Abstract
The late 1800s Louis Pasteur and Robert Koch introduced and popularized the germ theory of disease. At that time gastric cancer was the most common cause of cancer deaths in most countries making the stomach an early site of microbial research with a focus on gastric luminal and mucosal bacteria and the role of Boas-Oppler bacillus (Lactobacillus) in the diagnosis of gastric cancer. In the 1970’s the focus evolved to studies of the gastric microbiome in the production of nitrosamines and included development of the Correa cascade. Interest in nitrosamine production peaked in the late 1980s and was replaced by studies of the newly described Helicobacter pylori and studies of its role in gastritis, gastric atrophy, and gastric cancer. The last decade has witnessed a rebirth in interest in the gastric microbiota as part of worldwide interest in the human microbiome. Although fungi were prominent in the studies of gastric microbiology in the 19th century, their potential role in disease pathogenesis has yet to be addressed using modern techniques. Overall, current studies of the gastric bacterial microbiome do not provide convincing evidence to expand the role of the gastric microbiome in cancer pathogenesis beyond what is directly attributable to the oncogenic potential of H. pylori and its role in persisting acute-on-chronic inflammation.
Keywords: Helicobacter pylori, microbiome, gastric cancer, inflammation, culture, next generation sequencing
Introduction
Interest in the gastric microbiome began in the second half of the nineteenth century when Louis Pasteur and Robert Koch introduced and popularized the germ theory of disease. Their observations resulted in a virtual explosion of interest in bacteria and fungi as well as the role of microbes in disease. During that period gastric cancer was the most common cause of cancer deaths in most countries [1]. During that period early diagnosis of gastric cancer was impossible. In 1879 von den Velden linked achlorhydria with gastric cancer which greatly stimulated interest in gastritis, gastric acid secretion and early diagnosis of gastric cancer [2,3]. Physicians also began to examine gastric contents and their investigations included staining of the gastric contents for bacteria and fungi. In 1895 Izmar Isidor Boas and Bruno Oppler reported the association of gastric cancer with the presence of both lactic acid and a large amount of bacteria in the stomach [4]. Boas described “a string picture of the carcinomatous gastric contents is formed through immovable bacilli, first observed by myself. They are long, filiform, frequently joining one another at angles; they appear in numberless specimens in the field of vision. Particularly frequent, almost consistent, they are found in stagnant, achlorhydric gastric contents containing lactic acid” [5]. That same year, Fenton B. Turck, who stained microscopic from material collected both luminal and targeted scraping from the gastric mucosa also reported that the majority of the gastric samples he studied contained organisms [6,7]. He attempted to determine the predominant organism and reported that in “all cases of carcinoma of the stomach, numerous colonies of a large variety of germs are found. They seem to revel in this superb culture bed”. Truck insightfully noted that “It is one thing to find stray germs in the lumen of the stomach which may occur in health… and another thing to find colonization (original italics) of germs adherent to the mucous membrane of the walls of the stomach under changed conditions, is transformed into a large plate culture”.[6].
This period was also one of increasing interest in gastric bacteriology for the early diagnosis of gastric carcinoma especially the value of finding Boas-Oppler bacillus in clinical specimens. Boas’s Textbook of Gastroenterology [5] provides details of his experience as well as that of other authors such as Kellogg, who in 1900, reported 7000 examinations of gastric contents including those with hyperchlorhydria. He found mold-fungi 457 times including Oidium lactis, Aspergillus fumigatus and flavescens (page 233) [5] and included details of studies by Abelous [8] and Schmidt who reported changing his diagnosis of benign ulcer to gastric cancer on the basis of a stool examination showing numerous Boas-Oppler bacillii [9].
By 1916, interest in the Boas-Oppler bacillus waned and was summarized by Heinemann and Ecker who confirmed that the Boas-Oppler bacillus was a Lactobacillus, or several types of Lactobacilli that were able to overgrow in states associated with hypo- or achlorhydria. Their conclusion was that the Boas-Oppler bacillus was neither causative nor diagnostic of gastric cancer [10]. An editorial in JAMA supported the notion that high concentrations of Lactobacillii in the stomach was a consequence of reduced acid secretion coupled with the organism’s ability to survive in acid and concluded that its presence was independent of the presence of gastric ulcer, gastritis, pernicious anemia, or gastric carcinoma [10,11].
The gastric bactericidal barrier
Spallanzani (1783) is given credit for pointing out the antiseptic properties of gastric juice when he noted that “the flesh locked with the gastric juice are not subject to decay” [13,14] In 1887 Allen Macfadyen published studies on the effect of gastric acid (HCl) and pepsin on pathogenic bacteria including Staphylococcus aureus, the typhoid bacillus and the anthrax bacillus. He showed pepsin alone did not kill them and asked “Is the hydrochloric acid the actual antiseptic factor of the gastric juice, and, if so, does it act in natural quantities so powerfully as to kill the bacteria that find their way into the stomach, and so prevent their passage in a living state into the intestine? He tested a number of different organisms and concluded that “hydrochloric acid is the germicide of the gastric juice” but that certain germs were able to survive and that no boundary could be set, but each germ must be examined on its merits”. He went on to show that the addition of pepsin to acid was no more effective than acid alone. Finally, he showed that spores survived the action of gastric juice. He then went on to examine the effect of acid in vivo using a dog model and confirmed his in vitro experiments. [15]. Additional work by Gillespie [16] and a host of others culminated in Giannella et al. who conclusively showed that the gastric pH barrier was primarily pH-hydrochloric acid dependent [17].
Bacteria and gastric cancer: the nitrosamine hypothesis of gastric carcinogenesis
By 1950 the association of gastric cancer with atrophic gastritis/gastric atrophy had become firmly established and interest turned from confirming that association toward investigation of the causes and associations of chronic gastritis [3,12]. Interest in the gastric microbiome resurged in the last quarter of the 20th century based on the premise that intestinal and gastric bacteria might be a potential source of carcinogens (i.e., the nitrosamine hypothesis).
