Abstract
We describe a highly sensitive assay for quantitation of varicella-zoster virus (VZV) DNA in blood, involving PCR amplification, solution hybridization with Tris-(2,2′-bipyridine)-ruthenium(II) chelate-labeled probes, and measurement by electrochemiluminescence (ECL). Extraction and amplification efficiencies were monitored by the inclusion of internal control (IC) DNA, mimicking the VZV target, in the DNA extraction. Viral DNA load was calculated from the ratio of VZV and IC ECL signals. The lower limit of sensitivity was 20 VZV DNA copies/ml of plasma or serum and 80 copies/ml of whole blood. In reconstruction experiments, expected and calculated VZV DNA loads were in excellent accordance. Blood specimens from 42 VZV-infected patients were tested for the presence of VZV DNA and showed detection rates of 86% in patients with varicella and 81% in patients with herpes zoster. In specimens obtained during the first week after onset of the rash, detection rates were 100 and 89%, respectively. Viral DNA was detected in all immunocompromised patients with herpes zoster, emphasizing the risk of disseminated disease in this patient group. VZV DNA load was similar in patients with varicella and multidermatomal herpes zoster and lower in patients with unidermatomal zoster. Despite the cell-associated nature of the virus, VZV DNA was detected in serum and plasma at high copy numbers, and at similar frequencies compared to whole-blood specimens. Quantitation of VZV DNA in blood is of potential importance for diagnosis and clinical management of VZV-infected patients. Plasma and serum provide convenient matrices for this purpose.
Varicella-zoster virus (VZV) is a human alphaherpesvirus that causes chicken pox (varicella) during primary infection, following which the virus establishes latency in cells of the dorsal root ganglia. Reactivation of latent virus causes shingles (herpes zoster), a disease predominantly occurring in elderly or immunocompromised patients.
Varicella and herpes zoster are usually clinical diagnoses, based on the typical morphology and distribution of the skin lesions. Confirmation of the diagnosis can be obtained by immunofluorescence or immunoperoxidase staining of vesicle scrapings and viral culture of vesicle fluid. However, the sensitivities of these diagnostic methods are limited and highly dependent on the quality of scrapings, handling time of vesicle fluids, and stage of the skin lesions. Alternative sensitive diagnostic methods may thus be helpful.
In the pathogenesis of varicella, a viremic phase occurs, beginning during the incubation period of the disease, which allows the virus to disseminate to cutaneous epithelial cells and produce the characteristic varicella lesions (1). In herpes zoster, the virus is transmitted from the ganglion, via neuronal axons, to the epidermis of the corresponding dermatome, causing localized disease (1). Despite this apparently contained transmission pathway, viremia has also been reported to occur during localized VZV reactivation, as suggested by the detection of viral DNA in peripheral blood mononuclear cells (PBMC) of some patients with dermatomal zoster and zoster-associated pain (5, 8, 9, 11, 13, 14).
Detection and quantitation of the viremia during VZV infections are potentially useful for diagnostic, prognostic, and therapeutic monitoring purposes (9). However, it is generally assumed that, similar to in vitro infections, the viremia during VZV infections is highly cell associated (1). Possibly due to this assumption, reported studies of VZV viremia, as detected by PCR, have restricted their analyses to detection of viral DNA in PBMC, which require elaborate methods for isolation (5, 7–14). Until now, other, more convenient blood compartments, such as plasma or serum, have not been studied.
We have developed a sensitive assay for detection and quantitation of VZV DNA, which has the same format as a recently described quantitative assay for detection of cytomegalovirus DNA in plasma and serum (3). With this method, VZV DNA could be detected in substantial copy numbers, not only in whole blood but also in plasma and serum from a large proportion of patients with varicella as well as herpes zoster.
MATERIALS AND METHODS
VZV DNA.
Quantified VZV DNA, prepared from sucrose density gradient-purified VZV (strain rod) and quantified electron microscopically by direct particle count, which discriminates between full and empty particles, was obtained from Advanced Biotechnologies Inc. (Columbia, Md.). Results of limiting-dilution experiments followed by PCR indicated that the concentration of VZV DNA given by the manufacturer was accurate (data not shown).
Laboratory parameters for VZV infection.
Viral culture of vesicle fluids or scrapings was done by cocultivation with human embryo lung fibroblast cells and microscopic examination of VZV-specific cytopathological effects. Confirmation was done by immunofluorescent staining of cells with murine monoclonal anti-VZV immunoglobulin M (IgM) (BioWhittaker, Inc., Walkersville, Md.). VZV IgM in serum was detected by immunofluorescence (VZV IgM immunofluorescence assay; Gull Laboratories, 's-Hertogenbosch, The Netherlands).
