Abstract
Several studies are now underway as a worldwide response for the containment of the COVID-19 outbreak; unfortunately, none of them have resulted in an effective treatment. Salvadora persica L. (Salvadoraceae), commonly known as meswak, is one of the popular plants used by Muslims as an oral hygiene tool. It is documented that the meswak possesses antiviral activity, but no report discusses its use for coronavirus treatment. Herein, a mixture of 11 flavonoids prepared from the aqueous plant extract and its liposomal formulation were shown to inhibit SARS-CoV-2 in an in vitro A549 cell line culture and a RT-PCR test almost as well as the FDA-approved anti-COVID-19 agent, remdesivir. Encapsulation within liposomal formulation led to a highly significant increase in the percentage of inhibition of viral replication from 38.09 ± 0.83 to 85.56 ± 1.12% in a flavonoid mixture and its liposomal preparation, respectively, and this figure approached that obtained for remdesivir (91.20 ± 1.71%). Preliminary tests were also performed, including a total flavonoid assay, a molecular docking study, a 3CL-protease inhibition assay and a cytotoxicity study. It was worthy to find a cheap, readily available, safe natural source for promising anti-SARS-CoV-2 agents, that leak their phytochemicals into the aqueous saliva during regular use as a brushing agent.
Several studies are now underway as a worldwide response for the containment of the COVID-19 outbreak; unfortunately, none of them have resulted in an effective treatment.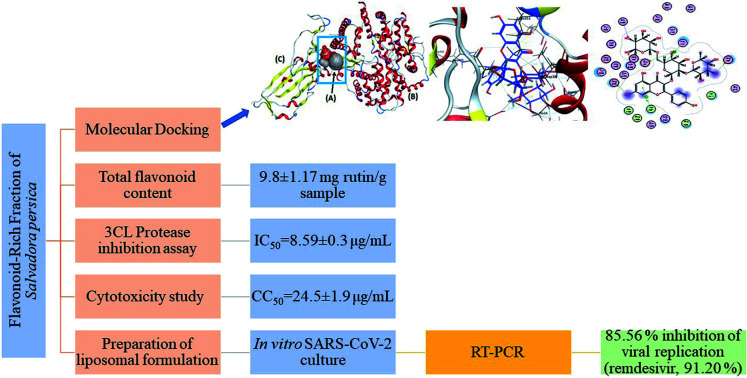
1. Introduction
At the end of 2019, an outbreak of coronavirus disease 2019 (COVID-19), caused by a novel severe acute respiratory syndrome corona virus-2 (SARS-CoV-2), emerged in China and rapidly spread across the world leading to a major global threat.1 The World Health Organization (WHO) declared SARS-CoV-2 as an emergency world health pandemic. The SARS-CoV-2 is an RNA-enveloped virus that encodes four structural proteins.2 Exceptionally, the S protein has attracted great attention because of its earlier role during the attachment of the virus to a host cell surface receptor called an angiotensin converting enzyme-2 (ACE-2) receptor.3 Scientists are engaged in trying to discover or repurpose drugs having the ability to affect the binding of the viral spike protein with the human ACE-2 receptor, e.g., Alexpandi et al. in 2020, who screened quinoline derivatives in silico for their spike-RBD-ACE-2 inhibitory properties.4 Also, the viral main protease Mpro is another important target, because upon infection of the host cell, the viral genome is translated into a large polyprotein which is cleaved by viral-encoded proteases (Mpro), releasing several non-structural proteins responsible for replicating the viral genome and generating nested transcripts that are used in the synthesis of viral proteins.5 Many researchers have reported their studies on Mpro drug repurposing.6–8 Marine products were also screened for in silico inhibition of Mpro.9 High throughput virtual screening to discover inhibitors of the Mpro of the SARS-CoV-2 have been reported and among millions of compounds, bioflavonoids were some of the top scoring compounds.10
A few protocols are being used to treat laboratory confirmed or suspected COVID-19 patients hospitalized with severe disease, for example, chloroquine derivatives,11 convalescent plasma,12 azithromycin13 and remdesivir, with limited known information about its safety on human cells.14 Natural products are considered as viable lead candidates that have a highly significant role in drug discovery. Salvadora persica L. (Salvadoraceae) or chewing stick, commonly known in Arabic as meswak, is one of the popular plants used by Muslims as an oral hygiene tool. Several scientific studies have worked on the pharmacological profile of S. persica, with a wealth of published literature now available. Besides the effects on oral health, its biological activities have been found to include antimicrobial, hypoglycemic, antioxidant, anticonvulsant, antiulcer, sedative, analgesic, antiosteoporosis, anti-inflammatory, hypolipidemic, antitumor and antiviral effects,15,16 but there are no reports dealing with coronavirus. A literature survey also revealed the presence of a variety of phytochemicals in S. persica extracts including alkaloids, tannins, glycosides, saponins, essential oils and flavonoids.15 Previous research on an aqueous extract of S. persica led to the isolation and identification of 11 flavonoid glycosides.17 Interestingly, 10 of them exhibited significant binding stability at the N3 binding site, found through a molecular docking study against the COVID-19 main protease (Mpro), suggesting their potential effect on blocking viral replication.14 Therefore, the objectives of this study were to prepare a flavonoid-rich fraction (FRF) from an aqueous extract of the aerial parts of S. persica, and investigate its anti-COVID-19 activity using the preliminary steps as a molecular docking study on the contact surface of hACE2-COVID 19 spike protein complex, a 3CL-protease inhibition assay, and cytotoxic activity. The final objective was to prepare a liposomal formulation (FRF-Lip) from this fraction to increase its viral inhibitory action and assess the potential ability of both fractions to affect human SARS-CoV-2 using an in vitro assay with remdesivir as reference. As far as is known, this is the first practical study dealing with the effect of S. persica on COVID-19.
