Abstract
We determined the nucleotide sequences (329 bp) of the rpoB DNAs from 22 reference strains of Borrelia. No insertions or deletions were observed. Deduced amino acid sequences of amplified rpoB DNA comprised 109 amino acid residues (N450 to M558 [Escherichia coli numbering]). All amino acid sequences were identical with the exception of those of Borrelia lusitaniae PotiB2 (T461→A) and B. bissettii DN127 (I498→V). Each species of B. burgdorferi sensu lato was differentiated as a distinct entity in the phylogenetic tree constructed by a neighbor-joining method. B. burgdorferi sensu lato could be distinguished from B. turicatae and B. hermsii, which are associated with relapsing fever. Seventeen Korean isolates could be identified by PCR-linked direct sequencing and restriction analysis of the rpoB DNA. These results suggest that rpoB DNA is useful for identification and characterization of Borrelia. In addition, we developed the rapid species identification method using the species-specific primer sets based on rpoB gene sequences.
Lyme disease is one of the most prevalent tick-borne infectious diseases in Europe and North America (30). Since the first isolation of Borrelia burgdorferi in 1982, the etiologic agent of Lyme disease, a large number of strains have been isolated and reported in all parts of the world (2). B. burgdorferi sensu lato is currently classified into 10 species: B. burgdorferi (19), B. afzelii (12), B. garinii (4), B. japonica (20), B. valaisiana (33), B. lusitaniae (24), B. andersonii (25), B. turdi and B. tanukii (15), and B. bissettii (29). Although B. burgdorferi sensu lato is present all over the world, most species have a limited geographical distribution. Among these species, B. burgdorferi and B. bissettii, which were also reported in Europe (31), and B. andersonii are mainly found in the United States (29), while B. garinii and B. afzelii are found in Eurasia (28). B. japonica, B. turdi, and B. tanukii are found in Japan (15), and B. valaisiana and B. lusitaniae are found in Europe (24, 33). Ixodes scapularis and I. pacificus ticks are the main vectors in the United States, and I. ricinus and I. persulcatus ticks are the main vectors in Europe and Asia, respectively (14).
B. burgdorferi sensu lato has been characterized conventionally and has been identified by protein analysis with monoclonal antibodies (7, 12), multilocus enzyme electrophoresis (3, 10), and plasmid profile analysis (6). Since the members of the genus Borrelia are usually fastidious microorganisms, however, these methods are time-consuming and expensive (28). As recent alternatives, 16S rRNA gene (rDNA) sequence analysis (26) and PCR-restriction fragment length polymorphism (PCR-RFLP) analysis of the 5S-23S intergenic spacer amplicons (28) are frequently used. However, unlike other bacteria, the hypervariable region for differentiation is inadequate, so 16S rDNA sequence analysis requires at least 800 nucleotide sequences (24, 33). Also, identification with the species-specific primers that target 16S rDNA has several problems. PCR with B. garinii-specific primers amplified the 16S rDNAs of strains of other species (15). Primers specific for several species could not be designed (24, 33). Also, various RFLP patterns in one species emerged, and these patterns were unlike those of the first report on PCR-RFLP analysis of 5S-23S intergenic spacer amplicons (27, 28, 29). In short, a new identification method that completes the 16S rDNA sequence analysis and PCR-RFLP analysis of the 5S-23S intergenic spacer amplicons was needed.
Our objective was to develop a new method for the identification of Borrelia species based on comparative sequence analysis and PCR-RFLP analysis of the rpoB DNA. rpoB, which encodes the β subunit of RNA polymerase, is related to the rifampin resistance of Mycobacterium tuberculosis (32). Recently, rpoB DNA was used as an alternative tool for the identification of mycobacteria (18, 21). The rpoB gene of B. burgdorferi was cloned and characterized (1). In this study, rpoB DNAs (369 bp) that comprised the sequence of a highly conserved region (11) were amplified from the 22 reference strains of Borrelia. Their nucleotide sequences (329 bp) were directly determined and compared. By comparison with reference strains and grouping of the strains into strain-specific clusters with low levels of sequence divergence, Korean isolates were identified. Also, we developed a rapid identification method using the primers specific for B. burgdorferi, B. garinii, B. afzelii, B. valaisiana, and B. lusitaniae.
MATERIALS AND METHODS
Bacterial strains and DNA extraction.
