Abstract
Patient‐derived cancer cells (PDCs) and patient‐derived xenografts (PDXs) are often used as tumor models, but have many shortcomings. PDCs not only lack diversity in terms of cell type, spatial organization, and microenvironment but also have adverse effects in stem cell cultures, whereas PDX are expensive with a low transplantation success rate and require a long culture time. In recent years, advances in three‐dimensional (3D) organoid culture technology have led to the development of novel physiological systems that model the tissues of origin more precisely than traditional culture methods. Patient‐derived cancer organoids bridge the conventional gaps in PDC and PDX models and closely reflect the pathophysiological features of natural tumorigenesis and metastasis, and have led to new patient‐specific drug screening techniques, development of individualized treatment regimens, and discovery of prognostic biomarkers and mechanisms of resistance. Synergistic combinations of cancer organoids with other technologies, for example, organ‐on‐a‐chip, 3D bio‐printing, and CRISPR‐Cas9‐mediated homology‐independent organoid transgenesis, and with treatments, such as immunotherapy, have been useful in overcoming their limitations and led to the development of more suitable model systems that recapitulate the complex stroma of cancer, inter‐organ and intra‐organ communications, and potentially multiorgan metastasis. In this review, we discuss various methods for the creation of organ‐specific cancer organoids and summarize organ‐specific advances and applications, synergistic technologies, and treatments as well as current limitations and future prospects for cancer organoids. Further advances will bring this novel 3D organoid culture technique closer to clinical practice in the future.
Keywords: cancer organoids, drug screening, personalized medicine, prognostic biomarker, organ‐on‐a‐chip, 3D bio‐printing, tumor microenvironment, cancer stroma
We discuss various methods for creation of organ‐specific cancer organoids and summarize organ‐specific advances and applications, synergistic technologies, and treatments as well as current limitations and future prospects for cancer organoids. Further advances will bring this novel 3D culture organoid technique closer to clinical practice in the future.
Abbreviations
- 3D
three‐dimensional
- ALI
air‐liquid interface
- CRC
colorectal cancer
- CRISPR‐HOT
CRISPR‐Cas9‐mediated homology‐independent organoid transgenesis
- CSCs
cancer stem cells
- DACH1
Dachshund homologue 1
- ECM
extracellular matrix
- ERBB3
Erb‐b2 receptor tyrosine kinase 3
- HCC
hepatocellular carcinoma
- MEK1/2
mitogen‐activated protein kinase kinase
- HNSCC
head and neck squamous cell carcinoma
- hPSCs
human pluripotent stem cells
- iPSCs
induced pluripotent stem cells
- NSCLC
non‐small cell lung cancer
- PD‐1
programmed cell death protein 1
- PDA
pancreatic ductal adenocarcinoma
- PDCs
patient‐derived cancer cells
- PD‐L1
programmed death‐ligand 1
- PDXs
patient‐derived xenografts
- PDOs
patient‐derived organoids
- SCCs
somatic stem cells
- TME
tumor microenvironment
- Wnt
Wingless‐related integration site
1. BACKGROUND
Patient‐derived cancer cells (PDCs) and patient‐derived xenografts (PDXs) are often used as tumor models, but have many shortcomings [1, 2]. PDCs not only lack diversity in terms of cell type, spatial organization, and microenvironment but also have adverse effects in stem cell cultures, whereas PDXs are expensive with a low transplantation success rate and require a long culture time [3, 4].
In recent years, a three‐dimensional (3D) cell culture technique known as “organoid” has been developed [5]. An organoid is a multi‐cell mass constructed from a 3D culture in vitro [3, 6] and can be generated from embryonic stem cells, induced pluripotent stem cells (iPSCs), and somatic stem cells (SSCs). With the capabilities of self‐renewal and self‐proliferation, while maintaining the physiological structure and function of their source tissues, organoids are useful for studying tumor models and stem cell biology [7, 8, 9]. Patient‐derived organoids bridge the conventional gaps in PDC and PDX models and have potential in clinical cancer research (Figure 1), particularly modeling of cancer [10, 11, 12], individualized therapy [13, 14], tumor drug screening [15, 16], tumor immunotherapy [17, 18], and translational medicine [19, 20]. Organoids have now been established for a wide range of tumor types [21, 22]. In this review, we elaborate on how the different types of cancer organoids were developed and discuss their applications, limitations, and future prospects.
FIGURE 1.
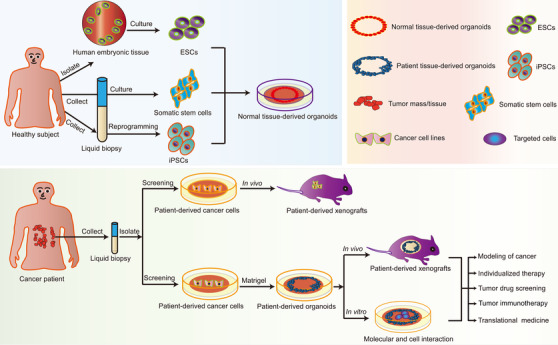
Summary of the procedures used to establish normal tissue and cancer organoids. Induced pluripotent stem cells, somatic stem cells, and embryonic stem cells can be used to establish normal tissue‐derived organoids. Patient‐derived cancer cells can be used to establish in vivo xenografts or can be propagated on an enriched Matrigel matrix and cultured into three‐dimensional tumor organoids that have in vivo and in vitro applications. Abbreviations: ESCs, embryonic stem cells; iPSCs, induced pluripotent stem cells
2. EVOLUTION AND ESTABLISHMENT OF ORGANOIDS
In 1907, Wilson [23] demonstrated that separated sponge cells could reassemble and self‐organize into a complete organism. Lindberg et al. [24] and Pellegrini et al. [25] then cultured limbal stem cells on 3T3 trophoblast cells and transplanted them into damaged eyes, which marked the start of the development of 3D organoid technology. A search on PubMed for the years 2000‐2020 using the terms “organoid” and “organoid and tumor/cancer” yielded 6864 items and 3006 items, respectively. Progress in organoid research was slow from 2000 to 2010 (Supplementary Figure S1). In 2009, Sato et al. [26] demonstrated that a single leucine‐rich repeat containing G protein‐coupled receptor 5 stem cells without a mesenchymal background could be used to construct small intestinal crypt‐villus structures in vitro. They used a novel Matrigel culture system that was able to replace the extracellular matrix (ECM) and contained growth factors, including epidermal growth factor, Noggin, R‐spondin 1, and Wingless‐related integration site (Wnt). The 3D mouse crypt structures were established in this culture system for 8 months [26]. Subsequently, in 2010, Unbekandt and Davies [27] used mouse embryonic stem cells to produce kidney organoids. In 2013, Lancaster et al. [28] showed that a brain organoid could be generated by growing human pluripotent stem cells (hPSCs) in Matrigel. Between 2012 and 2013, there was a rapid increase in the amount of published researches on general organoids and cancer organoids (from 84 items to 151 items) and cancer‐specific organoids (from 34 items to 66 items) (Supplementary Figure S1). After 2013, the number of organoid studies continued to increase year by year. By 2020, there were 2086 published items related to general organoids and cancer organoids, 907 (approximately 43%) of which were in the field of cancer organoids.
The 3D cancer organoids could serve as an important tool for unlocking the secrets of cancer. Given their ability to recapitulate the genotype, phenotype, and cellular features of their parent tissues, they can be used in the study of various tumors, including liver [29], breast [30], lung [31], and pancreatic cancers [32]. Standard tissue organoids for use in physiological studies can be derived from hPSCs or SSCs; hPSCs can be derived from embryonic stem cells or iPSCs, whereas SSCs are generally derived by the separation and purification of tissue samples after surgery [33, 34, 35]. A cancer/tumor organoid, also known as a “cancer surrogate,” uses the patient's tumor tissue for in vitro 3D cultures to simulate the biological characteristics of the tumor. However, organoids cultured from precancerous lesions are mainly used to simulate the occurrence and development of tumors and to analyze changes in tumor‐related omics. Oncolytic virotherapy is currently being considered as a potential treatment for cancer [36, 37]. Oncolytic viruses preferentially replicate in tumor cells; therefore, cytotoxicity can be enhanced by introducing target genes. Zhu et al. [38] found that ZIKV (a type of oncolytic virus) inhibited glioblastoma stem cells in organoids and showed promise as a treatment for glioblastoma. Furthermore, Raimondi et al. [39] found that pancreatic cancer patient‐derived organoids (PDOs) were suitable for preclinical exploration of oncolytic viruses. A tumor organoid encompasses the characteristics of the tissue of origin and retains the heterogeneity of individual cancers; thus, it could potentially be applied in precision medicine and translational medicine [14, 40, 41, 42].
3. APPLICATIONS OF ORGANOIDS IN VARIOUS TYPES OF CANCER
The lack of an in vitro tumor model that can mimic the heterogeneity of human cancers hinders our understanding of the pathogenesis of cancer and the efficacy and toxicity of treatment [43]. Three‐dimensional organoid culture models have shown considerable potential in the modeling of human cancers [44, 45, 46, 47]. A tumor organoid grows more slowly than its counterpart from normal epithelial organoid in living tissue, possibly because of the failure of mitosis and subsequent cell death [7, 48]. Therefore, pure tumor cell samples and selective culture conditions are required to avoid excessive growth of epithelioids and preserve normal tissues. Establishment of an organoid culture requires Matrigel or basement membrane extract as a substitute for ECM as well as a particular culture medium. The various components of the types of culture medium used for several different tumor organoids are listed in Table 1. The potential applications of tumor‐derived organoids in tumor modeling, drug screening, precision medicine, tumor immunotherapy, and gene profiling are presented in Figure 2.
TABLE 1.
Culture systems of different cancer organoid models
| Tumor type | Extracellular matrix | Culture media | Inhibitors | Ref |
|---|---|---|---|---|
| Colorectal cancer | Matrigel | DMEM/F12, Glutamax, HEPES, Primocin, recombinant human EGF, A83‐01, N‐acetylcysteine, recombinant human Noggin, Noggin‐conditioned media, R‐spondin‐1‐conditioned media | Y‐27632 dihydrochloride kinase inhibitor | [49] |
| Lung cancer | Matrigel | MBM, DMEM/F12, DNase, collagenase/dispase, penicillin/streptomycin, streptomycin, amphotericin B, bFGF, human EGF, N2, B27 | ROCK inhibitor | [67] |
| Pancreatic cancer | Matrigel | Wnt3a‐conditioned medium, B‐27, N‐acetyl‐L‐cysteine, nicotinamide, human EGF, human FGF10, prostaglandin E2, gastrin, R‐spondin, Noggin | A83‐01 | [79] |
| Breast cancer | Basement membrane extract | ADMEM/F12, penicillin/streptomycin, GlutaMAX, HEPES, B‐27, N‐acetylcysteine, R‐spondin‐1, FGF7, FGF10, nicotinamide, Noggin, primocin, and neuregulin 1 | A83‐01, Y‐27632 | [30] |
| Liver cancer | Basement membrane extract |
Classical human liver organoid isolation medium: ADMEM/F12, penicillin/streptomycin, GlutaMAX, HEPES, B‐27, N2, N‐acetyl‐1‐cysteine, nicotinamide, gastrin 1, EGF, FGF10, HGF, forskolin, R‐spondin‐1, Wnt3A, and Noggin Tumoroid‐specific isolation medium: Classical human liver organoid isolation medium with the elimination of R‐spondin‐1, Wnt3A, and Noggin as well as the addition of dexamethasone |
A83‐01, Y‐27632 | [29, 104] |
| Ovarian cancer | Matrigel | DMEM/F12, human EGF, R‐spondin1, Noggin, Jagged‐1, L‐glutamine solution, penicillin/streptomycin, amphotericin B | Y‐27632 | [108] |
| Bladder cancer | Matrigel | DMEM/F12, FGF10, FGF7, FGF2, B‐27, A83‐01, N‐acetyl‐1‐cysteine, nicotinamide | Y‐27632 | [115] |
| Prostate cancer | Matrigel | DMEM/F12, B‐27, Y‐27632‐HCl, nicotinamide, Rspondin, N‐acetyl‐cysteine, SB202190, Noggin, A83‐01, DHT, Wnt3a, HGF, EGF, FGF10, FGF2, PGE2 | Y‐27632 | [119, 121] |
| HNSCC | Matrigel | DMEM/F12, B‐27, BME type 2, CHIR‐99021, Forskolin, GlutaMAX, HEPES, N‐acetyl‐1‐cysteine, nicotinamide, Noggin‐Fc fusion protein‐conditioned medium, PGE2, hEGF, hFGF‐10, hFGF‐2 |
Y‐27632, A83‐01 |
[123, 124] |
| Gastric cancer | Matrigel | DMEM/F12, GlutaMAX, HEPES, B‐27, N‐acetylcysteine, human EGF, hFGF‐10, noggin‐conditioned medium, R‐spondin‐1‐conditioned medium, Wnt‐conditioned medium, gastrin, nicotinamide, IGF, PGE2 |
A83‐01, Y‐27632, SB202190, CHIR99021 |
[126] |
| Glioblastoma | Matrigel | DMEM/F12, GlutaMAX, hibernate A, antibiotic antimycotic, neurobasal medium, MEM‐NEAAs, N2, B‐27, human insulin solution | Y‐27632 | [129] |
DMEM/F12: Dulbecco's modified Eagle medium/F12; HEPES: N’‐a‐hydroxythylpiperazine‐N’‐ethanesulfanic acid; Wnt, Wingless‐related integration site; B‐27, a type of serum‐free supplement; BME, basement membrane extract; DHT, dihydrotestosterone; A‐83‐01: transforming growth factor; RHOK: Rho‐associated coiled coil forming protein serine/threonine kinase; SB202190: p38 inhibitor; CHIR99021: glykogen synthase kinase 3b inhibitor; IGF: insulin‐like growth factor; PGE: prostaglandin E; EGF: epidermal growth factor; FGF: fibroblast growth factor.