2. Materials and methods
2.1. Plant material and preparation of FRF
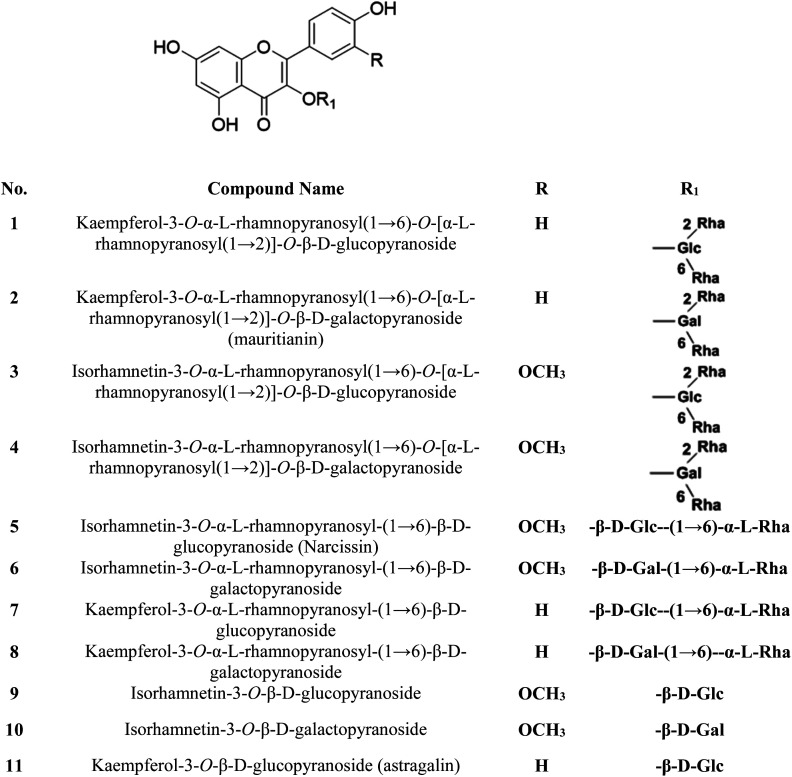
The S. persica stem and leaves were collected, as described by Owis et al.14 A mixture of 11 previously isolated and identified flavonoids (FRF) was prepared and identified according to the method adopted by Ali et al. and Owis et al.14,17 The chemical structures of the flavonoids are shown in Fig. 1.14,17
Fig. 1. Chemical structures of the major compounds present in FRF.
2.2. Determination of the total flavonoid contents
The total flavonoid content of the FRFs and the aqueous extracts was determined using microplate reader analysis with a FLUOstar® Omega multi-mode microplate reader (BMG Labtech) according to the method adopted by Herald et al.18 The total flavonoid contents were expressed as mg rutin equivalents per g dry sample (mg RU per g).
2.3. Docking and ADME studies
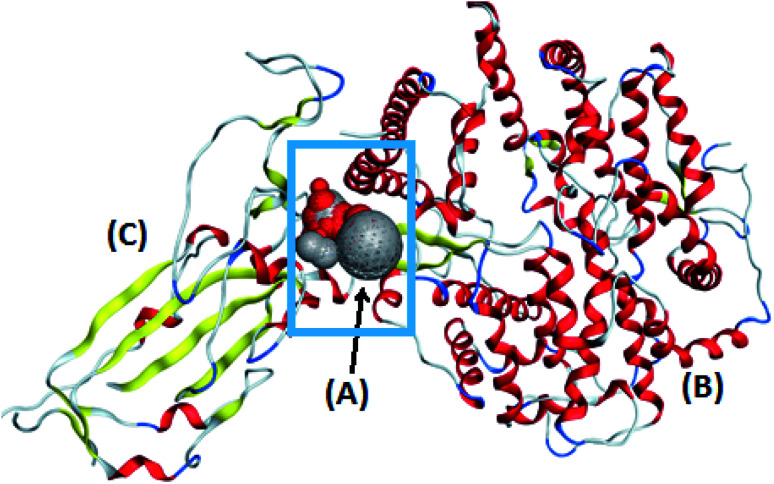
The target compounds were optimized according to the method adopted by Owis et al.14 The X-ray crystallographic structure of the hACE2 complex with a COVID-19 spike protein was obtained from the Protein Data Bank (http://www.rcsb.org, PDB code 6VW1).19 The proteins were prepared according to the method of Owis et al.14 Site Finder was used for active site search on the contact surface of the hACE2-COVID 19 spike protein complex and the biggest site of interaction (site 4) was selected (Fig. 2). A docking of conformation database of the target molecules to site 4 was prepared as described by Owis et al.14 The obtained poses were studied so that the poses that showed the best ligand-contact surface of the hACE2-COVID 19 spike protein complex interactions were selected and stored for use in the energy calculations. The absorption, distribution, metabolism and excretion (ADME) of the target flavonoids 1–11 and hesperidin (reference compound) were calculated using the free-to-use website: http://www.swissadme.ch (Swiss Institute of Bioinformatics).