This hypothesis was most popular in the pre-H. pylori era and relates the theory that reduction of dietary nitrates to nitrite could convert dietary amines into carcinogenic N-nitroso compounds (i.e., in vivo synthesis of carcinogens). The hypothesis was confirmed in numerous experimental studies in which inclusion of nitrites and secondary amines in rodent diets (food and water) was associated with an increased incidence of malignancies in a variety of organs [18]. The data in humans has been primarily based on epidemiology studies such as those that associated a high intake of processed meats or smoked fish and human cancers [19,20,20–22]. Overall, the data from epidemiologic studies relating nitrate ingestion and gastrointestinal cancer has proven inconclusive and as we will discuss below, in part because nitrate intake is not likely to be the ideal parameter to measure [20,20,23–26].
Research in the possible role of dietary nitrates in gastric cancer prominently occurred in the pre-H. pylori era and formed part of the basis of the Correa cascade [27–31]. However, some interest has continued up to the present time [21,24–28,32,33]. With the Boas-Oppler bacillus the focus was on the diagnosis of gastric cancer. In contrast, the nitrosamine hypothesis involved pathogenesis. Both theories are related to gastric hypochlorhydria or achlorhydria which results in overgrowth of intestinal bacteria in the stomach.
The concept is basically that carcinogenic N-nitroso compounds produced as noted above could cause progressive genetic instability that would eventually result in gastric cancer. One major problem that has haunted this hypothesis is that this conversion requires acid which is lacking in the achlorhydric stomach. A number of relative definitive studies have been done in humans that have not supported the hypothesis. For example, Hall et al. studied the formation of N-nitroso compounds in normal stomachs, those with achlorhydria, and after partial gastrectomy and reported that “although hypoacidity predisposes to bacterial overgrowth and nitrite generation, it does not enhance nitrosation. Instead, this is maximal at low pH, suggesting chemical rather than bacterial nitrosation, contrary to the nitrosamine hypothesis of gastric carcinogenesis” [34]. Koyayashi noted that formation of N-nitroso compounds is affected by diet, ingestion of antioxidants and gastric acidity including use of acid reducing drugs [26] with three proposed mechanisms: chemical (acid-catalyzed), bacterial, and inflammatory. The H. Pylori-infected hypochlorhydric stomach typically contains both acute and chronic inflammation and very low levels of ascorbic acid which favors formation of N-nitrosamine rather than S-nitrosothiol [26,35,36]. When directly examined Sobala et al. reported that total levels of N-nitroso compounds were not increased in such patients [37]. Despite the fact that these studies directly attempting to test the N-nitroso hypothesis in humans have invariably been negative and that autoimmune gastritis, another cause of achlorhydria, is rarely associated with gastric adenocarcinoma in the presence of H. pylori infection. Nontheless, finding of organisms potentially able to produce nitrosamines continues to be cited as the important finding of studies of the gastric microbiome and its role in gastric cancer (see below).
H. pylori as a carcinogen
Although a number of animal models of gastric cancer following H. pylori infection have been reported, invasive gastric cancers have not been described and whether the lesions in rodents actually represent cancer has been challenged [38,39]. Most recently the putative malignant lesions in rodents have been described instead as exuberantly proliferative metaplastic or reactive lesions [38]. Importantly, experiments with organoids produced from three mouse models of gastric cancer also failed to cause tumors in a xenograft model whereas the controls (carcinogen-treated gastric mucosa) produced tumors [38]. H. pylori-induced animal lesions and advanced dysplasia have also been noted to resolve following H. pylori therapy which would seem to make the diagnosis of intraepithelial neoplasia unlikely. Probably the most relevant animal experiments involve co-carcinogenesis in which chemical carcinogens plus inflammation such as that related to the H. pylori infection has been able to produce tumors [40–42] including in mice and ferrets [43]. Tsuda and coworkers examined the impact of gastric acid suppression and on the gastrointestinal tract microbiota by comparing PPI-users and controls not using PPI using modern sequencing [44]. Studies of gastric fluid showed that bacterial cell numbers identified by culture increased significantly in PPI users. The suggested that bacterial overgrowth in the stomach after PPI treatment may not be due to proliferation of bacteria but rather lack of bacterial killing in the acid-suppressed stomach.
The H. pylori, gastric microbiome, environment, host hypothesis of gastric cancer pathogenesis
Gastric cancer is an inflammation-associated cancer in which persistent H. pylori-related acute-on-chronic inflammation and direct H. pylori-related genetic damage is modulated by the host’s response to the inflammatory stimuli (i.e., polymorphisms in genes enhancing or reducing the inflammatory response) and by the environment of which food, especially the balance between the intake of cancer promoting agents such as salt and cancer preventing agents such as phyto-nutrients (eg, fresh fruits and vegetables) interact to predict gastric cancer risk [45,46]. H. pylori also vary in virulence which is largely reflected by the ability of the individual H. pylori strain to produce inflammation [46]. This bacterium-host-environment interplay can best be visualized in high cancer risk regions such as Japan where changes in diet and smoking has resulted in rapid and dramatic decline in rate of acquisition of atrophic gastritis and gastric cancer [47,48]. H. pylori also can directly cause genetic instability vis both epigenetic and direct methods including causing double-stranded DNA breaks [49,50]
The combination of persistent inflammation and H. pylori are associated with increased cancer risk and this risk accelerates after development of atrophic gastritis. Eradication of H. pylori results in a reduction in cancer risk irrespective of the degree of risk present at the time of H. pylori eradication including among those who have already experienced a resectable gastric cancer [45]. This is best illustrated by the reduction in the risk of metachronous gastric cancer following eradication of the infection after endoscopic removal of an early gastric cancer [51]. H. pylori eradication both removes H. pylori and its direct effects and H. pylori associated acute inflammation with a more gradual reduction in chronic inflammation. It also removes the inflammation-induced inhibition of any remaining parietal cells potentially allowing acid secretion to increase [39,45]. Elimination of ongoing damage may also allow some recovery of acid secreting mucosa [39,45]. As noted above, atrophic gastritis-associated reduced acid secretion removes the bacteriocidal acid barrier and allows colonization of the stomach with organism normally inhabiting the more distal intestine and potentially allows overgrowth of normal residents of the distal bowel that could enhance or stimulate inflammation or produce carcinogens that further accelerate carcinogenesis. The overall benefit of H. pylori eradication, even in advanced atrophy, has been to halt further progression of cancer risk and either stabilize or reduce subsequent risk.
