Abstract
The purpose of this study was to assess the usefulness of real-time automated PCR as a quantitative, highly reproducible, and sensitive method to detect cytomegalovirus (CMV) DNA in blood specimens. Intra- and interassay precision rates were 0.89% (small number of copies [L]), 1.43% (middle number of copies [M]), and 1.12% (high number of copies [H]), and 4.46% (L), 1.51% (M), and 2.28% (H), respectively. The linearity of this assay was obtained between 10 and 107 copies/well, with a minimum detection limit of 20 copies/well. Specimens from 55 of 70 healthy subjects were found to be positive for CMV antibody, but CMV DNA was not detected in any of them. In the qualitative assessment of each specimen, the results of the CMV antigenemia assay and those of the real-time PCR assay agreed in 80% (plasma specimens), 79% (all nucleated cells), and 86% (blood) of the cases examined. For eight patients diagnosed as having CMV infection or disease, no sample was positive in the antigenemia assay earlier than in the real-time PCR assay. Furthermore, the results of this assay could be obtained within 8 h. We concluded that the real-time PCR assay is useful for rapid diagnosis of CMV infection and monitoring of clinical courses.
Cytomegalovirus (CMV) disease is one of the fatal complications after bone marrow transplantation (BMT). Monitoring of its reactivation and preemptive or prophylactic treatment using ganciclovir are critical for BMT recipients. However, because of the myelotoxicity of ganciclovir and its prolongation of periods of neutropenia, the prognosis in patients at low risk of developing CMV disease is not necessarily improved. Identification of patients at high risk of developing CMV is therefore believed to be important in management of BMT recipients.
To date, surveillance bronchoscopy (9), surveillance blood culture (4), and a CMV antigenemia assay (3) have been used to identify patients at high risk of developing CMV disease; among them, the CMV antigenemia assay is, at present, widely used to monitor BMT recipients. This method aims to detect pp65 antigen expressed in CMV-infected all nucleated cells (ANC) using a monoclonal antibody (14). Because of its high sensitivity for CMV and the ease of blood sampling, it is quite useful and convenient for monitoring BMT recipients. However, this method has some problems. First, this assay shows false-negative results because of low-level expression of the pp65 antigen of white blood cells in a small number of patients with definite CMV disease (10). Second, it is difficult to perform the antigenemia assay before engraftment because of small leukocyte numbers.
Recently, a PCR method using CMV-specific primers has been used to diagnose CMV reactivation early after BMT. In addition to its high sensitivity and specificity for detecting CMV (11), this test is highly advantageous in that it is not influenced by the white blood cell count in peripheral blood or by pp65 antigen expression. PCR is definitely a useful diagnostic method for detecting CMV reactivation, but it may frequently be too sensitive for clinical use. That is, when the results of CMV PCR are positive, they do not necessarily indicate an imminent risk of CMV disease and the results obtained are frequently overestimates. In the case of PCR after transplantation, the results should be evaluated quantitatively. We have previously reported a semiquantitative PCR method to detect CMV (7), but it was not quantitative enough to detect small differences in viral burden.
Real-time automated PCR is a quantitative assay reported by Heid et al. in 1996 (5). This method measures PCR product accumulation by means of a dual-labeled fluorogenic probe (i.e., TaqMan probe), and it provides a very accurate and reproducible measure of gene copies (5). In this study, we evaluated the usefulness of this exonuclease-based PCR assay as an alternative to the conventional PCR to detect CMV reactivation after BMT.
MATERIALS AND METHODS
Extraction of viral DNA.
Two hundred microliters of whole blood or plasma was mixed with 20 μl of protease and 200 μl of lysis buffer, and the mixture was heated at 56°C for 10 min and then processed using a QIAamp Blood mini-kit (Qiagen, Valencia, Calif.). The DNA absorbed to the QIAamp spin column was eluted with 50 μl of distilled water and then subjected to PCR. ANC (including polymorphonuclear and mononuclear cells) were obtained by centrifugation of whole blood at 1,000 × g for 5 min, and red blood cells were destroyed with hypotonic solution (0.2% NaCl). The pellet of nucleated cells was suspended in 900 μl of DNAzol (Life Technology, Grand Island, N.Y.), mixed homogeneously by pipetting. DNA was recovered by adding 450 μl of ethanol (Wako Pure Chemicals, Osaka, Japan), followed by centrifugation at 17,360 × g for 5 min. After being washed twice with ethanol, the sedimented DNA was dissolved in 100 μl of distilled water.
