Abbreviations
- bp
Base pair
- CCND1
Cyclin D1
- FLI1
Friend leukemia integration 1 transcription factor
- hTERT
Human telomerase reverse transcriptase
- IgG
Immunoglobulin G
- MYC
MYC proto‐oncogene, BHLH transcription factor
- p27
Cyclin‐dependent kinase inhibitor 1B
- p53
Tumor protein p53
- Sp1
Sp1 transcription factor
- TERT
Telomerase reverse transcriptase
- TF
Transcription factor
- TFE3
Transcription factor binding to IGHM enhancer 3
- TSS
Transcription start site
- ZNF556
Zinc finger protein 556
Dear Editor,
Telomeres are located at the ends of chromosomes and are essential for chromosome stability and replication. They progressively get shorter with cell division, eventually resulting in cell proliferation arrest. The process is considered a barrier to tumor occurrence [1, 2]. Abnormally activated telomerase with constitutive telomerase reverse transcriptase (TERT) expression is a hallmark of almost all human tumors [3]. Some genomic alterations in the human TERT (hTERT) promoter are associated with activation of hTERT transcription in various cancer types [4].
The regulation of TERT expression can significantly affect tumor occurrence and progression. While lacking TATA and CAAT boxes, the hTERT promoter region contains an E‐box which interacts with the transcription factor (TF) MYC, and a GC‐box which is targeted by TF Sp1 [5]. The hTERT promoter also interacts with TFs as well as upstream stimulatory factors [6]. Furthermore, friend leukemia integration 1 (FLI1) specifically binds to novel transcription binding sites created by hTERT promoter mutations that frequently occur in tumors, thereby enhancing telomerase activity [7]. Thus, identification of other upstream regulators of hTERT could enrich our knowledge on the regulatory hTERT network involved in tumor biology.
To understand the molecular mechanisms underlying hTERT promoter activity, we screened, with the hTERT promoter sequence, a protein microarray made of a candidate subset of TFs selected from a larger microarray of 667 TFs [7, 8]. Two TFs – transcription factor binding to IGHM enhancer 3 (TFE3) and zinc finger protein 556 (ZNF556) – exhibited particularly strong binding to the hTERT promoter (Figure 1A). Since TFE3 is known to be associated with the occurrence and progression of several tumors [9], we focused our analyses on this molecule, leaving ZNF556 for future studies.
FIGURE 1.
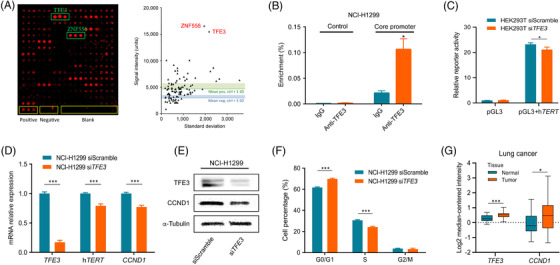
TFE3 is a regulator of hTERT. (A) Left panel: Screening of the binding of the hTERT promoter sequence to a protein microarray made of 96 TFs; a typical result is shown; pos. – positive control; neg. – negative control; blank – empty spots. Right panel: Quantification of the results; mean intensities, and the related standard deviations (SD) of the signals are given next to results of individual TFs. (B) The binding site prediction was confirmed in ChIP experiments with anti‐TFE3 antibody on chromatin isolated from NCI‐H1299 cells. As a negative control, rabbit IgGs were used instead of the anti‐TFE3 antibody. Next to the predicted binding sites at ‐170 bp and +29 bp, an unrelated DNA at position ‐5329 bp was used as a control. The degree of enrichment of the respective DNA segment was quantified by qPCR. (C) hTERT promoter activity is measured by a luciferase reporter assay in vector pGL3. HEK293T cells were subjected to knockdowns with an siRNA targeting TFE3 (siTFE3) and a control siRNA with scrambled, unspecific sequence. The relative fluorescence is shown that was produced by the luciferase activity. (D) siRNA‐mediated knockdown of TFE3 down‐regulates hTERT and CCND1 expression in NCI‐H1299. The effect of the TFE3 knockdown and the consequential reduction of hTERT and CCND1 transcript levels are shown. (E) The effect is also shown at the protein level by Western blot, comparing results obtained with an siRNA construct with scrambled sequence and a molecule targeting TFE3 (siTFE3). (F) Cell cycle analysis with NCI‐H1299 cells subjected to TFE3 knockdown or treated with a scrambled siRNA sequence. Cell percentages are shown. (G) Expression of TFE3 and CCND1 in lung cancer tissues in comparison to their levels in healthy control samples. The data were obtained from the Oncomine expression database. TFE3 and CCND1 were found up‐regulated in tumors in comparison to the respective healthy tissues (normal). * P < 0.05; ** P < 0.01; *** P < 0.001.
