Abstract
We recently analyzed 11 helicobacter isolates cultured from diarrhea patients in Canada. These isolates had been characterized biochemically by restriction fragment length polymorphism (RFLP; AluI, HhaI) analysis and by fatty-acid analysis as Helicobacter pullorum. However, four of the isolates differed biochemically from H. pullorum by their inability to hydrolyze indoxyl acetate and their resistance to nalidixic acid. Using complete 16S rRNA analysis, we determined that these four strains clustered near H. pullorum but had a sequence difference of 2% and therefore represent a novel helicobacter, Helicobacter canadensis. This novel helicobacter could also be distinguished from H. pullorum by RFLP analysis using ApaLI. The number of novel Helicobacter spp. associated with gastrointestinal disease in humans and animals is rapidly increasing. There are now six Helicobacter spp. isolated from diarrheic humans, the other five being H. pullorum, H. canis, “H. rappini,” H. fennelliae, and H. cinaedi. This finding highlights the importance of careful molecular analysis in addition to standard biochemical tests in identifying the increasing number of Helicobacter spp. isolated from humans and animals.
Enterohepatic Helicobacter spp. are increasingly recognized as microbial pathogens in humans and animals (6). Helicobacter pullorum was initially isolated from the feces and diseased livers of chickens (14). H. pullorum is now known to colonize many chicken flocks and is commonly isolated from the cecal contents and carcasses of slaughtered chickens (1, 2).
This microaerobe has been linked to a number of cases of gastrointestinal disease in humans (7, 14, 15; A. P. Burnens, J. Stanley, and J. Nicolet, Letter, Lancet 344:1569–1570, 1994). One case in particular was of interest because the organism was isolated from a male with chronic diarrhea who was also suspected of having liver disease based on elevated liver enzymes and an abdominal ultrasound examination (Burnens et al., letter). Reports subsequent to the original description of H. pullorum have cited the difficulty in correctly distinguishing H. pullorum from other enteric helicobacters as well as campylobacters (14, 15). H. pullorum, though clearly identifiable as a helicobacter by 16S rRNA analysis and biochemical features, differs from most other Helicobacter spp. in lacking sheathed flagella. H. pullorum is inert in most biochemical assays, and like most other enteric helicobacters isolated from humans, with the exception of “H. rappini,” H. pullorum is urease negative. However, this biochemical feature does not distinguish it from Campylobacter coli, so the inability of H. pullorum to hydrolyze indoxyl acetate was relied upon to differentiate between these two microaerophiles. Furthermore, H. pullorum had the same biochemical features as Campylobacter lari except its intolerance to 2% NaCl and its sensitivity to nalidixic acid.
It was of interest to us that several strains of H. pullorum previously confirmed using purported H. pullorum 16S rRNA-specific primers, fatty acid analysis, and restriction enzyme analysis with AluI and HhaI (7, 14) were indoxyl acetate positive, a biochemical feature not previously noted in H. pullorum (7). The purpose of this report is to describe indoxyl acetate-positive “H. pullorum” strains isolated from diarrheic humans based on 16S rRNA analysis and biochemical profiling as a novel Helicobacter species, Helicobacter canadensis.
MATERIALS AND METHODS
Case histories.
Four strains of indoxyl acetate-positive H. pullorum were isolated from diarrheic humans from 1994 to 1999. Unfortunately, any clinical information other than the patients' residency in Canada is not available. Two of the diarrheic individuals were 31-year-old females. The two males were 17 and 27 years old.
Biochemical and phenotypic characterization.
The four strains were collected and initially characterized by the National Laboratory for Enteric Pathogens, Laboratory Center for Disease Control (LCDC), Health Canada, as H. pullorum based on morphology, biochemical characteristics, and H. pullorum-specific PCR analysis (7, 9). The four indoxyl acetate-positive “H. pullorum” isolates (NLEP-16143, NLEP-16767, NLEP-17813, and NLEP-99-3017) shipped to the Massachusetts Institute of Technology were subjected to a detailed biochemical characterization as previously described by Shen et al. (13). The isolates were examined for catalase, oxidase, and urease activities. With the RapID NH System (Innovative Diagnostic Systems Inc., Norcross, Ga.), the isolates were examined for the presence of alkaline phosphatase hydrolysis and γ-glutamyl transpeptidase and for the hydrolysis of urea. Indoxyl acetate hydrolysis was determined by using indoxyl acetate discs (Remel, Lenexa, Kans.). The isolates were also tested for their ability to reduce nitrate by using nitrate broth (GIBCO Laboratories, Grand Island, N.Y.) (5). Growth at 37 and 42°C under microaerobic conditions was examined at 3- to 4-day intervals for up to 2 weeks. Susceptibility to cephalothin (30 μg/disc) and nalidixic acid (30 μg/disc) was determined by culturing the organisms in the presence of discs impregnated with the antibiotic in question (Difco Laboratories). The bacteria were also Gram stained and examined for motility in sterile phosphate-buffered saline by phase-contrast microscopy.