Antibody detection.
Serum samples were tested for anti-CMV immunoglobulin G (IgG) using commercial indirect enzyme immunoassays. Sera were tested by means of the Enzygnost anti-CMV IgG kit (Dade Behring Inc., Marburg, Germany) according to the manufacturer's recommendations and using the Behring enzyme-linked immunosorbent assay Processor III (Dade Behring Inc.).
Amplification and detection of viral DNA.
The sequences of the PCR primers and that of the probe were selected from the US17 region of CMV AD169 (1). The forward and reverse primers were 5′-GCGTGCTTTTTAGCCTCTGCA-3′ and 5′-AAAAGTTTGTGCCCCAACGGTA-3′, respectively. The TaqMan probe selected between both primers was fluorescence labeled with 6-carboxyfluorescein at the 5′ end as the reporter dye and 6-carboxytetramethylrhodamine at the 3′ end as the quencher (FAM-TgATCggCgTTATCgCgTTCTTgATC-TAMRA). The primers and the TaqMan probe were purchased from Greiner Japan (Tokyo, Japan). The PCR product was detected as an increase in fluorescence using ABI PRISM 7700 (PE Biosystems, Foster City, Calif.). The PCR was performed using TaqMan Universal PCR master mix (PE Biosystems). Five hundred nanograms of DNA from whole blood or ANC or the DNA extracted from 100 μl of plasma was mixed with 25 μl of PCR master mix, 15 pmol of each of the primers, and 10 pmol of the TaqMan probe, and then distilled water was added to a total volume of 50 μl. The concentration of the extracted DNA was quantified by spectrophotometric measurement at a wavelength of 260 nm using a U-2100 spectrophotometer (Hitachi, Tokyo, Japan). The PCR was performed in 96-well microtiter plates under the following conditions: 1 cycle at 50°C for 2 min and 95°C for 10 min and 50 cycles at 95°C for 15 s and 61°C for 1 min. DNA from CMV AD169 (obtained from the American Type Culture Collection, Rockville, Md.) was amplified using both primers and inserted into the pCR2.1 TA cloning vector (Invitrogen, Carlsbad, Calif.). This plasmid was used as a CMV standard in this study. CMV was quantified using serially diluted CMV standard within the range of 10 to 107 copies/well, and the number of CMV copies was calculated using Sequence Detection System version 1.6.3. software (PE Biosystems). Fluorescence intensity was measured in each well every 7 s, and the result was deemed to be positive when the fluorescence intensity exceeded 10 times the baseline fluorescence. The threshold cycle (Ct value) was defined as the PCR cycle number at that point. The normalized reporter (Rn) value was calculated by dividing the emission intensity of the reporter dye by the emission intensity of the reference dye included in the TaqMan Universal PCR master mix, which does not participate in the exonuclease-based PCR assay. ΔRn is the difference between the Rn+ value and the Rn− value, where Rn+ is the Rn value of a reaction containing all components including the template and Rn− is the Rn value obtained from early cycles prior to a detectable increase in fluorescence during real-time PCR.
In vitro examination of sensitivity and specificity of real-time automated PCR.
To determine the sensitivity of the method, standard DNA at a concentration of 100 copies/μl was serially diluted with a solution of salmon sperm DNA at a concentration of 200 μg/ml. The diluted DNA samples were measured in quintuplicate. For specificity, DNAs from human herpesviruses (Epstein-Barr virus P3HR-1, herpes simplex virus type 1 HF, herpes simplex virus type 2 UW-268, and varicella-zoster virus H-S1; all virus strains were obtained from the National Institute of Infectious Diseases, Tokyo, Japan) other than CMV were subjected to PCR.
Patients and hospitalization conditions.
Sixteen consecutive patients who underwent BMT in our institution were enrolled in this study. All of the patients gave their written informed consent. The characteristics of the patients and their hospitalization conditions are shown in Table 1. All of the patients were CMV seropositive before transplantation.
TABLE 1.
