Understanding the mechanisms by which specific genes are expressed in a temporal or tissue-specific manner is a central problem in biology. Transcription factors play a key role in this process. These proteins are modular and can be classified based on the structure of the domain that binds DNA. One type of DNA binding domain is defined by its requirement for zinc and has been designated a zinc finger motif (reviewed in reference 28). In this group, the C2H2 zinc fingers represent one of the most common types of DNA binding domains. The motif frequently occurs in tandem repeats and is defined by two cysteine and two histidine residues that coordinate a zinc ion and fold the domain into a finger-like projection that can interact with DNA (28). There are approximately 600 to 700 genes in the human genome encoding C2H2 motifs (7, 48a), suggesting that this class of transcription factors represents a substantial portion of the genes in the human genome.
Accompanying the zinc finger elements in the C2H2 class of zinc finger transcription factors are a variety of extended sequence motifs. These structural modules regulate subcellular localization, DNA binding, and gene expression by controlling selective association of the transcription factors with each other or with other cellular components. In the C2H2 class of zinc fingers, these associated modules include the poxvirus and zinc finger (POZ) domain (5), which is also known as the BTB domain (Broad-Complex, Tramtrack, and Bric-a-brac), the Kruppel-associated box (KRAB) (7), and the newly defined SCAN domain (51). These domains define subgroups and may provide insights into the functions of the members of this large family of zinc finger transcription factors. Here each of these domains will be introduced and representative members of the BTB/POZ and KRAB domain families will be used to place the more recently described SCAN domain family of transcription factors in perspective.
BTB/POZ DOMAIN
The BTB/POZ domain is an evolutionarily conserved protein-protein interaction domain that is found at the N terminus of some C2H2-type zinc finger transcription factors and in some actin binding proteins having a kelch motif (reviewed in reference 2). Examples of the BTB/POZ domain have been identified in organisms from yeast to human. With about 100 distinct BTB/POZ entries in available sequence databases (48a), it is estimated that 5 to 10% of the zinc finger proteins in humans contain these domains. Although there are many genes associated with this domain, two better-characterized members and their products will be overviewed to illustrate some of the interesting features of the family.
PLZF.
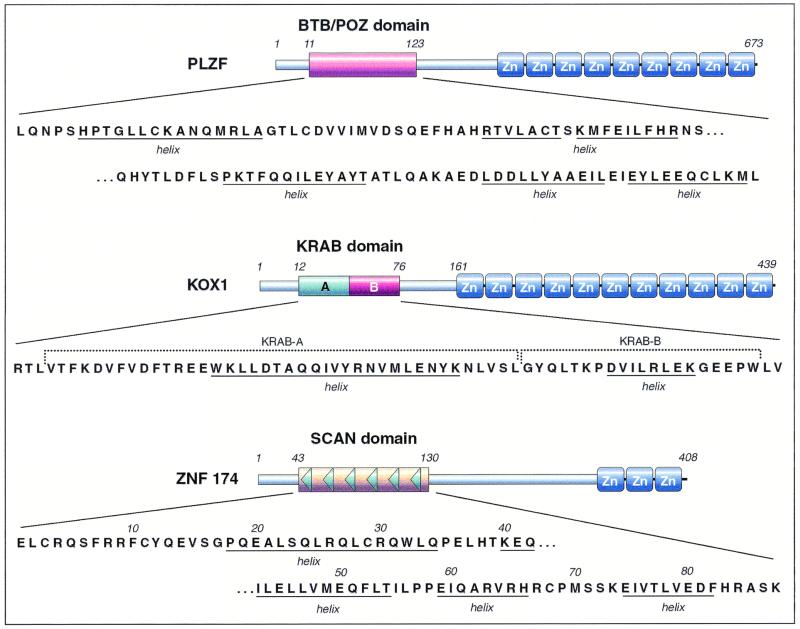
The organization of the promyelocytic leukemia zinc finger (PLZF) protein is typical of this group of proteins and consists of a single 120-amino-acid BTB/POZ domain found at the N terminus of the protein followed by a central region of several hundred amino acids and ending with nine C2H2 Kruppel-type zinc fingers (Fig. 1). PLZF has been implicated in embryonic development and hematopoiesis (for examples see references 6, 14, and 45). The BTB/POZ domain is associated with many of the biological functions of the PLZF protein, including oligomerization and transcriptional repression. The BTB/POZ domain of PLZF allows PLZF to self-associate and to form heteromeric complexes with other BTB/POZ domains, such as the Fanconi anemia zinc finger protein (17, 23). The crystal structure of the PLZF BTP/POZ domain reveals a tightly intertwined homodimer with an extensive dimer interface (1, 30). Detailed analysis of the PLZF domain demonstrated that a number of the evolutionarily conserved residues within the BTB/POZ domain are critical for dimerization. Mutations that disrupt the interface and block dimerization result in nonfunctional proteins. This supports the general concept that dimerization of the BTB/POZ domain is essential for the proper folding of the entire zinc finger protein (33).
FIG. 1.
Three types of C2H2-type zinc finger-associated domains. The structure of representatives of the BTB/POZ (PLZF) (1, 30), KRAB (KOX1) (37, 38), and the SCAN (ZNF174) (50, 51) domain families is drawn schematically. The primary amino acid sequences of each of the domains, as well as information about the actual (BTB/POZ) or predicted secondary structures of the domains, are provided. The A and B subregions of the KRAB domain are also indicated.
