Abstract
A simple molecular technique for rapid genotyping was developed to monitor the internal gene composition of currently circulating influenza A viruses. Sequence information from recent H1N1, H3N2, and H5N1 human virus isolates was used to identify conserved regions within each internal gene, and gene-specific PCR primers capable of amplifying all three virus subtypes were designed. Subtyping was based on subtype-specific restriction fragment length polymorphism (RFLP) patterns within the amplified regions. The strategy was tested in a blinded fashion using 10 control viruses of each subtype (total, 30) and was found to be very effective. Once standardized, the genotyping method was used to identify the origin of the internal genes of 51 influenza A viruses isolated from humans in Hong Kong during and immediately following the 1997–1998 H5N1 outbreak. No avian-human or H1-H3 reassortants were detected. Less than 2% (6 of 486) of the RFLP analyses were inconclusive; all were due to point mutations within a restriction site. The technique was also used to characterize the internal genes of two avian H9N2 viruses isolated from children in Hong Kong during 1999.
Influenza A viruses circulate worldwide and cause annual epidemics of human respiratory illness. Currently, two subtypes, H1N1 and H3N2, are in worldwide circulation within the human population. In addition, animal reservoirs of other antigenic subtypes exist, especially among aquatic bird species, from which 15 HA and 9 NA types have been isolated (19).
The segmented genome of influenza A viruses confers the ability for gene reassortment, and the coinfection of a single host by two distinct viruses may lead to the production of hybrid virions. Because the viruses responsible for the two most recent influenza pandemics, the Asian H2N2 of 1957 and Hong Kong H3N2 of 1968, arose through the reassortment of human and avian viruses (11), the isolation of avian influenza A H5N1 viruses from humans in Hong Kong in 1997 raised concerns about the emergence of a new pandemic strain. All eight genes from the 16 H5N1 viruses isolated from humans during the outbreak were of avian origin, indicating that these cases resulted from a complete viral host shift across the species barrier (7, 17). Similarly, in March 1999, two avian H9N2 viruses were isolated from children in Hong Kong (14). Although wholly avian viruses may not be highly transmissible from person to person, a reassortment event with cocirculating human H3N2 and H1N1 viruses could create a gene constellation that confers this trait (3, 11). The continued presence of these and other influenza viruses in avian populations suggests a need for continued monitoring to detect the inapparent movement of avian influenza virus genes into the human virus gene pool.
Previous studies have utilized a variety of molecular methods to detect naturally occurring and laboratory-generated reassortants (1, 6, 12, 15, 23). The purpose of the present study was to develop a simple genotyping method that could be used to monitor the internal gene composition of currently circulating human influenza A viruses. Our specific objective was to be able to rapidly genotype virus isolates and easily distinguish the internal genes of recent human H1N1 and H3N2 viruses from each other and from those of the recently isolated H5N1 avian viruses. An additional advantage of this strategy is that it can be used to rapidly assess the internal genes of other virus subtypes, such as the 1999 Hong Kong H9N2 isolates, to determine if they harbor H1N1, H3N2, or H5N1-like genes.
MATERIALS AND METHODS
RT-PCR and nucleotide sequencing.
