Abstract
Chlamydia pneumoniae has been associated with atherosclerosis and coronary artery disease (CAD), and its DNA has been detected in atheromatous lesions of the aorta, carotid, and coronary arteries by a variety of PCR assays. The objective of this study was to compare the performances of five published PCR assays in the detection of C. pneumoniae in peripheral blood mononuclear cells (PBMCs) from patients with coronary artery disease. The assays included two conventional PCRs, one targeting a cloned PstI fragment and one targeting the 16S rRNA gene; two nested PCRs, one targeting the 16S rRNA gene and one targeting ompA; and a touchdown enzyme time release (TETR) PCR, targeting the 16S rRNA gene. All PCRs had similar analytical sensitivities and detected a minimum of 0.005 inclusion-forming units (IFU) of C. pneumoniae; the ompA nested PCR and the TETR PCR were slightly more sensitive and detected 0.001 IFU. Assay reproducibility was examined by testing 10 replicates of C. pneumoniae DNA by each assay. All five assays showed excellent reproducibility at high levels of DNA, with scores of 10 out of 10 for 0.01 IFU, but exhibited decreased reproducibility for smaller numbers of C. pneumoniae IFU for all tests. Pairwise comparison of test results indicated that there was a significant difference between tests (Cochran Q = 32.0, P < 0.001), with the PstI fragment (P < 0.001) and 16S rRNA (P = 0.002) assays having lower reproducibility than the nested ompA and TETR assays. To further analyze assay sensitivity, C. pneumoniae-infected U-937 mononuclear cells were added to whole blood, and extracted mononuclear-cell DNA was tested by each assay. All five assays showed similar sensitivities, detecting 15 infected cells; three assays detected 3 infected cells, while all assays were negative at the next dilution (1.5 infected cells). A striking difference in performance of the five assays was seen, however, when PBMCs from CAD patients were tested for C. pneumoniae DNA. The ompA nested PCR detected C. pneumoniae DNA in 11 of 148 (7.4%) specimens, the 16S rRNA nested PCR detected 2 positives among the 148 specimens (1.4%) (P < 0.001), and the other 3 assays detected no positive specimens (P < 0.001, compared with the ompA assay). These results indicate that analytical sensitivity alone does not predict the ability of an assay to detect C. pneumoniae in whole-blood-derived PBMCs. Before standardized assays can be used in wide-scale epidemiological studies, further characterization of these assays will be required to improve our understanding of their performance in the detection of C. pneumoniae in clinical material.
Chlamydia pneumoniae has been associated with coronary artery disease (CAD) and myocardial infarction in several serological and pathological studies (6, 10, 20, 34, 37). C. pneumoniae has been detected in atherosclerotic plaques by immunocytochemical staining and PCR, and in limited studies it has been isolated in culture from atheromas (16, 18, 20, 33), providing evidence for an association between C. pneumoniae and atherogenesis without, however, demonstrating causality. Furthermore, animal models strongly implicate C. pneumoniae in the pathogenesis of atherosclerosis (8, 11, 12, 26, 28, 30). Although it has been shown that C. pneumoniae can replicate in human macrophages, endothelial cells, and smooth muscle cells (15), the exact role of C. pneumoniae in the pathogenesis of atherosclerosis is not known.
Chlamydiae are unique intracellular pathogens with a biphasic developmental cycle consisting of metabolically inactive and infectious elementary bodies and metabolically active but noninfectious reticulate bodies (29). Under certain conditions, including exposure to gamma interferon or antibiotics, chlamydia can become dormant and reside inside cells in a nonreplicating form that may escape immune detection and persist for long periods of time (1, 2). Induction of indoleamine-2,3-dioxygenase activity by gamma interferon and tryptophan catabolism is thought to be a mechanism for inducing persistence (2, 25). Persistently infected mononuclear cells and macrophages may facilitate the hematogenous dissemination of chlamydiae to other organ systems in an infected animal and may be important in atherogenesis (8, 27). There is increasing evidence that chronically infected mononuclear cells may play an important role in the development of chronic diseases such as atherosclerosis and coronary heart disease. In both the mouse and rabbit models of C. pneumoniae-induced atherosclerosis, C. pneumoniae has been detected in the blood prior to its appearance in atheromatous lesions of major blood vessels, including the aorta and coronary arteries (27; J. B. Mahony, I. W. Fong, E. Viira, and D. Jang, unpublished data).