PLZF functions as a transcriptional repressor by binding to the promoters of target genes involved in the regulation of the cell cycle, such as cyclin A (54). The consequences of PLZF expression in hematopoietic cells include growth suppression, cell cycle arrest in the G1/S phase, and differentiation blockade (41). The BTB/POZ domain is in part responsible for the transcriptional repression mediated by PLZF. Mutational analysis of the BTB/POZ domain suggests that the highly conserved groove or charge pocket formed at the dimer interface mediates transcriptional repression (33). In one model for transcriptional repression, the BTB/POZ domain interacts with Sin3A, SMRT (silencing mediator for retinoid and thyroid hormone receptors), and N-CoR (nuclear receptor corepressor) (15, 21, 24, 31). These corepressors, in turn, recruit a histone deacetylase (e.g., HDAC1) to the promoter of the target gene, resulting in local changes in chromatin structure that diminish transcription. The PLZF gene is disrupted in patients with t(11;17)(q23;q21)-associated acute promyelocytic leukemia. In this disorder, the N-terminal region of PLZF is fused to the DNA and hormone binding domains of retinoic acid receptor α (RARα). The PLZF-RARα fusion protein represses transcription at retinoic acid-sensitive sites through BTB/POZ-mediated recruitment of a histone deacetylase-containing complex (21, 31). This recruitment of histone deacetylases seems to be critical to the transforming potential of the acute promyelocytic leukemia fusion protein. Recruitment of similar repressive complexes to the BTB/POZ domain in other members of the family (e.g., BCL-6/LAZ3) supports the generality of the suppressive mechanism. However, it should be noted that this model is not universal and that some members of the BTB/POZ family can interact but are not associated with repression. The reason why some of the members of the family act as repressors and others do not is an open question for the field.
Trithorax-like.
A second interesting example of the BTB/POZ family is the Drosophila melanogaster GAGA transcription factor, which is encoded by the essential Trithorax-like gene (18). Normal expression of several developmental regulatory genes requires GAGA. Based on biochemical studies, GAGA activates transcription by counteracting chromatin repression. Natural target promoters for GAGA typically contain multiple, closely spaced, GAGA binding sites. The BTB/POZ domain in GAGA is responsible for oligomerization of the transcription factor into higher-order complexes and strong cooperative binding of GAGA to multiple DNA sites. An attractive possibility is that oligomerization of GAGA mediated by the BTB/POZ domain bends the target gene promoter. This may result in disruption of nucleosomes and binding of factors that are unable to overcome the nucleosome barriers by themselves (18, 26). The function of GAGA may not be restricted to the activation of specific target genes. Analysis of Trithorax-like mutations in the fly suggests that GAGA counteracts heterochromatic silencing (19). Interestingly, punctate nuclear staining patterns are frequently seen with BTB/POZ proteins in higher organisms. For example, the BTB/POZ domain is required for PLZF to localize to nuclear speckles, which may represent the accumulation of the protein on chromatin (2, 17, 41). The versatile BTB/POZ domain mediates oligomerization and interaction with cofactors, ultimately leading to chromatin remodeling and changes in gene expression.
KRAB DOMAIN
A second C2H2 zinc finger-associated protein-protein interaction motif is the KRAB domain. The KRAB box is a conserved amino acid sequence motif at the amino-terminal end of proteins that contain multiple C2H2 zinc fingers at their carboxy termini (7) (Fig. 1). The KRAB domain has been estimated to be found in about one-third of the human genes encoding Kruppel-type C2H2 zinc fingers. Like the BTB/POZ domain, the KRAB domain is a transcriptional repression module (32, 52). In contrast to the BTB/POZ domain, however, the KRAB box appears to be vertebrate specific (48a). The KRAB domain itself spans approximately 75 amino acids and is divided into A and B boxes, where the A box plays a key role in repression (Fig. 1). Surprisingly, little is known about the structure of the KRAB domain, although it is predicted to contain two charged amphipathic helices. KRAB-containing zinc finger transcription factors probably perform a wide range of functions during hematopoietic cell development and differentiation. For example, some KRAB-containing zinc finger proteins are downregulated during myeloid differentiation (7), while others are largely restricted to lymphoid cells and may play a specific role in lymphoid differentiation (8). Interestingly, a set of KRAB-encoding genes are clustered on chromosome 19q13, many of which exhibit hematopoietic specific expression (22). Recently a product of a member of the KRAB family, designated ZBRK1, was shown to interact with BRCA1, a protein that has been implicated in the transcriptional regulation of DNA damage-inducible genes that function in cell cycle arrest (55). ZBRK1 functions as a transcriptional repressor by binding to a specific sequence within the gene of GADD45, a gene that encodes a product involved in cell cycle arrest.
Transcriptional repression and the KRAB domain.
The KRAB domain functions to repress transcription by recruiting corepressors. At least in part, repression is mediated by binding of the transcriptional corepressor KRAB-associated protein-1 (KAP-1 [20]), which is also known as KRAB-A interacting protein (KRIP-1 [27]) or transcription intermediary factor 1β (TIF1β [34]). In one schema for KRAB-mediated repression, the KRAB zinc finger protein specifically binds DNA through its array of C2H2 zinc fingers. The DNA-bound KRAB domain then recruits a homotrimer of the KAP-1 corepressor (37, 38). The bound corepressor then interacts with downstream target genes, such as the protein HP1 family of nonhistone heterochromatin-associated proteins (42). This family of proteins has an established gene-silencing function in Drosophila. Thus, the KRAB-KAP complex results in the formation of a heterochromatin-like complex, ultimately leading to gene silencing.