FIGURE 2.
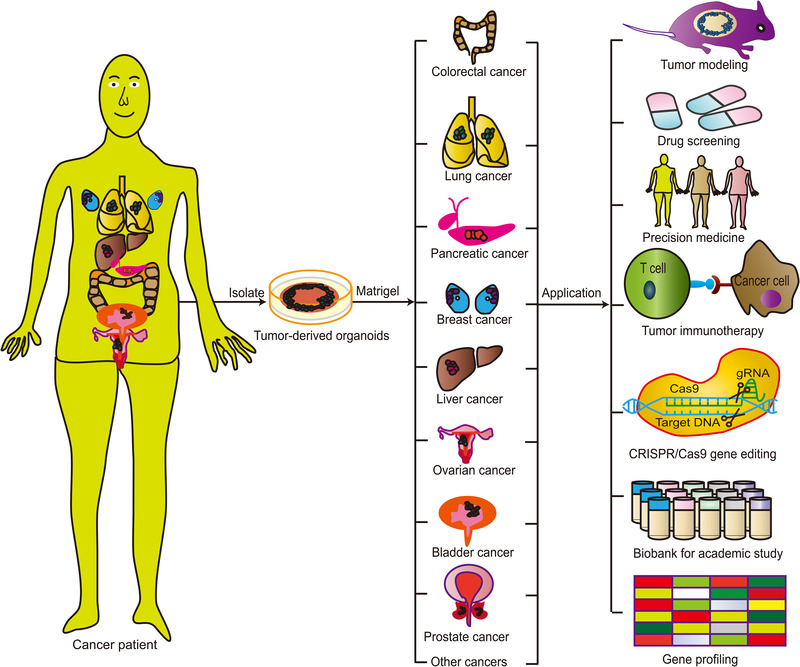
Various applications of tumor‐derived organoids in tumor modeling, drug screening, precision medicine, tumor immunotherapy, and gene profiling. Organoid technology can be used to model a variety of human cancers, to test drug efficacy and toxicity in precision medicine, and to develop novel targeted therapeutics. Furthermore, organoid biobanks can be used for academic studies and gene profiling of various cancers. Moreover, synergistic application with CRISPR/Cas9 gene editing can be used to further elucidate the pathophysiology of various cancers and to study the effects of specific genetic changes on tissue function and the development of disease
3.1. Colorectal cancer
Several research groups have successfully established colorectal cancer (CRC) organoids [49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59]. Patient‐derived CRC organoids have been used to find associations between stem cell markers, patient survival, and resistance to therapy [49, 60]. One study identified clusterin in patient‐derived tumor organoids resistant to 5‐fluorouracil and an association between the presence of clusterin and a low patient survival rate [49]. Organoids have also been used to model cancer stem cells (CSCs) and differentiated cancer cells (non‐CSCs). Zhao et al. [50] demonstrated that CSCs and non‐CSCs had distinctive metabolic profiles and that lactate derived from non‐CSCs promoted self‐renewal of CSCs, thereby, contributing to the progression of CRC.
Metastatic cancer can lead to abnormal functioning in other organs, which is often fatal [61]. Organoids have been used to study metastasis in CRC. Li et al. [51] modeled the progression of colorectal tumors and metastasis using paired PDOs and found that organoids from sites of metastasis showed more tumorigenesis and metastatic ability in comparison with those from the primary lesions. Organoids have also been used in the search for more effective treatments of CRC. Patients with CRC and peritoneal metastases have limited treatment options and a poor prognosis. Hyperthermic intraperitoneal chemotherapy is widely used to treat peritoneal metastases, and PDO models have been used to evaluate hyperthermic intraperitoneal chemotherapy regimens in the search for more effective and personalized treatment strategies [52]. Furthermore, Narasimhan et al. [53] used organoids to guide the precision treatment of CRC and peritoneal metastase in the clinical setting by isolating CRC and peritoneal metastase organoids from patients. Using parallel ex vivo application of next‐generation sequencing and medium‐throughput drug screening, they were able to identify patient‐specific drug sensitivities. Molecular tumor heterogeneity may play a role in the efficacy of targeted therapies on metastatic CRC. Bruun et al. [54] established a platform for ex vivo pharmacogenomic profiling using PDOs and found that it could be used for drug screening to find new therapeutic strategies for metastatic CRC.
CRCs have different gene expression patterns, and tissue‐derived organoids have been established to study these gene profiles. Costales‐Carrera et al. [55] used tissue‐derived organoids to compare the gene profiles of normal tissues with those of tumor tissues in the colon and rectum. Although organoids derived from normal colon and rectal tissues showed similar gene expression profiles, rectal tumor organoids displayed heterogeneous gene expression profiles that were different from those of colon tumor organoids. Therefore, studies on gene expression in organoids could help to identify useful prognostic markers and therapeutic targets in CRC. Hu et al. [56] revealed the potentially helpful role of Dachshund homologue 1 (DACH1) as a prognostic marker for CRC, and Nguyen et al. [57] demonstrated that inhibition of Erb‐b2 receptor tyrosine kinase 3 (ERBB3) could be a target for CRC therapy using organoid technology. Human CRC organoids have also been used to assess mutation‐targeted inhibitors and combination drug therapy [58].
Parallel application of other technologies with organoids can be advantageous. For example, mass spectrometry immunopeptidomics has been used to investigate neoantigen presentation and the effects of interferon‐gamma and MEK inhibitor therapy in CRC PDOs [59]. Schnalzger et al. [62] developed a platform to investigate the resistance of chimeric antigen receptor cells to cytotoxicity in a CRC‐derived PDO model using a confocal live imaging protocol for monitoring of effector cell recruitment and cytolytic activity at a single organoid level. PDO models of CRC have also been used to evaluate sensitivity to chemotherapy [63]. Therapeutic thresholds deduced from changes in the organoid growth rate and related optical metabolic imaging have similarly been used to predict therapeutic sensitivity in patients with CRC undergoing chemotherapy/radiotherapy [64].
3.2. Lung cancer
Lung cancer accounts for the majority of cancer‐related deaths [65]. Preclinical models that accurately recapitulate the biology of lung cancer are needed for further investigation of biomarkers and drug screening to allow more individualized therapy in these patients. Biobanks of lung cancer organoids and normal bronchial organoids have been established from primary lung cancer tissues, including adenocarcinoma [66], squamous cell carcinoma [67], small cell carcinoma [68], large cell carcinoma [31], and adenosquamous carcinoma [67], and from paired non‐neoplastic airway tissues [69]. Other researchers have also established PDO biobanks and organoid models for use in drug trials, elucidation of the mechanisms of metastasis and resistance, the pathophysiology of tumorigenesis, and personalized medicine [31, 66, 70, 71, 72, 73].
Most cancer organoids are established using tissue derived from patients. However, Mazzocchi et al. [74] successfully established lung cancer organoids using pleural effusion aspirate obtained from patients with lung cancer to compare the chemotherapeutic response with that in a 2D culture system. Although the 2D cultures were more chemosensitive than the 3D organoids, they concluded that the 3D system was more accurate for studying tumor progression and for drug screening.
In recent years, an increasing number of researchers have been using organoid technology to study the efficacy of immunotherapy. A system for evaluating immune checkpoint inhibitors in PDOs has been developed by researchers studying various classes of molecule‐targeted drugs [75]. Della Corte et al. [76] investigated the antitumor and immune effects of a combination of atezolizumab, anti‐programmed death‐ligand 1 (PD‐L1), and selumetinib in preclinical and clinical experiments using an organoid model of non‐small cell lung cancer (NSCLC).
Establishment of a pure organoid is important before these models can be used to tailor personalized medicine in the clinical setting. Dijkstra et al. [77] evaluated the purity of NSCLC tumors and reported that 80% of organoids from intrapulmonary lesions had a standard copy number profile and that the organoids did not reflect the characteristic mutations in the tumors of origin, which they attributed to an overgrowth of the normal airway organoids in culture. NSCLC organoids could be established in only 17% of cases. Therefore, the methods currently used to establish pure organoids must be improved before organoid technology could be used in clinical practice.
3.3. Pancreatic cancer
Pancreatic ductal adenocarcinoma (PDA) has a very poor prognosis because it is difficult to detect early, behaves aggressively, and is relatively resistant to conventional chemotherapy [78]. A biobank of 30 characterized pancreatic cancer organoids was created to explore the role of organoid technology in drug screening for PDA. These organoids retained the histological features of the primary tumors and carried genetic alterations commonly found in the tumors of origin. High‐throughput drug screening using a panel of 76 compounds identified a range of targeted therapies with efficacy in PDOs [79].
The time interval between biopsy and functional analysis of pancreatic cancer organoids is a major obstacle to precision oncology and individualized treatment. Generation of PDOs from cell‐free DNA can allow rapid molecular profiling and drug testing, which has implications for clinical practice [80]. The lack of predictive biomarkers hinders optimal systemic treatment in patients with PDA; therefore, rapid pharmacotyping of PDOs may guide postoperative treatment selection, which could mimic the response of primary tumors to drug treatment and therefore have the potential for use in translational research and drug discovery [81, 82].
Pancreatic cancer organoids can also be used to test immunomodulatory drugs. Immune checkpoint inhibitors, independent of cytotoxic immune responses, have direct cytotoxic effects on PDA cells. PDOs treated with immune checkpoint inhibitors have direct therapeutic effect on PDA, with the highest response induced by a combination of anti‐mitogen‐activated protein kinase kinase (MEK1/2) and anti‐PD‐L1 [83]. Moreover, organoid models have been utilized to study mechanisms of resistance and to predict the response of PDA to treatment [84].
The factors that contribute to the high recurrence rate of PDA (60%–80%) have yet to be elucidated [85, 86]. Organoid technology has shown potential in this regard. Braun et al. [87] established PDOs from patients with early and late recurrent PDA to investigate tumor metabolism and found that levels of nine metabolites were higher in PDOs from patients with early recurrence than in those with late recurrence. Another study employed an organoid model of PDA to determine the molecular alterations critical for invasion from 25 resected PDA samples and found a difference in molecular levels between mutant and wild‐type tumors [88]. These studies suggest that organoid models have value for exploring molecular mechanism of the tumor invasion programs in cancer research.
Organoids have also been used to study paraneoplastic syndromes, such as cachexia. A large percentage of patients with pancreatic cancer develop cachexia, but the underlying mechanism is not completely understood. Vaes et al. [89] established PDA organoids to identify the factors driving cancer‐related cachexia. Several cachexia‐related parameters, including physical performance, nutritional status, and inflammation, were assessed, and PDA organoids were found to express different levels of many common cachexia‐related genes.
3.4. Breast cancer
A few studies have described the procedure used to create breast cancer organoids from clinical samples. DeRose et al. [90] described applications related to organoid technology in depth. Moreover, Mazzucchelli et al. [91] designed a novel approach for constructing organoids derived from surgically resected tissue and biopsy samples in patients with breast cancer. Another study assessed the frequency of normal‐like organoids in primary breast carcinoma cultures using organoid methodology to establish a living biobank containing over 100 primary and metastatic breast cancer organoid lines [30].
