Abstract
Secreted extracellular vesicles (EVs) influence the tumor microenvironment and promote distal metastasis. Here, we analyzed the involvement of melanoma-secreted EVs in lymph node pre-metastatic niche formation in murine models. We found that small EVs (sEVs) derived from metastatic melanoma cell lines were enriched in nerve growth factor receptor (NGFR, p75NTR), spread through the lymphatic system and were taken up by lymphatic endothelial cells, reinforcing lymph node metastasis. Remarkably, sEVs enhanced lymphangiogenesis and tumor cell adhesion by inducing ERK kinase, nuclear factor (NF)-κB activation and intracellular adhesion molecule (ICAM)-1 expression in lymphatic endothelial cells. Importantly, ablation or inhibition of NGFR in sEVs reversed the lymphangiogenic phenotype, decreased lymph node metastasis and extended survival in pre-clinical models. Furthermore, NGFR expression was augmented in human lymph node metastases relative to that in matched primary tumors, and the frequency of NGFR+ metastatic melanoma cells in lymph nodes correlated with patient survival. In summary, we found that NGFR is secreted in melanoma-derived sEVs, reinforcing lymph node pre-metastatic niche formation and metastasis.
Keywords: small extracellular vesicles, lymph node metastasis, NGFR, p75NTR, CD271, pre-metastatic niche formation, lymphangiogenesis, melanoma metastasis, cell adhesion, metastasis mechanisms
Main
Melanoma preferentially disseminates to the lymph nodes (LNs)1. It has been established that tumors trigger changes in the sentinel LNs (sLNs), such as enhanced lymphangiogenesis2 and induction of an immunosuppressive environment3. These abnormalities precede metastatic colonization and contribute to the formation of the pre-metastatic niche (PMN) in the LNs4. Understanding the intricate mechanisms involved in LN metastasis is crucial to decipher the first steps of melanoma metastatic spread5. Secretion of VEGF-C and VEGF-A from melanoma primary tumors induce lymphangiogenesis in sLNs as well as LN metastasis and subsequent distal organ metastasis6-8. In addition to soluble factors, tumor-derived extracellular vesicles (EVs) reinforce metastasis in melanoma9-11. Exosomes are small extracellular vesicles (sEVs) 30 to 120 nm in diameter size12 that are key players in PMN formation at distal organ sites and in metastatic organotropism13-15. Melanoma-derived sEVs distribute through the mouse lymphatic system, facilitate tumor cell homing, and bind subcapsular sinus (SCS) CD169+ macrophages in tumor-draining LNs, eliciting enhanced immune responses against cancer cells that tumors usually overcome9, 10, 16. Furthermore, we and others have identified and characterized sEVs in the lymphatic drainage from melanoma patients and described their interactions with lymphatic endothelial cells (LECs)17, 18. However, the role of melanoma-secreted sEVs in LN PMN formation and the mechanisms involved are not yet known. In this work, we wanted to analyze the involvement of melanoma-secreted EVs in lymph node PMN formation to define the mechanisms involved in early metastatic cell dissemination.
Malignant melanomas originate from the oncogenic transformation of melanocytes, a cell type derived from multipotent neural crest stem cells1. Nerve growth factor receptor (NGFR; also known as p75NTR/CD271) belongs to the TNFR superfamily and is one of two types of neurotrophin receptors controlling survival and differentiation in the nervous system19, 20. NGFR expression during melanocyte development supports melanoma initiation21, 22, progression23, metastasis24, 25, chemoresistance26 and immune therapy resistance27. NGFR also regulates the invasive properties of metastatic melanoma cell lines, but to date, studies have mainly defined its intrinsic role in melanoma21, 22, 25.
Here, we show that melanoma-secreted sEVs spread through the lymphatic system and were taken up first by LECs followed by LN macrophages in murine models. Melanoma-derived sEVs induced lymphangiogenesis in LECs by promoting: 1) the expression of lymphangiogenic genes and VEGFR3 phosphorylation, 2) LEC proliferation, 3) activation of ERK and NF-κB pathways, and 4) ICAM-1 expression, reinforcing tumor cell adhesion. We found that NGFR was secreted in melanoma-derived sEVs and shuttled to LECs. Importantly, ablation of NGFR in sEVs or its pharmacological inhibition reversed the lymphangiogenic phenotype and decreased melanoma metastasis. Finally, the histological analysis of NGFR expression in human melanoma samples revealed that NGFR expression was higher in metastatic LNs compared to skin lesions and that the frequency of tumor cells expressing NGFR in metastatic LNs was correlated with decreased survival in melanoma patients.
RESULTS
sEV retention in lymphatics depends on metastatic behavior
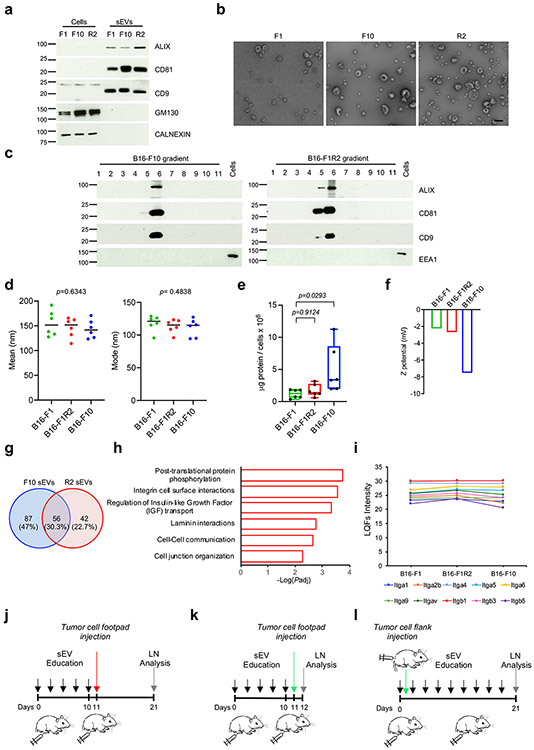
We purified sEVs from low and high metastatic mouse melanoma models (B16-F1 and B16-F10 cell lines, respectively) and a LN-metastatic model derived from B16-F1 (B16-F1R2)28. We confirmed the purity of our sEV preparations by analyzing exosomal markers (ALIX, CD81, CD9) and the absence of endoplasmic reticulum (CALNEXIN) and Golgi markers (GM130) (Extended Data Fig. 1a). We verified an sEV size <200nm by electron microscopy (Extended Data Fig. 1b) and purity by iodixanol density gradient (Extended Data Fig. 1c). We analyzed the biodistribution of sEVs derived from melanocytes (melan-a cell line) and from melanoma cell lines in the lymphatic system of naïve mice after intra-footpad injection29, a method used extensively to deliver material through the lymphatics into the sLN. B16-F10- and B16-F1R2-secreted sEVs had a broader distribution compared to B16-F1 and melan-a-derived sEVs that were mainly detected in the sLNs (Fig. 1a). Analysis of LNs after 3 weeks of sEV conditioning revealed that sEVs-associated fluorescence in the sLN was higher in animals treated with sEVs derived from B16-F10 and B16-F1R2 cells compared to parental B16-F1 cells (Fig. 1b, c). A time course analysis from 1 to 48 hours demonstrated that B16-F10-derived sEVs spread and accumulated in sLNs faster than B16-F1-secreted sEVs (Fig. 1d, e). We consistently observed that B16-F10-derived and B16-F1R2-derived sEV administration resulted in increased fluorescence signal in sLNs relative to B16-F1-derived sEVs 24 h after footpad injection and localized in LYVE-1-enriched cortical areas (Fig. 1f).
Figure 1. Melanoma-secreted sEVs are retained through the lymphatic system.
a, Representative images of sEV-associated signal in mice injected intra-footpad 3 weeks with the indicated NIR815-labeled sEVs (n = 2 mice per group). Po, popliteal LN; Il, Iliac LN; In, inguinal LN; Li, liver. b,c, Representative images and quantification of NIR815-associated signal in LNs from mice treated as above. Two independent experiments were performed (n = 4 LNs per group). L, lateral LN; CL, contralateral LN. d,e, Representative images and quantification of sEV-associated signal in mice 1, 4, 24 and 48 hours after footpad injection with NIR815-labeled sEVs. Data correspond to two independent experiments (n = 4 mice per group). Squares indicate popliteal LN area. f, Representative images of the distribution of melanoma DiD-labeled sEVs in popliteal LNs 16 h after intra-footpad injection (n = 4 LNs per group). Scale bar, 150 μm. g,h, Representative flow cytometry plots and quantification of B16-F1-GFP+ cells within the CD45− population in LNs educated with sEVs for 10 days. LN were analyzed 24 h post-injection of tumor cells (PBS and R2 n = 5 mice per group, F1 and F10 n = 4 mice per group). i,j, Representative images and quantification of B16-F1-mCherry+ cells in sections of popliteal LNs 10 days post tumor cell injection. Melanoma-derived sEVs or PBS were injected intra-footpad for 10 days prior to tumor inoculation. Two independent experiments were performed (CTRL, F1, F1R2 groups, n = 5 mice per group, F10 group, n = 4 mice). Scale bar, 20 μm. k,I, Representative images and quantification of metastatic area in inguinal LNs of animals bearing B16-F1-GFP flank tumors and educated with B16-F1-R2 sEVs injected intra-footpad for 21 days. Two independent experiments were performed (n = 12 mice per group). Scale bars, 500 μm and 200 μm. Boxes in the box plots in h and j define the IQR split by the median, with whiskers extending to the most extreme values within 1.5 × IQR beyond the box. All other data represent mean ± s.e.m. and p values were calculated by two-way ANOVA in c and e, by one-way ANOVA in h and j and by two-tailed Student’s t test in l.
To understand if changes in biophysical properties could explain differences in retention, we analyzed the size of sEVs from B16-F1, B16-F10, and B16-F1R2 cell lines and did not observe any significant variations in mean or mode sEV size (Extended Data Fig. 1d). The amount of protein packaged in sEVs from B16-F10-derived sEVs was increased when normalized by the number of cells, as previously described15 (Extended Data Fig. 1e). We measured the surface charge (Z-potential) of the different vesicles and observed that it was decreased in B16-F10-derived sEVs compared to B16-F1- and B16-F1R2-derived sEVs (Extended Data Fig. 1f). We then analyzed sEV surface proteins by mass spectrometry and found a total of 185 proteins upregulated in either B16-F1R2 or B16-F10 sEVs versus B16-F1 sEVs, with a shared group of 56 proteins (Extended Data Fig. 1g, Supplementary Table 1). Interestingly, pathway enrichment analysis of the upregulated proteins identified several processes related to cell adhesion as enriched in B16-F1R2 or B16-F10 sEVs compared to B16-F1 sEVs (Extended Data Fig. 1h), potentially explaining augmented retention in LNs. Analysis of sEV integrin levels showed that the integrin profile was similar for all the sEV types assessed (Extended Data Fig. 1i).
Next, we explored if prolonged exposure to sEVs (defined as education15) could have functional consequences in LN metastasis (see Extended Data Fig. 1j-l for diagrams of experimental set-up). We first assessed the effect of sEV education on B16-F1 tumor cell homing (see scheme in Extended Data Fig. 1k). We observed a significant increase in B16-F1-GFP cells when LNs were educated with sEVs derived from highly metastatic cell lines B16-F1R2 and B16-F10 (Fig. 1g,h). Moreover, education with B16-F1R2 and B16-F10 sEVs enhanced tumor colonization in experimental metastasis assays (see Extended Data Fig. 1j schematic), whereas education with PBS or B16-F1-derived sEVs did not (Fig. 1i,j) .
To study the effect of sEV education on spontaneous metastasis, mice were injected intra-footpad with B16-F1R2-derived EVs and, one day later, tumor cells were injected in the flank. Education with B16-F1R2-derived sEVs continued for 20 days (see Extended Data Fig. 1l schematic). We observed a significant increase in LN spontaneous metastases after B16-F1R2 sEV education (Fig. 1k, l). These data support the hypothesis that melanoma-derived sEVs circulate through the lymphatic system and reinforce LN metastasis.
Tumor sEVs are taken up by LECs and macrophages
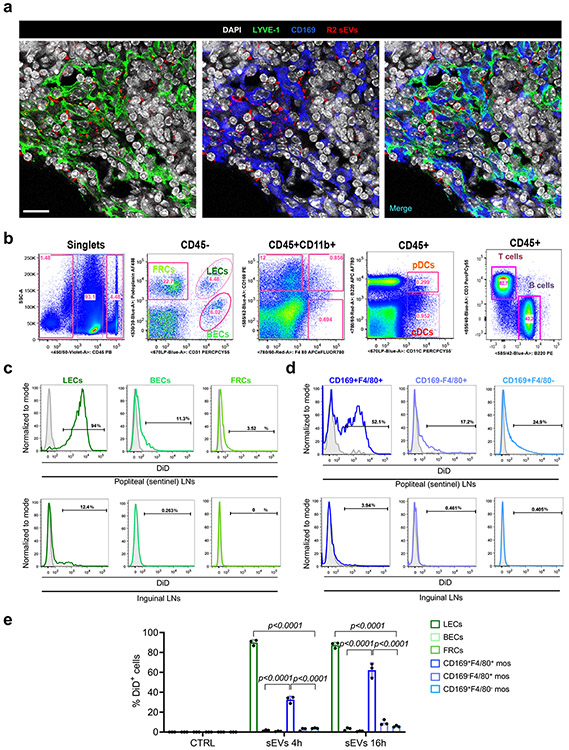
To identify the main LN cell types that incorporated melanoma-derived sEVs, we injected sEVs intradermally in the ear of Prox-1-GFP mice30. EVs were detected inside the lymphatic vasculature 1 hour post-injection (Fig. 2a). Analysis of cervical LNs 16 hours post-injection demonstrated that sEVs were retained in the paracortical area mainly by LYVE-1+ cells and CD169+ cells (Fig. 2b). Popliteal LN analysis after footpad injection of B16-F10- and B16-F1R2-derived sEVs showed the presence of melanoma-derived sEVs in LYVE-1+ cells as well as CD169+ cells (Fig. 2b, Extended Data Fig. 2a). Analysis of EVs in LNs by immunofluorescence showed that LYVE-1+ areas were mainly enriched in sEVs (Fig. 2c). For a more detailed assessment, we performed flow cytometry analysis of popliteal LNs 16 hours after footpad injection of B16-F10 fluorescently-labeled sEVs (See gating stategy in Extended Data Fig. 2b). We observed that around 95% of LECs (CD45−CD31+Podoplanin+), 60% of double-positive (DP)-macrophages (CD45+CD11b+CD169+F4/80+), 30% of F4/80+ macrophages (CD45+CD11b+CD169−F4/80+) and 20% of subcapsular macrophages (CD45+CD11b+CD169+F4/80−) displayed sEV-associated fluorescence after treatment with melanoma-derived sEVs (Fig. 2d,e and Extended Data Fig. 2c,d, upper panels). EV fluorescence signal was also detected in 10-15% of dendritic cells (DC, CD45+CD11c+B220+/−), 8% of blood endothelial cells (BECs, CD45−CD31+Podoplanin−) and 4.5% of fibroblastic reticular cells (FRCs, CD45−CD31−Podoplanin+) but not detected in T cells (CD45+CD3+B220−) or B cells (CD45+CD3−B220+) (Fig. 2d,e and Extended Data Fig. 2c,d, upper panels). The analysis of distal LNs (inguinal LNs) revealed that around 12% of LECs and less than 4% of (DP)-macrophages incorporated tumor-derived sEVs (Extended Data Fig. 2c,d, lower panels). At shorter time points (4 hours), the percentage of popliteal LECs with sEV-associated fluorescence was similar, but only around 30% of DP-macrophages were positive, with a median fluorescence intensity significantly lower than that of LECs (Fig. 2f,g and Extended Data Fig. 2e). These data suggest that LECs and macrophages were the main cells types taking up tumor-derived sEVs in the sLN and we subsequently focused on LECs.
