Abstract
Objective:
The larger sample sizes available from multi-site publicly available neuroimaging data repositories makes machine-learning based diagnostic classification of mental disorders more feasible by alleviating the curse of dimensionality. However, since multi-site data are aggregated post-hoc, i.e. they were acquired from different scanners with different acquisition parameters, non-neural inter-site variability may mask inter-group differences that are at least in part neural in origin. Hence, the advantages gained by the larger sample size in the context of machine-learning based diagnostic classification may not be realized.
Methods:
We address this issue using harmonization of multi-site neuroimaging data using the ComBat technique, which is based on an empirical Bayes formulation to remove inter-site differences in data distributions, to improve diagnostic classification accuracy. Specifically, we demonstrate this using ABIDE (Autism Brain Imaging Data Exchange) multisite data for classifying individuals with Autism from healthy controls using resting state fMRI-based functional connectivity data.
Results:
Our results show that higher classification accuracies across multiple classification models can be obtained (especially for models based on artificial neural networks) from multi-site data post harmonization with the ComBat technique as compared to without harmonization, outperforming earlier results from existing studies using ABIDE. Furthermore, our network ablation analysis facilitated important insights into autism spectrum disorder pathology and the connectivity in networks shown to be important for classification covaried with verbal communication impairments in Autism.
Conclusion:
Multi-site data harmonization using ComBat improves neuroimaging-based diagnostic classification of mental disorders.
Significance:
ComBat has the potential to make AI-based clinical decision-support systems more feasible in psychiatry.
Keywords: ABIDE, Autism, Data Harmonization, fMRI, Inter-site Diagnostic classification, Multi-site Data
I. INTRODUCTION
Autism Spectrum Disorder (ASD) is a developmental non-focal brain disorder that is clinically characterized by impaired social communication, restricted interests and repetitive behaviors and can be diagnosed in early years of life [1]. The diagnosis of ASD is typically performed using observation of behavior as well as clinical interviews and questionnaires of the child and the parents [2]. These techniques however may create disparities in diagnosis and therefore it has become crucial to identify objective pathological biomarkers of ASD that can support clinical diagnosis, especially in ambiguity, as well as an aid in predicting the risk of ASD before the manifestation of behavioral symptoms [3]–[5].
Earlier neuroimaging-based research in ASD relied on univariate analysis techniques such as voxel-based morphometry, region-of-interest based analysis etc. These studies indicated widespread brain abnormalities that include gray matter, white matter volume differences and atrophy in frontal, parietal, temporal and limbic regions mainly based on structural and diffusion MRI [6]–[10]. Resting state functional magnetic resonance imaging (rs-fMRI) probes the dynamic alterations in ASD by mapping connectivity and deviations in the activation patterns [11], [12]. Existing work has illustrated altered connectivity and activation patterns in various brain regions within the frontal, parietal, temporal, limbic and striatal regions using rs-fMRI. However, there is little agreement on the connectivity and activation patterns that could be attributed towards variability in populations, the pre-processing and analytic techniques employed, MRI acquisition protocol etc. Moreover, these studies that relied on statistical group analysis, could not provide a patient specific prediction or a quantifiable score that could serve as a pathophysiological signature of autism.
Recent studies have therefore focused on multivariate analysis based on machine learning (ML) algorithms that can facilitate patient specific quantifiers of pathology. These classifier-based techniques have illustrated reasonably high prediction accuracies using multiple imaging modalities that include structural MRI, diffusion MRI, magnetoencephalography and fMRI [13]–[19]. Furthermore, including the interactions between the brain regions via large scale structural and/or functional brain connectomes, ML has been able to additionally support in predicting ASD [20]–[22]. Connectomic multi-variate analysis methods have gained importance as these not only facilitate subject specific signatures but also offer a non-invasive means to understand the macroscopic regional interactions and anomalies that exist between these interactions. However, majority of these studies have been carried out on small locally scanned datasets and the reproducibility on other sites/scanners has not been evaluated thoroughly [14].
Autism Brain Imaging Data Exchange (ABIDE) is a large-scale imaging dataset (about 1000 subjects) of MRI data pooled from multiple sites. It has provided a platform to gain a deeper understanding of the pathological mechanisms underlying autism. Moreover, the dataset facilitates as an open-access benchmarking instrument for novel ML based algorithms that are being developed to identify autism from neuroimaging. Earlier work on ABIDE employed standard machine learning algorithms on functional connectomes which includes general linear model, supervised methods such as support vector machines, random forest (RF), logistic regression, naïve Bayes classifier and linear discriminant analysis, as well as unsupervised methods [23]–[29]. Recent developments in identifying ASD have focused on employing novel deep learning algorithms, which takes the classification problem to a new level by allowing better predictions than standard ML algorithms [30], [31]. These algorithms use complex data representations and have the capability to auto-extract the most relevant features [32]. Convolutional neural net (CNN), a deep learning technique, has been applied to discriminate ASD patients with superior accuracies [33]. However, effectively using connectomic data for purposes of classification is more complicated when compared to other conventional data representations such as images or signals.
For example, CNNs inherently may not perform optimally on graphs as graphs do not possess a smoothly varying neighborhood like images making it complicated for the convolution kernel to match the underlying pattern. Accounting for this problem with CNNs, Parisot et al. proposed to employ graph based convolutional neural nets that represents populations as sparse graphs where the nodes represent the subjects and edge weights represent the pair-wise similarity features computed from auxiliary phenotypic data [34]. Although the technique combines imaging and non-imaging data, it expects the phenotypic information to be available for each subject. Moreover, the entire connectome is compressed into a single number (similarity) which may not be the best representation of the complete connectome. Another deep learning technique applied to ABIDE that employs denoising autoencoders, has facilitated state-of-the-art accuracies. However, these do not exceed 70%, making these ML algorithms ineffective for clinical usage [35].
In general, prediction tasks on connectomes are non-trivial and require a cautious effort in engineering the most distinctive features. Moreover, with neuro-psychiatric disorders like autism, the differences are subtle and capturing these in a diverse population is challenging. Additionally, multi-center studies such as ABIDE are often afflicted with non-pathological variability emerging from scanner magnetic strength and vendor differences, inconsistencies in MR protocols and other intrinsic factors such as head motion etc. [36], [37].