Researchers have developed many models that combine organoids with other technologies [92]. CRISPR/Cas9 has been used in conjunction with human breast organoids to model breast cancer. Dekkers et al. [93] generated breast epithelial organoids using specimens obtained during reduction mammoplasty, and CRISPR/Cas9 was used for targeted knockout of genes mimicking neoplasia in organoids that were transplanted into mice. The response to therapy was recorded. This study highlighted how other technologies can be combined with organoids to create models that could be used to study molecular events and drug responses in breast cancer [93]. Another study engineered a 3D biochemicomimetic and mechanomimetic scaffold for culture of breast cancer organoids that allowed better conservation of the gene expression profile of the tumor of origin than the classic methods used to establish an organoid model. This 3D scaffold model recapitulated the behavior of the tumor and its response to drug therapy more realistically and has potential applications in the development of effective and personalized chemotherapy regimens [94]. Heterogeneous cell populations within a tumor display varying metabolic profiles that can cloud interpretation of the response to treatment. Organoids can help overcome this problem [30]. Sharick et al. [95] showed that the heterogeneity of cell metabolism could be captured accurately using optical metabolic imaging technology and attributed the increased metabolic heterogeneity seen after treatment to metabolic shifts within the tumor.
Metastasis is the most common cause of cancer‐related death; however, the value of organoid models of metastatic cancer has not been assessed using large‐scale genomic data. Liu et al. [96] detected genomic differences using RNA sequencing data from 26 patient‐derived breast cancer organoids and showed that these organoids had a higher degree of genomic similarity to their parent sample than to cell lines.
Organoids have also been used to study the more uncommon breast cancers, such as papillary carcinoma. Using surgical specimens to establish papillary carcinoma PDOs, Li et al. [97] performed drug screening and subsequently examined the roles of endocrine agents and targeted drugs in breast cancer.
3.5. Liver cancer
Three major types of liver cancer organoids, namely, hepatocellular carcinoma (HCC), cholangiocarcinoma, and HCC combined with cholangiocarcinoma, have been established [29] using typical human liver organoid culture medium and tumoroid‐specific culture medium (Table 1). Some studies have established and characterized organoids from primary mouse liver tumors [98]. These organoid models have significant potential to advance liver cancer research, aid in signaling networks interacting with specific cell types, and could be used as a tool in personalized medicine [99, 100]. However, there are still no models in which the initiation of liver cancer can be investigated. Sun et al. [101] established liver cancer organoids using reprogrammed human‐induced hepatocytes and demonstrated that human‐induced hepatocyte organoids can be genetically manipulated in a liver cancer model to show initiation of the cancer. Furthermore, Artegiani et al. [102] created human liver cancer organoids using CRISPR/Cas9 technology, which is a good example of a synergistic experimental platform for studying the underlying mechanisms of genes involved in the development of cancer.
Resistance to chemotherapy is common in all types of cancer, and liver cancer organoids have been shown to be helpful in identifying the mechanisms of resistance to anticancer drugs. For example, sorafenib is widely used to treat HCC; however, the underlying mechanism of resistance has yet to be elucidated. Emerging evidence suggests that tumor‐initiating cells are a crucial driving force in resistance, with CD44 and Hedgehog signaling playing a fundamental role in the properties of tumor‐initiating cells in HCC [103]. Wang et al. [104] used PDO models of HCC to explore the roles of CD44 and Hedgehog signaling and to investigate the therapeutic effects of and resistance to sorafenib. Organoids have also been used to demonstrate intra‐tumor genetic heterogeneity, which may contribute to the failure of chemotherapy in liver cancer. To evaluate heterogeneity in primary human liver cancer, Li et al. [105] established 27 liver cancer organoid lines that were tested with 129 cancer drugs. They subsequently generated 3483 datum points for cell survival, which showed that the liver cancer organoids displayed intra‐patient and inter‐patient functional heterogeneity. Liver transplantation is one of the treatment options for HCC. However, tumor recurrence due to immunosuppression is a major complication of liver transplantation. Liver cancer organoids derived from mice have been used to test the ability of mycophenolic acid, an immunosuppressant commonly used after liver transplantation, to inhibit tumor recurrence [106]. Overall, these studies show the promise of organoid technology in liver cancer modeling and as a platform for drug screening and point to the possibility of its clinical applications for individualized treatment of liver cancer.
3.6. Ovarian cancer
Ovarian cancer is a common malignant tumor of the female reproductive system [107]. Despite advances in chemotherapy, there has been no significant improvement in the overall survival of patients with ovarian cancer. Several studies have developed effective methods for establishing ovarian cancer organoids. Three‐dimensional cultures of gynecological tumor organoids using Matrigel bilayer organoid culture and checking the histopathology and sequencing of the targeted genome revealed that ovarian cancer organoids maintained various features of the tumor of origin [108]. However, different gynecological cancers may require different environments or different types of medium for the establishment of organoids. For example, stable expansion of high‐grade serous ovarian cancer organoids requires a low‐Wnt environment [109]. Nanki et al. [110] assessed the association between the in vitro response of ovarian cancer PDOs and patients’ responses to chemotherapeutic drugs and found an association between the response of the PDO and several clinical response measures. These data demonstrated that PDOs are physiologically suitable ex vivo cancer drug screening models that could allow the use of effective personalized drugs targeting ovarian cancer. PDOs have also been shown to display inter‐patient and intra‐patient heterogeneity in terms of the toxicity of chemotherapy and targeted drugs. This finding can be partly explained by genetic aberrations. PDO drug screening identified a high response rate to at least one drug in 88% of patients with ovarian cancer [111].
Ovarian cancer is a highly heterogeneous disease that is often diagnosed at a late stage. Experimental in vitro models capturing hallmarks of tumor heterogeneity are not limited but are difficult to establish. One study in which 56 organoids were established from 32 patients developed a protocol that was able to capture intra‐patient and inter‐patient tumor heterogeneity. Subsequently, these organoids were genetically manipulated for drug screening applications [112]. In summary, ovarian cancer organoids maintain the gene expression pattern of their tumor of origin and can be used to target genes and guide future research on targeted therapy [113]. Ovarian cancer organoids can also be cultured on a large scale to enable high‐throughput drug screening.
3.7. Bladder cancer
Bladder cancer is a common malignancy of the urinary tract. Encountered regularly in clinical practice, bladder cancer has a high risk of recurrence and a high mutation rate. Owing to the heterogeneity of this tumor, the prognosis is poor [114]. Culture systems for bladder cancer organoids have been mentioned in many studies [115, 116]. Mullenders et al. [115] created a biobank consisting of bladder cancer organoids derived from 53 patients for the use as a drug screening platform. Identification of biomarkers that can predict the response to anticancer drugs is imperative in the effort to improve therapeutic outcomes. Kong et al. [117] used pharmacogenomic data derived from 3D organoid culture models and a machine‐learning framework to identify biomarkers that accurately predicted the response to chemotherapy in 77 patients with bladder cancer. This novel network‐based approach combined with application of gene modules was able to predict the therapeutic response and has the potential to be applied in drug screening for personalized medicine and for pharmacogenomic research in the field of cancer.
3.8. Prostate cancer
Prostate cancer is a common malignant tumor of the genitourinary system in men [118]. The natural history of prostate cancer varies from person to person. Gao et al. [119] used engineered mutations to successfully generate prostate cancer organoids, suggesting a promising way to test antitumor drugs both in vivo and in vitro. Puca et al. [120] and Karkampouna et al. [121] developed organoid models of prostate cancer with specific biological and genetic landscapes that could be used to investigate tumor growth, metastasis, and drug resistance in the early stages of the disease and to evaluate the response to chemotherapy, with the aim of providing a basis for clinical decision‐making. Furthermore, Servant et al. [122] developed a biobank consisting of prostate cancer organoids derived from 81 patients that were used to construct stable cell lines.
3.9. Other types of cancer
There have been some researches on head and neck squamous cell carcinoma (HNSCC) organoids. Driehuis et al. [123] devised an in‐depth protocol for the creation of HNSCC organoids and their applications in semi‐automated drug screening. Another study established HNSCC organoid lines derived from patients’ tumors and characterized their growth and response to various pharmacological agents [124]. Furthermore, organoids have been used to assess the efficacy of targeted photodynamic therapy on HNSCC using 3D PDO models in vitro [125].
Human gastric cancer organoids have also been created. Bartfeld et al. [126] successfully developed a system for creating gastric organoids that could be used to study various gastric pathologies, and Seidlitz et al. [127] established gastric cancer organoids for assessment of responses to conventional chemotherapy. Moreover, Steele et al. [128] used gastric cancer organoids to predict the responses of tumors to treatment in individual patients.
A procedure for generating patient‐derived glioblastoma organoids was described by Jacob et al. [129]. Glioblastoma organoids maintain many key features of the tumor of origin, so can be used to model the disease and for rapid investigation of patient‐specific treatment strategies. Patient‐derived glioblastoma organoids were confirmed to recapitulate critical features of the tissue of origin, including its histological characteristics, cellular diversity, gene expression patterns, and mutation profiles [130].
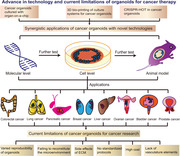
4. ADVANCES IN TECHNOLOGY WITH SYNERGISTIC APPLICATION OF CANCER ORGANOIDS
The applications of various cancer organoid models isolated from patients are described in Table 2. Although cancer organoids display various key features of cancer progression, there is room for improvement [131]. For example, cancer organoids usually include epithelial cells and progenitor cells but not non‐parenchymal cells, including fibroblasts and endothelial cells. Moreover, classic 3D organoid culture techniques do not allow for exact spatiotemporal control of various factors in the tumor microenvironment (TME) [17, 132]. Synergistic application of cancer organoids with other technologies, such as organ‐on‐a‐chip, 3D bio‐printing, and CRISPR‐Cas9‐mediated homology‐independent organoid transgenesis (CRISPR‐HOT), has allowed the development of highly precise cancer models for studying various mechanisms of tumor interactions, metastasis, and drug resistance (Figure 3).
TABLE 2.
