Abstract
“Norwalk-like viruses” (NLVs) are the most common cause of outbreaks of nonbacterial gastroenteritis worldwide. To date, the method most widely used for typing of NLV strains is sequencing and subsequent phylogenetic analysis of reverse transcription (RT)-PCR products, which has revealed the existence of stable distinct lineages (genotypes). This typing method is rather costly, not routinely used in clinical laboratories, and not very suitable for the analysis of large numbers of samples. Therefore, we have developed a rapid and simple method for genotyping of NLVs. The method, designated reverse line blot hybridization, is based on the nucleotide divergence of a region of the gene for RNA polymerase which can be used to classify NLVs into genotypes. NLV RNA was amplified by RT-PCR and then hybridized to 18 different membrane-bound oligonucleotides that were able to discriminate among 13 NLV genotypes. Application of the method to a panel of 132 positive stool samples from 34 outbreaks and 20 sporadic cases of gastroenteritis collected in a 6-year period (1994 to 1999) resulted in successful genotyping of 124 samples (94%), as confirmed by phylogenetic analysis. The nucleotide sequences of the remaning eight strains (6%) from three outbreaks did not cluster with the known NLV genotypes. Phylogenetic analysis of the complete and partial open reading frame 2 (capsid gene) sequences of these strains revealed the existence of one novel genotype (Alphatron) and one potentially novel genotype (Amsterdam). This novel method, which allows simultaneous detection and genotyping of NLVs, is useful in the diagnosis and typing of NLVs obtained from outbreaks and in large-scale epidemiological studies.
In recent years, “Norwalk-like viruses” (NLVs), members of the family Caliciviridae (14), have emerged as the single most common cause of outbreaks of acute nonbacterial gastroenteritis in people of all age groups (4, 9, 33). These outbreaks are most frequently recognized in adults in semiclosed institutions such as hospitals and nursing homes, where the infection rate is high due to extensive person-to-person spread (2, 9, 33). To date, the most widely used detection assays for NLVs are electron microscopy and sensitive generic reverse transcription (RT)-PCR assays with the RNA polymerase gene (pol) as the target (1, 6, 25, 32). Since NLVs, also known as small round-structured viruses, cannot be grown in cell culture, no serotype classification system exists. Therefore, until antigenic typing assays based on recombinant virus-like particles become available, genotyping of NLVs will be based on the sequences of complete capsid (open reading frame 2 [ORF2]) genes (8, 13). NLVs have a surprisingly high level of genetic diversity and can be phylogenetically divided into two genogroups (GG) that differ from each other by up to 50% of the nucleotides.
On the basis of complete ORF2 gene sequences, at least 15 different NLV capsid types have been described, 7 in GGI and 8 in GGII (8). In a previous study, we defined genotypes as capsid types having >80% amino acid similarity. Furthermore, genotypes should be represented by at least two different strains of which the branches in the dendrogram were supported by high bootstrap values regardless of the method used to determine phylogeny (34). Using these criteria, we recognized at the start of this study at least 13 genotypes (Table 1, genotypes 1 to 13) and two capsid types (Seacroft and Wortley) represented by a single strain (34). The virus position in phylogenetic trees is generally consistent across the ORF1, ORF2, and ORF3 genes, with few exceptions that are thought to represent recombinant strains (27, 34). Therefore, typing of strains in large-scale studies is now achieved by amplifying a relatively small region of ORF1 and subsequent sequencing and phylogenetic analysis of the RT-PCR products.
TABLE 1.
Division of NLVs into genotypes
| ORF1 cluster | Genotype
|
Genotype division based on sequence identity in:
|
Reference(s) | ||||
|---|---|---|---|---|---|---|---|
| Name | No. | Complete ORF2a | ORF1b | 5′ part of ORF2c | ORF3c | ||
| Norwalk | Norwalk | 1 | +d | + | + | + | 8, 34 |
| Desert Shield | Desert Shield | 2 | + | + | + | + | 8, 34 |
| Southampton | Southampton | 3 | + | + | + | + | 8, 34 |
| Queens Arms | Queens Arms | 4 | + | + | + | + | 8, 34 |
| Musgrove | Musgrove | 5 | + | + | + | + | 8, 34 |
| Winchester | Winchester | 6 | + | + | + | − | 8, 34 |
| Sindlesham | Sindlesham | 7 | + | + | + | − | 8, 34 |
| Hawaii | Hawaii | 8 | + | + | + | + | 8, 34 |
| Mexico | Mexico | 9 | + | + | + | + | 8, 34 |
| Lordsdale | Lordsdale | 10 | + | + | + | + | 8, 34 |
| Melksham | Melksham | 11 | + | + | + | − | 8, 34 |
| Hillingdon | Hillingdon | 12 | + | + | + | + | 8, 34 |
| Leeds | Leeds | 13 | + | + | + | − | 8, 34 |
| Alphatron | Alphatron | 14 | + | + | + | + | This study |
| Wortley | Hawaii | 8 | − | + | − | − | 8, 34 |
| Rotterdam | Mexico | 9 | − | + | − | − | This study |
| Amsterdam | —e | + | + | + | + | This study | |
| Seacroft | — | + | + | + | + | 8, 34 | |
>80% of the amino acid sequence (i.e., genotypes). Complete capsid sequences were obtained from GenBank (Norwalk virus, M87661; Desert Shield, U04469; Southampton, L07418; Hawaii, U07611; Mexico, U22498; Lordsdale, X86557; Melksham, X81879) and reference 8.
85% of the nucleotides (i.e., ORF1 clusters) and >90% for GGII strains (34).
>80% of the nucleotides.
+ indicates <80% amino acid (or nucleotide) identity with other known strains; − means >80% identity.
NG, No genotype has yet been determined because the cluster comprises only one strain.
In this report, we describe the development of a rapid, cheap, and simple method for the simultaneous detection and typing of NLVs. As a target for the generic RT-PCR (33), we used a region of the pol gene and determined the genotype by hybridization of the RT-PCR product to multiple genotype-specific oligonucleotide probes immobilized on a nylon membrane (36). This method, designated reverse line blot hybridization (RLB), allows easy detection and genotyping of NLV strains from outbreaks (OBs) and sporadic cases without sequencing of the PCR product. As transmission of NLVs is not limited to countries, discrimination between strains with the described method may form the basis of an international laboratory surveillance network for NLVs.