Figure 2. Small EVs are incorporated by LN stromal and immune cells.
a, Representative images of earlobe lymphatic vessels in Prox-1-GFP mice 1 h after intra-ear injection of PKH26-labeled B16-F10 sEVs. Top right inset displays the orthogonal projection corresponding to the white dashed line (n = 2 mice per group). Scale bars, 100 μm and 20 μm. b, Representative images of cervical LNs 16 h after intra-footpad injection of PKH26-labeled B16-F1 and B16-F10 sEVs. Arrows indicate areas of overlapping PKH26 and LYVE-1 staining (n = 3 mice per group). Scale bar, 200 μm and 20 μm. c, Representative images of two consecutive popliteal LN sections dissected 16 h after intra-footpad injection of DiD-labeled B16-F1-R2 sEVs. White lines delineate areas of lymphatic (upper panels) and macrophage (lower panels) stained networks (n = 3 LNs per group). Scale bar, 250 μm and 50 μm. d, e, Representative flow cytometry plots and quantification related to B16-F10 sEV uptake in the indicated stromal and immune populations. DiD-labeled sEVs were injected intra-footpad and LNs populations were analyzed 16 h later. Gates were depicted based on corresponding dye-only signal for each population. Gating strategy is described in Extended Data Fig 2b. Three independent experiments were performed (n = 4 LNs analyzed). BECs, blood endothelial cells; FRCs, fibroblastic reticular cells; DC, dendritic cells. f, Representative plots of B16-F1R2 sEV-associated fluorescence in LECs and CD169+F4/80+ macrophages 4 h after intra-footpad injection. Three independent experiments were performed (n = 3 LNs analyzed). g, Median DiD fluorescence intensity associated to B16-F1-R2 sEVs in LECs and CD169+F4/80+ macrophages (mos) at the indicated times. Two independent experiments were performed (n = 3 LNs analyzed). All data represent mean ± s.e.m. and p values were calculated by two-tailed Student’s t test in e and by two-way ANOVA in g.
Melanoma sEVs influence LEC phenotype promoting tumor cell adhesion
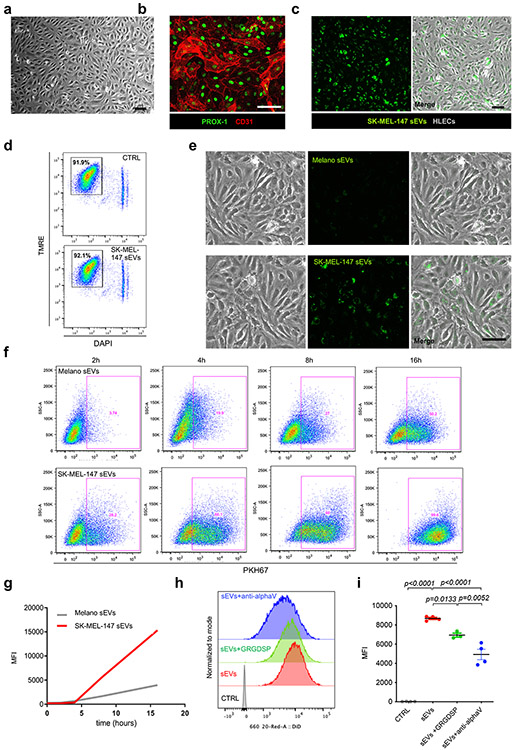
We hypothesized that melanoma-derived sEVs may influence LEC behavior. We analyzed sEV size distribution and integrity of human metastatic melanoma SK-MEL-147 cell-derived sEVs by electron microscopy (Extended Data Fig. 3a,b). We verified the purity of our sEV preparations by analyzing exosomal markers (ALIX, CD81, CD63) and the absence of CALNEXIN and GM130 (Extended Data Fig. 3c). We also analyzed the distribution of ALIX, CD81 and EEA1 after iodixanol density gradient (Extended Data Fig. 3d). We observed that human LECs (hLECs) expressing typical LEC markers (Extended Data Fig. 4a,b) incorporated SK-MEL-147-derived sEVs (Extended Data Fig. 4c) that did not induce apoptosis of hLECs (Extended Data Fig. 4d). Remarkably, SK-MEL-147-derived sEVs were taken up faster and in larger amounts than melanocyte-derived sEVs (Extended Data Fig. 4e-g). We tested several integrin blocking agents for their effect on sEV uptake by hLECs and found that GRGDSP peptide, a RGD-based peptide31, reduced sEV uptake by 20%, and a blocking antibody against integrin αv reduced sEV uptake by 50% (Extended Data Fig. 4h,i).
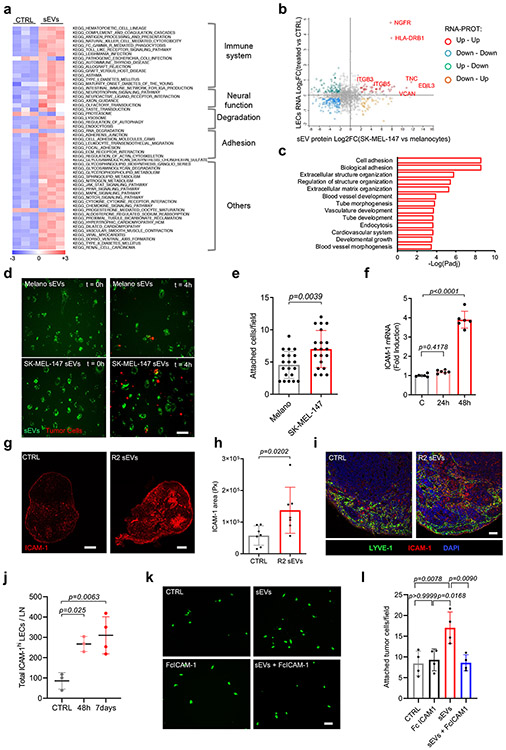
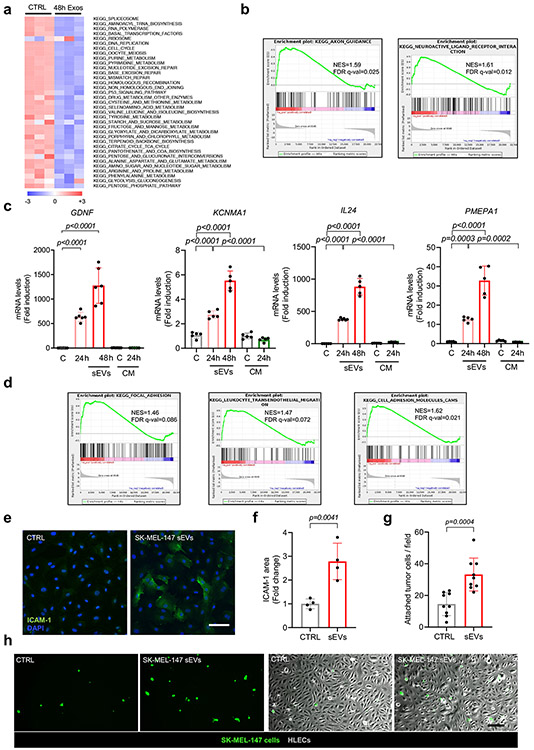
To study the effect of sEV uptake on LEC phenotype, we performed RNA sequencing in hLECs after 48 hours of treatment with SK-MEL-147-derived sEVs or conditioned medium from the same cultures and focused on genes whose expression was exclusively affected by tumor-derived sEVs. A total of 447 genes were upregulated and 318 genes were downregulated (FDR<0.05) in hLECs sEV-treated for 48 hours (Supplementary Table 2). Gene set enrichment analysis (GSEA) identified enrichment of signatures related to the immune system, neural function, cell adhesion, and cytokine signaling, among other gene functions (Fig. 3a, Extended Data Fig. 5a,b). We validated the expression of several genes that were upregulated in hLECs in response to melanoma-derived sEVs, such as GDNF, KCNMA1, IL24 and PMEPA1 (Extended Data Fig. 5c, Supplementary Table 2).
Figure 3. Melanoma-derived sEVs influence LEC transcriptional profile and promote adhesion.
a, Single sample GSEA analysis showing significantly enriched KEGG signatures in human LECs treated with SK-MEL-147 sEVs or PBS for 48 h (n = 3 samples per group). b, Correlation between RNAseq data in LECs and proteomic data in SK-MEL-147 sEVs. Color code indicates significantly regulated gene-protein pairs (FDR < 5%). c, Top pathways significantly enriched in the group of gene-proteins positively correlated shown in (b). Pathways were obtained using PANTHER overrepresentation analysis applying Fisher’s exact test and FDR correction. d,e, Representative images of PKH26-labelled SK-MEL-147 cells adhered to sEV-treated LECs in flow. LECs were previously exposed to PKH67-labeled sEVs from primary melanocytes (Melano) or SK-MEL-147 cells during 24 h. Plot in (e) shows quantification of attached tumor cells at t = 4 h. Two independent experiments performed (all groups, n = 20 fields from one representative experiment. Scale bar, 50 μm. f, ICAM-1 expression in hLECs treated with SK-MEL-147 sEVs for 48 h. Two independent experiments performed (n = 6 LNs per group). g,h, Representative images and quantification of ICAM-1 expression in LNs treated with B16-F1-R2 sEVs intra-footpad for 10 days. Two independent experiments were performed (n = 7 LNs per group). Scale bar, 100 μm and 200 μm. i, LYVE-1 and ICAM-1 staining in LNs treated with F1-R2 sEVs or PBS for 48h (n = 3 LNs per group). Scale bar, 50 μm. j, Quantification by flow cytometry of LECs expressing high levels of ICAM-1 in LNs of animals injected intra-footpad with B16-F1R2 sEVs for the indicated times. Two independent experiments were performed (CTRL and 48 h, n = 3 LNs per group and 7 days, n = 4 LNs per group). k,l, Representative images and quantification of the adhesion of SK-MEL-147 cells (green) to HLEC monolayers in the presence of Fc ICAM blocking molecule or vehicle. HLECs were previously exposed or not to SK-MEL-147 sEVs. Two independent experiments were performed (n = 4 samples per group). Scale bar, 20 μm. All data represent mean ± s.e.m. and p values were calculated by two-tailed Student’s t test in e and h and by one-way ANOVA in f, j and l.
We next integrated RNA expression data for hLECs treated with SK-MEL-147 sEVs with proteomic analyses of SK-MEL-147 sEVs. The analysis demonstrated that there was a fraction of positively correlated genes/proteins, including NGFR, HLA-DRB1, Tenascin C (TNC), and EDIL3 (Fig. 3b). Functional enrichment analysis indicated that this group of genes was associated with cell adhesion, extracellular matrix re-organization, and vasculature development (Fig. 3c). These findings were consistent with the enrichment in cell adhesion signatures, such as focal adhesion, leukocyte trans-endothelial migration, and cell adhesion molecules observed in the RNAseq data set (Extended Data Fig. 5d).
We tested whether treatment with sEVs may influence tumor cell adhesion to LECs. We observed enhanced adhesion of SK-MEL-147 tumor cells to hLECs in vitro after treatment with SK-MEL-147- derived sEV in flow conditions compared to treatment with melanocyte-derived sEVs during 4 hours (Fig. 3d,e). Since tumor cells normally mimic immune cell mechanisms to cross endothelial monolayers and ICAM-1 favors the transmigration of melanoma and colon cells through endothelial monolayers32, 33, we tested whether ICAM-1 was involved in tumor cell adhesion to LECs in our setting. LEC monolayers treated with SK-MEL-147 sEVs upregulated ICAM-1 mRNA (Fig. 3f) and protein levels (Extended Data Fig 5e,f) concurrent with an increased adhesion of SK-MEL-147 cells to hLECs in vitro (Extended Data Fig. 5g,h). In vivo analysis showed that sEV injection in the footpad enhanced the overall expression of ICAM-1 in LNs (Fig. 3g,h), with increased ICAM-1 expression in LYVE-1+ areas (Fig. 3i). B16-F1R2-derived sEVs increased the number of LECs expressing high levels of ICAM-1 in LNs exposed to melanoma sEVs (Fig. 3j). Finally, we performed adhesion experiments in the presence of ICAM-1-Fc, a molecule that impairs ICAM-1 binding function34. We found that soluble ICAM-1-Fc abrogated the adhesion of tumor cells promoted by melanoma-derived sEVs (Fig. 3k, l), supporting an important role for ICAM1 in tumor cell adhesion to LECs.
Melanoma sEVs promote lymphangiogenesis in LECs
Lymphangiogenesis is a hallmark of the LN PMN4. Thus, we next analyzed the genes involved in the development of vasculature upregulated in our RNAseq data. Several pro-lymph-/angiogenic genes were upregulated after 48 hours of treatment with melanoma sEVs (Fig. 4a). Accordingly, we observed an enrichment in signatures related with lymphangiogenesis and angiogenesis (Extended Data Fig. 6a). We validated a subset of genes, such as LYVE-1, VEGF-C, ENDOGLIN, VEGFR2, PDGFR and VEGF-A, by qPCR (Fig. 4b). In addition, we confirmed that tumor-derived sEVs promoted the activation of VEGFR3, a central regulator of vascular network formation in LECs by paracrine factors35 (Fig. 4c), suggesting the activation of a pro-lymphangiogenic program.