Patients.
Whole-blood, plasma, or serum specimens were tested from 42 patients presenting at the outpatient department of or admitted to the Academic Medical Center with a clinical diagnosis of varicella (21 persons) or herpes zoster (21 persons), which was confirmed by positive cultures of vesicle fluids or detection of VZV-specific IgM in serum. In addition, blood specimens from 30 individuals without evidence of VZV disease were tested.
Specimen processing and DNA purification.
After centrifugation (10 min, 125 × g), plasma was separated from EDTA-anticoagulated blood by pipetting, leaving approximately 1 cm of plasma untouched in the collection tube to prevent contamination by cells. In a subset of patients, separated plasma was again centrifuged for 30 min at 1,750 × g to remove any possible remaining cells. Since the PCR results were not influenced by this procedure (data not shown), this was not performed for all samples. Serum was separated from clotted whole blood after centrifugation for 10 min at 1,750 × g.
After the addition of 20 molecules of internal control (IC) DNA (see below), DNA was purified from 50 μl of EDTA-anticoagulated whole blood or 200 μl of plasma or serum, as described previously (3). DNA was eluted in 100 μl of TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8.0).
PCR.
Primers and probes were obtained from Perkin-Elmer B.V. (Nieuwerkerk a/d IJssel, The Netherlands). The primer pair used for amplification consisted of VZV-3 (5′-TCT TTC ACG GAG GCA AAC AC-3′) and Bio-VZV-4 (5′-TCC AAG GCG GGT GCA TAT CT-3′; 5′ biotinylated) (7). This primer pair amplifies a 161-bp DNA fragment from VZV gene 29 (nucleotide [nt] positions 51133 to 51293 according to numbering by Davison and Scott [4]), encoding the major DNA-binding protein, as well as a fragment of identical size and GC content from IC DNA (see below). Primers were diluted in TE buffer to 100 ng/μl. In the qualitative diagnostic PCR (D-PCR), 25 μl of the DNA eluate was used as input in the PCR. In the quantitative PCR (Q-PCR), 15 μl of DNA eluate and 10 μl of reference IC DNA (4 molecules/μl) were subjected to PCR. The uracil-N-glycosylase system (Perkin-Elmer) was used to prevent false-positive reactions due to carryover of amplimers.
The final reaction mixture (50 μl) contained 200 ng of each primer, 2.5 U of Ampli-Taq DNA polymerase (Perkin-Elmer); 0.5 U of uracil-N-glycosylase (Perkin-Elmer); 5 μg of bovine serum albumin (Boehringer-Mannheim B.V., Almere, The Netherlands); 10 mM Tris-HCl (pH 8.3); 50 mM KCl; 4 mM MgCl2; dATP, dGTP, and dCTP at concentrations of 200 μM each; and 400 μM dUTP (Perkin-Elmer). The PCRs were performed in a Perkin-Elmer 9600 thermocycler, as follows: 2 min at 50°C and 5 min at 95°C, followed by 35 cycles each consisting of 20 s at 95°C, 20 s at 63°C, and 1 min at 72°C, followed by 5 min at 72°C.
IC DNA.