MATERIALS AND METHODS
Clinical specimens.
Stool samples (n = 112) from 34 NLV-positive OBs of gastroenteritis in The Netherlands from 1994 to 1999 were used throughout this study. In addition, NLV-positive stool samples (n = 20) from sporadic cases were used that had been collected from a physician-based case-control study in 1996 to 1999, the details of which will be described elsewhere. To determine the sensitivity of the RLB assay, strains representing the 13 different NLV genotypes (34) and two unclassified strains (Rotterdam and Wortley) that were present either as stool samples (OB98-14 [Southampton], OB97-26 [Desert Shield], OB97-1 [Queens Arms], OB94-9 [Hawaii], OB97-2 [Mexico], OB98-11 [Lordsdale], OB94-1 [Hillingdon], OB97-6 [Rotterdam], and OB96-67 [Leeds]) or as cloned DNAs (Norwalk L-52 [Norwalk], LWymondley/95 [Winchester], Butlins/96 [Musgrove], Mikkeli/97 [Sindlesham], sewage/1-3 [Melksham], and SP630/90 [Wortley]) were used. Stool samples had been screened for the presence of NLVs using RT-PCR as previously described (6, 33). All stool samples had been stored at 4°C.
Extraction of viral RNA.
Viral RNA was extracted from a 10% stool suspension by binding to glass beads as previously described (32) and eluted in 20 μl of RNase-free water (Sigma Chemical Co., St. Louis, Mo.) containing 5 U of RNase inhibitor (Amersham, 's-Hertogenbosch, The Netherlands) and 3 mM dithiothreitol. RNA was either used directly in the RT-PCR or stored at −70°C.
Detection of NLVs by RT-PCR.
RT-PCR was performed with the generic primer pair JV12-JV13, which generates a 326-bp product (33). For RT, 5 μl of the extracted viral RNA was mixed with 4 μl of 1 μM 5′-biotin-labeled JV13 and heated for 2 min at 90°C. After quenching on ice for 2 min, 6 μl of RT buffer was added. The RT reaction was performed with a 15-μl reaction mixture consisting of 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 3.0 mM MgCl2, 1 mM each deoxynucleoside triphosphate (Boehringer Mannheim, Almere, The Netherlands), and 5 U of avian myeloblastoma virus reverse transcriptase (Promega, Leiden, The Netherlands). Reaction mixtures were incubated for 60 min at 42°C. After heat denaturation at 99°C for 5 min, 5 μl of the RT mixture was added to 45 μl of PCR mixture containing 10 mM Tris-HCl (pH 9.2), 75 mM KCl, 1.5 mM MgCl2, 0.2 mM deoxynucleoside triphosphates, 2.5 U of ampliTaq (Perkin Elmer, Nieuwerkerk a/d IJssel, The Netherlands) and each primer (JV12 and 5′-biotin-labeled JV13) at 0.3 μM. The mixture was overlaid with mineral oil (Sigma), and the samples were denatured for 3 min at 94°C and subjected to 40 cycles of 94°C for 1 min, 37°C for 1.5 min, and 74°C for 1 min. A final extension of 7 min at 74°C was then performed.
Design of GG- and genotype-specific oligonucleotides.
The ∼300 pol sequences available in the National Institute of Public Health and the Environment database, including strains from The Netherlands (32, 33, 34) and the United Kingdom (5, 6, 7, 28) and representing 15 different NLV capsid types (8), were aligned with 17 GGI and GGII NLV sequences submitted to GenBank using the GeneWorks software (Intelligenetics, Mountain View, Calif.). A region of 160 nt upstream of the JV13 primer region was used for selection of GG- and genotype-specific oligonucleotides. GGI and GGII sequences were aligned separately and used to calculate pairwise distances of the sequences. The probes were selected based on the following criteria: length, 18 to 20 nucleotides (nt); melting temperature at 54 to 60°C; and more than three mismatches between the probe and other genotypes, ideally evenly distributed over the oligonucleotide sequence. All oligonucleotides were analyzed with the OLIGO 5.0 software (National Biosciences, Inc., Plymouth, Minn.) for possible hairpins. The probes were 5′ hexylamine labeled (Isogen, Maarssenbroek, The Netherlands).
RLB.
Each amplified biotinylated product was hybridized to a set of 18 immobilized oligonucleotide probes (Table 2), each corresponding to a GG (I or II) or a genotype. The oligonucleotides were covalently bound to a nylon membrane (Biodyne C; Pall Biosupport, Portsmouth, Cambridge, United Kingdom) by the 5′ hexylamino group as previously described (18). Briefly, the carboxyl groups on the membrane were activated by incubation for 15 min in 16% (wt/vol) 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (Sigma). After being rinsed with water, the membrane was placed in a miniblotter system (MN45; Immunetics, Cambridge, Mass.). The slots were filled in parallel with 150 μl of different twofold dilutions of the 5′-hexylamine-labeled oligonucleotide (0.156 to 1.25 μM diluted in freshly prepared 500 mM NaHCO3 [pH 8.4]). The oligonucleotides were immobilized on the membrane through their amino group, and after 1 min of incubation at room temperature, the excess solution was aspirated and the membrane was removed from the miniblotter. The remaining active esters on the membrane were hydrolyzed by incubation in 0.1 M NaOH for 10 min. After a rinse with water, the membrane was washed for 10 min at 60°C in 2× SSPE (360 mM NaCl, 20 mM NaH2PO4, 2 mM EDTA [pH 7.2]) with 0.1% sodium dodecyl sulfate (SDS) (BDH, Poole, United Kingdom). After a rinse in 2× SSPE, the membrane was used immediately or stored at 4°C in a solution of 20 mM EDTA. Prior to use in hybridization, the membrane was washed for 5 min in 2× SSPE–0.1% SDS, placed in the miniblotter, and rotated 90° from the previous position, resulting in a perpendicular position of the slots on the lines which contained the oligonucleotides. The slots were filled with 150 μl of heat-denatured PCR products (5 μl of the PCR product diluted in 150 μl of 2× SSPE–0.1% SDS, heated at 99°C for 10 min, and chilled on ice) and incubated for 60 min at an empirically determined optimal hybridization temperature.