Figure 4. Melanoma-sEVs promote LN lymphangiogenesis.
a, Heatmap of angiogenesis and lymphangiogenesis-related genes based on RNAseq data (all groups, n = 3 samples per group b, Expression of lymph/angiogenesis-related genes in SK-MEL-147 sEVs-treated or control hLECs. Data were obtained from 2 independent experiments (n = 6 independent cell cultures per group). c, Representative WB of phospho-VEGFR3 and total VEGFR3 levels in HLECs treated with SK-MEL-147-derived sEVs. Three independent experiments were performed (n = 3 samples per group). d, Representative pictures of endothelial cell tube assays performed in HLECs growing on matrigel for 16 h after treatment with SK-MEL-147-derived sEVs. Data were obtained from 3 independent experiments (n = 15 samples per group). Scale bar, 100 μm. e,f, Representative images and quantification of LYVE-1+ cells in control (PBS) and SK-MEL-147 sEV-embedded matrigel plugs. Data were collected from two independent experiments (CTRL, n = 5 plugs and sEVs, n = 4 plugs). Scale bar, 50 μm. g, Representative images of consecutive sections of a sEV-embedded matrigel plugs stained with LYVE-1 and F4/80 (n = 4 plugs per group). Scale bar, 250 μm and 80 μm (inset). h,i, Representative images and quantification of luminescence associated to LNs of Vegfr3-EGFP-luc mice after 7 days of exposure to B16-F10 sEVs. Po, popliteal LN; In, inguinal LN; Ax, axillary LN. Data were obtained from three independent experiments (Popliteal CTRL, axilary CTRL and inguinal sEV groups n = 3 LNs; inguinal CTRL groups, n = 2 LNs; axillary sEVs group, n = 4 LNs; popliteal sEVs group, n = 5 LNs). j,k, Representative images and quantification of LYVE-1 staining in LNs after 48 h of intra-footpad injection with B16-F1-R2 sEVs. Two independent experiments were performed (n = 8 LNs per group). Scale bar 250 μm. l,m, Representative images and quantification of LEC proliferation in LNs after 24 h of intra-footpad injection with B16-F1-R2 sEVs (n = 5 LNs per group). Scale bar 40 μm. All data represent mean ± s.e.m. and p values were calculated by two-tailed Student’s t test in f, k and m and by two-way ANOVA in b and i.
To test if sEV treatment influenced LEC branching, we performed sprouting assays in human LECs (HLECs and HMVECs) (Fig. 4d, Extended Data Fig. 6b-d) and co-cultures of HMVECs with LN fibroblasts (Extended Data Fig. 6e,f) after sEV treatment. We found that the number and length of sprout/tube structures were significantly increased in both scenarios at 16 hours.
To analyze the effect of sEVs in lymphangiogenesis in vivo, we performed matrigel plug assays with embedded SK-MEL-147 sEVs. We observed that sEVs induced a significant increase in LYVE-1+ vessels compared to control matrigel plugs (Fig. 4e,f). We verified that LYVE-1+ cells were indeed LECs and not macrophages by staining consecutive sections of matrigel with LYVE-1 and the macrophage marker F4/80 (Fig. 4g). Additionally, we investigated the expression of lymphatic genes in LN LECs sorted 24 hours after sEV injection in Prox-1-tdTomato mice36. Footpad injection of B16-F10-derived sEVs induced Lyve-1 and Prox-1 mRNA expression 24 hours post-injection in sorted LECs positive for sEV-associated fluorescence (Extended Data Fig. 6g). Consistent with these findings Vegfr3 expression was significantly induced in popliteal and inguinal LNs of, Vegfr3/Flt4 EGFP-Luc lymphoreporter mice37 (Fig. 4h,i) and LYVE-1 area was significantly increased in animals treated with B16-F1R2-derived sEVs (Fig. 4j,k), suggesting increased lymphangiogenesis.
Since lymphangiogenesis is a complex mechanism involving lymphatic cell growth and sprouting38, we investigated LEC proliferation by measuring Ki67 levels in PROX-1+ and LYVE-1+ cells after sEV treatment and found that B16-F1R2 sEVs increased LEC proliferation by 3.5- and 2.5-fold after 24 h or 7 days, respectively (Fig. 4l,m; Extended Data Fig. 6h,i).
LN macrophages have been implicated in sEV-induced lymphangiogenesis and have been shown to secrete lymphangiogenic factors39. Therefore, we analyzed the expression of lymphangiogenic genes in F4/80+ CD169+ macrophages sorted from LNs after B16-F1R2-sEV footpad injection. We found that melanoma-derived sEVs did not influence the expression of lymphangiogenic genes in LN macrophages (Extended Data Fig. 7a). We also tested the effects of B16-F1R2 sEVs on the proliferation of popliteal LN LECs from animals depleted of CD169+ macrophages after diphtheria toxin injection (CD169DTR mice)40. These animals did not present changes in sEV uptake by LECs (Extended Data Fig. 7b). Interestingly, depletion of CD169+ macrophages stimulated LEC proliferation, but sEVs did not have any significant additional effect (Extended Data Fig. 7c), suggesting that CD169 macrophages may indeed be restricting LN LEC proliferation.
NGFR is shuttled into melanoma sEVs and induced in LECs
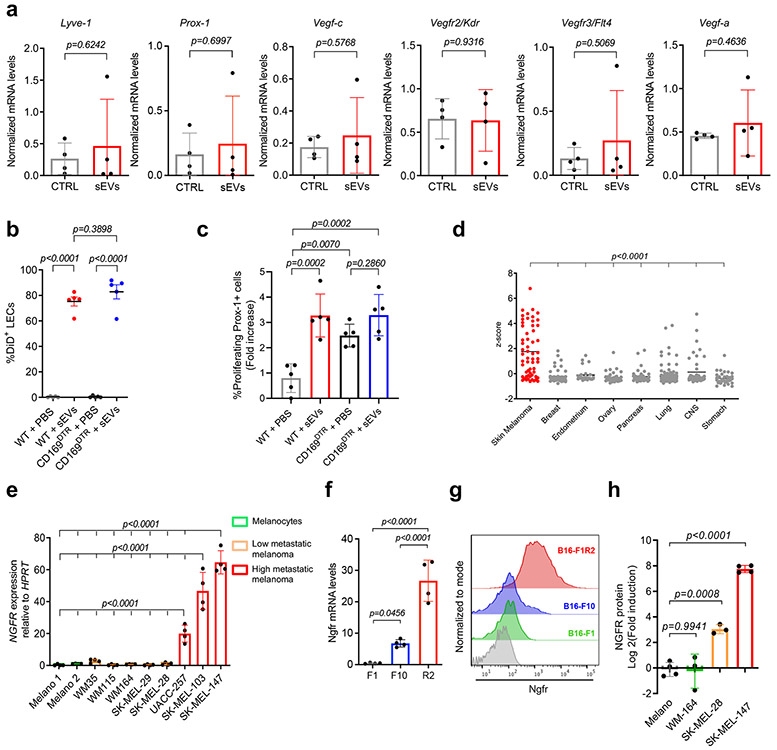
In order to investigate the sEV-dependent mechanism involved in LN PMN formation, we examined the most highly upregulated genes in LECs after treatment with SK-MEL-147 sEVs (Supplementary Table 2) that also positively correlated with sEV cargo (Fig. 3b). Among these, NGFR, a neurotrophin receptor previously related to melanoma metastasis22, 41 and lymphangiogenesis during corneal inflammation,42 was the top candidate. Study of NGFR expression in the Cancer Cell Line Encyclopedia (CCLE) database showed an increased expression of NGFR in human melanoma cell lines compared to all other cancer cell lines analyzed (Extended Data Fig. 7d). NGFR expression analysis in a panel of melanoma cell lines confirmed it was highly expressed in metastatic human (Extended Data Fig. 7e) and mouse (Extended Data Fig. 7f) melanoma cell lines compared to low/medium metastatic and non-invasive melanoma cell lines. Importantly, sEVs secreted by highly metastatic cell lines contained NGFR protein whereas this receptor was undetectable or nominally expressed at very low levels in low metastatic models or melanocytes (Fig. 5a,b and Extended Data Fig. 7g). In addition, analysis of mass spectrometry data17 showed that NGFR was highly expressed in tumor versus melanocyte-derived sEVs and NGFR levels increased in highly metastatic SK-MEL-147 sEVs in comparison to SK-MEL-28- and WM164-derived sEVs (Extended Data Fig. 7h).
Figure 5. Metastatic melanoma-derived sEVs contain NGFR and transfer it to LECs.
a, Representative WB of NGFR protein levels in sEVs from human primary melanocytes and melanoma cell lines (upper panels) or murine cell lines (lower panels). Two independent experiments were performed (n = 2 samples per group). b, Representative overlay flow cytometry plots of NGFR staining on SK-MEL-28 and SK-MEL-147 sEVs. Two independent experiments were performed (n = 2 samples per group). c, NGFR mRNA levels in human LECs treated for 24 h and 48 h with SK-MEL-147-derived sEVs or conditioned medium (CM). Data were obtained from two independent experiments (n = 4 independent cell cultures per group, except 24h sEVs, n = 6 and 48h sEVs and cCM, n = 5). d, Representative images of NGFR staining in HLECs exposed to SK-MEL-147 sEVs for 48 h. Three independent experiments were performed (n = 6 cell culture samples per group). Scale, 30 μm. e, NGFR mRNA levels in human LECs exposed to sEVs from melanocytes or different melanoma cell lines for 48 h. Data were collected from two independent experiments (n = 6 samples per group). f-h, Representative images of LYVE-1 and GFP-expression in LNs 16 h after the injection of B16-F1-NGFR-GFP sEVs. White lines delineate areas of LYVE-1+ lymphatic network. Quantification of GFP+ area and mean fluorescence in LYVE-1 regions are shown in (g) and (h), 4-3 whole sections per LN were analyzed. Data were obtained from 2 independent experiments (n = 6 LNs per group). Scale bar, 200μm (left images) and 50 μm (right image). i,j, Representative flow cytometry plots and quantification of NGFR+ cells in HLECs exposed to SK-MEL-147 sEVs for 4h. Two independent experiments were performed (n = 3 samples per group). All data represent mean ± s.e.m. and p values were calculated by two-tailed Student’s t test in g, h and j and by one-way ANOVA in c and e.
To investigate if other secreted factors could promote the induction of NGFR expression, we treated hLECs with SK-MEL-147 sEVs or sEV-depleted conditioned medium and observed that only the sEV fraction induced NGFR mRNA (Fig. 5c) and protein expression (Fig. 5d). Interestingly, NGFR expression was induced by SK-MEL-103 and SK-MEL-147-secreted sEVs but not by sEVs derived from less aggressive cell lines or primary melanocytes (Fig. 5e).
We tested the horizontal transfer of NGFR protein in vivo using sEVs from B16-F1 cells overexpressing NGFR-GFP fusion protein, injecting them intra-footpad, and found that they localized to LYVE-1+ areas (Fig. 5f). Quantification showed that the GFP percentage in LYVE-1+ areas and its fluorescence intensity increased 4- and 1.5-fold, respectively, in LNs exposed to sEVs derived from B16-F1-NGFR-GFP compared to B16-F1-GFP-derived sEVs (Fig. 5g,h), suggesting GFP-NGFR shuttling in vesicles and uptake by LN cells. We further confirmed the increase in NGFR levels by flow cytometry in vitro. The percentage of NGFR-expressing hLECs increased 4-fold when cells were exposed to SK-MEL-147-derived sEVs for 4 hours compared to untreated cells (Fig. 5i,j).
These findings show that NGFR expression is induced in LECs through two mechanisms: 1) small but significant horizontal transfer of NGFR protein by sEVs and 2) strong de novo induction of expression of NGFR mRNA.
sEV-loaded NGFR induces MAPK and NF-κB pathways in LECs
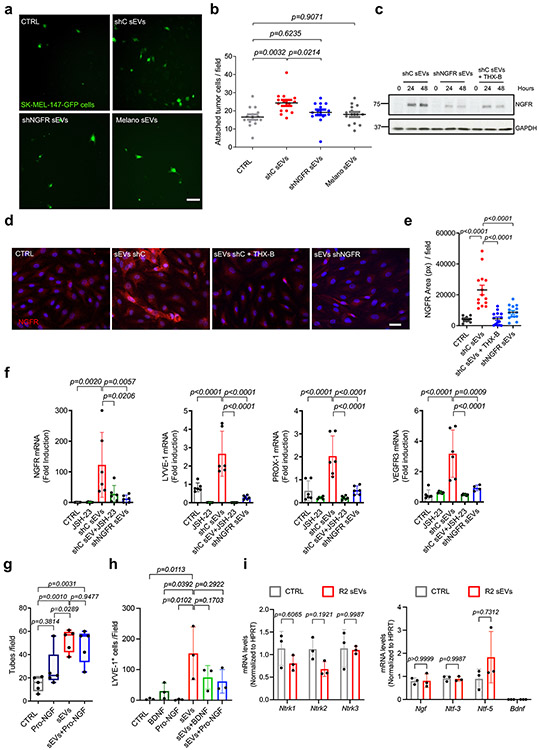
To investigate if the transcriptional and functional effects of sEVs on LECs were dependent on NGFR, we generated a human SK-MEL-147 NGFR knock-down cell line (shNGFR) and a B16-F1R2 Ngfr knock-out cell line (NGFR KO) using CRISPR technology (Extended Data Fig. 8a, b). NGFR depletion did not affect sEV number or protein abundance (Extended Data Fig. 8c) nor sEV uptake by LN cells (Extended Data Fig. 8d). Proteomic analysis of shNGFR versus control (shC) SK-MEL-147 sEVs revealed a group of 126 significantly downregulated proteins, including NGFR, and 281 other significantly upregulated proteins (Supplementary Table 3). Enrichment analysis showed that processes related to metabolism were overrepresented in the group of significantly downregulated proteins (Extended Data Fig. 8e).
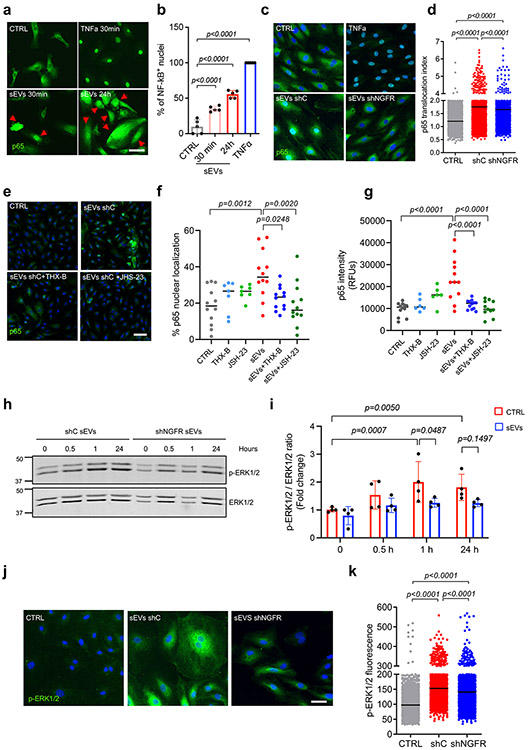
We next analyzed the signaling pathways affected by sEVs in LECs. We focused on NF-κB and MAPK pathways since these are well-known pathways activated downstream of NGFR19, 43, 44. Treatment with SK-MEL-147-derived sEVs promoted the nuclear translocation of p65 in hLECs after 30 minutes and 24 hours compared to untreated hLECs (Fig. 6a,b), TNF-α was used as positive control. Importantly, p65 translocation induced by melanoma-derived sEVs was significantly reduced by treatment with shNGFR sEVs (Fig. 6c,d). We corroborated the effect of NGFR on NF-κB activation by using the NGFR small molecule inhibitor THX-B45. Treatment with THX-B significantly reduced both p65 nuclear translocation and p65 expression induced by sEV treatment at 48 hours (Fig. 6e-g, dark blue dots vs red). The NF-κB inhibitor JSH-23 was used as a control (Fig. 6e-g, dark green dots vs red).