Restriction enzymes and T4 DNA ligase were obtained from Boehringer-Mannheim B.V. For construction of IC DNA, the above-mentioned 161-bp amplimer was generated by PCR from VZV strain rod DNA (Advanced Biotechnologies Inc.), using primers VZV-3 and nonbiotinylated VZV-4. The amplimer was digested with AvaI and visualized on an agarose gel containing 1 μg of ethidium bromide per ml, and a 70-bp fragment (nt 51133 to 51202) was purified from the gel. In addition, the amplimer was digested with DraII, followed by purification of a 58-bp fragment (nt 51234 to 51293) from the gel. From two in vitro-synthesized 5′-phosphorylated single-stranded DNAs (5′-TCG ACA CCT GTC GGA TCC GTA GTT GCT GTA AG-3′ and 5′-GCC CTT ACA GCA ACT ACG GAT CCG ACA GGT G-3′), a double-stranded DNA sequence was obtained by hybridization of the complementary sequences. This DNA sequence contained DNA overhangs and was ligated to the AvaI site of the 70-bp fragment and to the DraII site of the 58-bp fragment, generating a 161-bp fragment again. Relative to the wild-type VZV sequence, the AvaI site (5′-CTCGAG-3′, nt 51202 to 51207) was thus replaced by the sequence 5′-CTCGAC-3′, and the VZV sequence at nt positions 51208 to 51231 was replaced by the sequence 5′-ACC TGT CGG ATC CGT AGT TGC TGT-3′ to serve as a probe area. The latter sequence contains a BamHI restriction site and has the same, albeit shuffled, nucleotide content as the wild-type sequence. These modifications allow for discrimination between VZV and IC DNA amplimers by digestion with the restriction enzymes AvaI and BamHI or by hybridization with probes specific for either VZV DNA- or IC DNA-specific probe areas (see below). The constructed DNA fragment was amplified by PCR with primers VZV-3 and nonbiotinylated VZV-4. Purified amplimer was cloned into a plasmid vector (PCRII; Promega, Leiden, The Netherlands), resulting in a plasmid pVZV marker which served as IC DNA. The pVZV marker was purified from bacterial cultures as described previously, linearized by HindIII digestion, and purified by the Boom procedure (2, 3). The DNA was quantified by measuring UV absorption at 260 nm and was stored in TE buffer (at 10 μg/ml) at −70°C. Dilutions of linearized plasmid were made in TE buffer containing 20 ng of human placental DNA (Sigma Chemical Company, lot no. 160H3807) per μl to stabilize dilute DNA preparations during storage. A different batch of human placental DNA (lot no. 74H3848) was used as carrier DNA in initial experiments and yielded false-positive results at a low frequency (data not shown). These findings suggested the presence of VZV DNA at low copy number in this batch of placental DNA. No false-positive results have been observed since the use of the current batch of carrier DNA. Reference IC DNA contained four molecules of linearized plasmid, as determined by limiting dilution followed by PCR, and 20 ng of human placental DNA per μl.
Hybridization and measurement by ECL.
The probes used for hybridization were the VZV-specific probe TBR-VZV-1 [5′-AAC GGT TTG GGT TTT CAC GCT GCC-3′, 5′ labeled with Tris-(2,2′-bipyridine)-ruthenium(II) chelate (TBR)] and the IC DNA-specific probe TBR-VZV-2 (5′-ACC TGT CGG ATC CGT AGT TGC TGT-3′, 5′ labeled with TBR). The probes were diluted in 1× PCRII buffer (Perkin-Elmer) to 1 ng/μl. Prior to hybridization, excess primers were removed from the PCR products as described previously (3). For D-PCR, the purified PCR product was used directly for hybridization; for Q-PCR, the PCR product was diluted 15-fold in 1× PCRII buffer before hybridization. Hybridization with VZV DNA- and IC DNA-specific probes was performed in separate reactions by adding 20 μl of probe (either TBR-VZV-1 or TBR-VZV-2) to 30 μl of purified PCR product, followed by incubation of the mixtures for 2 min at 95°C and 5 min at 60°C in a 9600 thermocycler (Perkin-Elmer). Next, 10 μl of streptavidin-coated magnetic beads (Perkin-Elmer) was added, followed by incubation for another 15 min at 60°C. Forty microliters of the bead-hybrid suspension was added to 400 μl of Origen assay buffer (Biozym, Landgraaf, The Netherlands), supplemented with sodium azide to 0.05%, and the electrochemiluminescence (ECL) signal, expressed in luminosity units (LU), was measured in the Q-PCR System 5000 (Perkin-Elmer). In this system, streptavidin-coated beads are concentrated magnetically and subsequently washed with Origen washing buffer (Biozym), after which bound hybrids are detected by ECL.
Criteria for D-PCR.
Each test was run in duplicate with the inclusion of three controls in the DNA extraction: one positive control (12.5 copies of VZV rod DNA [Advanced Biotechnologies Inc.]) and two negative controls. The first negative control contained 100 ng of human placental DNA and 6.25 copies of IC DNA and served as a control for the complete procedure. The second negative control contained 100 ng of human placental DNA only and thus should give negative results with both probes.
A signal of >100 LU, equal to four times the mean background signal for either probe, was considered to be positive. If neither VZV nor IC DNA could be detected, the result was considered inconclusive, and the test was repeated. To date, inconclusive results have not been obtained.
Algorithm for quantitation in Q-PCR.
