Abstract
The purpose of this research was to develop a simple, sensitive, and accurate method for simultaneous determination of 35 free amino acids using ultra-performance liquid chromatography coupled with quadrupole time-of-flight mass spectrometry (UPLC-Q-TOF/MS). Tea samples were extracted with boiling water bath, and then separated by XBridge BEH Amide column by gradient elution. The exact mass and MS/MS spectra of the target compound was detected under the TOF–MS and Information dependent acquisition (IDA)–MS/MS mode. The results demonstrated good linearity (R2 > 0.9980) in the range of 0.5–1,000 ng/mL. The limits of detection (LODs) were 0.13–25.00 mg/kg and the limits of quantitation (LOQs) were 0.25–50.00 mg/kg. The recovery rate ranged from 70.1 to 105.1% with relative standard deviations (RSDs) <11% (n = 6). This research provides a targeted strategy for developing an analysis method for amino acids in tea.
Keywords: ultra-performance liquid chromatography, quadrupole-time of flight mass spectrometry, tea, free amino acids, detection
Introduction
Tea, which originated in China, is one of the most popular and widely consumed beverage in the world because of its refreshing taste, attractive aroma, and potential health benefits (1). The production and processing of tea have a significant impact on the economic and social development in China due to the huge economic benefits associated with it. In recent years, there has been extensive research on the valorization and health benefits of medicinal plants (in general) and teas (in particular), but systematic and in-depth research on the chemical components of flavor in tea is still weak in China (2). Amino acids and other molecules such as carotenoids, fatty acids, glycosides, and sugars contribute to the aroma in teas (3, 4). Free amino acids play an important role in the chemical composition of tea (5), which is one of the important evaluation factors of tea quality (6). Amino acids are the basic units of proteins, which are also the important components of active peptidases and other bioactive molecules (7–9). The composition and content of amino acids and their degradation products and transformation products also directly affect the quality of tea. Amino acids participate in the formation of aroma in tea processing, that is because amino acids converted into volatile aldehydes or other products during tea processing, forming tea aroma. In addition, some amino acids themselves have a certain fragrance and are among the most important ingredients that contribute to the quality of tea. For example, theanine, which is a unique amino acid in tea, can relieve the bitterness and astringency of the tea itself due to its fresh smell and caramel aroma, and is the main ingredient that produces sweetness in tea (10). Alanine and glutamic acid have floral aroma characteristics. Serine and tyrosine have wine aroma characteristics (11). The free amino acids in tea participate in the formation of tea color, aroma, and taste through a variety of ways (12). The content and combination of ingredients have a direct impact on the taste of tea soup and the quality of tea (13), which requires accurate and rapid amino acid detection technology.
In recent years, many scholars have devoted themselves to the research on the detection technology of amino acids in tea, and various analytical methods have been proposed including ninhydrin colorimetric method (14–16), amino acid automatic analyzer method (17), high performance liquid chromatography (HPLC) (18–20), and high performance liquid chromatography–triple quadrupole mass spectrometry (HPLC-QQQ-MS) (21–25). Traditionally, the determination of the total amount of free amino acids in tea has been conducted by the ninhydrin colorimetric method, which is stipulated in the national standard (26). However, the limitation of this strategy is that it can only measure the total amount of amino acids and cannot accurately measure the content of individual amino acid components (27). The determination using automated amino acid analyzer is a traditional method for the analysis of free amino acids. The disadvantage of the method is post-column derivatization with long analysis times and low sensitivity (28). At present, high performance liquid chromatography method mostly involves pre-column derivatization technology, in which the measuring procedures are complicated and greatly affected by derivatization reagents (29–32). The accuracy and operability of derivatization need to be further improved. Liquid chromatography–mass spectrometry (LC–MS) is a powerful analytical technique, and depending on the mass analyzer coupled to the separation technique offers other advantages (33). LC-QQQ-MS and LC-QTrap-MS have high specificity but low resolution, and there are problems such as serious matrix interference and little information about compound fragmentation. On the other hand, LC-Orbitrap-MS and LC-Q-TOF/MS approaches have high resolution and accuracy, and have strong anti-interference ability and wide scanning range (34, 35). In this study, UPLC-Q-TOF/MS was used for the first time to detect 35 free amino acids in tea. This method provides technical support for quality control and research and development of tea, and promotes the sustainable development of the industry.
Materials and Methods
Reagents and Standards
Acetonitrile was HPLC-MS grade and purchased from Fisher Scientific (Loughborough, UK). Water was HPLC grade and supplied by Watson's Food & Beverage Co., Ltd. (Guangzhou, China). Formic acid was HPLC grade supplied by Sigma–Aldrich (St. Louis, Missouri, USA). Magnesium oxide was purchased from Kemiou Chemical Reagent Co., Ltd. (Tianjin, China). The origin of congou black tea was Hangzhou, Zhejiang Province, China. The origin of Yellow mountain fuzz tip was Huangshan City, Anhui Province, China. The origin of rizhao green tea was Yantai City, Shandong Province, China. The origin of green tea 1 was Suzhou City, Jiangsu Province, China. The origin of green tea 2 was Suzhou City, Jiangsu Province, China. The origin of maojian tea was Hangzhou City, Zhejiang Province, China. The origin of yacca tea was Hangzhou, Zhejiang Province, China. The origin of taiping houkui tea was Huangshan City, Anhui Province, China. The origin of sword-shaped green tea was Shaoxing City, Zhejiang Province, China. All dry tea samples were purchased from local tea shops in Shijiazhuang, China. All samples were stored in dark and dry places at room temperature.
L-Arginine (purity ≥ 99.0%), L-aspartic acid (purity ≥ 99.0%), L-glutamic acid (purity ≥ 99.0%), glycine (purity ≥ 99.5%), L-histidine (purity ≥ 98.0%), L-isoleucine (purity ≥ 99.0%), L-leucine (purity ≥ 99.0%), L-lysine (purity ≥ 98.0%), L-methionine (purity ≥ 99.0%), L-phenylalanine (purity ≥ 99.0%), L-proline (purity ≥ 99.0%), L-serine (purity ≥ 99.0%), L-threonine (purity ≥ 99.0%), L-tyrosine (purity ≥ 99.0%), L-valine (purity ≥ 99.0%), and L-alanine (purity ≥ 99.0%) were supplied from J&K Scientific Ltd. (Shanghai, China). L-Cysteine (purity ≥ 99.4%) was supplied from Dr. Ehrenstorfer (Augsburg, Germany). Theanine (purity ≥ 98.0%), L-tryptophan (purity ≥ 99.0%), L-asparagine (purity ≥ 98.7%), and L-glutamine (purity ≥ 99.5%) were supplied from ANPEL Laboratory Technologies (Shanghai, China). L-cystine (purity ≥ 99.3%), aminoadipic acid (purity ≥ 98.0%), sarcosine (purity ≥ 98.0%), L-pipecolic acid (purity ≥ 98.0%), α-aminobutyric acid (purity ≥ 98.0%), β-aminobutyric acid (purity ≥ 98.0%), citrulline (purity ≥ 98.0%), hydroxylysine (purity ≥ 98.0%), 1-methyl-L-histidine (purity ≥ 98.0%), and 3-methyl-L-histidine (purity ≥ 98.0%) were purchased from Sigma (California, USA). Hydroxyproline (purity ≥ 98.0%), γ-aminobutyric acid (purity ≥ 98.5%), DL-homocysteine (purity ≥ 98.0%), and L-ornithine (purity ≥ 98.0%) were purchased from Chromadex Trading Company (California, USA).