Electron microscopy.
Isolate NLEP-16143 was examined by electron microscopy. Cells grown on blood agar plates were centrifuged and gently suspended in 10 mM Tris-HCl buffer (pH 7.4) at a concentration of about 108 cells per ml. Samples were negatively stained with 1% (wt/vol) phosphotungstic acid (pH 6.5) for 20 to 30 s. The specimens were examined with a JEOL model JEM-1200EX transmission electron microscope.
DNA extraction.
The High Pure PCR template preparation kit (Roche Molecular Biochemicals, Indianapolis, Ind.) was used to extract DNA from bacterial pellets as outlined by the manufacturer and previously described (12).
Helicobacter-specific PCR amplification.
A 16S rRNA-based primer set that is genus specific for all Helicobacter spp. was used for PCR amplification. Primer C97 (5′GCT ATG ACG GGT ATC C) and primer C05 (5′ACT TCA CCC CAG TCG CTG) produced a 1,200-bp product. Amplification PCRs were performed with a thermal cycler and an Expand high-fidelity PCR system (Roche). The reaction mixture (100 μl) contained 1× polymerase buffer (supplied by the manufacturer but supplemented with 1 M MgCl2 to a final concentration of 2.25 mM), a 0.5 μM concentration of each of the two primers, a 200 μM concentration of each deoxyribonucleotide, and bovine serum albumin (200 μg/ml). Samples were heated at 94°C for 4 min, briefly centrifuged, and cooled to 58°C. Polymerase (2.5 U) was then added, and this was followed by an overlay of 100 μl of mineral oil. Amplification was achieved by denaturation at 94°C for 1 min, annealing at 58°C for 2 min, and elongation at 72°C for 3 min for 35 cycles. A 15-μl portion of the sample was then electrophoresed through a 6% Visigel separation matrix (Stratagene, La Jolla, Calif.).
16S rRNA gene sequencing.
The sequences of the 16S rRNA genes of the four strains NLEP-16143, -16767, -17813, and -99-3017 were determined. 16S rRNA gene amplification, sequencing, and data analysis were performed as previously described by Dewhirst et al. (4). In brief, PCR-purified DNA was sequenced using an ABI Prism cycle sequencing kit (BigDye terminator cycle sequencing ready reaction kit with AmpliTaq DNA polymerase, FS; PE Applied Biosystems). The sequencing primers and methods were as listed previously (4). 16S rRNA sequence data were entered and aligned using the program RNA, which is set for data entry, editing, sequencing alignment, secondary structure comparison, similarity matrix generation, and dendrogram construction and is written in Microsoft QuickBasic for use with PCs (10).
The database used contains approximately 400 Helicobacter, Wolinella, Arcobacter, and Campylobacter sequences and more than 1,000 sequences for other bacteria. Similarity matrices were constructed from the aligned sequences by using only those base positions for which data were available for 90% of the strains and were corrected for multiple base changes by the method of Jukes and Cantor (8). Phylogenetic trees were constructed by the neighbor-joining method (11).
Restriction fragment length polymorphism (RFLP) of the 16S rRNA gene.
PCR-amplified DNA (20 μl) was digested with 10 U of enzyme in the appropriate buffer recommended by the enzyme manufacturer at 37°C for 3 h. Restriction patterns were compared after the digested PCR products were separated on a 6% Visigel separation matrix. Restriction enzymes HhaI, AluI, and ApaLI were used for digestion.
Nucleotide sequence accession number.
The 16S rRNA sequence for NLEP-16143 has been deposited in GenBank under accession no. AF262037.
RESULTS
Biochemical characterization.
All four strains grew as a spreading film on blood agar at 37 and 42°C and were oxidase and catalase positive and urease, alkaline phosphatase, and γ-glutamyl transpeptidase negative. Unlike other reported H. pullorum strains, the organisms were indoxyl acetate positive and nalidixic acid (30 mg) and cephalothin (30 mg) resistant. Nitrate reduction was variable (two of four) (Table 1).