TABLE 2.
Sequences of the GG- and genotype-specific oligonucleotides used in this study
| Target specificity | GG | Nucleotide sequencea | Genomic locationb |
|---|---|---|---|
| GGIa | I | 5′-ATG GAT GTA GGT GAY TAY GT | 4685–4704 |
| GGIb | I | 5′-ATG GAY GTT GGY GAY TAT GT | 4685–4704 |
| GGII | II | 5′-GGA AYT CCA TYR CMC AYT G | 4494–4512 |
| Norwalk | I | 5′-GAG GCC ACT GGT TTA TCA C | 4800–4818 |
| Desert Shield | I | 5′-CAC TGG ATC CTA ACT CTA TG | 4770–4789 |
| Southampton | I | 5′-TGT CAT AAG AGT GAA GGG AG | 4703–4722 |
| Queens Arms | I | 5′-CAC MTT GTG TGC CAT GTC T | 4781–4799 |
| Musgrove | I | 5′-GCT CTC TCC TGA TGT AAT AC | 4811–4830 |
| Winchester | I | 5′-TTT ACA CTC TGT GTG CTC TG | 4777–4796 |
| Sindlesham | I | 5′-CGG CTT TCC ATG CAC ATC T | 4733–4751 |
| Hawaii | II | 5′-ACG AGG GTC TCC CAT CCG | 4455–4472 |
| Mexico | II | 5′-TTC TGA AGT GAC AGG ACT AG | 4534–4553 |
| Lordsdale | II | 5′-TAC AAA CTT GTC CCC TGA CA | 4543–4562 |
| Melksham | II | 5′-TGA CAT TGT GCA GGC CAA C | 4558–4576 |
| Hillingdon | II | 5′-GAA ACC ACC ACC CTG TCT C | 4538–4556 |
| Leeds | II | 5′-CTY TCA CCA GAT GTT GTC CA | 4550–4569 |
| Wortley | II | 5′-GGT TAC AAA TCT ATC CCC TA | 4540–4559 |
| Rotterdam | II | 5′-CAC TCT CTG AAA CTA CAA AT | 4530–4549 |
After hybridization, the membrane was washed twice for 10 min (each time) at the hybridization temperature with 2× SSPE–0.5% SDS prewarmed at 42°C and incubated in 10 ml of streptavidin-peroxidase conjugate (Boehringer Mannheim) diluted 1/4,000 in 2× SSPE–0.5% SDS for 45 min at 42°C. The membrane was washed twice with 2× SSPE–0.5% SDS at 42°C and with 2× SSPE at room temperature. The bound PCR products were detected by chemiluminescence assay using ECL detection liquid (Amersham) and visualized by exposure of the blot for 15 min to an X-ray film (Hyperfilm; Amersham). For repeated use, the membranes were stripped by being washed twice, for 30 min each time, in 0.1% SDS at 80°C. After incubation for 15 min at room temperature in 20 mM EDTA solution, the membranes were sealed and stored at 4°C until further use. In our laboratory, these membranes have been stored for up to 1 year and stripped at least 15 times without detectable loss of sensitivity.
Sequence analysis and data analysis.
To confirm the RLB typing, NLV-positive RT-PCR products were sequenced using an ABI PRISM BigDye Terminator Cycle Sequencing Reaction Kit (Perkin Elmer) on an automated sequencer (Applied Biosystems model 373; Applied Biosystems, Nieuwerkerk a/d IJssel, The Netherlands). DNA sequences were edited using Seq Ed (version 1.03; Applied Biosystems), converted to the Geneworks format (version 2.45; Intelligenetics), and aligned using the unweighted pair group method with arithmetic mean (26). Multiple-sequence alignments were subsequently imported into the Treecon software package (31) for distance calculation using the Jukes and Cantor correction for evolutionary rate (16). A dendrogram was constructed using the unweighted pair group method with arithmetic mean, and the confidence values of the internal lineages within the dendrogram were assessed by repeating the analysis after resampling of the data to reduce the influence of sequence input order (bootstrapping, 100 rounds).
ORF2 and ORF3 sequencing.
The distinctness of the lineages (genotypes) had been confirmed by sequencing and clustering in phylogenetic trees of ORF1, ORF2, and ORF3 for all clusters (34), except for the Rotterdam strains. Since in ORF1 these viruses clustered separately from other GGII NLVs (>10% of the nucleotides), we sequenced the 5′ end of ORF2 and part of ORF3 from two strains belonging to this Rotterdam cluster. In addition, the complete capsid sequences of two strains representing OB98-15 and OB98-18 that were nontypeable by RLB were determined. The primers and RT-PCR protocols used to amplify partial or complete NLV capsid genes and parts of ORF3 have been described elsewhere (8, 34).
Nucleotide sequence accession numbers.
Sequence data of the complete Alphatron and Amsterdam capsid genes have been submitted to GenBank and assigned accession numbers AF195847 and AF195848, respectively.
RESULTS
Development of genotype- and GG-specific probes.