Despite the promise that neuroimaging markers facilitate, translating these to the clinic is still infeasible. Multiple fMRI studies have reported systemic scanner differences [37]. However, these are not accounted for during post-processing and analysis, that in turn may perturb the multi-variate model. This is illustrated in a study by Lanka et al. where leave-site-out type of analysis was carried out using 18 different conventional machine learning classifiers (in ABIDE as well as other multi-site data such as ADHD-200) revealing a substantial drop in accuracy on test data [26], [27]. Although merging data from multiple sites may facilitate more generalizability to the multi-variate model, it is crucial to test the robustness and/or uncertainty about the adaptability to unseen datasets. Recent work in site-harmonization, that statistically removes the scanner effects, has demonstrated exceptional results on diffusion imaging, structural imaging as well as on functional connectivity (FC) analysis [38], [39]. Applying such techniques on the 18-site ABIDE-I data may facilitate promising classification results as well as support in gaining insights into the discriminative connectivity patterns that emerge after harmonization. Our work harmonizes the ABIDE-I connectivity matrices using the state-of-the-art ComBat technique and employs a simple ANN-based architecture for classification of typically developing kids from autism [39]. Our sub-network-based ablation analysis on the harmonized data extracts the most significant sub-networks that are finally correlated with clinical markers in autism.
II. MATERIALS AND METHODS
A. Participants
The present study was carried out using the rs-fMRI data from the ABIDE-I dataset [40]. ABIDE is an open access, multisite image repository comprising structural and functional scans of ASD and matched typically developing (TD) controls [41]. We included 432 ASD and 556 TDs in our dataset. Note that ABIDE I data release contains 1112 subjects. We excluded some subjects based on the following criteria: (i) 36 PDD-NOS subjects since this disorder has been removed in DSM-V, (ii) 6 subjects with a diagnosis of “Asperger’s or PDD NOS”, (iii) 10 subjects from the UCLA (University of California, Los Angeles) site with partial data missing, and (iv) 72 subjects from Stanford and OHSU (Oregon Health and Science University) sites who did not have a DSM-IV diagnosis. This left us with a total of 988 subjects for our analysis.
B. Imaging
ABIDE-I encompasses rs-fMRI and T1 structural brain images that were acquired at 18 sites. The image acquisition parameters and protocol information can be found at https://fcon_1000.projects.nitrc.org/indi/abide.
C. Data Preprocessing
The ABIDE I fMRI dataset’s preprocessing followed a standard pipeline using Data Processing Assistant for Resting-State fMRI Toolbox (DPARSF) which is based on SPM (Statistical Parametric Mapping) [42], [43]. The pipeline consisted of first five volumes removal, slice time correction and motion correction. T1-weighted anatomical images were co-registered to the mean functional images, using which the fMRI images were spatially registered to a standard MNI152 template. Nuisance variables such as low-frequency drifts and motion parameters were regressed out. Unwanted physiological fluctuations (white-matter and cerebrospinal fluid signals) were removed using aCOMPCor (anatomical component-based noise correction). The fMRI time series from every voxel in the brain was deconvolved by estimating the voxel-specific hemodynamic response function (HRF) using a blind deconvolution procedure to obtain the latent neural signals [44], [45]. This is necessitated by the fact that HRF variability across brain regions and individuals has been shown to corrupt functional connectivity estimates [46], [47]. Further, HRFs have been shown to vary considerably in mental disorders including Autism [48]–[50]. Following deconvolution, fMRI data was then temporally band-pass filtered (0.01–0.1 Hz) using a 30th order finite impulse response filter.
D. Functional Connectivity computation
We used FC as the feature to classify the ASD group from the TD group. FC matrix is a weighted adjacency matrix, which indicates the level of co-activation between paired regions of interest in the brain during resting state. To construct the FC matrix, 200 homogeneous regions of interest (ROIs) were defined using the Craddock CC200 functional parcellation atlas [51]. The corresponding mean time series were extracted from these 200 regions for each subject. Each value in the FC matrix was calculated using the Pearson correlation coefficient of two corresponding time series. The range of each value in the FC matrix is from −1 to 1. The 200×200 correlation matrix across 988 subjects was vectorized by removing the upper triangle owing to the symmetric nature of the matrix. The diagonal elements which correspond to self-correlation were omitted. Thus, a column array of 19900 values was obtained for analysis.
Further, the fMRI FC matrices were Fischer-Z transformed. Absolute values of the matrices were used. Twelve sub-networks namely sensory/somatomotor hand, sensory/somatomotor mouth, cingulo-opercular task control, auditory, default mode, cingulo-parietal, visual, fronto-parietal task control, salience, subcortical, ventral attention and dorsal attention were identified among the ROIs used. In addition, we extracted seven global graph theoretical measures for each sub-network. The graph measures obtained included density, modularity, transitivity, global efficiency, assortativity, characteristic path-length and clustering coefficient. Associations were assessed between global measures of each sub-network and symptom-severity/behavioral measures of the subjects’ ADIR-Social and Verbal scores. Multiple comparisons were corrected using the false discovery rate (FDR) method.
E. ComBat Harmonization
To evaluate whether site-harmonization impacts the power of classification models in predicting diagnostic status using the ABIDE dataset, we employed ComBat named for ‘combating batch effects when combining batches’ [52]. ComBat technique was initially developed for adjustment of batch effects in genomic microarray analysis and has since been applied to harmonization of diffusion tensor imaging data [39] and brain connectivity data [53]. ComBat is based on the empirical Bayes method; it assumes that the errors introduced in the imaging features can be standardized by adjusting the location (means) and scale (variances) across the batches. This location and scale (L/S) model is defined as:
| (1) |
Where, for every feature v, in our case, this is the connectivity between two given brain regions, yijv represents the actual value of the feature which in our case, it is the actual value of FC between the given brain regions for the scan j at site i. αv corresponds to the overall feature value for the feature v. X is a design matrix for the covariates of interest (e.g. age and gender). βv is the feature specific vector of regression coefficients corresponding to X. The terms γiv and δiv represent the additive and multiplicative site effects of site i for feature v respectively and εijv is the error term which is assumed to follow a normal distribution with mean zero and variance . ComBat improves the variance of the parameter estimates and by estimating an empirical statistical distribution for each of those parameters. It assumes that all the features share the same common distribution, and site-effect parameters have the parametric prior distributions. The hyperparameters are estimated empirically from the data as described in [52]. The final ComBat-harmonized values are defined as:
| (2) |
F. Classification
We employed three classification techniques that include an artificial neural network (ANN) architecture, random forest (RF) classification as well as state of art auto-encoders as proposed by Heinsfeld et al. on harmonized and non-harmonized connectivity matrices [24], [35], [54]. For all the methods, classification was implemented in leave-one-site-out (LOSO) manner. Of the 18 available scanner sites, the training set included subjects belonging to 17 sites and the remaining site was used for testing.