The changes that then occur in the intragastric milieu would also be expected to possibly be reflected in changes in the microbiome. Overall, the outcome favors the hypothesis that sum of the effects is beneficial and cancer risk is reduced. Current investigations have been unable to take the interactions between ongoing inflammation and the microbiome into account. The fact that adenocarcinoma rarely accompanies autoimmune gastritis suggests that microbiome changes associated achlorhydria are in themselves insufficient to play a dominant role in carcinogenesis.
Autoimmune gastritis and the microbiome
Autoimmune gastritis results in potentially reversible atrophy of the gastric corpus mucosa, achlorhydria, and a microbiome characteristic of overgrown of intestinal bacteria and fungi. The antrum remains normal or near normal, inflammation is primarily chronic and in the later stage sparse [52]. Gastric adenocarcinoma is rare whereas neuroendocrine neoplasms may occur due to the over expression of the hormone gastrin [52]. The difference in outcome likely relates to the difference in the type, extent, and duration of inflammation and damage by reactive oxygen and nitrogen species which with the lack of acid would likely reduce the local production of carcinogens as well as the proper soil for them to act.
The gastric microbiome in the culture-dependent era
The ability to culture bacteria from the stomach is dependent largely on the acidity, or lack thereof, in the stomach at the time the culture was made and the methods used for culture (eg, media type, composition, conditions -aerobic, anaerobic, specific microaerophilic, etc) and whether organisms are cultivatable using current techniques. One major limitation is that the microflora sampled has most often been largely been limited to luminal contents rather than mucosa-associated organisms. In 1946 Barber and Franklin published their attempts to culture bacteria from the gastric mucosal swabs and reviewed studies prior to that time [13]. At that time the main two research issues were the role of the gastric microbiome in post-operative infections and whether an infection was casually related to peptic ulcers [13]. Older observations that cultures are most frequently positive in diseases and conditions where acidity is low and is most likely in the presence of achlorhydria were confirmed [53,54]. Positive anaerobic culture was also more likely in gastric cancer patients many of which are associated with culture of Enterobacteriaceae.
Microbial colonization of the upper gastrointestinal-tract using culture was also studied by Sjöstedt et al. in the mid-1980s [55,56]. They investigated luminal bacteria by aspiration of the intraluminal content at different locations in oropharynx, esophagus, stomach and duodenum together with gastric biopsies obtained from the gastric tumor and from the normal appearing gastric mucosa [55]. As expected, counts of microorganisms isolated from gastric fluid showed that patients with gastric carcinoma were colonized with highest numbers of different microorganisms compared to patients with gastric or duodenal ulcers [56]. All tumors were heavily colonized with a dominance of anaerobic bacteria in the cancer tissue. The bacteria obtained from the cancer tissue were not found in the normal oropharyngeal flora.
The pre-H. pylori era was also one with considerable interest in the gastric microflora in relation to surgical and intensive care complications. Many studies were done in patients in the intensive care unit, especially in intubated patients and included studies on the effects of antacids, H2-receptor antagonists and proton pump inhibitors on the upper gastrointestinal microbiology [44,57–63]. The discovery of H. pylori in the early 1980s led to increased interest in gastric histology and microbiology and many of the older observations such as the effect of reduced acid secretion on promoting a diverse gastric flora were relearned or reemphasized [64]. The declaration that H. pylori was a type I human carcinogen and agreement regarding its role as the most common cause of gastric cancer subsequently refocused interest on the possible relationship between the gastric microbiome and carcinogenesis.
The gastric microbiome in the era of genome sequencing
The introduction of DNA sequencing to study the gastric microbiota has markedly enhanced study of the profile the bacterial flora in the gastrointestinal tract. Until today, the 16S rRNA gene has remained the most common target for such studies. However, such methods exclusively provide information about the bacterial community but no information about gene functions such as resistance to antibiotics or potential toxin and virulence genes in the microbial community. Consequently, bacterial profiling targeting the 16S rRNA gene only tell us what bacteria in there – not what they are doing. Neither will viruses, protozoa or fungi be identified. As noted above, early investigations of gastric contents in achlorhydria and hypochlorhydria stressed the importance of the fungal microbiome. Lack of study of these highly metabolically active, highly prevalent residents of hypochlorhydric stomachs remains a major impediment to understanding the relative importance of the microbiome in various gastric diseases.
Methods relying on sequence-specific separation of equal sized 16S rDNA PCR amplified fragments such as temporal temperature gradient gel-electrophoresis (TTGE) and terminal restriction fragment length polymorphism (T-RFLP) were introduced in 2000 [65]. However, these studies lack the resolution to define sequence differences at the species level and often relied on short 16S rDNA amplified fragments. A study with a similar cloning and sequencing approach for characterization of bacterial diversity within the gastric mucosa was published in 2006 [66]. A diverse community of 128 phylotypes was identified in the stomach with 10 percent of the phylotypes having not previously been described. In the 19 biopsies where H. pylori was identified only 12 subjects also tested positive for H. pylori by conventional methods such as culture, serology or urea breath test. The authors did not consider the possibility of false positive results and concluded that the sensitivity of molecular identification was superior to traditional culture methods. These studies, however, further focused attention on the role of the gastric microbiota in human health and disease. A similar lack of resolution was reported in a study of the stomach microbiota in cancer patients using T-RFLP in combination with cloning and sequencing of 16S rRNA genes to assess the composition of the gastric microbiota [67]. That study reported relatively low abundance of H. pylori with the gastric cancer microbiota being dominated by different species of the genera Streptococcus, Lactobacillus, Veillonella and Prevotella. No major differences were seen compared to H. pylori negative control patients.