The application of cancer organoid models
| Tumor organoid model | Date | Cell derived | Sample size | Research type | Achievement | Ref |
|---|---|---|---|---|---|---|
| Colorectal cancer | 2020.01 | Patient | 11 | Drug resistance | Clusterin, a drug resistance marker used to detect colorectal cancer progression | [49] |
| Colorectal cancer | 2020.11 | Patient | 15 | Tumor metabolic properties and phenotypes | Established a basis for the development of new treatments which target metabolic parameters in colorectal cancer | [50] |
| Colorectal cancer | 2020.09 | Patient | 2 | Tumor metastasis | Development of an experimental model for investigating colorectal cancer progression | [51] |
| Colorectal cancer | 2019.09 | Patient | 40 | Tumor metastasis | Developed an evaluation method to assess existing hyperthermic intraperitoneal chemotherapy regimens on an individual patient level | [52] |
| Colorectal cancer | 2020.07 | Patient | 28 | Personalized therapy | Displayed the use of organoids in guiding precision treatment for patients with CRC and peritoneal metastases | [53] |
| Colorectal cancer | 2020.08 | Patient | 22 | Drug screening and gene profiling | The use of ex vivo drug screening to identify novel treatment options for metastatic colorectal cancer | [54] |
| Colorectal cancer | 2020.08 | Patient | 50 | Tumor gene profile | Distinguished genetic profiles of rectal and colon tumors using organoids | [55] |
| Colorectal cancer | 2020.06 | Patient and cells | 22 | Tumor biomarker | DACH1 as a potential prognostic marker and therapeutic target for colorectal cancer | [56] |
| Colorectal cancer | 2016.11 | Patient | NA | Drug screening | Demonstrating the potential of colorectal cancer organoid libraries in drug screening | [58] |
| Colorectal cancer | 2019.11 | Patient | 5 | Neoantigen presentation | Identified novel approaches to increase neoantigen presentation | [59] |
| Colorectal cancer | 2019.06 | Patient | NA | CAR‐mediated cytotoxicity | Colorectal cancer organoids successfully evaluate CAR efficacy and tumor specificity in a personalized manner | [62] |
| Colorectal cancer | 2019.09 | Patient | 90 | Chemotherapy and/or radiotherapy sensitivity | Predict treatment sensitivity for patients with cancer undergoing chemotherapy and/or radiotherapy | [64] |
| Lung cancer | 2019.09 | Patient | 80 | Biobank of lung cancer organoids construction | Successfully construct biobank of lung cancer organoids | [67] |
| Lung cancer | 2020.03 | Patient | 30 | Tumor modeling | Successfully construct NSCLC organoid for drug testing | [31] |
| Lung cancer | 2020.08 | Patient | 12 | Drug screening | To identify new therapeutic targets and advanced personalized medicine | [66] |
| Lung cancer | 2020.08 | Patient | 12 | Genomic characteristics and drug screening | PDOs are highly credible models for personalized precision medicine | [71] |
| Lung cancer | 2020.03 | Patient | 4 | Drug screening | PDOs were relatively more sensitive to CF10 | [72] |
| Lung cancer | 2020.03 | Patient | 10 | Drug screening | To identify the anticancer activity of chelerythrine chloride, cantharidin, and harmine in PDOs | [73] |
| Lung cancer | 2019.04 | Pleural effusion aspirate from patient | 2 | Drug response | Serve as more accurate disease models for the study of tumor progression and drug development | [74] |
| Lung cancer | 2019.05 | PDOs | 3 | Evaluating molecular targeted drugs | PDOs are suitable for evaluation molecular targeted drugs | [75] |
| Lung cancer | 2019.06 | Patient | 11 | Immunotherapy | Combining PD‐L1 with MEK‐I in 3D‐culture model, useful to predict sensitivity of patients to immunotherapy | [76] |
| Pancreatic cancer | 2019.12 | Patient | 30 | Personalized drug screening | Development of a platform for identification of novel therapeutics for pancreatic cancer using PDOs | [79] |
| Pancreatic cancer | 2020.08 | Patient | 10 | Personalized therapy | Generation of PDOs from a limited sample can allow molecular profiling and drug testing | [80] |
| Pancreatic cancer | 2020.09 | Patient | 76 | Precision medicine | To guide postoperative adjuvant chemotherapeutic selection | [82] |
| Pancreatic cancer | 2019.09 | PDOX models | NA | Drug sensitivity and resistance | Development of PDOX‐derived organoid system for use in prediction of treatment response in advanced pancreatic cancer | [81] |
| Pancreatic cancer | 2019.01 | Patient | NA | Immunotherapy | Exploring the role of PD‐L1 in pancreatic cancer organoids | [83] |
| Pancreatic cancer | 2019.11 | Patient | NA | Tumor resistance | Pan‐ERBB kinase inhibitor resulted in suppression of cell viability and tumor regressions when combined with MEK inhibition | [84] |
| Pancreatic cancer | 2020.06 | Patient | 6 | Investigate the metabolism in PDOs | A therapeutic intervention could delay PDA recurrence and prolong the survival of affected patients | [87] |
| Pancreatic cancer | 2020.07 | Patient | 25 | Study the pattern of invasion in PDA | Invasion programs in SMAD4‐mutant and SMAD4 wild‐type tumors are different in both morphology and molecular mechanism | [88] |
| Pancreatic cancer | 2020.12 | Patient | 8 | Study human PDA induced cachexia | To further understand the mechanisms driving cancer cachexia | [89] |
| Breast cancer | 2020.05 | Patient | 12 | Using CRISPR/Cas9 to model tumor organoids | Modeling breast cancer using CRISPR/Cas9‐mediated engineering of human breast cancer organoids | [93] |
| Breast cancer | 2019.08 | Primary patient‐derived breast cancer cells | NA | Personalized chemotherapy | Development of a new platform for culturing primary cells for developing personalized chemotherapy regimens | [94] |
| Breast cancer | 2019.06 | Genetically engineered mouse model | NA | Cellular metabolic heterogeneity | Found that metabolic heterogeneity after upon treatment is attributed to heterogeneous metabolic shifts within tumor cells | [95] |
| Breast cancer | 2019.05 | Patient | 26 | Metastasis cancer related translational research | Demonstrated metastatic breast cancer organoids closely resemble the transcriptome of their parent lesion | [96] |
| Breast cancer | 2020.03 | Patient | 1 | Drug screening | Identified possible treatments in patients with breast papillary carcinoma | [97] |
| Liver cancer | 2019.03 | Primary mouse liver tumors | 129 | Drug development and personalized medicine | The antitumor drug can be successfully used in the organoids from primary mouse liver tumors | [100] |
| Liver cancer | 2019.08 | Reprogrammed human hepatocytes | NA | Modeling liver cancer | Showed human‐induced hepatocyte organoids can be genetically manipulated to model cancer initiation | [101] |
| Liver cancer | 2019.06 | Patient | NA | CRISPR/Cas9 engineer human liver organoids | Demonstrate combination of organoid technology with CRISPR/Cas9 can serve as an experimental platform for mechanistic studies of human cancer gene function | [102] |
| Liver cancer | 2020.01 | Patient | 4 | Tumor resistance | Combination of sorafenib and Hedgehog signaling inhibitors might be effective in HCC patients with high CD44 levels as a personalized‐medicine approach | [104] |
| Liver cancer | 2019.01 | Patient | 5 | Drug response heterogeneity | This study lay the foundation for functional personalized oncology approaches | [105] |
| Liver cancer | 2019.05 | Primary mouse liver tumors | NA | Tumor growth | Mycophenolic acid inhibits liver tumor organoids initiation and growth | [106] |
| Ovarian cancer | 2020.07 | Patient | 7 | Drug sensitivity and resistance testing | PDOs are suitable cancer models that can be used to screen effective personalized ovarian cancer drugs | [110] |
| Ovarian cancer | 2020.06 | Patient | 23 | Tumor heterogeneity | Increase our knowledge of genetic and drug response heterogeneity | [111] |
| Ovarian cancer | 2019.05 | Patient | 32 | Genetic manipulations and drug screening | Ovarian cancer organoids illustrating intra‐ and inter‐patient heterogeneity to use for drug‐screening assays | [112] |
| Bladder cancer | 2019.3 | Patient | 53 | Construct a bladder organoids biobank | Bladder organoids biobank for drug testing in the future | [115] |
| Bladder cancer | 2020.10 | Patient | 77 | Predict cancer patient drug responses | Used pharmacogenomic data derived from organoids and developed a novel machine learning framework to identify biomarkers and predict drug response in bladder cancer | [117] |
| Prostate cancer | 2014.09 | Patient | 32 | Predict cancer patient drug responses |
Enable the generation of a large repertoire of patient‐derived prostate cancer lines amenable to genetic and pharmacologic studies |
[119] |
| Prostate cancer | 2021.08 | Patient | 81 | Explores determinants of outcome |
Ensure the reliable establishment of organoids derived from specific prostate cancer molecular subtypes |
[122] |
| HNSCC | 2018.12 | Patient | 43 | Predict drug sensitivity | Show organoids can predict drug sensitivity and potential of organoids in the development of precision treatments for HNSCC | [124] |
| HNSCC | 2019.11 | Patient | 7 | For PDT | Demonstrated HNSCC organoid as a useful model for in‐vitro testing of targeted PDT | [125] |
| Gastric cancer | 2019.02 | Patient | 20 | Modeling gastric cancer | Modeled human gastric cancer using organoids | [127] |
| Gastric cancer | 2019.01 | Patient | 7 | Personalized treatment | To predict individual therapy response and patient outcome | [128] |
| Glioblastoma organoids | 2020.01 | Patient | 53 | Personalized treatment | Establishment of a glioblastoma organoid biobank for testing personalized therapies | [130] |
DACH1, Dachshund homolog 1; NA, Not available; CAR, Chimeric antigen receptor; PDOs, Patient‐derived organoids; CF10, fluoropyrimidine polymer F10; PD‐L1, Programmed cell death ligand 1; MEK‐I, MAP‐ERK kinase inhibitor; PDOX, Patient‐derived orthotropic xenograft; ERBB, Receptor tyrosine‐protein kinase; PDA, Pancreatic ductal adenocarcinoma; SMAD4, Mothers against decapentaplegic homolog 4; HCC, Hepatocellular carcinoma; HNSCC, Head and neck squamous cell carcinoma; PDT, Photodynamic therapy.
FIGURE 3.
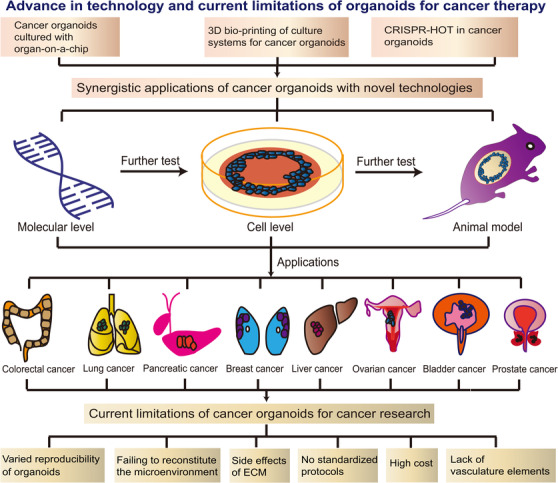
Advance in technology and current limitations of organoids for cancer therapy. Synergistic applications of cancer organoids with novel technologies include organ‐on‐a‐chip, 3D bio‐printing, and CRISPR‐HOT. These synergistic technologies with cancer organoids underwent testing at the molecular and cellular levels and in animal models and were ultimately investigated in various cancers. At present, the main limitations of cancer organoids for cancer research are their variable reproducibility, failure to reconstitute the microenvironment, the unwanted effects of the ECM, lack of standardized protocols, high cost, and absence of vascular elements. ECM, extracellular matrix; CRISPR‐HOT, CRISPR‐Cas9‐mediated homology‐independent organoid transgenesis
4.1. Cancer organoids cultured with organ‐on‐a‐chip
Organoids are incomplete in the sense that they lack vasculature, stromal components, and tissue‐resident immune cells [133]. Furthermore, the random configuration in conventional 3D organoid culture does not allow for accurate spatiotemporal control of biophysical and biochemical factors in the TME. Moreover, 3D organoid culture systems are unable to replicate the microenvironment of the organ and lack the signaling that prompts organogenesis.
To advance in vitro technology, researchers have integrated organoid culture systems with “organ‐on‐a‐chip” technology [134]. An organ‐on‐a‐chip is a microfabricated device for integrated culture of living cells, ECM, and microstructures that mimic organs or tissues. To create an organ‐on‐a‐chip, elements in an organ essential for physiological function are identified, after which the biochemical and physical microenvironments of the organ‐specific functional unit can be assessed. A cell culture device was designed to retain these features and produced using microfabrication techniques [135]. Integration of living human cells with a synthetically generated microenvironment creates a model that can mimic not only physiological homeostasis but also complex disease processes that benefit medical end‐users [136].
Although the fundamental concepts of organoids and organ‐on‐a‐chip are different, they share the same goal of recapitulating complex human organs in vitro. Combining features from both concepts allows for a more powerful in vitro technology. As mentioned above, traditional 3D cultures do not allow for precise spatiotemporal control of the microenvironment. Researchers have tackled this issue by using microengineering techniques to create a microfluidic system [137]. Similar methods were used to develop a human brain organoid‐on‐a‐chip model derived from human iPSCs, which facilitated 3D culture, in situ neural differentiation, and self‐organization of brain organoids in a controlled manner [138].
Conventional organoid cultures also do not have a vasculature, relying entirely on their culture medium for nutrients, and therefore can only grow to a certain size because diffusion alone cannot support their metabolic requirements. Because organs‐on‐a‐chip can mimic perfusable blood vessels, integration of this technology with organoids can help overcome the obstacle of nutrient supply. Shirure et al. [139] successfully demonstrated this integration by developing a tumor‐on‐a‐chip microfluidic platform that was used to investigate the progression and response to chemotherapy and antiangiogenic therapy in cell lines and PDOs.
Another challenge is the inability of organoids to model interactions between different tissues and organs. Interactions between different tissues within the organ of interest and other organs play an essential role in organ development and homeostasis. Organs‐on‐a‐chip provides a more controllable environment for culturing different types of cells and tissues in organoid systems, as seen in the vascularized liver organ‐on‐a‐chip model developed by Jin et al. [140]. The same team also created a multiorgan model for coculture with stem cell‐derived liver, intestinal, and stomach organoids, which were able to display features of inter‐organ crosstalk. Together, these studies illustrate how a synergistic combination of organoids and organ‐on‐a‐chip technology can help achieve a level of cell maturity that is not possible when either technology is applied alone.
4.2. 3D bio‐printing of culture systems for cancer organoids
Three‐dimensional bio‐printing allows the production of biomaterials through designed structures joined by hydrogels with the properties of printability, crosslinking, biocompatibility, and controllability [141, 142]. These features can provide researchers with accurate control over spatial heterogeneity within the TME by spatially depositing predefined biobanks [143, 144]. The ECM plays an important role in controlling the fate of cells and has important implications for developmental biology, tissue engineering, regenerative medicine, and tumor therapy; however, classic organoid cultures lack tissue‐specific ECM, making it difficult to study cell interactions. The synergistic application of organoids and 3D bio‐printing can allow for the development of a more sophisticated cancer model with structures that are more cell‐specific and well‐separated properties that are more suited for the growth and maturation of organoids [145, 146].