SCAN DOMAIN
A third type of extended sequence motif found in some zinc finger transcription factors is the SCAN domain. The name for this domain, which is also called LeR because it is a leucine-rich region, was derived from the first letters of the names of four proteins initially found to contain this domain [SRE-ZBP, CTfin51, AW-1 (ZNF174), and Number 18 cDNA or ZnF20] (51). A screen of both public and private (Celera) human genome databases with the SCAN domain suggests that there may be more than 60 members of the SCAN family in the human genome. Like the KRAB box, the SCAN domain appears to be vertebrate specific (48a). To date, all of the members of the SCAN family encode a single SCAN domain. The SCAN domain was originally defined to consist of about 80 highly conserved contiguous amino acids (51) (Fig. 1). As more members of the family are defined, a more detailed comparison of the residues in the domain becomes possible. Based on this new information, two residues have been added to the N-terminal end and five residues to the C-terminal end of the domain, defining the SCAN box as at least 87 residues in length (Fig. 2). Little is known about the structure of the SCAN domain, although three conserved proline residues probably serve to divide the SCAN domain into at least three predicted amphipathic α-helices (51). Like the BTB/POZ domain, the SCAN box is capable of mediating homo- and heterodimerization between specific members of the SCAN domain family of zinc finger transcription factors (43, 44, 50). An attractive possibility is that the domain generates combinatorial diversity within the SCAN family of proteins. However, the mechanisms involved in dimerization and partner selection are unknown. In striking contrast to the BTB/POZ and KRAB domains, the isolated SCAN domain is not associated with either transcriptional activation or repression (50, 51). Like the distribution of the genes for other zinc finger proteins, at least some of the genes for the members of the SCAN family are clustered in the human genome. For example, a disproportionate density of SCAN family members is found on 3p21-22 (e.g., ZnF20 and TRFA), 6p21-22 (e.g., ZNF192 and ZNF193), 16p13.3 (e.g., ZNF174, ZNF213, and ZNF315), and 17p12-13 (e.g., ZNF18/KOX11, ZNF29/KOX26, ZNF62, and ZFP3). Some of these sites are of particular interest because the locations are associated with cytogenetic abnormalities. In fact, several of the SCAN family members were cloned as a result of attempts to identify candidate tumor suppressor genes that lie within these chromosomal regions.
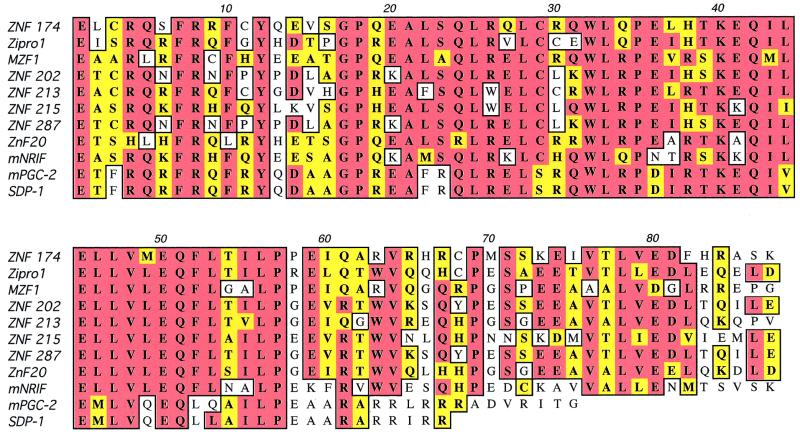
FIG. 2.
Alignment of the amino acid sequences of selected members of the SCAN domain family. To illustrate the remarkable homology characteristic of the SCAN domain, the amino acid sequences of the following SCAN domains are aligned: ZNF174 (GenBank accession number U31248); Zipro1 (also known as Ctfin51 [D10630], Ru49 [U41671], or Zfp38 [X63747]); MZF1 (also known as ZNF42 [AF161886]); ZNF202 (AF027219); ZNF213 (AF017433, also known as CR53); ZNF215 (AF056618); ZNF287 (AF217227); ZnF20 (AF011573, also known as p18 [Z21707]); murine NRIF (AJ242914); and murine PGC-2 (AF220501, also known as Leap1) and its human homolog, SDP1 (AF204271).
The SCAN family: one system of classification.
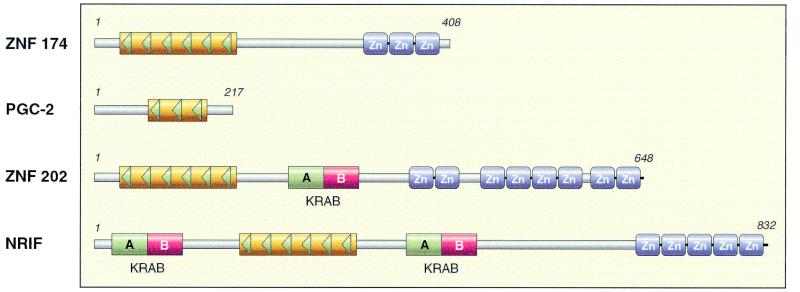
The members of the SCAN family of transcription factors can be divided into classes based on the presence of other modular elements (Fig. 3). Most SCAN domains are found in transcription factors with a variable number of C-terminal (C2H2)x-type zinc fingers [SCAN-(C2H2)x]. A second class of SCAN family members consists of the recently discovered group of isolated SCAN domain proteins. A third group of SCAN family members is composed of proteins predicted to have a SCAN-KRAB-(C2H2)x domain alignment, and another category consists of a protein with a predicted domain composition of KRAB-SCAN-KRAB-(C2H2)x. An overview of representatives from each of these families demonstrates that the SCAN domain-containing transcription factors perform a wide range of functions important in cell development or differentiation.
FIG. 3.
