherlands in the Period 2009-2016. Eur. J. Cancer 137, 214–223. 10.1016/j.ejca.2020.07.008 [DOI] [PubMed] [Google Scholar]
- Rimassa L., Wörns M. A. (2020). Navigating the New Landscape of Second‐line Treatment in Advanced Hepatocellular Carcinoma. Liver Int. 40, 1800–1811. 10.1111/liv.14533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salehan M. R., Morse H. R. (2013). DNA Damage Repair and Tolerance: a Role in Chemotherapeutic Drug Resistance. Br. J. Biomed. Sci. 70, 31–40. 10.1080/09674845.2013.11669927 [DOI] [PubMed] [Google Scholar]
- Schwartz S., Agarwala S. D., Mumbach M. R., Jovanovic M., Mertins P., Shishkin A., et al. (2013). High-resolution Mapping Reveals a Conserved, Widespread, Dynamic mRNA Methylation Program in Yeast Meiosis. Cell 155, 1409–1421. 10.1016/j.cell.2013.10.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X., Nie F., Wang Z., Sun M. (2016). Pseudogene-expressed RNAs: a New Frontier in Cancers. Tumor Biol. 37, 1471–1478. 10.1007/s13277-015-4482-z [DOI] [PubMed] [Google Scholar]
- Statello L., Guo C.-J., Chen L.-L., Huarte M. (2021). Gene Regulation by Long Non-coding RNAs and its Biological Functions. Nat. Rev. Mol. Cel Biol. 22, 96–118. 10.1038/s41580-020-00315-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M., Nie F.-q., Zang C., Wang Y., Hou J., Wei C., et al. (2017). The Pseudogene DUXAP8 Promotes Non-small-cell Lung Cancer Cell Proliferation and Invasion by Epigenetically Silencing EGR1 and RHOB. Mol. Ther. 25, 739–751. 10.1016/j.ymthe.2016.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Tay Y., Rinn J., Pandolfi P. P. (2014). The Multilayered Complexity of ceRNA Crosstalk and Competition. Nature 505, 344–352. 10.1038/nature12986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvanathan A., Patil V., Arora A., Hegde A. S., Arivazhagan A., Santosh V., et al. (2018). Essential Role of METTL3-Mediated m6A Modification in Glioma Stem-like Cells Maintenance and Radioresistance. Oncogene 37, 522–533. 10.1038/onc.2017.351 [DOI] [PubMed] [Google Scholar]
- Wang X.-K., Liao X.-W., Huang R., Huang J.-L., Chen Z.-J., Zhou X., et al. (2020). Clinical Significance of Long Non-coding RNA DUXAP8 and its Protein Coding Genes in Hepatocellular Carcinoma. J. Cancer 11, 6140–6156. 10.7150/jca.47902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei C.-M., Gershowitz A., Moss B. (1975). Methylated Nucleotides Block 5′ Terminus of HeLa Cell Messenger RNA. Cell 4, 379–386. 10.1016/0092-8674(75)90158-0 [DOI] [PubMed] [Google Scholar]
- Wei L., Wang X., Lv L., Liu J., Xing H., Song Y., et al. (2019). The Emerging Role of microRNAs and Long Noncoding RNAs in Drug Resistance of Hepatocellular Carcinoma. Mol. Cancer 18, 147. 10.1186/s12943-019-1086-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., Cao K., Ni Z., Wang S., Li W., Liu X., et al. (2019). Rhein Reverses Doxorubicin Resistance in SMMC-7721 Liver Cancer Cells by Inhibiting Energy Metabolism and Inducing Mitochondrial Permeability Transition Pore Opening. Biofactors 45, 85–96. 10.1002/biof.1462 [DOI] [PubMed] [Google Scholar]
- Wu L. K., Liu Y. C., Ma G., Shi L. L., He X. M. (2016). High Levels of Glucose Promote the Activation of Hepatic Stellate Cells via the P38-Mitogen-Activated Protein Kinase Signal Pathway. Genet. Mol. Res. 15. 10.4238/gmr.15038419 [DOI] [PubMed] [Google Scholar]