Patient characteristicsa
| Patient no. | Age (yr) | Sex | Disease | Status | Treatment | GVHD prophylaxis | Stem cell source | Acute GVHD grade | Outcome | Cause of death |
|---|---|---|---|---|---|---|---|---|---|---|
| 82 | 20 | F | ALL | First complete remission | VP-16/CY/TBI | sMTX and CsA | HLA identical sibling | II | Alive | |
| 84 | 31 | M | NHL | Second relapse | VP-16/CY/TBI | sMTX and CsA | MUD | I | Alive | |
| 85 | 44 | F | CML | Accelerated phase | CA/BU/CY | sMTX and CsA | HLA identical sibling | I | Dead | Aspergillus tracheobronchitis |
| 87 | 46 | M | CML | First chronic phase | CY/TBI | sMTX and CsA | HLA identical sibling | 0 | Alive | |
| 89 | 23 | F | ANLL | First relapse | CA/CY/TBI | sMTX and CsA | MUD | 0 | Alive | |
| 90 | 48 | M | ANLL | Induction failure | CA/CY/TBI | sMTX and CsA | HLA identical sibling | I | Alive | |
| 91 | 47 | F | CML | First chronic phase | BU/CY | sMTX and CsA | HLA identical sibling | I | Alive | |
| 93 | 28 | M | ALL | Third relapse | VP-16/CY/TBI | sMTX and FK506 and T-cell depletion | Two-locus mismatch sibling | III | Dead | Acute GVHD |
| 95 | 33 | M | ANLL | First relapse | CA/CY/TBI | sMTX and CsA | HLA identical sibling | I | Alive | |
| 96 | 34 | M | ALL | First complete remission | VP-16/CY/TBI | sMTX and CsA | MUD | I | Alive | |
| 97 | 20 | F | ALL | Second relapse | CY/TBI | sMTX and CsA | Two-locus mismatch unrelated donor | 0 | Alive | |
| 98 | 23 | F | ALL | Second relapse | VP-16/CY/TBI | sMTX and CsA | One-locus mismatch unrelated donor | 0 | Alive | |
| 99 | 18 | M | ALL | Second relapse | VP-16/CY/TBI | sMTX and CsA | HLA identical sibling | 0 | Alive | |
| 101 | 50 | F | CML | Accelerated phase | CY/TBI | sMTX and CsA | MUD | I | Alive | |
| 102 | 40 | M | ANLL | Induction failure | BU/CY | sMTX and CsA | HLA identical sibling | 0 | Alive |
ALL, acute lymphoblastic leukemia; NHL, non-Hodgkin's lymphoma; CML, chronic myelocytic leukemia; ANLL, acute non-lymphoblastic leukemia; VP-16, etoposide; CY, cyclophosphamide; TBI, total body irradiation; BU, busulfan; CA, cytosine arabinoside; sMTX, short-term methotrexate; CsA, cyclosporine A; MUD, matched unrelated donor.
We administered acyclovir at a dosage of 750 mg/day intravenously from days −5 to 28. However, CMV-specific immunoglobulin was not prophylactically administered, and all of the patients received blood transfusions using products that had not been tested for CMV antibody.
Criteria for diagnosis of CMV infection and disease and management of CMV infection.
A patient was considered to be infected with CMV when CMV was detected in the blood by shell-vial culture and/or by the CMV antigenemia assay, and a patient was considered to have CMV disease when CMV was demonstrated in biopsy specimens by immunohistochemical analysis and this was accompanied by clinical signs and symptoms. For diagnosis of CMV retinitis, histopathological identification of CMV was not necessarily required; the criterion was retinal opacity with a perivascular distribution and associated hemorrhage.
We started the patient on ganciclovir either when more than 10 cells were found to be positive by the antigenemia assay in the case of transplantation from related donors or when a single cell was found to be positive in the case of transplantation from unrelated donors.
Blood collection and examination of CMV infection.
In this study, 136 and 70 blood samples were drawn from BMT recipients and healthy volunteers, respectively. All of the samples were treated with EDTA-ACD anticoagulant. The samples drawn from BMT recipients once a week from the time of admission until either discharge or death were subjected to real-time automated PCR and were also tested by the CMV antigenemia assay and by shell-vial culture.
The CMV antigenemia assay was started after engraftment. The CMV antigenemia assay was performed using the monoclonal antibody against the pp65 antigen of CMV according to the method described by Tanabe et al. (14). Briefly, 1.5 × 105 peripheral blood leukocytes were attached to slides using a cytocentrifuge and fixed with formaldehyde. The cells were sequentially incubated with Clonab CMV monoclonal antibody C10/11 (Biotest), reacted with goat anti-mouse IgG antibody coupled to alkaline phosphatase (DAKO Japan), and stained using new fuchsin as the substrate. A total of 105 cells were analyzed per sample, counterstained with hematoxylin, and the results were expressed as the number of positive cells per 50,000 leukocytes.