TABLE 1.
Characteristics which differentiate H. canadensis strains from other Helicobacter speciesa
| Taxon | Catalase production | Nitrate reduction | Alkaline phosphatase hydrolysis | Urease activity | Indoxyl acetate hydrolysis | γ-Glutamyl transpeptidase activity | Growth at 42°C | Growth with 1% glycine | Susceptibility to:
|
Periplasmic fibers | No. of flagella | Distribution of flagella | G+C content (mol%) | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Nalidixic acid (30-μg disc) | Cephalothin (30-μg disc) | |||||||||||||
| H. canadensis | + (4/4)b | + (2/4) | − (4/4) | − (4/4) | + (4/4) | − (4/4) | + (4/4) | + (4/4) | R (4/4) | R (4/4) | − | 1–2 | Bipolar | ND |
| H. pullorum | + | + | − | − | − | ND | + | ND | S | R | − | 1 | Unipolar | 34–35 |
| H. rodentium | + | + | − | − | − | − | + | + | R | R | − | 2 | Bipolar | ND |
| Helicobacter sp. strain CLO-3 | + | − | + | − | + | − | + | + | I | R | − | 45 | ||
| H. pylori | + | − | + | + | − | + | − | − | R | S | − | 4–8 | Bipolar | 35–37 |
| H. nemestrinae | + | − | + | + | − | ND | + | − | R | S | − | 4–8 | Bipolar | 24 |
| H. acinonychis | + | − | + | + | − | + | − | − | R | S | − | 2–5 | Bipolar | 30 |
| H. felis | + | + | + | + | − | + | + | − | R | S | + | 14–20 | Bipolar | 42 |
| H. fennelliae | + | − | + | − | + | − | − | + | S | S | − | 2 | Bipolar | 35 |
| H. trogontum | + | + | − | + | ND | + | + | ND | R | R | + | 5–7 | Bipolar | ND |
| H. muridarum | + | − | + | + | + | + | − | − | R | R | + | 10–14 | Bipolar | 34 |
| H. hepaticus | + | + | ND | + | + | ND | − | + | R | R | − | 2 | Bipolar | ND |
| H. canis | − | − | + | − | + | ND | + | ND | S | I | − | 2 | Bipolar | 48 |
| H. bilis | + | + | ND | + | − | ND | + | + | R | R | + | 3–14 | Bipolar | ND |
| “H. rappini” | + | − | − | + | ND | + | + | − | R | R | + | 10–20 | Bipolar | 34 |
| H. cinaedi | + | + | − | − | − | − | − | + | S | I | − | 1–2 | Bipolar | 37–38 |
| H. pametensis | + | + | + | − | − | − | + | + | S | S | − | 2 | Bipolar | 38 |
| Helicobacter sp. strain Bird-C | + | + | + | + | + | − | + | + | S | R | − | 2 | Bipolar | 30 |
| Helicobacter sp. strain Bird-B | + | + | + | + | − | + | + | + | S | R | − | 2 | Bipolar | 31 |
| H. mustelae | + | + | + | + | + | + | + | − | S | R | − | 4–8 | Peritrichous | 36 |
Ultrastructure.
Cells had a spiral shape and measured approximately 0.3 by 1 to 4 μm. The bacterium was also characterized by nonsheathed flagella located singly or bipolarly.
Phylogenetic analysis.