Schematic diagram of four classes of SCAN domain family members. One representative of each of the four classes of SCAN family members discussed in the text is provided for comparison. The first class has an N-terminal SCAN domain and a variable number of C-terminal (C2H2)x-type zinc fingers [SCAN-(C2H2)x]. Found in this class are ZNF174 (51), and as described in the text, MZF1B (40) and Zipro1 (53). A second class of SCAN family members consists of the recently discovered group of isolated SCAN domain proteins. This group includes the mouse PGC-2, which has a partial SCAN domain (11) as well as the corresponding human homologs (43, 44). A third group of SCAN family members is composed of proteins predicted to have a SCAN-KRAB-(C2H2)x domain alignment, such as that encoded by the hypoalphalipoproteinemia susceptibility gene, ZNF202 (49); a BWS-associated gene, ZNF215 (3); ZNF213; which is linked with familial Mediterranean fever (13); and ZNF287, which has been associated with a deletion found in Smith-Magenis syndrome (NM020653). A final category consists of proteins with a predicted domain composition of KRAB-SCAN-KRAB-(C2H2)x, represented by the mouse protein NRIF, which interacts with the neurotrophin receptor (10).
SCAN-(C2H2)x.
The first group of SCAN domain proteins consists of family members encoding a SCAN domain and a variable number of C-terminal C2H2 zinc fingers. There are a number of genes, in the recent release of the human DNA database, but only a few have been characterized. ZNF174, the member of the SCAN family identified by our group, encodes a single SCAN domain and three C2H2 zinc fingers. It is typical of the SCAN family in that it is broadly expressed and interacts with itself and other members of the family (50, 51). Application of the proposed DNA-zinc finger recognition rules (16, 47) suggests that the zinc fingers of ZNF174, like some of the fingers of other members of the SCAN family, can participate in optimal DNA binding. ZNF174 appears to be a transcriptional repressor, although little is known about the mechanism of repression or its authentic target genes. Other products of this group of SCAN family members include myeloid zinc finger protein (MZF1). MZF1 has 13 zinc fingers and is expressed preferentially in hematopoietic progenitor cells of the myeloid lineage (reviewed in references 25 and 48). MZF1 has been implicated in hematopoiesis and has been found to be essential for granulopoiesis. DNA binding sites of MZF1 were determined, and computer searches of databases for promoter sequences revealed potential matches in the genes for CD34, c-myb, myeloperoxidase, and lactoferrin (35, 36, 39). Interestingly, MZF1 is a bifunctional transcriptional regulator in that it activates transcription in cells of hematopoietic origin and represses transcription in nonhematopoietic cells (36). Alternative splicing of the MZF1 gene generates a SCAN box-containing zinc finger protein, MZF1B (40). MZF1B shares identity to the carboxy terminus of MZF1, including the 13 C2H2 zinc finger modules, but MZF1B has an additional amino-terminal extension that contains a SCAN box. Thus, while MZF1 and MZF1B may bind to the same DNA target, it is likely that they exert distinct regulatory effects because of their unique amino termini. Additionally, it is possible that when MZF1 is bound to a target gene it may act as a dominant-negative inhibitor of MZF1B function. This phenomenon is not unique to MZF1 in that other SCAN family members are alternatively spliced, generating forms that lack the SCAN domain.
Another member of this class of the SCAN box family, Zipro1 (which is also known as Ctfin51, Ru49, or Zfp38) may also be important in lineage determination. In the cerebellar cortex, this SCAN box family member is a marker for the cerebellar granule neuronal lineage and may play a role in the proliferation of granule cell precursors in the developing cerebellum (53). Additionally, the gene product is expressed in skin and increased dosage results in a hair loss phenotype associated with increased epithelial cell proliferation and abnormal hair follicle development.
Isolated SCAN domain proteins.
The second group in the SCAN family consists of a protein with a SCAN domain without associated zinc fingers. A murine SCAN domain-containing protein was identified as an adipogenic cofactor bound by the differentiation domain of peroxisome proliferator-activated receptor γ (PPARγ) (11). This protein, termed PPARγ coactivator 2 (PGC-2, or its human homolog, SDP1 or RAZ1), can potentiate PPARγ-dependent gene expression, presumably by facilitating the assembly of a coactivation complex and by enhancing fat cell formation. PGC-2 does not contain zinc fingers but does contain a partial SCAN box that consists of the N-terminal 60 residues of the “authentic” SCAN domain (Fig. 2). The functional significance of the shortened SCAN-like domain in PGC-2 remains to be determined.
SCAN-KRAB-(C2H2)x.
A third class of SCAN family members consists of a SCAN-KRAB-(C2H2)x modular architecture, where the KRAB domain may be complete, or more frequently, consist of only the A domain. In this group the SCAN domain is N terminal to the KRAB domain (Fig. 3), although the significance of the placement is uncertain. It appears as though the KRAB domain can still function as a repression module when associated with the SCAN box, although nothing is known about how one domain influences the function of the other. An interesting member of this subgroup of SCAN family members is the hypoalphalipoproteinemia susceptibility gene, ZNF202. This member of the family resides in a chromosomal region linked genetically to low high-density lipoprotein cholesterol (29). The gene encodes a protein predicted to contain a SCAN box, an intact KRAB domain, and eight C2H2 zinc finger motifs. The ZNF202 gene product is a transcriptional repressor that binds to elements found predominantly in genes that participate in lipid metabolism, such as the structural components of lipoprotein particles (apolipoproteins AIV, CII, and E) and enzymes involved in lipid processing (lipoprotein lipase, lecithin cholesteryl ester transferase) (49).