In the Q-PCR, DNA was purified from 200 μl of plasma or serum, or 50 μl of whole blood, together with 20 molecules of IC DNA, and DNA was eluted in 100 μl of TE buffer. Fifteen microliters of DNA was subjected to Q-PCR in the presence of an additional 40 molecules of IC DNA present in the PCR master mixture, and the ECL signals obtained after hybridization were determined as described above. After correction for the background, the ratio of VZV-specific signal to IC DNA-specific signal (R) was determined, and the virus load, expressed as VZV DNA copies per milliliter, was calculated by amplifying R by a factor of 1,433 for plasma or serum, and by a factor of 5,733 for whole-blood specimens. This factor was determined from two separate sets of factors: (3 + 40) × 33.33 or 133.33. The factor (3 + 40) represents the number of IC DNA molecules present during the PCR, and the factor 33.33 or 133.33 is required to calculate the copy number per milliliter of plasma or serum and of whole blood, respectively.
Statistical analysis.
VZV DNA load was analyzed after log transformation. The lower limit of the assay was used in the case of negative PCR results. For group comparisons, the independent-sample t test or Mann-Whitney U test was performed where appropriate. Correlation coefficients (r) and slopes were obtained by least-squares linear regression. All analyses were performed with SPSS 8.0 for Windows software (SPSS, Inc., Chicago, Ill.).
RESULTS
Specificity and lower limit of detection.
No signals were obtained when DNAs of other herpesviruses were used at PCR inputs of up to 25,000 copies, including herpes simplex virus type 1 MacIntyre, herpes simplex virus type 2 G, Epstein-Barr virus B95-8, human herpesvirus 6 Z-29, and cytomegalovirus AD169 (Advanced Biotechnologies Inc.). From 33 Dutch patients with varicella or herpes zoster, VZV isolates obtained after culture of vesicle fluids and stored at −70°C were tested and were all positive (data not shown).
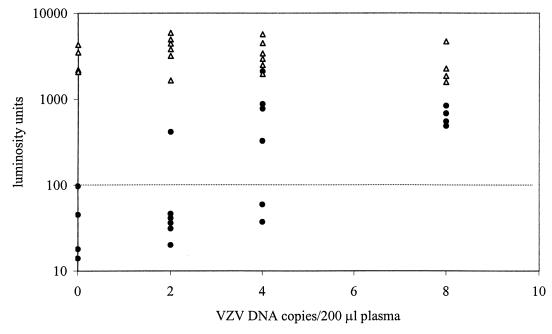
As previously reported, the Boom method for purification of DNA is characterized by a nearly 100% recovery of DNA from plasma, even in the case of very low DNA input (3). To study the lower limit of detection of the assay, VZV-negative plasma was supplemented with VZV DNA to a concentration of 40 copies/ml and serial twofold dilutions thereof. Figure 1 shows that, with an extraction input as low as four copies per 200 μl of plasma, resulting in one copy of VZV DNA in the PCR, the assay was positive in four of six reactions. With 100% extraction efficiency, this is in accordance with the number of expected positive reactions according to the Poisson distribution in the case of the presence of a single molecule of target DNA per PCR (6). Negative results were truly negative since the ECL signals for coextracted IC DNA were all positive. Extraction inputs of eight copies per 200 μl of plasma all yielded positive results. Together, these data indicate that the assay can detect virus loads as low as 20 copies per ml of plasma or serum (extraction input, 200 μl) and 80 copies per ml of whole blood (extraction input, 50 μl).
FIG. 1.
Lower limit of detection of VZV DNA in plasma. DNA was purified from 200 μl of plasma containing 20 molecules of IC DNA and zero, two, four, or eight VZV DNA copies. Extractions were done fourfold for specimens containing zero and eight VZV DNA copies and sixfold for specimens containing two and four copies. One-quarter of extracted DNA was subjected to PCR, and the amounts of VZV amplimers (circles) and IC amplimers (triangles) were determined by ECL. The cutoff level is indicated by the dotted line.
Quantitative assay.