Twenty-two reference strains of the genus Borrelia and two Haenam strains found in the Haenam area of Korea (strains HN-6 and HN-19), which were isolated from Ixodes granulatus and Apodemus agrarius (23), respectively, were used in this study (Table 1). These were cultivated at 32°C in BSKII medium (5). DNA was extracted by modification of a previously described method (8). Briefly, a pellet from a 5-ml culture was suspended in 650 μl of TE (10 mM Tris [pH 8.0], 100 mM EDTA) solution. Ten microliters of RNase (10 mg/ml) and 20 μl of lysozyme (10 mg/ml) were added to the suspension. After 30 min of incubation at 37°C, sodium dodecyl sulfate (0.5%) was added, and the suspension was incubated at 65°C for 10 min. The DNA was extracted four times with equal volumes of phenol and once with an equal volume of chloroform. The DNA was precipitated by adding 0.1 volume of 3 M sodium acetate and 2 volumes of absolute ethanol, washed with 70% ethanol, and resuspended in TE (pH 8.0).
TABLE 1.
Borrelia strains used in this study
| Species | Strain | Accession no.a |
|---|---|---|
| B. burgdorferi | B31T | L48488 |
| IP2 | AF191588 | |
| B. garinii | HP13 | AF164217 |
| Sika1 | AF164218 | |
| Sika2 | AF164219 | |
| IP90 | AF164220 | |
| Pbi | AF164221 | |
| G2 | AF164222 | |
| G25 | AF164223 | |
| PD89 | AF164224 | |
| IP89 | AF164225 | |
| B. afzelii | IPer3 | AF164229 |
| PKo-85 | AF164230 | |
| VS461T | AF164231 | |
| M7 | AF164232 | |
| B. valaisiana | VS116T | AF164226 |
| B. japonica | HO14T | AF164227 |
| B. lusitaniae | PotiB2T | AF164228 |
| B. bissettii | DN127T | AF164233 |
| B. andersonii | 21123 | AF164234 |
| Haenam strains | HN-6 | AF191589 |
| HN-19 | AF191592 | |
| B. hermsii | HS1T | AF164235 |
| B. turicatae | M2007 | AF164236 |
GenBank accession number for the rpoB sequence.
Nucleotide sequencing.
PCR was performed with a set of primers (primer BF [5′-GATGATATTGACCATTTAGG-3′] and primer BR [5′-TTCAGGGGTTTCAATAGGAC-3′]) to amplify rpoB DNA (369 bp). Template DNA (50 ng) and 20 pmol of each primer were added to a PCR mixture tube (AccuPower PCR PreMix; Bioneer, Chungbuk, Korea), which contained 1 U of Taq DNA polymerase, each deoxynucleoside triphosphate at a concentration of 250 μM, 50 mM Tris-HCl (pH 8.3), 40 mM KCl, 1.5 mM MgCl2, and gel loading dye. The volume was adjusted with distilled water to 20 μl. The reaction mixture was subjected to 30 cycles of amplification (30 s at 94°C, 30 s at 59°C, and 45 s at 72°C), followed by a 5-min extension at 72°C (model 9600 thermocycler; Perkin-Elmer Cetus). The PCR products were electrophoresed on a 1.5% agarose gel and were purified with a QIAEX II gel extraction kit (QIAGEN, Hilden, Germany).
Nucleotide sequences (329 bp) were determined from the purified PCR product (369 bp) with forward and reverse primers by using an Applied Biosystems 373A automatic sequencer and BigDye Terminator Cycle Sequencing kit (PE Applied Biosystems, Warrington, United Kingdom). For the sequencing reaction, 60 ng of PCR-amplified DNA, 3.2 pmol of either the forward or the reverse primer, and 8 μl of BigDye Terminator RR mixture (part no. 4303153; PE Applied Biosystems) were mixed and adjusted to a final volume of 20 μl by adding distilled water. The reaction was run with 5% (vol/vol) dimethyl sulfoxide for 30 cycles of 15 s at 95°C, 10 s at 50°C, and 4 min at 60°C. Both strands were sequenced as a cross-check.
Sequence analysis.
The sequences were aligned with the multiple alignment algorithm in the MegAlign package (Windows version 3.12e; DNASTAR, Madison, Wis.). A phylogenetic tree of the Borrelia spp. was constructed by the neighbor-joining method with the MEGA (molecular evolutionary genetics analysis) program (22). A bootstrap analysis (100 repeats) was performed to evaluate the topology of the phylogenetic tree.
Identification of Korean isolates by PCR-linked DNA sequencing and RFLP analysis.
PCR-linked DNA sequencing was performed to determine the rpoB DNA sequences of 15 Borrelia strains isolated from ticks or mice in Korea as described above. The sequences determined were compared to those of reference strains. For the PCR-RFLP analysis of rpoB DNA, endonuclease Tsp509-I (New England Biolabs, Beverly, Mass.) was used as recommended by the manufacturer to cleave the PCR products. The restriction fragments were electrophoresed on a 3% wide-range–standard (3:1) agarose (Sigma, St. Louis, Mo.) gel.
PCR for species-specific identification of B. burgdorferi sensu lato strains.