1). ANN
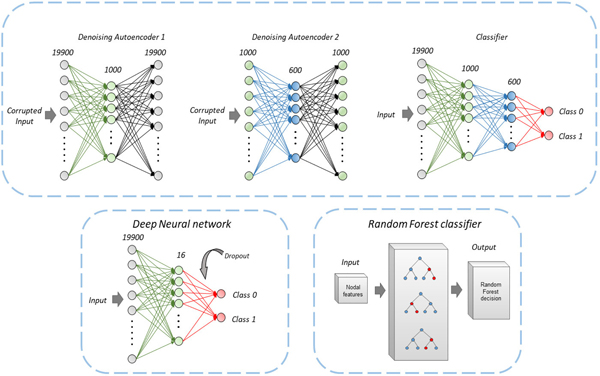
The ANN approach comprised of a shallow set of dense layers for the classification task. The architecture for the ANN model included a dense layer at the input. This dense layer consisted of 19900 neurons corresponding to the vectorized array of input from the connectivity matrices. A hidden layer with 16 neurons was employed to scale down the dimensionality and learn the classification encodings followed by a ReLU activation function. Batch normalization was used between layers and dropout of 0.5 was applied before the final layer [55]. The final output dense layer had 2 neurons corresponding to the autistic and typically developing class with a SoftMax activation function yielding probability values for every class. Training was performed with Adam optimizer using binary cross-entropy as the loss function. An illustration of the ANN architecture is provided in Fig. 1.
Fig. 1.
A schematic diagram of all the classification methods used. An artificial neural network (ANN) based classifier was implemented along with a Random forest (RF) of classification trees. Architecture for classification involving denoising autoencoders based on Heinsfeld et. al. has been shown.
2). Autoencoders
This network architecture involved denoising autoencoders to scale down the dimensionality of data from 19900 features to 1000 features using a first autoencoder setup. Input features were corrupted by adding noise with data corruption of 20% (binomial distribution: n=1, p=0.8). Dimensionality was further reduced from 1000 features to 600 features using a second encoder-decoder model with corruption of 30% this time (binomial distribution: n=1, p=0.7). The classifier comprised a model which takes its shape based on the first two auto-encoders with 19900 neurons at the input dense layer followed by 1000 and 600 neurons in intermediate hidden layers. Finally, a SoftMax based dense layer with two neurons was employed for the two classes. The weights of the classifier were initialized with the weights of autoencoders used for dimensionality reduction to utilize the knowledge extracted from the autoencoders. For details, please refer to Heinsfeld et al. 2019 [35]. An illustration of the auto-encoder architecture is provided in Fig. 1.
3). Random forests
The random forest (RF) classification involved an ensemble of classification trees for predictive modeling of the connectivity matrix [54], [56]. It uses ‘bagging’ (bootstrap-aggregating) and feature randomness when building each individual tree to try to create an uncorrelated ‘forest’ of trees whose combined prediction is more accurate than that of any individual tree.
The number of estimators was set to 100 trees with minimum samples per leaf equal to 1. The data samples were split based on the condition of minimum samples equal 100. Maximum of square root of the total features was chosen at a time for the input per estimator. Gini impurity which measures the likelihood of an incorrect classification of a new instance of a random variable was used as the loss function and bootstrapping was set to True to train the RF classifier. An illustration of the RF architecture is provided in Fig. 1.
G. Ablation analysis
Gaining deeper understanding about the discriminative connectivity sub-networks is crucial for clinical interpretability. To this end, we performed ablation analysis between 12 well-defined sub-networks (please refer to Table S1 in the supplement for ROI names, MNI coordinates and corresponding sub-networks). The sub-networks included sensory/somatomotor hand (15 ROIs), sensory/somatomotor mouth (3 ROIs), cingulo-opercular task control (12 ROIs), auditory (9 ROIs), default mode (41 ROIs), cingulo-parietal (3 ROIs), visual (20 ROIs), fronto-parietal task control (24 ROIs), salience (10 ROIs), subcortical (25 ROIs), ventral attention (8 ROIs), dorsal attention sub-networks (13 ROIs) and uncertain (17 ROIs; regions that did not belong to any other defined network). One of these network assignments were made to each of the 200 ROIs (the number of ROIs in each network is mentioned above). The networks, originally derived in Power et al. were adopted to the CC200 atlas by matching each CC200 ROI with an ROI from the Power264 atlas, and then assigning the network based on Power264’s network assignment [57]. The criteria for matching was the minimum Euclidean distance between the CC200 and Power264 ROIs. The assignments were further manually checked to ensure that the ROIs from the two atlases were visually and numerically close by.
For ablation analysis, the fMRI connectivity matrix was separated based on the sub-networks assigned to every node. A zero-valued mask was generated for each sub-network. Each mask was multiplied with the input array of 19900 elements separately to obtain a vector consisting of feature values belonging to all other subgroups except the one to be ablated. The resultant output was fed as input to the ANN based classifier for inferencing. Ablated sub-network inferencing was performed by occluding each sub-network at the input by multiplying with a zero-valued mask corresponding to the ablated sub-network. The accuracy with the ablated region was calculated and compared for a drop or increase with the baseline accuracy without ablation. The train-test split was based on LOSO where one site was tested with a trained model on rest of the 17 scanner sites. In ablation analysis, the sub-networks were ranked based on the drop-in accuracy for every site. Maximum drop indicated most significant sub-network for the classification. Frequency for every sub-network being the most significant was calculated.