Fig. 2. The alpha sphere (A) of the main binding pocket between hACE2 (B) and the COVID-19 spike protein (C).
2.4. The 3CL-protease inhibition assay of FRF
The 3CL-protease inhibition assay was used to assess the effect of the FRFs on the inhibition of the main protease (Mpro) enzyme which is responsible for viral replication and transcription, and consequently affects human SARS-CoV-2. The assay was performed using a 3CL Protease (SARS-CoV-2) Assay Kit (BPS Bioscience), according to the manufacturer's protocol. The fluorescence intensity was measured in a microtiter plate-reading fluorimeter (TECAN) capable of excitation at a wavelength of 360 nm and detection of emission at a wavelength of 460 nm.
2.5. Preparation and evaluation of liposomal formulation of FRF
A liposomal formulation of FRF (FRF-Lip) was prepared using a spraying technique adopted by Refaat et al.20 The particle size and polydispersity index (PDI) were determined using laser light diffraction techniques, and transmission electron microscopy (TEM) was used to evaluate the prepared liposomes. The entrapment efficiency percentage (% EE) was determined using the method of Refaat et al.20
2.6. In vitro study on human coronavirus
A human lung adenocarcinoma epithelial cell line (A549, American Type Culture Collection, USA) and human coronavirus (COV 19) were kindly supplied by VACSERA (Egypt). Prior to assessment of the antiviral activity of the FRFs, a cytotoxicity study was performed using the MTT assay as described by Mosmann,21 and the absorbance was measured at a wavelength of 530 nm. To evaluate the antiviral potential of the FRFs and the FRF-Lip against SARS-CoV-2, a classical cell culture was performed.22 The percentage of inhibition of SARS-CoV-2 replication was determined by estimation of the viral RNA concentration using a quantitative real time-polymerase chain reaction (RT-PCR) assay performed according to the manufacturer's protocol (genesig® Real-Time PCR Coronavirus COVID-19 (CE IVD), Primerdesign Ltd., UK) and remdesivir was used as a reference standard.
2.7. Statistical analysis
The previous experiments were performed in triplicate and the data were presented as mean ± standard error (SE). To study the significance of the differences between the results, a one-way analysis of variance (ANOVA) was used.
3. Results and discussion
