Abstract
Epidemiological studies have yet to identify a single cause for the most common late-onset form of Alzheimer's disease. The common respiratory pathogen Chlamydia pneumoniae recently has been implicated as a risk factor for this form of Alzheimer's disease. Were this true, there would be a dramatic shift in current paradigms of Alzheimer's disease research and treatment. In the absence of published confirmation, we obtained postmortem brain tissue from late-onset Alzheimer's disease patients (n = 15) and representative controls (n = 5) and extracted DNA from up to six separate brain regions in each instance, including those areas particularly relevant to Alzheimer's disease neuropathology. Each sample of DNA (n = 101) was assayed five times or more for the presence of C. pneumoniae DNA using a nested-PCR protocol targeting a species-specific gene sequence coding for the major outer membrane protein of this organism. We were unable unequivocally to detect C. pneumoniae in any of the 101 samples tested by PCR and failed to culture the organism from tissue samples. We conclude that C. pneumoniae is neither strongly nor uniquely associated with the neuropathology seen in late-onset Alzheimer's disease.
Alzheimer's disease (AD) is a neurodegenerative disorder characterized behaviorally by the progressive loss of memory and disorder of other cognitive functions. Most cases are of the nonfamilial, late-onset type for which a specific cause(s) has yet to be established, although a number of risk factors have been identified (8). A recent report has raised the intriguing possibility that Chlamydia pneumoniae, a common human respiratory pathogen, may be a risk factor in late-onset AD (2). C. pneumoniae was reported to be detectable by PCR in 17 of the 19 (90%) late-onset AD brains examined, compared to 1 of 19 (5%) controls. These results were unequivocal in two assays and were confirmed in selected subsets of these samples by electron microscopy, immunoelectron microscopy, immunohistochemistry, reverse transcription-PCR, and culture analysis. Were this to be the case, there would be a dramatic shift from current paradigms employed in AD research and treatment.
Since the presence of C. pneumoniae in AD brains has not been confirmed, an independent investigation was performed to examine the extent of C. pneumoniae presence in the brains of late-onset AD patients. Postmortem brain tissue from late-onset AD and appropriate controls was screened for evidence of C. pneumoniae DNA using PCR targeting a species-specific gene sequence of the major outer membrane protein (MOMP), as previously published (4), and modified since publication by the original authors (personal communication). Attempts were also made to culture the organism from tissue homogenates.
MATERIALS AND METHODS
Clinical tissue specimens.
Postmortem tissue samples from the brains of late-onset AD patients and appropriate controls were obtained from the Alzheimer's Disease Research Center Neuropathology Core, USC School of Medicine. All cases fulfilled the neuropathologic criteria for definite AD as defined by the Consortium to Establish a Registry for Alzheimer's Disease. Samples were frozen following autopsy (the average postmortem intervals for controls and AD samples were 6.3 and 5.9 h, respectively) and stored at −70°C until DNA extraction. Average ages of control and AD patients at time of death were 86.2 and 77.0 years, respectively. Stringent precautions were employed during the processing of clinical specimens to avoid the possibility of contamination while performing dissections, nucleic acid extractions, and culturing from clinical samples in separate laboratories at different times.
Extraction of DNA from tissue.
DNA was extracted from tissue using hot buffered phenol. Briefly, 50 mg of frozen central nervous system (CNS) tissue was placed in a 7-ml Dounce glass homogenizer containing 0.5 ml of lysis buffer (100 mM NaCl, 10 mM Tris-HCl, 25 mM EDTA, and 0.5% sodium dodecyl sulfate; final pH of 8.0) and homogenized briefly before adding 1.0 ml of phenol (65°C, pH 8.0). The sample was then homogenized for another 2 min, transferred to a sterile 1.7-ml tube, and centrifuged at 12,000 rpm for 20 min. Aqueous phase was carefully removed and washed twice with phenol at room temperature, which was followed by two washes with chloroform. DNA was precipitated with three volumes of ethanol (−20°C) and centrifuged at 12,000 rpm for 20 min. Pellets were washed twice with 70% ethanol, air dried, and resuspended in Tris-EDTA (pH 8). This method differs from the reference cited in the original paper describing the method (4). This modification was conveyed to us to be the currently preferred procedure by the senior author of the original paper (personal communication). Our own comparison of the two methods confirmed there to be improvement in DNA yield with the above modification. Spectrophotometric analysis was used for quantitative determination and standardization of DNA concentrations.