The sustainability of these bio‐printed organoid models has been demonstrated in several studies. Mollica et al. [147] and Sandercock et al. [148] generated 3D hydrogels composed solely of human or mouse mammary ECM and demonstrated that their novel 3D culture substrates effectively maintained large 3D bio‐printed organoids and formation of tumoroids. Reid et al. [149] found that bio‐printing significantly increased the formation of tumoroids in 3D collagen gels and allowed accurate production of tumoroid arrays when studying the tumorigenesis of breast cancer. They also demonstrated that bio‐printed organoids can accurately mimic in vivo conditions, showing that the capacity of 3D bio‐printing served as a mediator to investigate tumorigenesis and the TME for the control of cancer cells. Bio‐printing can also allow the production of multiple scales of vasculature, enabling the growth of larger organoids [150]. Furthermore, bio‐printing techniques can also increase the throughput of 3D drug screening, as shown by Maloney et al. [151]. Ideally, 3D bio‐printed organoids would be representative of an in vitro functional organ containing different cell types, a vasculature, and even nervous and immune system components, aiding the investigation of potential mechanisms of tumor‐stroma interactions, multiorgan metastasis, TME interactions for multiple tumor types, and drug screening.
4.3. CRISPR‐HOT in cancer organoids
Although CRISPR‐Cas9 has helped to simplify genetic engineering, there is considerable room for error during this process. In 2020, Artegiani et al. [152] developed CRISPR‐HOT, a new genetic tool for labeling specific genes in human organoids, which they used initially to investigate modes of division in human hepatocytes. CRISPR‐HOT was applied to fluorescently tag and visualize subcellular structural molecules for rare intestinal cell types by generating reporters. Some cell types are rare and difficult to study, but CRISPR‐HOT can easily tag and allow visualization of these cells. Another report from the same group indicated that it took only 2‐3 months to produce genetically engineered human fetal hepatocyte organoids using CRISPR‐HOT [153], which is highly efficient. This novel tool can be applied in the study of cell fate and differentiation and in developmental diseases, and can be used to visualize any type of gene or cell. CRISPR‐HOT also shows promise for the advancement of cancer research.
5. CANCER ORGANOIDS AND IMMUNOTHERAPY
Cancer immunotherapy uses the host's own immune system to combat tumors. In recent years, there has been a series of major breakthroughs in cancer immunotherapy. Owing to its advantages of relatively minor toxicity and long‐lasting efficacy, immunotherapy has become a hot topic in cancer research [154, 155]. The use of human organoid cultures is a novel approach in the study of tumor immunobiology. Organoids are commonly used as cancer models and are created by genetically engineering iPSCs to bear oncogenes or tumor suppressor genes or by propagating biopsied cells in vitro to create PDOs. This has led to the creation of many tumor biobanks, as discussed in many of the studies mentioned in the earlier sections. However, due to their genomic instability, there may be genetic variations between PDO models and their parent tumor tissues. Furthermore, 3D organoids require artificial reconstitution to recapitulate the TME, although various strategies to overcome this problem have been described. Yuki et al. [156] showed that tumor‐only 3D PDO systems can be reconstituted by the addition of exogenous immune components. Whole air‐liquid interface (ALI) cancer organoid culture and microfluidic culture can reproduce the TME by retaining endogenous matrix components (including multiple immune cells) without recombination. Recent developments in tumor organoid/immune cell cocultures and ALI systems have allowed new insights into the interactions between epithelial cells and immune cells. Organoids that incorporate cells (lymphocytes/immune cells/fibroblasts) from the TME that combat cancers have been extensively studied [157, 158, 159]. For example, Scognamiglio et al. [157] found that programmed cell death protein 1(PD‐1)/CD8‐positive lymphocytes isolated from PD‐L1‐positive organoids were a promising tool. Similarly, Holokai et al. [158] reported that co‐cultured mouse and human‐derived autologous PDA organoids and immune cells effectively inhibited the effector functions of T cells in the TME. Nakamura et al. [159] also used a cancer organoid model to evaluate the effect of cancer‐associated fibroblasts and found it to be useful for the assessment of the TME. Finally, Neal et al. [17] used ALI to establish a PDO system and modeled immune checkpoint blockade with anti‐PD‐1/anti‐PD‐L1. They successfully demonstrated that immuno‐oncology investigations could be carried out successfully using organoid models and that these organoids may facilitate personalized immunotherapy testing.
Tumor organoids have several potential applications, including translational immuno‐oncology research, immunotherapy modeling, and personalized medicine [6, 22, 160, 161]. Combination of different technologies, such as organ‐on‐a‐chip, CRISPR‐Cas9, and microfluidics, has helped model immunotherapeutic responses within organoids [162]. For example, Hai et al. [163] showed that the CRISPR‐Cas9 system can efficiently and rapidly generate lung squamous cell carcinomas that closely mimic human disease at the genomic and phenotypic levels.
Overall, these results demonstrate that cancer organoids are an ideal model for investigating immunotherapy and have the potential to allow personalized treatment strategies. Moreover, the combination of different strategies with organoids has allowed for advancement of models of immunological disease and aided in further investigation of the link between immunity and cancer [164]. Further combinations of organoids and other technologies in the future could help to overcome the current challenges of organoid technology.
6. CURRENT LIMITATIONS OF TUMOR ORGANOIDS FOR CANCER THERAPY
Although organoids have shown strong potential in cancer research and clinical applications, there are still many drawbacks [165, 166]. A summary of the current limitations with regard to cancer organoids is provided in Figure 3. Currently, successful creation of organoid models is highly variable, with success rates ranging between 15% and 90% [108, 116]. The conditions for organoid culture require optimization. Only when the same samples can produce very similar phenotypic characteristics, including organ‐like diameter, cell composition, 3D structure, and gene expression, under the same experimental conditions can the organoid formation process be accurately repeated and the organoid culture system be considered mature and applied in various studies [167]. The reproducibility of organoids can vary under the same conditions, as demonstrated by Velasco et al. [168]. Therefore, determinants for the generation of reproducible organoids must be identified. This variation limits large‐scale reproducibility and inhibits the potential application of organoids in high‐throughput drug screening.
Furthermore, in vitro organoids cannot completely recapitulate all the cell types and maturity of the organ of origin owing to the dynamic process and pulsatile nature of many growth factors. Moreover, most PDOs lack stromal cells. Therefore, they fail to reconstitute the microenvironment, which includes fibroblasts, endothelial cells, immune cells, and ECM, and lack the signaling that prompts organogenesis, hindering the ability to accurately predict clinical outcomes. As discussed in the above sections, cancer organoids have the potential to serve as an in vitro testing platform to predict the response to personalized immunotherapy. However, the lack of these fundamental components can affect the response to treatment and make it more difficult to assess the pharmacological effects of immunomodulatory agents. Furthermore, these models can only be used for a short term because the numbers of fibroblasts and immune cells they contain decline over 1‐2 months. Moreover, the effects of the composition of the ECM on human organoid cultures remain uncertain and could influence the outcomes of various chemical and genetic screening processes. To make further progress in organoid technology, all materials, protocols, and quality control procedures used to establish organoids must be completely defined and standardized.
Current organoid models have a limited size owing to the lack of vascular elements to allow for adequate absorption of nutrients. Recently, considerable progress has been made with vascularized organoids and application of tumor‐on‐a‐chip platforms, as discussed earlier. However, vascularization remains a difficult obstacle to overcome. Current organoid models lack the ability to model interactions and crosstalk between different tissues and organs; thus, they cannot completely reflect the developmental and pathological characteristics of their in vivo origins. Moreover, human organoids have limited tissue‐specific micro‐physiology. Research efforts to overcome this limitation are underway.
Finally, there are some practical issues worthy of mention. First, the production of PDOs is very expensive. Therefore, this technology cannot be incorporated into existing health care systems. Second, owing to the complex 3D culture system, production cannot be conducted on a large scale to reduce costs. Finally, standardized protocols must be established for reliable assessment of drug screening outcomes in both preclinical models and clinical research.
7. CONCLUSIONS
Patient‐derived tumor organoids are physiologically and clinically more advanced than classic cancer cell lines and animal cancer models. Furthermore, compared with classic cancer cell lines and PDX models, patient‐derived tumor organoids can better capture and retain the molecular, cellular, genetic, and histological phenotypes of the tumor of origin while also maintaining patient‐specific tumor heterogeneity. Despite the remaining challenges, human organoids have considerable potential in the treatment of cancer. With the rapid development of other technologies, we believe that synergistic applications that include organoids can help narrow the gap between ex vivo organoids and their in vivo counterparts and help pave the way for novel cancer treatments. Correct application of these remarkable 3D cultures could allow for comprehensive development of high‐throughput drug screening for better prediction of drug responses and personalized medicine that can guide optimized therapeutic strategies for individual patients. Advances in the future will undoubtedly bring this novel 3D culture technique closer to clinical use.
DECLARATIONS
DECLARATION OF CONFLICTING INTERESTS
The authors declare there is no conflict of interest.
AUTHORS’ CONTRIBUTIONS
JQ and FSK wrote the manuscript; LC, LL, and TC collected the references and modified the manuscript; LC and FSK designed the manuscript and approved the final manuscript for publication.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
CONSENT FOR PUBLICATION
Not applicable.
AVAILABILITY OF DATA AND MATERIALS
Please contact the corresponding author for data requests.
Supporting information
Supporting Information
ACKNOWLEDGMENTS
The authors thank Prof. Nong Yang from the Lung Cancer and Gastroenterology Department, Hunan Cancer Hospital, Affiliated Tumor Hospital of Xiangya Medical School of Central South University for writing help with this manuscript. Additionally, we would like to thank Editage (www.editage.cn) for English language editing. This work was supported by the National Natural Science Foundation of China (81802278 and 81900563) and the Natural Science Foundation of Hunan Province (2019JJ50361 and 2020JJ4418).
Qu J, Kalyani FS, Liu L, Cheng T, Chen L. Tumor organoids: synergistic applications, current challenges, and future prospects in cancer therapy. Cancer Commun. 2021;41:1331–1353. 10.1002/cac2.12224
Contributor Information
Farhin Shaheed Kalyani, Email: farhin@zju.edu.cn.
Lijun Chen, Email: chenlijun@zju.edu.cn.