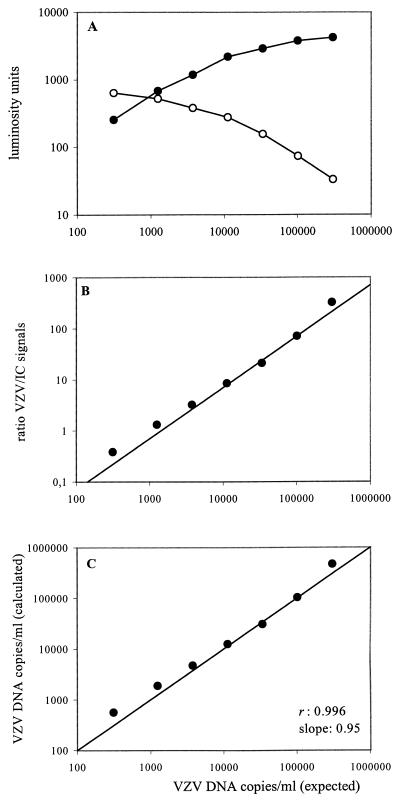
VZV-negative plasma was supplemented with VZV DNA to a concentration of 607,500 copies/ml and threefold dilutions thereof and was subjected to Q-PCR. Figure 2A represents the ECL signals obtained by Q-PCR after hybridization and shows that the amount of VZV amplimers increased with increasing virus load until a plateau was reached, while IC DNA signals decreased with higher virus loads. The latter observation was due to the fact that the plateau for IC DNA was reached earlier with increasing virus loads. When the plateau is reached during PCR, IC DNA amplification also enters a plateau phase at a level determined by the virus load. Therefore, the initial ratio of VZV DNA to IC DNA present at the start of the PCR will be maintained throughout the PCR, including the plateau phase. Figure 2B shows that, with increasing virus loads, the VZV DNA/IC DNA ratios (R) followed a straight line, the slope of which corresponded with the expected ratios. To calculate the virus load, expressed as the number of VZV DNA copies per milliliter, the ratio was amplified by a constant factor (see Materials and Methods for the algorithm). As shown in Fig. 2C, the expected and calculated virus loads were in excellent accordance for copy numbers ranging from 625 to 607, 500 VZV DNA copies/ml.
FIG. 2.
Q-PCR. VZV-negative plasma was supplemented with VZV DNA to a concentration of 607,500 copies/ml and threefold dilutions thereof and subjected to Q-PCR. (A) ECL signals (LU) obtained with the VZV-specific probe (closed circles) and the IC-specific probe (open circles). (B) Values of the VZV DNA/IC DNA ratio (R) after background correction. (C) VZV DNA loads as calculated by amplifying R by a constant factor (1,433). The correlation coefficient (r) and slope were obtained by least-squares linear regression analysis.
Detection rate of VZV DNA in clinical specimens.
Forty-two patients with culture-proven or serologically proven varicella (21 patients) or herpes zoster (21 patients) were tested for the presence of VZV DNA in whole blood, plasma, or serum. When available, more than one matrix was tested (Table 1). The time between onset of the rash and blood sampling was similar in both groups of patients (median, 2 days, and range, 0 to 10 days, in both groups; P = 0.51). In the herpes zoster group, 15 patients were immunocompromised due to immunosuppressive therapy or human immunodeficiency virus infection. Six patients with herpes zoster had multidermatomal or disseminated disease. In the varicella group, six patients were immunocompromised.
TABLE 1.
Detection rate and quantity of VZV DNA in blood from patients with VZV infections
| Infection type | No. of patients | % VZV DNA positive (no. of patients positive/total no. of patients)
|
VZV DNA load [mean log copies/ml (SD)]
|
|||
|---|---|---|---|---|---|---|
| Total | Whole blood | Plasma or serum | Whole blood | Plasma or serum | ||
| Varicella | 21 | 85.7 (18/21) | 87.5 (7/8) | 88.9 (16/18) | 3.2 (0.7) | 3.3 (1.4) |
| Herpes zoster | ||||||
| Dermatomal | 15 | 73 (11/15) | 90 (9/10) | 66.7 (6/9) | 2.3 (0.4) | 2.0 (0.6) |
| Disseminated or multidermatomal | 6 | 100 (6/6) | 100 (5/5) | 100 (6/6) | 3.7 (1.0) | 3.6 (1.2) |
| No VZV disease | 30 | 0 (0/30) | 0 (0/20) | 0 (0/10) | ||
As shown in Table 1, VZV DNA could be detected in the blood of 86% of patients with varicella and 81% of patients with herpes zoster. In specimens obtained within the first week after onset of the rash, the detection rates were 100 and 89%, respectively. In herpes zoster patients, negative results were observed only with immunocompetent patients. Blood specimens from individuals without symptoms of VZV infection were invariably VZV negative. Detection rates of VZV DNA were similar in whole blood, plasma, and serum. No discrepant results between whole blood and plasma or serum were observed for patients from whom both matrices were tested. Of note, VZV DNA could also be detected in the blood of seven patients with a clinical diagnosis of varicella (four patients) or herpes zoster (three patients), for whom cultures of vesicle fluids were negative (data not shown).
Quantitation of VZV DNA in clinical specimens.