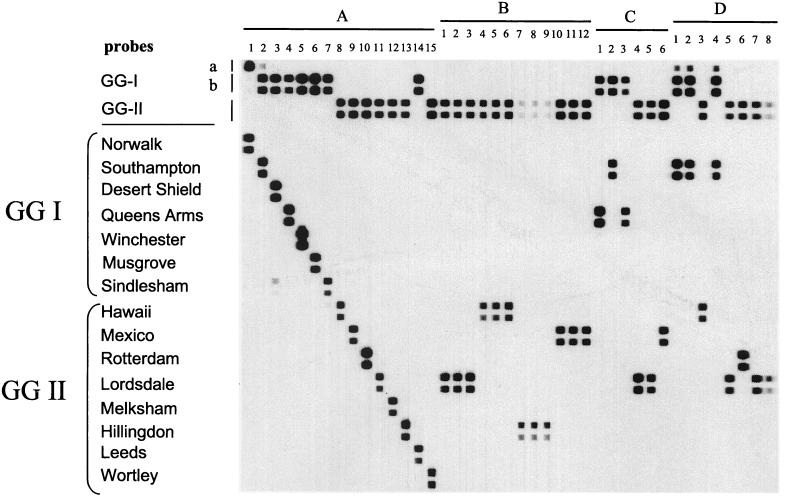
For the selection of probes, we used a GGI alignment consisting of 18 pol sequences covering all genotypes. For GGII, 27 Pol sequences were used to create a multiple-sequence alignment, again including representatives of each (putative) genotype. For GGI viruses, areas on which to construct a consensus probe could not be identified within the 145-nt region of the pol gene used for phylogenetic analysis of NLVs and could only be developed when at least a 160-nt fragment upstream of the JV13 primer was used. This probe (GGIb; Fig. 1, rows 2 and 3) detected all of the GGI viruses in this study, excluding Norwalk virus. For viruses within the Norwalk virus genotype, a second GGI probe was developed (GGIa; Fig. 1, row 1) based on the same target area in the alignment. All of the GGI NLVs tested in this study reacted with one of the GGI probes. In addition, viruses of the Leeds genotype belonging to GGII NLVs also reacted with the GGIb probe (Fig. 1A, lane 14) although they clearly clustered with the GGII viruses based on complete capsid sequences. For GGII viruses, a single probe could be developed which detects all of the known lineages within the GGII NLVs except for the Leeds genotype, which reacts with the GGIb probe. For the differentiation of the 13 established NLV genotypes, a single probe for each genotype could be developed (Fig. 1A, lanes 1 to 9 and 11 to 14). In addition, type-specific probes for the Wortley and Rotterdam genotypes were developed (Fig. 1A, lanes 10 and 15). Because no Seacroft-positive sample was available for analysis, a specific probe for this potential genotype could not be evaluated.
FIG. 1.
Detection and simultaneous typing of NLV strains from OBs (20 strains in panels B and D) and sporadic cases (6 strains in panel C) of gastroenteritis in The Netherlands. The probes used to detect GGIa and GGIb strains, GGII strains, 13 different genotypes (Table 1, genotypes 1 to 13), and strains Wortley and Rotterdam are indicated. For reference, the hybridization patterns of these 15 strains are shown in panel A. In panel B, strains belonging to the same OB have identical hybridization patterns (lanes 1 to 3, 4 to 6, 7 to 9, and 10 to 12), whereas panel D shows the hybridization patterns of two OBs in which two different strains were detected (lanes 1 to 4 and 5 to 8). Hybridization patterns of six different sporadic cases are shown in panel C.
Optimal probe dilutions were determined empirically by binding twofold dilutions of each individual probe (one dilution for the GGIa probe), starting at a concentration of 1.25 μM, to a membrane. Except for the Leeds and Rotterdam probes (optimal concentration, 0.156 μM), the optimal concentration was 0.625 μM. To optimize the hybridization temperature, three membranes to which the same panel of probes was bound were hybridized with nine biotin-labeled RT-PCR products representing six different genotypes at three hybridization temperatures (45, 50, and 55°C). At 42°C, all of the samples reacted with the corresponding GG- and genotype-specific probe. However, cross-reactions of several RT-PCR products with heterologous probes were observed. These cross-reactions were low in level or absent from the membranes incubated at 50 and 55°C, but at the latter temperature, the sensitivity of the homologous reactions dropped significantly (data not shown). In the present format, we use membranes with all of the probes (except the GGIa probe) in two twofold dilutions and the hybridization is done at 50°C (Fig. 1).
Since Rotterdam viruses segregated separately from the other GGII NLVs in ORF1, (>10% of the nucleotides), these viruses formed a potential distinct genotype, which required confirmation by sequencing of the 5′ end of ORF2 of these viruses (see below).
Specificity, detection limit, and sensitivity of RLB.
The specificity of the RT-PCR assay has been determined previously; no products were detected when RNAs from 16 different enteric RNA viruses were subjected to the RT-PCR (32). The detection limit, which has previously been determined to be 3 to 30 RNA-containing particles (19), was not different when a biotin-labeled primer was used. The sensitivity of the NLV RLB assay was determined utilizing 15 NLV-positive fecal samples representing 13 different genotypes and two strains possibly representing new genotypes (Rotterdam and Wortley) (Fig. 1A, lanes 1 to 15). They were amplified using the generic NLV RT-PCR with one 5′-biotinylated primer (JV13). All 15 samples could be confirmed as NLV while reacting with at least one of the GG-specific probes. Seven samples reacted with the GGIb probe (Fig. 1A, lanes 2 to 7 and 14), and seven reacted with the GGII probe (Fig. 1A, lanes 8 to 13 and 15). As expected, Norwalk virus reacted with the GGIa probe (Fig. 1A, lane 1). For genotyping, each sample reacted with the homologous probe (Fig. 1A, lanes 1 to 15). Some low-level cross-reactivity was observed, both between the GGIa and GGIb probes (Fig. 1A, lanes 1 and 2, and D, lanes 1, 2, and 4) and between different genotypes (Fig. 1A, lane 3 between the Desert Shield and Sindlesham genotypes and lane 7 between the Sindlesham and Hawaii genotypes). In all cases, however, the strongest hybridization signal was obtained with the homologous probe.
The detection limit for NLV RNA using the RLB method was compared with that of the Southern hybridization used in the routine NLV RT-PCR (33). RNAs from four different NLV strains (two GGI [Southampton and Queens Arms genotypes] and two GGII [Mexico and Lordsdale genotypes]) were tested in 10-fold serial dilutions with both assays. RLB was equally sensitive for genogrouping but slightly less sensitive (maximally, 10-fold) than Southern hybridization for genotyping (data not shown).
Evaluation of the RLB assay.
One hundred thirty-two NLV-positive stool samples from 34 OBs (112 samples) and 20 sporadic cases were used for validation of the RLB assay. Using the NLV RT-PCR followed by RLB, we were able to confirm the presence of NLV in all of the 132 samples analyzed (positive for one of the GG-specific probes). Of the 132 samples positive by GG probe, 124 (94%) could be genotyped. A selection of samples (n = 26) is shown in Fig. 1B, C, and D). Panel B contains samples from four OBs (three samples each) in which all of the specimens had a strain of the same genotype (Fig. 1B, lanes 1 to 12). In addition, in our collection (34 OBs), 2 OBs (four samples each) were found to contain two different NLV genotypes. In one OB (Fig. 1D, lanes 1 to 4), three samples tested positive for a strain with the Southampton genotype while one sample (Fig. 1D, lane 3) clearly had the Hawaii genotype. The other OB consisted of three Lordsdale-like viruses and one virus in the Rotterdam cluster (Fig. 1D, lane 6). Panel C shows samples from sporadic cases of gastroenteritis that were typed into three GGI and three GGII NLV strains (Fig. 1C, lanes 1 to 6).