Provocatively, another member of this class of the SCAN family (ZNF287, NM020653) was found recently in the 17p11.2 interstitial deletion associated with Smith-Magenis syndrome. This disorder is associated with mental retardation, facial dysmorphism, and behavioral problems such as aggressive and self-injurious behavior (reviewed in reference 12). In a similar correlation, ZNF213 is in strong linkage disequilibrium with the gene for familial Mediterranean fever. This febrile disorder is found in people originating in the Near East and is characterized by brief episodes of fever accompanied by a tissue-damaging inflammatory response of serosal surfaces, such as the peritoneum, pleura, and synovial membranes (reviewed in reference 4). ZNF213 encodes a SCAN box, a KRAB A domain, and three zinc fingers (13). Although this member of the SCAN family is strongly linked with the disorder, both genetic and mutational analyses have excluded the gene as being directly responsible for familial Mediterranean fever. The adjacent pyrin gene has been designated as the gene for familial Mediterranean fever because of the presence of missense mutations in affected patients but not in normal individuals. Interestingly, the pyrin gene product may be a transcription factor, possibly regulating the expression of target genes involved in the suppression of inflammation.
Another member of this class of the SCAN family may play a role in Beckwith-Wiedemann syndrome (BWS), a disorder characterized by a wide variety of growth abnormalities, including enlargement of many of the body organs and gigantism. The molecular basis of this syndrome involves multiple genes and genomic imprinting. Inheritance of two functional copies of the primarily paternally expressed insulin-like growth factor 2 (IGF2) gene results in overproduction of this fetal growth factor and changes in development. Evidence for the role of IGF2 in the disorder comes from the observation that the majority of BWS patients show loss of imprinting of this gene and from studies with transgenic mice in which overexpression of IGF2 results in mice with BWS-like morphologic features (46). In addition to the IGF2-containing region of chromosome 11p15 (BWSCR1), it is known that two other regions of 11p15 play a role in this syndrome. Found within one of these regions (BWSCR2) is ZNF215, a member of the SCAN family that encodes four zinc fingers, as well as an amino-terminal SCAN domain, a KRAB A domain, and a region with similarities to a KRAB B domain (3). The two chromosomal rearrangements that define BWSCR2 both disrupt ZNF215, thus implicating the gene in the etiology of BWS. The disrupted transcripts encode a truncated protein lacking the zinc fingers but retaining the SCAN and KRAB domains. It is tempting to speculate that ZNF215 may interact with the IGF2 system and influence the BWS phenotype (3).
KRAB-SCAN-KRAB-(C2H2)x.
The fourth group of SCAN domain-containing proteins consists of one family member with a KRAB-SCAN-KRAB-(C2H2)x composition. Remarkably, a SCAN domain in a recently cloned murine zinc finger protein may provide new insights into the function of this class of SCAN box-containing proteins. The zinc finger protein-designated neurotrophin receptor interacting factor (NRIF) binds to the cytoplasmic domain of the neurotrophin receptor p75NTR, a member of the tumor necrosis factor receptor family (reviewed in reference 9). The retinae of mice with a targeted mutation in the NRIF gene show reduced cell death, and this reduction is similar to that seen in both p75 and nerve growth factor-null mice (10). These findings suggest that the gene encoding NRIF is a SCAN family member that is an essential component downstream of nerve growth factor and p75 in the cell death pathway. NRIF contains five zinc fingers, a putative nuclear localization sequence, and two N-terminal KRAB domains as well as the SCAN box (Fig. 3). Although overexpression of NRIF alone localizes to the nucleus, coexpression of both p75 and NRIF changes the pattern of expression such that NRIF localizes to both the cytoplasm and the nucleus. The SCAN motif does not appear to participate in the interaction between the transcription factor and the cell surface receptor. A provocative model suggests that upon receptor activation, NRIF translocates to the nucleus and regulates the expression of genes related to cell viability in the developing nervous system. This raises the exciting general possibility that upon ligand-induced activation of some cell surface receptors, SCAN box-containing transcription factors are released from a cell surface receptor and translocate to the nucleus, where they oligomerize with themselves or with other members of the SCAN family and regulate transcription.
Collectively, these examples illustrate that members of the SCAN family of C2H2-type zinc finger proteins are involved in a diverse set of biological functions. A number of interesting questions remain about the SCAN domain, including a detailed understanding of its structure, mechanism of partner choice, and the nature of interacting proteins. Additionally, identifying the target genes regulated by this family will be useful in determining the function of this group of zinc finger transcription factors. Like the BTB/POZ and KRAB domains, the presence of a SCAN domain may provide some mechanistic insights into a large number of C2H2-type zinc finger proteins.
ACKNOWLEDGMENTS
This work was supported by NIH grant R37 HL35716.
We thank Simos Simionidis and Christopher Leo for helpful suggestions as well as Jenny Maki for enthusiastic technical support.