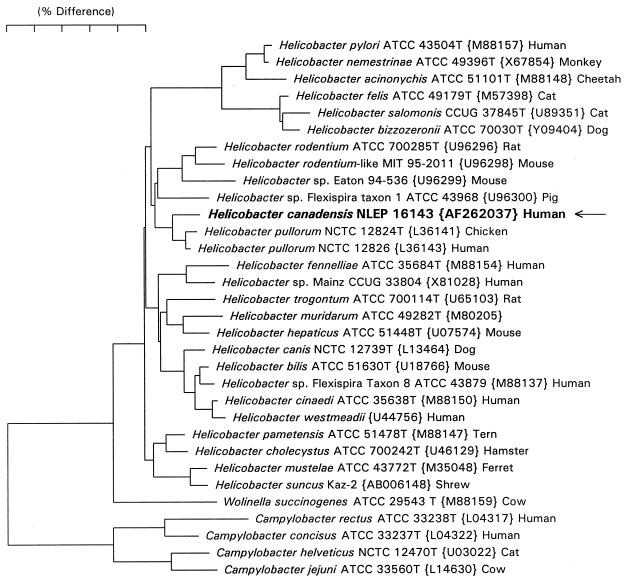
Full 16S rRNA sequences (∼1,500 bases) were determined for all four strains: NLEP-16143, -16767, -99-3017 and -17813. Two H. canadensis strains, NLEP-16143 and -16767, differ by a single base (C or U at position 1137 [Escherichia coli numbering]); NLEP-99-3017 differs by one base from NLEP-16143 and two bases from NLEP-16767; NLEP-17813 differs by seven bases from NLEP-16143 but still clusters with H. canadensis. H. canadensis strains NLEP-16143 and -16767 differ from H. pullorum strains NCTC 12824 (L36141) and NCTC 12826 (L36143) by 28 and 23 bases, respectively (Fig. 1).
FIG. 1.
Dendrogram depicting the taxonomic location of H. canadensis, based on 16S rRNA sequence similarity values. Scale bar = 5% difference in the nucleotide sequence as determined by measuring the lengths of the horizontal lines connecting any two species.
RFLP analysis.
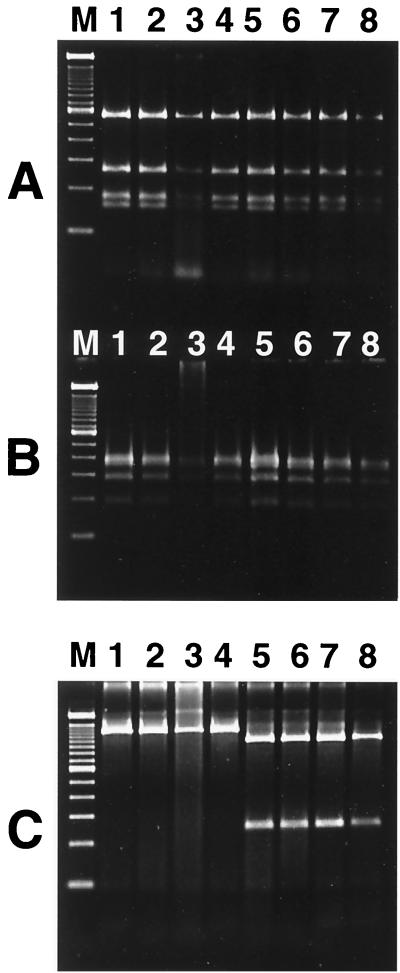
The 1.2-kb Helicobacter-specific PCR products of the 16S rRNA gene were subjected to digestion by AluI, HhaI, and ApaLI. H. pullorum and H. canadensis had similar RFLP patterns when digested by AluI and HhaI; however, the two helicobacters could be distinguished by ApaLI digestion. H. canadensis has an ApaLI site at position 1040 of the 16S rRNA gene, whereas H. pullorum does not; H. canadensis was therefore digested into two fragments, one of 250 bp and the other of 950 bp (Fig. 2).
FIG. 2.
PCR-RFLP patterns for H. pullorum and H. canadensis of 1.2-kb 16S rRNA Helicobacter-specific PCR products. (A) AluI digestion; (B) HhaI digestion; (C) ApaLI digestion. The 1.2-kb PCR product of the Helicobacter 16S rRNA gene was digested for 3 h with selected endonucleases and then separated by electrophoresis on a 6% Visigel matrix. Lane M, 100-bp DNA ladder; lane 1, H. pullorum NCTC 12824; lane 2, H. pullorum NCTC 12827; lane 3, H. pullorum human isolate MIT 98-5493; lane 4, H. pullorum human isolate MIT 98-5494; lanes 5 to 8, H. canadensis isolates NLEP-16143, NLEP-16767, NLEP-17813, and NLEP-99-3017.