In this research, molecular docking in the contact surface of the hACE2-COVID 19 spike protein complex with reference to hesperidin was performed to investigate the capability of the 11 flavonoids (Fig. 1) to prevent the virus entry to the host cells, thus protecting against infection or the release of virus from the hACE2 after its intracellular entry. Hesperidin was selected from both Chinese and Indian medicinal plants as a competent natural product to treat COVID-19 due to its potent ability to bind at the S-protein-ACE-2 site.23
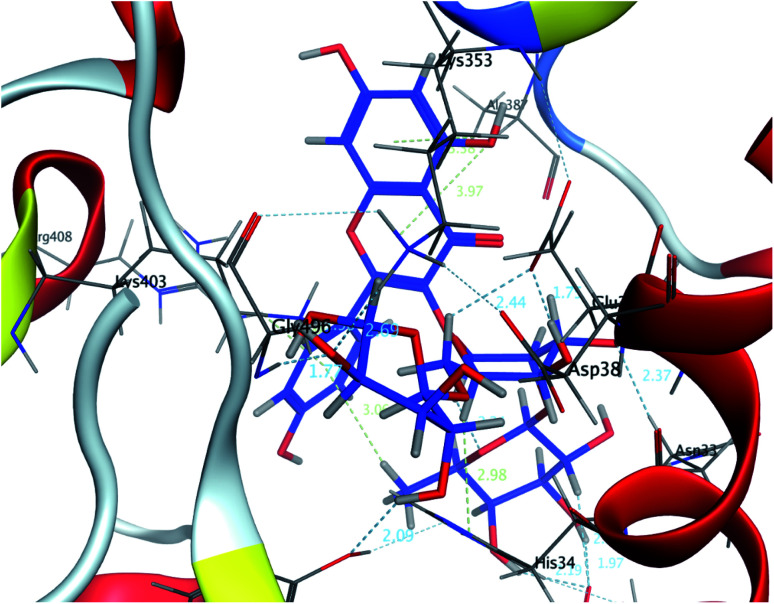
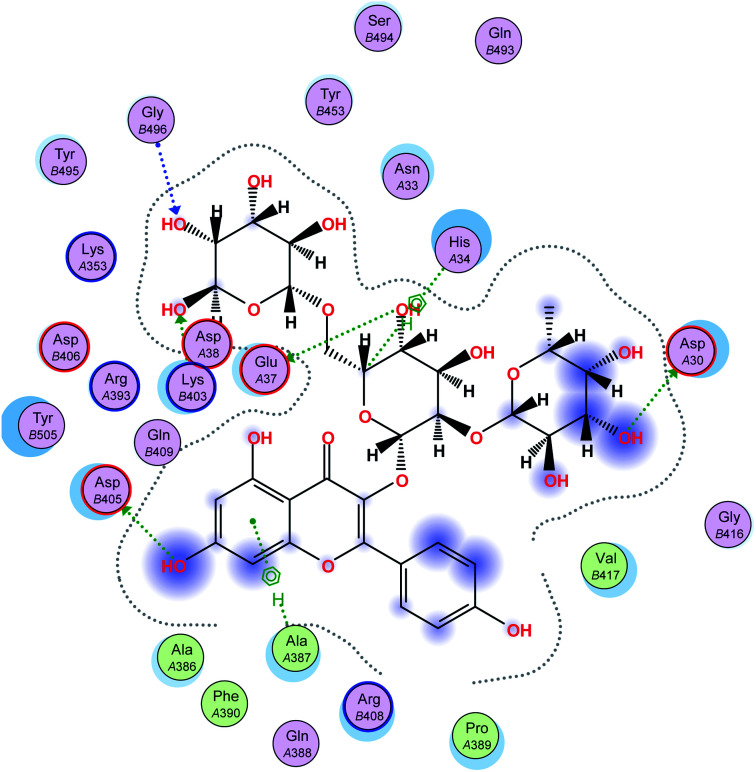
The interaction energies of compounds 1–11 revealed that most of the compounds showed a prominent interaction with both hACE2 and the viral spike protein (Table 1, Fig. 3 and 4). The most promising compound, with the highest binding energy (−9.4799 kcal per mole), was compound 2. Compounds 5, 1, 3, 4 and 7, with binding energies of −7.6404, −7.3869, −7.3769 and −7.2929 kcal per mole, respectively, showed higher binding energies than hesperidin. Whereas compounds 9, 8, 10, 11 and 6 showed moderate binding energies which were less than that of hesperidin (−6.6629 kcal per kmole). The six compounds (1–6) with the highest binding energies were able to bind to the spike protein with similar hydrogen bonding with Asp 405 or Asp 406, and Lys 403. Compounds 1, 2 and 5 were like hesperidin and were able to bind to hACE2 with hydrogen bonding with Glu 37. Compounds 1, 2, 6 and 7 had similar binding with His 34, either by hydrogen bonding or H–pi bonding. It is worth noting that all the compounds showed many other types of hydrogen bonding in addition to pi–pi and other hydrophobic bonds with the spike protein and hACE2. Interestingly, compound 2 could be used as a starting point for developing more potent derivatives with better therapeutic properties. Flavonols are the most studied, and documented, compounds tested against SARS-CoV-2.24. Pandey et al. reported that kaempferol and isorhamnetin interacted with the S2 domain of the S protein with high binding affinities when compared to hydroxychloroquine (positive control).25 Mouffouk et al. proved that the presence of ortho di-hydroxyl groups in kaempferol created stronger hydrogen bonds and a stable hACE2-COVID 19 spike protein complex than those of isorhamnetin.24 These data were consistent with the results obtained in the research reported here.
Receptor interaction of compounds 1–11 and hesperidin.