REFERENCES
- 1.Ahmad K F, Engel C K, Prive G G. Crystal structure of the BTB domain from PLZF. Proc Natl Acad Sci USA. 1998;95:12123–12128. doi: 10.1073/pnas.95.21.12123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albagli O, Dhordain P, Deweindt C, Lecocq G, Leprince D. The BTB/POZ domain: a new protein-protein interaction motif common to DNA- and actin-binding proteins. Cell Growth Differ. 1995;6:1193–1198. [PubMed] [Google Scholar]
- 3.Alders M, Ryan A, Hodges M, Bliek J, Feinberg A P, Privitera O, Westerveld A, Little P F R, Mannens M. Disruption of a novel imprinted zinc-finger gene, ZNF215, in Beckwith-Wiedemann syndrome. Am J Hum Genet. 2000;66:1473–1484. doi: 10.1086/302892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Babior B M, Matzner Y. The familial Mediterranean fever gene—cloned at last. N Engl J Med. 1997;337:1548–1549. doi: 10.1056/NEJM199711203372112. [DOI] [PubMed] [Google Scholar]
- 5.Bardwell V J, Treisman R. The POZ domain: a conserved protein-protein interaction motif. Genes Dev. 1994;8:1664–1677. doi: 10.1101/gad.8.14.1664. [DOI] [PubMed] [Google Scholar]
- 6.Barna M, Hawe N, Niswander L, Pandolfi P P. Plzf regulates limb and axial skeletal patterning. Nat Genet. 2000;25:166–172. doi: 10.1038/76014. [DOI] [PubMed] [Google Scholar]
- 7.Bellefroid E J, Poncelet D A, Lecocq P J, Relevant O, Martial J A. The evolutionarily conserved Kruppel-associated box domain defines a subfamily of eukaryotic multifingered proteins. Proc Natl Acad Sci USA. 1991;88:3608–3612. doi: 10.1073/pnas.88.9.3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bellefroid E J, Marine J C, Ried T, Lecocq P J, Riviere M, Amemiya C, Poncelet D A, Coulie P G, de Jong P, Szpirer C. Clustered organization of homolgous KRAB zinc-finger genes with enhanced expression in human T cells. EMBO J. 1993;12:1363–1374. doi: 10.1002/j.1460-2075.1993.tb05781.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bibel M, Barde Y A. Neurotrophins: key regulators of cell fate and cell shape in the vertebrate nervous system. Genes Dev. 2000;14:2919–2937. doi: 10.1101/gad.841400. [DOI] [PubMed] [Google Scholar]
- 10.Casademunt E, Carter B D, Benzel I, Frade J M, Dechant G, Barde Y A. The zinc finger protein NRIF interacts with the neutrophin receptor p75NTR and participates in programmed cell death. EMBO J. 1999;18:6050–6061. doi: 10.1093/emboj/18.21.6050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Castillo G, Brun R P, Rosenfield J K, Hauser S, Park C W, Troy A E, Wright M E, Spiegelman B M. An adipogenic cofactor bound by the differentiation domain of PPARγ. EMBO J. 1999;18:3676–3687. doi: 10.1093/emboj/18.13.3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen K S, Potocki L, Lupski J R. The Smith-Magenis syndrome [del(17)p11.2]: clinical review and molecular advances. Ment Retard Dev Disabil Res Rev. 1996;2:122–129. [Google Scholar]
- 13.Chen X, Hamon M, Deng Z, Centola M, Sood R, Taylor K, Kastner D L, Fischel-Ghodsian N. Identification and characterization of a zinc finger gene (ZNF213) from 16p13.3. Biochim Biophys Acta. 1999;1444:218–230. doi: 10.1016/s0167-4781(98)00273-5. [DOI] [PubMed] [Google Scholar]
- 14.Cook M, Gould A, Brand N, Davies J, Strutt P, Shaknovich R, Licht J, Waxman S, Chen Z, Gluecksohn-Waelsch S. Expression of the zinc-finger gene PLZF at rhombomere boundaries in the vertebrate hindbrain. Proc Natl Acad Sci USA. 1995;92:2249–2253. doi: 10.1073/pnas.92.6.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.David G, Alland L, Hong S H, Wong C W, DePinho R A, Dejean A. Histone deacetylase associated with mSin3A mediates represion by the acute promyelocytic leukemia-associated PLZF protein. Oncogene. 1998;16:2549–2556. doi: 10.1038/sj.onc.1202043. [DOI] [PubMed] [Google Scholar]
- 16.Desjarlais J R, Berg J M. Toward rules relating zinc finger protein sequences and DNA binding site preferences. Proc Natl Acad Sci USA. 1992;89:7345–7349. doi: 10.1073/pnas.89.16.7345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dong S, Zhu J, Reid A, Strutt P, Guidez F, Zhong H J, Wang Z Y, Licht J, Waxman S, Chomienne C. Amino-terminal protein-protein interaction motif (POZ-domain) is responsible for activities of the promyelocytic leukemia zinc finger-retinoic acid receptor-alpha fusion protein. Proc Natl Acad Sci USA. 1996;93:3624–3629. doi: 10.1073/pnas.93.8.3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Espinas M L, Jimenez-Garcia E, Vaquero A, Canudas S, Bernues J, Azorin F. The N-terminal POZ domain of GAGA mediates the formation of oligomers that bind DNA with high affinity and specificity. J Biol Chem. 1999;274:16461–16469. doi: 10.1074/jbc.274.23.16461. [DOI] [PubMed] [Google Scholar]
- 19.Farkas G, Gausz J, Galloni M, Reuter G, Gyurkovics H, Karch F. The trithorax-like gene encodes the Drosophila GAGA factor. Nature. 1994;371:806–808. doi: 10.1038/371806a0. [DOI] [PubMed] [Google Scholar]
- 20.Friedman J R, Fredericks W J, Jensen D E, Speicher D W, Huang X P, Neilson E G, Rauscher F J., III KAP-1, a novel corepressor for the highly conserved KRAB repression domain. Genes Dev. 1996;10:2067–2078. doi: 10.1101/gad.10.16.2067. [DOI] [PubMed] [Google Scholar]
