Abstract
The stability of IS6110 restriction fragment length polymorphism (RFLP) patterns of Mycobacterium tuberculosis strains in actual transmission chains has been assessed by analyzing the variability of IS6110 RFLP patterns of strains in fingerprint clusters that have been confirmed by classical epidemiological data. Forty susceptible and 35 drug-resistant (including 17 multidrug-resistant) M. tuberculosis strains obtained from 75 patients living in Germany have been analyzed. The epidemiological relationship among strains within the fingerprint clusters has been verified by family contacts (14 clusters) or by contact tracing of the public health offices (7 clusters). The time spans between the first and the last isolate of one cluster ranged from less than 1 to 29 months. Of the 75 strains only 1 showed a one-band variation when compared to the other nine isolates grouped in the same cluster, corresponding with a rate of change of ∼1.9% per possible transmission (one index patient per cluster was subtracted from the total number of isolates). These results confirm a high degree of stability of IS6110 RFLP patterns of transmitted M. tuberculosis strains. Furthermore, the data presented indicate that isolates with identical IS6110 DNA fingerprint patterns are a good indicator for the rate of recent transmission in a study population.
DNA fingerprinting using the insertion sequence IS6110 as a probe has become the standard technique for the comparison of Mycobacterium tuberculosis isolates on the strain level (3, 10, 11). IS6110, a transposable sequence belonging to the IS3 family, is found in virtually all members of the M. tuberculosis complex and is apparently restricted to this group of organisms (3). It has been widely used in epidemiological studies, e.g., to detect the rate of recent transmission in a study population or to identify outbreaks of M. tuberculosis strains (e.g., see references 2 and 6). These studies are based on the assumption that the DNA polymorphism of IS6110 patterns among unrelated clinical isolates is high, whereas epidemiologically related M. tuberculosis strains show identical or similar (one- or two-band variations) fingerprint patterns (11).
For the correct interpretation of the results obtained by IS6110 typing, it is necessary to know the degree of stability of M. tuberculosis IS6110 restriction fragment length polymorphism (RFLP) patterns. Recently, three studies have analyzed the rate of change of M. tuberculosis IS6110 RFLP patterns in serial patient isolates (4, 7, 12). Two of these publications confirmed a high degree of stability (4, 7), whereas the study of Yeh and coworkers (12) indicates a high variability of IS6110 patterns. A high rate of change was also found by Alito et al. (1) for one cluster of a multidrug-resistant (MDR) outbreak strain. These studies indicate that the degree of stability of IS6110 RFLP patterns might vary for different families of M. tuberculosis strains or for strains that are isolated in different geographical regions.
Since the factors inducing transposition or mutation activity in M. tuberculosis remain uncertain so far, the mobility of IS6110 in M. tuberculosis strains remaining in the body of the patient generally may differ from that in strains that undergo a person-to-person transmission. Hence, the stability of IS6110 RFLP patterns in serial patient isolates might be different from that of isolates in actual transmission chains (4).
In this study we analyzed the stability of IS6110 RFLP patterns of M. tuberculosis strains grouped in well-defined fingerprint clusters. All these clusters have been confirmed by classical epidemiological data; thus, isolates of any given cluster are part of a recent chain of transmission.