PCR.
A nested PCR was performed five or more times using DNA samples extracted from each patient, with 1 μg of DNA per reaction mixture. The primers targeted a species-specific sequence of the MOMP gene of C. pneumoniae (13) and were synthesized by the City of Hope DNA Synthesis Facility according to sequences provided by Alan Hudson. The primers used were outer forward (5′-TATTATCCGCCGCATTTG), corresponding to bases 26 to 43 of the MOMP gene; outer reverse (5′-GAGGTGTCTGTGTAAAGTTC), corresponding to bases 560 to 541; nested forward (5′-TACAATATGGGAAGGTGCTGC), corresponding to bases 114 to 134; and nested reverse (5′-TGAACGCTGTAGAGTTTCC), corresponding to bases 457 to 439. These sequences differ from those published in the original paper, but represent those currently used in Dr. Hudson's laboratory (personal communication).
The PCR cocktail used contained 10 mM Tris-HCl, pH 8.3; 50 mM KCl; 1.5 mM MgCl2; 0.001% (wt/vol) gelatin; and 0.25 U of AmpliTaq Gold DNA polymerase (catalog no. N808-0241; Perkin-Elmer). Thermocycling was performed on an MJ Research PT-100 device with a hot bonnet. The PCR program consisted of 95°C for a 10-min hot start; 35 cycles of denaturation at 95°C, annealing at 55°C, and extension at 72°C (each for 1 min); and a final extension at 72°C for 5 min. A 2.5-μl aliquot from the first reaction mixture was added to a second reaction cocktail and was identical except for the substitution of nested primers. Products were run on 2% agarose gel and Tris-borate-EDTA running buffer and were visualized by ethidium bromide staining. The PCR assay of each sample was repeated a minimum of five times. Products were isolated, and sequences were verified on an ABI prism sequencer. Reaction setup, thermocycling, and product analysis were performed in separate laboratories to avoid problems associated with amplimer contamination.
Propagation of Chlamydia strains.
Several strains of Chlamydia were propagated and enumerated for use in control experiments. C. pneumoniae AR39 (ATCC 53592), C. pneumoniae CM-1 (ATCC VR-1360), Chlamydia trachomatis serotype D (ATCC VR-885) and Chlamydia psittaci (ATCC VR-351) were propagated and titrated in cyclohexamide (1 μg/ml)-inhibited HEp-2 cells (ATCC CCL-23) grown in 20 mM HEPES buffered and glutamine containing (1 mM) Eagle's minimal essential medium supplemented with 10% heat-inactivated fetal bovine serum and 0.45% glucose. C. pneumoniae inclusions were detected and enumerated using standard indirect enzyme-linked immunosorbent assay techniques that employed a species-specific anti-C. pneumoniae monoclonal antibody (DAKO Corp. M660001).
Culture of C. pneumoniae from brain tissue.
Homogenates from PCR-positive brain samples and identical regions from control brains were cultured for C. pneumoniae in the human monocyte cell line THP-1 (ATCC TIB-202) by the method reported in the original paper (4) with slight modification. Approximately 0.5 g of brain tissue, in 2.0 ml of THP-1 growth medium (RPMI 1640 medium [Irvine Scientific, Santa Ana, Calif.] supplemented with 10% heat-inactivated fetal bovine serum [Omega Scientific, Tarzana, Calif.], 10 mM HEPES, 1 mM sodium pyruvate, 1 mM glutamine [all from Irvine Scientific], and 0.45% glucose [Mallinckrodt Baker, Paris, Ky.]), were subjected to three freeze-thaw cycles with tissues homogenized and sonicated between cycles. Four hundred microliters of homogenate was mixed with 3 × 106 THP-1 cells suspended in 2 ml of fresh growth medium, centrifuged for 30 min at 500 × g, diluted to a 10-ml volume with medium, transferred to 25-cm2 tissue culture flasks, and incubated for 72 h at 37°C in 5% CO2. Following initial culture, 1 ml of culture was used for secondary passage in 3 × 106 THP-1 cells as described above, and 4 ml was frozen at −70°C. The remaining 5 ml of primary culture material was centrifuged at 500 × g for 30 min, separated into pellet and supernatant, and frozen at −70°C until analyzed by PCR. Secondary culture material was processed as described above, with one half frozen neat and the other half processed for PCR.