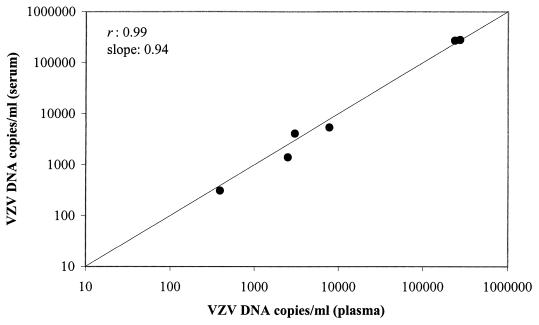
Paired plasma and serum specimens, both obtained at the same time, were available from six patients. Quantitation of VZV DNA in these specimens revealed highly similar viral DNA loads (Fig. 3). For this reason, results from plasma and serum specimens were combined in subsequent analyses of VZV DNA load.
FIG. 3.
VZV DNA loads in paired plasma and serum specimens from six VZV-infected patients. The correlation coefficient (r) and slope were obtained by least-squares linear regression analysis.
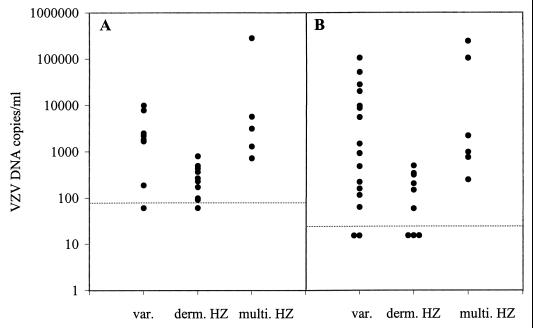
Quantitative analyses of viral DNA load in the blood of the above-mentioned 42 patients with VZV disease are shown in Table 1 and Fig. 4. VZV DNA load in both whole blood and plasma or serum was significantly higher in varicella patients than in patients with unidermatomal herpes zoster (whole blood, P = 0.014; plasma or serum, P = 0.012). A similar difference in virus load was observed between patients with multidermatomal or disseminated zoster and patients with unidermatomal zoster (whole blood, P = 0.002; plasma or serum, P = 0.003). The virus loads in varicella patients and in patients with multidermatomal or disseminated zoster did not differ significantly. The numbers of patients were too small to allow for comparisons between immunocompetent and immunocompromised patients. However, virus loads in excess of 1 million copies/ml were observed only for immunocompromised patients.
FIG. 4.
VZV DNA loads in whole blood (A) and plasma or serum (B) from patients with varicella (var.), unidermatomal zoster (derm. HZ), and disseminated or multidermatomal zoster (multi. HZ). The lower limits of detection are indicated by the dotted lines.
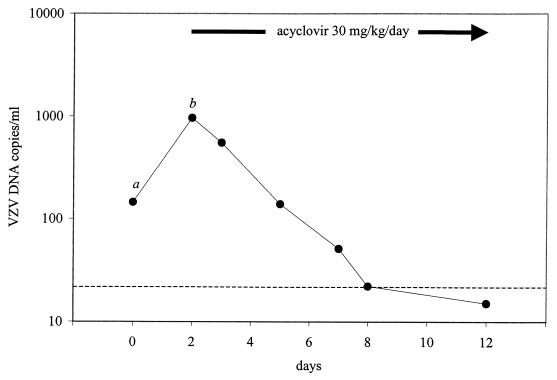
Figure 5 depicts the results of Q-PCR in seven consecutive plasma specimens from an immunocompromised patient who presented with a unidermatomal zoster involving S2, which was not correctly diagnosed at that time. Two days later, she returned with progression of the skin lesions to S3, which coincided with an 0.8 log increase in VZV DNA load. After the institution of antiviral treatment, VZV DNA load decreased sharply, nearing the lower limit of the assay within 6 days.
FIG. 5.
Q-PCR of seven consecutive plasma specimens from a patient who progressed from unidermatomal herpes zoster (a) to multidermatomal disease (b). Antiviral treatment was initiated at day 2. The dashed line signifies the lower limit of detection.
A large fraction of whole-blood VZV DNA is present in plasma or serum.
A striking observation was the detection of VZV DNA in serum and plasma at high copy numbers and similar frequencies compared to whole-blood specimens. Assuming a maximum fraction of plasma or serum in blood of 50%, which will represent an underestimation in most cases, the ratio of virus load in plasma or serum to virus load in whole blood could be calculated for 10 patients in whom it was measured in both matrices. The median ratio, which provides an estimation of the fraction of viral DNA in blood that is present in serum or plasma, was 0.39 (range, 0.06 to 1.78).