Shell-vial culture was performed as previously described. CMV was isolated from blood by rapid immunoperoxidase staining for the major immediate-early protein in infected human embryonic fibroblasts using a previously described technique (13).
Statistical analysis.
Statistical analysis was performed using Pearson's correlation coefficient, and a P value of <0.05 was accepted as statistically significant.
RESULTS
Sensitivity of PCR.
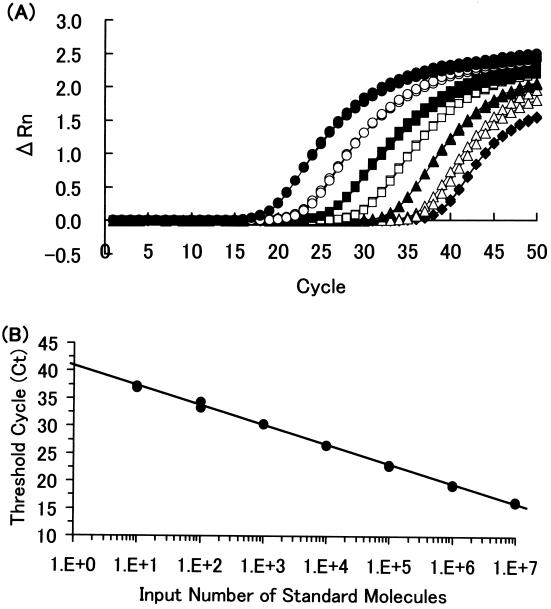
Ct values of the serially diluted standard were plotted on the y axis, and copy numbers were plotted on the x axis on a logarithmic scale. The plot showed good linearity (Fig. 1). One hundred copies of CMV standard DNA per μl were serially diluted with salmon sperm DNA (200 μg/ml). Each diluted standard sample was measured in quintuplicate. The coefficient of variation (CV) values were less than 5% from 20 to 100 copies/well (1.0 to 3.1%) and 7.1% at 10 copies/well. Therefore, the sensitivity limit of this assay was defined as 20 copies/well.
FIG. 1.
(A) Amplification profile of standards for CMV real-time PCR. Standard DNA at 107 (solid circles), 106 (open circles), 105 (solid squares), 104 (open squares), 103 (solid triangles), 102 (open triangles), and 10 (solid diamonds) copies/well in duplicate was amplified for 50 cycles. (B) Standard curve for CMV real-time PCR. Ct values were plotted against various numbers of copies of standard DNA. The slope was −3.54 cycles/log decade. The correlation coefficient was 0.988.
Reproducibility of PCR.
The CV values obtained for control low (L, 100 copies/well), middle (M, 10,000 copies/well), and high (H, 106 copies/well) viral load samples in the test of intra-assay variation were 0.89 ± 0.28, 1.43 ± 0.37, and 1.12 ± 0.21 (values are means ± standard deviations), respectively, with 10 replicates each, and in the test of interassay variation the CV values were 4.46 ± 1.43, 1.51 ± 0.40, and 2.28 ± 0.44, respectively, with measurement in quintuplicate.
Specificity of PCR.
To confirm the specificity of this assay, several virus strains other than CMV were tested. No cross-reactivity between Epstein-Barr virus, herpes simplex virus, varicella-zoster virus, and CMV was observed (data not shown).
CMV antibody and CMV copy number in normal subjects.
Although specimens from 55 of 70 volunteers had CMV antibodies, CMV DNA was not detectable in any of them using this assay (data not shown).
Patient characteristics.
Sixteen patients who received BMT between September 1998 and February 1999 were examined. There were nine males and seven females, with a median age of 34.9 years (range, 18 to 54). T-cell depletion was performed in one case (UPN93). The patients' characteristics are shown in Table 1. Acute graft versus host disease (GVHD) grading was done as previously described (8).
Analysis of each patient.
Seven patients were diagnosed as having CMV infection, and one patient (UPN84) was diagnosed as having CMV disease (CMV retinitis). CMV retinitis responded to systemic and intraocular administration of ganciclovir. No sign of CMV disease was seen in any other organ in this patient. Table 2 shows the results of laboratory tests for each patient. Samples from 12 (80%), 10 (67%), 9 (60%), 8 (53%), and 1 (7%) patient became positive more than once in the ANC PCR assay, plasma PCR assay, blood PCR assay, antigenemia assay, and shell-vial culture, respectively.
TABLE 2.
