Abstract
Coronavirus disease 2019 (COVID-19) has been recognized as a global pandemic outbreak, opening the most severe socio-economic crisis since World War II. Different scientific activities have been emerged in this global scenario, including the development of innovative analytical tools to measure nucleic acid, antibodies, and antigens in the nasopharyngeal swab, serum, and saliva for prompt identification of COVID-19 patients and to evaluate the immune response to the vaccine. The detection of SARS-CoV-2 in saliva remains a challenge for the lack of sufficient sensitivity. To address this issue, we developed a novel paper-based immunoassay using magnetic beads to support the immunological chain and 96-well wax-printed paper plate as a platform for color visualization by using a smartphone combined with Spotxel free-charge app. To assess the reliability of the measurement of SARS-CoV-2 in saliva, untreated saliva was used as a specimen and the calibration curve demonstrated a dynamic range up to 10 μg/mL, with a detection limit equal to 0.1 μg/mL. The effectiveness of this sustainable analytical tool in saliva was evaluated by comparing the data with the nasopharyngeal swab specimens sampled by the same patients and tested with Real-Time PCR reference method, founding 100% of agreement, even in the case of high Cycle Threshold (CT) numbers (low viral load). Furthermore, the positive saliva samples were characterized by the next-generation sequencing method, demonstrating the capability to detect the Delta variant, which is actually (July 2021) the most relevant variant of concern.
Keywords: Antigen detection, Delta variant, Paper-based immunoassay, Biological specimens
1. Introduction
European Centre for Disease Prevention and Control (https://www.ecdc.europa.eu/en/covid-19/variants-concern) and Centers for Disease Control and Prevention (https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-info.html) have highlighted the emerging circulation of genetic variants of SARS-CoV-2 around the world throughout the COVID-19 pandemic. They established a variant classification scheme that includes the variant of concern, the variant of interest, and the variant under monitoring. For the variant of concern, there is clear evidence of a significant impact on transmissibility, severity, and/or immunity that is likely to have an impact on the epidemiological situation, while the variant of interest could imply a significant impact on transmissibility, severity, and/or immunity, realistically having an impact on the epidemiological situation. Variants under monitoring are the additional variants of SARS-CoV-2 detected as signals through epidemic intelligence, rules-based genomic variant screening, or preliminary scientific evidence, and they could have properties similar to those of a variant of concern although the evidence is weak or has not yet been assessed. WHO proposed labels by using letters of the Greek alphabet for global SARS-CoV-2 variants of concern and variants of interest to be used alongside the scientific nomenclature in communications about variants to the public (https://www.who.int/news/item/31-05-2021-who-announces-simple-easy-to-say-labels-for-sars-cov-2-variants-of-interest-and-concern). In this period (July 2021), the Delta variant is the one that is quickly spreading, triggering a new phase in COVID-19. For instance, the Centers for Disease Control and Prevention declared Delta as a variant of concern on 15 June in the United States, being its prevalence estimated at least at 14%, while in the United Kingdom Delta variant was found in more than 90% of all infections (Kupferschmidt and Wadman, 2021). Despite the vaccination campaign, it seems that COVID-19 will continue to be part of our lives for many years to come. Recently (April 2021) the CEO of Pfizer Albert Bourla reported that people may need to get vaccinated against the coronavirus annually (https://www.cnbc.com/2021/04/15/pfizer-ceo-says-third-covid-vaccine-dose-likely-needed-within-12-months.html). This means that testing for antigens and antibodies will continue to be of vital importance to ensure that we remain fully protected. For reliable evaluation of infectious people, the antibody test is not recommended because antibodies are produced by human beings in 7–15 days after the exposure to the virus, thus it is not suitable for identifying the infected people in a timely fashion as well as for an early diagnosis of COVID-19 (Ma et al., 2020; Mekonnen et al., 2021). For RNA detection several analytical devices have been developed (Wang et al., 2020; Zhu et al., 2020), including the use of CRISPR-Technology (Ning et al., 2021), which is a vanguard technology as demonstrated by the Nobel Prize in Chemistry 2020 to Emmanuelle Charpentier and Jennifer A. Doudna, having discovered the CRISPR/Cas9 genetic scissor (https://www.nobelprize.org/prizes/chemistry/2020/press-release/). In the case of antigen detection (Fabiani et al., 2021a), the immunosensors developed are based on the detection of i) nucleocapsid (N) protein, which is the protein responsible for genome packaging (Masters, 2019; Laude and Masters, 1995); ii) Spike (S) protein, which is the protein present of the surface of the SARS-CoV-2 virus with affinity with the human angiotensin-converting enzyme 2 (hACE2), exploited to infect human cells (Verdecchia et al., 2020; Ward et al., 2020; Xia et al., 2020). In detail, the S protein of SARS-CoV-2 is composed of two subunits, S1 and S2, where the S1 subunit contains a receptor-binding domain that recognizes and binds the host receptor, while the S2 subunit umpires the viral cell membrane fusion (Huang et al., 2020). Among N, S1, and S2, the S1 subunit is less conserved and more highly specific to SARS-CoV-2, thus its detection could improve the selectivity of the analytical system (Ward et al., 2020). For S protein detection, the first immunosensor was reported by Seo et al. which developed a field-effect transistor device for detecting SARS-CoV-2 in nasopharyngeal swab specimens by using antibody for the S protein immobilized on the graphene sheets coating the field-effect transistor (Seo et al., 2020). This sensor was able to detect S protein and SARS-CoV-2 with a detection limit of 100 fg/mL and 1.6 × 101 PFU/mL, respectively for clinical trasport medium and culture medium . Even if the nasopharyngeal swab specimen is one of the most used sample tested in the biosensing field (Shao et al., 2021; Kim et al., 2021, Raziq et al., 2021), this sampling requires an invasive procedure and skilled personnel, hampering the accurate analysis with sampling carried out by the end-users. Another useful specimen is saliva because saliva has the advantage of non-invasive sampling and delivers accurate data, avoiding the variability of nasopharyngeal specimens due to the complex sampling (Wyllie et al., 2020). To this regard, we developed the first immunosensing system for the S and N proteins in saliva by exploiting magnetic beads (MBs) as support of the immunological chain as well as for a sensitive sampling of target analyte in saliva, and nano-modified screen-printed electrodes combined with a portable instrument for the measurement, obtaining a detection limit equal to 19 ng/mL and 8 ng/mL in untreated saliva, respectively for S and N protein (Fabiani et al., 2021b). The suitability and advantages of saliva for SARS-CoV-2 detection by smart biosensors were successively reported in the literature by other relevant groups (Hall's, Kelley's, and Gao's groups) (Singh et al., 2021; Yousefi et al., 2021; Torrente-Rodríguez et al., 2020).