- 21.Grignani F, DeMatteis S, Nervi C, Tomassoni L, Gelmetti V, Cioce M, Fanelli M, Ruthardt M, Ferrara F F, Zamir I, Seiser C, Grignani F, Lazar M A, Minucci S, Pelicci P G. Fusion proteins of the retinoic acid receptor-alpha recruit histone deacetylase in promyelocytic leukemia. Nature. 1998;391:815–818. doi: 10.1038/35901. [DOI] [PubMed] [Google Scholar]
- 22.Han Z G, Zhang Q H, Ye M, Kan L X, Gu B W, He K L, Shi S L, Zhou J, Fu G, Mao M, Chen S J, Yu L, Chen Z. Molecular cloning of six novel Kruppel-like zinc finger genes from hematopoetic cells and identification of a novel transregulator domain KRNB. J Biol Chem. 1999;274:35741–35748. doi: 10.1074/jbc.274.50.35741. [DOI] [PubMed] [Google Scholar]
- 23.Hoatlin M E, Zhi Y, Ball H, Silvey K, Melnick A, Stone S, Arai S, Hawe N, Owen G, Zelent A, Licht J D. A novel BTB/POZ transcriptional repressor protein interacts with the Fanconi anemia group C protein and PLZF. Blood. 1999;94:3737–3747. [PubMed] [Google Scholar]
- 24.Hong S H, David G, Wong C W, Dejean A, Privalsky M L. SMRT corepressor protein interacts with PLZF and with the PML-retinoic acid receptor alpha (PARalpha) and PLZF-RARalpha oncoproteins associated with acute promyelocytic leukemia. Proc Natl Acad Sci USA. 1997;95:9028–9033. doi: 10.1073/pnas.94.17.9028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hromas R, Davis B, Rauscher F J, Klemsz M, Tenen D, Hoffman S, Xu D, Morris J F. Hematopoietic transcriptional regulation by the myeloid zinc finger gene MZF-1. Curr Top Microbiol Immunol. 1996;211:159–164. doi: 10.1007/978-3-642-85232-9_16. [DOI] [PubMed] [Google Scholar]
- 26.Katsani K R, Hajibagheri M A N, Verrijzer C P. Co-operative DNA binding by GAGA transcription factor requires the conserved BTB/POZ domain and reorganizes promoter topology. EMBO J. 1999;18:698–708. doi: 10.1093/emboj/18.3.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim S S, Chen Y M, O'Leary E, Witzgall R, Vidal M, Bonventre J V. A novel member of the RING finger family, KRIP-1, associates with the KRAB-A transcriptional repressor domain of zinc finger proteins. Proc Natl Acad Sci USA. 1996;93:15299–15304. doi: 10.1073/pnas.93.26.15299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klug A, Schwabe J W. Protein motifs 5. Zinc fingers. FASEB J. 1995;9:597–604. [PubMed] [Google Scholar]
- 29.Kort E N, Ballinger D G, Ding W, Hunt S C, Bowen B R, Abkevich V, Bulka K, Campbell B, Capener C, Gutin A, Harshman K, McDermott M, Thorne T, Wang H, Wardell B, Wong J, Hopkins P N, Skolnick S, Samuels M. Evidence of linkage of familial hypoalphalipoproteinemia to a novel locus on chromosome 11q23. Am J Hum Genet. 2000;66:1845–1856. doi: 10.1086/302945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li X, Peng H, Schultz D C, Lopez-Guisa J M, Rauscher III F J, Marmorstein R. Structure-function studies of the BTB/POZ transcriptional repression domain from the promyelocytic leukemia zinc finger oncoprotein. Cancer Res. 1999;59:5275–5282. [PubMed] [Google Scholar]
- 31.Lin R J, Nagy L, Inoue S, Shao W, Miler W H, Evans R M. Role of the histone deacetylase complex in acute promyelocytic leukemia. Nature. 1998;391:811–814. doi: 10.1038/35895. [DOI] [PubMed] [Google Scholar]
- 32.Margolin J F, Friedman J R, Meyer W K, Vissing H, Thiesen H J, Rauscher F J., III Kruppel-associated boxes are potent transcriptional repression domains. Proc Natl Acad Sci USA. 1994;91:4509–4513. doi: 10.1073/pnas.91.10.4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Melnick A, Ahmad K F, Arai S, Polinger A, Ball H, Borden K L, Carlile G W, Prive G G, Licht J D. In-depth mutational analysis of the promyelocytic leukemia zinc finger BTB/POZ domain reveals motifs and residues required for biological and transcriptional functions. Mol Cell Biol. 2000;20:6550–6567. doi: 10.1128/mcb.20.17.6550-6567.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moosmann P, Georgiev O, Le Douarin B, Bourquin J P, Schaffner W. Transcriptional repression by RING finger protein TIF1 beta that interacts with the KRAB repressor domain of KOX1. Nucleic Acids Res. 1996;24:4859–4867. doi: 10.1093/nar/24.24.4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morris J F, Hromas R, Rauscher F J. Characterization of the DNA-binding properties of the myeloid zinc finger protein MZF1: two independent DNA-binding domains recognize two DNA consensus sequences with a common G-rich core. Mol Cell Biol. 1994;14:1786–1795. doi: 10.1128/mcb.14.3.1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morris J F, Rauscher F J, Davis B, Klemsz M, Xu D, Tenen D, Hromas R. The myeloid zinc finger gene MZF-1 regulates the CD34 promoter in vitro. Blood. 1995;186:3640–3647. [PubMed] [Google Scholar]
- 37.Peng H, Begg G E, Harper S L, Friedman J R, Speicher D W, Rauscher F J., III Biochemical analysis of the Kruppel-associated box (KRAB) transcriptional repression domain. J Biol Chem. 2000;275:18000–18010. doi: 10.1074/jbc.M001499200. [DOI] [PubMed] [Google Scholar]