Instruments
High-speed refrigerated centrifuge (3K13, Sigma, USA), vortex mixer (Vortex Genius 3, IKA, Germany), and magnetic stirrer with heating (RET CV S025, IKA, Germany) were used in the procedure of sample preparation. The separation of compounds were carried out on an UPLC system (LC-30AD, Shimadzu, Japan). Quantitative analysis of 35 free amino acids in tea was conducted on a Q-TOF/MS (TripleTOFTM 5600+, Sciex, USA). OS software (Version 1.5.0, Sciex, USA) was used for data processing.
Methods
Sample Preparation
Refer to GB/T 8312-2013 (36) for sample preparation: 0.5 g crushed tea (dry leaves) was weighed into a 500 mL breaker followed by addition of 4.5 g magnesium oxide and 250 mL boiling water. The solution was extracted for 20 min in a boiling water bath. The prepared tea soup was transferred to a 50 mL centrifuge tube and centrifuged for 5 min at 2,128 g at room temperature. The supernatant was filtered through a 0.22 μm filter membrane. When the sample content was too high, it was diluted to the linear range with ultrapure water according to the actual concentration.
UPLC Conditions
The chromatographic separation was carried out on a Waters XBridge BEH Amide column (2.1 × 150 mm, 2.5 μm) with a flow rate of 0.4 mL/min. The mobile phases were 0.2% formic acid in water (A) and 0.2% formic acid in acetonitrile (B). The elution gradient was carried out for 20 min as follows: 0–3.00 min, 90% B; 3.0–13.0 min, 90% to 48% B; 13.0–15.0 min, 48% B; 15.0–16.0 min, 48% to 90% B; and 16.0–20.0 min, 90% B. The injection volume was 5.0 μL.
Q-TOF/MS Conditions
For the MS analysis, the Triple TOFTM5600+ equipped with a DuoSprayTM ion source was used. In this study, the electrospray ionization (ESI) source was used for detection and atmospheric pressure chemical ionization (APCI) source was used for calibration. The ionization voltage was set at 5.5 kV and the source temperature at 400°C under positive mode. The curtain gas pressure was 35 psi, the nebulizer gas pressure was 50 psi, and the auxiliary gas pressure was 55 psi. The TOF MS data were collected between m/z 20 and m/z 500 with a duration time of 20 min, and accumulation time was 0.15 s. The information dependent acquisition (IDA)-MS/MS conditions were as follows: accumulation time was 0.05 s, isotopes within 4 Da were excluded, the declustering potential was 80 V, and the collision energy was 40 ± 20 V. Automatic batch calibration was performed to ensure the accuracy and reproducibility.
Database Establishment
Under the instrument conditions given in sections UPLC Conditions and Q-TOF/MS Conditions, 35 amino acid standard solutions were analyzed, and the retention time, accurate mass, isotope distribution, and MS/MS spectrum of each compound were obtained (Table 1). In the Product Ion mode of Triple TOF, when the response value of the target in the retention time window exceeded the threshold, it automatically triggered the acquisition and superimposition of the MS/MS spectrums under different collision energies (20, 40, and 60 eV), thereby establishing a library of MS/MS spectrum of the compound. Import the MS/MS spectrums into the Library module of OS 1.5.0 software to create a mass spectrum database of 35 amino acids.
Table 1.
Mass spectral parameters for the 35 amino acids.
| Compound name | Formula | Retention time (min) |
Adduct/
charge |
Theoretical mass (m/z) | Experimental mass (m/z) | Mass error (ppm) | Isotope ratio difference (%) |
|---|---|---|---|---|---|---|---|
| L-Arginine | C6H14N4O2 | 10.190 | [M+H]+ | 175.11895 | 175.11968 | 4.2 | 0.4 |
| L-Aspartic acid | C4H7NO4 | 7.876 | [M+H]+ | 134.04478 | 134.04471 | −0.6 | 0.5 |
| L-Cysteine | C3H7NO2S | 6.302 | [M+H]+ | 122.02703 | 122.02692 | −0.9 | 1.5 |
| L-Cystine | C6H12N2O4S2 | 10.789 | [M+H]+ | 241.03113 | 241.03148 | 1.5 | 2.4 |
| L-Glutamic acid | C5H9NO4 | 7.557 | [M+H]+ | 148.06043 | 148.06055 | 0.8 | 0.8 |
| Glycine | C2H5NO2 | 6.716 | [M+H]+ | 76.03930 | 76.03962 | 4.2 | 1.2 |
| L-Histidine | C6H9N3O2 | 10.282 | [M+H]+ | 156.07675 | 156.07688 | 0.8 | 0.3 |
| L-Isoleucine | C6H13NO2 | 9.908 | [M+H]+ | 132.10191 | 132.10202 | 0.9 | 0.1 |
| L-Leucine | C6H13NO2 | 10.239 | [M+H]+ | 132.10191 | 132.10202 | 0.9 | 0.1 |
| L-Lysine | C6H14N2O2 | 10.415 | [M+H]+ | 147.11280 | 147.11278 | −0.1 | 0.8 |
| L-Methionine | C5H11NO2S | 4.421 | [M+H]+ | 150.05833 | 150.05836 | 0.2 | 1.7 |
| L-Phenylalanine | C9H11NO2 | 2.420 | [M+H]+ | 166.08626 | 166.08689 | 3.8 | 2.4 |
| L-Proline | C5H9NO2 | 4.620 | [M+H]+ | 116.07060 | 116.07063 | 0.2 | 1.6 |
| L-Serine | C3H7NO3 | 8.149 | [M+H]+ | 106.04987 | 106.04996 | 0.8 | 1.4 |
| L-Threonine | C4H9NO3 | 7.457 | [M+H]+ | 120.06552 | 120.06562 | 0.8 | 0.1 |
| L-Tyrosine | C9H11NO3 | 5.626 | [M+H]+ | 182.08117 | 182.08129 | 0.6 | 0.9 |
| L-Valine | C5H11NO2 | 4.204 | [M+H]+ | 118.08626 | 118.08631 | 0.4 | 0.6 |
| L-Alanine | C3H7NO2 | 6.031 | [M+H]+ | 90.05495 | 90.05502 | 0.7 | 1.3 |
| Theanine | C7H14N2O3 | 6.039 | [M+H]+ | 175.10772 | 175.10757 | −0.8 | 0.4 |
| L-Tryptophan | C11H12N2O2 | 4.113 | [M+H]+ | 205.09715 | 205.09657 | −2.9 | 0.6 |