In the era of sustainability, the use of paper-based devices and smartphone-assisted biosensors has established one of the main pillars in the diagnostics field. The use of paper in their design allows for: i) the fabrication of the plastic-free devices; ii) the loading of the reagents onto the cellulose network delivering a reagent-free measure; iii) the customization of microfluidics with an equipment-free system exploiting the capillarity of the paper; iv) a treatment-free measurement thanks to the filter paper which can treat the sample during the analysis, v) the possibility to be incinerated, rendering more sustainable the management of waste in the case of infected samples (Cate et al., 2015; Hamedi et al., 2016; Caratelli et al., 2020, Grant et al., 2020; Antiochia, 2021, Zhu et al., 2021, Zhang et al., 2020, Noviana et al., 2021). At the same level, smartphone-assisted devices can boost the acquisition of the signal by smartphone, avoiding the use of a dedicated instrument. Furthermore, an easy management is carried out by using a dedicated app or Internet of Things (Roda et al., 2016). For the antigen detection, to our knowledge, the first paper-based device is the one developed in 2021 by Yakoh et al. (2021). In detail, they developed a paper-based electrochemical platform as a screening tool to detect SARS-CoV-2 immunoglobulins (represented by IgG and IgM) as well as S protein using a portable potentiostat (Emstat3 Blue wireless potentiostat). In the case of S protein, the antibody was immobilized onto the paper-based working electrode surface and the presence of S protein was evaluated by measuring after 45 min the decrease of the response in the presence of 5 mM [Fe(CN)6]3-/4- as the electrochemical probe. This device was tested only in standard solution, obtaining a dynamic range comprised between 1 and 1000 ng/mL with a detection limit of 0.11 ng/mL.
Herein, we report a novel immunoassay based on:
-
i)
the use of MBs as support for the immunological chain and for an easy way for pre-concentration;
-
ii)
a 96-well wax-printed paper-based plate as a platform to load MBs and to contain the reagent for colorimetric detection without the addition of enzymatic substrate;
-
iii)
the smartphone combined with Spotxel free-charge app for an equipment-free and accurate evaluation of colored signal.
The suitability of the paper-based platform was evaluated by testing S protein in standard solutions and in saliva samples of COVID-19 patients affected also by Delta variant, demonstrating the ability to identify the infectious patients at low viral load, e.g. Cycle Threshold (CT) values higher than 30 in RT-PCR measures, overcoming the drawbacks of the commercial available immunochromatographic antigen kit able to identify only patients with high viral load.
2. Materials and methods
2.1. Reagents and equipment
The target SARS-CoV Spike (subunit S1) protein (0.5 mg/mL), monoclonal antibody anti-SARS-CoV-2 (produced in mouse, 1 mg/mL), SARS-CoV-2 Spike antibody-HRP Chimeric MAb (MAb-HRP, 100 μg), were purchased from Sinobiological (Germany). The target SARS Coronavirus 2019 Spike Recombinant protein 100 μg from Sinobiological (Germany), the bovine serum albumin (BSA) was used as blocking agent for covered magnetic beads to reduce the aspecific adsorptions, Tween 20, sodium azide, 3,3',5,5’-tetramethylbenzidine (TMB) liquid substrate, Supersensitive, for ELISA, ready to use solution, and all other reagents were obtained from Sigma (USA). Dynabeads® Pan Mouse IgG pre-coated with anti-Mouse IgG able to bind monoclonal antibody (supplied as a suspension containing 4 × 108 beads/mL in phosphate buffered saline, pH 7.4) was from Life Technologies (USA). A rotary shaker and a magnetic rack/particle concentrator were from Dynal Biotech (USA). Phosphate saline buffer (PBS) = 0.0015 M KH2PO4, 0.0081 M Na2HPO4, 0.137 M NaCl, 0.0027 M KCl, pH 7.4; Buffer = PBS+0.05% Tween 20; Storage buffer = PBS + 0.02% NaN3 were used as buffer solution. For specificity test, the SARS-CoV-2 Nucleocapside protein was from Acrobiosystem (USA, 0.6 mg/mL), Legionella pneumophila NCTC 12821 Lenticule discs from Sigma (USA), Botulinum neurotoxin C was kindly supplied by Prof. Ornella Rossetto and Prof. Marco Pirazzini, Department of Biomedical Sciences, University of Padova. Micrographs of the 96-well paper platform before and after the addition of MBs were acquired through electron microscopy FEI Quanta 400. The monitoring of the colorimetric response was carried out using a Huawei smartphone assisted with Spotxel free-charge app. This app is able to process colorimetric data. It is important to acquire the image of the plate by photographing it, respecting the guide wells drawn in the app. Once the image has been captured, the app starts processing the data and provides numeric values.
2.2. Production of the 96-well wax-printed paper plate
The 96-well paper-based platform was designed using a drawing software (Adobe Illustrator). We designed the 96-well paper-based platform with a 7 mm-diameter well, being the same diameter as the conventional ELISA plate. Successively, the designed template was printed onto filter paper (67 g/m2, Cordenons, Italy) with a solid-ink printer (ColorQube 8580, Xerox, USA). The 96-well paper-based platform was then cured in an oven for 2 min at 100 °C. For delivering a reagent-free 96-well paper-based platform, each well was loaded with 10 μL of TMB solution to ask the end-user only the addition of MBs. The pre-loaded 96-well wax-printed paper-based platform stored under vacuum at room temperature was stable up to 10 days, after 15 days a decrease of response was observed.
2.3. Immunoassay
The MBs-based assay involves sequential procedures:
-
i)
a preliminary blocking-coating procedure of the Dynabeads® Pan Mouse IgG (to store them at 4 °C until use for several months), where 250 μL of MBs was pipetted into 2 mL tube and washed twice in 1 mL of PBS pH 7.4. Then, MBs were blocked by incubating them in 1 mL of PBS pH 7.4 + 3% (w/v) BSA for 30 min at room temperature (RT) with slow tilt rotation (using Dynal sample mixer). After, the supernatant was discarded and 500 μL of PBS containing 10 μg MAb were added to the MBs suspension and incubated for 30 min at RT with slow tilt rotation. Finally, the supernatant was discarded and the MBs were resuspended in 250 μL of PBS +0.02% NaN3 and stored up to several months at 4 °C;
-
ii)
Immunoassay procedure:
-
1.