MATERIALS AND METHODS
All isolates analyzed were identified as M. tuberculosis using gene probes (ACCUProbe; GenProbe, San Diego, Calif.) and by standard biochemical procedures (5). Drug susceptibility was determined by the proportion method on Löwenstein-Jensen medium and/or the modified proportion method in the BACTEC 460TB system (Becton Dickinson Microbiology Systems, Cockeysville, Md.). DNA fingerprinting using IS6110 as a probe was performed according to a standardized protocol as described elsewhere (6, 9). Briefly, for isolation of genomic DNA M. tuberculosis strains were grown on Löwenstein-Jensen slants for 3 to 5 weeks. All bacterial cells from one slant were transferred in 400 μl of TE buffer (0.01 M Tris-HCl, 0.001 M EDTA, pH 8), and the solution was heated at 80°C for 20 min to kill the bacteria. Fifty microliters of lysozyme (10 mg/ml) was added, and the tube was incubated for 1 h at 37°C. Seventy microliters of sodium dodecyl sulfate (10%) and 6 μl of proteinase K (10 mg/ml) were added, and the mixture was incubated for 10 min at 65°C. A 100-μl volume of 5 M NaCl and of an N-cetyl-N,N,N-trimethylammonium bromide (CTAB)-NaCl solution (4.1 g of NaCl and 10 g of CTAB per 100 ml) was added. The cups were vortexed and incubated for 10 min at 65°C. An equal volume of chloroform-isoamylalcohol (24:1) was added, the mixture was centrifuged for 5 min at 12,000 × g, and the aqueous supernatant was carefully transferred to a fresh tube. The total DNA was precipitated using isopropanol and redissolved in an appropriate volume of double-distilled water. For fingerprinting, PvuII-digested total DNA was separated using horizontal 1% agarose gels in Tris-acetate buffer and vacuum blotted onto a nylon membrane. Hybridization of the DNA was carried out with a 254-bp internal PCR fragment of IS6110 (amplified with the primer pair INS1 and INS2) as a probe using the ECL system (Amersham, Little Chalfont Buckinghamshire, United Kingdom). PvuII-digested total DNA of reference strain Mt. 14323 was used in each Southern blot experiment as an external size standard.
IS6110 fingerprint patterns of mycobacterial strains were analyzed using the Gelcompar software (Windows 95, version 4.0; Applied Maths, Kortrijk, Belgium) as described previously (9). Autoradiograms were digitized using a scanner with an optical resolution of 220 dots per inch (ScanMaker E6; Microtek Europe B. V., Rotterdam, The Netherlands). The fingerprint patterns were analyzed for similarity using the Dice coefficient, and a dendrogram was calculated with the unweighted pair group method using average linkage according to the supplier's instruction.
The analysis of clusters that are initially identified by IS6110 fingerprinting might lead to the exclusion of isolates with changed IS6110 RFLP patterns, which in turn may result in falsely low rates of change of IS6110 RFLP patterns. For this reason, in this study we have mainly selected clusters that were initially based on family contacts (family outbreaks [clusters 1, 3, 4, 9, 10, and 11b–19) or on epidemiologically certified data of the public health office (clusters 11a and 20) that have since been confirmed by IS6110 fingerprinting. Hence, the preselection of isolates with identical IS6110 RFLP patterns can be excluded in these cases. Clusters 2 and 5 to 8 were initially identified by IS6110 DNA fingerprint analysis, but so far no other isolates with a one- or two-band variation has been identified in a DNA fingerprint database that cover more than 75% of all culture-confirmed M. tuberculosis cases in the respective geographical region for a study period of 3 years. The epidemiological relationship of the strains within these clusters was verified by contact tracing performed by the public health offices. The fingerprint data were partially published in two previous studies (6, 8).
RESULTS AND DISCUSSION
A total of 40 susceptible and 35 drug-resistant (including 17 MDR) M. tuberculosis isolates obtained from 75 patients with pulmonary tuberculosis living in Germany have been analyzed. The isolates were grouped in 21 clusters, of which 14 were family outbreaks and 7 were community outbreaks confirmed by contact tracing of the public health offices (Table 1). The size of the clusters varied from 2 to 11 isolates obtained over time spans ranging from less than 1 to 29 months between the first and last isolate within a cluster. The number of IS6110 copies per isolate varied from 7 to 16 bands, with a mean of 10 bands. Drug resistances of isolates ranged from fully susceptible to MDR with resistance to six drugs (Table 1). Seventeen MDR isolates have been included.
TABLE 1.