REFERENCES
- 1. Lee JK, Liu Z, Sa JK, Shin S, Wang J, Bordyuh M, et al. Pharmacogenomic landscape of patient‐derived tumor cells informs precision oncology therapy. Nat Genet. 2018;50(10):1399‐1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Invrea F, Rovito R, Torchiaro E, Petti C, Isella C, Medico E. Patient‐derived xenografts (PDXs) as model systems for human cancer. Curr Opin Biotechnol. 2020;63:151‐156. [DOI] [PubMed] [Google Scholar]
- 3. Lancaster MA, Knoblich JA. Organogenesis in a dish: modeling development and disease using organoid technologies. Science. 2014;345(6194):1247125. [DOI] [PubMed] [Google Scholar]
- 4. Lai Y, Wei X, Lin S, Qin L, Cheng L, Li P. Current status and perspectives of patient‐derived xenograft models in cancer research. J Hematol Oncol. 2017;10(1):106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yan Q, Dong H, Su J, Han J, Song B, Wei Q, et al. A Review of 3D Printing Technology for Medical Applications. Engineering. 2018;4(5):729‐742. [Google Scholar]
- 6. Fatehullah A, Tan SH, Barker N. Organoids as an in vitro model of human development and disease. Nat Cell Biol. 2016;18(3):246‐254. [DOI] [PubMed] [Google Scholar]
- 7. Xu H, Lyu X, Yi M, Zhao W, Song Y, Wu K. Organoid technology and applications in cancer research. J Hematol Oncol. 2018;11(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rauth S, Karmakar S, Batra SK, Ponnusamy MP. Recent advances in organoid development and applications in disease modeling. Biochim Biophys Acta Rev Cancer. 2021;1875(2):188527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang X. Stem cells in tissues, organoids, and cancers. Cell Mol Life Sci. 2019;76(20):4043‐4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tuveson D, Clevers H. Cancer modeling meets human organoid technology. Science. 2019;364(6444):952‐955. [DOI] [PubMed] [Google Scholar]
- 11. Xu Y, Chen J, Huang Y, Luo Y, Hsieh AC, Chen J, et al. Patient‐derived organoids in cellulosic sponge model chemotherapy response of metastatic colorectal cancer. Clin Transl Med. 2021;11(1):e285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zanoni M, Cortesi M, Zamagni A, Arienti C, Pignatta S, Tesei A. Modeling neoplastic disease with spheroids and organoids. J Hematol Oncol. 2020;13(1):97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Aboulkheyr Es H, Montazeri L, Aref AR, Vosough M, Baharvand H. Personalized Cancer Medicine: An Organoid Approach. Trends Biotechnol. 2018;36(4):358‐371. [DOI] [PubMed] [Google Scholar]
- 14. Xu R, Zhou X, Wang S, Trinkle C. Tumor organoid models in precision medicine and investigating cancer‐stromal interactions. Pharmacol Ther. 2021;218:107668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kondo J, Inoue M. Application of Cancer Organoid Model for Drug Screening and Personalized Therapy. Cells. 2019;8(5):470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Boretto M, Maenhoudt N, Luo X, Hennes A, Boeckx B, Bui B, et al. Patient‐derived organoids from endometrial disease capture clinical heterogeneity and are amenable to drug screening. Nat Cell Biol. 2019;21(8):1041‐1051. [DOI] [PubMed] [Google Scholar]
- 17. Neal JT, Li X, Zhu J, Giangarra V, Grzeskowiak CL, Ju J, et al. Organoid Modeling of the Tumor Immune Microenvironment. Cell. 2018;175(7):1972‐1988.e1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wagar LE, Salahudeen A, Constantz CM, Wendel BS, Lyons MM, Mallajosyula V, et al. Modeling human adaptive immune responses with tonsil organoids. Nat Med. 2021;27(1):125‐135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tatullo M, Marrelli B, Benincasa C, Aiello E, Makeeva I, Zavan B, et al. Organoids in Translational Oncology. J Clin Med. 2020;9(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu L, Yu L, Li Z, Li W, Huang W. Patient‐derived organoid (PDO) platforms to facilitate clinical decision making. J Transl Med. 2021;19(1):40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fan H, Demirci U, Chen P. Emerging organoid models: leaping forward in cancer research. J Hematol Oncol. 2019;12(1):142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lo Y‐H, Karlsson K, Kuo CJ. Applications of organoids for cancer biology and precision medicine. Nature Cancer. 2020;1(8):761‐773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wilson HV. A New Method by Which Sponges May Be Artificially Reared. Science. 1907;25(649):912‐915. [DOI] [PubMed] [Google Scholar]
- 24. Lindberg K, Brown ME, Chaves HV, Kenyon KR, Rheinwald JG. In vitro propagation of human ocular surface epithelial cells for transplantation. Invest Ophthalmol Vis Sci. 1993;34(9):2672‐2679. [PubMed] [Google Scholar]
- 25. Pellegrini G, Traverso CE, Franzi AT, Zingirian M, Cancedda R, De Luca M. Long‐term restoration of damaged corneal surfaces with autologous cultivated corneal epithelium. Lancet. 1997;349(9057):990‐993. [DOI] [PubMed] [Google Scholar]
- 26. Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, et al. Single Lgr5 stem cells build crypt‐villus structures in vitro without a mesenchymal niche. Nature. 2009;459(7244):262‐265. [DOI] [PubMed] [Google Scholar]
- 27. Unbekandt M, Davies JA. Dissociation of embryonic kidneys followed by reaggregation allows the formation of renal tissues. Kidney Int. 2010;77(5):407‐416. [DOI] [PubMed] [Google Scholar]
- 28. Lancaster MA, Renner M, Martin CA, Wenzel D, Bicknell LS, Hurles ME, et al. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501(7467):373‐379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Broutier L, Mastrogiovanni G, Verstegen MM, Francies HE, Gavarro LM, Bradshaw CR, et al. Human primary liver cancer‐derived organoid cultures for disease modeling and drug screening. Nat Med. 2017;23(12):1424‐1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sachs N, de Ligt J, Kopper O, Gogola E, Bounova G, Weeber F, et al. A Living Biobank of Breast Cancer Organoids Captures Disease Heterogeneity. Cell. 2018;172(1‐2):373‐386 e310. [DOI] [PubMed] [Google Scholar]
- 31. Shi R, Radulovich N, Ng C, Liu N, Notsuda H, Cabanero M, et al. Organoid Cultures as Preclinical Models of Non‐Small Cell Lung Cancer. Clin Cancer Res. 2020;26(5):1162‐1174. [DOI] [PubMed] [Google Scholar]
- 32. Tiriac H, Plenker D, Baker LA, Tuveson DA. Organoid models for translational pancreatic cancer research. Curr Opin Genet Dev. 2019;54:7‐11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Barker N, Huch M, Kujala P, van de Wetering M, Snippert HJ, van Es JH, et al. Lgr5(+ve) stem cells drive self‐renewal in the stomach and build long‐lived gastric units in vitro. Cell Stem Cell. 2010;6(1):25‐36. [DOI] [PubMed] [Google Scholar]
- 34. Huch M, Gehart H, van Boxtel R, Hamer K, Blokzijl F, Verstegen MM, et al. Long‐term culture of genome‐stable bipotent stem cells from adult human liver. Cell. 2015;160(1‐2):299‐312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Takebe T, Zhang RR, Koike H, Kimura M, Yoshizawa E, Enomura M, et al. Generation of a vascularized and functional human liver from an iPSC‐derived organ bud transplant. Nat Protoc. 2014;9(2):396‐409. [DOI] [PubMed] [Google Scholar]
- 36. Russell SJ, Peng KW, Bell JC. Oncolytic virotherapy. Nat Biotechnol. 2012;30(7):658‐670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Li L, Liu S, Han D, Tang B, Ma J. Delivery and Biosafety of Oncolytic Virotherapy. Front Oncol. 2020;10:475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhu Z, Gorman MJ, McKenzie LD, Chai JN, Hubert CG, Prager BC, et al. Zika virus has oncolytic activity against glioblastoma stem cells. J Exp Med. 2017;214(10):2843‐2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Raimondi G, Mato‐Berciano A, Pascual‐Sabater S, Rovira‐Rigau M, Cuatrecasas M, Fondevila C, et al. Patient‐derived pancreatic tumour organoids identify therapeutic responses to oncolytic adenoviruses. EBioMedicine. 2020;56:102786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. van de Wetering M, Francies HE, Francis JM, Bounova G, Iorio F, Pronk A, et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell. 2015;161(4):933‐945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Calandrini C, Schutgens F, Oka R, Margaritis T, Candelli T, Mathijsen L, et al. An organoid biobank for childhood kidney cancers that captures disease and tissue heterogeneity. Nat Commun. 2020;11(1):1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jiang S, Zhao H, Zhang W, Wang J, Liu Y, Cao Y, et al. An Automated Organoid Platform with Inter‐organoid Homogeneity and Inter‐patient Heterogeneity. Cell Rep Med. 2020;1(9):100161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Marusyk A, Janiszewska M, Polyak K. Intratumor Heterogeneity: The Rosetta Stone of Therapy Resistance. Cancer Cell. 2020;37(4):471‐484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kuo CJ, Curtis C. Organoids reveal cancer dynamics. Nature. 2018;556(7702):441‐442. [DOI] [PubMed] [Google Scholar]
- 45. Muthuswamy SK. Organoid Models of Cancer Explode with Possibilities. Cell Stem Cell. 2018;22(3):290‐291. [DOI] [PubMed] [Google Scholar]
- 46. Crespo M, Vilar E, Tsai SY, Chang K, Amin S, Srinivasan T, et al. Colonic organoids derived from human induced pluripotent stem cells for modeling colorectal cancer and drug testing. Nat Med. 2017;23(7):878‐884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Di Modugno F, Colosi C, Trono P, Antonacci G, Ruocco G, Nistico P. 3D models in the new era of immune oncology: focus on T cells, CAF and ECM. J Exp Clin Cancer Res. 2019;38(1):117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Drost J, van Jaarsveld RH, Ponsioen B, Zimberlin C, van Boxtel R, Buijs A, et al. Sequential cancer mutations in cultured human intestinal stem cells. Nature. 2015;521(7550):43‐47. [DOI] [PubMed] [Google Scholar]
- 49. Engel RM, Chan WH, Nickless D, Hlavca S, Richards E, Kerr G, et al. Patient‐Derived Colorectal Cancer Organoids Upregulate Revival Stem Cell Marker Genes following Chemotherapeutic Treatment. J Clin Med. 2020;9(1):128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhao H, Yan C, Hu Y, Mu L, Liu S, Huang K, et al. Differentiated cancer cell‐originated lactate promotes the self‐renewal of cancer stem cells in patient‐derived colorectal cancer organoids. Cancer Lett. 2020;493:236‐244. [DOI] [PubMed] [Google Scholar]
- 51. Li H, Dai W, Xia X, Wang R, Zhao J, Han L, et al. Modeling tumor development and metastasis using paired organoids derived from patients with colorectal cancer liver metastases. J Hematol Oncol. 2020;13(1):119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ubink I, Bolhaqueiro ACF, Elias SG, Raats DAE, Constantinides A, Peters NA, et al. Organoids from colorectal peritoneal metastases as a platform for improving hyperthermic intraperitoneal chemotherapy. Br J Surg. 2019;106(10):1404‐1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Narasimhan V, Wright JA, Churchill M, Wang T, Rosati R, Lannagan TRM, et al. Medium‐throughput Drug Screening of Patient‐derived Organoids from Colorectal Peritoneal Metastases to Direct Personalized Therapy. Clin Cancer Res. 2020;26(14):3662‐3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bruun J, Kryeziu K, Eide PW, Moosavi SH, Eilertsen IA, Langerud J, et al. Patient‐Derived Organoids from Multiple Colorectal Cancer Liver Metastases Reveal Moderate Intra‐patient Pharmacotranscriptomic Heterogeneity. Clin Cancer Res. 2020;26(15):4107‐4119. [DOI] [PubMed] [Google Scholar]
- 55. Costales‐Carrera A, Fernandez‐Barral A, Bustamante‐Madrid P, Dominguez O, Guerra‐Pastrian L, Cantero R, et al. Comparative Study of Organoids from Patient‐Derived Normal and Tumor Colon and Rectal Tissue. Cancers (Basel). 2020;12(8):2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hu X, Zhang L, Li Y, Ma X, Dai W, Gao X, et al. Organoid modelling identifies that DACH1 functions as a tumour promoter in colorectal cancer by modulating BMP signalling. EBioMedicine. 2020;56:102800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Nguyen AT, Lee SY, Chin HJ, Le QV, Lee D. Kinase activity of ERBB3 contributes to intestinal organoids growth and intestinal tumorigenesis. Cancer Sci. 2020;111(1):137‐147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Verissimo CS, Overmeer RM, Ponsioen B, Drost J, Mertens S, Verlaan‐Klink I, et al. Targeting mutant RAS in patient‐derived colorectal cancer organoids by combinatorial drug screening. Elife. 2016;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Newey A, Griffiths B, Michaux J, Pak HS, Stevenson BJ, Woolston A, et al. Immunopeptidomics of colorectal cancer organoids reveals a sparse HLA class I neoantigen landscape and no increase in neoantigens with interferon or MEK‐inhibitor treatment. J Immunother Cancer. 2019;7(1):309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Reidy E, Leonard NA, Treacy O, Ryan AE. A 3D View of Colorectal Cancer Models in Predicting Therapeutic Responses and Resistance. Cancers (Basel). 2021;13(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ganesh K, Massague J. Targeting metastatic cancer. Nat Med. 2021;27(1):34‐44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Schnalzger TE, de Groot MH, Zhang C, Mosa MH, Michels BE, Roder J, et al. 3D model for CAR‐mediated cytotoxicity using patient‐derived colorectal cancer organoids. EMBO J. 2019;38(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ooft SN, Weeber F, Dijkstra KK, McLean CM, Kaing S, van Werkhoven E, et al. Patient‐derived organoids can predict response to chemotherapy in metastatic colorectal cancer patients. Sci Transl Med. 2019;11(513). [DOI] [PubMed] [Google Scholar]