Primer sets specific for B. burgdorferi, B. garinii, B. afzelii, B. lusitaniae, and B. valaisiana were designed on the basis of the rpoB DNA sequences determined (Table 2). Template DNA (50 ng) and 20 pmol of each primer were added to a PCR mixture tube (AccuPower PCR PreMix; Bioneer), and the volume was adjusted with distilled water to 20 μl. PCR amplification conditions are shown in Table 2.
TABLE 2.
The oligonucleotide sequences of species-specific primer sets
| Species | Primera | PCR conditions | DNA size (bp) |
|---|---|---|---|
| B. burgdorferi | F, CTGTTGGTGAGCTTCTTACT | 94°C for 30 s, 58°C for 30 s, and 72°C for 30 s for 30 cycles | 308 |
| R, GCATTATACTCATTATGGTAGA | |||
| B. garinii | F, GTGCGTTCTGTTGGGGAG | 94°C for 10 s, 62°C for 5 s, and 72°C for 20 s for 20 cycles | 257 |
| R, CCTTGGACCAGGGGGACT | |||
| B. afzelii | F, AGAGTGCGTTCTGTTGGC | 94°C for 30 s, 56°C for 30 s, and 72°C for 30 s for 25 cycles | 318 |
| R, ACATTATACTCATTATGGTAGA | |||
| B. valaisiana | F, AGGAGAGTACGTTCTGTTGGA | 94°C for 10 s, 62°C for 5 s, and 72°C for 30 s for 25 cycles | 306 |
| R, TGAAGTAAGAGACGTCCATTAT | |||
| B. lusitaniae | F, AGAGCTTCTTGCTAATATATATA | 94°C for 30 s, 60°C for 30 s, and 72°C for 30 s for 30 cycles | 250 |
| R, GCCTGGGGGACTTTCAAGA |
F, forward primer; R, reverse primer.
Nucleotide sequence accession numbers.
The rpoB DNA sequences determined for the Borrelia strains have been deposited in GenBank (accession no. AF164217 to AF164236, AF191588, AF191589, and AF191592). The rpoB sequence of B. burgdorferi B31T (GenBank accession no. L48488), available from GenBank, was used for comparison (Table 1).
RESULTS
rpoB sequences of reference strains.
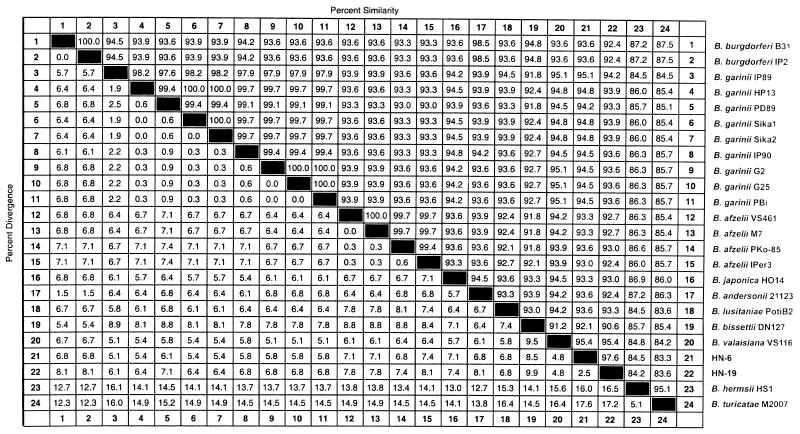
rpoB DNAs (369 bp) were successfully amplified from 22 reference strains of Borrelia. The nucleotide sequences (329 bp) of the amplified DNAs were determined. No insertions or deletions were observed. The nucleotide sequences determined were compared for pairwise similarity (Fig. 1). More than 91.2% similarity was observed among B. burgdorferi sensu lato strains. The sequence similarity among B. garinii strains was 97.6 to 100%. B. garinii IP89 showed the lowest level of similarity (97.6 to 98.2%) with the other B. garinii strains tested. The sequence similarity of B. burgdorferi sensu lato to Borrelia hermsii and Borrelia turicatae, which are associated with relapsing fever, was 83.6 to 87.5%.
FIG. 1.
Similarity matrix for the rpoB DNA sequences of B. burgdorferi sensu lato reference strains and two Haenam strains. Similarities were determined by using the Clustal program with the weighted residue weight table (MegAlign package [Windows version 3.12e]; DNASTAR).
Deduced amino acid sequences.
The deduced amino acid sequences of amplified rpoB DNA comprised 109 amino acid residues (N450 to M558 [Escherichia coli numbering]). The amino acid sequences among the Borrelia species were highly conserved. All amino acid sequences were identical with the exception of those of B. lusitaniae PotiB2T (T461→A) and B. bissettii DN127T (I498→V). Borrelia is known to be naturally resistant to rifampin, which was possibly explained by the S531→N amino acid change (1). This amino acid change was also observed in all of the strains of Borrelia.