DISCUSSION
We developed a highly sensitive assay for detection and quantitation of VZV DNA, using the same format as a recently described quantitative method for detection of cytomegalovirus DNA (3). The assay involved PCR amplification, followed by solution hybridization with nonradioactive TBR-labeled probes, and measurement by ECL. IC DNA, mimicking the VZV target, was included in the DNA extraction of clinical specimens. IC DNA and VZV were amplified with the same primer pair, yielding amplimers with identical sizes and GC contents. The only difference was a shuffled nucleotide sequence at the probe area of the IC DNA, which enabled discrimination between IC DNA and VZV amplimers. The inclusion of IC DNA controls for variations in extraction and amplification efficiency, e.g., due to inhibitory substances, thereby excluding false-negative results. The uracil-N-glycosylase system was used to prevent false-positive results due to amplimer carryover.
The lower limit of detection of the D-PCR was 20 VZV DNA copies/ml of plasma or serum and 80 copies/ml of whole blood. Since 20 molecules of IC DNA were present during DNA extraction, a clinical specimen negative for VZV DNA but positive for IC DNA should formally be interpreted as containing less than 100 copies of VZV DNA/ml of plasma or serum and less than 400 VZV DNA copies/ml of whole blood.
Since VZV and IC DNA were amplified with similar efficiencies using the same primer pair, the initial ratio of target to IC DNA remained constant throughout amplification. Therefore, the viral DNA load could be calculated by a simple algorithm from the ratio of ECL signals of VZV DNA to those of IC DNA, after solution hybridization with VZV- and IC-specific probes. In our quantitative format (Q-PCR), the dynamic range of measurement was extended by including an additional amount of IC DNA during PCR and diluting the PCR products prior to hybridization. In reconstruction experiments, expected and calculated VZV DNA loads were in excellent accordance up to 600,000 copies/ml.
We tested 42 patients with culture-proven or serologically proven VZV infections for the presence of VZV DNA in blood specimens and found detection rates which are higher than previously reported (9–12). In patients with varicella, VZV DNA could be detected in 100% of blood specimens obtained within 1 week after the onset of the skin rash. Strikingly, in immunocompromised patients with herpes zoster, most of whom did not show signs of disseminated or multidermatomal disease, VZV DNA was also detected in all cases. This suggests that, rather than being an exception, the occurrence of viremia during herpes zoster is a rule in immunocompromised patients and emphasizes the potential risk of disseminated disease in this patient group. Quantitative analyses showed that viral DNA loads were similar in patients with varicella and patients with disseminated or multidermatomal zoster and significantly lower in individuals with unidermatomal zoster. Prospective studies are needed to evaluate whether the development of disseminated zoster can be predicted by the level of VZV DNA in blood. Likewise, the potential use of quantitation of VZV DNA load for the purpose of therapeutic monitoring requires further study. Interestingly, quantitative analysis of sequential plasma specimens from a patient who progressed from unidermatomal to multidermatomal herpes zoster showed a coincident marked increase in VZV DNA load, followed by a sharp decline upon the institution of antiviral treatment.
The viremia during VZV infections is thought to be highly cell associated (1). This assumption seems largely based on the cell-associated nature of the virus when grown in vitro. Perhaps due to this assumption, attempts to detect viral DNA in blood have been restricted to analyses of isolated PBMC (5, 7–14). In the present study, VZV DNA could be detected not only in whole blood but also, at similar frequencies and in high copy numbers, in plasma and serum of patients with varicella and herpes zoster. Viral DNA loads were equal in plasma and serum. It was estimated that a considerable proportion (median, 39%) of VZV DNA in blood is present in the cell-free compartment. This proportion seemed to exhibit a substantial degree of variability, the source of which requires further research. At present, the precise origin of the viral DNA in plasma and serum is unclear, and we cannot exclude the possibility that the VZV DNA detected in these specimens was derived from lysed infected cells. Alternatively, VZV DNA as detected in plasma and serum may originate from circulating infectious virus, implying that VZV viremia is less cell associated than currently assumed. Further investigations are needed to elucidate the relative contributions of both possibilities. Until more insight has been gained into the nature and clinical relevance of detectable VZV DNA in the respective blood compartments, as well as into the source of variability in the plasma fraction of VZV DNA, whole blood probably represents the preferred matrix for diagnosis.
Irrespective of the origin of VZV DNA, whole blood, plasma, or serum is a more convenient biological matrix for diagnosis of VZV viremia than PBMC, which require elaborate methods for isolation. Furthermore, the detection in plasma and serum enables retrospective analysis of stored specimens. In our view, for diagnostic, prognostic, and therapeutic monitoring purposes, quantitation of viremia is of potential importance in the clinical management of patients with VZV infections. For this reason, the availability of sensitive quantitative methods and convenient biological matrices for diagnosis of VZV viremia is important.