Next generation sequencing and molecular analysis of the gut microbiota
The relatively poor resolution fingerprinting methods described above where successively replaced in mid-2000 by high-throughput DNA sequencing techniques (e.g. 454 pyrosequencing]. These technologies allowed researchers to analyze the bacterial composition rapidly in many samples with greater sequencing depth and sequence coverage. However, even here the analyses are based on the 16S rRNA gene restricting the analyses to studies of the composition of bacterial communities. The large amounts of sequence data obtained from next generation sequencing platforms also required development of novel bioinformatics tools which fortunately have now been developed and access shared with the research community as open access [68,69]. The first analyses of the human gastric mucosa microbiota using massive parallel sequencing (e.g. 454 pyrosequencing] was published 2008 [70]. A hyper-variable region of the 16S rRNA gene in combination with sample-specific barcode sequences enabled parallel in-depth analysis of hundreds of samples with limited sample processing. The method describes microbial communities down to phylotypes below the genus level. Microbial communities in throat and stomach biopsies and fecal samples from humans were analyzed and confirmed the applicability of barcoded pyrosequencing as a high-throughput method for bacterial community studies and were not limited to the gastrointestinal tract. The fecal samples formed a distinct cluster while the throat and stomach samples grouped more closely. The gastric microbiota analysis revealed diverse microbial communities in the H. pylori-negative stomachs which harbored 262 phylotypes representing 13 phyla. Compared to the cloning and sequencing approach used in the past the new technology provided substantially more information about the composition of bacterial communities. Next generation sequencing was soon adopted by almost all researchers in the microbiota field. Massive parallel sequencing of the 16S rRNA gene has remained the gold standard for microbiota studies for more than a decade following the initial publication. One major advantage is that this technology does not need to take into account the presence of human DNA in the sample since 16S RNA is only present in prokaryotic cells. Consequently, samples where human DNA dominates, such as in biopsy material, can be analyzed only for its bacterial content.
The gastric microbiota in H. pyloripositive and negative gastric cancer tissues
The introduction of high-throughput sequencing technologies has confirmed and extending our knowledge of the diverse microbial community the human stomach harbors which differs slightly from that of the oral cavity and markedly from the more distal bowel. Current data is largely restricted to the bacterial microbiome which is predominated by Proteobacteria and Firmicutes. A larger bacterial load, diversity and abundance of Firmicutes and Bacteroidetes is found in the lower gastrointestinal tract [71].
Gastric cancer is most often the end result of a cascade of events arising in patients colonized with H. pylori. As noted previously, multiple host and environmental factors also influence disease development with gastric cancer typically appearing after the development of H. pylori-induced gastric mucosal atrophy with achlorhydria or hypochlorhydria. These changes in the intragastric environment result in alterations to the composition and function of the normal gastric microbiota. The relative contribution of the bacteria vs. components other than H. pylori in the development of gastric cancer remain unclear. Although, the critical role of H. pylori is universally recognized, there is no consensus on whether or which specific non-H. pylori bacteria might play important roles in the pathogenesis of gastric cancer. Furthermore, microbial changes within the tumor itself may be important especially in tumor behavior and have not been thoroughly investigated. For example, a recent study evaluated variations in the composition, interaction network and functions of gastric microbiota in cancerous and matched non-cancerous tissues in the same patient [72]. They reported that the composition of the microbiota in the non-cancerous mucosal samples was significantly influenced by whether H. pylori was also present. In contrast to the results of the culture-based study performed by Sjöstedt et al. [55] they reported that bacterial taxa that were enriched in the cancer tissue mostly consisted of oral bacteria such as Peptostreptococcus, Streptococcus, and Fusobacterium whereas the adjacent non-tumor tissues had abundant lactic acid-producing bacteria such as L. lactis, and L. brevis.
H. pylori and gastric microbiota
Almost all sequencing-based studies of the gastric microbiota have resulted in describing the composition of the bacterial communities within the stomach. When H. pylori is present it dominates the gastric niche such as in patients with gastritis and ulcers (i.e. in individuals with an acidic gastric milieu). H. pylori-negative subjects contain a diverse stomach microbiota [73,74]. Alterations in gastric microbiota following H. pylori eradication have also been reported in a number of studies [58,72,75].
Characterization of the gastric bacterial communities has also been performed in patients with gastric cancer, for example in one study the gastric bacterial communities were described in 135 gastric carcinoma cases and chronic gastritis controls by massive parallel sequencing of the 16S rRNA gene [76]. Patients with gastric cancer had a significant decrease in H. pylori abundance with several taxa being significantly more abundant including intestinal commensals such as Achromobacter, Lactobacillus, Citrobacter, Clostridium and Rhodococcus. Hearkening back to the studies described decades ago, they used functional analyses of the commensal community in cancer tissues and noted that these bacteria had increased nitrosating functions which has consistently been shown in any condition associated with reduced acid secretion. Importantly, functional capabilities of microbial communities can be predicted using an algorithm that estimates the functional potential of microbial communities based on 16S rRNA marker gene sequences [77]. As noted above, the drawback is that only indirect evidence for such functions in the community can be achieved since the algorithm is based on 16S rRNA gene sequencing.
The presence of a significant mucosa microbial dysbiosis in intestinal metaplasia and gastric carcinoma patients was confirmed by Coker et al. [78] who noted enrichment of 21 and depletion of 10 bacterial taxa in stomachs with gastric cancer compared with superficial gastritis. They followed the differences in bacterial interactions and metagenomic functions across the stages of gastric carcinogenesis using algorithm tools and reported that oral bacteria were enriched in cancer tissue which again contrasts with the previous culture and sequencing based studies where bacterial genera from intestinal commensals were observed.
Discussion
Despite more than a century of interest and increasing detailed studies, a definite role for the non-H .pylori microbiota in the development of gastric carcinogenesis is not yet been established. Staining and culture of gastric contents has given way to culture-independent technologies allowing for characterization of the bacterial communities in the stomach (e.g. high-throughput DNA sequencing techniques). Prospective and retrospective case-controls studies have been performed [72,78–83]. However, differences in sequencing platforms, sequencing depth, 16S rDNA targets for PCR amplification, DNA extraction kits and logistics for obtaining, transport and storage of samples all lead to caution with regard to interpretation of results. Currently, there is no consensus how to analyze the microbiota samples obtained from different sites inside and on the surface of humans. The fecal microbiota community has proven to be a poor proxy for the mucosa-associated communities present as has gastric fluid as a surrogate for gastric mucosal biopsies [84–86]. Because host-microbial cross-talk is both present and critically importance for the understanding of microbial communities at the local level where such cross-talks take place [87]. The ideal study will be one able to adjust for confounding factors to ensure lack of a severe selection bias in choice of cases and/or controls. Furthermore, statistical power in the analyses requires adequate sample size and study material. Most available studies are not population-based and often used a case-control study design apt to introduce selection-bias problems that are difficult or impossible to adjust for. Finally, and most importantly, associations cannot provide good evidence regarding causality and mechanistic studies are needed to make confident predictions regarding the role of the gastric microbiota in development of gastric cancer.