Phylogenetic tree.
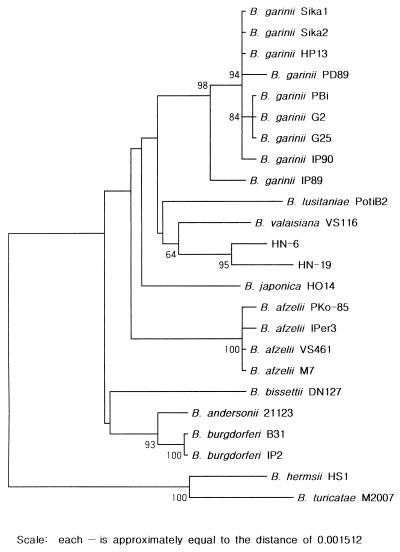
A phylogenetic tree was constructed by the neighbor-joining method to investigate the relationships among B. burgdorferi sensu lato strains (Fig. 2). rpoB DNA sequence analysis could differentiate Borrelia species as well as analysis of 16S rDNA and the flagellin gene could (16, 29). B. burgdorferi sensu lato could be distinguished from B. turicatae and B. hermsii, which are associated with relapsing fever. Two B. burgdorferi strains (strains B31T and IP2) and four B. afzelii strains (strains IPer3, PKo-85, VS461T, and M7) formed a cluster separated from the other Borrelia strains. Also, nine B. garinii strains (strains Sika1, Sika2, HP13, PD89, IP90, PBi, G2, G25, and IP89) formed a cluster separated from the other Borrelia strains, while strain IP89 was deeply branched from the other B. garinii strains. B. valaisiana, B. lusitaniae, B. japonica, B. bissettii, and B. andersonii were branched independently. Two Haenam strains (strains HN-6 and HN-19) formed a distinctive cluster that was clearly separated from the other members of B. burgdorferi sensu lato. B. bissettii DN127T and B. andersonii 21123 were more closely related to B. burgdorferi B31T. Similar results were shown by phylogenetic analysis of 16S rDNA and the flagellin gene (16, 29).
FIG. 2.
Phylogenetic tree of B. burgdorferi sensu lato strains and two Haenam strains on the basis of the rpoB DNA sequences. The tree was constructed by a neighbor-joining method. Topology was also evaluated by bootstrap analysis. The values in the tree represent bootstrap results.
Identification of Korean isolates by PCR-linked DNA sequencing and RFLP analysis.
Among the 10 species currently classified in the genus Borrelia, B. afzelii and B. garinii were isolated in Korea. In addition, unclassified Haenam strains (strains HN-6 and HN-19) were recently characterized (23). On the basis of the rpoB data for reference strains, 15 strains were analyzed by PCR-linked DNA sequencing and restriction analysis. They were identified as either B. afzelii (six strains identical to B. afzelii VS461), B. garinii (one strain identical to B. garinii IP89), or Haenam strains (seven strains identical to HN-6 and 1 strain with 99.7% similarity to HN-19). By referring to the phylogenetic tree together with the use of the reference strains, we could identify those isolates. The tree was quite similar to that based on the 16S rDNA sequences (23) (data not shown).
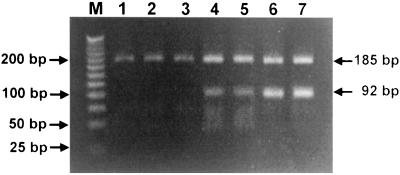
PCR-RFLP analysis is a simple and rapid method for the identification of isolated bacterial strains in clinical laboratories. Reference strains of Borrelia could be differentiated by restriction analysis of rpoB DNA with Tsp509-I (Table 3). By use of the most discernible bands Korean isolates could be identified as either B. afzelii (more than three bands; 185, 92, 53, and 39 bp), B. garinii (two bands; 185 and 92 bp) or Haenam strains (single band; 185 or 184 bp) (Fig. 3).
TABLE 3.
Tsp509-I restriction fragments of amplified rpoB DNAs (369 bp)
| Species | Restriction fragment sizes (bp)a |
|---|---|
| Haenam strains | 185, 184 |
| B. afzelii, B. burgdorferi, B. japonica, B. valaisiana, and B. andersonii | 185, 92, 53, 39 |
| B. garinii | 185, 92 |
| B. lusitaniae | 185, 92, 55, 37 |
| B. bissettii | 185, 92, 37, 35, 16, 4 |
| B. hermsii | 128, 92, 53, 39, 30, 27 |
| B. turicatae | 128, 53, 46, 39, 30, 27 |
Exact restriction fragment sizes were determined from the sequences.