III. RESULTS
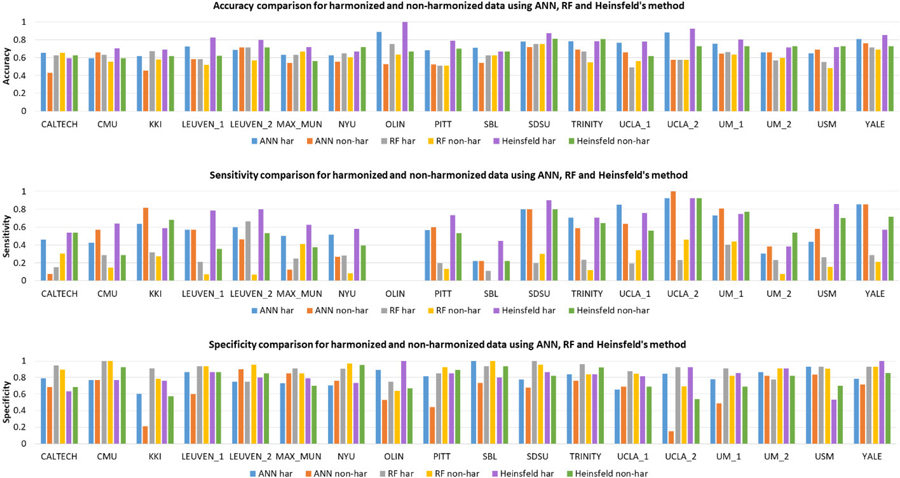
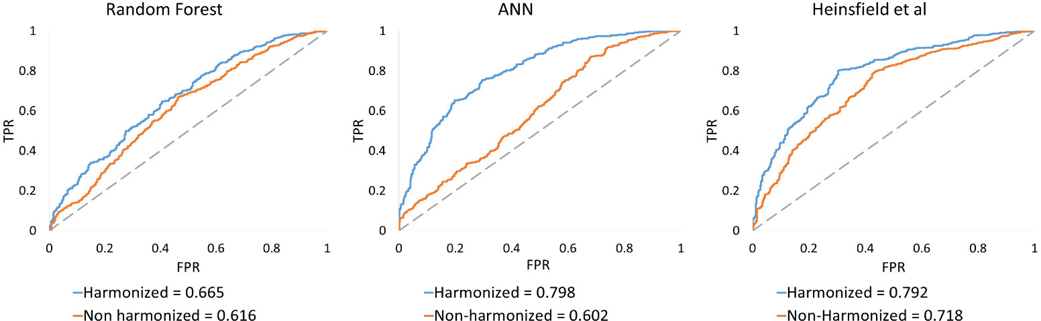
Table I shows the comparison of classification metrics obtained from harmonized and non-harmonized data for each of the methods employed for a 10-fold LOSO cross-validation procedure. While harmonization did not have much effect on accuracy obtained from RF, the accuracy improved with harmonized data for auto-encoders (~2.5%) and ANNs (4.5%). In fact, ANN with harmonized data provided highest accuracy of 71.35%. Fig. 2 illustrates the site-specific classification metrics obtained from harmonized and non-harmonized data for each of the methods employed. The general trend across sites replicates trends observed in the entire dataset shown in Table I, i.e. ANN and Heinsfeld methods perform better than RF and that performance metrics for harmonized data outperform those obtained from non-harmonized data at almost every single site. The area under receiver operator characteristic curve (AUROC) over the complete harmonized dataset and non-harmonized dataset is shown in Fig. 3. For the RF classifier, AUROC of harmonized data was 0.665 whereas AUROC of non-harmonized data was 0.616. Substantial increase in AUROC was observed for the ANN classifier where AUROC for harmonized data was 0.798 compared to AUROC for non-harmonized that was 0.602. Heinsfeld’s approach of using denoising autoencoders gave an AUROC of 0.792 for harmonized data and an AUROC of 0.718 for non-harmonized data.
TABLE I.
10-FOLD CROSS-VALIDATION PERFORMANCE OF RANDOM FOREST (RF), ARTIFICIAL NEURAL NETWORK (ANN) AND AUTO-ENCODER (FROM HEINSFELD ET AL.) USING HARMONIZED AND NON-HARMONIZED DATASETS
| RF | ANN | Heinsfeld et al | |
|---|---|---|---|
|
| |||
| Non-harmonized data | |||
| Accuracy (%) | 60.93 | 65.99 | 67.61 |
| Sensitivity | 0.247 | 0.593 | 0.787 |
| Specificity | 0.890 | 0.712 | 0.532 |
|
| |||
| Harmonized data | |||
| Accuracy (%) | 60.63 | 71.35 | 69.93 |
| Sensitivity | 0.245 | 0.595 | 0.750 |
| Specificity | 0.886 | 0.806 | 0.634 |
Fig. 2.
Bar chart showing the site-specific accuracy, sensitivity and specificity obtained from harmonized as well as non-harmonized data for the three methods (random forests, artificial neural networks and Heinsfeld’s auto-encoders) employed.
Fig. 3.
Comparison of Area under receiver-operating characteristic (AU-ROC) between harmonized and non-harmonized datasets for all the classification methods used.
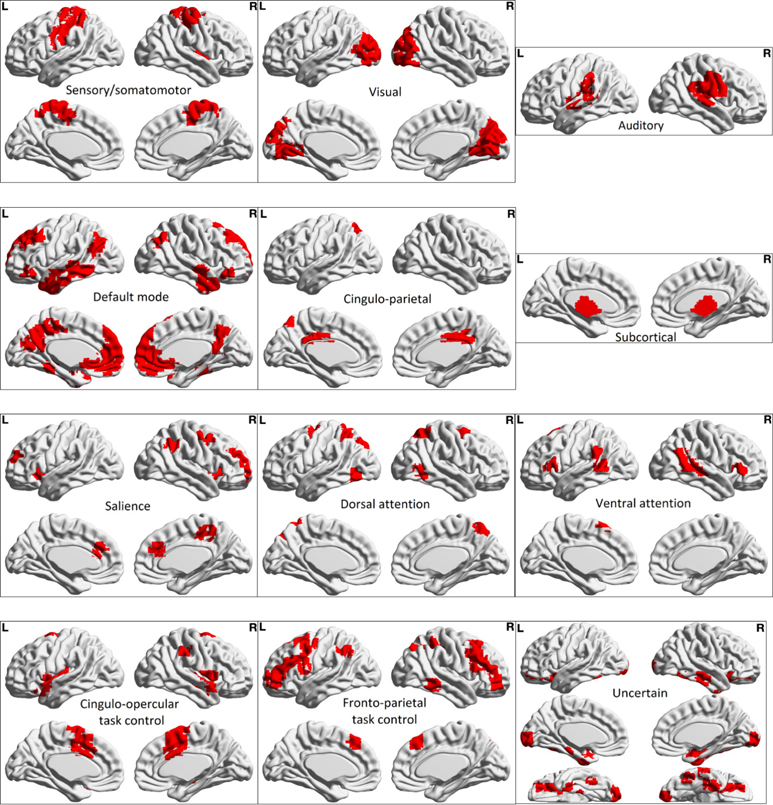
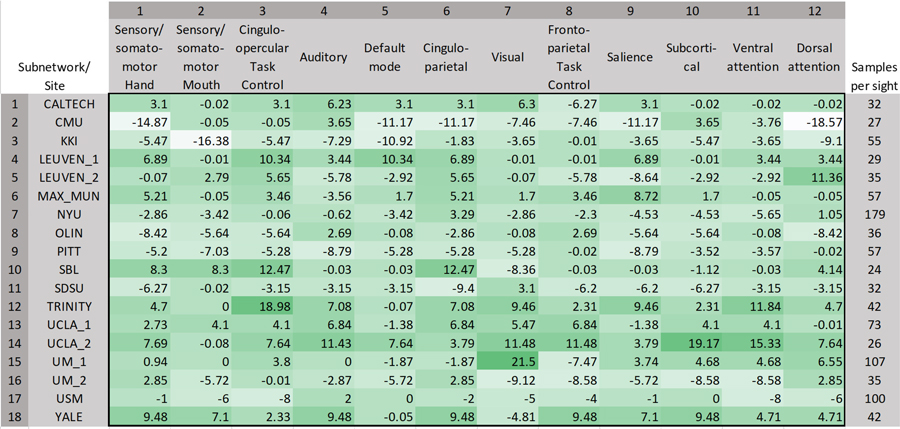
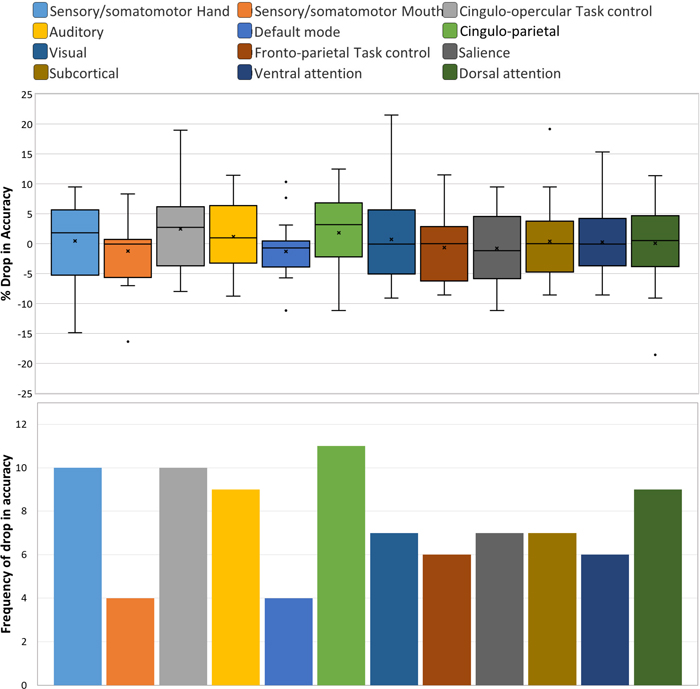
The brain regions representing the 12 sub-networks used in ablation analysis of harmonized data with the ANN classifier is shown in Fig. 4. The corresponding results are illustrated in Fig. 5, which shows the drop in accuracy when any of the given 12 sub-networks are occluded from the analysis. The drop in accuracy (indicated as positive values) is shown for individual sites in the LOSO framework. It can be seen that there is some variability across sites. Therefore, we assessed the frequency of drop in accuracy, i.e. the number of sites where in a drop in accuracy is observed, for occlusion of each of the sub-networks in ablation analysis (Fig. 6). The percentage drop in accuracy (the median and range is shown) across all sites when each of the sub-networks are occluded in the ablation analysis is also shown in Fig. 6. Both in terms of frequency and absolute percentage drop in accuracy, sensory/somatomotor (drop in 10 sites), auditory (drop in 9 sites), cingulo-opercular task control (drop in 10 sites) and cingulo-parietal (drop in 11 sites) networks stand out.