Shaking and transfer 10 μL of coated and blocked MBs suspension (stored at 4 °C) into 2 mL tube (in the number required by the samples to analyze);
-
2.
Addition of 200 μL of chimeric MAb-HRP anti-SARS-CoV-2 2 μg/mL in PBS +0.05% Tween 20 and 300 μL of saliva sample;
-
4.
Incubation for 30 min at RT;
-
5.
Washing by adding 1 mL of PBS +0.05% Tween 20 into the tube containing the MBs, followed by shaking and inserting the magnet to concentrate the MBs. After, the supernatant was discarded and the procedure was repeated twice to eliminate the excess of unreacted labeled antibodies;
-
6.
Resuspension of the MBs in 100 μL of PBS;
-
7.
For the measurement, 10 μL of this suspension (three replicates for each sample) were cast onto the wells of the printed plate, preloaded with 10 μL TMB solution, and the colorimetric response was revealed using Huawei smartphone-assisted with Spotxel free-charge app.
In this immunoassay procedure, the classical sequential incubations for the immuno-recognition events are merged in a single incubation of 30 min, for delivering an easy-to-use device.
2.4. SARS-CoV-2 virus propagation
SARS-CoV-2 was passaged once in Vero cells to generate a virus master stock used to produce a virus working stock. The virus was propagated in Vero cells cultured in minimum essential medium (MEM) containing 2% (w/v) fetal bovine serum (Euroclone S. p.A.). After infection, virus stock was collected by centrifuging the culture supernatants of infected Vero cells at 600 g for 5 min. The clarified supernatant was supplemented to 20% with fetal bovine serum (w/v), frozen, and kept at −80 °C until use. The concentration of infectious virus was determined by plaque-forming titer assay. Virus propagation, virus isolation or neutralization assays of SARS-CoV-2 needs to be conducted in a bio-safety Level-3 facility according to WHO laboratory biosafety guidance (https://www.who.int/publications/i/item/laboratory-biosafety-guidance-related-to-coronavirus-disease-(covid-19)).
2.5. SARS-CoV-2 quantification by Real-Time PCR
Nasopharyngeal swabs were collected and tested from the Italian Scientific Department of Army Medical Centre for SARS-CoV-2. Viral RNA was extracted from 300 μL of swab medium using a Maxwell RSC Viral Total Nucleic Acid Purification Kit on Maxwell RSC Instrument (Promega, USA). Total nucleic acid was eluted in a final volume of 50 μL of nuclease-free water. Then, one step Real-Time Reverse Transcription-PCR was performed using TaqPath TM COVID-19 CE-IVD RT-PCR Kit (ThermoFisher Scientific, USA) on QuantStudio 5 Real-Time PCR Instrument (ThermoFisher Scientific, USA). ORF1ab, N, and S genes were selected as target regions plus an internal control MS2 to verify the efficacy of the sample preparation and the absence of inhibitors in the PCR reaction. RT-PCR reactions were performed aliquoting 15 μL of reaction mix (5.0 μL TaqPath TM 1-Step Multiplex Master Mix, 1.0 μL COVID-19 Real-Time PCR Assay Multiplex, 4.0 μL Nuclease-free Water) plus 10 μL of RNA sample using the following thermal cycling protocol: incubation 25 °C for 2 min, reverse transcription 53 °C for 10 min, activation 95 °C for 2 min, and 40 cycles of PCR denaturation 95 °C for 3 s and annealing/extension 60 °C for 30 s. Reporter dye detectors were FAM, VIC, ABY, and JUN for ORF1ab, N gene, S gene, and MS2, respectively.
2.6. Next generation sequencing
The viral RNA was extracted from nasopharyngeal swabs using the RNeasy Mini Kit (Qiagen, Hilden, Germany). The obtained RNAs were retro-transcribed using the SuperScript III Reverse Transcriptase kit (Invitrogen, US) and double-stranded DNAs were subsequently obtained by Klenow enzyme according to the instructions of the manufacturer (Roche, Switzerland). The Nextera XT kit was used for library preparations and whole genome sequencing was performed using the Illumina NextSeq 500 High Output Kit V 2.5 (2 × 150) on the NextSeq 500 sequencer (Illumina, San Diego, US). The reads were trimmed for quality (q score≥25) and minimum length (= 100) using BBDuk trimmer. High-quality reads were assembled by mapping to the reference genome from Wuhan, China (GenBank an. NC_045512.2) with a bowtie 2 mapping algorithm. All software was integrated into Geneious Prime (www.geneious.com). Then, all viral sequences were deposited in GISAID, a dedicated database (Global Initiative on Sharing All Influenza Data, https://www.gisaid.org).
2.7. 2009 H1N1 influenza pandemic virus propagation for selectivity test
2009 H1N1 influenza pandemic (2009 pH1N1) virus master stock was passaged in Madin-Darby canine kidney (MDCK) to generate virus working stocks. The viruses were propagated in MDCK cultured in minimum essential medium (MEM) containing 2% (w/v) fetal bovine serum (Euroclone S. p.A.), 2 μg/ml trypsin-TPCK (Merck, Germany). After infection, virus stock was collected by centrifuging the culture supernatants of infected MDCK cells at 600 g for 5 min. The clarified supernatants were frozen and kept at −80 °C until use. The concentration of infectious virus was equal to 7 × 104 PFU/mL.
3. Results and discussions
3.1. Description and characterization of immunoassay based on the use of MBs, 96-well wax-printed paper plate, and smartphone
The use of paper for the development of cost-effective and easy-to-use diagnostic devices for low resource conditions has been boosted in the last years for the diagnosis of diseases. COVID-19 outbreak has further enlarged their application due to the unravel features of paper-based devices including multiplexing capabilities, high sensitivity, high selectivity, easiness to use, cost-effectiveness, mass manufacturing, rapidity, and single-use, all characteristics that match the ones highlighted in the case of an ideal device for effective use in pandemics (Bhalla et al., 2020).