Serving as positive controls, companion aliquots of brain tissue homogenates were seeded with approximately 100 infection-forming units (IFU) of C. pneumoniae (AR39) prior to initial centrifugation with THP-1 cells and were processed alongside the test aliquots.
RESULTS
Specificity and sensitivity of PCR.
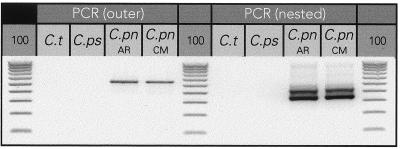
The PCR assay employed distinguished two separate strains of C. pneumoniae from C. trachomatis and C. psittaci as shown in Fig. 1. Both the outer and nested PCR products are shown. The product of the first (outer) amplification is 538 bp, and the second (nested) amplification produced a product of 344 bp. Each product was verified by sequence analysis (data not shown).
FIG. 1.
Species specificity of PCR assay. Both the outer and nested primer pairs specifically amplify two strains of C. pneumoniae (C.pn) DNA but do not amplify DNA from C. trachomatis (C.t) or C. psittaci (C.ps). A 100-bp ladder was run (lanes labeled 100) alongside PCR products for size determination.
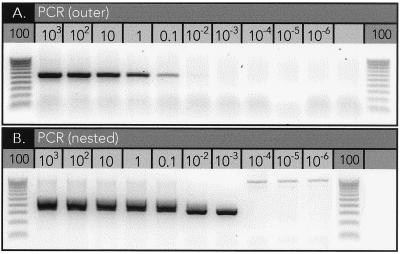
To estimate the sensitivity of our PCR assay, a dilution series of C. pneumoniae DNA (104 to 10−4 IFU) was added to 1 μg of human genomic DNA prior to performing nested PCR (Fig. 2). In five separate PCRs we were consistently able to detect between 0.01 and 0.001 IFU within this 1-μg background of nontarget nucleic acids. This degree of sensitivity is in agreement with previously reported assays (5, 10, 11). The detection of less than 1 IFU demonstrates the ability of the assay to detect both cultivatable and replicating forms of C. pneumoniae.
FIG. 2.
Range of detection. Known amounts of C. pneumoniae DNA (103 to 10−6 IFU) were spiked into a constant background of human genomic DNA (1 μg) and assayed by PCR. (A) The first round of amplifications with the outer primer pair demonstrates the ability of the assay to detect as few as 0.1 IFU. (B) The second round of amplifications using the nested primer pair extends this range to 0.001 to 0.0001 IFU. A 100-bp ladder was run (lanes labeled 100) alongside PCR products for size determination.
To obviate the potential sampling errors associated with a low-copy-number amplification of C. pneumoniae from within our DNA samples, PCRs were repeated separately a minimum of five times. The DNA used in each PCR (∼1 μg) was derived from roughly 150,000 cells (9), bringing the maximum sensitivity of the combined five PCRs down to 0.01 to 0.001 IFU per 750,000 cells.
Analysis of extraction.
Extraction of DNA from clinical samples for use in PCR assays can be problematic because of the historically recognized problems generally associated with recovery efficiencies of different DNA extraction methods as well as the copurification of PCR inhibitors (e.g., lipids, Ca2+, acidic polysaccharides, etc.) (10, 15).
To determine the efficiency of extraction we added a dilution series of C. pneumoniae (104 to 10−4 IFU) to control brain tissue prior to homogenization and extraction of DNA and then assayed 1 μg of the recovered nucleic acids by PCR for C. pneumoniae (data not shown). We were able to detect a minimum of 1 IFU of C. pneumoniae following extraction. This sensitivity of the assay is actually much greater, because the 1 μg of input DNA represented the entire DNA sample recovered. The average yield of total DNA per extraction was roughly 30 μg, bringing the range of detection following extraction down to approximately 0.03 IFU. This is similar to the detection sensitivity of the PCR assay reported above and indicates that there is minimal loss of DNA or that significant copurification of inhibitors had not occurred during our extraction.
PCR of patient samples.