Results of real-time automated PCR, antigenemia assay, and shell-vial culturea
| Patient no. | Status | Shell-vial culture result | Maximum no. of CMV DNA copies detected (day post-BMT)
|
Antigenemia | ||
|---|---|---|---|---|---|---|
| ANC PCR | Plasma PCR | Blood PCR | ||||
| 82 | Control | Negative | 22 (102) | NDb | ND | ND |
| 84 | Disease | Negative | 865 (94) | 133 (94) | 739 (94) | 4 (94) |
| 85 | Infection | Positive | 2,998 (88) | 10,604 (95) | 633 (88) | 3 (95) |
| 87 | Control | Negative | 57 (87) | ND | ND | ND |
| 89 | Control | Negative | ND | 32 (69) | ND | ND |
| 90 | Infection | Negative | 1,097 (47) | 246 (47) | 577 (47) | 4 (47) |
| 91 | Control | Negative | 33 (53) | 111 (53) | ND | ND |
| 93 | Infection | Negative | 3,795 (17) | 4,137 (10) | 1,978 (17) | 10 (31) |
| 95 | Infection | Negative | 1,129 (137) | 78 (137) | 255 (130) | 1 (137) |
| 96 | Infection | Negative | 20 (85) | ND | 41 (32) | 1 (32) |
| 97 | Control | Negative | ND | ND | ND | ND |
| 98 | Control | Negative | ND | ND | ND | ND |
| 99 | Infection | Negative | 1,939 (46) | 508 (53) | 652 (46) | 4 (53) |
| 101 | Infection | Negative | 355 (31) | 87 (31) | 161 (24) | 2 (31) |
| 102 | Control | Negative | ND | ND | ND | ND |
| 103 | Control | Negative | ND | ND | ND | —c |
Day post-BMT, the day posttransplantation when the first positive result was obtained. Results are no. of copies per 500 ng of DNA (ANC and blood PCR), per 100 μl of plasma (plasma PCR), or per 50,000 cells (antigenemia).
ND, not detected.
—, CMV antigenemia assay not performed because the patient died of cardiac tamponade before engraftment.
For seven patients, all of the assays except shell-vial culture became positive during their clinical course. The day on which each test became positive is shown in Table 2. Among these seven patients, none was positive in the antigenemia assay earlier than in the real-time PCR assay. There were time lags of 5.4, 4.7, and 3.0 days on average between the occurrence of positive findings in the antigenemia assay and blood, ANC, and plasma PCR, respectively.
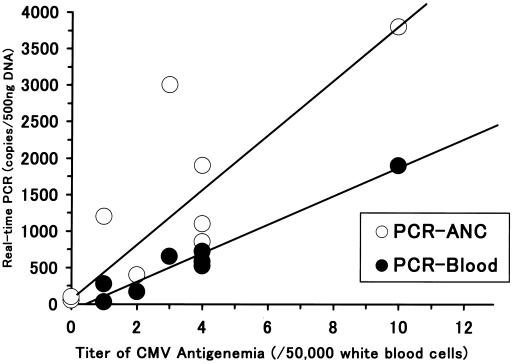
In Fig. 2, we show the association between the maximum level of CMV antigenemia and the maximum CMV DNA level as determined by real-time PCR for each patient. Samples with DNA levels below 20 copies were excluded from this analysis. A statistically significant correlation was observed between the CMV antigenemia assay and ANC PCR (correlation coefficient, 0.840; P = 0.0006), between the CMV antigenemia assay and blood PCR (correlation coefficient, 0.983; P < 0.0001), between ANC PCR and blood PCR (correlation coefficient, 0.838, P = 0.0066), and between ANC PCR and plasma PCR (correlation coefficient, 0.728; P = 0.0235). On the other hand, there was no significant correlation between the antigenemia assay and plasma PCR (correlation coefficient, 0.339; P = 0.3507) or between blood PCR and plasma PCR (correlation coefficient, 0.351; P = 0.4123).
FIG. 2.
Association between maximum level of CMV antigenemia and maximum level of CMV DNA as determined by real-time PCR for each patient. There was a statistically significant correlation between the CMV antigenemia assay and ANC PCR (correlation coefficient, 0.873; P < 0.001) and between the CMV antigenemia assay and blood PCR.
Analysis of each specimen.
Among the 55 samples obtained before transplantation, only one specimen became positive in one of the five different assays (ANC PCR, blood PCR, plasma PCR, CMV antigenemia assay, and shell-vial culture). The sample obtained from patient UPN93 became positive by plasma PCR (22 copies).