Figure 6. Melanoma sEVs activate NGFR, MAPK and NF-kB signaling pathways in LECs.
a,b, Representative images and quantification of p65 staining in HLECs after exposure to TNFα or SK-MEL-147-derived sEVs for the indicated times. Arrows indicate cells with p65 nuclear staining (n = 5 samples per group). Scale bar, 50 μm. c,d, Representative confocal images and quantification of p65 staining in HLECs in basal conditions or after the addition of TNFα, control shRNA or NGFR shRNA SK-MEL-147 sEVs for 30 min. p65 translocation was analyzed using a confocal high content screening system. Plot shows data from one representative experiment out of two (CTRL, n = 5325 cells, CTRL shC, n = 3355 cells and shNGFR, n = 3005 cells). Scale bar, 40 μm. e-g, Representative confocal images and quantification of nuclear area (f) and average fluorescence intensity (g) for p65 staining in HLECS exposed to of SK-MEL-147-derived sEVs in the presence or absence of NF-kB inhibitor JSH-23 or NGFR inhibitor THX-B. Data were collected from two independent experiments (n = 12 samples per group except THX-B and JSH-23 groups, n = 7 samples for f and n = 6 samples for g). h,i, Representative WB of ERK1/2 phosphorylation levels in HLECs treated with shControl (shC) and shNGFR SK-MEL-147-derived sEVs for the indicated times. Data were collected from four independent experiments (n = 4 samples per group). j,k, Representative confocal images and quantification of phospho-ERK1/2 in HLECs in basal conditions or after the addition of shControl (shC) or shNGFR SK-MEL-147-derived sEVs for 30 min. Phospho-ERK1/2-associated cell fluorescence was analyzed using a confocal high content screening system. Plot shows data from one representative experiment out of two (CTRL, n = 4777 cells, CTRL shC, n = 3419 cells and shNGFR, n = 2323 cells). Scale bar, 50 μm. All data represent mean ± s.e.m. and p values were calculated by one-way ANOVA except by two-way ANOVA in i.
In addition to NF-κB activation, SK-MEL-147-derived sEVs also induced phosphorylation of ERK1/2, which was reduced after NGFR depletion in sEVs (Fig. 6h-k). MEK inhibition abolished sEV-dependent ERK1/2 activation (Extended Data Fig. 8f). Since the AKT/mTOR pathway is a mediator of tumor-induced lymphangiogenic signals46, we also investigated if phosphorylation of AKT and/or mTOR was activated upon sEV treatment. However, we did not observe significant changes (Extended Data Fig. 8g). These data support that activation of NF-κB and MEK in LECs by melanoma-derived sEVs depend on NGFR expression or activity in sEVs.
sEV-secreted NGFR promotes adhesion and lymphangiogenesis
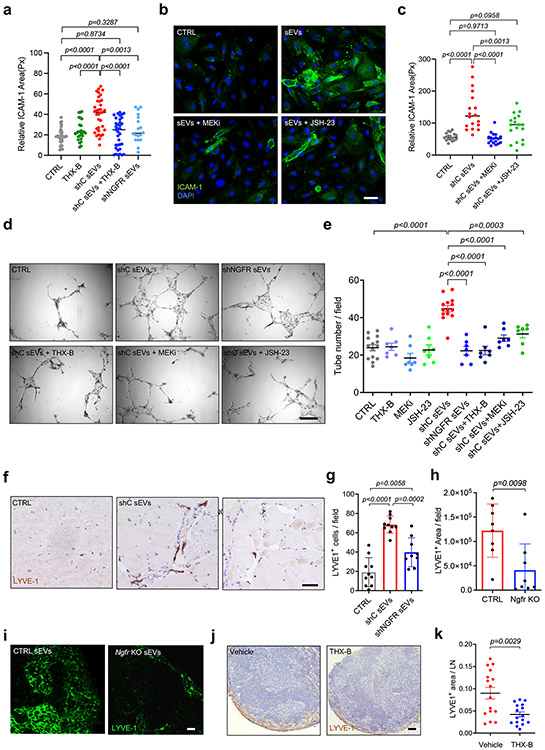
We next studied if the phenotypic changes induced in LECs by melanoma-derived sEVs were dependent on NGFR. We found that ICAM-1 overexpression induced in hLECs by shC sEVs in LECs was significantly reduced when sEVs from NGFR knock-down cells (shNGFR sEVs) or control sEVs plus THX-B were administered (Fig. 7a). Similarly, MEK or NF-κB inhibitors abolished ICAM-1 induction in hLECs (Fig. 7b,c). Consequently, melanoma cell adhesion to a hLEC monolayer was also reduced after treatment with sEVs derived from shNGFR cells (Extended Data Fig. 9a,b). Intriguingly, we noticed that the induction of NGFR gene by sEVs in hLECs was in part due to an autoregulatory loop, since it was reduced by depleting NGFR from sEVs or administering THX-B in combination with shC sEVs (Extended Data Fig. 9c-e). Induction of NGFR and lymphatic/lymphangiogenic genes by sEVs was also controlled by NGFR and the NF-κB pathway, as their expressions were reduced by shNGFR sEVs or shC sEVs in combination with JSH-23 inhibitor (Extended Data Fig. 9f).
Figure 7. sEVs induced ICAM-1 expression and lymphangiogenesis through NGFR pathway.
a, Quantification of ICAM-1 fluorescence area in HLECs treated with shControl (shC) or shNGFR SK-MEL-147-derived sEVs for 48 h with or without THX-B. Data were collected from three independent experiments (CTRL, n = 36, THX-B, n = 26, shC sEVs and shC sEVs+THX-B, n = 35, shNGFR sEVs, n = 18 fields per group). b,c, Representative images and quantification of ICAM-1 fluorescence of HLECs treated with SK-MEL-147-derived sEVs with or without MEK inhibitor or JSH-23. Data were collected from two independent experiments (n = 18 fields per group). Scale bar, 40 μm. d,e, Representative images and quantification of endothelial tubbing assays performed with HLECs previously exposed to shControl (shC) or shNGFR SK-MEL-147-derived sEVs for 48 h with or without THX-B, JSH-23 or MEK inhibitors (CTRL and shC sEVs groups, n = 14 samples and rest of groups, n = 7 samples per group). Scale bar, 100 μm. f,g, Representative images and quantification of LYVE-1+ cells in PBS, shControl (shC) or shNGFR sEVs-embedded matrigel plugs 15 days post-injection. Scale bar, μm 150. Two independent experiments were performed (CTRL and shC sEVs groups, n = 10 plugs analyzed and shNGFR sEVs group, n = 9 plugs analyzed). h,i, Quantification of LYVE-1 area and representative images of popliteal LNs from mice educated with intra-footpad injections of B16-F1R2 Ngfr KO or control (CTRL) sEVs for 13 days. (n = 8 LNs analyzed). Scale bar, 40 μm. j,k, Representative LYVE-1 histological staining in inguinal LNs of mice with B16-F1R2 flank tumors and corresponding quantification. Animals were treated or not intraperitoneally with 2.5 mg/Kg of THX-B twice a week, starting on day 7 post-injection of tumor cells. Animals were sacrificed at 21 days (n = 16 LNs analyzed). Scale bar, 200 μm. All data represent mean ± s.e.m. and p values were calculated by one-way ANOVA in a- g and by two-tailed Student’s t test in h and k.
In agreement with this finding, depletion of NGFR in sEVs or use of NGFR, MEK, and NF-κB inhibitors impaired lymphatic branching formation induced by melanoma sEVs in in vitro tube formation assays (Fig. 7d-e). To analyze if LEC lymphangiogenic behavior induced by NGFR was dependent on ligands (neurotrophins), we investigated the possible effect of pro-NGF, one of the main ligands of NGFR19. We found that treatment with pro-NGF did not influence tube formation nor changed the effect of SK-MEL-147 sEVs in vitro in LECs (Extended Data Fig. 9g). In vivo analysis showed that SK-MEL-147-derived sEVs efficiently increased the number of LYVE-1+ cells in matrigel plugs; however pro-NGF and brain-derived neurotrophic factor (BDNF) did not influence LYVE-1+ cell number nor modified the effect of sEVs (Extended Data Fig. 9h). Finally, education with B16-F1R2 sEVs LNs did not influence the expression of neurotrophin receptor genes Ntrk1, Ntrk2 and Ntrk3 nor their ligands Ngf, Ntf-3, Ntf-5 and Bdnf in popliteal LN (Extended Data Fig. 9i). Together, these data suggest that LEC lymphangiogenesis induced by melanoma-derived sEVs is driven by NGFR through a ligand-independent mechanism.
Analysis of lymphangiogenesis in vivo using matrigel plugs embedded with shNGFR SK-MEL-147-derived sEVs showed that reduction of NGFR in sEVs decreased the number of LYVE-1+ cells compared to shC sEVs (Fig. 7f,g). Importantly, we also observed that lymphatic vessel area, as measured by LYVE-1 staining, was decreased in LNs educated with NGFR KO B16-F1R2 sEVs compared to control sEVs (Fig. 7h,i). Consistent with these findings, THX-B treatment of mice bearing B16-F1R2 flank tumors decreased LN lymphangiogenesis, as measured by LYVE-1 network area (Fig. 7j,k).
sEVs-loaded NGFR promotes LN metastasis and reduces survival
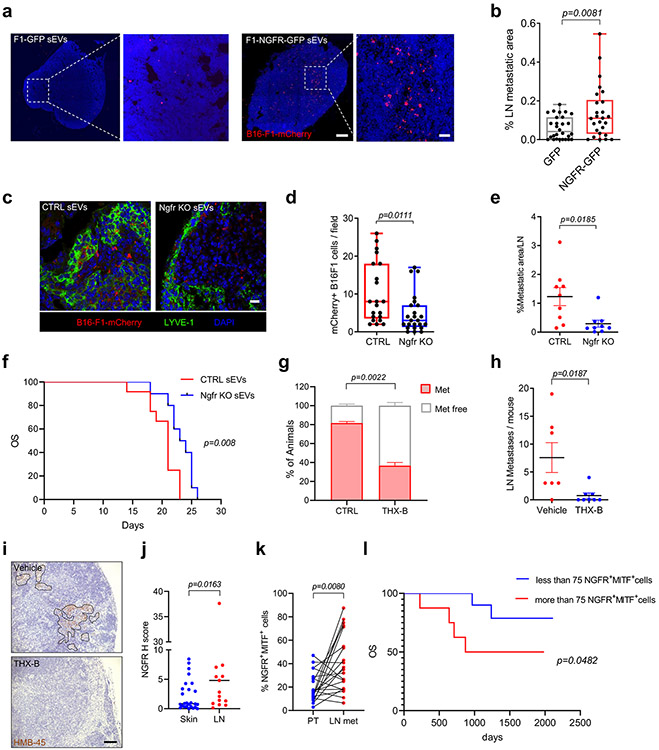
We investigated if sEV-shed NGFR could be responsible for the enhanced pro-metastatic capacity of melanoma-derived sEVs. We performed education experiments using sEVs overexpressing or depleted of NGFR. To determine the requirement for NGFR in metastasis, we injected sEVs intra-footpad twice a week for 10 days and, one day after the last injection of sEVs, we injected B16-F1-mCherry intra-footpad (see schematic diagram in Extended Data Fig. 1j). We found that education with NGFR over-expressing sEVs increased popliteal LN metastasis (Fig. 8a, b). Conversely, education with NGFR KO sEVs significantly reduced the number of metastases in mice compared to control sEV conditioning (Fig. 8c,d). We further performed spontaneous LN metastasis assays (see scheme in Extended Data Fig. 1l). In this setting, we observed that education with NGFR KO sEVs significantly reduced spontaneous LN metastasis (Fig. 8e). Furthermore, animals treated with NGFR KO sEVs had significantly prolonged survival compared to those exposed to control sEVs (Fig. 8f).
Figure 8. EV-shed NGFR favors LN metastasis and influences survival.
a,b, Representative images and quantification of B16-F1-mCherry+ cells in popliteal LNs. Mice were educated with B16-F1-GFP and B16-F1-NGFR-GFP sEVs as in Fig. 1i. Data from two independent experiments (n =10 mice per group, GFP group 28 LN sections and NGFR-GFP group 26 LN sections). Scale bar, 200 μm and 40 μm. c,d, Representative images and quantification of B16-F1-mCherry+ cells in popliteal LNs. Animals were educated with control or Ngfr KO B16-F1R2 sEVs as in Fig. 1i (n = 5 mice per group; CTRL group, n = 21 LN sections and Ngfr KO group, n = 24 LN sections). Scale bar, 20 μm. e, Metastatic area in mice educated with control and Ngfr KO B16-F1R2 sEVs as described in Fig. 1k. Two independent experiments were performed (n = 9 mice per group). f, Survival of animals educated with control and Ngfr KO B16-F1R2-secreted sEVs as indicated in (e) (CTRL sEV, n = 12 mice and Ngfr KO sEV, n = 10 mice). g-i, Percentage of animals (g) number of LN metastases (h) and representative images (i) in animals bearing B16F1-R2 flank tumors treated or not with THX-B as in Figure 7j (vehicle n = 7 mice and THX-B n = 8 mice). j, NGFR h Score in skin and LN sections from melanoma patients (n = 26 skin/soft tissue samples and n = 17 LN samples). k, Percentage of NGFR+MITF+ tumor cells in skin and LN samples from melanoma patients (n = 21 matched samples). l, Overall survival (OS) of stage II/III melanoma patients according to NGFR+MITF+ cell number in LN biopsies (less than 75 NGFR+MITF+ cells, n = 13 patients, more than 75 NGFR+MITF+ cells, n = 12 patients). Boxes and whiskers in the box plots in b and d are defined as in Fig. 1). All other data represent mean ± s.e.m f, k and l. and p values were calculated by two-tailed Student’s t test with Welch’s correction b, d and e, by two-sided Mann-Whitney test in j, by two-sided paired Wilcoxon matched-pairs signed rank test in k and two-sided log rank test in f and l.
Analysis of the effect of NGFR loss on tumor growth and metastasis showed that although B16-F1R2 NGFR KO primary tumors grew slightly faster to control tumors (Extended Data Fig. 10a), the number of LN metastases was significantly reduced (Extended Data Fig. 10b,c). Likewise, the number of metastatic lesions in popliteal LNs was significantly diminished among mice injected intra-footpad with NGFR KO B16-F1R2 cells than among those injected with control cells (Extended Data Fig. 10d). In agreement with these results, THX-B treatment after flank tumor initiation by B16-F1R2 cells reduced the number of animals with LN metastases as well as the average number of metastatic lesions per LN (Fig. 8g-i).