Laboratory work with the highly pathogenic H5N1 viruses was carried out under biosafety level 3+ containment conditions. Viral RNA was extracted from cell culture supernatants or allantoic fluid suspensions that contained from 8 to 512 hemagglutinating units using the QIAmp viral RNA extraction kit (Qiagen, Chatsworth, Calif.) in accordance with the manufacturer's instructions. RNA was extracted from 140 μl of each specimen and eluted in a final volume of 50 μl. The amount of RNA recovered was not routinely quantified. Amplification of viral genes was accomplished by using a previously standardized two-step reverse transcriptase (RT)-PCR protocol. First-step RT reaction mixtures containing 2.5 mM each of the deoxynucleoside triphosphates (dNTPs) dATP, dCTP, dGTP, and dTTP, 10 μl of 10× PCR buffer (Roche Diagnostics, Indianapolis, Ind.), 7 μl of water, 1 μl of the forward primer at a concentration of 0.5 μg/μl, and 3 μl of viral RNA were incubated for 90 s at 90°C before being quenched on ice. Twenty-five units of avian myeloblastosis virus reverse transcriptase and 40 U of RNase inhibitor were then added, and the mixture was incubated at 42°C for 60 min. Subsequent PCR amplification of cDNA was accomplished in the same tube through the addition of a second mixture containing an additional 10 mM dNTPs, 5 U of Taq DNA polymerase (Roche Diagnostics), 1 μl of the reverse primer at a concentration of 0.5 μg/μl, and enough water to give a final reaction mixture of 100 μl. Thermocycling conditions for the PCR step consisted of 95°C for 5 min followed by 30 to 35 cycles of 94°C for 1 min, 50°C for 2 min, 70°C for 3 min, and a final extension step of 72°C for 7 min. Prior to sequencing, PCR products were purified by using the QIAquick PCR purification kit (Qiagen). Automated sequencing of the PCR products was accomplished by using the Prism Ready Dye Deoxy Terminator Cycle sequencing kit on an ABI model 373A automated DNA sequencer (Perkin-Elmer, Applied Biosystems Division, Foster City, Calif.). Computerized analyses of virus sequence data were performed with the Pretty, Pileup, and Mapsort programs of the University of Wisconsin Genetics Computer Group software package (Genetics Computer Group, 1994).
Primer design and location of subtype-specific restriction sites.
In order to design broadly cross-reactive PCR primers, we first determined the complete nucleotide sequences of the M, NS, NP, PA, PB1, and PB2 genes of two 1997 human H3N2 and H1N1 viruses isolated from Hong Kong (Table 1). The sequences of amplifying PCR and sequencing primers used for this purpose are available upon request. These data, along with those from two of the human H5N1 viruses, were used to locate conserved primer sites within each gene. Highly conserved regions were identified and used to design gene-specific PCR primers capable of amplifying all three virus subtypes. Rapid subtyping was based on the detection of restriction fragment length polymorphism (RFLP) patterns within these amplified regions. Specifically, complete digest maps that included restriction sites for all commercially available restriction enzymes were generated for each internal gene amplicon. Genotyping strategies for individual gene segments were designed based on the following criteria: three enzymes were selected per gene segment; each enzyme cut only one of the three subtypes at a single location within the region amplified by the conserved primers; and each enzyme generated a visually distinct restriction pattern. The designated primer pairs used for each gene and the subtype-specific restriction enzymes used are shown in Table 2. The nucleotide sequences of the conserved PCR primers are as follows: NSF1, 5′-AGCAAAAGCAGGGTGACAAAGACA-3′; NSR890, 5′-AGTAGAAACAAGGGTGTTTTTTAT-3′; MF149, 5′-CTCATGGAATGGCTAAAGACA-3′; MR847, 5′-CGATCAAG/TAATCCACAATATC-3′; NPF528, 5′-CAGA/GATGTGCTCTC/TTGATGCA-3′; NPR1506, 5′-ATAAGATCCTTCATTACTCAT-3′; PAF29, 5′-AAGAT/CTTTGTGCGACAATGCT-3′; PAR773, 5′-GACATTTGAGAAAGCTTGCC-3′; PB1F22, 5′-TTGAATGGATGTCAATCCGA-3′; PB1R715, 5′-CATCTTTIGTCATC/TGTGTTCA-3′; PB2F160, 5′-CTTAGA/GATGAAATGGATGAT-3′; and PB2 R1007, 5′-CCICCAAAIG/CTGAAGGATGA-3′. Primer sequences correspond to the viral RNA sequences beginning at the designated nucleotide. Degenerate sites are noted with a forward slash mark; I stands for inosine.
TABLE 1.
Human influenza A viruses sequenced
| Strain designation (subtype) | GenBank accession no.
|
|||||
|---|---|---|---|---|---|---|
| NS | M | NP | PA | PB1 | PB2 | |
| A/Hong Kong/470/97 (H1N1) | AF258520 | AF258522 | AF258517 | AF258518 | AF258527 | AF258524 |
| A/Hong Kong/427/98 (H1N1) | AF258521 | AF258523 | AF258516 | AF258519 | AF258526 | AF258525 |
| A/Hong Kong/497/97 (H3N2) | AF256182 | AF255369 | AF255748 | AF257197 | AF258822 | AF258841 |
| A/Hong Kong/498/97 (H3N2) | AF256183 | AF255370 | AF255749 | AF257198 | AF258823 | AF258842 |
TABLE 2.