Amplification of potentially low-copy-number targets within a disproportionately larger background of host DNA requires modifications of traditional PCR in order to ensure specificity and efficiency of the reaction (9). A nested PCR following the first round of amplification was employed to increase the efficiency of identifying C. pneumoniae. Nested PCR involves a second round of amplification with primers internal to the original primer pair. This technique gives a built-in confirmation of the outer products and is an advantage for low-copy-number amplifications because of its ability to dilute the original material along with any inhibitory substances (1). The use of AmpliTaq Gold DNA polymerase allowed the integration of a hot start into the amplification (3). A hot start can significantly improve the specificity, sensitivity, and yield in PCR by eliminating the formation of misprimed products and primer-oligomers often formed during pre-PCR at temperatures lower than the optimal annealing temperature (6).
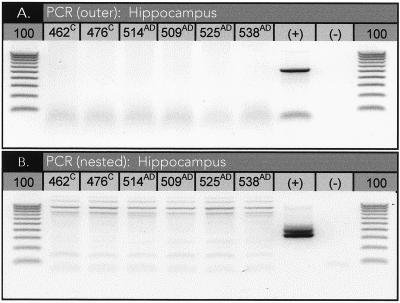
DNA samples from six separate brain regions of each patient were assayed for C. pneumoniae by PCR five or more times. These regions included areas relevant to AD (e.g., the hippocampus, frontal cortex, and parietal cortex). For a case to be considered positive, the PCR results had to be unequivocally positive for at least one of the regions sampled. Our PCR results were uniformly negative for C. pneumoniae in both control and AD samples (Table 1; Fig. 3). Although three samples among all studied returned at least one PCR-positive result for C. pneumoniae (sample 509AD, one of seven; sample 526AD, two of five; and sample 462C, one of six [where the superscripts AD and C indicate samples from AD patients and controls, respectively]), confirmation in each instance was not observed in subsequent reactions and these samples were not considered truly positive according to the above criteria.
TABLE 1.
PCR resultsa
| Patientb | Sexc | Age (yr) | No. of positive results/no. of assays performed
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Hippocampus | Frontal cortex 6 | Frontal cortex 9 | Frontal cortex 10 | Parietal cortex | Cerebellum | Basal ganglia | Cochlear nucleus | |||
| 401C | F | 71 | 0/6 | 0/6 | 0/6 | 0/5 | 0/5 | |||
| 405C | M | 88 | 0/5 | 0/7 | 0/5 | 0/5 | 0/5 | 0/5 | ||
| 462C | F | 92 | 0/6 | 0/6 | 0/6 | 0/5 | 1/6 | |||
| 476C | M | 88 | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | ||
| 529C | F | 92 | 0/6 | 0/6 | 0/6 | 0/6 | 0/5 | 0/6 | ||
| 434AD | M | 63 | 0/5 | 0/5 | ||||||
| 435AD | M | 70 | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | |||
| 453AD | M | 68 | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | |||
| 474AD | M | 74 | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | |||
| 485AD | M | 64 | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | |||
| 504AD | M | 72 | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | |||
| 508AD | M | 88 | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | |||
| 509AD | F | 97 | 0/5 | 0/5 | 0/7 | 1/7 | 0/5 | 0/5 | ||
| 514AD | F | 82 | 0/5 | 0/7 | 0/7 | 0/5 | 0/5 | 0/5 | ||
| 517AD | M | 75 | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | |||
| 519AD | M | 66 | 0/5 | 0/5 | 0/5 | 0/5 | ||||
| 525AD | F | 94 | 0/5 | 0/7 | 0/5 | 0/7 | 0/5 | 0/5 | ||
| 526AD | F | 77 | 2/7 | 0/7 | 0/5 | 0/7 | 0/5 | 0/5 | ||
| 531AD | M | 87 | 0/5 | 0/5 | 0/5 | 0/5 | ||||
| 538AD | F | 77 | 0/5 | 0/7 | 0/7 | 0/5 | 0/5 | 0/5 | ||
Boldface type indicate the three samples that returned at least one positive PCR result. Different areas of the frontal cortex are referred to by their numbered position on Brodmann's cytoarchitectonic map of the human cortex (1909).
PCR results for C. pneumoniae DNA in brain regions of control cases (n = 28) are indicated with a superscript C; PCR results for C. pneumoniae DNA in brain regions of AD patients (n = 73) are indicated by a superscript AD.
F, female; M, male.
FIG. 3.