| L-Asparagine | C4H8N2O3 | 8.421 | [M+H]+ | 133.06077 | 133.06076 | −0.1 | 1.7 |
| L-Glutamine | C5H10N2O3 | 8.107 | [M+H]+ | 147.07642 | 147.07622 | −1.4 | 1.1 |
| Aminoadipic acid | C6H11NO4 | 7.144 | [M+H]+ | 162.07608 | 162.07594 | −0.9 | 1.0 |
| α-Aminobutyric acid | C4H9NO2 | 4.751 | [M+H]+ | 104.07060 | 104.07050 | −1.0 | 3.3 |
| β-Aminobutyric acid | C4H9NO2 | 4.095 | [M+H]+ | 104.07060 | 104.07061 | 0.1 | 3.9 |
| γ-Aminobutyric acid | C4H9NO2 | 4.427 | [M+H]+ | 104.07060 | 104.07050 | −1.0 | 3.3 |
| Citrulline | C6H13N3O3 | 8.555 | [M+H]+ | 176.10297 | 176.10302 | 0.3 | 0.6 |
| Hydroxyproline | C5H9NO3 | 6.961 | [M+H]+ | 132.06552 | 132.06550 | −0.2 | 0.5 |
| Hydroxylysine | C6H14N2O3 | 10.953 | [M+H]+ | 163.10772 | 163.10748 | −1.5 | 1.0 |
| 1-Methyl-L-histidine | C7H11N3O2 | 8.023 | [M+H]+ | 170.09240 | 170.09261 | 1.2 | 3.4 |
| 3-Methyl-L-histidine | C7H11N3O2 | 8.293 | [M+H]+ | 170.09240 | 170.09272 | 1.8 | 2.5 |
| L-Ornithine | C5H12N2O2 | 10.571 | [M+H]+ | 133.09715 | 133.09692 | −1.8 | 1.0 |
| Sarcosine | C3H7NO2 | 4.188 | [M+H]+ | 90.05495 | 90.05502 | 0.7 | 1.3 |
| DL-Homocysteine | C4H9NO2S | 4.821 | [M+H]+ | 136.04268 | 136.04254 | −1.0 | 1.1 |
| L-Pipecolic acid | C6H11NO2 | 4.344 | [M+H]+ | 130.08626 | 130.08633 | 0.6 | 1.0 |
Results and Discussion
Optimization of UPLC-Q-TOF/MS Conditions
Liquid chromatography–mass spectrometry spectra were studied in both positive and negative modes. The results showed that 35 amino acids had higher response in positive mode, and so the positive mode was used for detection in this study. The experiment investigated the response of the 35 amino acids under different declustering potentials (50, 80, 100, 120, and 150 V), and it was found that the amino acid responses were the highest at the declustering potential of 80 V. A lower declustering potential was not conductive to ion transmission, and excessively high declustering potential caused the target compound to fragment within the source. The accurate mass deviations of target compounds were <5.0 × 10−6 (Table 1). The MS/MS spectra of the 35 amino acids (Supplementary Figure 1) was used for the final confirmation of the initial screening results.
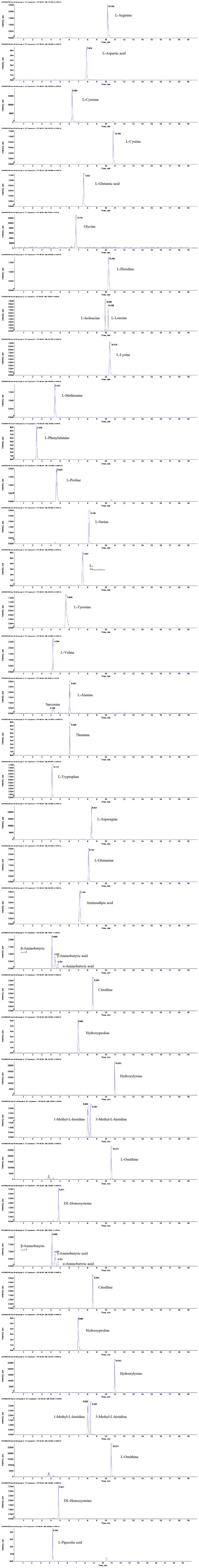
A series of preliminary experiments was carried out using different chromatographic columns including Waters Acquity UPLC HSS T3 (2.1 × 100 mm, 1.8 μm), Waters XBridge BEH C18 (2.1 × 100 mm, 2.5 μm), Waters Cortecs UPLC HILIC (2.1 × 100 mm, 1.6 μm), and Waters XBridge BEH Amide (2.1 × 150 mm, 2.5 μm). The results showed (Supplementary Figure 2) that the 35 amino acids were not retained on Waters Acquity UPLC HSS T3 and Waters XBridge BEH C18 chromatographic columns. The peak shape obtained on Waters Cortecs UPLC HILIC chromatographic column was poor and wide. The extracted ion chromatograms of the 35 amino acids precursor ions are illustrated in Figure 1. The results showed that the peak shape of the analyte obtained by the Waters XBridge BEH Amide column was the best, and could effectively separate the four groups of isomers. L-alanine and sarcosine displayed the same protonated ion [M+H]+ at m/z 90.05502 and 90.05502, respectively, with the same molecular formula C3H7NO2. α-Aminobutyric acid, β-aminobutyric acid, and γ-aminobutyric acid displayed a similar protonated ion [M+H]+ at m/z 104.07050, 104.07061, and 104.07050, respectively, with the same molecular formula C4H9NO2. L-isoleucine and L-leucine displayed the same protonated ion [M+H]+ at m/z 132.10202 with the same molecular formula C6H13NO2. 1-Methyl-L-histidine and 3-methyl-L-histidine displayed a similar protonated ion [M+H]+ at m/z 170.09261 and 170.09272, respectively, with the same molecular formula C7H11N3O2. Therefore, the Waters XBridge BEH Amide column was selected as the analytical column.
Figure 1.
Extracted ion chromatograms of the 35 amino acids precursor ions.
This experiment also compared different mobile phase systems (water–acetonitrile, water–methanol, 0.1% formic acid water−0.1% formic acid acetonitrile, and 0.2% formic acid water−0.2% formic acid acetonitrile) to obtain better resolution and chromatographic separation ability. In the positive mode, the addition of formic acid provided more H+, which helped to enhance the response intensity of the target compound. When the organic phase of the mobile phase system was methanol and acetonitrile, the chromatographic response and peak shape of the target compound were not much different, but when methanol was used as the mobile phase, a longer equilibration time was required and the system pressure fluctuated greatly. Therefore, water containing 0.2% (v/v) formic acid and acetonitrile containing 0.2% (v/v) formic acid were selected as the mobile phase. Three different flow rates (0.3 mL/min, 0.4 mL/min, and 0.5 mL/min) were tested to obtain a fast and reliable separation. The results indicated that 0.4 mL/min was the optimal flow rate considering the resolution, run time, and column pressure. Under the optimal UPLC conditions, the extracted ion chromatograms of the 35 amino acids are presented in Figure 1.