We collected 136 samples after transplantation, of which 83 samples (61%) became negative in all of the assays. Of 136 specimens, 46, 40, 31, 19, and 1 sample were positive in ANC PCR, blood PCR, plasma PCR, the CMV antigenemia assay, and shell-vial culture, respectively. Relative sensitivities, specificities, and predictive values for detecting CMV DNA and CMV antigen in blood were calculated for all four assays. These methods of analysis were compared to that of Boeckh et al. (2). Concordant results were observed in 80% of sample pairs between plasma PCR and the antigenemia assay, 79% of sample pairs between ANC PCR and the antigenemia assay, 86% of sample pairs between blood PCR and the antigenemia assay, 84% of sample pairs between plasma PCR and ANC PCR, 89% of sample pairs between ANC PCR and blood PCR, and 85% of sample pairs between plasma PCR and blood PCR.
Typical clinical course.
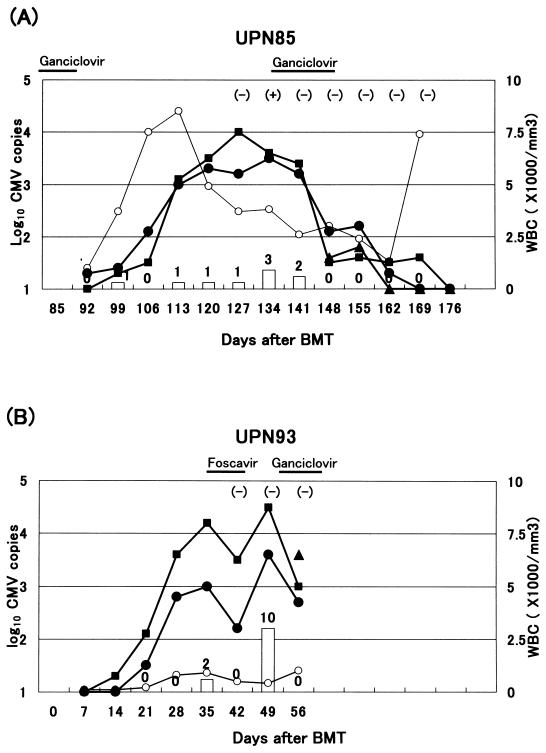
The clinical courses of patients UPN85 and UPN93 are shown in Fig. 3. Patient UPN85 became positive in the shell-vial culture of peripheral blood. This patient also developed Aspergillus tracheobronchitis concomitantly under long-term steroid therapy for the treatment of acute GVHD and died on day 176. Patient UPN93 died of grade III acute GVHD on day 60. Both patients were administered ganciclovir starting on the day they became positive for CMV in the antigenemia assay.
FIG. 3.
Clinical course of patients UPN85 and UPN93. (A) Patient UPN85 became positive in the shell-vial culture of peripheral blood. She also developed Aspergillus tracheobronchitis concomitantly under long-term steroid therapy for the treatment of acute GVHD and died of it on day 176. + and −, positive and negative in shell-vial culture, respectively. Open circles, white blood cells (WBC); solid circles, ANC; solid squares, plasma; solid triangles, whole blood. Bars and numbers show the number of CMV detected by the antigenemia assay. (B) Patient UPN93 died of grade III acute GVHD on day 60. Both patients responded well to ganciclovir. Results of real-time PCR for CMV accurately reflect their responses to the anti-CMV treatment.
DISCUSSION
We have established a new method of diagnosis of CMV infection using real-time automated PCR. This is a modification of conventional PCR, and it possesses the advantages of this assay. Although real-time automated PCR might be less sensitive in detection of CMV DNA than conventional PCR, we could detect as few as 20 copies of CMV DNA using this method. However, it should be noted that none of eight diagnosed as having CMV infection or disease were positive in the antigenemia assay earlier than in the real-time PCR assay. There was a time lag of 5.4, 4.7, or 3.0 days, on average, between the occurrence of positive findings in the antigenemia assay and blood, ANC, or plasma PCR, respectively. Using real-time automated PCR, it was possible to detect CMV infection at a very early stage compared with conventional detection methods. In addition, real-time automated PCR proved to be more time-saving than the antigenemia assay.
PCR is a very useful method for early diagnosis of CMV disease, but it may frequently be too sensitive for clinical use. That is, CMV-positive results obtained by PCR do not necessarily indicate an imminent risk of developing CMV disease, and these results are frequently overestimates. However, we overcame this problem by applying real-time automated PCR to the diagnosis of CMV infection. This method has some advantages over the conventional PCR assay.