Results of drug susceptibility tests and IS6110 DNA fingerprinting of the 75 isolates analyzed
| Isolate no. | Cluster | Date culture obtained (mo/yr) | Time spana (mo) | Drug resistanceb | No. of IS6110 bands | Epidemiological method of cluster confirmation |
|---|---|---|---|---|---|---|
| 1 | 1 | 10/97 | 8 | Susc | 8 | Family contacts |
| 2 | 1 | 11/97 | 8 | Susc | 8 | Family contacts |
| 3 | 1 | 6/98 | 8 | Susc | 8 | Family contacts |
| 4 | 2 | 11/96 | 21 | Susc | 11 | Contact tracing |
| 5 | 2 | 4/97 | 21 | Susc | 11 | Contact tracing |
| 6 | 2 | 3/98 | 21 | Susc | 11 | Contact tracing |
| 7 | 2 | 8/98 | 21 | Susc | 11 | Contact tracing |
| 8 | 3 | 3/98 | 6 | Susc | 13 | Family contacts |
| 9 | 3 | 3/98 | 6 | Susc | 13 | Family contacts |
| 10 | 3 | 6/98 | 6 | Susc | 13 | Family contacts |
| 11 | 3 | 9/98 | 6 | Susc | 13 | Family contacts |
| 12 | 4 | 11/97 | 12 | Susc | 8 | Family contacts |
| 13 | 4 | 1/98 | 12 | Susc | 8 | Family contacts |
| 14 | 4 | 7/98 | 12 | Susc | 8 | Family contacts |
| 15 | 4 | 11/98 | 12 | Susc | 8 | Family contacts |
| 16 | 5 | 3/97 | 12 | Susc | 9 | Contact tracing |
| 17 | 5 | 4/97 | 12 | Susc | 9 | Contact tracing |
| 18 | 5 | 6/97 | 12 | Susc | 9 | Contact tracing |
| 19 | 5 | 10/97 | 12 | Susc | 9 | Contact tracing |
| 20 | 5 | 3/98 | 12 | Susc | 9 | Contact tracing |
| 21 | 6 | 6/97 | 14 | Susc | 7 | Contact tracing |
| 22 | 6 | 7/97 | 14 | Susc | 7 | Contact tracing |
| 23 | 6 | 3/98 | 14 | Susc | 7 | Contact tracing |
| 24 | 6 | 7/98 | 14 | Susc | 7 | Contact tracing |
| 25 | 6 | 8/98 | 14 | Susc | 7 | Contact tracing |
| 26 | 7 | 4/97 | 20 | Susc | 7 | Contact tracing |
| 27 | 7 | 6/97 | 20 | Susc | 7 | Contact tracing |
| 28 | 7 | 10/97 | 20 | Susc | 7 | Contact tracing |
| 29 | 7 | 10/97 | 20 | Susc | 7 | Contact tracing |
| 30 | 7 | 12/97 | 20 | Susc | 7 | Contact tracing |
| 31 | 7 | 1/98 | 20 | Susc | 7 | Contact tracing |
| 32 | 7 | 4/98 | 20 | Susc | 7 | Contact tracing |
| 33 | 7 | 7/98 | 20 | Susc | 7 | Contact tracing |
| 34 | 7 | 8/98 | 20 | Susc | 7 | Contact tracing |
| 35 | 7 | 12/98 | 20 | Susc | 7 | Contact tracing |
| 36 | 7 | 12/98 | 20 | Susc | 7 | Contact tracing |
| 37 | 8 | 6/97 | 17 | Susc | 8 | Contact tracing |
| 38 | 8 | 3/98 | 17 | Susc | 8 | Contact tracing |
| 39 | 8 | 9/98 | 17 | Susc | 8 | Contact tracing |
| 40 | 8 | 11/98 | 17 | Susc | 8 | Contact tracing |
| 41 | 9 | 7/95 | 3 | H, R, S, Z | 11 | Family contacts |
| 42 | 9 | 10/95 | 3 | H, R, S, Z | 11 | Family contacts |
| 43 | 10 | 7/95 | ∼1 | H, S | 18 | Family contacts |
| 44 | 10 | 7/95 | ∼1 | E, H, S | 18 | Family contacts |
| 45 | 11ac | 9/95 | 3 | E, H, P, R, S | 16 | Contact tracing |
| 46 | 11ac | 12/95 | 3 | E, H, P, R, S | 16 | Contact tracing |
| 47 | 11bc | 1/95 | 27 | H, R, S | 16 | Family contacts |
| 48 | 11bc | 4/97 | 27 | H, R, S | 16 | Family contacts |
| 49 | 12 | 11/95 | ∼1 | E, H, P, R, S, Z | 10 | Family contacts |