We next explored the relevance of NGFR expression in LN metastasis in melanoma patients. We first used TCGA data to compare NGFR mRNA levels in primary and metastatic melanoma tumors. NGFR expression was found to be significantly higher in metastatic tumors (Extended Data Fig. 10e). Similarly, this receptor was expressed at higher levels in metastatic tumors with LN involvement compared to primary tumors with LN involvement (Extended Data Fig. 10f). To corroborate these findings, we performed histological analyses of NGFR expression in a cohort of 44 Stage III/IV melanoma patients and found that NGFR expression was significantly higher in metastatic LN tissues than in skin lesions (Fig. 8j). Evaluation of another independent cohort of 25 Stage III melanoma patients with matched primary tumor and LN tissue biopsies uncovered a significant increase in the frequency of NGFR+ tumor cells (identified by co-staining with the melanoma marker MITF) in LN metastases compared to matched primary tumors (Fig. 8k). Remarkably, the number of NGFR+MITF+ cells in LN biopsies predicted patient survival (Fig. 8l). These results reveal a role for NGFR in the early steps of metastatic spreading in lymph nodes.
Discussion
We found from our research that, in LNs, sEVs are taken up mainly by LECs, followed by macrophages. Previous studies demonstrated the uptake of melanoma EVs by LN immune cells, including subcapsular macrophages and B cells16, but at later time points during the metastatic cascade. Our data support the hypothesis that LECs are the first cell type to encounter tumor-derived sEVs in LNs while macrophages can uptake these sEVs, but to a slower and lesser extent. Thus, it is plausible that there is a hierarchy in tumor-derived sEV uptake in LN that involves several sequential steps. Melanoma-derived EVs are known to distribute through the lymphatic system and LNs9, 10, 16. In addition, melanoma-secreted sEVs are detected in the lymphatic drainage from melanoma patients and described to interact with LECs17, 18. Our data support the idea that LECs are the primary barrier taking up melanoma-derived EVs, promoting lymphangiogenesis and tumor cell adhesion. Results using the CD169DTR model to deplete SCS macrophages suggest that they are important in restricting LEC basal proliferation and may be limiting the interaction of sEVs with LECs, as recently described with B cells16.
Whether sEVs can reach LNs by passive or active mechanisms is a matter of debate. Integrins have been shown to be involved in organotropic homing of exosomes in lung or liver13. Our data confirm a role for integrins in sEV uptake by LECs, since blocking of integrin αv or GRGDSP-binding integrins reduces their uptake by 50% and 20%, respectively. However, proteomic analysis did not reveal differences among sEV integrins between murine melanoma cell lines with distinct metastatic potentials and we have not determined the specific integrin combination involved or if additional molecules play a role in the uptake. We observed an increased protein cargo and reduced z-potential in sEVs derived from highly metastatic models. Our data support the hypothesis that the combination of biophysical properties and surface proteins in sEVs influences sEV dissemination, homing, and uptake by LECs in LNs. Nevertheless, it would be interesting to explore whether various subpopulations of sEVs may differ in functionality.
Changes in cell adhesion molecules in the lymphatic vasculature have been found to be associated with tumor-induced lymphovascular niches in LNs47. In our model, ICAM-1 upregulation in LECs facilitated the adhesion of tumor cells through a NGFR-dependent mechanism induced by melanoma-derived sEVs. Our data support the idea that melanoma cells adhere to ICAM-1 on the LEC surface in a manner akin to immune cells48, 49. Since ICAM-1 is upregulated in inflamed lymphatic capillaries regulating cell adhesion and transmigration of immune cells50, 51, our data suggest that tumor cells may hijack this molecular mechanism in order to metastasize to LNs. Moreover, there was an overall upregulation of ICAM-1 in other LN populations suggesting that this molecule could support PMN formation by favoring the adhesion of tumor cells to LECs and possibly to other cell types.
Our data also support the notion that tumor-derived sEVs promote lymphangiogenesis by inducing the expression of lymphangiogenic genes, such as VEGF-C or VEGF-A, and the activation of VEGFR3. In addition, we observed a sustained induction of lymphangiogenesis in lymphoreporter mice, indicating that tumor-derived sEVs are actively involved in LN remodeling through a mechanism dependent on NGFR. Recently, the induction of lymphangiogenesis by tumor-derived sEVs has been depicted in other cancer models39, 52, 53 validating the important role of sEVs, in addition to other soluble factors, in lymphangiogenesis6, 7.
Among the genes upregulated in hLECs by melanoma-derived sEVs, NGFR was the top candidate packaged in sEVs, horizontally transferred and expressed de novo in LECs. NGFR is involved in melanocyte development in the neural crest54. Our study suggests that, in addition to its known intrinsic functions in melanoma cells, NGFR secreted in melanoma-derived sEVs influences stromal cells in the tumor microenvironment21-27, 55. Interestingly, NGFR expression is mainly observed in a small population of melanoma stem cells with high metastatic abilities21, 25. The properties of melanoma-derived sEVs expressing NGFR in a heterogeneous population versus non-expressing sEVs as well as the impact of sEV subpopulations on metastatic behavior remain to be explored.
Importantly, the variability of NGFR levels in our cell/sEV models suggests that the combination of NGFR and additional cargo in melanoma-derived sEVs contribute to LN metastasis. Our results support a model in which NGFR shed in sEVs leads to the education of LECs and in turn reinforces lymphangiogenesis and LN metastasis along with other soluble factors secreted by tumors.
A previous study demonstrated that NGFR can be secreted in sEVs from neural models56, but NGFR secretion in sEVs has never been described in the context of tumor metastasis. We found that NGFR secreted in sEVs orchestrates local changes in LECs favoring lymphangiogenesis and LN metastasis in melanoma. We observed that NGFR secretion in sEVs promotes: 1) p65 nuclear translocation in LECs, 2) induction of phospho-ERK1/2 and endogenous NGFR expression, 3) lymphangiogenesis in vitro and in vivo, 4) tumor cell adhesion and 5) LN metastasis and decreased survival in preclinical models. Importantly, depletion of NGFR or its pharmacological inhibition in melanoma-derived sEVs not only reduced lymphangiogenesis and LN metastasis but also impaired the activation of NF-κB and MAPK signaling pathways, demonstrating that NGFR shedding and downstream signaling in LECs are crucial in metastatic outcome.
The NF-κB pathway activates VEGFR-3 promoter in LECs, ultimately driving lymphatic vessel formation57. Consistent with this prior observation, our findings demonstrated that melanoma-derived sEVs promoted the activation of VEGFR-3 in LECs. Importantly, inhibition of NF-κB signaling blocked LEC sprouting and pro-lymphangiogenic gene induction by NGFR-loaded sEVs, supporting the idea that NF-κΒ as one of the main downstream effectors of NGFR in LECs in the absence of NGFR ligands58-60.
Finally, analysis of NGFR in matched sLNs and primary tumors from melanoma patients showed that the number of melanoma cells expressing NGFR in LNs correlated with a worse patient survival. The fact that patients with a frequency of NGFR+ tumor cells in LNs above a threshold progress faster could imply that these initial steps in the metastatic cascade influence disease progression and points to this receptor as critical to early melanoma metastatic dissemination in LNs and a novel potential therapeutic target. Studies with larger cohorts are, however, required to assess their potential. Analyzing NGFR in biofluids of patients with pre-metastatic disease could help to verify if measuring sEV NGFR in liquid biopsies are useful to predict patient outcome.
Taken together, our findings offer clues to a better understanding of the earliest events in metastatic spread, demonstrating that tumors corrupt their surrounding microenvironment (e.g. LNs) and prepare favorable niches for metastasis via the uptake of sEVs. We also provide insights into how NGFR expression in LN metastatic cells could be useful in the clinical setting to predict melanoma patient outcome.
Methods
Ethical regulations
The research performed in the present study complies with all ethical regulations. For retrospective clinical studies, all patients signed written informed consent form and accepted or did not explicitly refuse the use of their data for research purposes. The protocols were approved by Kantonale Ethikkommission Zurich (protocol EK647-EK800) and by Maria Sklodowska-Curie National Research Institute of Oncology Ethical Committee (protocol 27/2018). Experiments using animals were performed in accordance with the protocol 709-666A approved by the Weill Cornell Medicine IACUC’s committee in USA or the protocols PROEX178/15 and PROEX225/17 approved by the ISCIII /Comunidad de Madrid Ethical Committee in Spain. Animal experiments were performed in accordance with the guidelines for Ethical Conduct in the Care and Use of Animals as stated in The International Guiding Principles for Biomedical Research involving Animals, developed by the Council for International Organizations of Medical Sciences (CIOMS). When required, tumor size did not exceed the maximum volume (1500mm3) allowed by the ISCIII Ethical Committee.
Patient samples
The study included two independent cohorts. Cohort 1 consisted of 44 stage III/IV melanoma patients for unpaired analysis of tumors and LN metastases and cohort 2 involved 25 Stage II/III melanoma patients with paired tumor/ regional LN biopsies. Additional information is provided in Supplementary Tables 4 and 5. Biopsies were obtained from the University Hospital Zurich in Switzerland (cohort 1) or the Maria Sklodowska-Curie National Research Institute of Oncology in Poland (cohort 2).
Cell lines and reagents
Detailed information about cell lines is included in Supplementary Table 6. All melanoma cell lines were grown in high glucose DMEM (Lonza) supplemented with 10% fetal bovine serum (Hyclone), 2 mM glutamine and 20 μg/mL gentamicin. Primary melanocytes were cultured in 254CF medium (Gibco) supplemented with human melanocyte growth supplement (Gibco); melan-a61 were cultured in RPMI (Gibco), supplemented with 5% fetal calf serum (Gibco) and 200 nM 12-o-tetradecanoyl phorbol-13-acetate (Sigma). Human dermal lymphatic microvascular endothelial cells HMVEC-dLyAd and human lymph node endothelial cells HLEC were cultivated in Clonetics EGM-2 MV BulletKit (Lonza) following the manufacturer’s instructions. Human lymphatic fibroblasts HLF were cultured in high glucose DMEM (Lonza) supplemented with 10% fetal bovine serum (Hyclone). All cells were grown at 37°C in a humidified 5% CO2 atmosphere.
When indicated, HLECs were treated with the following compounds: NGFR inhibitor THX-B (15 μM) and MEKi PD0325901 (1 μm) synthetized by CNIO Experimental Therapeutics Program; NF-kB inhibitor JSH-23 (5 μM, Merck), GRGDSP peptide (10 ng/mL, Merck), anti-integrin αv blocking antibody (0.1 μg/mL, Merck), Fc soluble ICAM-1 (10 μg/mL, R&D Systems), diphtheria toxin (10 μg/Kg mouse body weight, Sigma Aldrich), human Pro-NGF (50ng/mL, Alomone) and human BDNF (5 ng/mL, Peprotech).
Mice
8-12 weeks-old C57BL/6J males from Envigo and JAX were used unless otherwise specified for sEV in vivo distribution, sEV conditioning, lymphangiogenesis and analysis of metastatic outcome. 11-15 weeks Vegfr3/FLT4 EGFP-Luc lymphoreporter females37 (obtained from Dr. Sagrario Ortega (CNIO, Spain) were used for in vivo lymphangiogenesis assays. Prox-1-tdTomato and Prox-1-GFP reporter males were purchased from JAX and used for analysis of sEV uptake. CD169DTR males (obtained from Dr. Andres Hidalgo, CNIC, Spain) were described elsewhere40, 62 and were used for studying sEV-induced effects on LN macrophages. Animals were housed at 21°C+/−2°C and humidity was 50-60%. Light cycle was light:13 hours/dark: 11hours.
EV purification
Cells were cultured in medium supplemented with 10% sEV-reduced FBS (FBS, Hyclone). FBS was reduced of bovine sEVs by ultracentrifugation at 100,000 x g for 70 min. Supernatant fractions collected from 72 h cell cultures were pelleted by centrifugation at 500 x g for 10 min. The supernatant was centrifuged at 12,000 x g for 20 min. sEVs were then harvested by centrifugation at 100,000 x g for 70 min. The supernatant (conditioned medium) was collected and used as control. The sEV pellet was resuspended in 20 mL of PBS and collected by ultracentrifugation at 100,000 x g for 70 min. All spins were performed at 10 °C using a Beckman Optima X100 centrifuge with Beckman 70Ti rotor, sEVs were resuspended in PBS and the protein content was measured by bicinchoninic acid assay (Pierce). When indicated, sEVs were further purified by discontinuous density gradient layering 1.5 mL of each of the 40, 30, 20, 10 and 5% (w/v) iodixanol solutions prepared with Optiprep™ (60%w/v) in 0.25 M sucrose/1 mM EDTA/10 mM Tris-HCl, (pH 7.5). Samples were placed on top and ultracentrifuged at 100,000 x g for 16h at 10 °C. Sequential fractions of 0.75 mL were collected, washed with 2.5 mL of PBS and ultracentrifuged at 100,000 x g for 70 min. Pellets were resuspended in 40 μl of PBS.
When indicated, sEVs pellets were labeled with 3 μl of infrared fluorescent dye NIR815 (eBiosciences), DiD (Molecular Probes), CSFE (Thermo Fisher) or PKH26 (Sigma) in 1 mL of PBS. Labeled sEVs were washed twice in 20 mL of PBS, collected by ultracentrifugation and resuspended in PBS. For Z potential measurement, sEVs with a concentration of 108 particles/mL were acquired in a Zetasizer Ultra (Malvern Panalytical) and analysis was performed using the ZS xplorer v1.0 software (Malvern Panalytical) .
Electron Microscopy
For negative staining, 3 μl of each sample was applied onto glow-discharged, carbon-coated, electron microscopy grids and incubated for 30 s. After the sample was adsorbed and excess was blotted, grids were deposited successively on top of three different 50 μl drops of MilliQ water and blotted. Grids were laid on the top of two distinct 50 μl drops of 1% uranyl formate, stained for 1 min and air dried. Grids were visualized on a JEM-2200FS (JEOL Ltd.) field emission gun operated at 200 keV. Images were taken at nominal magnification x8,000 with K3 direct electron detector (Gatan).
Distribution analysis of sEVs through the lymphatic system
8-weeks old C57BL/6J female mice were injected intra-footpad or in the ear lobule to deliver sEVs directly into the lymphatic circulation as described. A total of 5 μg of NIR815-labeled sEVs were injected intra-footpad in a total volume of 20 μl PBS twice a week for 3 weeks. Mice were monitored using In-Vivo Multisprectral System FX(Kodak) or IVIS Spectrum imaging system (Perkin Elmer). For ear vessels and cervical LN analyses, a single dose of 5 μL containing 2 μg/μL of PKH-26-labeled sEVs in PBS was injected in the earlobe. After 2 h and 24 h later, ear skin, and draining LNs were collected, embedded in O.C.T., cryo-sectioned, stained and visualized by confocal microscopy.