Abbreviations: TFE3, transcription factor binding to IGHM enhancer 3; hTERT, human telomerase reverse transcriptase; CCND1, cyclin D1; bp, base pair; siRNA, small interfering RNA; IgG, immunoglobulin G; qPCR, quantitative polymerase chain reaction; ChIP, chromatin immunoprecipitation
In silico prediction of binding sites in the hTERT promoter region indicated two TFE3 binding sites (Supplementary Figure S1A, B), one at ‐170 bp upstream of the transcription start site (TSS) on the positive strand, and the other one at +29 bp downstream on the negative strand Supplementary Figure S2A). We performed chromatin immunoprecipitation with anti‐TFE3 antibody to confirm TFE3 binding. The core binding sequence was significantly enriched compared to the immunoglobulin G (IgG) control; a non‐promoter region acting as negative control produced an even weaker signal (Figure 1B and Supplementary Figure S2B).
To verify the role of TFE3 in hTERT activation, we measured the promoter activity with a luciferase reporter assay in HEK293T cells. After knocking down TFE3 (Supplementary Figure S2C), luciferase activity was decreased by about 10% (Figure 1C), indicating that TFE3 positively regulates the hTERT promoter activity by direct interaction. The relatively small effect of TFE3 knockdown is not surprising since several TFs have binding sites in the hTERT promoter (Supplementary Figure S2A). Nevertheless, there is a substantial contribution of TFE3 to the overall regulation of hTERT transcription.
To explore the effect of TFE3 on hTERT expression, we measured hTERT transcript levels after TFE3 knockdown in the lung cancer cell line (NCI‐H1299) and pancreatic cancer cell line (MiaPaCa‐2). In NCI‐H1299, TFE3 mRNA and protein levels were reduced significantly (Figure 1D‐E). The effect was even more pronounced in MiaPaCa‐2 cell line (Supplementary Figure S2D‐E). Concomitantly to TFE3 knockdown, hTERT mRNA levels decreased by 20% in NCI‐H1299 (Figure 1D) and dropped even more drastically in MiaPaCa‐2 (Supplementary Figure S2D). In both cell lines, the hTERT protein level was also reduced (Supplementary Figure S3A). The difference between the cells could partly be the result of different knockdown efficacy. Additionally, MiaPaCa‐2 has already a relatively lower endogenous hTERT expression. To confirm that TFE3 binds specifically to the promoter region and exerts its effect on hTERT, we simulated site‐directed mutagenesis in silico. A change of one highly conserved base of the TFE3 binding motif led to the loss of binding (data not shown). These results imply that TFE3 positively regulates the expression of hTERT.
Besides analyzing the changes to hTERT, we looked at the expression of cell cycle‐associated marker cyclin D1 (CCND1), since several regulators of hTERT activity have been reported to influence the cell cycle. Knockdown of TFE3 affected its binding partner hTERT and reduced CCND1 expression (Figure 1D‐E and Supplementary Figure S2D‐E), suggesting that TFE3 is involved in cell cycle regulation. Knockdown of TFE3 also elevated cyclin‐dependent kinase inhibitor 1B (p27) protein levels, an upstream regulator of CCND1, indicating that p27 is involved in stopping or slowing down of the cell division. There was no change in tumor protein p53 levels (Supplementary Figure S3A). To rule out that this effect was due to mutations in the hTERT promoter, we performed a dual luciferase reporter assay with wildtype hTERT promoter and a version with hotspot mutation G250A. There was no change in promoter activity, emphasizing that the effect was in fact due to TFE3 binding (Supplementary Figure S3B).
For confirmation of the functional consequence indicated by CCND1, we performed cell cycle analyses after TFE3 knockdown. In NCI‐H1299, the cell percentage in the G0/G1 phase increased by 8%, decreased by 6% in the S phase and did not change significantly in the G2/M phase (Figure 1F). MiaPaCa‐2 cell percentages increased by 9% in the G0/G1 phase, decreased by 5% in the S phase, and 3% in the G2/M phase (Supplementary Figure S2F) inferring that TFE3 moderately promotes cell cycle progression. In order to show that TFE3 promotes cancer development also through other hTERT related functions, we performed apoptosis and ROS assays. Apoptosis was increased upon TFE3 knockdown and no difference in ROS activity in both the cell lines was observed (Supplementary Figure S3C‐D).
To confirm that the relationship of TFE3 and CCND1 could also be observed in human tumor tissues rather than merely in cell lines, the transcript profiles of the Oncomine data repository were used. Both TFE3 and CCND1 were significantly up‐regulated not only in lung and pancreatic cancer tissues (Figure 1G and Supplementary Figure S2G), but also in kidney, breast, colorectal and gastric tumors compared to respective healthy control samples (Supplementary Figure S4) indicating a common feature across tumors. Due to the low expression level of hTERT, TFE3 and hTERT showed no or only weak correlation in pancreatic and lung cancers. TFE3 and CCND1 were weakly correlated in 4/6 datasets (data not shown). Given the known interactions and large number of predicted TF binding sites in hTERT, it is likely that several TFs orchestrate the regulation of hTERT, repressing or activating the gene in a concerted manner, which could be further affected by genetic and epigenetic factors.