| 50 | 12 | 11/95 | ∼1 | E, H, P, R, S, Z | 10 | Family contacts |
| 51 | 13 | 3/95 | 2 | H, R, Rb | 6 | Family contacts |
| 52 | 13 | 5/95 | 2 | H, R, Rb | 6 | Family contacts |
| 53 | 14 | 1/97 | ∼1 | E, H, P, S | 16 | Family contacts |
| 54 | 14 | 1/97 | ∼1 | E, H, P, R, S | 16 | Family contacts |
| 55 | 15 | 3/96 | 4 | H, R | 11 | Family contacts |
| 56 | 15 | 7/96 | 4 | E, H, R | 11 | Family contacts |
| 57 | 16 | 9/96 | 9 | H | 13 | Family contacts |
| 58 | 16 | 6/97 | 9 | H | 13 | Family contacts |
| 59 | 16 | 6/97 | 9 | H | 13 | Family contacts |
| 60 | 17 | 2/96 | 8 | E, H, R, S, Z | 10 | Family contacts |
| 61 | 17 | 10/96 | 8 | E, H, R, S, Z | 10 | Family contacts |
| 62 | 18 | 11/95 | 2 | E, H, R | 13 | Family contacts |
| 63 | 18 | 1/96 | 2 | E, H, R | 13 | Family contacts |
| 64 | 19 | 12/95 | 6 | E, H, R, Z | 8 | Family contacts |
| 65 | 19 | 6/96 | 6 | H | 8 | Family contacts |
| 66 | 20 | 10/96 | 29 | S, R | 7 | Contact tracing |
| 67 | 20 | 1/97 | 29 | S, R | 7 | Contact tracing |
| 68 | 20 | 2/97 | 29 | S, R | 7 | Contact tracing |
| 69 | 20 | 4/97 | 29 | S, R | 7 | Contact tracing |
| 70 | 20 | 6/97 | 29 | S, R | 7 + 1d | Contact tracing |
| 71 | 20 | 11/97 | 29 | S, R | 7 | Contact tracing |
| 72 | 20 | 12/97 | 29 | S, R | 7 | Contact tracing |
| 73 | 20 | 10/98 | 29 | S, R | 7 | Contact tracing |
| 74 | 20 | 11/98 | 29 | S, R | 7 | Contact tracing |
| 75 | 20 | 3/99 | 29 | S, R | 7 | Contact tracing |
Time span between the first and last isolate within one cluster.
Susc, susceptible. Other abbreviation indicate resistance as follows: E, ethambutol; H, isoniazid; P, protionamide; R, rifampin; Rb, rifabutin; S, streptomycin; Z, pyrazinamide.
These isolates showed identical IS6110 RFLP patterns, but no epidemiological relationship has been verified among the patients of cluster 11a and the patients of cluster 11b.
One additional band was found.
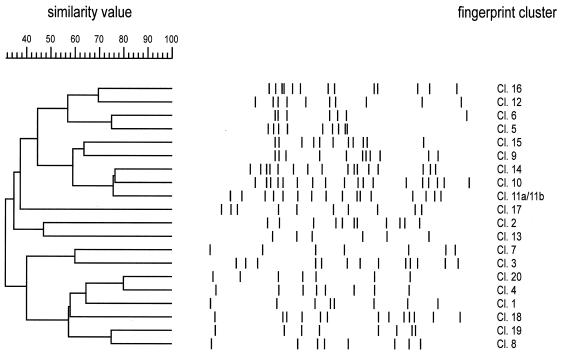
The IS6110 fingerprint patterns of each cluster have been analyzed for similarity to prove the identity of the clusters that were initially based on epidemiological data. Except for those of two clusters, the RFLP patterns showed a high variability (Fig. 1). The isolates of clusters 11a (isolates 45 and 46) and 11b (isolates 47 and 48) showed identical RFLP patterns, but no epidemiological relationship between the patients of cluster 11a and the patients of cluster 11b could be verified. Hence, they were further regarded as two different epidemiological clusters for the calculation of the rate of change of IS6110 RFLP patterns.
FIG. 1.
IS6110 banding patterns and dendrogram of IS6110 fingerprint clusters 1 to 20. One pattern is shown for each cluster. The positions of IS6110 bands have been normalized and are displayed as lanes. The scale depicts the similarity of patterns calculated as described in Materials and Methods.