Conserved PCR primer designations and subtype-specific restriction enzymes
| Gene | Forward primer | Reverse primer | Amplicon size (bp) | Subtype-specific restriction enzyme
|
||
|---|---|---|---|---|---|---|
| H1N1 | H3N2 | H5N1 | ||||
| NS | 1 | 890 | 890 | DraI | XbaI | BsrBI |
| M | 149 | 847 | 698 | HindIII | ScaI | AvaII |
| NP | 528 | 1506 | 978 | HaeII | SacII | BamHI |
| PA | 29 | 773 | 744 | BbsI | EcoNI | NsiI |
| PB1 | 22 | 715 | 693 | ScaI | XmnI | BsrBI |
| PB2 | 160 | 1007 | 847 | EcoRV | BstZ171 | BglII |
Aliquots of unpurified amplicon DNA were individually digested with all three enzymes for 1 h at 37°C in 96-well microtiter plates. The resulting products were electrophoresed through 1.5% agarose gels containing ethidium bromide and visualized under a UV light source.
Viruses.
To test our PCR primers and the feasibility of using the RFLP strategy for rapid genotyping, an initial test using a group of 30 human influenza viruses was conducted. Rapid subtyping was based on subtype-specific RFLP patterns within the amplified region. Ten each of H1N1 and H3N2 viruses, collected from different geographic regions around the world, as well as 10 H5N1 isolates from the Hong Kong outbreak were tested in a blinded fashion. Rapid subtyping of the resultant PCR amplicons was based on subtype-specific RFLP patterns within the amplified region. To test for possible reassortment between current human subtypes H3N2 and H1N1 and viruses of avian origin, the internal genes from 51 additional human viruses isolated in Hong Kong during the 1997 H5N1 outbreak were also genotyped (strain designations available upon request). These techniques were also used for the initial molecular characterization of two avian H9N2 viruses from Hong Kong. These viruses, A/Hong Kong/1073/99 and A/Hong Kong/1074/99, were isolated from children during March 1999.
Further analysis of nontypeable isolates.
A small number of virus isolates examined in the current study could not be genotyped completely by the PCR-RFLP method because of the presence of an amplicon with an atypical restriction pattern. Because the most likely cause was a base substitution within a restriction site, the genotypes of all such nontypeable amplicons were identified by nucleotide sequencing of the appropriate region, and the reason for the loss of the restriction site was determined.
RESULTS
PCR amplifications and subtype-specific restriction patterns.
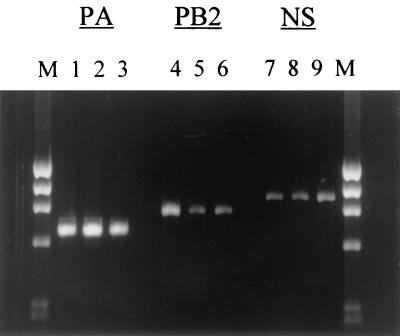
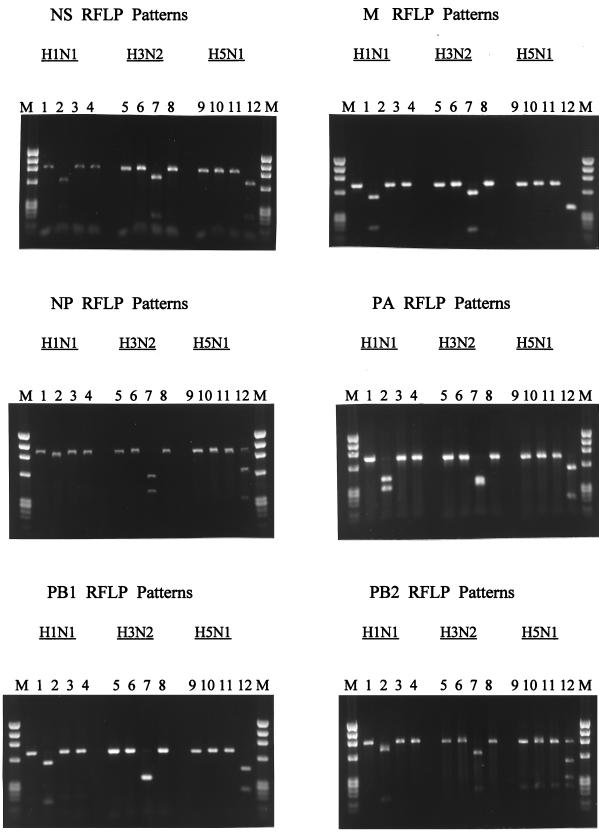
Data from an initial control group of 30 viruses indicated that the designed PCR primers performed well and produced gene-specific amplicons for these viruses, as well as for the 51 additional Hong Kong isolates that were tested later. Representative amplicons for three of the six internal genes are shown in Fig. 1. Typical subtype-specific restriction patterns for each of the six amplified gene regions are shown in Fig. 2. In all cases, visibly distinct restriction patterns were observed for each of the three virus subtypes. However, it should be noted that in several cases the relative sizes of the restriction products necessitated a longer period of electrophoresis for clear separation of the subtypes.