DISCUSSION
In this study we identified, based on 16S rRNA analysis and biochemical traits, a novel helicobacter, H. canadensis, previously isolated from the feces of diarrheic humans and classified as H. pullorum. H. canadensis strains are indoxyl acetate-positive helicobacters first described as H. pullorum based on 16S rRNA primers designed to be specific for H. pullorum (14), as well as on other phenotypic and biochemical features which characterize the organism. Subsequent analysis has revealed that errors in printed sequence and base positions occurred in the H. pullorum-specific primers described by Stanley et al. (14). The primers should be as follows: forward, positions 819 to 839, 5′ATG AAT GCT AGT TGT TGT GAG3′, and reverse, positions 1282 to 1265, 5′GAT TGG CTC CAC TTC ACA3′ (E. coli numbering). Under the conditions used, these primers amplify H. pullorum and H. canadensis sequences. The reverse primer, with a one-base mismatch near the 3′ end, is evidently not sufficient to prevent PCR amplification. Thus, primers developed for the early description of Helicobacter may not be specific and may misidentify newly described Helicobacter spp.
In support of our findings, Gibson et al. recently described genetic diversity in H. pullorum strains isolated from humans and chickens (7). Two of these strains, NLEP-16143 and NLEP-16767, which we have now characterized as H. canadensis, were included in their study (7). Interestingly, these two strains, when analyzed by an amplified RFLP technique, were 73% similar to each other but showed only 33% similarity to 18 other H. pullorum strains, all of which grouped at the 70% similarity level (7). The authors concluded that these two Canadian strains were distinct from the other Canadian H. pullorum isolates as well as from other strains isolated in other countries (7). These two strains also differed from other H. pullorum strains in their SacII pulsed-field gel electrophoresis profiles by not being digested by SmaI (7).
We have recently identified cytolethal distending toxin (CDT) in several enterohepatic helicobacters, including H. pullorum (3, 16, 17). H. pullorum from both human and avian sources has DNA sequence homology and cytotoxic activity which position it as a member of the CDT family of bacterial toxins. Interestingly, H. canadensis strains NLEP-16143 and -16767 were tested for the presence of the cdtB gene by PCR as well as for the production of the CDT cytopathic effect and cell cycle arrest and were negative, thus providing further evidence that these novel strains are distinct from H. pullorum (17).
In summary, we have identified and named a novel Helicobacter sp., H. canadensis, associated with diarrhea in humans. Our study reemphasizes that enteric helicobacters cannot always be reliably identified by biochemical reactions or PCR. Proper classification of these novel helicobacters will allow the more accurate description of their pathogenic potential as well as elucidation of key features of their epidemiology. For example, since its original isolation and description, H. pullorum has been isolated from diarrheic humans in North America and Europe (1, 7, 14; Burnens et al., letter). Because of its association with chicken feces and carcasses, studies have suggested that, as in the case of Campylobacter jejuni, a zoonotic link to chicken consumption may exist with H. pullorum infection in people as well (7, 14; Burnens et al., letter). Whether H. canadensis has similar reservoir hosts and zoonotic potential requires further study.
Description of H. canadensis sp. nov.
H. canadensis relating to the country of original isolation. Cells are slender, curved to spiral rods (0.3 by 1.5 to 4 μm), which have one to three spirals. The bacterium is gram negative and nonsporulating; it is motile by means of nonsheathed, single unipolar or bipolar flagella. Cultures grown on solid agar media appear as spreading layers. Cells exhibit microaerobic but not aerobic or anaerobic growth. Growth occurs at 37 and 42°C. The bacteria are urease, alkaline phosphatase, and γ-glutamyl transpeptidase negative but catalase and oxidase positive. The organism hydrolyzes indoxyl acetate, and some strains reduce nitrate to nitrite. Cells are resistent to nalidixic acid and cephalothin. Bacteria have been isolated from the feces of diarrheic humans. The type strain is NLEP-16143 (MIT 98-5491) (= ATCC 700968).
ACKNOWLEDGMENTS
We thank H. G. Trüper, Rleinische Friedrich-Wilhelms-Universität Bonn, for assistance in naming the organism.
This work is supported in part by NIH grants R01CA67529, R01DK52413, and RR-01046 (J.G.F.).