First, using real-time automated PCR, it is possible to determine the number of copies of viral DNA that are present quite accurately. In this study, we found a good positive correlation between the number of copies of CMV DNA present and the cycles at which the amplification curves became steep using samples containing from 20 to 107 copies of CMV DNA. In this range, we could accurately determine the number of copies of viral DNA present in the samples. In addition, the maximal levels observed in ANC and blood PCR were significantly correlated with those in the antigenemia assay (Fig. 2). Figure 3 shows the clinical course of patients at high risk of developing CMV disease. Both patients were administered anti-CMV drugs because they became positive in the antigenemia assay. After the start of treatment with these drugs, the antigenemia assay and real-time automated PCR both showed a rapid decrease in the number of viral copies. We suppose that real-time automated PCR reflected the effect of ganciclovir more sensitively than the antigenemia assay. PCR is frequently too sensitive, but we succeeded in overcoming this shortcoming by using real-time automated PCR, which may prove useful to evaluate BMT recipients with CMV infection who are receiving ganciclovir therapy.
Second, the results of real-time automated PCR were highly reproducible. In our study, the intra-assay variation was 0.89, 1.43, and 1.12% for samples containing 102, 104, and 106 copies, respectively. The interassay variation was 4.46, 1.51, and 2.28%, respectively. We consider that these variations will be negligible in clinical use. Furthermore, although PCR involves the potential risk of DNA contamination, this seemed to be minimal in this study, since the control samples employed in every assay were always negative. Our real-time automated PCR seemed to be stable.
In this study, we compared three sources of DNA, whole blood, ANC, and plasma. Previous studies have shown that detection of CMV by PCR can be applied to blood, ANC, plasma (12), and serum (6), but we do not know which is the best source for the diagnosis of CMV disease by PCR. In this study, we found a good positive correlation between the results of the antigenemia assay and blood or ANC PCR, and ANC PCR was most sensitive in detecting the CMV DNA, which was comparable to that in the previous report (2). In contrast, the results of plasma PCR did not seem to be correlated with those of the antigenemia assay. We suppose that plasma PCR may be less sensitive in detecting of CMV infection because CMV infects leukocytes and a small number of cell-free CMV usually exist in peripheral blood. Since Woo et al. reported that the results of leukocyte PCR became positive prior to those of the plasma CMV DNA assay (15), it is possible that plasma PCR becomes positive for patients with advanced CMV infection, and this method may be less sensitive than the other PCR methods. However, a positive result by plasma PCR indicates the presence of cell-free CMV, which may be a predictor of subsequent CMV disease. At present, we cannot conclude which is the best type of specimen for detecting CMV infection by PCR.
Although we believe that the real-time PCR assay is promising, this study has some limitations to be discussed. First, this was a pilot study involving only a limited number of patients. Definite CMV disease was identified in only one patient, and we have not obtained enough data on the application of real-time automated PCR in the case of patients with definite CMV disease. Second, if it were possible to establish a threshold for this test between patients with CMV infection and those who are at a high risk of developing CMV disease, the threshold would be an important indicator of the time to initiate anti-CMV treatment, and this should be clarified in further studies.
In conclusion, the use of a quantitative PCR assay in the management of BMT recipients may serve to further define the population at high risk of developing CMV disease and lead to a method for long-term preemptive therapy.