Method Validation
Linearity and Sensitivity
The calibration curves were generated from the relationship between the peak area and the concentration of target compounds with a concentration range of 0.5–1,000 μg/L. LOD was defined as the lowest concentration of free amino acids in sample, which can be detected but not quantified. LOQ was determined as the lowest concentration of free amino acids in sample, which can be quantified with acceptable precision and accuracy under the normal operating conditions of the method. A series of spiked samples was prepared to determine the LOD and LOQ when the signal-to-noise ratios of the 35 amino acids were about 3 and 10, respectively. As shown in Table 2, the linearity of the 35 amino acids were satisfactory with correlation coefficients (R2) higher than 0.9980. The LODs and LOQs of the 35 amino acids were 0.13–25.00 and 0.25–50.0 mg/kg, respectively.
Table 2.
Linearity, LOD, and LOQ of 35 amino acids.
| Compound | Linear equation |
Linear range (μg/L) |
R 2 |
LOD (mg/kg) |
LOQ (mg/kg) |
Recovery (RSD)* | ||
|---|---|---|---|---|---|---|---|---|
| 50 mg/kg | 150 mg/kg | 500 mg/kg | ||||||
| L-Arginine | Y = 712.49596X+1855.66828 | 2.5–1,000 | 0.99914 | 0.50 | 1.25 | 77.3 (4.73) | 90.7 (3.66) | 87.4 (3.90) |
| L-Aspartic acid | Y = 245.91896X+44.40999 | 20–1,000 | 0.99963 | 5.00 | 10.00 | 80.3 (9.10) | 91.0 (7.52) | 86.2 (5.53) |
| L-Cysteine | Y = 128.74993X-2483.03825 | 50–1,000 | 0.99944 | 12.5 | 25.00 | 80.0 (4.47) | 89.4 (9.39) | 96.3 (4.18) |
| L-Cystine | Y = 371.23366X+5246.96481 | 2.5–1,000 | 0.99954 | 0.50 | 1.25 | 83.3 (9.56) | 91.8 (5.27) | 95.8 (5.01) |
| L-Glutamic acid | Y = 392.30557X+10.05360 | 5–1,000 | 0.99962 | 1.25 | 2.50 | 72.7 (9.80) | 92.9 (5.19) | 99.7 (2.47) |
| Glycine | Y = 42.62232X+758.58867 | 100–1,000 | 0.99828 | 25.0 | 50.00 | 84.3 (3.49) | 90.9 (6.06) | 94.0 (4.99) |
| L-Histidine | Y = 652.69714X+2906.99013 | 2.5–1,000 | 0.99916 | 0.50 | 1.25 | 81.0 (4.62) | 91.9 (8.96) | 93.3 (4.58) |
| L-Isoleucine | Y = 400.87005X+7.90515e4 | 20–1,000 | 0.99834 | 5.00 | 10.00 | 76.3 (8.99) | 90.4 (5.39) | 89.6 (8.52) |
| L-Leucine | Y = 246.71219X+25428.48128 | 20–1,000 | 0.99895 | 5.00 | 10.00 | 80.3 (6.57) | 88.6 (9.67) | 89.7 (5.60) |
| L-Lysine | Y = 388.81504X-9916.88241 | 5–1,000 | 0.99944 | 1.25 | 2.50 | 88.0 (6.89) | 89.2 (8.50) | 91.9 (6.71) |
| L-Methionine | Y = 794.93701X-1225.64686 | 5–1,000 | 0.99951 | 1.25 | 2.50 | 82.3 (4.18) | 95.0 (1.31) | 100.8 (0.95) |
| L-Phenylalanine | Y=1047.32901X+3.00590e4 | 0.5-1000 | 0.99810 | 0.13 | 0.25 | 71.0 (10.2) | 89.3 (7.17) | 87.8 (5.38) |
| L-Proline | Y = 2785.34443X+15998.35998 | 2.5–1,000 | 0.99995 | 0.50 | 1.25 | 77.7 (8.53) | 80.8 (8.97) | 89.8 (5.25) |
| L-Serine | Y = 175.19613X-459.03363 | 10–1,000 | 0.99862 | 2.50 | 5.00 | 77.3 (8.60) | 88.4 (8.94) | 89.2 (7.78) |
| L-Threonine | Y = 319.78661X-640.30454 | 10–1,000 | 0.99941 | 2.50 | 5.00 | 77.0 (9.40) | 82.3 (9.67) | 88.1 (6.18) |
| L-Tyrosine | Y = 695.20787X+1382.31684 | 5–1,000 | 0.99988 | 1.25 | 2.50 | 80.3 (4.57) | 105.1 (5.26) | 95.3 (3.94) |
| L-Valine | Y = 660.48179X+19098.57317 | 15–1,000 | 0.99959 | 3.75 | 7.50 | 90.0 (7.57) | 94.9 (2.50) | 102.0 (1.23) |
| L-Alanine | Y = 111.59111X-1462.13625 | 20–1,000 | 0.99864 | 5.00 | 10.00 | 86.0 (4.16) | 93.0 (2.21) | 96.3 (4.37) |
| Theanine | Y = 1776.92273X+6744.45179 | 5–1,000 | 0.99984 | 1.25 | 2.50 | 79.7 (9.45) | 94.6 (2.11) | 92.1 (8.12) |
| L-Tryptophan | Y = 798.22194X+5509.98570 | 5–1,000 | 0.99989 | 1.25 | 2.50 | 89.0 (7.35) | 92.7 (1.88) | 101.8 (0.45) |
| L-Asparagine | Y = 271.81905X-678.93195 | 10–1,000 | 0.99944 | 2.50 | 5.00 | 79.7 (10.1) | 90.0 (8.63) | 92.6 (4.49) |
| L-Glutamine | Y = 268.81006X+3744.16195 | 5–1,000 | 0.99916 | 1.25 | 2.50 | 79.0 (11.0) | 72.9 (7.47) | 88.3 (3.11) |
| Aminoadipic acid | Y = 252.66609X-1009.17764 | 10–1,000 | 0.99864 | 2.50 | 5.00 | 84.7 (5.10) | 91.4 (3.37) | 91.6 (6.44) |
| α-Aminobutyric acid | Y = 30.95211X+2875.25568 | 50–1,000 | 0.99856 | 12.5 | 25.00 | 86.0 (6.06) | 94.2 (4.28) | 89.3 (0.89) |
| β-Aminobutyric acid | Y = 304.38085X-7311.81303 | 10–1,000 | 0.99897 | 2.50 | 5.00 | 86.7 (5.40) | 92.4 (1.75) | 70.1 (3.59) |
| γ-Aminobutyric acid | Y = 237.63091X | 10–1,000 | 0.99920 | 2.50 | 5.00 | 73.7 (6.74) | 89.9 (6.56) | 89.1 (3.02) |
| Citrulline | Y = 326.91270X-359.40922 | 5–1,000 | 0.99919 | 1.25 | 2.50 | 96.3 (8.45) | 93.7 (1.33) | 93.6 (1.23) |
| Hydroxyproline | Y = 1188.51708X-10472.55309 | 5–1,000 | 0.99803 | 1.25 | 2.50 | 86.7 (4.30) | 90.9 (7.36) | 97.1 (2.20) |
| Hydroxylysine | Y = 147.61396X+158.38431 | 10–1,000 | 0.99949 | 2.50 | 5.00 | 88.3 (5.98) | 97.0 (1.29) | 95.1 (0.74) |
| 1-Methyl-L-histidine | Y = 1959.69840X-8.65412e4 | 5–1,000 | 0.99817 | 1.25 | 2.50 | 88.0 (4.77) | 97.9 (1.85) | 95.9 (4.59) |
| 3-Methyl-L-histidine | Y = 1096.39834X-18386.66480 | 5–1,000 | 0.99983 | 1.25 | 2.50 | 86.7 (9.53) | 93.0 (5.24) | 95.6 (1.88) |
| L-Ornithine | Y = 353.22736X-12881.75504 | 50–1,000 | 0.99975 | 12.5 | 25.00 | 82.7 (8.47) | 90.2 (8.50) | 88.5 (3.76) |
| Sarcosine | Y = 196.41758X+11684.55602 | 50–1,000 | 0.99953 | 12.5 | 25.00 | 86.7 (4.04) | 85.9 (7.09) | 85.0 (2.95) |
| DL-Homocysteine | Y = 142.21203X+6753.17341 | 50–1,000 | 0.99887 | 12.5 | 25.00 | 88.0 (5.75) | 86.7 (6.88) | 79.3 (8.59) |
| L-Pipecolic acid | Y = 4544.31048X-1525.44628 | 5–1,000 | 0.99857 | 1.25 | 2.50 | 73.0 (9.92) | 89.2 (6.22) | 92.0 (3.12) |
Accuracy expressed as average trueness of the calculated (quantified) concentration value, and relative standard deviation (RSD) of n = 6 replicate samples.