Among the 75 strains analyzed, only 1 showed a change of the IS6110 fingerprint pattern (one additional band) (Fig. 2) compared to the other 9 strains in the corresponding fingerprint cluster (cluster 20) (Table 1). The epidemiological relationship of the isolates within this cluster could be confirmed by contact tracing of the public health office and additionally by drug resistance to rifampin and streptomycin. Thus, the rate of change amounted to ∼1.9% per transmission when the possible number of transmissions was calculated by subtracting one patient per cluster (index patient) from the total number of patients (number of changed isolates/total number of isolates − total number of clusters) or 11% if only the corresponding cluster was regarded. The changed isolate was obtained 8 months after the retrieval of the first isolate of this cluster; however, five isolates with no changes in their fingerprint pattern were obtained up to 21 months later. Hence, the number of changes in the IS6110 RFLP patterns of M. tuberculosis isolates that belong to a particular outbreak may not be correlated to the length of the time span between the collection of the isolates, but changes may be induced by chance during infection of a new host.
FIG. 2.
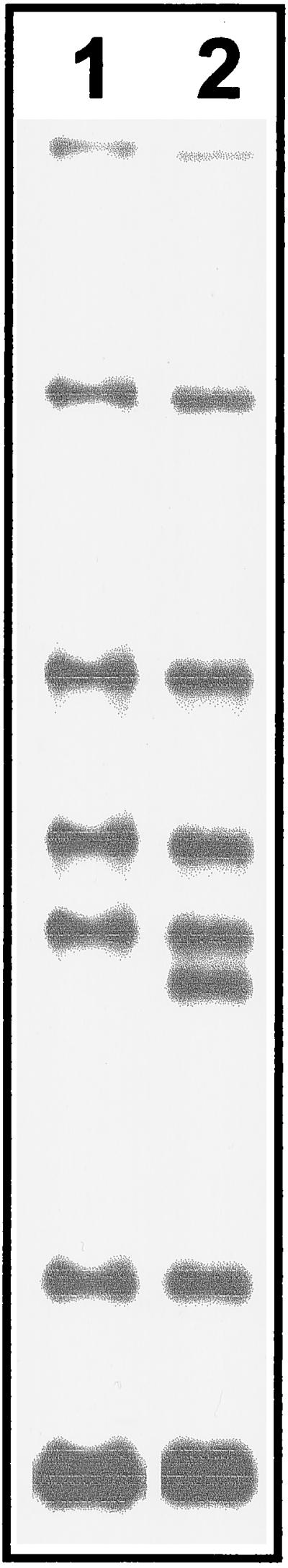
RFLP patterns of the M. tuberculosis strains obtained from patient 1 (lane 1) and patient 5 (lane 2) of cluster 20. The isolate from patient 5 shows a clearly visible additional IS6110 RFLP band.
As reported previously (7, 12), changes in the drug resistance pattern of the isolates in one cluster were not accompanied by changes of the IS6110 RFLP pattern (see clusters 10, 15, and 19). Moreover, we did not observe the instability of IS6110 RFLP patterns in clusters of MDR M. tuberculosis isolates to be greater than that of susceptible strains as reported by Alito et al. (1).
In the present study, a high degree of stability of M. tuberculosis IS6110 RFLP patterns in actual transmission chains could be demonstrated, indicating that the rate of change of IS6110 RFLP patterns in the situation of person-to-person transmission of M. tuberculosis strains is low. Compared to studies analyzing serial patient isolates (4, 7, 12), our study revealed a remarkably higher stability of IS6110 RFLP patterns in clusters of transmitted M. tuberculosis strains. Even considering the one cluster with a changed IS6110 RFLP pattern, the rate of change (11%) still was less than half that observed by Yeh et al. (25% [12]) when analyzing serial isolates of patients living in the San Francisco area.
In conclusion, M. tuberculosis IS6110 RFLP patterns in actual transmission chains showed a high degree of stability that appears to be higher than that observed for serial patient isolates. The isolates within a cluster in the present study were obtained over time spans of up to 29 months, indicating that for periods of more than 2 years IS6110 fingerprint clusters based on identical isolates can account for nearly all cases of recent transmission in a study population. Hence, clustering rates that are based on IS6110 fingerprint clusters including strains with one or two band changes may result in an overestimation of the rate of recent transmission.