FIG. 1.
Representative amplicons for the PA, PB2, and NS genes of influenza A H1N1, H3N2, and H5N1 viruses. Lanes 1, 4, and 7, H1N1 virus; lanes 2, 5, and 8, H3N2 isolate; lanes 3, 6, and 9, H5N1 virus isolate. Lanes M, molecular size markers.
FIG. 2.
RFLP patterns for the internal genes of H1N1, H3N2, and H5N1 influenza A viruses. In each instance, the samples were loaded (left to right) in the order of no enzyme, digest with the H1N1-specific enzyme, digest with the H3N2-specific enzyme, and digest with the H5N1-specific enzyme. Lanes M, molecular size markers.
To test for possible reassortment between the current human subtypes, H3N2 and H1N1, and viruses of avian origin, the internal genes from 51 additional human influenza A viruses isolated in Hong Kong during the 1997 H5N1 outbreak were also genotyped. No avian influenza virus genes were present in the 51 human viruses from Hong Kong, and no naturally occurring H1N1-H3N2 reassortants were detected among the viruses that were sampled. In all, 486 genes from 81 human H1N1, H3N2, and H5N1 viruses were examined.
The initial molecular characterization of the internal genes of the two H9N2 viruses, A/Hong Kong/1073/99 and A/Hong Kong/1074/99, by RFLP resulted in restriction patterns identical to those of the 1997 Hong Kong H5N1 viruses. The similarity of the internal genes from these two H9N2 isolates to those of the H5N1 viruses isolated in 1997 was confirmed by subsequent sequencing of the PCR amplicons (data not presented).
Sequence analysis of nontypeable amplicons.
A small number of PCR amplicons did not generate typical restriction profiles and therefore could not be subtyped by their RFLP patterns. An overview of these untypeable viruses is shown in Table 3. Six (1.2%) of the 486 PCR amplicons examined fell into this category. Further sequence analysis of these six genes revealed that in each case, the RFLP strategy failed because of a single point mutation within a restriction site. The facts that these six untypeable amplicons included H1N1 and H3N2 subtypes, occurred in four different gene segments, and were observed for virus isolates spanning a wide geographic distribution support the notion that the mutations that resulted in the loss of a restriction site were random events. However, it should be noted that the same point mutation was detected in the PA genes of three Asian H3N2 viruses. This may be of concern in later surveys if this mutation becomes fixed within the H3N2 lineage.
TABLE 3.