The current studies do not provide convincing evidence to expand the role of cancer pathogenesis beyond that is directly attributable to the presence of chronic inflammation and the oncogenic potential of H. pylori. The large volume of information available regarding the gastric microbiome likely does not mean nothing but currently we find it impossible to identifies areas we are confident that more investigation will lead to better understanding of the pathogenesis of H. pylori-associated gastric cancer.
Gastric acidity serves as a major barrier limiting entry of microbes into the gastrointestinal tract. Dysrupton of the acid barrier by drugs or damage to the acid secreting mucosa changes the stomach from a being a hostile environment for microbial colonization to a site where many species can thrive either as intraluminal or mucosal-associated microbiota.
H. pylorican survive with the stomach but its ability to colonize gastric pits in the gastric corpus is restricted by the high concentration of acid that exits from those pits during secretion. Progressive gastric inflammation/damage increases the regions of the stomach affected by progressive inflammatory damage resulting in atrophy. Both inflammation and H. pylori can damage host genes resulting in genetic instability and eventually malignancy
There is currently no solid evidence that the non-H. pylori bacterial community in the stomach is directly involved in gastric carcinogenesis.
Longitudinal studies that track the changes in the gastric microbiome over the evolution of the disease including H. pylori gastritis, autoimmune gastritis, and autoimmune gastritis with concomitant H. pylori infection are needed. The studies should include changes in the fungal and virus microbiomes.
Different underlying methods, standards and operating procedures for studies of the human microbiome makes it difficult to properly compare results from prospective and retrospective studies in the field. There is an urgent need for initiatives to strengthen international cooperation to increase data comparability and to agree common standards, procedures and methods.
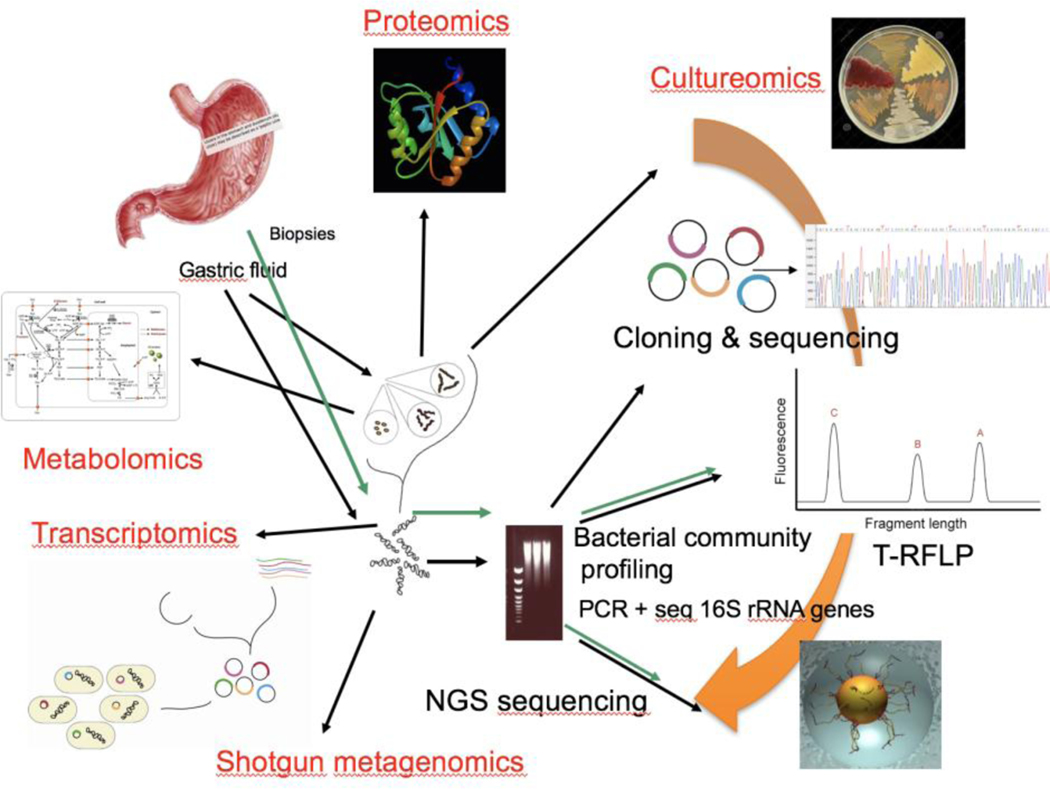
Draft Figure 1:
Overview of the major “omics” approaches using culture, DNA, RNA, metabolites and proteins for studies of the gastric microbiome. The traditional culture-based methods have been used for more than 100 years. Culture was replaced by DNA-based techniques 20 years ago and future investigations of the gastric microbiome will include transcriptomics, metabolomics and proteomics that will provide more opportunities for functional studies of microorganisms in the gastric niche.
Draft Figure 2. Evolution of the gastric microbiome following H. pylori infection.
This shows the relation between the mucosal histology, acidity, and gastric microbiome over the progression from antral predominant gastritis with predominant H pylori to gastric atrophy when the microbial diversity, number, and pH all increase. Representative culture plates are also shown. (needs to be drawn)
Changing pH such as in color across the time interval
Changing (increasing number and diversity of microbiota shown as Different shaped and colored organisms
Changing Histology from normal stomach to cancer
Acknowledgments
Support: Dr. Engstrand is supported in part byFerring Phamaceuticals (public-private partnership) and grants from European Union (Horizon 2020) and the Swedish research council. Dr. Graham is supported in part by the Office of Research and Development Medical Research Service Department of Veterans Affairs, Public Health Service grant DK56338 which funds the Texas Medical Center Digestive Diseases Center.