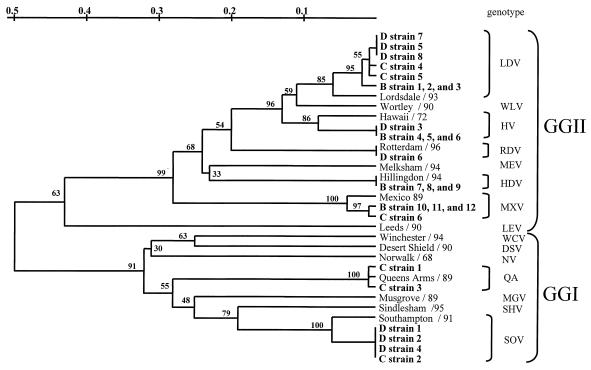
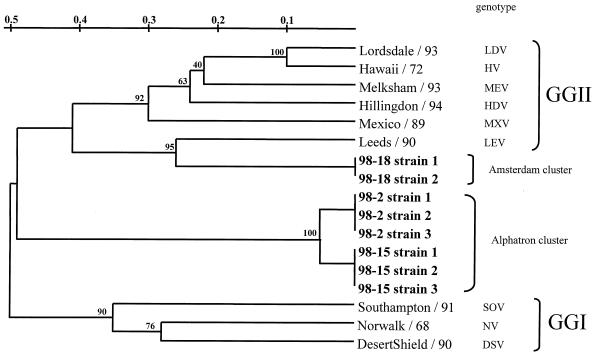
To confirm the clustering of strains typed by RLB, the corresponding RT-PCR products of all 132 stool samples were sequenced. These sequences were analyzed for their phylogenetic relationship in the 145-nt region of ORF1. In addition, strain sequences from a previous study (34) were added to the analysis. The nucleotide sequence identity ranged from 90 to 100% for viruses with the Alphatron genotype to 98 to 100% for viruses with the Southampton genotype, compared with the reference strains indicated in Table 3. All of the strains were in GGI and GGII (Fig. 2). The RLB results of 124 strains were all confirmed by sequencing, as shown in Fig. 2 for the 26 field strains in Fig. 1B, C, and D. Sequencing of the eight nontypeable strains and subsequent phylogenetic analysis revealed that these strains did not cluster with any of the known genotypes and therefore could represent potentially new genotypes (Fig. 3). Note that the sequence diversity of strains from OB98-2 and OB98-15 was approximately 50% with GGI and GGII viruses (Fig. 3).
TABLE 3.
Range of nucleotide sequence identities of NLV strains analyzed in this study
| No. of strainsa | Genotype with closest sequence identity | Range of nucleotide sequence identity (%) with reference strain | Reference strain (reference) |
|---|---|---|---|
| 23 | Lordsdale | 97–100 | Grimsby (34) |
| 11 | Mexico | 96–100 | Mexico (34) |
| 4 | Hawaii | 97–100 | Hawaii (34) |
| 5 | Rotterdam | 97–100 | Rotterdam (this study) |
| 2 | Hillingdon | 99–100 | Hillingdon (34) |
| 3 | Queens Arms | 96–100 | Queens Arms (34) |
| 3 | Southampton | 98–100 | Southampton (20) |
| 1 | Desert Shield | 98 | Desert Shield (21) |
| 1 | Leeds | 97 | Leeds (34) |
| 1 | Amsterdamb | 100 | OB98-18 (this study) |
| 2 | Alphatronb | 90–100 | OB98-2 (this study) |
Strains within an OB with identical sequences are counted as a single strain.
Potential novel NLV genotype.
FIG. 2.
Phylogenetic relationships based on a 145-bp region of the RNA polymerase-encoding gene showing the relationships among representative strains of NLV genotypes from GGI (Norwalk/68 [NV], Southampton/91 [SOV], Desert Shield/90 [DSV], Queens Arms/89 [QA], Sindlesham/95 [SHV], Musgrove/89 [MGV], and Winchester/94 [WCV]) and GGII (Lordsdale/93 [LDV], Mexico/89 [MXV], Melksham/94 [MEV], Hawaii/72 [HV], Hillingdon/94 [HDV], Leeds/90 [LEV], viruses in the Rotterdam cluster [RDV], and Wortley/90 [WLV]), as well as the 26 strains previously typed by the RLB assay shown in Fig. 1 (in bold). Designations of RLB-typed strains refer to lanes in Fig. 1. Bootstrap values of the internal nodes are indicated.
FIG. 3.
Phylogenetic relationships based on a 145-bp region of the RNA polymerase-encoding gene showing the relationships among NLV genotypes from GGI (Norwalk/68 [NV], Southampton/91 [SOV], and Desert Shield/90 [DSV]), GGII (Lordsdale/93 [LDV], Mexico/89 [MXV], Melksham/93 [MEV], Hawaii/72 [HV], Hillingdon/94 [HDV], and Leeds/90 [LEV]), and eight strains previously nontypeable by RLB (in bold). Bootstrap values of the internal nodes are indicated.
ORF2 and ORF3 sequencing.
To identify whether viruses from the Rotterdam cluster and the clusters of the nontyped RLB strains (Fig. 3) represent new genotypes, the nucleotide sequence of the complete capsid gene (Alphatron and Amsterdam) or the 5′ end of the capsid gene (Rotterdam) of one strain of each cluster was determined. In addition, partial ORF3 sequences of these strains were determined and pairwise distances from other ORF3 sequences were calculated (Table 3). To our surprise, the 5′ end of the capsid region of the Rotterdam strain was almost identical to that of the Mexico virus (96% amino acid identity). The complete capsid sequences of nontypeable strains from OB98-15 (Alphatron) and OB98-18 (Amsterdam) had maximum amino acid identities of 52 and 76%, respectively, with all of the NLV capsid sequences in the Genank and National Institute of Public Health and the Environment databases and therefore represent potential new genotypes.