Characterization of viruses with unusual RFLP patterns
| Virus designation | Subtype | Gene | Mutation making amplicon untypeable |
|---|---|---|---|
| A/Russia/01/98 | H1N1 | M | T to G |
| A/Paris/0453/97 | H3N2 | NP | C to T |
| A/Caen/1273/98 | H1N1 | PB1 | G to A |
| A/Hong Kong/523/97 | H3N2 | PA | C to A |
| A/Hong Kong/546/97 | H3N2 | PA | C to A |
| A/Singapore/21/97 | H3N2 | PA | C to A |
DISCUSSION
A rapid PCR-RFLP genotyping method capable of distinguishing the internal genes of human H1N1, H3N2, and avian H5N1 influenza A viruses was developed and used to screen viruses from clinical samples isolated in Hong Kong during and immediately following the 1997 H5N1 avian influenza outbreak. No avian-human virus reassortants were detected among these samples. The technique worked with human H1N1 and H3N2 viruses isolated from many locations and successfully identified the H5N1-like internal genes of two H9N2 viruses. Based on these results, the procedure will be of great future use in the genetic screening of new influenza A virus isolates. No new human cases of H5N1 or H9N2 influenza have recently been reported. However, similar avian viruses continue to circulate within the bird population of Asia (8) and may infect humans again. The isolation of an avian H6N1 virus, A/teal/Hong Kong/W312/97, in which all six internal genes are highly homologous to those of the 1997 H5N1 viruses, has recently been reported (E. Hoffman, J. Stech, S. Krauss, K. F. Shortridge, and R. G. Webster, 18th Annu. Meet. Am. Soc. Virol., abstr. W3-10). The detection of this internal gene constellation in three avian influenza A virus subtypes, two of which have crossed the species barrier to infect humans, is cause for concern and further illustrates the need for rapid diagnostic tests to detect their presence.
Toward this end, it is important to note that our current molecular strategies may need to be periodically reassessed. The technique was designed to detect and distinguish among three well-defined cocirculating subtypes and is based on the current sequences of human H1N1 and human H3N2 viruses and those of the avian H5N1 viruses isolated from patients in Hong Kong. Although the rate of evolution for influenza virus internal genes is substantially lower than those of the HA and NA genes (19), the sequences of these genes do change over time. For this reason it may be necessary to update both the primer sequences and the choice of restriction enzymes as needed to compensate for genetic drift.
Another minor concern is that because our primers are complementary to highly conserved genetic regions, they may be capable of amplifying the internal genes of other animal influenza viruses, such as those from classical swine viruses (Cooper, unpublished data). Although the ability of our primers to amplify a wide range of viruses is advantageous for the rapid identification of unknown isolates through sequencing of the PCR amplicons, in rare cases the presence of zoonotic genes could be overlooked if the tested regions not only amplify but, by chance alone, also have restriction patterns that are similar to those of human influenza A viruses. While such false-positives may be exceptionally rare events, they must be considered if this technique is to be used for routine viral screening, and it would be prudent to spot-check the results of future genotyping surveys by sequence analysis. Such sequence-based confirmation would be especially important when there is a high index of suspicion that reassortment between human and animal viruses has occurred.
The true extent of genetic reassortment between cocirculating influenza viruses is hard to assess, and only in the last two decades have advances in molecular biology allowed detailed genetic characterizations of influenza isolates. Without routine genotyping, even the best-informed estimates of genetic exchange between virus lineages may not accurately reflect the true amount of genetic mixing. Shu et al. (16) analyzed the internal genes of 122 viruses isolated from humans during interpandemic periods spanning 1933 to 1992 and found no reassortant genotypes. Yet clearly gene flow between divergent influenza A virus lineages does occur. The best-known examples are the avian and human virus reassortment events that give rise to the last two influenza pandemic strains in 1957 and 1968, but many other examples of cross-species movement of influenza virus genes exist. A short list of such events includes the infection of humans with avian and swine influenza viruses, sea mammals with avian strains, and camels, equines, and pigs with human and/or avian strains (2, 4, 5, 9, 10, 20, 22).
Likewise, genetic exchanges between strictly human viruses have been documented. Since 1977, human H1N1 and H3N2 viruses have cocirculated within the human population, and naturally occurring reassortants have been isolated on several occasions (13, 21). In a review of the published literature, Xu et al. (21) concluded that genetic reassortment of human influenza viruses occurs frequently in nature. Even the best surveillance systems sample only a portion of all naturally occurring infections. For example, our study detected no reassortants, but because it examined only a fraction of the total number of currently circulating influenza viruses, the existence of such hybrids was not ruled out. Viable reassortants may arise almost continuously but may not persist because they are at a selective disadvantage or may be lost from the circulating virus population by chance alone.