Footnotes
Potential conflicts: Dr. Graham is a consultant for RedHill Biopharma and Phathom Pharmaceuticals regarding novel H. pylori therapies and has received research support for culture of Helicobacter pylori and is the PI of an international study of the use of antimycobacterial therapy for Crohn’s disease. Dr. Engstrand has no relevant conflicts.
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
References
- (1).Tan MC, Balakrishnan M, Graham DY. Gastric cancer worldwide except Japan. In: Gastric cancer with special focus on studies from Japan. In: Shiotani A, editor. Gastric cancer with special focus on studies from Japan. Singapore: Springer, 2018: 17–28. [Google Scholar]
- (2).von den Velden R. Ueber vorkommen und mandgel der freien salzsaure in magensaft bei gastrektasie. Deutsches Arch F Klin Med 1879;23:369–99. [Google Scholar]
- (3).Graham DY, Asaka M. Eradication of gastric cancer and more efficient gastric cancer surveillance in Japan: two peas in a pod. J Gastroenterol 2010;45(1):1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Boas II, Oppler B. Zur kenntniss des mageninhalts beim carcinoma ventriculi. Deutsch Med Wchnsch 1895;21:73–5. [Google Scholar]
- (5).Boas I. Cancer of the Stomach. In: Boas I, Bernheim A, editors. Diseases of the stomach. Philadelphia: F.A. Davis Company, 1907: 561–604. [Google Scholar]
- (6).Turck FB. The early diagnosis of carcinoma of the stomach, with bacteriology of the stomach contents. JAMA 1895;24:317–9. [Google Scholar]
- (7).Turck FB. Combined gastroscope and gyromele for diagnostic and therapeutic purposes. JAMA 1903;23:1412–3. [Google Scholar]
- (8).Abelous. Recherches sur les microbes de l’estomac a L’etat normal et leur action sur les substances alimentares. In: Masson G, editor. Comptes rendus hebdomadaires des séances et mémoires de la Société de biologie. Paris: Library of the Academy of Medicine, 1889: 86–9. [Google Scholar]
- (9).Schmidt R. Demonstration bakteriologischer Fazespraparte. Munch Med Wochenschr 1903;50(2):2165. [Google Scholar]
- (10).Heinemann PG, Ecker EE. A Study of the Boas-Oppler bacillus. J Bacteriol 1916;1(4):435–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Anonymous. The Boas-Oppler Bacillus. JAMA 1917;69(5):377. [Google Scholar]
- (12).Comfort MW. Gastric acidity before and after development of gastric cancer: its etiologic, diagnostic and prognostic significance. Ann Intern Med 1951;36(6):1331–48. [DOI] [PubMed] [Google Scholar]
- (13).Barber M, Franklin RH. Bacteriology of stomach and duodenum in cases of peptic ulcer and gastric carcinoma. Br Med J 1946;1:951–3. [PubMed] [Google Scholar]
- (14).Spallanzani L. Experiences sur la digestion de L’homme et de diferentes especes d’animaux. Geneva: Chez Barthelemi Chirol, 1783. [Google Scholar]
- (15).Macfadyen A. Behavior of bacteria in the digestive tract. J Anat Physiol 1887;21(Pt 3):413–37. [PMC free article] [PubMed] [Google Scholar]
- (16).Gillespie AL. The bacteria of the stomach. In German S.Woodhead, editor. The journal of pathology and bacteriology. Edinburgh: Young J. Pentland, 1893: 279–302. [Google Scholar]
- (17).Giannella RA, Broitman SA, Zamcheck N. Gastric acid barrier to ingested microorganisms in man: studies in vivo and in vitro.Gut 1972;13(4):251–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Bartsch H. O’Neill IK and Shulte-Hermann R. The Relevance of N-nitroso compounds to human cancer: Exposures and mechanisms. 1–658. 1987. Baden, Austria, IARC Scientific Publication No. 84. Proceedings of the IXth International Symposium on N-Nitroso Compounds. [PubMed] [Google Scholar]
- (19).Jakszyn P, Bingham S, Pera G et al. Endogenous versus exogenous exposure to N-nitroso compounds and gastric cancer risk in the European Prospective Investigation into Cancer and Nutrition (EPIC-EURGAST) study. Carcinogenesis 2006;27(7):1497–501. [DOI] [PubMed] [Google Scholar]
- (20).Tricker AR, Preussmann R. Carcinogenic N-nitrosamines in the diet: occurrence, formation, mechanisms and carcinogenic potential. Mutat Res 1991;259(3–4):277–89. [DOI] [PubMed] [Google Scholar]
- (21).Xu L, Qu YH, Chu XD et al. Urinary levels of N-nitroso compounds in relation to risk of gastric cancer: findings from the Shanghai cohort study. PLoS One 2015;10(2):e0117326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Loh YH, Jakszyn P, Luben RN et al. N-nitroso compounds and cancer incidence: the European Prospective Investigation into Cancer and Nutrition (EPIC)-Norfolk Study. Am J Clin Nutr 2011;93(5):1053–61. [DOI] [PubMed] [Google Scholar]
- (23).Gilchrist M, Winyard PG, Benjamin N. Dietary nitrate--good or bad? Nitric Oxide 2010;22(2):104–9. [DOI] [PubMed] [Google Scholar]
- (24).Ward MH, Heineman EF, Markin RS et al. Adenocarcinoma of the stomach and esophagus and drinking water and dietary sources of nitrate and nitrite. Int J Occup Environ Health 2008;14(3):193–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Keszei AP, Goldbohm RA, Schouten LJ et al. Dietary N-nitroso compounds, endogenous nitrosation, and the risk of esophageal and gastric cancer subtypes in the Netherlands Cohort Study. Am J Clin Nutr 2013;97(1):135–46. [DOI] [PubMed] [Google Scholar]
- (26).Kobayashi J. Effect of diet and gut environment on the gastrointestinal formation of N-nitroso compounds: A review. Nitric Oxide 2018;73:66–73. [DOI] [PubMed] [Google Scholar]
- (27).Correa P, Haenszel W, Cuello C et al. A model for gastric cancer epidemiology. Lancet 1975;2:58–60. [DOI] [PubMed] [Google Scholar]
- (28).Correa P, Cuello C, Gordillo G et al. The gastric microenvironment in populations at high risk to stomach cancer. Natl Cancer Inst Monogr 1979;167–70. [PubMed] [Google Scholar]
- (29).Montes G, Cuello C, Gordillo G et al. Mutagenic activity of gastric juice. Cancer Lett 1979;7:307–12. [DOI] [PubMed] [Google Scholar]
- (30).Ruddell WS, Bone ES, Hill MJ et al. Gastric-juice nitrite. A risk factor for cancer in the hypochlorhydric stomach? Lancet 1976;2(7994):1037–9. [DOI] [PubMed] [Google Scholar]