Fig. 4.
Brain maps showing ROIs associated with each of the 12 sub-networks used in the ablation analysis. Table S1 in the supplement provides further information about each sub-network, such as ROIs in each sub-network, their names and MNI centroids.
Fig. 5.
Results from the ablation analysis of harmonized data with the ANN classifier. The drop in accuracy due to occluding every sub-network can be observed per test site (LOSO). Positive values indicate a drop in accuracy due to ablation.
Fig. 6.
The percentage drop in accuracy (the median and range is shown) across all sites when each of the sub-networks are occluded in the ablation analysis (top). The frequency of drop in accuracy, i.e. the number of sites where in a drop in accuracy is observed, for occlusion of each of the sub-networks in ablation analysis (bottom).
Although the percentage drop in accuracy was not very high, the dorsal attention network showed a drop in 9 sites. T-test performed between baseline accuracies and accuracies obtained after ablation showed near-significant p-values for Cingulo-opercular task control sub-network (p = 0.07) and Cingulo-parietal network (p = 0.1).
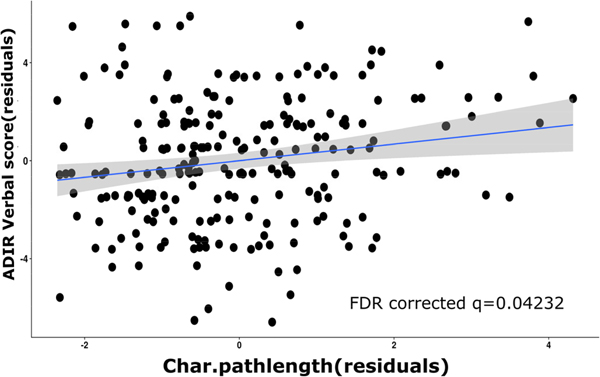
Only the characteristic path-length of the auditory sub-network showed a significant positive correlation with ADIR verbal scores in the Autism group alone (n=234) as shown in Fig. 7 (FDR-corrected q=0.04). Subsequently, sub-networks identified as important in the ablation analysis (sensory/somatomotor, auditory, cingulo-opercular task control and cingulo-parietal sub-networks) were combined and graph measures obtained from them and correlated with non-imaging measures controlling for gender and age in the Autism group. For this, Global Efficiency and ADIR-Verbal scores were found to have a negative correlation with a p-value of 0.01. However, this did not survive FDR correction for multiple comparisons (FDR-corrected q=0.08).
Fig. 7.
Correlation between characteristic path length obtained from the FC matrices of the auditory network in Autism subjects with the ADIR verbal scores in those subjects.
IV. DISCUSSION
With the advent of multi-site publicly available neuroimaging data repositories, there has been renewed interest in leveraging the larger sample size for predicting diagnostic status of subjects within a machine learning framework [58]. The larger sample size makes machine learning more feasible by alleviating the curse of dimensionality. However, many of these multi-site data are aggregated post-hoc, i.e. they were acquired from different scanners with different acquisition parameters. Consequently, inter-site variability may mask inter-group differences, and hence, the advantages gained by the larger sample size in the context of machine learning based diagnostic classification may not be realized. Our work focused on addressing this issue using harmonization of multi-site neuroimaging data to improve diagnostic classification accuracy. Specifically, we demonstrate this using ABIDE multi-site data for classifying individuals with Autism from healthy controls using resting state fMRI-based functional connectivity data. We demonstrate that consistently higher classification accuracies across multiple classification models can be obtained from multi-site data post harmonization with the ComBat technique as compared to without harmonization, outperforming earlier results from existing studies using ABIDE. Furthermore, our sub-network ablation analysis facilitated important insights into ASD pathology by determining the importance of each sub-network based on their drop in accuracy across all the sites.