Recovery and Precision
The recovery and precision experiments were carried out by adding 35 amino acid standard solutions to the known amino acid content of tea sample. Samples spiked at three different concentrations (50, 150, and 500 mg/kg) for each of the amino acids, and each concentration was repeated six times. As shown in Table 2, the average recovery rate ranged from 70.1 to 105.1%, and the RSDs ranged from 0.45 to 11.0%. The recoveries were considered acceptable data to the method, and the results indicated that the precision was reasonable.
Application to Actual Samples
In order to investigate the content of the 35 amino acids in tea, nine tea samples from local tea shops were analyzed using the established method in this research, and each sample was repeated three times. The results (Table 3) showed that the compositions and contents of 35 amino acids were different in the nine tea samples. Except for L-cysteine, L-cystine, glycine, aminoadipic acid, α-aminobutyric acid, β-aminobutyric acid, hydroxylysine, L-ornithine, and DL-homocysteine, the content of amino acids were higher than LOQs. There was little difference in the types of amino acids in different teas. Theanine was the dominating amino acid in tea infusions of different cultivars, followed by L-arginine, L-aspartic acid, L-glutamine, L-valine, L-glutamic acid, and L-asparagine. L-Phenylalanine is associated with the intensity of bitter and astringent taste of the tea. The content of L-phenylalanine in green tea was higher than in black tea. It showed that the quantity of phenylalanine could lay a foundation to assess the bitter and astringent taste of the tea. For green tea, theanine content varied from 3.844 to 11.638 mg/g, which is consistent with the literature results (37, 38). The theanine content in green tea was higher than that in black tea (2.787 mg/g). Furthermore, a clear corresponding relationship between the content of amino acid and the elaboration process of tea could be observed. The total amount of amino acids in black tea was lower than that in green tea, which may be due to the degradation of amino acids during the fermentation process. Green tea (non-fermented) and black tea (fully fermented) are the two major commercial types of tea. Green tea is derived directly from inactivating by steaming or microwave and drying the fresh tea leaves. For black tea, most of tea catechins are enzymatic oxidized and polymerized during the fermentation process. Accompanying the fermentation process, the free amino acids in tea leaves may take great changes. Accordingly, black teas contain less free amino acids compared with green tea (39).
Table 3.
Contents* of free amino acids in different types of tea.
| Compound | Congou black tea | Yellow mountain fuzz tip | Rizhao green tea | Green tea 1 | Green tea 2 | Maojian tea | Yacca tea | Taiping houkui tea | Sword shaped green tea |
|---|---|---|---|---|---|---|---|---|---|
| L-Arginine | 0.203 ± 0.002 | 1.213 ± 0.039 | 0.482 ± 0.009 | 1.339 ± 0.034 | 1.466 ± 0.035 | 0.704 ± 0.020 | 0.875 ± 0.028 | 0.908 ± 0.009 | 1.091 ± 0.010 |
| L-Aspartic acid | 0.812 ± 0.018 | 2.373 ± 0.001 | 2.380 ± 0.012 | 2.302 ± 0.026 | 2.565 ± 0.009 | 2.435 ± 0.005 | 1.951 ± 0.025 | 2.218 ± 0.001 | 3.343 ± 0.037 |
| L-Cysteine | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| L-Cystine | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| L-Glutamic acid | 0.507 ± 0.003 | 2.592 ± 0.002 | 1.712 ± 0.016 | 2.953 ± 0.000 | 2.492 ± 0.033 | 2.003 ± 0.005 | 4.170 ± 0.018 | 3.024 ± 0.014 | 2.948 ± 0.025 |
| Glycine | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| L-Histidine | 0.036 ± 0.002 | 0.149 ± 0.006 | 0.146 ± 0.003 | 0.092 ± 0.003 | 0.209 ± 0.001 | 0.143 ± 0.000 | 0.449 ± 0.010 | 0.120 ± 0.004 | 0.376 ± 0.006 |
| L-Isoleucine | 0.180 ± 0.011 | 0.284 ± 0.008 | 0.369 ± 0.007 | 0.326 ± 0.009 | 0.577 ± 0.007 | 0.427 ± 0.004 | 0.770 ± 0.002 | 0.217 ± 0.004 | 0.735 ± 0.006 |
| L-Leucine | 0.232 ± 0.011 | 0.363 ± 0.007 | 0.343 ± 0.014 | 0.324 ± 0.007 | 0.486 ± 0.006 | 0.333 ± 0.005 | 0.581 ± 0.005 | 0.323 ± 0.006 | 0.649 ± 0.003 |
| L-Lysine | 0.126 ± 0.001 | 0.240 ± 0.002 | 0.334 ± 0.006 | 0.341 ± 0.009 | 0.528 ± 0.001 | 0.361 ± 0.004 | 0.786 ± 0.009 | 0.228 ± 0.006 | 0.766 ± 0.013 |
| L-Methionine | 0.038 ± 0.005 | 0.036 ± 0.001 | 0.022 ± 0.001 | 0.022 ± 0.000 | 0.031 ± 0.001 | 0.021 ± 0.001 | 0.039 ± 0.001 | 0.037 ± 0.001 | 0.033 ± 0.000 |
| L-Phenylalanine | 0.290 ± 0.002 | 0.523 ± 0.005 | 0.557 ± 0.116 | 0.336 ± 0.015 | 0.410 ± 0.010 | 0.640 ± 0.006 | 0.610 ± 0.024 | 0.379 ± 0.009 | 0.437 ± 0.003 |