- (31).Ruddell WS, Blends LM, Walters CL. Proceedings: Nitrite and thiocyanate in gastric juice. Gut 1976;17(5):401. [PubMed] [Google Scholar]
- (32).Jakszyn P, Gonzalez CA. Nitrosamine and related food intake and gastric and oesophageal cancer risk: a systematic review of the epidemiological evidence. World J Gastroenterol 2006;12(27):4296–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Choi NW, Miller AB, Fodor JG et al. Consumption of precursors of N-nitroso compounds and human gastric cancer. IARC Sci Publ 1987;(84):492–6. [PubMed] [Google Scholar]
- (34).Hall CN, Darkin D, Brimblecombe R et al. Evaluation of the nitrosamine hypothesis of gastric carcinogenesis in precancerous conditions. Gut 1986;27(5):491–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Aditi A, Graham DY. Vitamin C, gastritis, and gastric disease: A historical review and update. Dig Dis Sci 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Sobala GM, Schorah CJ, Sanderson M et al. Ascorbic acid in the human stomach. Gastroenterology 1989;97(2):357–63. [DOI] [PubMed] [Google Scholar]
- (37).Sobala GM, Pignatelli B, Schorah CJ et al. Levels of nitrite, nitrate, N-nitroso compounds, ascorbic acid and total bile acids in gastric juice of patients with and without precancerous conditions of the stomach. Carcinogenesis 1991;12:193–8. [DOI] [PubMed] [Google Scholar]
- (38).Petersen CP, Mills JC, Goldenring JR. Murine models of gastric corpus preneoplasia. Cell Mol Gastroenterol Hepatol 2017;3(1):11–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Graham DY, Zou WY. Guilt by association: intestinal metaplasia does not progress to gastric cancer. Curr Opin Gastroenterol 2018;34(6):458–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Nozaki K, Shimizu N, Ikehara Y et al. Effect of early eradication on Helicobacter pylori-related gastric carcinogenesis in Mongolian gerbils. Cancer Sci 2003;94(3):235–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Shimizu N, Ikehara Y, Inada K et al. Eradication diminishes enhancing effects of Helicobacter pylori infection on glandular stomach carcinogenesis in Mongolian gerbils. Cancer Res 2000;60(6):1512–4. [PubMed] [Google Scholar]
- (42).Shimizu N, Inada K, Nakanishi H et al. Helicobacter pylori infection enhances glandular stomach carcinogenesis in Mongolian gerbils treated with chemical carcinogens. Carcinogenesis 1999;20(4):669–76. [DOI] [PubMed] [Google Scholar]
- (43).Hayakawa Y, Fox JG, Gonda T et al. Mouse models of gastric cancer. Cancers (Basel) 2013;5(1):92–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Tsuda A, Suda W, Morita H et al. Influence of proton-pump inhibitors on the luminal microbiota in the gastrointestinal tract. Clin Transl Gastroenterol 2015;6:e89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Graham DY. Helicobacter pylori update: Gastric cancer, reliable therapy, and possible benefits. Gastroenterology 2015;148(4):719–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Miftahussurur M, Yamaoka Y, Graham DY. Helicobacter pylori as an oncogenic pathogen, revisited. Expert Rev Mol Med 2017;19:e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Kamada T, Haruma K, Ito M et al. Time trends in Helicobacter pylori infection and atrophic gastritis over 40 years in Japan. Helicobacter 2015;20(3):192–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Wang C, Weber A, Graham DY. Age, period, and cohort effects on gastric cancer mortality. Dig Dis Sci 2015;60(2):514–23. [DOI] [PubMed] [Google Scholar]
- (49).Hanada K, Graham DY. Helicobacter pylori and the molecular pathogenesis of intestinal-type gastric carcinoma. Expert Rev Anticancer Ther 2014;14(8):947–54. [DOI] [PubMed] [Google Scholar]
- (50).Kidane D. Molecular mechanisms of H. pylori-induced DNA double-strand breaks. Int J Mol Sci 2018;19(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Choi IJ, Kook MC, Kim YI et al. Helicobacter pylori therapy for the prevention of metachronous gastric cancer. N Engl J Med 2018;378(12):1085–95. [DOI] [PubMed] [Google Scholar]
- (52).Coati I, Fassan M, Farinati F et al. Autoimmune gastritis: Pathologist’s viewpoint. World J Gastroenterol 2015;21(42):12179–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Marne C, Pallares R, Casanova A et al. Gastric juice microflora in patients with gastric ulcer and gastric cancer. Eur J Clin Microbiol 1985;4(4):426–7. [DOI] [PubMed] [Google Scholar]
- (54).Gray JD, Shiner M. Influence of gastric pH on gastric and jejunal flora. Gut 1967;8(6):574–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Sjostedt S, Kager L, Heimdahl A et al. Microbial colonization of tumors in relation to the upper gastrointestinal tract in patients with gastric carcinoma. Ann Surg 1988;207(3):341–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Sjostedt S, Heimdahl A, Kager L et al. Microbial colonization of the oropharynx, esophagus and stomach in patients with gastric diseases. Eur J Clin Microbiol 1985;4(1):49–51. [DOI] [PubMed] [Google Scholar]
- (57).Minalyan A, Gabrielyan L, Scott D et al. The gastric and intestinal microbiome: Role of proton pump inhibitors. Curr Gastroenterol Rep 2017;19(8):42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Li TH, Qin Y, Sham PC et al. Alterations in gastric microbiota after H. pylori eradication and in different histological stages of gastric carcinogenesis. Sci Rep 2017;7:44935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Massarrat S, Saniee P, Siavoshi F et al. The Effect of Helicobacter pyloriinfection, aging, and consumption of proton pump inhibitor on fungal colonization in the stomach of dyspeptic patients. Front Microbiol 2016;7:801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Azab M, Doo L, Doo DH et al. Comparison of the hospital-acquired Clostridium difficileinfection risk of using proton pump inhibitors versus histamine-2 receptor antagonists for prophylaxis and treatment of stress ulcers: A systematic review and meta-analysis. Gut Liver 2017;11(6):781–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Rosen R, Hu L, Amirault J et al. 16S community profiling identifies proton pump inhibitor related differences in gastric, lung, and oropharyngeal microflora. J Pediatr 2015;166(4):917–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Wang K, Lin HJ, Perng CL et al. The effect of H2-receptor antagonist and proton pump inhibitor on microbial proliferation in the stomach. Hepatogastroenterology 2004;51(59):1540–3. [PubMed] [Google Scholar]