ACKNOWLEDGMENTS
We thank I. Radzio, B. Schlüter, and A. Zyzik, Forschungszentrum Borstel, Borstel, Germany, for excellent technical assistance, and T. Goldmann, Forschungszentrum Borstel, for critical reading of the manuscript.
Parts of this work were supported by the Robert Koch-Institut, Berlin, Germany.
REFERENCES
- 1.Alito A, Morcillo N, Scipioni S, Dolmann A, Romano M I, Cataldi A, van Soolingen D. The IS6110 restriction fragment length polymorphism in particular multidrug-resistant Mycobacterium tuberculosis strains may evolve too fast for reliable use in outbreak investigation. J Clin Microbiol. 1999;37:788–791. doi: 10.1128/jcm.37.3.788-791.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bauer J, Yang Z, Poulsen S, Anderson Å B. Results from 5 years of nationwide DNA fingerprinting of Mycobacterium tuberculosis complex isolates in a country with a low incidence of M. tuberculosis infection. J Clin Microbiol. 1998;36:305–308. doi: 10.1128/jcm.36.1.305-308.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cave M D, Eisenach K D, McDermott P F, Bates J H, Crawford J T. IS6110: conservation of sequence in the Mycobacterium tuberculosis complex and its utilization in DNA fingerprinting. Mol Cell Probes. 1991;5:73–80. doi: 10.1016/0890-8508(91)90040-q. [DOI] [PubMed] [Google Scholar]
- 4.De Boer A S, Borgdorff M W, de Haas P E, Nagelkerke N J, van Embden J D A, van Soolingen D. Analysis of rate of change of IS6110 RFLP patterns of Mycobacterium tuberculosis based on serial patient isolates. J Infect Dis. 1999;180:1238–1244. doi: 10.1086/314979. [DOI] [PubMed] [Google Scholar]
- 5.Kent P T, Kubica G P. Public health mycobacteriology. A guide for the level III laboratory. U.S. Atlanta, Ga: Department of Health and Human Services, Centers for Disease Control; 1985. [Google Scholar]
- 6.Niemann S, Rüsch-Gerdes S, Richter E. IS6110 fingerprinting of drug-resistant Mycobacterium tuberculosis strains isolated in Germany during 1995. J Clin Microbiol. 1997;35:3015–3020. doi: 10.1128/jcm.35.12.3015-3020.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Niemann S, Richter E, Rüsch-Gerdes S. Stability of Mycobacterium tuberculosis IS6110 restriction fragment length polymorphism patterns and spoligotypes determined by analyzing serial isolates from patients with drug-resistant tuberculosis. J Clin Microbiol. 1999;37:409–412. doi: 10.1128/jcm.37.2.409-412.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Niemann S, Richter E, Rüsch-Gerdes S, Thielen H, Heykes-Uden H. Outbreak of rifampin and streptomycin resistant tuberculosis among homeless in Germany. Int J Tuberc Lung Dis. 1999;3:1146–1147. [PubMed] [Google Scholar]
- 9.Van Embden J D A, Cave M D, Crawford J T, Dale J W, Eisenach K D, Gicquel B, Hermans P, Martin C, McAdam R, Shinnick T M, Small P M. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J Clin Microbiol. 1993;31:406–409. doi: 10.1128/jcm.31.2.406-409.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van Soolingen D, de Haas P E W, Hermans P W M, Groenen P M A, van Embden J D A. Comparison of various repetitive DNA elements as genetic markers for strain differentiation and epidemiology of Mycobacterium tuberculosis. J Clin Microbiol. 1993;31:1987–1995. doi: 10.1128/jcm.31.8.1987-1995.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Soolingen D, Hermans P W M. Epidemiology of tuberculosis by DNA fingerprinting. Eur Respir J. 1995;8(Suppl. 20):649s–656s. [PubMed] [Google Scholar]
- 12.Yeh R W, Ponce de Leon A, Agasino C B, Hahn J A, Daley C L, Hopewell P C, Small P M. Stability of Mycobacterium tuberculosis DNA Genotypes. J Infect Dis. 1998;177:1107–1111. doi: 10.1086/517406. [DOI] [PubMed] [Google Scholar]