Education and LN colonization studies
To analyze the role of sEVs in LN tumor metastasis, 5 μg of sEVs in 30 μL of PBS or PBS alone were injected in the footpad of 8- to 10-week-old C57BL/6J males every 3 days for 10 days. On day 11, 5 x 104 B16-F1-mCherry cells in 30 μL of PBS were injected intra-footpad. Animals were sacrificed 10 days later and LN were dissected, fixed in 4 % PFA and frozen at −80 °C in O.C.T. To study the influence of sEVs in spontaneous metastasis, animals were injected with 5 μg of sEVs in 30 μL of PBS in the footpad every 3 days for 21 days. On day 2, 1 x 105 B16-F1-GFP cells were injected in the flank of educated animals. Mice were sacrificed 20 days later and LN were dissected, fixed in 4 % PFA and paraffin-embedded. For quantification of mCherry+ area, Whole LN sections were analyzed capturing mosaic images (6-20 per LN) using a 20X objective with an Eclipse E800 microscope (Nikon) or a TCS SP5 confocal laser scanning microscope (Leica). FIJI software was used for calculating fluorescence area. For quantification of HMB-45 area at least 4 whole sections of each LN were analyzed. Images were captured with an Olympus AX70 microscope applying CellSens 1.18 software (Olympus). FIJI 2.0 software was used for color deconvolution and staining area measurement.
Immunofluorescence
6 μm O.C.T. LN sections were treated with glycine 100 mM for 10 mins. After 3 washes with PBS, sections were incubated with PBS-0.3% Triton X-100 for 15 min. Samples were blocked with PBS containing 1% BSA, 5% Donkey Serum and 0.05% Triton X-100 for 1 h at RT. After 3 washes, tissues were incubated with primary antibodies at 4 °C overnight. Antibodies are listed in Supplementary Table 7. After another 3 washes, sections were incubated with secondary antibodies from the Alexa Fluor series (dil:1/500, Molecular Probes) for 1h and washed again. Samples were mounted with Prolong-DAPI (Thermo) and images were obtained using an Eclipse TE2000U confocal microscope (Nikon) and analyzed using Nikon software (EZ-C1 3.6) or using TCS-SP-5 Leica confocal microscope and Fiji software.
Proteomic analysis
Samples were lysed in urea and digested with Lys-C/trypsin using the standard FASP protocol. Peptides were analyzed by LC-MS/MS analysis using an LTQ Orbitrap Velos mass spectrometer (Thermo Scientific). Raw files were analyzed with MaxQuant against a mouse or human protein database and the MaxLFQ algorithm was used for label-free protein quantification. Proteomic analysis of shNGFR vs shControl SK-MEL-147-derived sEVs was performed as previously described63.
For integration of RNAseq performed in hLECs treated with SK-MEL-147 sEVs and the proteomic profile of SK-MEL-147 sEVs, 1311 RNA-protein pairs were considered. The total number of significantly co-regulated pairs was 322 (FDR < 5%) applying a protein fold change >2 or <−2.
Flow cytometry
LNs were prepared for flow cytometry as described65. Each sample consisted of a pool of 4 popliteal LNs. LNs were incubated in freshly prepared RPMI-1640 containing 0.8 mg/mL dispase, 0.2 mg/mL collagenase P and 0.1 mg/mL DNase at 37°C for 20 min. After incubation, LNs were disrupted by pipetting. Cell suspension was transferred to new tubes and the enzymatic mix was removed by centrifugation. Afterwards, another cycle of incubation and disruption with fresh enzymatic mix was applied for 10 min. Single cell suspensions were filtered and washed in PBS, 2 mM EDTA and 1% BSA and incubated with the primary antibodies described in Supplementary Table 8. To define cell viability DAPI (Thermo Fisher) was used. For sEV analysis, 15 μg of sEVs were incubated with aldehyde/sulphate latex beads (4 μm, Thermo) for 1 h at RT and subsequently blocked with 0.1M Glycine during 30min. After wash with PBS twice, beads were incubated with the indicated antibodies for 30 min on ice. Data were acquired on BD FACSCanto II and Fortessa LSR cytometers (BD), at least 10,000 single live events were acquired using FACS Diva 6.1.3 and 9.0 (BD) and data were analyzed using FlowJo software v10 (TreeStar).
CRISPR editing and shRNA tools
The sequences and commercial plasmids used for CRISPR editing and shRNA knock-down are described in Supplementary Table 9. Lentiviruses were prepared as previously described64. Briefly, 9 μg of target lentiviral vector, prepared with the EndoFree Plasmid Maxi Kit (QIAGEN, USA) was transfected in HEK293T cells together with 3 μg of pMD2.G (Addgene plasmid #12259), 5 μg of psPAX2 (Addgene plasmid #12260,) and 62.5 μl of Lipofectamine™ 2000 (Life Technologies) in 1 10-cm dish plate, 48 hours later supernatant containing lentiviruses was recovered and filtered through a 0.45 um low-binding protein filter (Millipore). Melanoma cells were transduced with lentiviral supernantants at low MOI.
RNA extraction, sequencing and data analysis
Total RNA was extracted using TRIzol Reagent (Invitrogen). After the recovery of the aqueous phase using chloroform, a DNase treatment and further purification were performed using the Mini RNeasy kit (Qiagen) following manufacturer instructions.
Total RNA (1 μg) from the samples was used. cDNA libraries were generated using the Illumina TruSeq RNA Sample Preparation Kit, and sequenced on an Illumina HiSeq1500 sequencer. 50bp single-end sequenced reads were analyzed with the nextpresso pipeline66, as follows: sequencing quality was checked with FastQC v0.10.1. Reads were aligned to the human genome (GRCh37/hg19) with TopHat-2.0.10 using Bowtie 1.0.0 and Samtools 0.1.1.9, allowing two mismatches and 5 multi hits. Transcript quantification and differential expression were calculated with Cufflinks 2.2.1, using the human GRCh37/hg19 transcript annotations from https://ccb.jhu.edu/software/tophat/igenomes.shtmL. GSEAPreranked was used to perform gene set enrichment analysis of the KEGG pathways database signatures on a pre-ranked gene list, setting 1,000 gene set permutations. Only those gene sets with significant enrichment levels (FDR q-value < 0.15) were finally considered. A single-sample GSEA was further obtained from the GSEA significant signatures.
Quantitative PCR
Total RNA was extracted using the RNeasy kit (Qiagen) and reverse transcription was performed using Superscript III reverse transcriptase kit (Invitrogen). Samples were analyzed for specific gene expression using indicated pre-designed TaqMan® assays or designed oligos (Supplementary Table 9.) Quantitative real-time PCR was performed on a 7500 Fast Real Time PCR System (Applied Biosystems), using TaqMan Universal PCR (Thermo) or SYBR® Green Master Mix (Applied Biosystems). Relative expression was calculated following delta delta Ct calculation method. HPRT/Hrpt and 18S genes were used as housekeeping genes.
NGFR Analysis in human clinical samples
Immunohistochemistry was performed in 5 μm sections. Tissues were stained with hematoxylin and eosin (H&E). Antibodies against human NGFR and MITF described in Supplementary Table 10 were used for staining. NGFR was scored as negative or positive taking into account expression detection in tumor cells. Image analysis was performed with QuPath v0.1.2 and Zen. Survival curves were estimated with the Kaplan–Meier product-limit method and compared using Mantel-Cox log-rank test.
Apoptosis assays
HLECs cells were growing until confluence. 5 μg/mL of SK-MEL-147 sEVs were added to the plate. After 48h, cells were harvested and stained first with TMRE (Thermo Fisher) for 30 min at 37°C and subsequently with DAPI. Samples were analyzed in a Fortessa LSR cytometer (BD), 30,000 single live events were acquired and data were analyzed using FlowJo software v10 (TreeStar).
Adhesion assays
HLECs were grown to form a monolayer on gelatin-coated M6 plates and treated with 5 μg/ml of sEVs for 72 h. CSFE-labeled 50,000 SK-MEL-147 cells were then added to each well. Cells were gently washed twice with medium 3 h later and fixed in 4% PFA. Images were taken with a Leica LED microscope. For analysis in flow conditions, HMVECs plated in μ-Slide IV 0.4 flow chambers (Ibidi) were incubated during 24 h with 5 μg/ml PKH-67-labeled sEVs. Subsequently a flow of SK-MEL-147 cells (500,000 cells/mL) at 0.0625 mL/min passed through the flow chambers during 4 h. Images were taken on a DM16000B Widefield microscope (Leica Microsystems) and analyzed with FIJI 2.0 software.
Endothelial cell tube formation assays
HLECs or HMVECs were incubated with 5 μg/mL of SK-MEL-147-derived sEVs or PBS. The inhibitors JHS-23 (5 μM), MEKi (1 μM) or THX-B (15 μM) were added simultaneously with the sEV treatment. After 48 h, the medium was removed and cells were starved in EBM-2 basal medium for 3 h. Cells were harvested and 25,000 cells in 200 μL of EBM-2 basal medium were added to each well of a 96-well plate coated with growth factor-reduced matrigel (Corning). Tube formation was evaluated 16 h later using a Leica LED microscope (Leica microsystems).
Lymphangiogenic assay in 2D co-cultures of human LECs and fibroblasts.
Co-cultures of human LECs and fibroblasts were prepared by mixing 75,000 human lymphatic fibroblasts HLF and 15,000 HMVECs in EBM-2 basal medium and were added to each well of gelatin-coated 48-well plates. 5 μg/mL of SK-MEL-147-derived sEVs were added 8 h later to the co-cultures. Medium and sEVs were changed every 2 days. On day 7, cultures were fixed with 4% paraformaldehyde for 15 min. After washing and blocking with 1% BSA, 5% Donkey Serum and 0.05% triton X-100 for 1 h at room temperature, cultures were incubated with rabbit anti-CD31-FITC (Supplementary Table 7) as indicated above. Images were taken with a Leica LED microscope (Leica microsystems).
Matrigel plug assays
SK-MEL-147-derived sEVs (5 μg/mL) or identical volume of PBS were embedded in growth factor reduced/phenol free matrigel (Corning). A total volume of 150 μL was injected subcutaneously into the ventral region of the animals. After 15 days, animals were sacrificed and plugs were fixed in formalin and embedded in paraffin for histological analysis. Rabbit anti-LYVE-1 (ab14917, Abcam) was used. Images were captured with an Olympus AX70 microscope using CellSens software.
Lymphoreporter experiments
Vegfr3/FLT4 EGFP-Luc nu/nu immunodeficient female mice received 3 doses of B16-F10-derived sEVs (10 μg) in the footpad for a week. Animals were anesthetized with isoflurane 4%, injected with 150 mg/kg luciferin (PerkinElmer) and then LNs were collected. Luciferase bioluminescence imaging was performed using an IVIS-SPECTRUM imaging system (PerkinElmer).
Immunoblotting
sEVs (10 μg) were lysed in Laemmli buffer at 95 °C for 5 min and the sEV protein extracts and RIPA whole cell lysates were resolved by SDS-PAGE and probed using the antibodies listed in Supplementary Table 10. Signals were detected using ECL Western Blotting Substrate kit (GE Healthcare). Western blots were cropped in Adobe Photoshop CC 2017 and GIMP 2.8.
High content screen imaging
HLECs were grown in EGM-2 MV medium on gelatin-coated 96-well plates. 5 μg/mL of SK-MEL-147 sEVs were added to wells and cells were fixed 30 min later in 4% PFA. Immunofluorescence procedure was performed as indicated above. Around 40 confocal fields were automatically acquired from each well by the High Content Screening (HCS) platform Opera LX (Perkin Elmer). Image analysis was performed using Acapela Software V2.0. Nuclei identification was performed using the DAPI channel and cytoplasmic regions were determined through the specific antibody signal. Translocation index was calculated as the nucleus versus cytoplasm antibody signal ratio. At least, 2,000 cells were analyzed per condition.
Statistics and reproducibility
Sample size for in vitro and in vivo experiments was selected based on pilot studies and previous similar studies 13, 15, 46, as well as on availability of transgenic mice. Animals were randomly allocated for exosome injection studies. For studies concerning exosome injection and subsequent tumor cell inoculation, animals were also randomly allocated prior to exosome injection. For all other experiments, no specific randomization method was followed as this was irrelevant to in vitro assays. The researchers were blinded during final outcome assessment of mouse experiments (in vivo imaging , fixed image capturing, FACS data collection, etc) but not during allocation and during treatments for mouse experiments as it was neccesary to know which were the groups to treat with sEVs. For the rest of in vitro experiments researchers were not blinded but experiments were performed by different individuals. The error bars in the graphical data represent the means ± s.e.m unless otherwise specified. For in vivo and in vitro assays, specifics regarding number of independent experiments and biological replicates are stated in the figure legends unless when only one experiment was performed. In those cases only the number of replicates was stated. When appropriate, the statistical significance was determined applying the two-tailed unpaired or paired Student’s t test or One-way or Two-way ANOVA or non-parametric tests using GraphPad Prism software version 9.1.0.
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Extended Data
Extended Data Fig. 1. Biophysical properties and cargo of melanoma-derived sEVs.
a, Analysis of sEV (ALIX, CD81 and CD9) and non-sEV markers (GM130 and CALNEXIN) in sEVs from the indicated mouse melanoma cell lines purified by ultracentrifugation. Two independent experiments were performed (n = 2 samples per group). b, Electron microscopy images of the indicated sEVs after iodixanol density gradient. Two independent experiments were performed (n = 2 samples per group). Scale bar, 200 μm. c, Analysis of sEV markers in iodixanol density gradient fractions obtained for B16F10 and B16-F1R2 sEVs. EEA-1 was included as non-sEV marker. Two independent experiments were performed (n = 2 samples per group). d,e, Measurement of the average and mode diameter size and protein content of sEVs purified by ultracentrifugation from the indicated cell lines. Two independent experiments were performed (n = 6 samples per group) f, Z-potential measurements of B16-F1, B16-F10 and B16-F1R2-derived sEVs. N = 2 samples per group. g, Venn diagram for significant upregulated proteins in B16-F10 and B16-F1R2-secreted sEVs compared to B16-F1-derived sEVs. Statistically significant changes were defined using a Student’s t test (FDR < 5%, p value < 0.05). Fold change was set at 1.32 (log2). h, Significantly enriched pathways associated with the upregulated proteins found in B16-F10 and B16-F1R2-secreted sEVs compared to B16-F1-derived sEVs. The enrichment analysis was performed using ClueGO pluggin and used a two-sided test for p value calculation followed by Bonferroni step down correction for p adjusted value. Padj, p adjusted value. i, Quantification of integrins detected by mass spectrometry in the indicated sEVs (n = 2 samples per group). j-l, Schemes showing the experimental planning for LN education experiments. j, Doses of 5 μg of melanoma-derived sEVs were injected intra-footpad every 2 days for the indicated days. At indicated time points, 50,000 B16-F1mCherry or B16-F1-GFP cells were injected intra-footpad (j, k) or in the flank (l). Data represent mean ± s.e.m. and p values were calculated by two-sided Kruskal-Wallis test in d and by one-way ANOVA in e and f.