- 64. Pasch CA, Favreau PF, Yueh AE, Babiarz CP, Gillette AA, Sharick JT, et al. Patient‐Derived Cancer Organoid Cultures to Predict Sensitivity to Chemotherapy and Radiation. Clin Cancer Res. 2019;25(17):5376‐5387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Duruisseaux M, Esteller M. Lung cancer epigenetics: From knowledge to applications. Semin Cancer Biol. 2018;51:116‐128. [DOI] [PubMed] [Google Scholar]
- 66. Li Z, Qian Y, Li W, Liu L, Yu L, Liu X, et al. Human Lung Adenocarcinoma‐Derived Organoid Models for Drug Screening. iScience. 2020;23(8):101411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kim M, Mun H, Sung CO, Cho EJ, Jeon HJ, Chun SM, et al. Patient‐derived lung cancer organoids as in vitro cancer models for therapeutic screening. Nat Commun. 2019;10(1):3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Choi SY, Cho YH, Kim DS, Ji W, Choi CM, Lee JC, et al. Establishment and Long‐Term Expansion of Small Cell Lung Cancer Patient‐Derived Tumor Organoids. Int J Mol Sci. 2021;22(3):1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Lu T, Cao Y, Zhao P, Shen S, Xi Y. Organoid: a powerful tool to study lung regeneration and disease. Cell Regen. 2021;10(1):21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Wang J, Li X, Chen H. Organoid models in lung regeneration and cancer. Cancer Lett. 2020;475:129‐135. [DOI] [PubMed] [Google Scholar]
- 71. Chen JH, Chu XP, Zhang JT, Nie Q, Tang WF, Su J, et al. Genomic characteristics and drug screening among organoids derived from non‐small cell lung cancer patients. Thorac Cancer. 2020;11(8):2279‐2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Gmeiner WH, Miller LD, Chou JW, Dominijanni A, Mutkus L, Marini F, et al. Dysregulated Pyrimidine Biosynthesis Contributes to 5‐FU Resistance in SCLC Patient‐Derived Organoids but Response to a Novel Polymeric Fluoropyrimidine, CF10. Cancers (Basel). 2020;12(4):788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Li YF, Gao Y, Liang BW, Cao XQ, Sun ZJ, Yu JH, et al. Patient‐derived organoids of non‐small cells lung cancer and their application for drug screening. Neoplasma. 2020;67(2):430‐437. [DOI] [PubMed] [Google Scholar]
- 74. Mazzocchi A, Devarasetty M, Herberg S, Petty WJ, Marini F, Miller L, et al. Pleural Effusion Aspirate for use in 3D Lung Cancer Modeling and Chemotherapy Screening. ACS Biomater Sci Eng. 2019;5(4):1937‐1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Takahashi N, Hoshi H, Higa A, Hiyama G, Tamura H, Ogawa M, et al. An In Vitro System for Evaluating Molecular Targeted Drugs Using Lung Patient‐Derived Tumor Organoids. Cells. 2019;8(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Della Corte CM, Barra G, Ciaramella V, Di Liello R, Vicidomini G, Zappavigna S, et al. Antitumor activity of dual blockade of PD‐L1 and MEK in NSCLC patients derived three‐dimensional spheroid cultures. J Exp Clin Cancer Res. 2019;38(1):253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Dijkstra KK, Monkhorst K, Schipper LJ, Hartemink KJ, Smit EF, Kaing S, et al. Challenges in Establishing Pure Lung Cancer Organoids Limit Their Utility for Personalized Medicine. Cell Rep. 2020;31(5):107588. [DOI] [PubMed] [Google Scholar]
- 78. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7‐34. [DOI] [PubMed] [Google Scholar]
- 79. Driehuis E, van Hoeck A, Moore K, Kolders S, Francies HE, Gulersonmez MC, et al. Pancreatic cancer organoids recapitulate disease and allow personalized drug screening. Proc Natl Acad Sci U S A. 2019;116(52):26580‐26590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Dantes Z, Yen H‐Y, Pfarr N, Winter C, Steiger K, Muckenhuber A, et al. Implementing cell‐free DNA of pancreatic cancer patient–derived organoids for personalized oncology. JCI Insight. 2020;5(15):e137809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Choi SI, Jeon AR, Kim MK, Lee YS, Im JE, Koh JW, et al. Development of Patient‐Derived Preclinical Platform for Metastatic Pancreatic Cancer: PDOX and a Subsequent Organoid Model System Using Percutaneous Biopsy Samples. Front Oncol. 2019;9:875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Seppala TT, Zimmerman JW, Sereni E, Plenker D, Suri R, Rozich N, et al. Patient‐derived Organoid Pharmacotyping is a Clinically Tractable Strategy for Precision Medicine in Pancreatic Cancer. Ann Surg. 2020;272(3):427‐435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Gao M, Lin M, Moffitt RA, Salazar MA, Park J, Vacirca J, et al. Direct therapeutic targeting of immune checkpoint PD‐1 in pancreatic cancer. Br J Cancer. 2019;120(1):88‐96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Ponz‐Sarvise M, Corbo V, Tiriac H, Engle DD, Frese KK, Oni TE, et al. Identification of Resistance Pathways Specific to Malignancy Using Organoid Models of Pancreatic Cancer. Clin Cancer Res. 2019;25(22):6742‐6755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Yoo HJ, You MW, Han DY, Hwang JH, Park SJ. Tumor conspicuity significantly correlates with postoperative recurrence in patients with pancreatic cancer: a retrospective observational study. Cancer Imaging. 2020;20(1):46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Moletta L, Serafini S, Valmasoni M, Pierobon ES, Ponzoni A, Sperti C. Surgery for Recurrent Pancreatic Cancer: Is It Effective? Cancers (Basel). 2019;11(7):991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Braun LM, Lagies S, Klar RFU, Hussung S, Fritsch RM, Kammerer B, et al. Metabolic Profiling of Early and Late Recurrent Pancreatic Ductal Adenocarcinoma Using Patient‐Derived Organoid Cultures. Cancers (Basel). 2020;12(6):1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Huang W, Navarro‐Serer B, Jeong YJ, Chianchiano P, Xia L, Luchini C, et al. Pattern of Invasion in Human Pancreatic Cancer Organoids Is Associated with Loss of SMAD4 and Clinical Outcome. Cancer Res. 2020;80(13):2804‐2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Vaes RDW, van Dijk DPJ, Welbers TTJ, Blok MJ, Aberle MR, Heij L, et al. Generation and initial characterization of novel tumour organoid models to study human pancreatic cancer‐induced cachexia. J Cachexia Sarcopenia Muscle. 2020;11(6):1509‐1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. DeRose YS, Gligorich KM, Wang G, Georgelas A, Bowman P, Courdy SJ, et al. Patient‐derived models of human breast cancer: protocols for in vitro and in vivo applications in tumor biology and translational medicine. Curr Protoc Pharmacol. 2013;Chapter 14:Unit14 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Mazzucchelli S, Piccotti F, Allevi R, Truffi M, Sorrentino L, Russo L, et al. Establishment and Morphological Characterization of Patient‐Derived Organoids from Breast Cancer. Biol Proced Online. 2019;21:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Kim J, Koo BK, Knoblich JA. Human organoids: model systems for human biology and medicine. Nat Rev Mol Cell Biol. 2020;21(10):571‐584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Dekkers JF, Whittle JR, Vaillant F, Chen HR, Dawson C, Liu K, et al. Modeling breast cancer using CRISPR/Cas9‐mediated engineering of human breast organoids. J Natl Cancer Inst. 2020;112(5):540‐544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Nayak B, Balachander GM, Manjunath S, Rangarajan A, Chatterjee K. Tissue mimetic 3D scaffold for breast tumor‐derived organoid culture toward personalized chemotherapy. Colloids Surf B Biointerfaces. 2019;180:334‐343. [DOI] [PubMed] [Google Scholar]
- 95. Sharick JT, Jeffery JJ, Karim MR, Walsh CM, Esbona K, Cook RS, et al. Cellular Metabolic Heterogeneity In Vivo Is Recapitulated in Tumor Organoids. Neoplasia. 2019;21(6):615‐626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Liu K, Newbury PA, Glicksberg BS, Zeng WZD, Paithankar S, Andrechek ER, et al. Evaluating cell lines as models for metastatic breast cancer through integrative analysis of genomic data. Nat Commun. 2019;10(1):2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Li X, Pan B, Song X, Li N, Zhao D, Li M, et al. Breast cancer organoids from a patient with giant papillary carcinoma as a high‐fidelity model. Cancer Cell Int. 2020;20:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Kruger M, Oosterhoff LA, van Wolferen ME, Schiele SA, Walther A, Geijsen N, et al. Cellulose Nanofibril Hydrogel Promotes Hepatic Differentiation of Human Liver Organoids. Adv Healthc Mater. 2020;9(6):e1901658. [DOI] [PubMed] [Google Scholar]
- 99. Qin X, Sufi J, Vlckova P, Kyriakidou P, Acton SE, Li VSW, et al. Cell‐type‐specific signaling networks in heterocellular organoids. Nat Methods. 2020;17(3):335‐342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Cao W, Liu J, Wang L, Li M, Verstegen MMA, Yin Y, et al. Modeling liver cancer and therapy responsiveness using organoids derived from primary mouse liver tumors. Carcinogenesis. 2019;40(1):145‐154. [DOI] [PubMed] [Google Scholar]
- 101. Sun L, Wang Y, Cen J, Ma X, Cui L, Qiu Z, et al. Modelling liver cancer initiation with organoids derived from directly reprogrammed human hepatocytes. Nat Cell Biol. 2019;21(8):1015‐1026. [DOI] [PubMed] [Google Scholar]
- 102. Artegiani B, van Voorthuijsen L, Lindeboom RGH, Seinstra D, Heo I, Tapia P, et al. Probing the Tumor Suppressor Function of BAP1 in CRISPR‐Engineered Human Liver Organoids. Cell Stem Cell. 2019;24(6):927‐943 e926. [DOI] [PubMed] [Google Scholar]
- 103. Vidal SJ, Rodriguez‐Bravo V, Galsky M, Cordon‐Cardo C, Domingo‐Domenech J. Targeting cancer stem cells to suppress acquired chemotherapy resistance. Oncogene. 2014;33(36):4451‐4463. [DOI] [PubMed] [Google Scholar]
- 104. Wang S, Wang Y, Xun X, Zhang C, Xiang X, Cheng Q, et al. Hedgehog signaling promotes sorafenib resistance in hepatocellular carcinoma patient‐derived organoids. J Exp Clin Cancer Res. 2020;39(1):22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Li L, Knutsdottir H, Hui K, Weiss MJ, He J, Philosophe B, et al. Human primary liver cancer organoids reveal intratumor and interpatient drug response heterogeneity. JCI Insight. 2019;4(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Chen K, Sheng J, Ma B, Cao W, Hernanda PY, Liu J, et al. Suppression of Hepatocellular Carcinoma by Mycophenolic Acid in Experimental Models and in Patients. Transplantation. 2019;103(5):929‐937. [DOI] [PubMed] [Google Scholar]
- 107. Lheureux S, Braunstein M, Oza AM. Epithelial ovarian cancer: Evolution of management in the era of precision medicine. CA Cancer J Clin. 2019;69(4):280‐304. [DOI] [PubMed] [Google Scholar]
- 108. Maru Y, Tanaka N, Itami M, Hippo Y. Efficient use of patient‐derived organoids as a preclinical model for gynecologic tumors. Gynecol Oncol. 2019;154(1):189‐198. [DOI] [PubMed] [Google Scholar]
- 109. Hoffmann K, Berger H, Kulbe H, Thillainadarasan S, Mollenkopf HJ, Zemojtel T, et al. Stable expansion of high‐grade serous ovarian cancer organoids requires a low‐Wnt environment. EMBO J. 2020;39(6):e104013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Nanki Y, Chiyoda T, Hirasawa A, Ookubo A, Itoh M, Ueno M, et al. Patient‐derived ovarian cancer organoids capture the genomic profiles of primary tumours applicable for drug sensitivity and resistance testing. Sci Rep. 2020;10(1):12581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. de Witte CJ, Espejo Valle‐Inclan J, Hami N, Lohmussaar K, Kopper O, Vreuls CPH, et al. Patient‐Derived Ovarian Cancer Organoids Mimic Clinical Response and Exhibit Heterogeneous Inter‐ and Intrapatient Drug Responses. Cell Rep. 2020;31(11):107762. [DOI] [PubMed] [Google Scholar]
- 112. Kopper O, de Witte CJ, Lohmussaar K, Valle‐Inclan JE, Hami N, Kester L, et al. An organoid platform for ovarian cancer captures intra‐ and interpatient heterogeneity. Nat Med. 2019;25(5):838‐849. [DOI] [PubMed] [Google Scholar]