- Xiang Q.-F., Zhan M.-X., Li Y., Liang H., Hu C., Huang Y.-M., et al. (2019). Activation of MET Promotes Resistance to Sorafenib in Hepatocellular Carcinoma Cells via the AKT/ERK1/2-EGR1 Pathway. Artif. Cell Nanomedicine, Biotechnol. 47, 83–89. 10.1080/21691401.2018.1543195 [DOI] [PubMed] [Google Scholar]
- Xiang X., Mei H., Qu H., Zhao X., Li D., Song H., et al. (2015). miRNA-584-5p Exerts Tumor Suppressive Functions in Human Neuroblastoma through Repressing Transcription of Matrix Metalloproteinase 14. Biochim. Biophys. Acta (Bba) - Mol. Basis Dis. 1852, 1743–1754. 10.1016/j.bbadis.2015.06.002 [DOI] [PubMed] [Google Scholar]
- Yang M., Parikh N. D., Liu H., Wu E., Rao H., Feng B., et al. (2020). Incidence and Risk Factors of Hepatocellular Carcinoma in Patients with Hepatitis C in China and the United States. Sci. Rep. 10, 20922. 10.1038/s41598-020-77515-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J.-h., Yang F., Wang F., Ma J.-z., Guo Y.-j., Tao Q.-f., et al. (2014). A Long Noncoding RNA Activated by TGF-β Promotes the Invasion-Metastasis Cascade in Hepatocellular Carcinoma. Cancer Cell 25, 666–681. 10.1016/j.ccr.2014.03.010 [DOI] [PubMed] [Google Scholar]
- Yuan S.-x., Wang J., Yang F., Tao Q.-f., Zhang J., Wang L.-l., et al. (2016). Long Noncoding RNADANCRincreases Stemness Features of Hepatocellular Carcinoma by Derepression ofCTNNB1. Hepatology 63, 499–511. 10.1002/hep.27893 [DOI] [PubMed] [Google Scholar]
- Zhang M., Weng W., Zhang Q., Wu Y., Ni S., Tan C., et al. (2018). The lncRNA NEAT1 Activates Wnt/β-Catenin Signaling and Promotes Colorectal Cancer Progression via Interacting with DDX5. J. Hematol. Oncol. 11, 113. 10.1186/s13045-018-0656-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Wang H., Li C., Gao L., Zheng Y., Chang W., et al. (2020). CircSMYD4 Regulates Proliferation, Migration and Apoptosis of Hepatocellular Carcinoma Cells by Sponging miR-584-5p. Cancer Cel Int. 20, 556. 10.1186/s12935-020-01648-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B. S., Roundtree I. A., He C. (2017). Post-transcriptional Gene Regulation by mRNA Modifications. Nat. Rev. Mol. Cel Biol. 18, 31–42. 10.1038/nrm.2016.132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Hao S., Wang M., Xing D., Wang C. (2019). Knockdown of pseudogene DUXAP8 Expression in Glioma Suppresses Tumor Cell Proliferation. Oncol. Lett. 17, 3511–3516. 10.3892/ol.2019.9994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L., Li X., Gu Y., Lv X., Xi T. (2015). The 3′UTR of the Pseudogene CYP4Z2P Promotes Tumor Angiogenesis in Breast Cancer by Acting as a ceRNA for CYP4Z1. Breast Cancer Res. Treat. 150, 105–118. 10.1007/s10549-015-3298-2 [DOI] [PubMed] [Google Scholar]
- Zhu P., Wang Y., Huang G., Ye B., Liu B., Wu J., et al. (2016). Lnc-β-Catm Elicits EZH2-dependent β-catenin Stabilization and Sustains Liver CSC Self-Renewal. Nat. Struct. Mol. Biol. 23, 631–639. 10.1038/nsmb.3235 [DOI] [PubMed] [Google Scholar]
- Zhu Y.-J., Zheng B., Luo G.-J., Ma X.-K., Lu X.-Y., Lin X.-M., et al. (2019). Circular RNAs Negatively Regulate Cancer Stem Cells by Physically Binding FMRP against CCAR1 Complex in Hepatocellular Carcinoma. Theranostics 9, 3526–3540. 10.7150/thno.32796 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://doi.org/10.6084/m9.figshare.17049722.v1, https://doi.org/10.6084/m9.figshare.17049716.v1, https://doi.org/10.6084/m9.figshare.17049686.v1.