FIG. 3.
Identification of Korean isolates (HN-6, HN-14, HN-19, KK1, and KW3) of B. burgdorferi sensu lato strains by PCR-RFLP analysis of the rpoB DNAs (369 bp) Lanes: M, marker DNA (25 bp ladder); 1, HN-6; 2, HN-14; 3, HN-19; 4, B. afzelii VS461; 5, KK1; 6, B. garinii IP89; 7, KW3.
Species-specific PCR for identification of B. burgdorferi sensu lato strains.
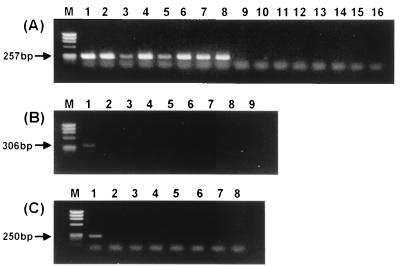
PCR with species-specific primers sufficiently differentiated the Borrelia species (Fig. 4). B. garinii-specific primers could amplify only the strains of B. garinii (Fig. 4A), while primers specific for 16S rDNA amplified the DNAs of other species (15). Also, B. burgdorferi- and B. afzelii-specific primers amplified the rpoB DNA only from the strains B. burgdorferi and B. afzelii, respectively (data not shown). Previously, PCRs specific for B. valaisiana and B. lusitaniae 16S rDNAs could not be performed (24, 33), but rpoB DNAs were specifically amplified from B. valaisiana and B. lusitaniae (Fig. 4B and C).
FIG. 4.
Amplification of rpoB DNA from B. burgdorferi sensu lato strains with the primers specific for B. garinii (A), B. valaisiana (B), and B. lusitaniae (C). (A) Lanes: 1, B. garinii HP13; 2, B. garinii Sika1; 3, B. garinii IP90; 4, B. garinii PBi; 5, B. garinii G2; 6, B. garinii G25; 7, B. garinii PD89; 8, B. garinii IP89; 9, B. burgdorferi B31T; 10, B. afzelii VS461T; 11, B. japonica HO14T; 12, B. valaisiana VS116T; 13, B. lusitaniae PotiB2T; 14, B. bissettii DN127T; 15, B. andersonii 21123; 16, B. hermsii HS1T. (B) Lanes: 1, B. valaisiana VS116T; 2, B. burgdorferi B31T; 3, B. garinii HP13; 4, B. afzelii VS461T; 5, B. japonica HO14T; 6, B. lusitaniae PotiB2T; 7, B. bissettii DN127T; 8, B. andersonii 21123; 9, B. hermsii HST. (C) Lanes: 1, B. lusitaniae PotiB2T; 2, B. burgdorferi B31T; 3, B. garinii HP13; 4, B. afzelii VS461T; 5, B. japonica HO14T; 6, B. valaisiana VS116T; 7, B. bissettii DN127T; 8, B. andersonii 21123.
DISCUSSION
Since the first isolation and description of B. burgdorferi, many strains have been isolated (2, 30) and classified as B. burgdorferi, B. garinii, and B. afzelii. However, since the description of B. japonica as a new species in 1993 (20), B. burgdorferi strains have been reclassified such that several new species were recently described (15, 24, 25, 29, 33).
Characterization and identification of B. burgdorferi sensu lato by conventional methods are time-consuming and expensive (3, 6, 7, 10, 12). Recently, 16S rDNA sequence analysis (26) and PCR-RFLP analysis of 5S-23S intergenic spacer amplicons (28) have frequently been used. Analysis of the hypervariable region of 16S rDNA nucleotide sequences provides a rapid means of identification and characterization. For example, Treponema identification by 16S rDNA sequence analysis requires only 500 nucleotides (13). Similarly, analysis of the hypervariable region of mycobacteria with species-specific probes provides a rapid means of identification (9), but 16S rDNA sequence analysis of B. burgdorferi sensu lato requires at least 800 nucleotide sequences due to a nonexistent hypervariable region. Also, species-specific PCR was not possible (24, 33). Furthermore, various RFLP patterns in one species emerged, unlike in the first report on PCR-RFLP analysis of 5S-23S intergenic spacer amplicons (27, 28, 29).
We used rpoB DNA to differentiate the species of B. burgdorferi sensu lato. rpoB, which encodes the β subunit of RNA polymerase, was used as an alternative tool to identify mycobacteria (21). Forty-four reference strains of mycobacteria were successfully differentiated by comparative sequence analysis of rpoB DNA. In particular, this analysis could differentiate Mycobacterium kansasii from M. gastri and could differentiate M. szulgai from M. malmoense, which could not be distinguished by 16S rDNA sequence analysis.