DISCUSSION
To date, the method most widely used for typing of NLV strains is sequencing and subsequent phylogenetic analysis of RT-PCR products (6, 27, 33). This typing method is rather costly, not routinely used in clinical laboratories, and not very suitable for the analysis of large numbers of samples. Therefore, we have adapted a method previously developed for the detection and typing of bacteria (17, 29). As the target region for RT-PCR and typing, a region of the pol gene was chosen that is used in most diagnostic NLV RT-PCR assays. Important for optimal performance of this RLB assay is the size of the RT-PCR product that is generated. GG- and genotype-specific oligonucleotide probes were developed based on a 160-nt fragment and immobilized to a membrane. This length was necessary only to find areas on which to construct probes (e.g., GGIa and GGIb probes) to detect and discriminate all of the strains used in this study. Biotin-labeled RT-PCR products were generated by a generic NLV RT-PCR (33) and hybridized to the membrane. Using this RLB assay, we were able to detect all and differentiate 94% of the strains in this study. In our laboratory, we now use the RLB assay for the routine detection and differentiation of NLV in stool samples obtained from large-scale epidemiological studies and from OB investigations. In contrast to sequencing, RLB can never resolve if strains within an OB are identical. However, common-source exposure can rapidly be excluded as different genotypes can easily be distinguished; allowing a reduction of the number of samples to be sequenced. All 132 NLV-positive strains were sequenced for confirmation and to identify potentially novel genotypes. In addition, the assay has been introduced in several diagnostic laboratories in different European countries to evaluate its performance in routine diagnostic laboratories.
Because of the genetic diversity in the target region of GGI, it was not possible to develop a probe that detects all GGI strains. Therefore, two probes were developed of which one is specific for strains related to Norwalk virus and the other is specific for all of the other GGI strains in our database. All of the strains that, by phylogenetic analysis of pol sequences, belong to the GGI viruses reacted with one of the two GGI probes. This includes the Leeds/Venlo-like strains, which are equidistant from the GGI and GGII strains in the pol region (33) but, on the basis of the capsid gene sequences, clearly belong to GGII (34). Similar strains (designated P1-B strains) have been described by other investigators (1, 27). Although the Leeds probe is able to detect most P1-B strains, these strains cannot be correctly genogrouped by the present probes and based on the expanding number of available NLV sequences, efforts should be undertaken to refine the current probes for viruses belonging to this cluster. GGII viruses that cluster differently when comparing phylogenetic trees based on the pol fragment with those based on the complete or partial capsid gene have been described (12, 13, 34). While the viruses in the Rotterdam cluster were clearly distinct from viruses in the Mexico cluster in the pol region, the capsid gene showed a high level of similarity to the capsid gene of Mexico virus. Viruses in the Rotterdam cluster were observed in several temporally different OBs, which means that they are viable viruses that seem to have evolved into a stable lineage. In theory, the discrepancies in clustering of the Rotterdam strain could be explained by artifacts of the phylogenetic trees based on pol sequences, since only a short fragment was used. Another explanation, however, could be the existence of recombinant NLV strains that have obtained the pol and capsid genes from different NLV parent strains. Indications for the circulation of such strains exist (34; X. Jiang, C. Espul, W. M. Zhong, H. Cuello, and D. O. Matson, Int. Workshop Human Caliciviruses 1999, P2-g), but it is unclear in what proportion. In any case, these findings stress the need for total capsid sequencing as a reference method for identification of novel genotypes.
Most RT-PCR-based detection assays are based on the pol region because this region is more conserved than the capsid region (10, 12, 35). Therefore, to date, most groups involved in molecular epidemiological studies have generated sequence data based on this region. Since for almost all strains phylogenetic grouping based on the capsid gene correlates well with that based on the pol regions (27, 34), we feel that pol typing still remains a valid method for these studies. Although RLB will never replace sequencing when detailed OB investigations (e.g., source tracing) are required, genotyping by RLB is a relatively simple method and will be a great advantage when RT-PCR products obtained are faint bands or present as a smear when analyzed on agarose gels and therefore cannot be confirmed by sequencing without cloning. While the RT-PCR has been optimized to minimize such effects, they still occur occasionally when clinical specimens or sewage extracts are tested. Potentially, multiple NLV genotypes in sewage samples (24) or even the presence of two different NLV strains in one stool sample (30) can be detected. By RLB, it was clearly shown that in fecal samples from two OBs, two different genotypes of NLV were present. This finding suggests either that the NLV infection was acquired from an overlap between an OB and a sporadic case or was acquired by water-borne transmission.
Comparison of the results obtained on genotyping by RLB and sequencing shows a 100% correlation. By sequence analysis of a region of the pol gene, we found that the eight strains not typed by RLB formed two clusters separate from all other currently known genotypes and therefore potentially represent new genotypes. Sequencing of the complete capsid gene of a representative strain from each cluster (OB98-15 and OB98-18) confirmed these findings. Moreover, the nucleotide sequence of the 5′ end of ORF2 of OB98-2 (Alphatron/98/NET) was determined and showed 93% identity with that of OB98-15. The novel capsid sequences have coding capacities for capsid proteins of 556 and 536 amino acids for Alphatron and Amsterdam, respectively. The tentative grouping with GGII viruses was supported by analyzing the sequence at the start of each ORF2 sequence. Like all of the GGII strains analyzed so far, both the Alphatron and Amsterdam ORF2 sequences start with the RNA sequence AUGAAGAUG (3, 8, 13, 22, 23).
Based on the current knowledge of sequence diversity of NLVs, we propose a provisional scheme for genotyping of NLVs (Table 1) that reflects most of the published NLV capsid types (3, 8, 11, 14, 15, 20, 21, 22, 23), the new ORF2 and ORF3 sequences determined in this study, and their correlation with ORF1 clustering and genotypes (34). When defined as >80% amino acid identity in the complete capsid gene confirmed by partial capsid sequences of at least one additional strain, NLVs can now be divided into at least 14 different genotypes (Table 1, genotypes 1 to 14). For these 14 genotypes, clustering was independent of the genomic region (5′ end of ORF2, >80% of the nucleotides; ORF1 region, >85% of the nucleotides for GGI and >90% of the nucleotides for GGII) (34). Based on these critiera, strains Rotterdam and Wortley, which could be considered potential novel genotypes based on their ORF1 sequences, are in fact ORF1 clusters belonging to the Mexico and Hawaii genotypes, respectively (Table 1). Two other potential genotypes (Seacroft and Amsterdam) await confirmation by sequence analysis of additional strains.