| L-Proline | 0.051 ± 0.000 | 0.029 ± 0.001 | 0.085 ± 0.001 | 0.059 ± 0.001 | 0.114 ± 0.002 | 0.079 ± 0.001 | 0.173 ± 0.000 | 0.037 ± 0.001 | 0.183 ± 0.002 |
| L-Serine | 0.305 ± 0.013 | 0.332 ± 0.007 | 0.539 ± 0.006 | 0.578 ± 0.008 | 0.684 ± 0.007 | 0.486 ± 0.004 | 0.746 ± 0.011 | 0.411 ± 0.004 | 0.784 ± 0.008 |
| L-Threonine | 0.164 ± 0.004 | 0.566 ± 0.002 | 0.610 ± 0.002 | 0.508 ± 0.000 | 0.740 ± 0.012 | 0.578 ± 0.004 | 0.965 ± 0.002 | 0.536 ± 0.001 | 0.951 ± 0.007 |
| L-Tyrosine | 0.051 ± 0.003 | 0.145 ± 0.002 | 0.074 ± 0.031 | 0.133 ± 0.003 | 0.185 ± 0.003 | 0.151 ± 0.003 | 0.279 ± 0.003 | 0.127 ± 0.001 | 0.165 ± 0.058 |
| L-Valine | 3.495 ± 0.013 | 3.182 ± 0.047 | 5.153 ± 0.013 | 3.235 ± 0.035 | 3.216 ± 0.001 | 5.009 ± 0.031 | 5.848 ± 0.038 | 2.233 ± 0.042 | 7.403 ± 0.078 |
| L-Alanine | 0.217 ± 0.005 | 0.150 ± 0.000 | 0.111 ± 0.001 | 0.190 ± 0.003 | 0.194 ± 0.002 | 0.183 ± 0.003 | 0.212 ± 0.007 | 0.163 ± 0.010 | 0.181 ± 0.005 |
| Theanine | 2.787 ± 0.009 | 11.638 ± 0.198 | 5.330 ± 0.020 | 5.775 ± 0.170 | 5.843 ± 0.023 | 4.499 ± 0.024 | 5.745 ± 0.040 | 9.073 ± 0.008 | 3.844 ± 0.010 |
| L-Tryptophan | 0.102 ± 0.001 | 0.258 ± 0.005 | 0.188 ± 0.001 | 0.160 ± 0.001 | 0.131 ± 0.042 | 0.236 ± 0.003 | 0.202 ± 0.006 | 0.206 ± 0.009 | 0.195 ± 0.001 |
| L-Asparagine | 0.254 ± 0.001 | 0.629 ± 0.001 | 1.354 ± 0.004 | 0.607 ± 0.005 | 1.303 ± 0.001 | 1.154 ± 0.028 | 3.248 ± 0.025 | 0.401 ± 0.007 | 1.987 ± 0.001 |
| L-Glutamine | 0.311 ± 0.002 | 5.773 ± 0.098 | 0.793 ± 0.014 | 1.637 ± 0.003 | 2.647 ± 0.005 | 1.949 ± 0.002 | 2.265 ± 0.040 | 5.168 ± 0.028 | 1.931 ± 0.005 |
| Aminoadipic acid | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| α-Aminobutyric acid | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| β-Aminobutyric acid | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| γ-Aminobutyric acid | 0.059 ± 0.005 | 0.119 ± 0.004 | 0.017 ± 0.002 | 0.072 ± 0.003 | 0.073 ± 0.005 | 0.052 ± 0.003 | 0.051 ± 0.004 | 0.052 ± 0.005 | 0.045 ± 0.003 |
| Citrulline | 0.004 ± 0.000 | 0.029 ± 0.000 | 0.015 ± 0.000 | 0.023 ± 0.001 | 0.041 ± 0.000 | 0.011 ± 0.001 | 0.012 ± 0.000 | 0.026 ± 0.001 | 0.036 ± 0.001 |
| Hydroxyproline | 0.017 ± 0.000 | 0.020 ± 0.000 | 0.024 ± 0.000 | 0.029 ± 0.000 | 0.020 ± 0.000 | 0.021 ± 0.000 | 0.019 ± 0.000 | 0.022 ± 0.000 | 0.020 ± 0.000 |
| Hydroxylysine | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| 1-Methyl-L-histidine | 0.013 ± 0.000 | 0.015 ± 0.000 | 0.016 ± 0.000 | 0.014 ± 0.000 | 0.017 ± 0.000 | 0.015 ± 0.000 | 0.018 ± 0.000 | 0.016 ± 0.000 | 0.021 ± 0.000 |
| 3-Methyl-L-histidine | ND | 0.009 ± 0.000 | 0.011 ± 0.000 | 0.010 ± 0.001 | 0.012 ± 0.000 | 0.009 ± 0.000 | 0.016 ± 0.000 | 0.010 ± 0.001 | 0.031 ± 0.001 |
| L-Ornithine | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| Sarcosine | ND | 0.018 ± 0.000 | ND | ND | ND | ND | ND | ND | ND |
| DL-Homocysteine | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| L-Pipecolic acid | 0.045 ± 0.001 | 0.083 ± 0.001 | 0.117 ± 0.003 | 0.117 ± 0.001 | 0.182 ± 0.002 | 0.133 ± 0.003 | 0.281 ± 0.001 | 0.081 ± 0.003 | 0.269 ± 0.007 |
| Total | 10.299 ± 0.041 | 30.765 ± 0.371 | 20.782 ± 0.106 | 21.181 ± 0.307 | 24.163 ± 0.160 | 21.631 ± 0.092 | 30.311 ± 0.119 | 26.018 ± 0.105 | 28.422 ± 0.187 |
Value (mg/g) = mean ± SD (n = 3). ND, not detectable.
Compared to other available methods for the same analytical determination (37, 40–44), this method measured the largest number of amino acids. At the same time, the method did not need derivatization, the pretreatment method was simple, the analysis time was short, and the selectivity was high.
Conclusion
An accurate, simple, and sensitive method for direct determination of 35 free amino acids in tea based on UPLC-Q-TOF/MS has been established. One injection can achieve both TOF–MS and IDA–MS/MS detection, and obtain the accurate mass, isotope distribution, and secondary ion fragment information. The UPLC-Q-TOF/MS is not affected by matrix effect, and the qualitative results can be confirmed by using the secondary ion fragmentation spectrum. It can be widely used for the rapid determination of free amino acids in tea, and provides a reference for the determination of amino acids in other plant tissues. This method also provides technical support for tea quality analysis and research.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
YZ and LZ conceived and designed the experiments. JL, JM, QL, and SF performed the experiments. LF and HM analyzed the data. JL and JM wrote the original draft. All authors have read and approved the manuscript.