Herein, we have chosen to configure our sustainable platform by exploiting:
-
i)
the use of a 96-well wax-printed paper plate to perform 96 simultaneous measurements to deliver a multiplexing analysis;
-
ii)
MBs as support of the immunological chain, being able to load a high number of antibodies thanks to the high surface/volume ratio as well as the capability of MBs to preconcentrate the virus with the final result of improved sensitivity;
-
iii)
the use of monoclonal antibodies for their ability to recognize only one epitope as well as the selection of S protein as target analyte being the specific protein of only SARS-CoV-2, allowing for an analytical tool with high selectivity;
-
iv)
the exploitation of porosity of paper in 96-well wax-printed paper to load the enzymatic substrate. In addition, each well is able to work as a sensing element and as a reagent reservoir for rendering the analysis easy to perform. Moreover, the selection of saliva as a specimen does not require a health worker for the sampling;
-
v)
the use of smartphone-assisted measurement using a free-charge app, boosting the cost-effective management of the data by wireless transmission and overcoming the limitation of eye naked visualization, in which the sensitivity of the operator can affect the analysis;
-
vi)
the fabrication of a 96-well wax-printed paper-based plate by a commercially available wax printer, delivering a facile mass-manufacturing of the sensing device with the advantage to easily customize the pattern by a design software;
-
vii)
the analysis time of 30 min allowing for rapid analysis, when compared with the reference method (RT-PCR), which requires several hours.
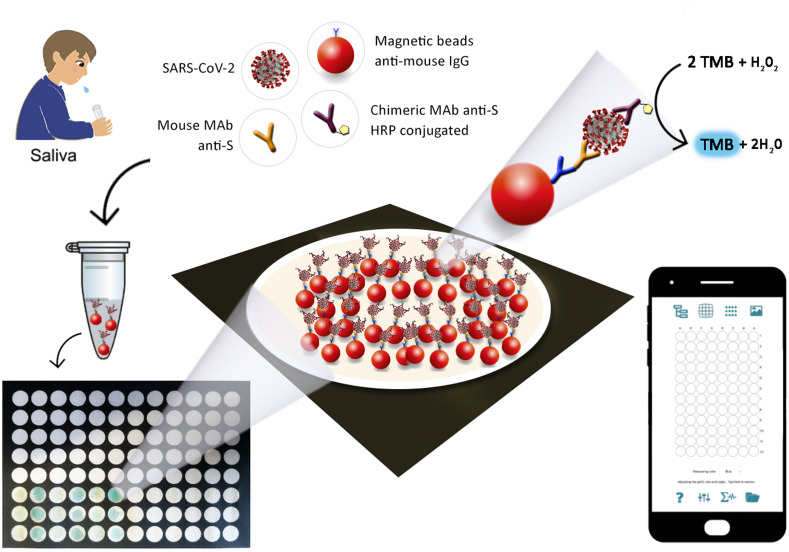
For the sampling, the end-user needs only to simply add the sputum in the tube in which all reagents are present. In 30 min, the immunological chain is constructed, and after washing steps, MBs were added to the wells which already contain the substrate, i.e. TMB supersensitive ready-to-use solution. The addition of the sample solubilizes the reagent entrapped in the paper, avoiding the further addition of the enzymatic substrate. After 5 min, the presence of SARS-CoV-2 in the saliva is attested by the blue color of the well, and the increase in the intensity of the blue color analyzed by the Spotxel free-charge app is correlated to the higher amount of the SARS-CoV-2 in the saliva tested, as schematized in Fig. 1 .
Fig. 1.
Scheme of the smartphone-assisted paper-based device.
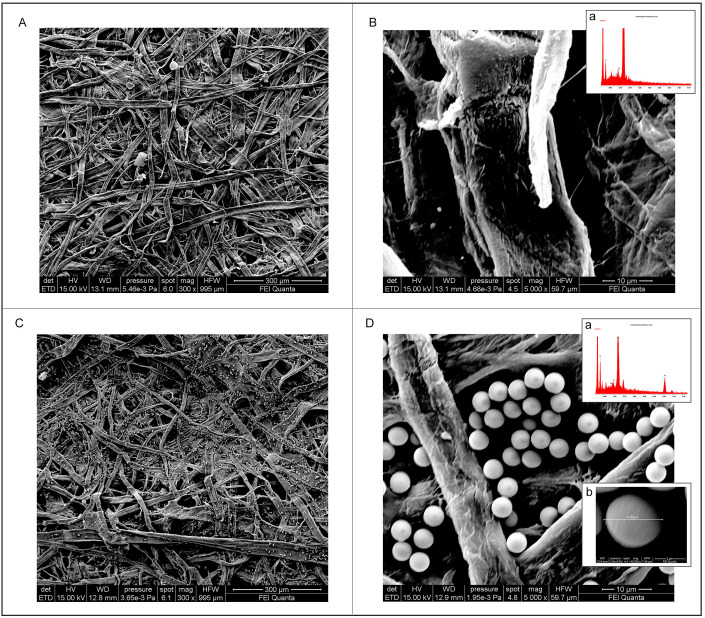
Because, to our knowledge, this is the first time that MBs are combined with a 96-well wax-printed paper, we have investigated the morphology of 96-well wax-printed paper-based platform before and after adding MBs onto the paper-based wells by scanning electron microscopy (SEM). The paper-based wells showed a porous network of cellulose fibers (Fig. 2 A/B), while when the MBs were added, it is well evident the presence of MBs decorating the cellulose fibers (Fig. 2C/D). In detail, MBs are present both on the fiber as well on the space between the fibers, allowing for a visible homogeneous blue color due to the formation of the enzymatic by-product, as reported in Fig. 1. In addition, we estimated the dimension of MBs (inset reported in Fig. 2D) equal to ca. 4 μm, which is a value in agreement with the one provided by the manufacturer (4.5 ± 0.5 μm). The presence of MBs was also confirmed by EDX, indeed the microanalysis confirmed the presence of Fe on the paper surface when MBs were added (Fig. 2D) when compared with the EDX analysis using paper without the addition of MBs (Fig. 2B).
Fig. 2.
SEM images of (A, B) bare paper-based well and relative EDS analysis (B, inset), (C, D) MBs-loaded paper-based well and relative EDS analysis (D, inset).