REFERENCES
- 1.Bankier A T, Beck S, Bohni R, Brown C M, Cerny R, Chee M S, Hutchison C A D, Kouzarides T, Martignetti J A, Preddie E. The DNA sequence of the human cytomegalovirus genome. DNA Sequences. 1991;2:1–12. doi: 10.3109/10425179109008433. [DOI] [PubMed] [Google Scholar]
- 2.Boeckh M, Gallez-Hawkins G M, Myerson D, Zaia J A, Bowden R A. Plasma polymerase chain reaction for cytomegalovirus DNA after allogeneic marrow transplantation: comparison with polymerase chain reaction using peripheral blood leukocytes, pp65 antigenemia, and viral culture. Transplantation. 1997;64:108–113. doi: 10.1097/00007890-199707150-00020. [DOI] [PubMed] [Google Scholar]
- 3.Boeckh M, Gooley T A, Myerson D, Cunningham T, Schoch G, Bowden R A. Cytomegalovirus pp65 antigenemia-guided early treatment with ganciclovir versus ganciclovir at engraftment after allogeneic marrow transplantation: a randomized double-blind study. Blood. 1996;88:4063–4071. [PubMed] [Google Scholar]
- 4.Goodrich J M, Mori M, Gleaves C A, Du Mond C, Cays M, Ebeling D F, Buhles W C, DeArmond B, Meyers J D. Early treatment with ganciclovir to prevent cytomegalovirus disease after allogeneic bone marrow transplantation. N Engl J Med. 1991;325:1601–1607. doi: 10.1056/NEJM199112053252303. [DOI] [PubMed] [Google Scholar]
- 5.Heid C A, Stevens J, Livak K J, Williams P M. Real time quantitative PCR. Genome Res. 1996;6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- 6.Ishigaki S, Takeda M, Kura T, Ban N, Saitoh T, Sakamaki S, Watanabe N, Kohgo Y, Niitsu Y. Cytomegalovirus DNA in the sera of patients with cytomegalovirus pneumonia. Br J Haematol. 1991;79:198–204. doi: 10.1111/j.1365-2141.1991.tb04522.x. [DOI] [PubMed] [Google Scholar]
- 7.Kanda Y, Chiba S, Suzuki T, Kami M, Yazaki Y, Hirai H. Time course analysis of semi-quantitative PCR and antigenaemia assay for prevention of cytomegalovirus disease after bone marrow transplantation. Br J Haematol. 1998;100:222–225. doi: 10.1046/j.1365-2141.1998.00518.x. [DOI] [PubMed] [Google Scholar]
- 8.Przepiorka D, Weisdorf D, Martin P, Klingemann H G, Beatty P, Hows J, Thomas E D. 1994 consensus conference on acute GVHD grading. Bone Marrow Transplant. 1995;15:825–828. [PubMed] [Google Scholar]
- 9.Schmidt G M, Horak D A, Niland J C, Duncan S R, Forman S J, Zaia J A. A randomized, controlled trial of prophylactic ganciclovir for cytomegalovirus pulmonary infection in recipients of allogeneic bone marrow transplants; the City of Hope-Stanford-Syntex CMV Study Group. N Engl J Med. 1991;324:1005–1011. doi: 10.1056/NEJM199104113241501. [DOI] [PubMed] [Google Scholar]
- 10.Seropian S, Ferguson D, Salloum E, Cooper D, Landry M L. Lack of reactivity to CMV pp65 antigenemia testing in a patient with CMV disease following allogeneic bone marrow transplant. Bone Marrow Transplant. 1998;22:507–509. doi: 10.1038/sj.bmt.1701354. [DOI] [PubMed] [Google Scholar]
- 11.Shibata D, Martin W J, Appleman M D, Causey D M, Leedom J M, Arnheim N. Detection of cytomegalovirus DNA in peripheral blood of patients infected with human immunodeficiency virus. J Infect Dis. 1988;158:1185–1192. doi: 10.1093/infdis/158.6.1185. [DOI] [PubMed] [Google Scholar]
- 12.Spector S A, Merrill R, Wolf D, Dankner W M. Detection of human cytomegalovirus in plasma of AIDS patients during acute visceral disease by DNA amplification. J Clin Microbiol. 1992;30:2359–2365. doi: 10.1128/jcm.30.9.2359-2365.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Swenson P D, Kaplan M H. Rapid detection of cytomegalovirus in cell culture by indirect immunoperoxidase staining with monoclonal antibody to an early nuclear antigen. J Clin Microbiol. 1985;21:669–673. doi: 10.1128/jcm.21.5.669-673.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tanabe K, Tokumoto T, Ishikawa N, Koyama I, Takahashi K, Fuchinoue S, Kawai T, Koga S, Yagisawa T, Toma H, Ota K, Nakajima H. Comparative study of cytomegalovirus (CMV) antigenemia assay, polymerase chain reaction, serology, and shell vial assay in the early diagnosis and monitoring of CMV infection after renal transplantation. Transplantation. 1997;64:1721–1725. doi: 10.1097/00007890-199712270-00016. [DOI] [PubMed] [Google Scholar]
- 15.Woo P C, Lo S K, Yuen K Y, Peiris J S, Siau H, Chiu E K, Liang R H, Chan T K. Detection of CMV DNA in bone marrow transplant recipients: plasma versus leucocyte polymerase chain reaction. J Clin Pathol. 1997;50:231–235. doi: 10.1136/jcp.50.3.231. [DOI] [PMC free article] [PubMed] [Google Scholar]