From a public health perspective, the ability to rapidly genotype the internal genes of influenza A virus isolates will be most valuable in outbreak situations in which there is concern about possible interspecies transmission and the emergence of novel reassortant viruses. The current study is valuable because it establishes a procedure by which this can be accomplished. Despite the ubiquity of influenza A viruses, the internal genes are rarely sequenced, and prior to our study few current human virus internal gene sequences were available through the public databases. Our PCR-RFLP methods are straightforward and can be performed by laboratories that do not have the ability to directly sequence virus isolates. If updated and kept current, the application of this strategy to the future monitoring of human H1N1 and H3N2 isolates could be of great value in the early detection of new gene combinations. Although our study was prompted by the recent emergence of H5N1 avian viruses, these techniques can easily be modified to monitor other avian or zoonotic strains and have broad applications within both public health and agricultural settings. They are therefore extremely valuable tools in the arsenal of techniques available for influenza virus molecular surveillance.
ACKNOWLEDGMENTS
This research was supported in part by an appointment to the Emerging Infectious Diseases Fellowship program (Lynn Cooper) administered by the Centers for Disease Control and Prevention and the Association of Public Health Laboratory Directors.
We thank Xiyan Xu and Yumiko Matsuoka for valuable scientific discussions, Miriam Laker for excellent laboratory assistance, and Nancy Cox and John O'Connor for their critical reviews of the manuscript.
REFERENCES
- 1.Adeyefa C A, Quayle O K, McCauley J W. A rapid method for the analysis of influenza virus genes: application to the reassortment of equine virus genes. Virus Res. 1994;32:391–409. doi: 10.1016/0168-1702(94)90087-6. [DOI] [PubMed] [Google Scholar]
- 2.Banks J, Speidel E, Alexander D J. Characterization of an avian influenza A virus isolated from a human—is an intermediate host necessary for the emergence of pandemic influenza viruses? Arch Virol. 1998;143:781–787. doi: 10.1007/s007050050329. [DOI] [PubMed] [Google Scholar]
- 3.Beare A S, Webster R G. Replication of avian influenza viruses in humans. Arch Virol. 1991;119:37–42. doi: 10.1007/BF01314321. [DOI] [PubMed] [Google Scholar]
- 4.Brown I H, Harris P A, McCauley J W, Alexander D J. Multiple genetic reassortment of avian and human influenza viruses in European pigs, resulting in the emergence of an H1N2 virus of novel genotype. J Gen Virol. 1998;12:2947–2955. doi: 10.1099/0022-1317-79-12-2947. [DOI] [PubMed] [Google Scholar]
- 5.Castrucci M R, Donatelli I, Sidoli L, Barigazzi G, Kawaoka Y, Webster R G. Genetic reassortment between avian and human influenza viruses in Italian pigs. Virology. 1993;193:503–506. doi: 10.1006/viro.1993.1155. [DOI] [PubMed] [Google Scholar]
- 6.Cha T, Zhao J, Lane E, Murray M, Stec D. Determination of the genome composition of influenza virus reassortants using multiplex reverse transcription-polymerase chain reaction followed by fluorescent single-strand conformation polymorphism analysis. Anal Biochem. 1997;252:24–32. doi: 10.1006/abio.1997.2269. [DOI] [PubMed] [Google Scholar]
- 7.Claas E C J, Osterhaus A D M E, van Beek R, de Jong J C, Rimmelzwann G F, Senne D A, Krauss S, Shortridge K F, Webster R G. Human influenza A H5N1 virus related to a highly pathogenic avian influenza virus. Lancet. 1998;351:472–477. doi: 10.1016/S0140-6736(97)11212-0. [DOI] [PubMed] [Google Scholar]