3.2. The evaluation of Tween 20 effect
One of the strategies to reduce the background in the immunoassay is the use of non-ionic detergents such as Tween 20 and Triton X-100 for washing steps, to avoid the aspecific adsorption (Steinitz, 2000). In the case of the immunoassay using MBs, the function of Tween 20 is not only the improvement of the washing step due to the different surface tension of the water-based solution with Tween 20, but also to interact with the surface of MBs for hindering the adsorption of the untargeted analyte. In our previous work (Fabiani et al., 2021b), we developed MBs-based immunoassay for SARS-CoV-2 detection with electrochemical transduction and alkaline phosphatase as immunological chain label, using Tween 20 at the concentration of 0.05% (w/v) as an additional reagent of buffer during the washing step. For improving sensitivity in the case of colorimetric detection, we have selected HRP as label enzyme, because the detection of blue color on paper gives a more sensitive measure in respect to yellow color (the color produced by the use of naphthyl phosphate as enzymatic substrate combined with alkaline phosphate as label enzyme). In the case of 96-well wax-printed paper-based plate, we investigated the suitability of HRP as a label, observing a high background signal (data not shown) due to the aspecific adsorption between MBs and HRP enzyme. To overcome this issue, we use Tween 20 0.05% (w/v) not only for the washing step (as in our previous work) but also as additive in the PBS buffer during the incubation step to avoid the aspecific adsorption. Because it is reported in the literature that non-ionic detergents have effects on the antigen (Hoffman and Jump, 1986), hampering the formation of immune complexes, we tested the concentration used in our test (0.05% (w/v)) to evaluate the possible inactivation of the SARS-CoV-2 by Tween 20. To study the effect of Tween 20 on the infectivity of SARS-CoV-2, we incubated SARS-CoV-2 (1.2 × 103 PFU/ml) for 30 min at RT with and without a solution consisting in 0.0015 M KH2PO4, 0.0081 M Na2HPO4, 0.137 M NaCl, 0,0027 M KCl, pH 7.4 + 0.05% Tween 20 (the solution used in the incubation step of the immunoassay).
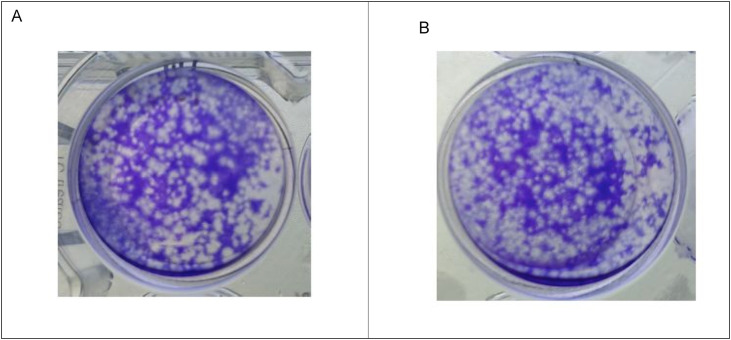
As reported in Fig. 3 no variation in the plaque forming has been observed when SARS-CoV-2 has been incubated with Tween 20, confirming any inactivation under our working condition, in agreement also with the literature (Patterson et al., 2020). Thus, we used Tween 20 as a reagent during the incubation step without any effect on the immunocomplex formation and on the immunoassay sensitivity.
Fig. 3.
Effect of Tween 20 on SARS-CoV-2 infectivity. Plaque formation in Vero E6 cells: (a) Virus incubated with Tween 20; b) Positive control (1.2 × 103 PFU/ml).
3.3. Suitability of the reagent-free approach using the 96-well wax-printed paper plate
With the final aim to deliver an easy-to-use device, we exploited the porosity of the paper to load the enzymatic substrate (TMB) in the well. In detail, we evaluated the response of the conceived paper-based immunoassay both i) when TMB was directly added in the well together with MAb-HRP or ii) when TMB was previously loaded onto the well and waited to dry for a reagent-free assay.
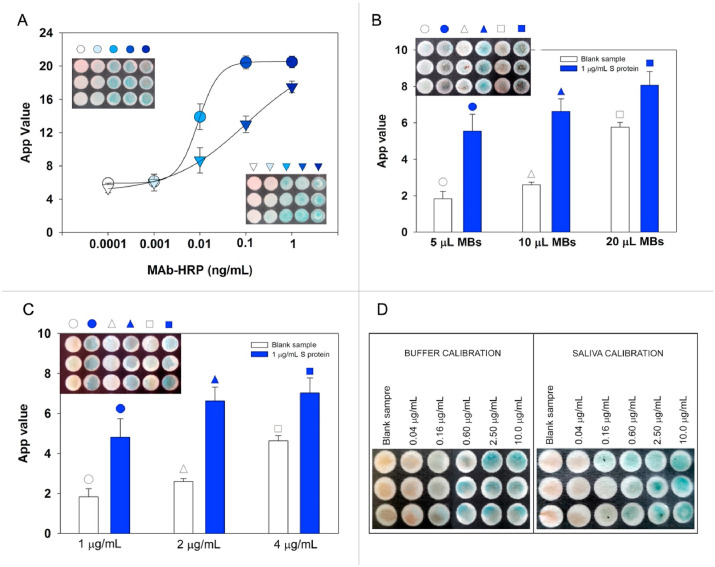
As depicted in Fig. 4 A, we observed that in the case of the simultaneous addition of both HRP-labeled antibody and TMB, a typical sigmoidal behavior was observed with a dynamic range of MAb-HRP from 0.0001 to 1 ng/mL. Otherwise, in the case of reagent-free approach a decrease of sensitivity was observed, due to the diffusion which is different when the reaction happens in drop or in the cellulose network. However, even if less sensitive, the reagent-free system allows for a useful detection of SARS-CoV-2, thanks to the high sensitivity of the system based on MBs used in the immunoassay.
Fig. 4.
A) Study of the suitability of the reagent-free approach: sigmoidal curve obtained in drop (•) by adding 10 μL of TMB + 10 μL MAb-HRP, sigmoidal curve obtained after pre-loaded 10 μL of TMB and by adding 10 μL of MAb-HRP (Δ); B) Selection of MBs volume. 5, 10, and 20 μL of MBs, 200 μL of MAb-HRP anti-SARS-CoV-2, 2 μg/mL (in PBS + 0.05% Tween 20) + 300 μL of sample tested (negative control or 1 μg/mL S protein; C) Study of MAb-HRP concentration. 10 μL of MBs, 200 μL of MAb-HRP anti SARS-CoV-2, 1, 2, and 4 μg/mL (in PBS + 0.05% Tween 20) + 300 μL of sample tested (negative control or 1 μg/mL S protein; D) Calibration images obtained using the chosen parameters for S protein detection in buffer and in untreated saliva. The mean value (n = 3) with the corresponding standard deviation and the image of paper plate were reported for each measurement.