- 38.Peng H, Begg G E, Schultz D C, Friedman J R, Jensen D E, Speicher D W, Rauscher F J., III Reconstitution of the KRAB-KAP-1 repressor complex: a model system for defining the molecular anatomy of RING-B box-coiled-coil domain-mediated protein-protein interactions. J Mol Biol. 2000;295:1139–1162. doi: 10.1006/jmbi.1999.3402. [DOI] [PubMed] [Google Scholar]
- 39.Perrotti D, Melotti P, Skorski T, Casella I, Peschle C, Calabretta B. Overexpression of the zinc finger protein MZF1 inhibits hematopoietic development from embryonic stem cells: correlation with negative regulation of CD34 and c-myb promoter activity. Mol Cell Biol. 1995;15:6075–6087. doi: 10.1128/mcb.15.11.6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peterson M J, Morris J F. Human myeloid zinc finger gene MZF produces multiple transcripts and encodes a SCAN box protein. Gene. 2000;22:105–118. doi: 10.1016/s0378-1119(00)00281-x. [DOI] [PubMed] [Google Scholar]
- 41.Read A, Gould A, Brand N, Cook M, Strutt P, Li J, Licht J, Waxman S, Krumlauf R, Zelent A. Leukemia translocation gene, PLZF, is expressed with a speckled nuclear pattern in early hematopoietic progenitors. Blood. 1995;83:4544–4552. [PubMed] [Google Scholar]
- 42.Ryan R F, Schultz D C, Ayyanathan K, Singh P B, Friedman J R, Fredericks W J, Rauscher F J., III KAP-1 corepressor protein interacts and colocalizes with heterochromatic and euchromatic HP1 proteins: a potential role for Kruppel-associated box-zinc finger proteins in heterochromatin-mediated gene silencing. Mol Cell Biol. 1999;19:4366–4378. doi: 10.1128/mcb.19.6.4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sander T A, Haas A L, Peterson M J, Morris J F. Identification of a novel SCAN box-related protein that interacts with MZF1B. J Biol Chem. 2000;275:12857–12867. doi: 10.1074/jbc.275.17.12857. [DOI] [PubMed] [Google Scholar]
- 44.Schumacher C, Wang H, Honer C, Ding W, Koehn J, Lawrence Q, Coulis C M, Wang L L, Ballinger D, Bowen B R, Wagner S. The SCAN domain mediates selective oligomerization. J Biol Chem. 2000;275:17173–17179. doi: 10.1074/jbc.M000119200. [DOI] [PubMed] [Google Scholar]
- 45.Shaknovich R, Yeyati P L, Ivins S, Melnick A, Lempert C, Waxman S, Zelent A, Licht J D. The promyelocytic leukemia zinc finger protein affects myeloid cell growth, differentiation and apoptosis. Mol Cell Biol. 1998;18:5533–5545. doi: 10.1128/mcb.18.9.5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sun F L, Dean W L, Kelsey G, Allen N D, Reik W. Transactivation of Igf2 in a mouse model of Beckwith-Wiedemann syndrome. Nature. 1997;389:809–815. doi: 10.1038/39797. [DOI] [PubMed] [Google Scholar]
- 47.Suzuki M, Gerstein M, Yagi N. Stereochemical basis of DNA recognition by Zn fingers. Nucleic Acids Res. 1994;22:3397–3405. doi: 10.1093/nar/22.16.3397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tenen G D, Hromas R, Licht J D, Zhag D. Transcription factors, normal myeloid development and leukemia. Blood. 1997;90:489–519. [PubMed] [Google Scholar]
- 48a.Venter J C, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 49.Wagner S, Hess M A, Ormonde-Hanson P, Malandro J, Hu H, Chen M, Kehrer R, Frodsham M, Schumacher C, Beluch M, Honer C, Skolnick M, Ballinger D, Bowen B R. A broad role for the zinc finger protein ZNF202 in human lipid metabolism. J Biol Chem. 2000;275:15685–15690. doi: 10.1074/jbc.M910152199. [DOI] [PubMed] [Google Scholar]
- 50.Williams A J, Blacklow S C, Collins T. The zinc finger-associated SCAN box is a conserved oligomerization domain. Mol Cell Biol. 1999;19:8526–8535. doi: 10.1128/mcb.19.12.8526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Williams A J, Khachigian L M, Shows T, Collins T. Isolation and characterization of a novel zinc-finger protein with transcriptional repressor activity. J Biol Chem. 1995;270:22143–22152. doi: 10.1074/jbc.270.38.22143. [DOI] [PubMed] [Google Scholar]
- 52.Witzgall R, O'Leary E, Leaf A, Onaldi D, Bonventre J V. The Kruppel-associated box-A (KRAB-A) domain of zinc finger proteins mediates transcriptional repression. Proc Natl Acad Sci USA. 1994;91:4514–4518. doi: 10.1073/pnas.91.10.4514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang X W, Wynder C, Doughty M L, Heintz N. BAC-mediated gene-dosage analysis reveals a role for Ziprol (Ru49/Zfp38) in progenitor cell proliferation in cerebellum and skin. Nat Genet. 1999;22:327–335. doi: 10.1038/11896. [DOI] [PubMed] [Google Scholar]
- 54.Yeyati P L, Shaknovich R, Boterashvili S, Li J, Ball H J, Waxman S, Nason-Burchenal K, Dmitrovsky E, Zelent A, Licht J D. Leukemia translocation protein PLZF inhibits cell growth and expression of cyclin A. Oncogene. 1999;18:925–934. doi: 10.1038/sj.onc.1202375. [DOI] [PubMed] [Google Scholar]
- 55.Zheng L, Pan H, Li S, Flesken-Nikitin A, Chen P, Boyer T G, Lee W-H. Sequence-specific transcriptional corepressor function for BRCA1 through a novel zinc finger protein, ZBRK1. Mol Cell. 2000;6:757–768. doi: 10.1016/s1097-2765(00)00075-7. [DOI] [PubMed] [Google Scholar]