Our study shows that TFE3 may act as a diagnostic marker in solid‐pseudo papillary neoplasms and granular cell tumors. Also, TFE3‐associated gene fusion and translocation is relatively common in some tumors and hTERT promoter rearrangements were detected in renal cell carcinoma subtypes. However, the effect of TFE3 on the hTERT promoter activity had not been reported before. TFE3 regulates hTERT activity positively and promotes cell cycle progression. As a functional partner of the E2F3 transcription factor, TFE3 is essential to DNA replication, which is another critical factor related to cell cycle control [10]. In conclusion, TFE3 directly triggers hTERT expression and is a newly identified part of the overall complex regulation of telomere biology, which is critical to tumor survival.
DECLARATIONS
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable
CONSENT FOR PUBLICATION
Not applicable
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest.
DATA AVAILABILITY STATEMENT
The datasets generated and/or analyzed during the current study are available from the corresponding author on reasonable request.
FUNDING
This research did not receive any specific grant from funding agencies in the public, commercial, or not‐for‐profit sectors.
AUTHOR'S CONTRIBUTIONS
BPM, JDH, ORB designed the study. BPM, CYZ, NS, LB; KH conducted the experiments and collected the data. BPM, ORB analyzed the data and interpreted the results. BPM, CYZ, JDH, ORB wrote the manuscript. All authors read and approved the final manuscript.
Supporting information
Supporting information
Supporting information
Supporting information
Supporting information
Supporting information
ACKNOWLEDGEMENTS
We thank Dr. Jing Yang (German Cancer Research Centre, DKFZ, Heidelberg) for her technical support in bioinformatic analysis and Cuncai Guo (University Hospital Heidelberg, Heidelberg) for technical support in Western blot experiments.
Open access funding enabled and organized by Projekt DEAL.
Contributor Information
Beiping Miao, Email: b.miao@kitz-heidelberg.de.
Obul Reddy Bandapalli, Email: bandapalli@gmail.com.
REFERENCES
- 1. Maciejowski J, de Lange T. Telomeres in cancer: tumour suppression and genome instability. Nat Rev Mol Cell Biol. 2017;18(3):175–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Shay JW. Role of Telomeres and Telomerase in Aging and Cancer. Cancer Discov. 2016;6(6):584–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Shay JW, Wright WE. Telomeres and telomerase: three decades of progress. Nat Rev Genet. 2019;20(5):299–309. [DOI] [PubMed] [Google Scholar]
- 4. Barthel FP, Wei W, Tang M, Martinez‐Ledesma E, Hu X, Amin SB, et al. Systematic analysis of telomere length and somatic alterations in 31 cancer types. Nat Genet. 2017;49(3):349–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Takakura M, Kyo S, Kanaya T, Hirano H, Takeda J, Yutsudo M, et al. Cloning of human telomerase catalytic subunit (hTERT) gene promoter and identification of proximal core promoter sequences essential for transcriptional activation in immortalized and cancer cells. Cancer Res. 1999;59(3):551–7. [PubMed] [Google Scholar]
- 6. Anderson CJ, Hoare SF, Ashcroft M, Bilsland AE, Keith WN. Hypoxic regulation of telomerase gene expression by transcriptional and post‐transcriptional mechanisms. Oncogene. 2006;25(1):61–9. [DOI] [PubMed] [Google Scholar]
- 7. Miao B, Bauer AS, Hufnagel K, Wu Y, Trajkovic‐Arsic M, Pirona AC, et al. The transcription factor FLI1 promotes cancer progression by affecting cell cycle regulation. Int J Cancer. 2020;147(1):189–201. [DOI] [PubMed] [Google Scholar]
- 8. Syafrizayanti, Lueong SS , Di C, Schaefer JV, Plückthun A, Hoheisel JD. Personalised proteome analysis by means of protein microarrays made from individual patient samples. Sci Rep. 2017;7:39756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jiang Y, Xie J, Wang B, Mu Y, Liu P. TFE3 is a diagnostic marker for solid pseudopapillary neoplasms of the pancreas. Hum Pathol. 2018;81:166–75. [DOI] [PubMed] [Google Scholar]
- 10. Giangrande PH, Hallstrom TC, Tunyaplin C, Calame K, Nevins JR. Identification of E‐box factor TFE3 as a functional partner for the E2F3 transcription factor. Mol Cell Biol. 2003;23(11):3707–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information
Supporting information
Supporting information
Supporting information
Supporting information
Data Availability Statement
The datasets generated and/or analyzed during the current study are available from the corresponding author on reasonable request.