| Compound no. | dG kcal per mole | Receptor | |
|---|---|---|---|
| Amino acid/type of bonding/distance (Å)/binding energy (kcal per mole) | |||
| hACE2 | Spike protein | ||
| 1 | −7.3869 | GLU 37/H–donor/3.06/−0.8 | ASP 405/H–donor/3.07/−1.3 |
| HIS 34/H–acceptor/2.96/−1.9 | LYS 403/H–acceptor/2.97/−5.2 | ||
| ARG 408/pi–cation/4.08/−1.3 | |||
| ARG 408/pi–cation/4.06/−0.6 | |||
| 2 | −9.4799 | GLU 37/H–donor/2.68/−3.1 | ASP 405/H–donor/3.01/−0.8 |
| ASP 30/H–donor/2.91/−3.8 | GLY 496/H–acceptor/2.76/−1.3 | ||
| HIS 34/H–pi/3.85/−0.9 | LYS 403/H–acceptor/2.90/−5.0 | ||
| ALA 387/pi–H/4.39/−0.6 | |||
| 3 | −7.3769 | ASP 405/H–donor/3.01/−2.3 | |
| ASP 406/H–donor/2.76/−4.0 | |||
| ASP 406/H–donor/3.11/−1.9 | |||
| ASP 406/H–donor/3.26/−1.0 | |||
| LYS 403/H–acceptor/2.96/−4.1 | |||
| LYS 403/H–acceptor/3.34/−0.9 | |||
| 4 | −7.2929 | ASP 30/H–donor/3.14/−1.1 | ASP 405/H–donor/2.76/−3.0 |
| LYS 403/H–acceptor/3.39/−0.6 | |||
| 5 | −7.6404 | GLU 37/H–donor/3.08/−1.1 | ASP 406/H–donor/2.67/−1.5 |
| GLU 37/H–donor/3.49/−0.8 | LYS 403/H–acceptor/2.76/−7.6 | ||
| ARG 393/H–acceptor/3.06/−1.4 | |||
| ASN 33/H–acceptor/2.91/−2.7 | |||
| 6 | −5.2061 | HIS 34/H–donor/2.98/−1.7 | ARG 408/H–acceptor/3.10/−2.5 |
| ARG 393/H–acceptor/2.87/−3.3 | |||
| HIS 34/pi–pi/3.90/−0.0 | |||
| 7 | −7.1293 | HIS 34/H–donor/2.78/−1.2 | TYR 453/H–donor/2.64/−1.0 |
| ARG 393/H–acceptor/3.15/−1.7 | LYS 403/H–acceptor/3.53/−1.0 | ||
| LYS 353/H–acceptor/2.97/−0.8 | ARG 408/pi–cation/3.85/−1.4 | ||
| HIS 34/H–pi/4.04/−1.8 | |||
| 8 | −5.9453 | ASP 30/H–donor/2.99/−3.1 | LYS 403/H–acceptor/2.91/−5.7 |
| ASN 33/H–acceptor/2.97/−2.5 | |||
| 9 | −6.2471 | ASP 30/H–donor/3.38/−0.8 | ASP 406/H–donor/2.94/−3.6 |
| LYS 403/H–acceptor/2.90/−3.3 | |||
| 10 | −5.7829 | ARG 408/H–acceptor/3.38/−0.9 | |
| ARG 408/H–acceptor/3.47/−0.8 | |||
| 11 | −5.3156 | SER 494/H–donor/2.80/−1.0 | |
| GLN 409/H–acceptor/2.75/−2.4 | |||
| ARG 408/H–acceptor/2.83/−4.1 | |||
| Hesperidin | −6.6629 | GLU 37/H–donor/2.75/−2.3 | GLN 409/H–acceptor/2.76 |
Fig. 3. A 3D representation of the docking poses of compound 2 on the COVID-19 spike binding site with hACE-2.
Fig. 4. A 2D representation of the docking of compound 2 on the COVID-19 spike binding site with hACE-2.
Studying the physicochemical and ADME properties revealed that all the compounds had a reasonable topological polar surface area (TPSA) between 20 Å2 and 130 Å2 except for the reference compound hesperidin (Table 2). Entries of the Moriguchi logarithm of the partition coefficient (MLOGP) < 4.15 were of reasonable lipophilicity to be taken orally. Compounds 5, 7, 9 and 2 were of reasonable lipophilicity. The predicted gastrointestinal (GI) absorption and blood–brain barrier (BBB) permeability showed that compounds 9, 2, 4, 3 and 1 were expected to be highly absorbed from the GI tract whereas compounds 9, 2, 4, and 1 can pass the BBB (Table 2). It is worth noting that all the tested compounds showed no pan assay interference compounds (PAINS) alerts. Fortunately, the possible deleterious effects on the CNS of COVID-19 infected patients26–31 could be treated or prevented by compounds 2, 1 and 4 as they were the top-scoring compounds with the capability of crossing the BBB.
Physicochemical parameters and ADME of compounds 1–11 and hesperidina.
| Compound no. | TPSA Å2 | log Po/w (MLOGP) | GI absorption | BBB permeant | PAINS alert |
|---|---|---|---|---|---|
| 1 | 37.30 | 4.38 | High | Yes | 0 |
| 2 | 54.37 | 3.59 | High | Yes | 0 |
| 3 | 37.30 | 4.75 | High | No | 0 |
| 4 | 37.30 | 4.29 | High | Yes | 0 |
| 5 | 110.02 | 3.99 | Low | No | 0 |
| 6 | 69.56 | 5.61 | Low | No | 0 |
| 7 | 110.02 | 3.88 | Low | No | 0 |
| 8 | 89.79 | 4.98 | Low | No | 0 |
| 9 | 37.30 | 3.81 | High | Yes | 0 |
| 10 | 110.02 | 4.71 | Low | No | 0 |
| 11 | 110.02 | 4.67 | Low | No | 0 |
| Hesperidin | 234.29 | −3.04 | Low | No | 0 |
TPSA: topological polar surface area, MLOGP: Moriguchi logarithm of the partition coefficient, BBB: blood–brain barrier, PAINS: pan assay interference compounds.