Extended Data Fig. 2. Analysis of sEV distribution in LNs.
a, Representative images of colocalization of labeled B16-F1R2-secreted sEVs with LYVE-1+ LECs and CD169+ macrophages in popliteal LNs. Nodules were analyzed 16 h after intra-footpad injection of DiD-labeled sEVs. Two experiments were performed (n = 4 mice per group). Scale bar, 20 μm. b, Gating strategy for the analysis of sEV uptake by LN populations. c, sEV-associated fluorescence in the indicated CD45− cell populations in popliteal and inguinal LNs 16 h after intra-footpad injection of DiD-labeled B16-F10-secreted sEVs determined by flow cytometry. Three experiments were performed (n = 3 samples per group). BECs, blood endothelial cells; FRCs, fibroblastic reticular cells. d, Representative plots showing sEV-associated fluorescence in the indicated LN macrophage populations treated in the same conditions as in (c). Three experiments were performed (n = 3 samples per group). e, Percentage of LECs and CD169+F4/80+ macrophages exhibiting sEV-associated fluorescence at the indicated times after intra-footpad injection of B16-F1R2 sEVs as determined by flow cytometry. Three experiments were performed (n = 3 samples per group). Data represent mean ± s.e.m and p values were calculated by two-way ANOVA.
Extended Data Fig. 3. Characterization of SK-MEL-147-derived sEVs.
a, Representative profile of nanoparticle size distribution analyzed by nanotracking analysis (NTA) of SK-MEL-147-derived sEVs. Three experiments were performed (n = 3 samples per group). b, Electron microscopy images of SK-MEL-147-secreted sEVs after iodixanol density gradient. Two experiments were performed (n = 2 samples per group). Scale bar, 200 μm. c, Analysis of sEV markers (ALIX, CD63 and CD81) and non-sEV markers (GM130 and CALNEXIN) in SK-MEL-147-derived sEVs purified by ultracentrifugation compared to whole SK-MEL-147 cell extracts. Two experiments were performed (n = 2 samples per group).d, Analysis of sEV markers in the sequential fractions obtained after iodixanol density gradient of a representative SK-MEL-147-derived sEV preparation. EEA-1 was included as non-sEV marker. Two experiments were performed (n = 2 samples per group).
Extended Data Fig. 4. Human LECs incorporate melanoma-secreted sEVs.
a, Representative bright field image of HLECs growing in monolayer, images were obtained from three independent experiments (n = 3 samples). Scale bar, 100 μm. b, Co-staining of lymphatic markers PROX-1 and CD31 in HLECs cultures (n = 3 samples). Scale bar 150 μm .c, Representative immunofluorescence (left panel) and bright field (right panel) images of cultured HLECs exposed to CSFE-labeled SK-MEL-147 sEVs for 16 h. Data were collected from two independent experiments (n = 4 samples per group). Scale bar, 100 μm. d, Representative flow cytometry plots showing apoptotic cell levels in HLECs treated with SK-MEL-147 sEVs for 48 h. Numbers on the gates show the percentage of live cells in each condition (n = 2 samples per group). e, Representative immunofluorescence and bright field images of cultures of HLECs 8 h after exposure to CSFE-labeled SK-MEL-147- or human primary melanocytes (Melano)-derived sEVs. Two experiments were performed (n = 3 samples per group). Scale bar, 100 μm. f, Representative flow cytometry plots for in vitro sEV uptake. Human LECs were treated with PKH67-labeled SK-MEL-147 or melanocyte sEVs and fluorescence was measured at the indicated time points (n = 2 samples per group). g, Median fluorescence intensity (MFI) signal obtained by flow cytometry analysis from measurements performed in (e), (n = 2 samples per group). h, i, Modal distribution and quantification of mean fluorescence intensity (MFI) of sEV-associated fluorescence in HLECs upon exposure to DiD-labeled SK-MEL-147 sEVs in combination with 10 μg/mL GRGDSP or 0.1 μg/mL anti-integrin αv antibody for 16 h obtained by flow cytometry. Control condition (CTRL) represented HLECs treated with the DiD dye alone. Data were collected from two independent experiments (CTRL, sEVs and sEVs+anti-αv groups, n = 4 samples, sEVs+GRGDSP group, n= 3 samples). Data represent mean ± s.e.m and p values were calculated by one-way ANOVA.
Extended Data Fig. 5. Melanoma sEVs promote transcriptional changes in LECs.
a, Single sample GSEA analysis showing KEGG significantly downregulated signatures obtained by RNAseq analysis in human LECs upon exposure to SK-MEL-147-derived sEVs during 48 h (n = 3 samples per group). b, GSEA plots of neural-related gene signatures exhibiting significant enrichment in sEV-treated LECs versus non treated cells according to RNAseq data. Nominal p value <0.0001. c, mRNA levels analyzed by qPCR of some of the most up-regulated genes obtained by RNAseq in hLECs. Cells were incubated with SK-MEL-147-derived sEVs (sEVs) or PBS for 24 h and 48 h or with sEV-depleted conditioned medium (CM) from SK-MEL-147 cells for 24 h. Data were collected from two independent experiments (n = 5 independent cell cultures per group) d, GSEAs showing positive enrichment of adhesion-related signatures in sEV-treated LECs versus non treated cells. Nominal p value <0.01. e,f, ICAM-1 expression in HLECs treated SK-MEL-147 sEVs during 48 h. Two experiments were performed (n = 4 samples per group). Scale bar, 25 μm. g,h Quantification and representative images of tumor adhesion on HLECs monolayer. HLECs were pre-treated with SK-MEL-147. sEVs during 48 and subsequently incubated with 5,263 tumor cells/cm2 for 3 h before fixation. Two independent experiments were performed (n = 9 samples per group). Scale bar, 50 μm. Data represent mean ± s.e.m and p values were calculated by one-way ANOVA in c and by two-tailed Student t-test in f and g.
Extended Data Fig. 6. Melanoma-derived sEVs promote lymphangiogenesis.
a, GSEA plots for lymphangiogenesis and angiogenesis-related gene sets positively enriched in HLECs treated with SK-MEL-147-derived sEVs for 48 h compared to control LECs (n = 3 samples per group). Nominal p value <0.01. b, Number and length of branches/tubes in endothelial cell tube assays in HLECs incubated for 48 h with SK-MEL-147 sEVs and subsequently plated on matrigel for 16 h. Data were collected from two independent experiments (n = 7 samples per group). c,d, Representative images and quantification of endothelial cell tube assays performed in HMVECs treated in the same conditions as described in (b). Data were collected from two independent experiments (n = 5 samples per group) Scale bar, 100 μm. e,f, Representative images and quantification of tubular structures in co-cultures of HMVECs (CD31+ cells) and HLF fibroblasts treated for 48 h with SK-MEL-147 sEVs and untreated (PBS) control. Two independent experiments were performed (CTRL group, n = 6 samples, SK-MEL-147 sEVs group, n = 8 samples). Scale bar, 50 μm. g, Quantitative PCR analysis of LYVE-1 and Prox-1 genes in sorted cell populations from popliteal LNs of Prox-1-tdTomato mice 24 h after injection of DiD-labeled B16-F10 sEVs or control dye (n = 4 samples per group). h,i, Representative histological images of popliteal LNs stained with LYVE-1 (magenta) and Ki67 (brown) and corresponding quantification of Ki67+Lyve+ cells. Animals were injected intra-footpad 3 times with B16-F1-R2 sEVs for 1 week (n = 5 mice per group, CTRL, n = 8 LN sections and R2 sEVs 10 LN sections). Scale bar, 150 μm. Boxes and whiskers in the box plots in b and d are defined as in Fig. 1. Data represent mean ± s.e.m and p values were calculated by two-tailed Student t-test in b, d and g or by two-tailed Student t test with Welch’s correction in f and i.
Extended Data Fig. 7. sEVs induce macrophage-independent changes in LECs.
a, Expression of pro-lymph-angiogenic genes in sorted CD169+F4/80+ macrophages. LNs were harvested 48h after intra-footpad injection of B16-F1-R2 sEVs. Data were collected from two independent experiments (n = 4 mice per group). b, Percentage of LECs with DiD-sEV associated fluorescence analyzed by flow cytometry. WT or CD169DTR mice were treated with diphtheria toxin and 48 h later, B16-F1-R2 sEVs were injected intra-footpad. Animals were sacrificed 24 h later. (n = 5 mice per group). c, Measurement of proliferating PROX-1+ LECs in popliteal LNs from WT and CD169DTR mice. Animals were treated with diphtheria toxin and 24h later, B16-F1-R2 sEVs were injected intra-footpad. Mice were sacrificed 48 h later (n = 5 mice per group). d, Comparison of NGFR mRNA levels (z-score) in cell lines from different tumor types obtained from the CCLE database (melanoma, n = 58; endometrium cancer, n = 25; breast cancer, n = 56; ovary cancer, n = 44; pancreatic cancer n = 42; lung cancer, n = 166 ; central nervous system (CNS) cancer, n = 47; and stomach cancer, n = 34). e, NGFR mRNA levels in a panel of primary melanocytes (Melano1 and Melano2) and human melanoma cell lines. Data were acquired from two independent experiments (n = 4 samples per group). f, Ngfr mRNA levels in mouse melanoma B16 cell lines. Data were collected from two independent experiments (all groups, n = 4 independent cell cultures per group. g, Ngfr fluorescence distribution analyzed by flow cytometry in mouse melanoma sEVs. Plot shows a representative analysis of two independent experiments (n = 2 samples per group). h, NGFR protein levels in human melanoma cell lines compared to primary melanocytes analyzed from a published mass spectrometry data set. (Melano and SK-MEL-147 groups, n = 4 samples, WM-164 and SK-MEL-28 groups, n = 3). Data represent mean ± s.e.m and p values were calculated by two-tailed Student t-test in a and by one-way ANOVA in b-f and h.
Extended Data Fig. 8. NGFR knock-down in metastatic melanoma cell lines.
a, Representative WB showing NGFR protein levels in control (shC) or NGFR shRNA (shNGFR) SK-MEL-147 whole cell lysates and secreted sEVs. Two independent experiments were performed (n = 2 samples per group). b, NGFR protein levels in whole cell lysates and sEVs from control (CTRL) and Ngfr KO B16-F1R2 cells. Two independent experiments were performed (n = 2 samples per group). c, Measurement by nanoparticle tracking analysis (NTA) of the number of particles (right plot) and protein content (left plot) after paired purification of sEVs from the indicated B16-F1R2 cell lines. Data were collected from six independent experiments (n = 6 samples per group). d, Percentage of LN cell types incorporating DiD-labeled sEVs 16 h after footpad injection of control (CTRL) and Ngfr KO B16-F1R2-derived sEVs (n = 3 mice per group). e, Overrepresented pathways associated with the significantly downregulated proteins found in shNGFR SK-MEL-147-derived sEVs compared to shC SK-MEL-147-derived sEVs (n = 3 samples per group). Pathways were obtained using PANTHER software overrepresentation tool applying Fisher’s exact test and FDR for multiple comparison correction. Padj,adjusted p value. f, Representative immunoblotting displaying ERK1/2 phosphorylation levels in HLECs treated with 5 μg/mL of SK-MEL-147-derived sEVs for the indicated times in the presence or absence of 1 μM MEK inhibitor PD0325901. Total ERK-1/2 levels are shown as loading control. Two independent experiments were performed (n = 2 cell samples per group). g, Quantification of phospho-AKT and phospho-mTOR staining in HLECs in basal conditions or after the addition of SK-MEL-147 sEVs for 30 min. Fluorescent signal was measured using Opera high content screening system. (Phosho-AKT fluorescence, CTRL cells n = 3598, sEV n = 3457. Phospho mTOR fluorescence, CTRL n = 517, sEV n =1509). Data represent mean ± s.e.m and p values were calculated by two-tailed paired Student t-test in c, by one-way ANOVA in d and by two-tailed unpaired Student t-test in g.
Extended Data Fig. 9. NGFR promotes tumor adhesion and lymphangiogenesis in LECs.
a,b, Images and quantification of SK-MEL-147-GFP tumor cells attached to a monolayer of human LECs after 3 h incubation. hLECs were pre-treated for 48 h with control shRNA (shC) or NGFR shRNA (shNGFR) SK-MEL-147 sEVs. Treatment with primary melanocytes-derived sEVs (Melano sEVs) was included as additional control. Two independent experiments were performed (n = 14 samples per group). Scale bar, 50 μm. c, NGFR protein levels in hLECs treated with control (shC) or NGFR shRNA (shNGFR) SK-MEL-147 sEVs for 24 h and 48 h. When indicated, cells were treated with THX-B 2 h before harvesting. Two independent experiments were performed (n = 2 samples per group). d,e, Images and quantification of NGFR staining in HLECs incubated with shControl or shNGFR SK-MEL-147 sEVs in the presence or absence of THX-B. Data were acquired from two independent experiments (n = 14 images per group). Scale bar, 25 μm. f, Expression of NGFR and pro-lymphangiogenic genes in HLECs treated with shControl or shNGFR SK-MEL-147 sEVs for 48 h in the presence or absence of JSH-23. Two independent experiments were performed (n = 6 samples per group). g, Quantification of tube structures in endothelial cell tube assays performed in HLECs incubated for 48 h with SK-MEL-147 sEVs alone or in combination with Pro-NGF (n = 5 samples per group). h, Quantification of LYVE-1+ cells in histological sections of matrigel plugs 15 days after implantation. SK-MEL-147-derived sEVs were embedded alone or in combination with BDNF or Pro-NGF in matrigel immediately prior to implantation (n = 3 plugs per group). i, Expression levels of neurotrophin receptors and NGFR ligands in popliteal LN exposed to B16-R2 sEVs for 48 h (n = 3 mice per group). Boxes and whiskers in the box plots in g are defined as in Fig. 1. All other data represent mean ± s.e.m and p values were calculated by one-way ANOVA in b-h and two-way ANOVA in i.
Extended Data Fig. 10. NGFR influences LN metastasis.
a, Growth curves of flank tumors in mice injected with 200,000 control or Ngfr KO B16-F1R2 cells (n = 6 mice per group). b, HMB-45 histological staining in inguinal LN sections in mice from experiment described in (a) (n = 6 mice per group). Scale bar 150 μm. c, Metastatic foci in inguinal LN sections of animals from experiment described in (a) (n = 6 mice per group). d, Metastatic lesions in LN of mice 15 days after intra-footpad injection of 50,000 control or NGFR KO B16-F1R2 cells (n = 6 mice per group). e, NGFR mRNA levels in primary tumors (PT) and metastatic tumors (Met) according to TCGA melanoma data set (PT group, n = 103 patients and Met group, n= 367 patients). f, NGFR mRNA levels in primary tumors (PT LN+) and metastatic tumors (Met LN+) with LN involvement. (PT LN+ group, n = 42 patients and Met LN+ group, n = 171 patients). Data represent mean ± s.e.m and p values were calculated by two-way ANOVA in a, by two-tailed Student t-test in c and d and by two-sided Mann-Whitney test in e and f.
Supplementary Material
Acknowledgements
We apologize those authors whose work could not be cited due to size restrictions. We thank Dr. MS. Soengas and the members of her lab for melanoma cells, primary melanocyte preparations and helpful discussions. We thank Dr. M. Detmar and Dr. S. Proulx for the mouse B16-F1-R2 cell line. We are grateful to Dr. M. Yañez-Mo and Dr. M. Valés for antibodies against sEV markers. We thank David Grela and Alberto Escobar from IESMAT for their support with the Zetasizer analysis. We thank Giovanna Roncador, Lorena Maestre and Jorge Luis Martinez Torrecuadrada for their help with the development and characterization of NGFR antibodies and Carolina Villarroya Beltri for her help in flow cytometry analysis.