Multi-scanner and multi-site studies such as ABIDE are significantly affected by inter-scanner variability. These variations usually arise from scanner hardware and calibration, magnetic homogeneity, acquisition parameters and reconstruction algorithms. These inter-site disparities can be detrimental to the study under consideration as these can limit the power to detect statistical differences and sometimes may also lead to erroneous findings. For functional MRI, previous work has illustrated existence of site differences that cannot be removed completely via ICA based techniques [59]. It is therefore crucial to employ harmonization techniques to remove the unsolicited site variations while retaining the underlying biological variability. Nonetheless, no studies on ABIDE till now have considered data harmonization on fMRI connectivity across sites before analyzing and interpreting ASD pathology.
We employed ComBat harmonization on ABIDE data, which earlier has been shown to eliminate site differences in functional connectivity while retaining the biological variability [53], [54]. Post harmonization, we employed standard classification models such as random forests and ANNs as well as the top performing model from earlier work by Heinsfeld et al. [35]. In all the classification models we observed that the overall accuracy increased significantly after harmonization (Table I). Validation was performed using leave one site out type analysis, where again we demonstrated that harmonized data could preserve and add power to detect subtle biological variations (17/18 sites using ANN, 14/18 using RF illustrated higher accuracies) in the absence of site-variations resulting in superior and robust classification between ASD from TD subjects which was also reflected in the AU-ROC analysis. Also, for the technique from Heinsfeld et al. which relied on de-noising auto-encoders for generalizability, harmonization was crucial for capturing sensitive variations (in 12 out of 18 sites (in 676 subjects)) and to boost the classification accuracy. Heinsfeld et al’s method used denoising auto-encoders which were able to eliminate noise from the connectivity matrices. The weight parameters used for denoising were further used to initialize the parameters for the classifier which resulted in better performance. The sites CALTECH and SBL match the sites where Heinsfeld et al. illustrated lower accuracies as well. The data acquired from these sites has high intra-site variability that is not present in data from other sites, a probable reason why harmonization did not help [35]. Novel AI-based data harmonization has illustrated superior results in general, however at the same time it is yet to be demonstrated in the context of neuroimaging-based classification (and specifically ABIDE) wherein we have high dimensionality and relatively small sample size [60].
To understand the neural patterns that were highly discriminative between ASD and TDs and contributed to the classification, we performed an ablation analysis where each sub-network was removed, and the ANN classifier was employed to find the drop in the accuracy. The results of this analysis (Figs 5 and 6) highlight the role of a few sub-networks namely sensory/somatomotor, auditory, cingulo-opercular task control and cingulo-parietal networks whose absolute drop in accuracy was generally higher than other networks. In addition, a drop in accuracy upon their ablation was found in 9–11 of the 18 sites. Thus, the most discerning brain regions which contributed to the classification were known. We could precisely quantify the contribution of every subnetwork based on the drop in accuracy.
Heightened sensitivity to sensory stimuli is a behavioral hallmark of Autism. Therefore, it is not surprising that neural abnormalities in sensory processing and alterations in sensory networks including somatomotor and auditory network has been observed before [61], [62]. Specifically, with respect to the auditory network, we found that Autistic subjects with higher characteristic path length also had a higher ADIR-Verbal score (Fig. 7). A larger value of characteristic path length indicates lower efficiency in the network and hence it is not surprising that it is associated with greater impairment in verbal communication. This also lends credence to the notion that verbal communication difficulties in Autism may be linked to impairments in auditory processing of sensory stimuli [63].
Cingulo-opercular and cingulo-parietal networks are basically control networks that are involved in a variety of executive functions including attention, salience, social cognition and communication [64]–[66]. Autism involves deficits in these domains and therefore, it is not surprising that we found these networks to be important for discrimination. In fact, our results are supported by previous studies that also found alterations in resting state networks anchored in the cingulate to be critical for identifying subjects with Autism [67]–[71].
V. CONCLUSION
In summary, our work illustrates the importance of site-harmonization in analysis of benchmark datasets such as ABIDE. Our classification using simple neural network models facilitated superior accuracy on harmonized data compared complex models that have been previously proposed. Ablation analysis was crucial for delineating the most discriminative sub-networks that were directly linked to the clinical markers of Autism.
Supplementary Material
Contributor Information
Madhura Ingalhalikar, Symbiosis Center for Medical Image Analysis, Symbiosis International University, Pune, India..
Sumeet Shinde, Symbiosis Center for Medical Image Analysis, Symbiosis International University, Pune, India..
Arnav Karmarkar, Symbiosis Center for Medical Image Analysis, Symbiosis International University, Pune, India..
Archith Rajan, Symbiosis Center for Medical Image Analysis, Symbiosis International University, Pune, India..
D. Rangaprakash, Athinoula A. Martinos Center for Biomedical Imaging, Massachusetts General Hospital, Charlestown, Massachusetts, USA Department of Radiology, Harvard Medical School, Boston, Massachusetts, USA; Division of Health Sciences and Technology, Harvard University and Massachusetts Institute of Technology, Cambridge, Massachusetts, USA..
Gopikrishna Deshpande, AU MRI Research Center, Department of Electrical and Computer Engineering, Auburn University, Auburn, Alabama, USA.; Department of Psychological Sciences, Auburn University, Auburn, Alabama, USA Alabama Advanced Imaging Consortium, University of Alabama Birmingham, Alabama, USA; Center for Neuroscience, Auburn University, Auburn, Alabama, USA; School of Psychology, Capital Normal University, Beijing, China; Key Laboratory for Learning and Cognition, Capital Normal University, Beijing, China; Department of Psychiatry, National Institute of Mental Health and Neurosciences, Bangalore, India..