The virtual protective and Mpro inhibitory role14 of the meswak flavonoids against SARS-CoV-2 was proved using in vitro experiments. Instead of using individual flavonoids, a mixture of 11 flavonoids (FRF) was used in this analysis, based on the synergism capability of natural products, which is thought to have more powerful antiviral activity.32 The FRF were successfully prepared from the aqueous extract of S. persica leaves and stems. The identity of the obtained fractions was confirmed by co-spot-TLC with previously isolated and characterized flavonoids.14,17 Total flavonoid content was determined on an aqueous extract and FRF with contents of 9.8 ± 1.17, and 7.4 ± 1.78 mg Ru per g sample, respectively. This confirmed that the FRFs contained most of the flavonoid content present in the aqueous extract.
According to virtual docking results, the FRFs had an inhibitory effect on both the Mpro enzyme14 and spike protein-ACE-2. The 3CL protease inhibition assay was performed as a preliminary practical evaluation. The FRFs exhibited a significant inhibitory action against 3CL-protease with an IC50 = 8.59 ± 0.3 μg mL−1 whereas that of the standard (tannic acid) was 2.1 ± 0.2 μM.33 This result was consistent with the stabilized binding of all the FRF-compounds (1–11) at the N3-binding site of Mpro as described in a previous docking study.14 The glycosylated flavonols with at least one rhamnose unit in their structure at the carbon 3 position can bind with a higher affinity to the active site of the Mpro than to the corresponding aglycones.34
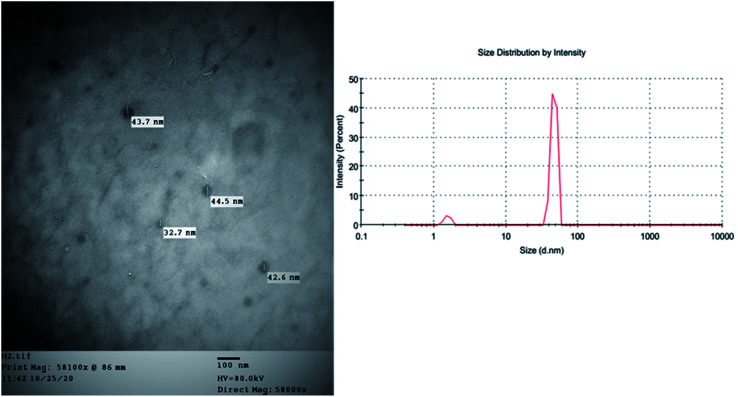
In an attempt to enhance the potential activity of FRF as an anti-SARS-CoV-2 agent, a nano-sized liposomal formulation (FRF-Lip) was prepared. This hypothesis was based upon the fact that nano-sized particles have a high surface area and possess the ability to introduce both water-soluble and lipophilic components, such as flavonoids, through the cellular membrane into the cells.20 The adopted spraying technique successfully produced a uniformly nanosized FRF-loaded liposomal formulation with a size of 43.7 ± 1.5 nm and a PDI of 0.32. The image obtained with TEM showed the vesicular multilayered structure of the prepared FRF-Lip as shown in Fig. 5 and the % EE y was 47.8 ± 2.1%.
Fig. 5. The TEM image and the particle size distribution of FRF-Lip.
Safety and possible secondary effects were considered as major requirements during the search for a new antiviral lead. The natural FRF obtained from meswak, a plant used regularly by Muslims, was screened for its cellular toxicity effect, and for the determination of the appropriate concentration using the in vitro inhibition assay. The estimated concentration associated with 50% cytotoxicity (CC50) was found to be 24.5 ± 1.9 μg mL−1.
The significant results of the effects of FRFs on 3CL-protease highlighted the need to investigate their behavior, using cell culture, on the human virus and to compare the obtained results with that of the FRF-Lip. The prospective anti-human coronavirus effect of FRF and FRF-Lip was investigated by in vitro RT-PCR testing which specifically detects SARS-CoV-2 RNA quantitatively. Encapsulation of FRF within the liposomal formulation led to a high significant increase in the percentage of inhibition in viral replication from 38.09 ± 0.83% to 85.56 ± 1.12% in FRF and FRF-Lip, respectively (p < 0.0001). This could be attributed to the higher surface area and better dispersibility of the liposomal formulation which was able to prevent precipitation and aggregation of the lipophilic components. Hence, it could optimize the expected poor cellular uptake of the hydrophilic components, and enhance the delivery of such challenging natural products.20 It was useful to find a natural product having the ability to inhibit SARS-CoV-2 replication in a value very close to that of the FDA-approved anti-COVID-19: remdesivir (91.20 ± 1.71%).