3.4. Evaluation of the experimental parameters
The paper-based ELISA using MBs was developed by using Dynabeads Pan Mouse IgG precoated with Anti-mouse IgG able to favor the correct orientation of the MAb Anti-S, improving the sensitivity of the system. The amount of MBs is a crucial parameter for highly sensitive detection, providing the best highest signal-to-noise ratio. We selected the amount of MBs employed by testing 5, 10, and 20 μL at a concentration of 4 × 108 beads/mL, added to a volume of 500 μL with a final concentration of 4 × 106, 8 × 106, 1.6 × 107 beads/mL, respectively. As shed light in Fig. 4B, the highest signal difference of the immunoassay in the absence and in the presence of 1 μg/mL of S protein was observed for 10 μL, being this value also in agreement with the value selected in our previous work (Fabiani et al., 2021b).
In this work, for the immunological chain, we have chosen a commercially available chimeric monoclonal antibody produced by combining the constant domains of the human IgG1 molecule with mouse variable regions. The variable region was obtained from a mouse immunized with purified, recombinant SARS-CoV Spike RBD Protein. The antibody was produced using recombinant antibody technology and conjugated with horseradish-peroxidase (HRP). We chose the concentration of MAb-HRP by testing S protein at a concentration of 1 μg/L by varying the MAb-HRP from 1 μg/L to 4 μg/L, observing the best sensitivity in terms of high ratio signal of the S protein to the blank, in the case of 2 μg/L (Fig. 4C).
3.5. Analytical features of 96-well wax-printed paper plate for S protein detection in standard solution and saliva
To assess the analytical features of the developed assay for measuring S protein in standard solutions, different concentrations of S protein diluted in PBS +0.05% Tween 20 pH = 7.4 ranging from 0.04 μg/mL to 10 μg/mL were tested in standard solution (Fig. 4D, left) obtaining a linear behavior described by the following equation y = (9.22 ± 0.03) + (2.63 ± 0.03) x, R2 = 0.999, with a detection limit equal to 0.01 μg/mL, calculated as blank signal + 3 standard deviation (SD). The matrix effect in the case of saliva specimen was evaluated by constructing the calibration curve in untreated saliva (Fig. 4D, right), observing a low matrix effect. Indeed, the calibration curve in saliva was described by the following equation: y = (5.62 ± 0.13) + (2.79 ± 0.15) x, R2 = 0.991, with a detection limit equal to 0.1 μg/mL. The repeatability was assessed at the concentration of 0.6 μg/mL for saliva (n = 3) observing an RSD% equal to 9.5%.
3.6. Specificity study
To assess the selectivity of the proposed 96-well wax-printed paper platform, a specificity test was performed analyzing:
-
i)
pandemic influenza A virus (pH1N1) (7 × 104 PFU/mL);
-
ii)
N protein (2 μg/mL), another target protein used for SARS-CoV-2 quantification
-
iii)
Legionella pneumophila (104 CFU/mL), bacteria that is transmitted by aerosol as in the case of SARS-CoV-2, being able to survive in this matrix for several months giving pneumonia disease
-
iv)
Botulinum toxin C (10−8 M), a toxin that can also be subjected to airborne transmission.
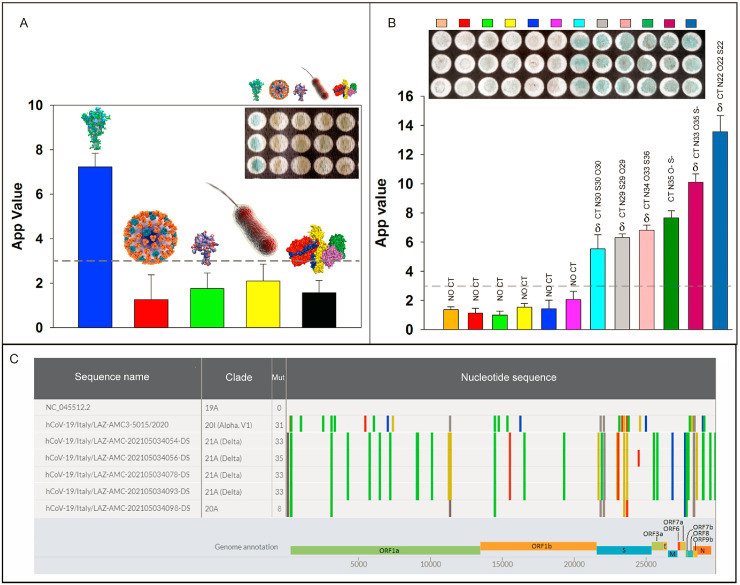
As reported in Fig. 5 A, the signal obtained for seasonal influenza virus A, N protein, Legionella pneumophila, and Botulinum toxin C gave a signal comparable to the blank signal, demonstrating the absence of the interference.
Fig. 5.
A) Specificity test with pandemic influenza A virus (pH1N1) (7 × 104 PFU/mL) (▀), N protein of SARS-CoV-2, 2 μg/mL (▀), Legionella pneumophila 104 CFU/mL (▀), Botulinum toxin C 10−8 M (▀),S protein of SARS-CoV-2 1 μg/mL (▀); B) Six negative (▀▀▀▀▀▀) and six positive (▀▀▀▀▀▀) clinical samples analyzed using 96-well wax-printed paper platform and confirmed by RT-PCR. The mean value (n = 3) with the corresponding standard deviation and the image of paper plate were reported for each measurement; C) Genomic comparison of SARS-CoV2 variants analyzed in this study vs reference strain isolated in Wuhan, China (Genbank acc. n. NC_045512.2).
3.7. Measurement of SARS-CoV-2 in clinical samples
To assess the effectiveness of 96-well wax-printed paper platform using clinical samples, saliva and nasopharyngeal swabs were tested using paper-based device and one-step Real-Time RT-PCR, respectively. In detail, we tested saliva samples using fresh samples because in the case of freezed samples we observed in our previous work a reduction of signal (Fabiani et al., 2021b). In detail, we used fresh saliva sampled after drinking a glass of water, with the aim of an easy sampling without any treatment, to match the requirement of a point of care system. As depicted in Fig. 5B, we observed a well-established difference in the case of positive and negative samples, and we selected the arbitrary value of 3 as the cut-off value, considering the variability of the different saliva samples. The agreement was obtained in 12/12 samples (Table S1), also in the case of low viral load, demonstrating the accuracy of the developed paper-based device. Furthermore, whole genome analysis was carried out on RT-PCR tested positive samples to characterize mutational variations and identify SARS-CoV-2 variants. Genomic comparison of SARS-CoV-2 samples analyzed in this study (Delta variants), an alpha genome (previously predominant variant) vs the reference strain isolated in Wuhan, China (Genbank acc. n. NC_045512.2) were found and are shown in Fig. 5C. All genome nucleotide mutations are highlighted in various colors, and the corresponding annotations are reported on the viral genome, which encodes two open reading frames (ORFs), ORF1a and ORF1b translated into 15 nonstructural proteins (nsps), four conserved structural proteins (spike [S], envelope [E], membrane [M], and nucleocapsid [N]), and at least six accessory proteins (3a, 6, 7a, 7 b, 8 and 9 b), identifying in the samples analyzed the Delta variant.