This work was funded by The Starr Cancer Consortium (B.J.M., D.L. and H.P.), US NIH (R01-CA169416), The Nancy C. and Daniel P. Paduano Foundation, Children’s Cancer and Blood Foundation (H.P. and D.L). The Melanoma Research Alliance, The Feldstein foundation, RETOS SAF2017-82924-R (AEI/10.13039/501100011033/FEDER-UE), Fundación Ramón Areces, Fundación Bancaria "la Caixa" (HR18-00256), ATRES-MEDIA AXA Foundation (CONSTANTES Y VITALES, una iniciativa de laSexta y Fundación AXA) and Fundación Científica AECC (LABAE19027PEIN, GCB15152978SOEN-HP) (H.P.). Malcolm Hewitt Wiener Foundation, The AHEPA Vth District Cancer Research Foundation, The Hartwell Foundation, The Manning Foundation (D.L.).
We are also grateful for the support of the Translational NeTwork for the CLinical application of Extracellular VesicleS, TeNTaCLES. RED2018-102411-T(AEI/10.13039/501100011033), Ramón y Cajal Programme, FERO Foundation, Comunidad of Madrid 2017-T2/BMD6026 (L.N.), La Caixa Foundation ID100010434, Fellowship LCF/BQ/ES17/11600007 (A.H.B.). The CNIO, certified as Severo Ochoa Excellence Centre, is supported by the Spanish Government through the Instituto de Salud Carlos III (ISCIII).
Footnotes
Competing Interests Statement
The authors have no conflict of interests.
Data availability
The RNA-Seq data generated have been deposited in the Gene Expression Omnibus under the accession number GSE135187. Public proteomic data set on human melanoma cell lines-derived sEVs used was obtained from the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD0009505. Proteomic data for the comparison between mouse B16 cell lines were uploaded to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD024108. Proteomic data for the comparison between human shControl and shNGFR SK-MEL-147-derived sEVs were uploaded to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD024852. Information on procedures and analysis of the described sEVs have been included in the EV-track database (EV-TRACK ID is EV210127).
Source data have been provided as Source Data files. All other data supporting the findings of this study are available from the corresponding author on reasonable request
References
- 1.Shain AH & Bastian BC From melanocytes to melanomas. Nat Rev Cancer (2016). [DOI] [PubMed] [Google Scholar]
- 2.van Akkooi AC, Verhoef C & Eggermont AM Importance of tumor load in the sentinel node in melanoma: clinical dilemmas. Nat Rev Clin Oncol 7, 446–454 (2010). [DOI] [PubMed] [Google Scholar]
- 3.Harrell MI, Iritani BM & Ruddell A Tumor-induced sentinel lymph node lymphangiogenesis and increased lymph flow precede melanoma metastasis. Am J Pathol 170, 774–786 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sleeman JP The lymph node pre-metastatic niche. Journal of Molecular Medicine 93, 1173–1184 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Pastushenko I et al. Blood microvessel density, lymphatic microvessel density and lymphatic invasion in predicting melanoma metastases: systematic review and meta-analysis. British Journal of Dermatology 170, 66–77 (2014). [DOI] [PubMed] [Google Scholar]
- 6.Hirakawa S et al. VEGF-C-induced lymphangiogenesis in sentinel lymph nodes promotes tumor metastasis to distant sites. Blood 109, 1010–1017 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hirakawa S et al. VEGF-A induces tumor and sentinel lymph node lymphangiogenesis and promotes lymphatic metastasis. J Exp Med 201, 1089–1099 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alitalo A & Detmar M Interaction of tumor cells and lymphatic vessels in cancer progression. Oncogene 31, 4499–4508 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Srinivasan S, Vannberg FO & Dixon JB Lymphatic transport of exosomes as a rapid route of information dissemination to the lymph node. Sci Rep 6, 24436 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hood JL, San RS & Wickline SA Exosomes released by melanoma cells prepare sentinel lymph nodes for tumor metastasis. Cancer Res 71, 3792–3801 (2011). [DOI] [PubMed] [Google Scholar]
- 11.Becker A et al. Extracellular Vesicles in Cancer: Cell-to-Cell Mediators of Metastasis. Cancer cell 30, 836–848 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Witwer KW & Thery C Extracellular vesicles or exosomes? On primacy, precision, and popularity influencing a choice of nomenclature. J Extracell Vesicles 8, 1648167 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoshino A et al. Tumour exosome integrins determine organotropic metastasis. Nature 527, 329–335 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Costa-Silva B et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nature cell biology 17, 816–826 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peinado H et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med 18, 883–891 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pucci F et al. SCS macrophages suppress melanoma by restricting tumor-derived vesicle-B cell interactions. Science 352, 242–246 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garcia-Silva S et al. Use of extracellular vesicles from lymphatic drainage as surrogate markers of melanoma progression and BRAF (V600E) mutation. J Exp Med 216, 1061–1070 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Broggi MAS et al. Tumor-associated factors are enriched in lymphatic exudate compared to plasma in metastatic melanoma patients. J Exp Med 216, 1091–1107 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hempstead BL The many faces of p75NTR. Curr Opin Neurobiol 12, 260–267 (2002). [DOI] [PubMed] [Google Scholar]
- 20.Chesa PG, Rettig WJ, Thomson TM, Old LJ & Melamed MR Immunohistochemical analysis of nerve growth factor receptor expression in normal and malignant human tissues. J Histochem Cytochem 36, 383–389 (1988). [DOI] [PubMed] [Google Scholar]
- 21.Boiko AD et al. Human melanoma-initiating cells express neural crest nerve growth factor receptor CD271. Nature 466, 133–137 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Restivo G et al. low neurotrophin receptor CD271 regulates phenotype switching in melanoma. Nat Commun 8, 1988 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nielsen PS, Riber-Hansen R & Steiniche T Immunohistochemical CD271 expression correlates with melanoma progress in a case-control study. Pathology 50, 402–410 (2018). [DOI] [PubMed] [Google Scholar]
- 24.Guo R et al. Increased expression of melanoma stem cell marker CD271 in metastatic melanoma to the brain. Int J Clin Exp Pathol 7, 8947–8951 (2014). [PMC free article] [PubMed] [Google Scholar]
- 25.Civenni G et al. Human CD271-positive melanoma stem cells associated with metastasis establish tumor heterogeneity and long-term growth. Cancer Res 71, 3098–3109 (2011). [DOI] [PubMed] [Google Scholar]
- 26.Li S et al. Epigenetic regulation of CD271, a potential cancer stem cell marker associated with chemoresistance and metastatic capacity. Oncol Rep 33, 425–432 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boshuizen J et al. Reversal of pre-existing NGFR-driven tumor and immune therapy resistance. Nat Commun 11, 3946 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liersch R et al. Analysis of a novel highly metastatic melanoma cell line identifies osteopontin as a new lymphangiogenic factor. Int J Oncol 41, 1455–1463 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Proulx ST, Ma Q, Andina D, Leroux JC & Detmar M Quantitative measurement of lymphatic function in mice by noninvasive near-infrared imaging of a peripheral vein. JCI Insight 2, e90861 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Choi I et al. Visualization of lymphatic vessels by Prox1-promoter directed GFP reporter in a bacterial artificial chromosome-based transgenic mouse. Blood 117, 362–365 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu MH, Ustinova E & Granger HJ Integrin binding to fibronectin and vitronectin maintains the barrier function of isolated porcine coronary venules. J Physiol 532, 785–791 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ghislin S et al. LFA-1 and ICAM-1 expression induced during melanoma-endothelial cell co-culture favors the transendothelial migration of melanoma cell lines in vitro. BMC Cancer 12, 455 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnson LA & Jackson DG Cell traffic and the lymphatic endothelium. Ann N Y Acad Sci 1131, 119–133 (2008). [DOI] [PubMed] [Google Scholar]
- 34.Sigal A et al. The LFA-1 integrin supports rolling adhesions on ICAM-1 under physiological shear flow in a permissive cellular environment. J Immunol 165, 442–452 (2000). [DOI] [PubMed] [Google Scholar]
- 35.Vaahtomeri K, Karaman S, Makinen T & Alitalo K Lymphangiogenesis guidance by paracrine and pericellular factors. Genes Dev 31, 1615–1634 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Truman LA et al. ProxTom lymphatic vessel reporter mice reveal Prox1 expression in the adrenal medulla, megakaryocytes, and platelets. The American journal of pathology 180, 1715–1725 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martínez-Corral I et al. In vivo imaging of lymphatic vessels in development, wound healing, inflammation, and tumor metastasis. Proceedings of the National Academy of Sciences of the United States of America 109, 6223–6228 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alitalo K The lymphatic vasculature in disease. Nat Med 17, 1371–1380 (2011). [DOI] [PubMed] [Google Scholar]
- 39.Sun B et al. Colorectal cancer exosomes induce lymphatic network remodeling in lymph nodes. Int J Cancer 145, 1648–1659 (2019). [DOI] [PubMed] [Google Scholar]
- 40.Ballesteros I et al. Co-option of Neutrophil Fates by Tissue Environments. Cell 183, 1282–1297 e1218 (2020). [DOI] [PubMed] [Google Scholar]
- 41.Redmer T et al. The nerve growth factor receptor CD271 is crucial to maintain tumorigenicity and stem-like properties of melanoma cells. PLoS One 9, e92596 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fink DM et al. Nerve growth factor regulates neurolymphatic remodeling during corneal inflammation and resolution. PLoS One 9, e112737 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lehraiki A et al. Increased CD271 expression by the NF-kB pathway promotes melanoma cell survival and drives acquired resistance to BRAF inhibitor vemurafenib. Cell Discov 1, 15030 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Richard G et al. ZEB1-mediated melanoma cell plasticity enhances resistance to MAPK inhibitors. EMBO Mol Med 8, 1143–1161 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bai Y et al. Chronic and acute models of retinal neurodegeneration TrkA activity are neuroprotective whereas p75NTR activity is neurotoxic through a paracrine mechanism. The Journal of biological chemistry 285, 39392–39400 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Olmeda D et al. Whole-body imaging of lymphovascular niches identifies pre-metastatic roles of midkine. Nature 546, 676–680 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Commerford CD et al. Mechanisms of Tumor-Induced Lymphovascular Niche Formation in Draining Lymph Nodes. Cell reports 25, 3554–3563.e3554 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reymond N, d'Água BB & Ridley AJ Crossing the endothelial barrier during metastasis. Nature Reviews Cancer 13, 858 (2013). [DOI] [PubMed] [Google Scholar]
- 49.Madri J, Graesser D & Haas T The roles of adhesion molecules and proteinases in lymphocyte transendothelial migration. Biochemistry and cell biology = Biochimie et biologie cellulaire 74, 749–757 (1996). [DOI] [PubMed] [Google Scholar]
- 50.Johnson LA et al. An inflammation-induced mechanism for leukocyte transmigration across lymphatic vessel endothelium. J Exp Med 203, 2763–2777 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Teijeira A et al. Lymphatic endothelium forms integrin-engaging 3D structures during DC transit across inflamed lymphatic vessels. J Invest Dermatol 133, 2276–2285 (2013). [DOI] [PubMed] [Google Scholar]
- 52.Li M et al. Horizontal transfer of exosomal CXCR4 promotes murine hepatocarcinoma cell migration, invasion and lymphangiogenesis. Gene 676, 101–109 (2018). [DOI] [PubMed] [Google Scholar]
- 53.Zhou C-F et al. Cervical squamous cell carcinoma-secreted exosomal miR-221-3p promotes lymphangiogenesis and lymphatic metastasis by targeting VASH1. Oncogene 38, 1256–1268 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kasemeier-Kulesa JC & Kulesa PM The convergent roles of CD271/p75 in neural crest-derived melanoma plasticity. Dev Biol 444 Suppl 1, S352–S355 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mohamed A, Gonzalez RS, Lawson D, Wang J & Cohen C Tumor stem cells (CD271, c-kit, SOX10) in Melanomas: prognostic and outcome implications. Appl Immunohistochem Mol Morphol 22, 142–145 (2014). [DOI] [PubMed] [Google Scholar]
- 56.Escudero CA et al. The p75 neurotrophin receptor evades the endolysosomal route in neuronal cells, favouring multivesicular bodies specialised for exosomal release. J Cell Sci 127, 1966–1979 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Flister MJ et al. Inflammation induces lymphangiogenesis through up-regulation of VEGFR-3 mediated by NF-kappaB and Prox1. Blood 115, 418–429 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Foehr ED et al. NF-κB Signaling Promotes Both Cell Survival and Neurite Process Formation in Nerve Growth Factor-Stimulated PC12 Cells. The Journal of Neuroscience 20, 7556 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Carter BD et al. Selective Activation of NF-κB by Nerve Growth Factor Through the Neurotrophin Receptor p75. Science 272, 542 (1996). [DOI] [PubMed] [Google Scholar]
- 60.Vilar M et al. Ligand-independent signaling by disulfide-crosslinked dimers of the p75 neurotrophin receptor. Journal of cell science 122, 3351–3357 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
References (methods)
- 61.Bennett DC, Cooper PJ & Hart IR A line of non-tumorigenic mouse melanocytes, syngeneic with the B16 melanoma and requiring a tumour promoter for growth. Int J Cancer 39, 414–418 (1987). [DOI] [PubMed] [Google Scholar]
- 62.Miyake Y et al. Critical role of macrophages in the marginal zone in the suppression of immune responses to apoptotic cell-associated antigens. J Clin Invest 117, 2268–2278 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hoshino A et al. Extracellular Vesicle and Particle Biomarkers Define Multiple Human Cancers. Cell 182, 1044–1061.e1018 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Torres-Ruiz R et al. Efficient Recreation of t(11;22) EWSR1-FLI1+ in Human Stem Cells Using CRISPR/Cas9. Stem Cell Reports 8, 1408–1420 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fletcher AL et al. Reproducible isolation of lymph node stromal cells reveals site-dependent differences in fibroblastic reticular cells. Front Immunol 2, 35 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Graña O, Rubio-Camarillo M, Fdez-Riverola F, Pisano DG & Glez-Peña D. Nextpresso: Next Generation Sequencing Expression Analysis Pipeline. Current Bioinformatics 13, 583–591 (2018). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-Seq data generated have been deposited in the Gene Expression Omnibus under the accession number GSE135187. Public proteomic data set on human melanoma cell lines-derived sEVs used was obtained from the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD0009505. Proteomic data for the comparison between mouse B16 cell lines were uploaded to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD024108. Proteomic data for the comparison between human shControl and shNGFR SK-MEL-147-derived sEVs were uploaded to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD024852. Information on procedures and analysis of the described sEVs have been included in the EV-track database (EV-TRACK ID is EV210127).
Source data have been provided as Source Data files. All other data supporting the findings of this study are available from the corresponding author on reasonable request