- 8.Guan Y, Shortridge K F, Krauss S, Webster R G. Molecular characterization of H9N2 influenza viruses: were they the donors of the “internal” genes of H5N1 viruses in Hong Kong? Proc Natl Acad Sci USA. 1999;96:9363–9367. doi: 10.1073/pnas.96.16.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo Y, Yang M, Kawaoka Y, Gorman O, Ito T, Webster R G. Characterization of a new avian-like influenza A virus from horses in China. Virology. 1992;188:245–255. doi: 10.1016/0042-6822(92)90754-d. [DOI] [PubMed] [Google Scholar]
- 10.Hinshaw V S, Webster R G, Easterday B C, Bean W J. Replication of avian influenza A viruses in mammals. Infect Immun. 1981;34:354–361. doi: 10.1128/iai.34.2.354-361.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kawaoka Y, Krauss S, Webster R G. Avian-to-human transmission of the PB1 gene of influenza A virus in the 1957 and 1968 pandemics. J Virol. 1989;63:4603–4608. doi: 10.1128/jvi.63.11.4603-4608.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klimov A J, Cox N J. PCR restriction analysis of genome composition and stability of cold-adapted reassortant live influenza vaccines. J Virol Methods. 1995;52:41–49. doi: 10.1016/0166-0934(94)00133-2. [DOI] [PubMed] [Google Scholar]
- 13.Lindstrom S E, Hiromoto Y, Nerome R, Omoe K, Sugita S, Yamazaki Y, Takahashi R, Nerome K. Phylogenetic analysis of the entire genome of influenza A (H3N2) viruses from Japan: evidence for genetic reassortment of the six internal genes. J Virol. 1998;72:8021–8031. doi: 10.1128/jvi.72.10.8021-8031.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peiris M, Yuen K Y, Leung C W, Chan K H, Ip P L S, Lai R W M, Orr W K, Shortridge K F. Human infection with influenza H9N2. Lancet. 1999;354:916–917. doi: 10.1016/s0140-6736(99)03311-5. [DOI] [PubMed] [Google Scholar]
- 15.Sakamoto S, Kino Y, Oka T, Herlocher M L, Maassab H. Gene analysis of reassortant influenza virus by RT-PCR followed by restriction enzyme digestion. J Virol Methods. 1996;56:161–171. doi: 10.1016/0166-0934(95)01909-x. [DOI] [PubMed] [Google Scholar]
- 16.Shu L P, Sharp G B, Lin Y P, Claas E C J, Krauss S L, Shortridge K F, Webster R G. Genetic reassortment in pandemic and interpandemic influenza viruses. Eur J Epidemiol. 1996;12:63–70. doi: 10.1007/BF00144430. [DOI] [PubMed] [Google Scholar]
- 17.Subbarao K, Klimov A, Katz J, Regnery H, Lim W, Hall H, Perdue M, Swayne D, Bender C, Huang J, Hemphill M, Rowe T, Shaw M, Xu X, Fukuda K, Cox N. Characterization of an avian influenza (H5N1) virus isolated from a child with a fatal respiratory illness. Science. 1998;279:393–396. doi: 10.1126/science.279.5349.393. [DOI] [PubMed] [Google Scholar]
- 18.Webster R G. Influenza virus: transmission between species and relevance to emergence of the next human pandemic. Arch Virol. 1997;13:105–113. doi: 10.1007/978-3-7091-6534-8_11. [DOI] [PubMed] [Google Scholar]
- 19.Webster R G, Bean W J, Gorman O T, Chambers T M, Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiol Rev. 1992;56:152–179. doi: 10.1128/mr.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wentworth D E, Thompson B L, Xu X, Regnery H L, Cooley A J, McGregor M W, Cox N J, Hinshaw V S. An influenza (H1N1) virus, closely related to swine influenza virus, responsible for a fatal case of human influenza. J Virol. 1994;68:2051–2058. doi: 10.1128/jvi.68.4.2051-2058.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu X, Guo Y, Rota P, Hemphill M, Kendal A, Cox N. Genetic reassortment of human influenza virus in nature. In: Hannoun C, Kendal A P, Klenk H D, Ruben F L, editors. Options for the control of influenza II. Amsterdam, The Netherlands: Excerpta Medica; 1993. pp. 203–207. [Google Scholar]
- 22.Yamnikova S S, Mandler J, Bekh-Ochir Z H, Dachtzeren P, Ludwig S, Lvov D K, Scholtissek C. A reassortant H1N1 influenza A virus caused fatal epizootics among camels in Mongolia. Virology. 1993;197:558–563. doi: 10.1006/viro.1993.1629. [DOI] [PubMed] [Google Scholar]
- 23.Zou S. A practical approach to genetic screening for influenza virus variants. J Clin Microbiol. 1997;35:2623–2627. doi: 10.1128/jcm.35.10.2623-2627.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]