REFERENCES
- [1].Association, A.P., Diagnostic and statistical manual of mental disorders (DSM-5®). 2013: American Psychiatric Pub. [DOI] [PubMed] [Google Scholar]
- [2].Eaves LC, et al. , Screening for autism spectrum disorders with the social communication questionnaire. Journal of Developmental & Behavioral Pediatrics, 2006. 27(2): p. S95–S103. [DOI] [PubMed] [Google Scholar]
- [3].Aggarwal S and Angus B, Misdiagnosis versus missed diagnosis: diagnosing autism spectrum disorder in adolescents. Australas Psychiatry, 2015. 23(2): p. 120–3. [DOI] [PubMed] [Google Scholar]
- [4].Beljan P, et al. , Misdiagnosis and Dual Diagnoses of Gifted Children and Adults: ADHD, Bipolar, OCD, Asperger’s, Depression, and Other Disorders. Gifted and Talented International, 2006. 21(2): p. 83–86. [Google Scholar]
- [5].Retico A, et al. , Neuroimaging-based methods for autism identification: a possible translational application? Functional neurology, 2014. 29(4): p. 231. [PMC free article] [PubMed] [Google Scholar]
- [6].Brambilla P, et al. , Brain anatomy and development in autism: review of structural MRI studies. Brain Res Bull, 2003. 61(6): p. 557–69. [DOI] [PubMed] [Google Scholar]
- [7].McAlonan GM, et al. , Mapping the brain in autism. A voxel-based MRI study of volumetric differences and intercorrelations in autism. Brain, 2005. 128(Pt 2): p. 268–76. [DOI] [PubMed] [Google Scholar]
- [8].Waiter GD, et al. , A voxel-based investigation of brain structure in male adolescents with autistic spectrum disorder. NeuroImage, 2004. 22(2): p. 619–625. [DOI] [PubMed] [Google Scholar]
- [9].Hazlett HC, et al. , Cortical gray and white brain tissue volume in adolescents and adults with autism. Biological psychiatry, 2006. 59(1): p. 1–6. [DOI] [PubMed] [Google Scholar]
- [10].Stigler KA, et al. , Structural and functional magnetic resonance imaging of autism spectrum disorders. Brain research, 2011. 1380: p. 146–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].von dem Hagen EAH, et al. , Reduced functional connectivity within and between ‘social’ resting state networks in autism spectrum conditions. Social cognitive and affective neuroscience, 2013. 8(6): p. 694–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Aoki Y, Cortese S, and Tansella M, Neural bases of atypical emotional face processing in autism: A meta-analysis of fMRI studies. World J Biol Psychiatry, 2015. 16(5): p. 291–300. [DOI] [PubMed] [Google Scholar]
- [13].Pagnozzi AM, et al. , A systematic review of structural MRI biomarkers in autism spectrum disorder: A machine learning perspective. International Journal of Developmental Neuroscience, 2018. 71: p. 68–82. [DOI] [PubMed] [Google Scholar]
- [14].Hyde KK, et al. , Applications of supervised machine learning in autism spectrum disorder research: a review. Review Journal of Autism and Developmental Disorders, 2019. 6(2): p. 128–146. [Google Scholar]
- [15].Ingalhalikar M, et al. , Diffusion based abnormality markers of pathology: toward learned diagnostic prediction of ASD. Neuroimage, 2011. 57(3): p. 918–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Libero LE, et al. , Multimodal neuroimaging based classification of autism spectrum disorder using anatomical, neurochemical, and white matter correlates. Cortex, 2015. 66: p. 46–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Barik K, et al. Classification of Autism in Young Children by Phase Angle Clustering in Magnetoencephalogram Signals. in 2020 National Conference on Communications (NCC). 2020. IEEE. [Google Scholar]
- [18].Ingalhalikar M, et al. , Creating multimodal predictors using missing data: Classifying and subtyping autism spectrum disorder. Journal of neuroscience methods, 2014. 235: p. 1–9. [DOI] [PubMed] [Google Scholar]
- [19].Deshpande G, et al. , Identification of neural connectivity signatures of autism using machine learning. Frontiers in human neuroscience, 2013. 7: p. 670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Hull JV, et al. , Resting-state functional connectivity in autism spectrum disorders: A review. Frontiers in psychiatry, 2017. 7: p. 205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Chaitra N, Vijaya P, and Deshpande G, Diagnostic prediction of autism spectrum disorder using complex network measures in a machine learning framework. Biomedical Signal Processing and Control, 2020. 62: p. 102099. [Google Scholar]
- [22].Syed MA, et al. , Investigating brain connectomic alterations in autism using the reproducibility of independent components derived from resting state functional MRI data. Frontiers in Neuroscience, 2017. 11: p. 459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Nielsen JA, et al. , Multisite functional connectivity MRI classification of autism: ABIDE results. Frontiers in human neuroscience, 2013. 7: p. 599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Plitt M, Barnes KA, and Martin A, Functional connectivity classification of autism identifies highly predictive brain features but falls short of biomarker standards. NeuroImage: Clinical, 2015. 7: p. 359–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Abraham A, et al. , Deriving reproducible biomarkers from multi-site resting-state data: An Autism-based example. NeuroImage, 2017. 147: p. 736–745. [DOI] [PubMed] [Google Scholar]
- [26].Lanka P, et al. , MALINI (Machine Learning in NeuroImaging): A MATLAB toolbox for aiding clinical diagnostics using resting-state fMRI data. Data in Brief, 2020. 29: p. 105213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Lanka P, et al. , Supervised machine learning for diagnostic classification from large-scale neuroimaging datasets. Brain Imaging Behav, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Zhao X, et al. , Investigating the correspondence of clinical diagnostic grouping with underlying neurobiological and phenotypic clusters using unsupervised machine learning. Frontiers in applied mathematics and statistics, 2018. 4: p. 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Zhao X, et al. , Identifying neuropsychiatric disorders using unsupervised clustering methods: data and code. Data in brief, 2019. 22: p. 570–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Parikh MN, Li H, and He L, Enhancing diagnosis of autism with optimized machine learning models and personal characteristic data. Frontiers in computational neuroscience, 2019. 13: p. 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Kassraian-Fard P, et al. , Promises, pitfalls, and basic guidelines for applying machine learning classifiers to psychiatric imaging data, with autism as an example. Frontiers in psychiatry, 2016. 7: p. 177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Li H, Parikh NA, and He L, A novel transfer learning approach to enhance deep neural network classification of brain functional connectomes. Frontiers in neuroscience, 2018. 12: p. 491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Khosla M, et al. , 3D convolutional neural networks for classification of functional connectomes, in Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support. 2018, Springer. p. 137–145. [Google Scholar]
- [34].Parisot S, et al. , Disease Prediction using Graph Convolutional Networks: Application to Autism Spectrum Disorder and Alzheimer’s Disease. Medical Image Analysis, 2018. 48. [DOI] [PubMed] [Google Scholar]