ACKNOWLEDGMENTS
We thank the clinicians of the Departments of Pediatrics and Internal Medicine, in particular Taco Kuijpers, Joep Lange, and Jan van der Meer, for providing clinical specimens and all coworkers of the Laboratory of Clinical Virology for their technical support.
REFERENCES
- 1.Arvin A A, Moffat J F, Redman R. Varicella-zoster virus: aspects of pathogenesis and host response to natural infection and varicella vaccine. Adv Virus Res. 1996;46:263–309. doi: 10.1016/s0065-3527(08)60074-3. [DOI] [PubMed] [Google Scholar]
- 2.Boom R, Sol C, Salimans M M M, Janssen C L, Wertheim-van Dillen P, Van der Noordaa J. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boom R, Sol C, Weel J, Gerrits Y, De Boer M, Wertheim-van Dillen P. A highly sensitive assay for detection and quantitation of human cytomegalovirus DNA in serum and plasma by PCR and electrochemiluminescence. J Clin Microbiol. 1999;37:1489–1497. doi: 10.1128/jcm.37.5.1489-1497.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davison A J, Scott J E. The complete DNA sequence of varicella-zoster virus. J Gen Virol. 1986;67:1759–1816. doi: 10.1099/0022-1317-67-9-1759. [DOI] [PubMed] [Google Scholar]
- 5.Devlin M E, Gilden D H, Mahalingam R, Dueland A N, Cohrs R. Peripheral blood mononuclear cells of the elderly contain varicella-zoster virus DNA. J Infect Dis. 1992;165:619–622. doi: 10.1093/infdis/165.4.619. [DOI] [PubMed] [Google Scholar]
- 6.Diaco R. Practical considerations for the design of quantitative PCR assays. In: Innis M A, Gelfland D A, Sninsky J J, editors. PCR strategies. New York, N.Y: Academic Press, Inc.; 1995. pp. 84–108. [Google Scholar]
- 7.Furuta Y, Fukuda S, Suzuki S, Takasu T, Inuyama Y, Nagashima K. Detection of varicella-zoster virus DNA in patients with acute peripheral facial palsy by the polymerase chain reaction, and its use for early diagnosis of zoster sine herpete. J Virol Methods. 1997;52:316–319. [PubMed] [Google Scholar]
- 8.Gilden D H, Devlin M E, Wellish M, Mahalingam R, Huff J C, Hayward A R, Vafai A. Persistence of varicella-zoster virus DNA in blood mononuclear cells of patients with varicella or zoster. Virus Genes. 1989;2:299–305. doi: 10.1007/BF00684037. [DOI] [PubMed] [Google Scholar]
- 9.Hawrami K, Breuer J. Development of a fluorogenic polymerase chain reaction assay (TaqMan) for the detection and quantitation of varicella zoster virus. J Virol Methods. 1999;79:33–40. doi: 10.1016/s0166-0934(98)00176-1. [DOI] [PubMed] [Google Scholar]
- 10.Koropchak C M, Graham G, Palmer J, Winsberg M, Ting S F, Wallace M, Prober C G, Arvin A A. Investigation of varicella-zoster virus infections by polymerase chain reaction in the immunocompetent host with acute varicella. J Infect Dis. 1991;163:1016–1022. doi: 10.1093/infdis/163.5.1016. [DOI] [PubMed] [Google Scholar]
- 11.Mainka C, Fuss B, Geiger H, Hofelmayr H, Wolff M H. Characterization of viremia at different stages of varicella-zoster virus infection. J Med Virol. 1998;56:91–98. doi: 10.1002/(sici)1096-9071(199809)56:1<91::aid-jmv15>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 12.Sawyer M M, Wu Y N, Chamberlin C J, et al. Detection of varicella-zoster virus DNA in the oropharynx and blood of patients with varicella. J Infect Dis. 1992;166:885–888. doi: 10.1093/infdis/166.4.885. [DOI] [PubMed] [Google Scholar]
- 13.Vafai A, Wellish M, Gilden D H. Expression of varicella-zoster virus in blood mononuclear cells of patients with postherpetic neuralgia. Proc Natl Acad Sci USA. 1988;85:2767–2770. doi: 10.1073/pnas.85.8.2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vonsover A, Leventon-Kriss S, Langer D, et al. Detection of varicella-zoster virus in lymphocytes by DNA hybridization. J Med Virol. 1987;21:57–66. doi: 10.1002/jmv.1890210108. [DOI] [PubMed] [Google Scholar]