- (63).Peterson WL, Mackowiak PA, Barnett CC et al. The human gastric bactericidal barrier: mechanisms of action, relative antibacterial activity, and dietary influences. J Infect Dis 1989;159(5):979–83. [DOI] [PubMed] [Google Scholar]
- (64).Osato MS, Gutierrez O, Kim JG et al. Microflora of gastric biopsies from patients with duodenal ulcer and gastric cancer: a comparative study of patients from Korea, Colombia, and the United States. Dig Dis Sci 1998;43(10):2291–5. [DOI] [PubMed] [Google Scholar]
- (65).Monstein HJ, Tiveljung A, Kraft CH et al. Profiling of bacterial flora in gastric biopsies from patients with Helicobacter pylori-associated gastritis and histologically normal control individuals by temperature gradient gel electrophoresis and 16S rDNA sequence analysis. J Med Microbiol 2000;49(9):817–22. [DOI] [PubMed] [Google Scholar]
- (66).Bik EM, Eckburg PB, Gill SR et al. Molecular analysis of the bacterial microbiota in the human stomach. Proc Natl Acad Sci U S A 2006;103(3):732–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Dicksved J, Lindberg M, Rosenquist M et al. Molecular characterization of the stomach microbiota in patients with gastric cancer and in controls. J Med Microbiol 2009;58(Pt 4):509–16. [DOI] [PubMed] [Google Scholar]
- (68).Delong EF. Preface. Microbial metagenomics, metatranscriptomics, and metaproteomics. Methods Enzymol 2013;531:xxi. [DOI] [PubMed] [Google Scholar]
- (69).Zhu Z, Ren J, Michail S et al. Correction to: MicroPro: using metagenomic unmapped reads to provide insights into human microbiota and disease associations. Genome Biol 2019;20(1):214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Andersson AF, Lindberg M, Jakobsson H et al. Comparative analysis of human gut microbiota by barcoded pyrosequencing. PLoS One 2008;3(7):e2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Vuik F, Dicksved J, Lam SY et al. Composition of the mucosa-associated microbiota along the entire gastrointestinal tract of human individuals. United European Gastroenterol J 2019;7(7):897–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Chen L, Xu W, Lee A et al. The impact of Helicobacter pylori infection, eradication therapy and probiotic supplementation on gut microenvironment homeostasis: An open-label, randomized clinical trial. EBioMedicine 2018;35:87–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Maldonado-Contreras A, Goldfarb KC, Godoy-Vitorino F et al. Structure of the human gastric bacterial community in relation to Helicobacter pylori status. ISME J 2011;5(4):574–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Aviles-Jimenez F, Vazquez-Jimenez F, Medrano-Guzman R et al. Stomach microbiota composition varies between patients with non-atrophic gastritis and patients with intestinal type of gastric cancer. Sci Rep 2014;4:4202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (75).Liu X, Shao L, Liu X et al. Alterations of gastric mucosal microbiota across different stomach microhabitats in a cohort of 276 patients with gastric cancer. EBioMedicine 2019;40:336–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (76).Ferreira RM, Pereira-Marques J, Pinto-Ribeiro I et al. Gastric microbial community profiling reveals a dysbiotic cancer-associated microbiota. Gut 2018;67(2):226–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (77).Langille MG, Zaneveld J, Caporaso JG et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 2013;31(9):814–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (78).Coker OO, Dai Z, Nie Y et al. Mucosal microbiome dysbiosis in gastric carcinogenesis. Gut 2018;67(6):1024–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (79).Noto JM, Peek RM Jr. The gastric microbiome, its interaction with Helicobacter pylori, and its potential role in the progression to stomach cancer. PLoS Pathog 2017;13(10):e1006573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (80).Castano-Rodriguez N, Goh KL, Fock KM et al. Dysbiosis of the microbiome in gastric carcinogenesis. Sci Rep 2017;7(1):15957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (81).Mailhe M, Ricaboni D, Vitton V et al. Repertoire of the gut microbiota from stomach to colon using culturomics and next-generation sequencing. BMC Microbiol 2018;18(1):157. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- (82).Ferreira RM, Pereira-Marques J, Pinto-Ribeiro I et al. Gastric microbial community profiling reveals a dysbiotic cancer-associated microbiota. Gut 2018;67(2):226–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (83).Park CH, Lee AR, Lee YR et al. Evaluation of gastric microbiome and metagenomic function in patients with intestinal metaplasia using 16S rRNA gene sequencing. Helicobacter 2019;24(1):e12547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (84).Sung J, Kim N, Kim J et al. Comparison of gastric microbiota between gastric juice and mucosa by next generation sequencing method. J Cancer Prev 2016;21(1):60–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (85).Gevers D, Kugathasan S, Denson LA et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe 2014;15(3):382–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (86).Carstens A, Roos A, Andreasson A et al. Differential clustering of fecal and mucosa-associated microbiota in ‘healthy’ individuals. J Dig Dis 2018;19(12):745–52. [DOI] [PubMed] [Google Scholar]
- (87).Saffarian A, Mulet C, Regnault B et al. Crypt- and mucosa-associated core microbiotas in humans and their alteration in colon cancer patients. MBio 2019;10(4). [DOI] [PMC free article] [PubMed] [Google Scholar]