The primers, which were selected from the previously known sequence of B. burgdorferi B31T (1), could successfully amplify rpoB DNAs (369 bp) from 22 reference strains of Borrelia. The primers corresponded to the highly conserved regions (HCR5 and HCR6) on the basis of known rpoB sequences of Bacillus subtilis (11). The sequence similarity of the rpoB DNAs of B. burgdorferi sensu lato strains was more than 91.2%. The intraspecies divergence of the rpoB DNA was narrow, as in mycobacteria (21). For example, two B. burgdorferi strains, B31T and IP2, had identical sequences, and the sequence similarities of the four B. afzelii strains tested (strains IPer3, PKo-85, VS461T, and M7) were 99.4 to 100%. However, the sequence similarities of the B. garinii strains were 97.6 to 100%, possibly indicating the heterogeneity of B. garinii, which was already explained by previous reports (16, 28). It is interesting that B. garinii IP89 showed the lowest level of similarity (97.6 to 98.2%) to other B. garinii strains. Previously, this strain was classified as a group different from B. garinii by multilocus enzyme electrophoresis (3). Also, the RFLP pattern of the rrf-rrl intergenic spacer amplicons of this strain were different from those of the other B. garinii strains tested (28). In the phylogenetic tree based on the rpoB sequences, B. garinii IP89 was separate from the other B. garinii strains tested.
In the phylogenetic tree based on the rpoB sequences, B. burgdorferi sensu lato could be distinguished from B. turicatae and B. hermsii, which are associated with relapsing fever. Each species of B. burgdorferi sensu lato was differentiated as a distinct entity. Two Haenam strains (strains HN-6 and HN-19) formed a distinctive cluster, clearly separated from the other members of B. burgdorferi sensu lato. These strains were closer to B. valaisiana VS116, as indicated by 16S rDNA analysis (23). Also, the MseI and DraI restriction patterns of the 5S-23S intergenic spacer amplicons of these strains differed from those of the other B. burgdorferi sensu lato strains tested (23). These results showed that the rpoB DNA is useful for the differentiation of the strains or species of B. burgdorferi sensu lato.
The aim of this study was not the phylogenetic analysis of Borrelia with rpoB DNA but the differentiation of Borrelia species that might be encountered in the clinical laboratory. It is difficult to show the phylogenetic relationships of many species because only a small portion of the whole rpoB gene was used in this study. However, as shown from the results, analysis of rpoB DNA (369 bp) could distinguish many of the strains or species of Borrelia, as was the case for 16S rDNA analysis, which, however, required a larger portion (800 bp) of 16S rDNA for the same procedure. Furthermore, signature nucleotides in that rpoB DNA enabled us to design primers which can be used for the rapid and specific identification and differentiation of B. burgdorferi sensu lato by PCR.
Borrelia is naturally resistant to rifampin, which was possibly explained by an amino acid change (S531→N) (1, 17). In our study these changes were observed in all strains of Borrelia. Similar findings on natural resistance to rifampin due to the primary amino acid sequence of the β subunit of RNA polymerase are known in Spiroplasma citri (S531→T) and Mycobacterium celatum (S531→N) (1, 17, 21).
In conclusion, rpoB DNA is a very useful marker for the differentiation of the Borrelia species. B. burgdorferi sensu lato could be successfully identified and differentiated by comparative sequence analysis, restriction analysis, and species-specific PCR.