PCR results. Example of PCR results for six hippocampal DNA samples. (A) First round of amplifications with outer primer pair; (B) second round of amplifications using the nested primer. A 100-bp ladder was run (lanes labeled 100) alongside PCR products for size determination.
Culture of PCR-positive samples.
A positive PCR result together with the ability to culture C. pneumoniae from clinical samples is considered a “gold standard” for its identification. We attempted to culture C. pneumoniae from our three PCR-positive samples along with PCR-negative samples from congruent control regions. Following two passages, we were unable to detect C. pneumoniae by PCR in either PCR-positive or -negative cultures. Control cultures of brain tissues seeded with C. pneumoniae during processing for culture consistently gave positive cultures.
CONCLUSION
Based on the results of this study, we conclude that C. pneumoniae is neither strongly nor uniquely associated with the neuropathology seen in late-onset AD. After a thorough examination of AD and control brains, employing previously described methods and confirmed in personal communications with the senior author of the first and only report, C. pneumoniae could not be unequivocally detected by PCR or culture methods in any CNS region from AD cases or controls that we assayed. This observation differs significantly from the original study, which claimed that C. pneumoniae DNA was extremely common in areas of AD neuropathology, being detectable by PCR in 90% (17 of 19) of AD brains compared to 5% (1 of 19) of control brains.
Notwithstanding the questionable detection of C. pneumoniae DNA in three samples, the difference between the two sets of specimens is even more striking considering both the number of brain areas affected and the apparent levels of C. pneumoniae DNA present in the PCR-positive specimens. It was reported previously that C. pneumoniae was detected in more than one CNS location in brain tissue from 8 of the 17 AD cases (2). In this study samples were considered positive if PCR results were positive in all aliquots tested.
In the present study, every effort was made to use the same conditions and methods as originally reported and subsequently modified. It is unlikely that the differences in findings are attributable to technical differences. Inadequate sample size also does not appear to be an issue, given both the high frequency of PCR-positive samples reported and the exhaustive testing of specimens in our study. This leaves characteristics of the patient populations as a possible source of the discrepancy. Of particular significance in this regard would be the season of death as well as the institutional history of each patient prior to death, as both of these circumstances could possibly contribute to differences in the recentness of exposure to C. pneumoniae. All of the tissue samples analyzed in the original study were obtained from three tissue banks associated with hospitals on the East Coast, while our samples were obtained from a single bank located in Southern California. These are different geographical regions possibly differing in institutional practices and social support systems.
C. pneumoniae possesses many attributes that would make it an attractive contributory factor to the chronic and progressively degenerative neuropathological patterns seen in AD. Multiple respiratory infections with chronic sequelae are very common, with seroprevalence (immunoglobulin G) increasing throughout life, reaching more than 75% among the elderly (7). C. pneumoniae is an obligate intracellular bacterium that can infect a wide range of cell types. C. pneumoniae can chronically infect and activate monocytes, which could traffic across the blood-brain barrier. It is a gram-negative bacterium and thus produces endotoxin. In short, it is the type of organism that could both directly and indirectly stimulate the kind of inflammation seen within AD lesions. Numerous studies have demonstrated conclusively its presence within atherosclerotic plaques and in the synovial tissue of some patients with reactive arthritis following respiratory infection (P. Saikku, presented at the Proceedings of the Ninth International Symposium on Human Chlamydial Infections, 1998). Most recently, C. pneumoniae was detected by PCR in the cerebrospinal fluid of 96% of multiple sclerosis patients, while being detected in only 18% of controls (14). Although the latter observation requires confirmation, what is common to all these diseases, as well as AD, is a chronic inflammatory process in which activated macrophages participate. Knowing when and where activation occurs and the types of responses chronically infected and activated macrophages can make when circulating will help establish the nature of any causal relationship between the presence of C. pneumoniae within inflammatory lesions and the diseases which these lesions define. During such chronic processes, the original of at least some of these cells might prove to be a site of infection within the lung. However, the almost universal and unequivocal association of C. pneumoniae with the neurodegenerative process occurring during the terminal phase of AD reported previously (2) was not confirmed in the present study.