Line markers represent nucleotide mutations: A (▀), C (▀), G (▀), T (▀), N (▀) and deletion (▀).
4. Conclusions
Herein, we reported a novel paper-based device for an easy, accurate, and fast detection of SARS-CoV-2 detection in saliva specimens using a smartphone as a reader (Table S2). The sensing scheme relies on the construction of the immunological chain onto magnetic beads in an untreated saliva sample in 30 min; after the washing steps, the magnetic beads are loaded onto the paper-based wells containing the enzymatic substrate, giving a colorimetric signal in the presence of SARS-CoV-2. The use of a smartphone, combined with Spotxel free-charge app, allowed for colorimetric detection, overcoming the limitation of eye-naked visualization, in which the sensitivity of the operator can affect the analysis. Furthermore, the use of paper allows for quantifying the target analyte by the only addition of the magnetic beads suspension onto the wells, because the enzymatic substrate has been already loaded on the cellulose network. In addition, the use of 96-wells wax-printed paper is able for the multiplexing analyses, boosting the rapid response and easy management of multiple samples. The specimens of patients analyzed namely saliva using the immunoassay and nasopharyngeal swab using RT-PCR, demonstrated the agreement of the results obtained with the advantages in terms of time analysis (30 min vs 3 h) and cost (ca. € 3 vs € 20). The possibility to test the samples with the next-generation sequencing method demonstrated the capability to detect also the Delta variant. Therefore, this novel paper-based assay provides a new family of paper-based devices, characterized by an equipment-free approach, boosting the application of this type of paper-based smart analytical tools for multiplexing analyses also in limited resources countries.
CRediT authorship contribution statement
Laura Fabiani, Riccardo De Santis, Silvia Fillo: Experimental investigation, methodology, data analysis, Writing - review & editing, Vincenzo Mazzaracchio, Anella Monte, Donatella Amatore: Experimental investigation, Danila Moscone,Review & editing, Florigio Lista and Fabiana Arduini: Conceptualization, data analysis, Writing - review & editing
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
Prof. Letizia Di Bella, Dr. Marco Albano and Dr. Tania Ruspandini (Laboratorio di Microscopia Elettronica e Microanalisi del Dipartimento di Scienze delle Terra, Università di Roma la Sapienza) for SEM analysis, Prof. Ornella Rossetto and Prof. Marco Pirazzini, Department of Biomedical Sciences, University of Padova for supplying Botulinum neurotoxin C used for selectivity study.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bios.2021.113909.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Antiochia R. Biosensors. 2021;11:110. doi: 10.3390/bios11040110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalla N., Pan Y., Yang Z., Payam A.F. ACS Nano. 2020;14:7783–7807. doi: 10.1021/acsnano.0c04421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caratelli V., Ciampaglia A., Guiducci J., Sancesario G., Moscone D., Arduini F. Biosens. Bioelectron. 2020;165:112411. doi: 10.1016/j.bios.2020.112411. [DOI] [PubMed] [Google Scholar]
- Cate D.M., Adkins J.A., Mettakoonpitak J., Henry C.S. Anal. Chem. 2015;87:19–41. doi: 10.1021/ac503968p. [DOI] [PubMed] [Google Scholar]
- Fabiani L., Caratelli V., Fiore L., Scognamiglio V., Antonacci A., Fillo S., De Santis R., Monte A., Bortone M., Moscone D., Lista F., Arduini F. Biosensors. 2021;11:310. doi: 10.3390/bios11090310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabiani L., Saroglia M., Galatà G., De Santis R., Fillo S., Luca V., Faggioni G., D'Amore N., Regalbuto E., Salvatori P., Terova G., Moscone D., Lista F., Arduini F. Biosens. Bioelectron. 2021;171:112686. doi: 10.1016/j.bios.2020.112686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant B.D., Anderson C.E., Williford J.R., Alonzo L.F., Glukhova V.A., Boyle D.S., Weigl B.H., Nichols K.P. Anal. Chem. 2020;92:11305–11309. doi: 10.1021/acs.analchem.0c01975. [DOI] [PubMed] [Google Scholar]
- Hamedi M.M., Ainla A., Güder F., Christodouleas D.C., Fernández-Abedul M.T., Whitesides G.M. Adv. Mater. 2016;28:5054–5063. doi: 10.1002/adma.201505823. [DOI] [PubMed] [Google Scholar]
- Hoffman W.L., Jump A.A. J. Immunol. Methods. 1986;94:191–196. doi: 10.1016/0022-1759(86)90232-2. [DOI] [PubMed] [Google Scholar]
- Huang Y., Yang C., Xu X., Xu W., Liu S. Acta Pharmacol. Sin. 2020;41:1141–1149. doi: 10.1038/s41401-020-0485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.-Y., Lee J.-H., Kim M.J., Park S.C., Choi M., Lee W., Ku K.B., Kim B.T., Changkyun Park E., Kim H.G., Kim S.I. Biosens. Bioelectron. 2021;175:112868. doi: 10.1016/j.bios.2020.112868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupferschmidt K., Wadman M. Science. 2021;372:1375–1376. [Google Scholar]
- Laude H., Masters P.S. In: The Coronaviridae. The Viruses. Siddell S.G., editor. Springer US.; Boston: 1995. The coronavirus nucleocapsid protein; pp. 141–163. [Google Scholar]