Funding
This work was supported by the National Key Research and Development Plan of China (2016YFD0401104).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnut.2021.767801/full#supplementary-material
References
- 1.Yukio Y, Nobuyuki K, Koichi K. The role of glutamine in neurogenesis promoted by the green tea amino acid theanine in neural progenitor cells for brain health. Neurochem Int. (2019) 129:104505. 10.1016/j.neuint.2019.104505 [DOI] [PubMed] [Google Scholar]
- 2.Naffa R, Holmes G, Zhang WK, Maidment C, Shehadi I, Norris G. Comparison of liquid chromatography with fluorescence detection to liquid chromatography-mass spectrometry for amino acid analysis with derivatisation by 6-aminoquinolyl-N-hydroxysuccinimidyl- carbamate: applications for analysis of amino acids in skin. Arabian J Chem. (2020) 13:3997–4008. 10.1016/j.arabjc.2019.05.002 [DOI] [Google Scholar]
- 3.Ni H, Jiang QX, Zhang T, Huang GL, Li LH, Chen F. Characterization of the aroma of an instant white tea dried by freeze drying. Molecules. (2020) 25:3625. 10.3390/molecules25163628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feng ZH, Li YF, Li M, Wang YJ, Zhang L, Wang XC, et al. Tea aroma formation from six model manufacturing processes. Food Chem. (2019) 285:347–54. 10.1016/j.foodchem.2019.01.174 [DOI] [PubMed] [Google Scholar]
- 5.Bi W, He C, Ma YY, Shen J, Zhang LH, Peng Y, et al. Investigation of free amino acid, total phenolics, antioxidant activity and purine alkaloids to assess the health properties of non-camellia tea. Acta Pharmaceutica Sinica B. (2016) 6:170–81. 10.1016/j.apsb.2015.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang JH, Wang YF, Cheng JJ, Wang J, Sun XD, Sun S, et al. Enhanced cross-category models for predicting the total polyphenols, caffeine and free amino acids contents in Chinese tea using NIR spectroscopy. LWT-Food Sci Technol. (2018) 96:90–7. 10.1016/j.lwt.2018.05.012 [DOI] [Google Scholar]
- 7.Azevedo MS, Seraglio SKT, Rocha G, Balderas CB, Piovezan M, Gonzaga LV, et al. Free amino acid determination by GC-MS combined with a chemometric approach for geographical classification of bracatinga honeydew honey (Mimosa scabrella Bentham). Food Control. (2017) 78:383–92. 10.1016/j.foodcont 2017.03.008 [DOI] [Google Scholar]
- 8.Duan JY, Li ZY, Li J, Song HB, Fu JS, Xie BG, et al. Comparison of nutritional and flavor characteristics between four edible fungi and four fruits and vegetables based on components and characteristics of free amino acids. Mycosystema. (2020) 39:1077–89. 10.13346/j.mycosystema.20062 [DOI] [Google Scholar]
- 9.Liang YX, Zhang L, Gao FH, Zhang XM, Li X, Zhang HH, et al. Changes in NaCl, reducing sugar and amino acids and formation of tastes in Chongqing Shuidouchi during fermentation. Food Fermentation Indust. (2019) 45:27–34. 10.13995/j.cnki.11-1802/ts021057 [DOI] [Google Scholar]
- 10.Zhou ZY, Cao YL, Wang YH, Chen DJ, Mi J, Li XY, et al. Analysis of chemical compositions and antioxidant activities of Lycium barbarum bud and leaf teas. Sci Technol Food Indus. (2017) 38:129–34+145. 10.13386/j.issn1002-0306.2017.10.017 [DOI] [Google Scholar]
- 11.Yilmaz C, Zdemir F, Gkmen V. Investigation of free amino acids, bioactive and neuroactive compounds in different types of tea and effect of black tea processing. LWT-Food Sci Technol. (2020) 117:108655. 10.1016/j.lwt.2019.108655 [DOI] [Google Scholar]
- 12.Scharbert S, Hofmann T. Molecular definition of black tea taste by means of quantitative studies, taste reconstitution, and omission experiments. J Agri Food Chem. (2005) 53:5377–84. 10.1021/jf050294d [DOI] [PubMed] [Google Scholar]
- 13.Wang X, Li XS, Chen XL, He B, Han H. Analysis and evaluation of amino acids types and contents of Hanzhong Congou Black Tea. Food Res Dev. (2017) 38:162–8. 10.3969/j.issn.1005-6521.2017.05.034 [DOI] [Google Scholar]
- 14.He XW, Gao QY, Huang JJ, Li YX, Zhang XW, Ma C. Simultaneous determination of 39 free amino acids in green tea beverages by ion exchange chromatography with ninhydrin post-column derivatization. Beverage Industry. (2020) 23:35–40. [Google Scholar]
- 15.Mendel F. Applications of the ninhydrin reaction for analysis of amino acids, peptides, and proteins to agricultural and biomedical sciences. J Agri Food Chem. (2004) 52:385–406. 10.1021/jf030490p [DOI] [PubMed] [Google Scholar]
- 16.Erica B, Crystal H, Anh ML, Lenka H, Juliana A, Jan H. New horizons for ninhydrin: colorimetric determination of gender from fingerprints. Analyt Chem. (2016) 88:2413–20. 10.1021/acs analchem.5b04473 [DOI] [PubMed] [Google Scholar]
- 17.Zhang H, Song Q, Lin J, Fei ZX. Determination of 16 amino acids in fresh Morchella esculenta of Yunnan province by amino acid automatic analyzer. J Food Safety Q. (2019) 10:7564–9. 10.19812/j.cnki.jfsq11-5956/ts.2019.22.021 [DOI] [Google Scholar]
- 18.Kazan RM, Seddik HA, Marstani ZM, Elsutohy MM, Yasri NG. Determination of amino acids content in tea species using liquid chromatography via pre-column fluorescence derivatization. Microchem J. (2019) 150:104103. 10.1016/j.microc.2019.104103 [DOI] [Google Scholar]
- 19.Yang T, Yang XY, Chen WJ, Chen BQ, Zhang GX, Deng JJ, et al. Assay of 18 amino acids in Chuanmingshen violaceum roots by pre-column derivative HPLC. China J Chin Materia Med. (2020) 45:1316–22. 10.19540/j.cnki.cjcmm.20200106.101 [DOI] [PubMed] [Google Scholar]
- 20.Cao JY, Zhang LX, Fan Y, Wang ZY, Liu YQ, Wei Y. Determination of seventeen free amino acids in Gynostemma pentaphyllum tea by reversed-phase high performance liquid chromatography with precolumn derivatization. Amino Acids Biotic Resources. (2016) 38:16–20. 10.14188/j.ajsh.2016.03.004 [DOI] [Google Scholar]
- 21.How ZT, Busetti F, Linge KL, Kristiana I, Joll CA, Charrois JWA. Analysis of free amino acids in natural waters by liquid chromatography-tandem mass spectrometry. J Chromatogr A. (2014) 1370:135–46. 10.1016/j.chroma.2014.10.040 [DOI] [PubMed] [Google Scholar]