A special advantage is that RLB membranes can be reused without substantial loss of sensitivity. The RLB assays for typing of streptococci have been reused up to 40 times (L. Schouls, personal communication). This offers the possibility of standardization throughout a study. The RLB membranes could be produced at a central facility and serve as a standard method for international collaborative studies.
In conclusion, we have developed an RLB method for the simultaneous detection and genotyping of NLV, based on current knowledge of NLV GGs and genotypes. The method is rapid, reproducible, cheap, and easy to perform with a high throughput of samples. The possibility of adding a probe specific for a newly emerging NLV genotype and the potential to develop a multiplex RT-PCR for other RNA viruses associated with gastroenteritis (e.g., Sapporo-like caliciviruses and astroviruses) make this method ideal for the early detection and investigation of multinational common-source OBs.
ACKNOWLEDGMENTS
We thank Hanneke Deijl, Petra de Bree, and Chantal Rison for excellent technical assistance; the NIVEL team, the SENSOR team, the physicians, and the epidemiologists of the municipal health services for their help; and Erwin Duizer for critical reading of the manuscript.
REFERENCES
- 1.Ando T, Monroe S S, Gentsch J R, Jin Q, Lewis D C, Glass R I. Detection and differentiation of antigenically distinct small round-structured viruses (Norwalk-like viruses) by reverse transcription-PCR and Southern hybridization. J Clin Microbiol. 1995;33:64–71. doi: 10.1128/jcm.33.1.64-71.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clarke I N, Lambden P R. Viral zoonoses and food of animal origin: calicivirus and human disease. Arch Virol Suppl. 1997;13:141–152. doi: 10.1007/978-3-7091-6534-8_14. [DOI] [PubMed] [Google Scholar]
- 3.Dingle K E, Lambden P R, Caul E O, Clarke I N. Human enteric caliciviridae: the complete genome sequence and expression of virus-like particles from a genetic group II small round-structured virus. J Gen Virol. 1995;76:2349–2355. doi: 10.1099/0022-1317-76-9-2349. [DOI] [PubMed] [Google Scholar]
- 4.Djuretic T, Wall P G, Ryan M J, Evans H S, Adak G K, Cowden J M. General outbreaks of infectious intestinal disease in England and Wales 1992 to 1994. Commun Dis Rep CDR Rev. 1996;6:R57–R63. [PubMed] [Google Scholar]
- 5.Green J, Norcott J P, Lewis D, Arnold C, Brown D W G. Norwalk-like viruses in the UK: demonstration of genomic diversity by PCR sequencing. J Clin Microbiol. 1993;31:3007–3012. doi: 10.1128/jcm.31.11.3007-3012.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Green J, Gallimore C I, Norcott J P, Lewis D, Brown D W G. Broadly reactive reverse transcriptase polymerase chain reaction for the diagnosis of SRSV-associated gastroenteritis. J Med Virol. 1995;47:392–398. doi: 10.1002/jmv.1890470416. [DOI] [PubMed] [Google Scholar]
- 7.Green J, Henshilwood K, Gallimore C I, Brown D W G, Lees D N. A nested reverse transcriptase PCR assay for detection of small round-structured viruses in environmentally contaminated molluscan shellfish. Appl Environ Microbiol. 1998;64:858–863. doi: 10.1128/aem.64.3.858-863.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Green, J., J. Vinjé, C. I. Gallimore, M. P. G. Koopmans, A. Hale, and D. W. G. Brown. Capsid protein diversity among small round-structured viruses. Virus Genes, in press. [DOI] [PubMed]
- 9.Green K Y. The role of human caliciviruses in epidemic gastroenteritis. Arch Virol Suppl. 1997;13:153–165. doi: 10.1007/978-3-7091-6534-8_15. [DOI] [PubMed] [Google Scholar]
- 10.Green S M, Dingle K E, Lambden P R, Caul E O, Ashley C R, Clarke I N. Human enteric Caliciviridae: a new prevalent SRSV group defined by RNA-dependent RNA polymerase and capsid diversity. J Gen Virol. 1994;75:1883–1888. doi: 10.1099/0022-1317-75-8-1883. [DOI] [PubMed] [Google Scholar]
- 11.Green S M, Lambden P R, Caul E O, Ashley C R, Clarke I N. Capsid diversity in small round-structured viruses: molecular characterization of an antigenically distinct human enteric calicivirus. Virus Res. 1995;37:271–283. doi: 10.1016/0168-1702(95)00041-n. [DOI] [PubMed] [Google Scholar]
- 12.Green S M, Lambden P R, Caul E O, Clarke I N. Capsid sequence diversity in small round structured viruses from recent UK outbreaks of gastroenteritis. J Med Virol. 1997;52:14–19. [PubMed] [Google Scholar]
- 13.Hardy M E, Kramer S F, Treanor J J, Estes M K. Human calicivirus genogroup II capsid diversity revealed by analysis of the prototype Snow Mountain agent. Arch Virol. 1997;142:1469–1479. doi: 10.1007/s007050050173. [DOI] [PubMed] [Google Scholar]
- 14.Jiang X, Wang M, Wang K, Estes M K. Sequence and genomic organization of Norwalk virus. Virology. 1993;195:51–61. doi: 10.1006/viro.1993.1345. [DOI] [PubMed] [Google Scholar]
- 15.Jiang X, Matson D O, Ruiz-Palacios G M, Hu J, Treanor J, Pickering L K. Expression, self-assembly, and antigenicity of a Snow Mountain agent-like calicivirus capsid protein. J Clin Microbiol. 1995;33:1452–1455. doi: 10.1128/jcm.33.6.1452-1455.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jukes T H, Cantor C R. Evolution of protein molecules. In: Munro H H, editor. Mammalian protein metabolism. New York, N.Y: Academic Press, Inc.; 1969. pp. 21–132. [Google Scholar]