- Ma H., Zeng W., He H., Zhao D., Jiang D., Zhou P., Cheng L., Li Y., Ma X., Jin T. Cell. Mol. Immunol. 2020;17:773–775. doi: 10.1038/s41423-020-0474-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters P.S. Virology. 2019;537:198–207. doi: 10.1016/j.virol.2019.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mekonnen D., Mengist H.M., Derbie A., Nibret E., Munshea A., He H., Li B., Jin T. Rev. Med. Virol. 2021;31 doi: 10.1002/rmv.2181. [DOI] [PubMed] [Google Scholar]
- Ning B., Yu T., Zhang S., Huang Z., Tian D., Lin Z., Niu A., Golden N., Hensley K., Threeton B., Lyon C.J., Yin X.-M., Roy C.J., Saba N.S., Rappaport J., Wei Q., Hu T.Y. Sci. Adv. 2021;7(2):eabe3703. doi: 10.1126/sciadv.abe3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noviana E., Ozer T., Carrell C.S., Link J.S., McMahon C., Jang I., Henry C.S. Chem. Rev. 2021;121(19):11835–11885. doi: 10.1021/acs.chemrev.0c01335. [DOI] [PubMed] [Google Scholar]
- Patterson E.I., Prince T., Anderson E.R., Casas-Sanchez A., Smith S.L., Cansado-Utrilla C., Solomon T., Griffiths M.J., Acosta-Serrano Á., Turtle L., Hughes G.L. J. Infect. Dis. 2020;222:1462–1467. doi: 10.1093/infdis/jiaa507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raziq A., Kidakova A., Boroznjak R., Reut J., Öpik A., Syritski V. Biosens. Bioelectron. 2021;178:113029. doi: 10.1016/j.bios.2021.113029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roda A., Michelini E., Zangheri M., Di Fusco M., Calabria D., Simoni P. Trac. Trends Anal. Chem. 2016;79:317–325. [Google Scholar]
- Seo G., Lee G., Kim M.J., Baek S.H., Choi M., Ku K.B., Lee C.S., Jun S., Park D., Kim H.G., Kim S.J., Lee J.O., Kim B.T., Park E.C.S. KimACS Nano. 2020;14:5135–5142. doi: 10.1021/acsnano.0c02823. [DOI] [PubMed] [Google Scholar]
- Shao W., Shurin M.R., Wheeler S.E., He X., Star A. ACS Appl. Mater. Interfaces. 2021;13:10321–10327. doi: 10.1021/acsami.0c22589. [DOI] [PubMed] [Google Scholar]
- Singh N.K., Ray P., Carlin A.F., Magallanes C., Morgan S.C., Laurent L.C., Aronoff-Spencer E.S., Hall D.A. Biosens. Bioelectron. 2021;180:113111. doi: 10.1016/j.bios.2021.113111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinitz M. Anal. Biochem. 2000;282:232–238. doi: 10.1006/abio.2000.4602. [DOI] [PubMed] [Google Scholar]
- Torrente-Rodríguez R.M., Lukas H., Tu J., Min J., Yang Y., Xu C., Rossiter H.B., Gao W. Matter. 2020;3:1981–1998. doi: 10.1016/j.matt.2020.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdecchia P., Cavallini C., Spanevello A., Angeli F. Eur. J. Intern. Med. 2020;76:14–20. doi: 10.1016/j.ejim.2020.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., He S., Wang X., Yan Y., Liu J., Wu S., Liu S., Lei Y., Chen M., Li L., Zhang J., Zhang L., Hu X., Zheng X., Bai J., Zhang Yulong, Zhang Yitong, Song M., Tang Y. Nat. Biomed. Eng. 2020;4:1150–1158. doi: 10.1038/s41551-020-00655-z. [DOI] [PubMed] [Google Scholar]
- Ward S., Lindsley A., Courter J., Assa’ad A. J. Allergy Clin. Immunol. 2020;146:23–34. doi: 10.1016/j.jaci.2020.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyllie A.L., Fournier J., Casanovas-Massana A., Campbell M., Tokuyama M., Vijayakumar P., Warren J.L., Geng B., Muenker M.C., Moore A.J., Vogels C.B.F., Petrone M.E., Ott I.M., Lu P., Venkataraman A., Lu-Culligan A., Klein J., Earnest R., Simonov M., Datta R., Handoko R., Naushad N., Sewanan L.R., Valdez J., White E.B., Lapidus S., Kalinich C.C., Jiang X., Kim D.J., Kudo E., Linehan M., Mao T., Moriyama M., Oh J.E., Park A., Silva J., Song E., Takahashi T., Taura M., Weizman O.-E., Wong P., Yang Y., Bermejo S., Odio C.D., Omer S.B., Dela Cruz C.S., Farhadian S., Martinello R.A., Iwasaki A., Grubaugh N.D., Ko A.I. N. Engl. J. Med. 2020;383:1283–1286. doi: 10.1056/NEJMc2016359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S., Liu M., Wang C., Xu W., Lan Q., Feng S., Qi F., Bao L., Du L., Liu S., Qin C., Sun F., Shi Z., Zhu Y., Jiang S., Lu L. Cell Res. 2020;30:343–355. doi: 10.1038/s41422-020-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakoh A., Pimpitak U., Rengpipat S., Hirankarn N., Chailapakul O., Chaiyo S. Biosens. Bioelectron. 2021;176:112912. doi: 10.1016/j.bios.2020.112912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousefi H., Mahmud A., Chang D., Das J., Gomis S., Chen J.B., Wang H., Been T., Yip L., Coomes E., Li Z., Mubareka S., McGeer A., Christie N., Gray-Owen S., Cochrane A., Rini J.M., Sargent E.H., Kelley S.O. J. Am. Chem. Soc. 2021;143:1722–1727. doi: 10.1021/jacs.0c10810. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Xu J., Zhou S., Zhu L., Lv X., Zhang J., Zhang L., Zhu P., Yu J. Anal. Chem. 2020;92:3874–3881. doi: 10.1021/acs.analchem.9b05343. [DOI] [PubMed] [Google Scholar]
- Zhu X., Wang X., Han L., Chen T., Wang L., Li H., Li S., He L., Fu X., Chen S., Xing M., Chen H., Wang Y. Biosens. Bioelectron. 2020;166:112437. doi: 10.1016/j.bios.2020.112437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L., Lv X., Li Z., Shi H., Zhang Y., Zhang L., Yu J. Biosens. Bioelectron. 2021;192:113524. doi: 10.1016/j.bios.2021.113524. [DOI] [PubMed] [Google Scholar]
- https://www.ecdc.europa.eu/en/covid-19/variants-concern
- https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-info.html
- https://www.nobelprize.org/prizes/chemistry/2020/press-release/
- https://www.cnbc.com/2021/04/15/pfizer-ceo-says-third-covid-vaccine-dose-likely-needed-within-12-months.html
- https://www.who.int/news/item/31-05-2021-who-announces-simple-easy-to-say-labels-for-sars-cov-2-variants-of-interest-and-concern
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.