- [35].Heinsfeld AS, et al. , Identification of autism spectrum disorder using deep learning and the ABIDE dataset. NeuroImage. Clinical, 2017. 17: p. 16–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Grech-Sollars M, et al. , Multi-centre reproducibility of diffusion MRI parameters for clinical sequences in the brain. NMR in Biomedicine, 2015. 28(4): p. 468–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Friedman L, et al. , Test–retest and between-site reliability in a multicenter fMRI study. Human brain mapping, 2008. 29(8): p. 958–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Fortin J-P, et al. , Harmonization of cortical thickness measurements across scanners and sites. Neuroimage, 2018. 167: p. 104–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Fortin J-P, et al. , Harmonization of multi-site diffusion tensor imaging data. NeuroImage, 2017. 161: p. 149–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Di Martino A, et al. , The autism brain imaging data exchange: towards a large-scale evaluation of the intrinsic brain architecture in autism. Molecular Psychiatry, 2014. 19(6): p. 659–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Di Martino A, et al. , Enhancing studies of the connectome in autism using the autism brain imaging data exchange II. Scientific data, 2017. 4(1): p. 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Chao-Gan Y and Yu-Feng Z, DPARSF: A MATLAB Toolbox for “Pipeline” Data Analysis of Resting-State fMRI. Front Syst Neurosci, 2010. 4: p. 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Penny WD, et al. , Statistical parametric mapping: the analysis of functional brain images. 2011: Elsevier. [Google Scholar]
- [44].Wu G-R, et al. , A blind deconvolution approach to recover effective connectivity brain networks from resting state fMRI data. Medical image analysis, 2013. 17(3): p. 365–374. [DOI] [PubMed] [Google Scholar]
- [45].Sreenivasan KR, Havlicek M, and Deshpande G, Nonparametric hemodynamic deconvolution of FMRI using homomorphic filtering. IEEE transactions on medical imaging, 2014. 34(5): p. 1155–1163. [DOI] [PubMed] [Google Scholar]
- [46].Rangaprakash D, et al. , Hemodynamic response function (HRF) variability confounds resting-state fMRI functional connectivity. Magnetic resonance in medicine, 2018. 80(4): p. 1697–1713. [DOI] [PubMed] [Google Scholar]
- [47].Rangaprakash D, et al. , Parameterized hemodynamic response function data of healthy individuals obtained from resting-state functional MRI in a 7T MRI scanner. Data in brief, 2018. 17: p. 1175–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Rangaprakash D, et al. , Hemodynamic variability in soldiers with trauma: implications for functional MRI connectivity studies. NeuroImage: Clinical, 2017. 16: p. 409–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Yan W, Rangaprakash D, and Deshpande G, Hemodynamic Response Function Parameters Obtained from Resting State BOLD fMRI. 2018. [DOI] [PMC free article] [PubMed]
- [50].Yan W, Rangaprakash D, and Deshpande G, Aberrant hemodynamic responses in autism: implications for resting state fMRI functional connectivity studies. NeuroImage: Clinical, 2018. 19: p. 320–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Craddock RC, et al. , A whole brain fMRI atlas generated via spatially constrained spectral clustering. Hum Brain Mapp, 2012. 33(8): p. 1914–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Johnson WE, Li C, and Rabinovic A, Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics, 2006. 8(1): p. 118–127. [DOI] [PubMed] [Google Scholar]
- [53].Yu M, et al. , Statistical harmonization corrects site effects in functional connectivity measurements from multi-site fMRI data. Hum Brain Mapp, 2018. 39(11): p. 4213–4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Breiman L, Random forests. Machine learning, 2001. 45(1): p. 5–32. [Google Scholar]
- [55].Xu B, Wang N, Chen T and Li M, 2015. Empirical evaluation of rectified activations in convolutional network. arXiv preprint arXiv:1505.00853. [Google Scholar]
- [56].Fredo J, et al. , Diagnostic Classification of Autism using Resting-State fMRI Data and Conditional Random Forest. Conference proceedings: Annual International Conference of the IEEE Engineering in Medicine and Biology Society. IEEE Engineering in Medicine and Biology Society. Conference, 2018. 2018: p. 1148–1151. [Google Scholar]
- [57].Power JD, et al. , Functional network organization of the human brain. Neuron, 2011. 72(4): p. 665–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Chaitra N, Vijaya PA, Deshpande G, “Diagnostic Prediction of Autism Spectrum Disorder Using Complex Network Measures in a Machine Learning Framework”, Biomedical Signal Processing and Control, vol. 62, pp. 102099, 2020. [Google Scholar]
- [59].Feis RA, et al. , ICA-based artifact removal diminishes scan site differences in multi-center resting-state fMRI. Frontiers in Neuroscience, 2015. 9(395). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].O’Sullivan S, Heinsen H, Grinberg LT et al. The role of artificial intelligence and machine learning in harmonization of high-resolution post-mortem MRI (virtopsy) with respect to brain microstructure. Brain Inf. 6, 3 (2019). 10.1186/s40708-019-0096-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Kozhemiako N, et al. , Alterations in Local Connectivity and Their Developmental Trajectories in Autism Spectrum Disorder: Does Being Female Matter? Cereb Cortex, 2020. 30(9): p. 5166–5179. [DOI] [PubMed] [Google Scholar]
- [62].Aggarwal P and Gupta A, Multivariate graph learning for detecting aberrant connectivity of dynamic brain networks in autism. Med Image Anal, 2019. 56: p. 11–25. [DOI] [PubMed] [Google Scholar]
- [63].Demopoulos C and Lewine JD, Audiometric Profiles in Autism Spectrum Disorders: Does Subclinical Hearing Loss Impact Communication? Autism Res, 2016. 9(1): p. 107–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Gratton C, Sun H, and Petersen SE, Control networks and hubs. Psychophysiology, 2018. 55(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Sestieri C, et al. , Domain-general signals in the cingulo-opercular network for visuospatial attention and episodic memory. J Cogn Neurosci, 2014. 26(3): p. 551–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Apps M, Lockwood P, and Balsters J, The role of the midcingulate cortex in monitoring others’ decisions. Frontiers in Neuroscience, 2013. 7(251). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Agastinose Ronicko J.F., et al. , Diagnostic classification of autism using resting-state fMRI data improves with full correlation functional brain connectivity compared to partial correlation. J Neurosci Methods, 2020. 345: p. 108884. [DOI] [PubMed] [Google Scholar]
- [68].Guo X, et al. , Diagnosing Autism Spectrum Disorder from Brain Resting-State Functional Connectivity Patterns Using a Deep Neural Network with a Novel Feature Selection Method. Front Neurosci, 2017. 11: p. 460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Zhou Y, et al. , Functional Connectivity of the Caudal Anterior Cingulate Cortex Is Decreased in Autism. PLoS One, 2016. 11(3): p. e0151879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Keown CL, et al. , Network organization is globally atypical in autism: A graph theory study of intrinsic functional connectivity. Biol Psychiatry Cogn Neurosci Neuroimaging, 2017. 2(1): p. 66–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Chen H, et al. , Multivariate classification of autism spectrum disorder using frequency-specific resting-state functional connectivity--A multi-center study. Prog Neuropsychopharmacol Biol Psychiatry, 2016. 64: p. 1–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.