- 22.Li DQ, Zhang XW. Direct determination of multiple amino acids in the tea using high performance liquid chromatography-mass spectrometry. J Anhui Agri University. (2019) 46:8–12. 10.13610/j.cnki.1672-352x.20190314.00416155232 [DOI] [Google Scholar]
- 23.Qiu X, Reynolds R, Johanningsmeier S, Truong VD. Determination of free amino acids in five commercial sweet potato cultivars by hydrophilic interaction liquid chromatography-mass spectrometry. J Food Composition Analysis. (2020) 92:103522. 10.1016/j.jfca.2020.103522 [DOI] [Google Scholar]
- 24.Zhou P, Zhao F, Chen MJ, Ye NX, Lin Q, Quyang LQ, et al. Determination of 21 free amino acids in 5 types of tea by ultra-high performance liquid chromatography coupled with tandem mass spectrometry (UHPLC–MS/MS) using a modified 6-aminoquinolyl-N-hydroxysuccinimidyl carbamate (AQC) method. J Food Composition Analy. (2019) 81:46–54. 10.1016/j.jfca.2019.05.007 [DOI] [Google Scholar]
- 25.Zhou J, Wu Y, Long PP, Ho CT, Wang YJ, Kan ZP, et al. LC-MS-based metabolomics reveals the chemical changes of polyphenols during high-temperature roasting of large-leaf yellow tea. J Agri Food Chem. (2019) 67:5405–12. 10.1021/acs.jafc.8b05062 [DOI] [PubMed] [Google Scholar]
- 26.General Administration of Quality Supervision Inspection and Quarantine of the People's Republic of China . GB/T 8314-2013. Tea—Determination of free amino acids content[S]. Beijing: Standards Press of China; (2014). [Google Scholar]
- 27.Chen Y, Zhang XQ, Li YX, Guo HP. Comparative study of determination of amino acid content in tea by two different methods. J Xinyang Agri Forestry University. (2016) 26:93–5+99. 10.16593/j.cnki.41-1433/s.2016.01.03028598509 [DOI] [Google Scholar]
- 28.Li F, Cai M, Chen B, Liu HL, Zhu F. Determination of γ-aminobutyric acid in health food by amino acid analyzer. Chinese J Analy Lab. (2016) 35:1012–5. 10.16593/j.cnki.411433/s 2016.01.030 [DOI] [Google Scholar]
- 29.Fleury MO, Ashley DV. High-performance liquid chromatographic analysis of amino acids in physiological fluids: on-line precolumn derivatization with o-phthaldialdehyde. Analy Biochem. (1983) 133:330–5. 10.1016/0003-2697(83)90092-1 [DOI] [PubMed] [Google Scholar]
- 30.Huang MY, Chen SL, Lin Y, Tao H, E WW, Hu YL, et al. Analysis of amino acids by ultra high performance liquid chromatography-electrospray ion tandem mass spectrometry using 4-nitrobenzoyl chloride as precolumn derivatization. Chin J Analytical Chem. (2019) 47:1857–65. 10.19756/j.issn.0253-3820.191334 [DOI] [Google Scholar]
- 31.Bidlingmeyer BA, Cohen SA, Tarvin TL. Rapid analysis of amino acids using pre-column derivatization. J Chromatography A. (1984) 336:93–104. [DOI] [PubMed] [Google Scholar]
- 32.Marino R, Iammarino M, Santillo A, Muscarella M, Caroprese M, Albenzio M. Technical note: Rapid method for determination of amino acids in milk. J Dairy Sci. (2010) 93:2367–70. 10.3168/jds.2009-3017 [DOI] [PubMed] [Google Scholar]
- 33.Guo S, Duan JA, Qian DW, Tang YP, Qian YF, Wu DW, et al. Rapid determination of amino acids in fruits of ziziphus jujubaby hydrophilic interaction ultra-high-performance liquid chromatography coupled with triple-quadrupole mass spectrometry. J Agri Food Chem. (2013) 61:2709–19. 10.1021/jf305497r [DOI] [PubMed] [Google Scholar]
- 34.Violi JP, Bishop DP, Padula MP, Steele JR, Rodgers KJ. Considerations for amino acid analysis by liquid chromatography-tandem mass spectrometry: a tutorial review. TrAC Trends Analyt Chem. (2020) 131:116018. 10.1016/j.trac.2020.116018 [DOI] [Google Scholar]
- 35.Das PR, Islam MT, Lee SH, Lee MK, Kim JR. UPLC-DAD-QTOF/MS analysis of green tea phenolic metabolites in their free, esterified, glycosylated, and cell wall-bound forms by ultra-sonication, agitation, and conventional extraction techniques. LWT-Food Sci Technol. (2020) 127:109440. 10.1016/j.lwt.2020.109440 [DOI] [Google Scholar]
- 36.General Administration of Quality Supervision Inspection and Quarantine of the People's Republic of China . GB/T 8312-2013. Tea—Determination of Caffeine Content[S]. Beijing: Standards Press of China; (2014). [Google Scholar]
- 37.Wang L, Xu RJ, Hu B, Li W, Sun Y, Tu YY, et al. Analysis of free amino acids in Chinese teas and flower of tea plant by high performance liquid chromatography combined with solid-phase extraction. Food Chem. (2010) 123:1259–66. 10.1016/j.foodchem.2010.05.063 [DOI] [Google Scholar]
- 38.Gao J, Xia T, Zhu B, Dai QY, Gao KJ. Simultaneous determination of theanine, catechins and alkaloids in tea by HPLC-PDA. J Anhui Agri University. (2008) 35:324–8. 10.13610/j.cnki.1672-352x.2008.03.013 [DOI] [Google Scholar]
- 39.Wang FH. Analysis and determination of free amini acids in different tea by HPLC. Food Res Dev. (2018) 39:141–6. 10.3969/j.issn.1005-6521.2018.01.02821875374 [DOI] [Google Scholar]
- 40.Li M, Li DX, Tai YL, Gu CC, Song YS, Jiao WT, et al. Determination of free amino acids in tea by a novel method of reversed-phase high performance liquid chromatography applying 6-Aminoquinolyl-N-Hydroxysuccinimidyl carbamate reagent. J Food Sci Technol. (2018) 55:4276–86. 10.1007/s13197-018-3366-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Horanni R, Engelhardt UH. Determination of amino acids in white, green, black, oolong, pu-erh teas and tea products. J Food Composit Analysis. (2013) 31:94–100. 10.1016/j.jfca.2013.03.005 [DOI] [Google Scholar]
- 42.Xu Y, Liu ZY, Liu ZY, Feng ZH, Zhang L, Wang XC, et al. Identification of d-amino acids in tea leaves. Food Chem. (2020) 317:126428. 10.1016/j.foodchem.2020.126428 [DOI] [PubMed] [Google Scholar]
- 43.Li NN, Liu Y, Zhao Y, Zheng XQ, Lu JL, Liang YR. Simultaneous HPLC determination of amino acids in tea infusion coupled to pre-column derivatization with 2,4-dinitrofluorobenzene. Food Anal Methods. (2016) 9:1307–14. 10.1007/s12161-015-0310-8 [DOI] [Google Scholar]
- 44.Dong SW, Liu TF, Dong MH. Determination of 18 kinds of aminoacids in fresh tea leaves by HPLC coupled with pre-column derivatization. Asian Agri Res. (2018) 10:55–63. 10.22004/ag.econ.273100 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.