The 11 compounds in the FRF are flavonols having a 3,4′-dihydroxy-2-phenylchromen-4-one backbone, namely kaempferol and its 3′-methoxylated derivative: isorhamnetin (Fig. 1). They contain O-sugar groups attached at position 3 of the carbon ring. These 3-O-glycosylated kaempferol analogs were previously documented to have antiviral activity generally and anti-SARS-CoV activity specifically.35
In addition to all the previous results together, the COVID-19 disease is characterized by an excessive inflammatory response leading to a major lung illness and consequent possible mortality. Anti-inflammatory agents are likely to be effective against the subsequent elevated cytokine levels, such as γ-interferon, typically associated with COVID-19 patients.36 The use of S. persica aqueous extract and its flavonoids as anti-inflammatory natural agents37 is not only likely to reduce SARS-CoV-2 viral infectivity, but also likely to reduce host inflammatory response. Moreover, S. persica (meswak) itself, as a natural famous plant used by Muslims, has the advantages of being a cheap, readily available, and safe drug source. Interestingly, human watery saliva can act as an aqueous extractor for its various innate phytochemicals during regular traditional use for oral hygiene by a mechanical cleaning process.
4. Conclusions
COVID-19 represents one of the pandemic diseases that play a crucial part in public health, when antiviral therapies are still not available. In this study, a mixture of 11 flavonol glycosides prepared from S. persica were studied hypothetically with a molecular docking study and found to affect SARS-CoV-2 by inhibiting Mpro and blocking the contact surface of the hACE2-COVID 19 spike protein complex. This postulation was proven by results of preliminary studies in addition to in vitro culture accompanied by RT-PCR testing. The 3CL-protease inhibition assay revealed that FRF produced a significant inhibitory effect with an IC50 of 8.59 ± 0.3 μg mL−1. Moreover, for FRF a safety profile was established with a CC50 = 24.5 ± 1.9 μg mL−1 for human use. Encapsulation in the form of a liposomal formulation resulted in a significant improvement in human coronavirus inhibition by 2.25-fold, when compared to the un-encapsulated fraction, which approached that of the FDA-approved anti-COVID-19 agent, remdesivir. The in vitro anti-COVID-19 effects of plant extracts in general and meswak specifically have not been studied before. This study provides a basis for the development of antiviral for SARS-CoV-2, which if administered during the outbreak, could help to protect people from person-to-person transmission, prevent disease progression, and limit viral load. Clinical studies will be continued for further investigation of the anti-viral and the anti-inflammatory effects of FRF-Lip on the treatment of COVID-19 patients.
Conflicts of interest
The authors declare that there is no conflict of interest.
Supplementary Material
Acknowledgments
The authors thank Prof. Esam Rashwan, Head of the Confirmatory Diagnostic Unit, VACSERA, Egypt, for carrying out the in vitro study on the human coronavirus.
Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ra00142f
References
- Zhang X. Tan Y. Ling Y. Lu G. Liu F. Yi Z. Jia X. Wu M. Shi B. Xu S. Nature. 2020:1–7. doi: 10.1038/s41586-020-2355-0. [DOI] [PubMed] [Google Scholar]
- Luan J. Lu Y. Jin X. Zhang L. Biochem. Biophys. Res. Commun. 2020;526:165–169. doi: 10.1016/j.bbrc.2020.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V. Jung Y.-S. Liang P.-H. Expert Opin. Ther. Pat. 2013;23:1337–1348. doi: 10.1517/13543776.2013.823159. [DOI] [PubMed] [Google Scholar]
- Alexpandi R. De Mesquita J. F. Pandian S. K. Ravi A. V. Front. Microbiol. 2020;11:1796–1810. doi: 10.3389/fmicb.2020.01796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra M. A. Jones S. J. Astell C. R. Holt R. A. Brooks-Wilson A. Butterfield Y. S. Khattra J. Asano J. K. Barber S. A. Chan S. Y. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- Ferraz W. R. Gomes R. A. S Novaes A. L. Goulart Trossini G. H. Future Med. Chem. 2020;12:1815–1828. doi: 10.4155/fmc-2020-0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh A. K. Brindisi M. Shahabi D. Chapman M. E. Mesecar A. D. ChemMedChem. 2020;15(11):907–932. doi: 10.1002/cmdc.202000223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagar M. Ahmed H. A. Aljohani G. Alhaddad O. A. Int. J. Mol. Sci. 2020;21:3922–3934. doi: 10.3390/ijms21113922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentile D. Patamia V. Scala A. Sciortino M. T. Piperno A. Rescifina A. Mar. Drugs. 2020;18:225–243. doi: 10.3390/md18040225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olubiyi O. O. Olagunju M. Keutmann M. Loschwitz J. Strodel B. Molecules. 2020;25:3193–3213. doi: 10.3390/molecules25143193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J. Tian Z. Yang X. BioSci. Trends. 2020;14:72–73. doi: 10.5582/bst.2020.01047. [DOI] [PubMed] [Google Scholar]