REFERENCES
- 1.Backman A, Johansson B, Olcen P. Nested PCR optimized for detection of Bordetella pertussis in clinical nasopharyngeal samples. J Clin Microbiol. 1994;32:2544–2548. doi: 10.1128/jcm.32.10.2544-2548.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balin B J, Gerard H C, Arking E J, Appelt D M, Branigan P J, Abrams J T, Whittum-Hudson J A, Hudson A P. Identification and localization of Chlamydia pneumoniae in the Alzheimer's brain. Med Microbiol Immunol (Berlin) 1998;187:23–42. doi: 10.1007/s004300050071. [DOI] [PubMed] [Google Scholar]
- 3.Birch D E. Simplified hot start PCR. Nature. 1996;381:445–446. doi: 10.1038/381445a0. [DOI] [PubMed] [Google Scholar]
- 4.Branigan P J, Gerard H C, Hudson A P, Schumacher H R, Jr, Pando J. Comparison of synovial tissue and synovial fluid as the source of nucleic acids for detection of Chlamydia trachomatis by polymerase chain reaction. Arthritis Rheum. 1996;39:1740–1746. doi: 10.1002/art.1780391018. . (Errata, 4D:387, 782, 1997.) [DOI] [PubMed] [Google Scholar]
- 5.Campbell L A, Perez Melgosa M, Hamilton D J, Kuo C C, Grayston J T. Detection of Chlamydia pneumoniae by polymerase chain reaction. J Clin Microbiol. 1992;30:434–439. doi: 10.1128/jcm.30.2.434-439.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.D'Aquila R T, Bechtel L J, Videler J A, Eron J J, Gorczyca P, Kaplan J C. Maximizing sensitivity and specificity of PCR by pre-amplification heating. Nucleic Acids Res. 1991;19:3749. doi: 10.1093/nar/19.13.3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grayston J T. Infections caused by Chlamydia pneumoniae strain TWAR. Clin Infect Dis. 1992;15:757–761. doi: 10.1093/clind/15.5.757. [DOI] [PubMed] [Google Scholar]
- 8.Lendon C L, Ashall F, Goate A M. Exploring the etiology of Alzheimer disease using molecular genetics. JAMA. 1997;277:825–831. [PubMed] [Google Scholar]
- 9.Lo Y M. Detection of minority nucleic acid populations by PCR. J Pathol. 1994;174:1–6. doi: 10.1002/path.1711740102. [DOI] [PubMed] [Google Scholar]
- 10.Maass M, Dalhoff K. Comparison of sample preparation methods for detection of Chlamydia pneumoniae in bronchoalveolar lavage fluid by PCR. J Clin Microbiol. 1994;32:2616–2619. doi: 10.1128/jcm.32.10.2616-2619.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meijer A, van der Vliet J A, Schouls L M, de Vries A, Roholl P J, Ossewaarde J M. Detection of microorganisms in vessel wall specimens of the abdominal aorta: development of a PCR assay in the absence of a gold standard. Res Microbiol. 1998;149:577–583. doi: 10.1016/s0923-2508(99)80005-9. [DOI] [PubMed] [Google Scholar]
- 12.Mirra S S, Heyman A, McKeel D, Sumi S M, Crain B J, Brownlee L M, Vogel F S, Hughes J P, van Belle G, Berg L. The Consortium to Establish a Registry for Alzheimer's Disease (CERAD). Part II. Standardization of the neuropathologic assessment of Alzheimer's disease. Neurology. 1991;41:479–486. doi: 10.1212/wnl.41.4.479. [DOI] [PubMed] [Google Scholar]
- 13.Perez Melgosa M, Kuo C C, Campbell L A. Sequence analysis of the major outer membrane protein gene of Chlamydia pneumoniae. Infect Immun. 1991;59:2195–2199. doi: 10.1128/iai.59.6.2195-2199.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sriram S, Stratton C W, Yao S, Tharp A, Ding L, Bannan J D, Mitchell W M. Chlamydia pneumoniae infection of the central nervous system in multiple sclerosis. Ann Neurol. 1999;46:6–14. [PubMed] [Google Scholar]
- 15.Thomas M, Wong Y, Thomas D, Ajaz M, Tsang V, Gallagher P J, Ward M E. Relation between direct detection of Chlamydia pneumoniae DNA in human coronary arteries at postmortem examination and histological severity (Stary grading) of associated atherosclerotic plaque. Circulation. 1999;99:2733–2736. doi: 10.1161/01.cir.99.21.2733. [DOI] [PubMed] [Google Scholar]