REFERENCES
- 1.Atabay I, Corry J E, On S L. Identification of unusual Campylobacter-like isolates from poultry products as Helicobacter pullorum. J Appl Microbiol. 1998;84:1017–1024. doi: 10.1046/j.1365-2672.1998.00438.x. [DOI] [PubMed] [Google Scholar]
- 2.Burnens A P, Stanley J, Nicolet J. Possible association of Helicobacter pullorum with lesions of vibrionic hepatitis in poultry. In: Newell D G, Ketley J M, Feldman R A, editors. Campylobacters, helicobacters, and related organisms. New York, N.Y: Plenum Press; 1996. [Google Scholar]
- 3.Chien, C. C., N. S. Taylor, Z. Ge, D. B. Schauer, V. B. Young, and J. G. Fox. Identification of cdtB homologues and cytolethal distending toxin activity in enterohepatic Helicobacter spp. J. Med. Microbiol., in press. [DOI] [PubMed]
- 4.Dewhirst F E, Chien C-C, Paster B J, Ericson R L, Orcutt R P, Schauer D B, Fox J G. Phylogeny of the defined murine microbiota: altered Schaedler flora. Appl Environ Microbiol. 1999;65:3287–3292. doi: 10.1128/aem.65.8.3287-3292.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fox J G, Dewhirst F E, Tully J G, Paster B J, Yan L, Taylor N S, Collins M J, Jr, Gorelick P L, Ward J M. Helicobacter hepaticus sp. nov., a microaerophilic bacterium isolated from livers and intestinal mucosal scrapings from mice. J Clin Microbiol. 1994;32:1238–1245. doi: 10.1128/jcm.32.5.1238-1245.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fox J G, Lee A. The role of Helicobacter species in newly recognized gastrointestinal tract diseases of animals. Lab Anim Sci. 1997;47:222–255. [PubMed] [Google Scholar]
- 7.Gibson J R, Ferrus M A, Woodward D, Xerry J, Owen R J. Genetic diversity in Helicobacter pullorum from human and poultry sources identified by an amplified fragment length polymorphism technique and pulsed-field gel electrophoresis. J Appl Microbiol. 1999;87:602–610. doi: 10.1046/j.1365-2672.1999.00858.x. [DOI] [PubMed] [Google Scholar]
- 8.Jukes T H, Cantor C R. Evolution of protein molecules. In: Munro H N, editor. Mammalian protein metabolism. New York, N.Y: Academic Press; 1969. pp. 21–132. [Google Scholar]
- 9.Melito P L, Woodward D L, Price L J, Khakhria R, Mulvey M R, Bernard K, Rodgers F G, Johnson W M. Helicobacter pullorum: an emerging pathogen. Gut. 1999;45:A63. . (Abstract.) [Google Scholar]
- 10.Paster B J, Dewhirst F E. Phylogeny of campylobacters, wolinellas, Bacteroides gracilis, and Bacteroides ureolyticus by 16S ribosomal ribonucleic acid sequencing. Int J Syst Bacteriol. 1988;38:56–62. [Google Scholar]
- 11.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 12.Shames B, Fox J G, Dewhirst F, Yan L, Shen Z, Taylor N S. Identification of widespread Helicobacter hepaticus infection in feces in commercial mouse colonies by culture and PCR assay. J Clin Microbiol. 1995;33:2968–2972. doi: 10.1128/jcm.33.11.2968-2972.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shen Z, Fox J G, Dewhirst F E, Paster B J, Foltz C J, Yan L, Shames B, Perry L. Helicobacter rodentium sp. nov., a urease-negative Helicobacter species isolated from laboratory mice. Int J Syst Bacteriol. 1997;47:627–634. doi: 10.1099/00207713-47-3-627. [DOI] [PubMed] [Google Scholar]
- 14.Stanley J, Linton D, Burens A P, Dewhirst F E, On S L W, Porter A, Owen R J, Costas M. Helicobacter pullorum sp. nov.—genotype and phenotype of a new species isolated from poultry and from human patients with gastroenteritis. Microbiology. 1994;140:3441–3449. doi: 10.1099/13500872-140-12-3441. [DOI] [PubMed] [Google Scholar]
- 15.Steinbrueckner B, Haerter G, Pelz K, Weiner S, Rump J A, Deissler W, Bereswill S V, Kist M. Isolation of Helicobacter pullorum from patients with enteritis. Scand J Infect Dis. 1997;29:315–318. doi: 10.3109/00365549709019053. [DOI] [PubMed] [Google Scholar]
- 16.Young V B, Knox K A, Schauer D B. Cytolethal distending toxin sequence and activity in the enterohepatic pathogen Helicobacter hepaticus. Infect Immun. 2000;68:184–191. doi: 10.1128/iai.68.1.184-191.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Young, V. B., C. C. Chien, N. S. Taylor, K. A. Knox, D. B. Schauer, and J. G. Fox. Cytolethal distending toxin in avian and human isolates of H. pullorum. J. Infect. Dis., in press. [DOI] [PubMed]