REFERENCES
- 1.Alekshun M, Kashlev M, Schwartz I. Molecular cloning and characterization of Borrelia burgdorferi rpoB. Gene. 1997;186:227–235. doi: 10.1016/s0378-1119(96)00714-7. [DOI] [PubMed] [Google Scholar]
- 2.Anderson J F. Epizootiology of Borrelia in Ixodes tick vectors and reservoir hosts. Rev Infect Dis. 1989;11:S1451–S1459. doi: 10.1093/clinids/11.supplement_6.s1451. [DOI] [PubMed] [Google Scholar]
- 3.Balmelli T, Piffaretti J C. Analysis of the genetic polymorphism of Borrelia burgdorferi sensu lato by multilocus enzyme electrophoresis. Int J Syst Bacteriol. 1996;46:167–172. doi: 10.1099/00207713-46-1-167. [DOI] [PubMed] [Google Scholar]
- 4.Baranton G, Postic D, Saint Girons I, Boerlin P, Piffaretti J C, Assous M, Grimont P A. Delineation of Borrelia burgdorferi sensu stricto, Borrelia garinii sp. nov., and group VS461 associated with Lyme borreliosis. Int J Syst Bacteriol. 1992;42:378–383. doi: 10.1099/00207713-42-3-378. [DOI] [PubMed] [Google Scholar]
- 5.Barbour A G. Isolation and cultivation of Lyme disease spirochetes. Yale J Biol Med. 1984;57:521–525. [PMC free article] [PubMed] [Google Scholar]
- 6.Barbour A G. Plasmid analysis of Borrelia burgdorferi, the Lyme disease agent. J Clin Microbiol. 1988;26:475–478. doi: 10.1128/jcm.26.3.475-478.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barbour A G, Heiland R A, Howe T R. Heterogeneity of major proteins in Lyme disease borreliae: a molecular analysis of North American and European isolates. J Infect Dis. 1985;152:478–484. doi: 10.1093/infdis/152.3.478. [DOI] [PubMed] [Google Scholar]
- 8.Barbour A G, Garon C F. Linear plasmids of the bacterium Borrelia burgdorferi have covalently closed ends. Science. 1987;237:409–411. doi: 10.1126/science.3603026. [DOI] [PubMed] [Google Scholar]
- 9.Böddinghaus B, Rogall T, Flohr T, Blöcker H, Böttger E C. Detection and identification of mycobacteria by amplification of rRNA. J Clin Microbiol. 1990;28:1751–1759. doi: 10.1128/jcm.28.8.1751-1759.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boerlin P, Peter O, Bretz A G, Postic D, Baranton G, Piffaretti J C. Population genetic analysis of Borrelia burgdorferi isolates by multilocus enzyme electrophoresis. Infect Immun. 1992;60:1677–1683. doi: 10.1128/iai.60.4.1677-1683.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boor K J, Dunkan M L, Price C W. Genetic and transcriptional organization of the region encoding the β subunit of Bacillus subtilis RNA polymerase. J Biol Chem. 1995;270:20329–20336. doi: 10.1074/jbc.270.35.20329. [DOI] [PubMed] [Google Scholar]
- 12.Canica M M, Nato F, du Merle L, Mazie J C, Baranton G, Postic D. Monoclonal antibodies for identificaton of Borrelia afzelii sp. nov. associated with late cutaneous manifestations of Lyme borreliosis. Scand J Infect Dis. 1993;25:441–448. doi: 10.3109/00365549309008525. [DOI] [PubMed] [Google Scholar]
- 13.Choi B-K, Wyss C, Göbel U B. Phylogenetic analysis of pathogen-related oral spirochetes. J Clin Microbiol. 1996;34:1922–1925. doi: 10.1128/jcm.34.8.1922-1925.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fukunaga M, Hamase A, Okada K, Inoue H, Tsuruta Y, Miyamoto K, Nakao M. Characterization of spirochetes isolated from ticks (Ixodes tanuki, Ixodes turdus, and Ixodes columnae) and comparison of the sequences with those of Borrelia burgdorferi sensu lato strains. Appl Environ Microbiol. 1996;62:2338–2344. doi: 10.1128/aem.62.7.2338-2344.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fukunaga M, Hamase A, Okada K, Nakao M. Borrelia tanukii sp. nov. and Borrelia turdae sp. nov. found from ixodid ticks in Japan: rapid species identification by 16S rRNA gene-targeted PCR analysis. Microbiol Immunol. 1996;40:877–881. doi: 10.1111/j.1348-0421.1996.tb01154.x. [DOI] [PubMed] [Google Scholar]
- 16.Fukunaga M, Okada K, Nakao M, Konishi T, Sato Y. Phylogenetic analysis of Borrelia species based on flagellin gene sequences and its application for molecular typing of Lyme disease borreliae. Int J Syst Bacteriol. 1996;46:898–905. doi: 10.1099/00207713-46-4-898. [DOI] [PubMed] [Google Scholar]
- 17.Gaurivaud P, Laigret F, Bove J M. Insusceptibility of members of the class Mollicutes to rifampin: studies of the Spiroplasma citri RNA polymerase beta-subunit gene. Antimicrob Agents Chemother. 1996;40:858–862. doi: 10.1128/aac.40.4.858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gingeras T R, Ghandour G, Wang E, Berno A, Small P M, Drobniewski F, Alland D, Desmond E, Holodniy M, Drenkow J. Simultaneous genotyping and species identification using hybridization pattern recognition analysis of generic Mycobacterium DNA arrays. Genome Res. 1998;8:435–448. doi: 10.1101/gr.8.5.435. [DOI] [PubMed] [Google Scholar]