- 113. Nero C, Vizzielli G, Lorusso D, Cesari E, Daniele G, Loverro M, et al. Patient‐derived organoids and high grade serous ovarian cancer: from disease modeling to personalized medicine. J Exp Clin Cancer Res. 2021;40(1):116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Huang HM, Li HX. Tumor heterogeneity and the potential role of liquid biopsy in bladder cancer. Cancer Commun (Lond). 2021;41(2):91‐108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Mullenders J, de Jongh E, Brousali A, Roosen M, Blom JPA, Begthel H, et al. Mouse and human urothelial cancer organoids: A tool for bladder cancer research. Proc Natl Acad Sci U S A. 2019;116(10):4567‐4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Lee SH, Hu W, Matulay JT, Silva MV, Owczarek TB, Kim K, et al. Tumor Evolution and Drug Response in Patient‐Derived Organoid Models of Bladder Cancer. Cell. 2018;173(2):515‐528 e517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Kong J, Lee H, Kim D, Han SK, Ha D, Shin K, et al. Network‐based machine learning in colorectal and bladder organoid models predicts anti‐cancer drug efficacy in patients. Nat Commun. 2020;11(1):5485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Ge R, Wang Z, Montironi R, Jiang Z, Cheng M, Santoni M, et al. Epigenetic modulations and lineage plasticity in advanced prostate cancer. Ann Oncol. 2020;31(4):470‐479. [DOI] [PubMed] [Google Scholar]
- 119. Gao D, Vela I, Sboner A, Iaquinta PJ, Karthaus WR, Gopalan A, et al. Organoid cultures derived from patients with advanced prostate cancer. Cell. 2014;159(1):176‐187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Puca L, Bareja R, Prandi D, Shaw R, Benelli M, Karthaus WR, et al. Patient derived organoids to model rare prostate cancer phenotypes. Nat Commun. 2018;9(1):2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Karkampouna S, La Manna F, Benjak A, Kiener M, De Menna M, Zoni E, et al. Patient‐derived xenografts and organoids model therapy response in prostate cancer. Nat Commun. 2021;12(1):1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Servant R, Garioni M, Vlajnic T, Blind M, Pueschel H, Muller DC, et al. Prostate cancer patient‐derived organoids: detailed outcome from a prospective cohort of 81 clinical specimens. J Pathol. 2021;254(5):543‐555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Driehuis E, Kretzschmar K, Clevers H. Establishment of patient‐derived cancer organoids for drug‐screening applications. Nat Protoc. 2020;15(10):3380‐3409. [DOI] [PubMed] [Google Scholar]
- 124. Tanaka N, Osman AA, Takahashi Y, Lindemann A, Patel AA, Zhao M, et al. Head and neck cancer organoids established by modification of the CTOS method can be used to predict in vivo drug sensitivity. Oral Oncol. 2018;87:49‐57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Driehuis E, Spelier S, Beltran Hernandez I, de Bree R, S MW, Clevers H, et al. Patient‐Derived Head and Neck Cancer Organoids Recapitulate EGFR Expression Levels of Respective Tissues and Are Responsive to EGFR‐Targeted Photodynamic Therapy. J Clin Med. 2019;8(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Bartfeld S, Bayram T, van de Wetering M, Huch M, Begthel H, Kujala P, et al. In vitro expansion of human gastric epithelial stem cells and their responses to bacterial infection. Gastroenterology. 2015;148(1):126‐136 e126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Seidlitz T, Merker SR, Rothe A, Zakrzewski F, von Neubeck C, Grutzmann K, et al. Human gastric cancer modelling using organoids. Gut. 2019;68(2):207‐217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Steele NG, Chakrabarti J, Wang J, Biesiada J, Holokai L, Chang J, et al. An Organoid‐Based Preclinical Model of Human Gastric Cancer. Cell Mol Gastroenterol Hepatol. 2019;7(1):161‐184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Jacob F, Ming GL, Song H. Generation and biobanking of patient‐derived glioblastoma organoids and their application in CAR T cell testing. Nat Protoc. 2020;15(12):4000‐4033. [DOI] [PubMed] [Google Scholar]
- 130. Jacob F, Salinas RD, Zhang DY, Nguyen PTT, Schnoll JG, Wong SZH, et al. A Patient‐Derived Glioblastoma Organoid Model and Biobank Recapitulates Inter‐ and Intra‐tumoral Heterogeneity. Cell. 2020;180(1):188‐204 e122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Wensink GE, Elias SG, Mullenders J, Koopman M, Boj SF, Kranenburg OW, et al. Patient‐derived organoids as a predictive biomarker for treatment response in cancer patients. NPJ Precis Oncol. 2021;5(1):30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Homicsko K. Organoid technology and applications in cancer immunotherapy and precision medicine. Curr Opin Biotechnol. 2020;65:242‐247. [DOI] [PubMed] [Google Scholar]
- 133. Worsdorfer P, I T, Asahina I, Sumita Y, Ergun S. Do not keep it simple: recent advances in the generation of complex organoids. J Neural Transm (Vienna). 2020;127(11):1569‐1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Wang Z, He X, Qiao H, Chen P. Global Trends of Organoid and Organ‐On‐a‐Chip in the Past Decade: A Bibliometric and Comparative Study. Tissue Eng Part A. 2020;26(11‐12):656‐671. [DOI] [PubMed] [Google Scholar]
- 135. Park SE, Georgescu A, Huh D. Organoids‐on‐a‐chip. Science. 2019;364(6444):960‐965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Ma C, Peng Y, Li H, Chen W. Organ‐on‐a‐Chip: A New Paradigm for Drug Development. Trends Pharmacol Sci. 2021;42(2):119‐133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Demers CJ, Soundararajan P, Chennampally P, Cox GA, Briscoe J, Collins SD, et al. Development‐on‐chip: in vitro neural tube patterning with a microfluidic device. Development. 2016;143(11):1884‐1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Wang Y, Wang L, Zhu Y, Qin J. Human brain organoid‐on‐a‐chip to model prenatal nicotine exposure. Lab Chip. 2018;18(6):851‐860. [DOI] [PubMed] [Google Scholar]
- 139. Shirure VS, Bi Y, Curtis MB, Lezia A, Goedegebuure MM, Goedegebuure SP, et al. Tumor‐on‐a‐chip platform to investigate progression and drug sensitivity in cell lines and patient‐derived organoids. Lab Chip. 2018;18(23):3687‐3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Jin Y, Kim J, Lee JS, Min S, Kim S, Ahn D‐H, et al. Vascularized Liver Organoids Generated Using Induced Hepatic Tissue and Dynamic Liver‐Specific Microenvironment as a Drug Testing Platform. Adv Funct Mater. 2018;28(37):1801954. [Google Scholar]
- 141. Murphy SV, De Coppi P, Atala A. Opportunities and challenges of translational 3D bioprinting. Nat Biomed Eng. 2020;4(4):370‐380. [DOI] [PubMed] [Google Scholar]
- 142. Dabbagh SR, Sarabi MR, Rahbarghazi R, Sokullu E, Yetisen AK, Tasoglu S. 3D‐printed microneedles in biomedical applications. iScience. 2021;24(1):102012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Peng W, Unutmaz D, Ozbolat IT. Bioprinting towards Physiologically Relevant Tissue Models for Pharmaceutics. Trends Biotechnol. 2016;34(9):722‐732. [DOI] [PubMed] [Google Scholar]
- 144. Lee H, Cho DW. One‐step fabrication of an organ‐on‐a‐chip with spatial heterogeneity using a 3D bioprinting technology. Lab Chip. 2016;16(14):2618‐2625. [DOI] [PubMed] [Google Scholar]
- 145. Correia Carreira S, Begum R, Perriman AW. 3D Bioprinting: The Emergence of Programmable Biodesign. Adv Healthc Mater. 2020;9(15):e1900554. [DOI] [PubMed] [Google Scholar]
- 146. Datta P, Dey M, Ataie Z, Unutmaz D, Ozbolat IT. 3D bioprinting for reconstituting the cancer microenvironment. NPJ Precis Oncol. 2020;4:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Mollica PA, Booth‐Creech EN, Reid JA, Zamponi M, Sullivan SM, Palmer XL, et al. 3D bioprinted mammary organoids and tumoroids in human mammary derived ECM hydrogels. Acta Biomater. 2019;95:201‐213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Sandercock AM, Rust S, Guillard S, Sachsenmeier KF, Holoweckyj N, Hay C, et al. Identification of anti‐tumour biologics using primary tumour models, 3‐D phenotypic screening and image‐based multi‐parametric profiling. Mol Cancer. 2015;14:147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Reid JA, Palmer XL, Mollica PA, Northam N, Sachs PC, Bruno RD. A 3D bioprinter platform for mechanistic analysis of tumoroids and chimeric mammary organoids. Sci Rep. 2019;9(1):7466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Lee A, Hudson AR, Shiwarski DJ, Tashman JW, Hinton TJ, Yerneni S, et al. 3D bioprinting of collagen to rebuild components of the human heart. Science. 2019;365(6452):482‐487. [DOI] [PubMed] [Google Scholar]
- 151. Maloney E, Clark C, Sivakumar H, Yoo K, Aleman J, Rajan SAP, et al. Immersion Bioprinting of Tumor Organoids in Multi‐Well Plates for Increasing Chemotherapy Screening Throughput. Micromachines (Basel). 2020;11(2):208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Artegiani B, Hendriks D, Beumer J, Kok R, Zheng X, Joore I, et al. Fast and efficient generation of knock‐in human organoids using homology‐independent CRISPR‐Cas9 precision genome editing. Nat Cell Biol. 2020;22(3):321‐331. [DOI] [PubMed] [Google Scholar]
- 153. Hendriks D, Artegiani B, Hu H, Chuva de Sousa Lopes S, Clevers H. Establishment of human fetal hepatocyte organoids and CRISPR‐Cas9‐based gene knockin and knockout in organoid cultures from human liver. Nat Protoc. 2021;16(1):182‐217. [DOI] [PubMed] [Google Scholar]
- 154. Waldman AD, Fritz JM, Lenardo MJ. A guide to cancer immunotherapy: from T cell basic science to clinical practice. Nat Rev Immunol. 2020;20(11):651‐668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Wang Y, Wang M, Wu HX, Xu RH. Advancing to the era of cancer immunotherapy. Cancer Commun (Lond). 2021;41(9):803‐829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Yuki K, Cheng N, Nakano M, Kuo CJ. Organoid Models of Tumor Immunology. Trends Immunol. 2020;41(8):652‐664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Scognamiglio G, De Chiara A, Parafioriti A, Armiraglio E, Fazioli F, Gallo M, et al. Patient‐derived organoids as a potential model to predict response to PD‐1/PD‐L1 checkpoint inhibitors. Br J Cancer. 2019;121(11):979‐982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Holokai L, Chakrabarti J, Lundy J, Croagh D, Adhikary P, Richards SS, et al. Murine‐ and Human‐Derived Autologous Organoid/Immune Cell Co‐Cultures as Pre‐Clinical Models of Pancreatic Ductal Adenocarcinoma. Cancers (Basel). 2020;12(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Nakamura H, Sugano M, Miyashita T, Hashimoto H, Ochiai A, Suzuki K, et al. Organoid culture containing cancer cells and stromal cells reveals that podoplanin‐positive cancer‐associated fibroblasts enhance proliferation of lung cancer cells. Lung Cancer. 2019;134:100‐107. [DOI] [PubMed] [Google Scholar]
- 160. Xia X, Li F, He J, Aji R, Gao D. Organoid technology in cancer precision medicine. Cancer Lett. 2019;457:20‐27. [DOI] [PubMed] [Google Scholar]
- 161. Yang L, Yang S, Li X, Li B, Li Y, Zhang X, et al. Tumor organoids: From inception to future in cancer research. Cancer Lett. 2019;454:120‐133. [DOI] [PubMed] [Google Scholar]
- 162. Bock C, Boutros M, Camp JG, Clarke L, Clevers H, Knoblich JA, et al. The Organoid Cell Atlas. Nat Biotechnol. 2021;39(1):13‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Hai J, Zhang H, Zhou J, Wu Z, Chen T, Papadopoulos E, et al. Generation of Genetically Engineered Mouse Lung Organoid Models for Squamous Cell Lung Cancers Allows for the Study of Combinatorial Immunotherapy. Clin Cancer Res. 2020;26(13):3431‐3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Gronholm M, Feodoroff M, Antignani G, Martins B, Hamdan F, Cerullo V. Patient‐Derived Organoids for Precision Cancer Immunotherapy. Cancer Res. 2021;81(12):3149‐3155. [DOI] [PubMed] [Google Scholar]
- 165. Granat LM, Kambhampati O, Klosek S, Niedzwecki B, Parsa K, Zhang D. The promises and challenges of patient‐derived tumor organoids in drug development and precision oncology. Animal Model Exp Med. 2019;2(3):150‐161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Huch M, Knoblich JA, Lutolf MP, Martinez‐Arias A. The hope and the hype of organoid research. Development. 2017;144(6):938‐941. [DOI] [PubMed] [Google Scholar]
- 167. Dutta D, Clevers H. Organoid culture systems to study host‐pathogen interactions. Curr Opin Immunol. 2017;48:15‐22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Velasco S, Kedaigle AJ, Simmons SK, Nash A, Rocha M, Quadrato G, et al. Individual brain organoids reproducibly form cell diversity of the human cerebral cortex. Nature. 2019;570(7762):523‐527. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information
Data Availability Statement
Please contact the corresponding author for data requests.