- 17.Kamerbeek J, Schouls L, Kolk A, van Agterveld M, van Soolingen D, Kuijper S, Bunschoten A, Molhuizen H, Shaw R, Goyal M, van Embden J. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J Clin Microbiol. 1997;35:907–914. doi: 10.1128/jcm.35.4.907-914.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaufhold A, Podbielski A, Baumgarten G, Bolkpoel M, Top J, Schouls L M. Rapid typing of group A streptococci by the use of DNA amplification and nonradioactive allele specific oligonucleotide probes. FEMS Microbiol Lett. 1994;119:19–26. doi: 10.1111/j.1574-6968.1994.tb06861.x. [DOI] [PubMed] [Google Scholar]
- 19.Koopmans M P G, Vinjé J, de Wit M, Leenen I, van der Poel W, van Duijnhoven Y. Molecular epidemiology of human enteric caliciviruses in The Netherlands. J Infect Dis. 2000;181:S262–S269. doi: 10.1086/315573. [DOI] [PubMed] [Google Scholar]
- 20.Lambden P R, Caul E O, Ashley C R, Clarke I N. Sequence and genome organization of a human small round-structured (Norwalk-like) virus. Science. 1993;259:516–519. doi: 10.1126/science.8380940. [DOI] [PubMed] [Google Scholar]
- 21.Lew J F, Kapikian A Z, Jiang X, Estes M K, Green K Y. Molecular characterization and expression of the capsid protein of a Norwalk-like virus recovered from a Desert Shield troop with gastroenteritis. Virology. 1994;200:319–325. doi: 10.1006/viro.1994.1194. [DOI] [PubMed] [Google Scholar]
- 22.Lew J F, Petric M, Kapikian A Z, Jiang X, Estes M K, Green K Y. Identification of minireovirus as a Norwalk-like virus in pediatric patients with gastroenteritis. J Virol. 1994;68:3391–3396. doi: 10.1128/jvi.68.5.3391-3396.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lew J F, Kapikian A Z, Valdesuso J, Green K Y. Molecular characterization of Hawaii virus and other Norwalk-like viruses: evidence for genetic polymorphism among human caliciviruses. J Infect Dis. 1994;170:535–542. doi: 10.1093/infdis/170.3.535. [DOI] [PubMed] [Google Scholar]
- 24.Lodder W L, Vinjé J, van der Heide R, Roda Husman A-M, Leenen E J T M, Koopmans M P G. Molecular detection of “Norwalk-like viruses” in sewage. Appl Environ Microbiol. 1999;65:5624–5627. doi: 10.1128/aem.65.12.5624-5627.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakayama M, Ueda Y, Kawamoto H, Han-jun Y, Saito K, Nishio O, Ushijima H. Detection and sequencing of Norwalk-like viruses from stool samples in Japan using RT-PCR amplification. Microbiol Immunol. 1996;40:317–320. doi: 10.1111/j.1348-0421.1996.tb03343.x. [DOI] [PubMed] [Google Scholar]
- 26.Nei M. Distance matrix methods. In: Nei M, editor. Molecular evolutionary genetics. New York, N.Y: Columbia University Press; 1987. pp. 293–298. [Google Scholar]
- 27.Noel J S, Ando T, Leite J P, Green K Y, Dingle K E, Estes M K, Seto Y, Monroe S S, Glass R I. Correlation of patient immune responses with genetically characterized small round-structured viruses involved in outbreaks of nonbacterial acute gastroenteritis in the United States, 1990 to 1995. J Med Virol. 1997;53:372–383. doi: 10.1002/(sici)1096-9071(199712)53:4<372::aid-jmv10>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 28.Norcott J P, Green J, Lewis D, Estes M K, Barlow K L, Brown D W G. Genomic diversity of small round structured viruses in the United Kingdom. J Med Virol. 1994;44:280–286. doi: 10.1002/jmv.1890440312. [DOI] [PubMed] [Google Scholar]
- 29.Rijpkema S G T, Molkenboer M J C H, Schouls L M, Jongejan F, Schellekens J F P. Simultaneous detection and genotyping of three genomic groups of Borrelia burgdorferi sensu lato in Dutch Ixodes ricinus ticks by characterization of the amplified intergenic spacer region between 5S and 23S rRNA genes. J Clin Microbiol. 1995;33:3091–3095. doi: 10.1128/jcm.33.12.3091-3095.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sugieda M, Nakajima K, Nakajima S. Outbreaks of Norwalk-like virus-associated gastroenteritis traced to shellfish: coexistence of two genotypes in one specimen. Epidemiol Infect. 1996;116:339–346. doi: 10.1017/s0950268800052663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van der Peer Y, de Wachter R. TREECON for windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput Appl Biosci. 1994;10:569–570. doi: 10.1093/bioinformatics/10.5.569. [DOI] [PubMed] [Google Scholar]
- 32.Vinjé J, Koopmans M P G. Molecular detection and epidemiology of small round-structured viruses in outbreaks of gastroenteritis in The Netherlands. J Infect Dis. 1996;174:610–615. doi: 10.1093/infdis/174.3.610. [DOI] [PubMed] [Google Scholar]
- 33.Vinjé J, Altena S A, Koopmans M P G. The incidence and genetic variability of small round-structured viruses in outbreaks of gastroenteritis in The Netherlands. J Infect Dis. 1997;176:1374–1378. doi: 10.1086/517325. [DOI] [PubMed] [Google Scholar]
- 34.Vinjé J, Green J, Lewis D C, Gallimore C I, Brown D W G, Koopmans M P G. Genetic polymorphism across regions of the three open reading frames of “Norwalk-like viruses.”. Arch Virol. 2000;145:1–19. doi: 10.1007/s007050050020. [DOI] [PubMed] [Google Scholar]
- 35.Wang X, Jiang X, Madore H P, Gray J, Desselberger U, Ando T, Seto Y, Oishi I, Lew J F, Green K Y, Estes M K. Sequence diversity of small, round-structured viruses in the Norwalk virus group. J Virol. 1994;68:5982–5990. doi: 10.1128/jvi.68.9.5982-5990.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y, Coyne M Y, Will S G, Levenson C H, Kawasaki E S. Single-base mutational analysis of cancer and genetic diseases using membrane bound modified oligonucleotides. Nucleic Acids Res. 1991;19:3929–3933. doi: 10.1093/nar/19.14.3929. [DOI] [PMC free article] [PubMed] [Google Scholar]