- 19.Johnson R C, Schmid G P, Hyde F W, Steingerwalt A G, Brenner D J. Borrelia burgdorferi sp. nov.: etiologic agent of Lyme disease. Int J Syst Bacteriol. 1984;34:496–497. [Google Scholar]
- 20.Kawabata H, Masuzawa T, Yanagihara Y. Genomic analysis of Borrelia japonica sp. nov. isolated from Ixodes ovatus in Japan. Microbiol Immunol. 1993;37:843–848. doi: 10.1111/j.1348-0421.1993.tb01714.x. [DOI] [PubMed] [Google Scholar]
- 21.Kim B-J, Lee S-H, Lyu M-A, Kim S-J, Bai G-H, Chae G-T, Kim E-C, Cha C-Y, Kook Y-H. Identification of mycobacterial species by comparative sequence analysis of the RNA polymerase gene. J Clin Microbiol. 1999;37:1714–1720. doi: 10.1128/jcm.37.6.1714-1720.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumar S, Tamura K, Masatoshi N. MEGA: molecular evolutionary genetics analysis, version 1.01. University Park: The Pennsylvania State University; 1993. [Google Scholar]
- 23.Lee S-H, Kim B-J, Kim J-H, Park K-H, Yeo S-J, Kim S-J, Kook Y-H. Characterization of Borrelia sp. isolated from Korea by 16S rDNA sequence analysis and PCR-RFLP analysis of rrf (5S)-rrl (23S) intergenic spacer amplicons. Int J Syst Evol Microbiol. 2000;50:857–863. doi: 10.1099/00207713-50-2-857. [DOI] [PubMed] [Google Scholar]
- 24.Le Fleche A, Postic D, Girardet K, Peter O, Baranton G. Characterization of Borrelia lusitaniae sp. nov. by 16S ribosomal DNA sequence analysis. Int J Syst Bacteriol. 1997;47:921–925. doi: 10.1099/00207713-47-4-921. [DOI] [PubMed] [Google Scholar]
- 25.Marconi R T, Liveris D, Schwartz I. Identification of novel insertion elements, restriction fragment length polymorphism patterns, and discontinuous 23S rRNA in Lyme disease spirochetes: phylogenetic analyses of rRNA genes and their intergenic spacers in Borrelia japonica sp. nov. and genomic group 21038 (Borrelia andersonii sp. nov.) isolates. J Clin Microbiol. 1995;33:2427–2434. doi: 10.1128/jcm.33.9.2427-2434.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marconi R T, Garon C F. Development of polymerase chain reaction primer sets for diagnosis of Lyme disease and for species-specific identification of Lyme disease isolates by 16S rRNA signature nucleotide analysis. J Clin Microbiol. 1992;30:2830–2834. doi: 10.1128/jcm.30.11.2830-2834.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Masuzawa T, Komikado T, Iwaki A, Suzuki H, Kaneda K, Yanagihara Y. Characterization of Borrelia sp. isolated from Ixodes tanuki, I. turdus, and I. columnae in Japan by restriction fragment length polymorphism of rrf (5S)-rrl (23S) intergenic spacer amplicons. FEMS Microbiol Lett. 1996;142:77–83. doi: 10.1111/j.1574-6968.1996.tb08411.x. [DOI] [PubMed] [Google Scholar]
- 28.Postic D, Assous M V, Grimont P A, Baranton G. Diversity of Borrelia burgdorferi sensu lato evidenced by restriction fragment length polymorphism of rrf (5S)-rrl (23S) intergenic spacer amplicons. Int J Syst Bacteriol. 1994;44:743–752. doi: 10.1099/00207713-44-4-743. [DOI] [PubMed] [Google Scholar]
- 29.Postic D, Ras N M, Lane R S, Hendson M, Baranton G. Expanded diversity among Californian Borrelia isolates and description of Borrelia bissettii sp. nov. (formerly Borrelia group DN127) J Clin Microbiol. 1998;36:3497–3504. doi: 10.1128/jcm.36.12.3497-3504.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Steere A C. Lyme disease. N Engl J Med. 1989;321:586–596. doi: 10.1056/NEJM198908313210906. [DOI] [PubMed] [Google Scholar]
- 31.Strle F. Lyme borreliosis in Slovenia. Int J Med Microbiol Virol Parasitol Infect Dis. 1999;289:643–652. doi: 10.1016/s0934-8840(99)80023-1. [DOI] [PubMed] [Google Scholar]
- 32.Telenti A, Imboden P, Marchesi F, Lowrie D, Cole S, Colston M J, Matter L, Schopfer K, Bodmer T. Detection of rifampin-resistance mutations in Mycobacterium tuberculosis. Lancet. 1993;341:647–650. doi: 10.1016/0140-6736(93)90417-f. [DOI] [PubMed] [Google Scholar]
- 33.Wang G, van Dam A P, Le Fleche A, Postic D, Peter O, Baranton G, de Boer R, Spanjaard L, Dankert J. Genetic and phenotypic analysis of Borrelia valaisiana sp. nov. (Borrelia genomic groups VS116 and M19) Int J Syst Bacteriol. 1997;47:926–932. doi: 10.1099/00207713-47-4-926. [DOI] [PubMed] [Google Scholar]