- Chen L. Xiong J. Bao L. Shi Y. Lancet Infect. Dis. 2020;20:398–400. doi: 10.1016/S1473-3099(20)30141-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautret P. Lagier J.-C. Parola P. Meddeb L. Mailhe M. Doudier B. Courjon J. Giordanengo V. Vieira V. E. Dupont H. T. Int. J. Antimicrob. Agents. 2020;56:105949. doi: 10.1016/j.ijantimicag.2020.105949. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Owis A. I. El-Hawary M. S. El Amir D. Aly O. M. Abdelmohsen U. R. Kamel M. S. RSC Adv. 2020;10:19570–19575. doi: 10.1039/D0RA03582C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aumeeruddy M. Z. Zengin G. Mahomoodally M. F. J. Ethnopharmacol. 2018;213:409–444. doi: 10.1016/j.jep.2017.11.030. [DOI] [PubMed] [Google Scholar]
- Taha M. Y. RDENTJ. 2008;8:50–55. doi: 10.33899/rden.2007.9041. [DOI] [Google Scholar]
- Ali A. Assaf M. El-Shanawany M. Kamel M. Bull. Pharm. Sci., Assiut Univ. 1997;20:181–186. doi: 10.21608/bfsa.1997.68759. [DOI] [Google Scholar]
- Herald T. J. Gadgil P. Tilley M. J. Sci. Food Agric. 2012;92:2326–2331. doi: 10.1002/jsfa.5633. [DOI] [PubMed] [Google Scholar]
- Shang J. Ye G. Shi K. Wan Y. Luo C. Aihara H. Geng Q. Auerbach A. Li F. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Refaat H. Naguib Y. W. Elsayed M. Sarhan H. A. Alaaeldin E. Pharmaceutics. 2019;11:558. doi: 10.3390/pharmaceutics11110558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosmann T. J. Immunol. Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- Günther S. Asper M. Röser C. Luna L. K. Drosten C. Becker-Ziaja B. Borowski P. Chen H.-M. Hosmane R. S. Antiviral Res. 2004;63:209–215. doi: 10.1016/j.antiviral.2004.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu A. Sarkar A. Maulik U. Sci. Rep. 2020;10:1–15. doi: 10.1038/s41598-019-56847-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouffouk C. Mouffouk S. Mouffouk S. Hambaba L. Haba H. Eur. J. Pharmacol. 2021;891:173759. doi: 10.1016/j.ejphar.2020.173759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey P. Rane J. S. Chatterjee A. Kumar A. Khan R. Prakash A. Ray S. J. Biomol. Struct. Dyn. 2020:1–11. doi: 10.1080/07391102.2020.1794969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson R. W. Brown R. L. Benjamin L. Nortley R. Wiethoff S. Bharucha T. Jayaseelan D. L. Kumar G. Raftopoulos R. E. Zambreanu L. Brain. 2020;143:3104–3120. doi: 10.1093/brain/awaa240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotfis K. Williams Roberson S. Wilson J. E. Dabrowski W. Pun B. T. Ely E. W. Crit. Care. 2020;24:1–9. doi: 10.1186/s13054-020-02882-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriguchi T. Harii N. Goto J. Harada D. Sugawara H. Takamino J. Ueno M. Sakata H. Kondo K. Myose N. Int. J. Infect. Dis. 2020;94:55–58. doi: 10.1016/j.ijid.2020.03.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanin L. Saraceno G. Panciani P. P. Renisi G. Signorini L. Migliorati K. Fontanella M. M. Acta Neurochir. 2020;162:1491–1494. doi: 10.1007/s00701-020-04374-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varatharaj A. Thomas N. Ellul M. A. Davies N. W. Pollak T. A. Tenorio E. L. Sultan M. Easton A. Breen G. Zandi M. Lancet Psychiatry. 2020;7:875–882. doi: 10.1016/S2215-0366(20)30287-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellul M. A. Benjamin L. Singh B. Lant S. Michael B. D. Easton A. Kneen R. Defres S. Sejvar J. Solomon T. Lancet Neurol. 2020;19:767–783. doi: 10.1016/S1474-4422(20)30221-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani J. S. Johnson J. B. Steel J. C. Broszczak D. A. Neilsen P. M. Walsh K. B. Naiker M. Virus Res. 2020;284:197989–198005. doi: 10.1016/j.virusres.2020.197989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelho C. Gallo G. Campos C. B. Hardy L. Würtele M. PLoS One. 2020;15:e0240079. doi: 10.1371/journal.pone.0240079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherrak S. A. Merzouk H. Mokhtari-Soulimane N. PLoS One. 2020;15:e0240653. doi: 10.1371/journal.pone.0240653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayed A. M. Khattab A. R. AboulMagd A. M. Hassan H. M. Rateb M. E. Zaid H. Abdelmohsen U. R. RSC Adv. 2020;10:19790–19802. doi: 10.1039/D0RA04199H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbing J. Phelan A. Griffin I. Tucker C. Oechsle O. Smith D. Richardson P. Lancet Infect. Dis. 2020;20:400–402. doi: 10.1016/S1473-3099(20)30132-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim A. Y. El-Gengaihi S. E. Motawea H. M. Sleem A. A. Not. Sci. Biol. 2011;3:22–28. doi: 10.15835/nsb346378. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.