C. pneumoniae infections have been difficult to diagnose either by serological methods or by isolation of the organism from infected tissue. Serological assays for C. pneumoniae have not yet been well standardized, and the use of methods such as the microimmunofluorescence test for large-scale epidemiological studies has been hampered by the poor correlation between serological results and tissue detection of this organism as well as by interlaboratory variation (20). C. pneumoniae has been difficult to recover from tissues, with special tissue culture techniques being required for its isolation (19, 32). Although numerous studies involving large numbers of patients have been performed, there are only three reports describing the isolation of C. pneumoniae from patients with vascular artery disease (16, 20, 33). Nucleic acid amplification (NAA) tests have therefore emerged as an important method of detecting C. pneumoniae in diseased tissues and have contributed significantly to the determination of an association of C. pneumoniae with atherosclerotic heart disease. C. pneumoniae DNA has been detected in large vessels, including the aortic, coronary, carotid, and femoral arteries, as well as in circulating mononuclear cells from a variety of patients with vascular diseases such as CAD, aortic aneurysm, and stroke (3, 6, 17, 31). Several different targets for amplification have been used, including a cloned PstI fragment (7), the gene encoding the major outer membrane protein OmpA (36), and the 16S rRNA gene (13, 14). These assays have involved a variety of formats, including conventional (nonnested) and nested PCR assays as well as a novel touchdown enzyme time release (TETR) PCR. Madico et al. recently reported that the clinical sensitivity of the TETR PCR equals that of the nested PCR for the 16S rRNA gene (21). As yet there are no commercially available NAA tests for C. pneumoniae. Although several publications document the use of these various assays, to our knowledge there have been no studies that have compared their performances on clinical material.
We report here a comparison of the performances of five different PCR assays, including two nested PCRs, two nonnested PCRs, and a TETR PCR, in the detection of C. pneumoniae DNA. Assay performances were compared by (i) determining the analytical sensitivity for detecting C. pneumoniae DNA, (ii) determining the reproducibility of the assays by testing replicates of serial DNA dilutions, (iii) testing peripheral blood mononuclear cells (PBMCs) spiked with C. pneumoniae-infected human mononuclear U-937 cells, and (iv) testing PBMCs from patients undergoing coronary angiography. Although these assays have similar analytical sensitivities, their abilities to detect C. pneumoniae in PBMCs differ significantly.
MATERIALS AND METHODS
Cell culture.
HEp2 cells (ATCC CCL-23) were maintained in minimal essential medium (Gibco BRL, Gaithersburg, Md.) containing Earle's salts and supplemented with 10% heat-inactivated fetal bovine serum (Gibco BRL) and 2 mM l-glutamine (Gibco BRL). HEp2 cells were grown in 25-cm2 culture flasks at 37°C and in a 5% CO2 atmosphere.
The promonocytic cell line U-937 was provided by Davis Taub (National Institute on Aging, National Institutes of Health, Bethesda, Md.) and maintained in RPMI 1640 medium (Gibco BRL) supplemented with 10% fetal bovine serum. U-937 cells were grown in 75-cm2 culture flasks at 37°C in a 5% CO2 atmosphere. Prior to infection, U-937 cells were harvested by centrifugation, counted, and seeded into 25-cm2 flasks to achieve the desired cell concentration.
C. pneumoniae propagation.
C. pneumoniae VR-1310 (ATCC 1310-VR) was propagated in HEp2 cells as described by Roblin et al. (33a) with slight modifications. C. pneumoniae was inoculated onto preformed HEp2 cell monolayers in 25-cm2 flasks, which were centrifuged for 60 min at 1,000 × g and 25°C and then incubated at 37°C for 1 h. The inoculum was removed and replaced with growth medium consisting of minimal essential medium containing 1 μg of cycloheximide/ml. Infected cultures were incubated 72 h at 37°C in a 5% CO2 atmosphere. C. pneumoniae was harvested by disrupting HEp2 cells with glass beads followed by sonication and then centrifugation at 250 × g to remove cellular debris. Supernatants containing C. pneumoniae were aliquoted and frozen at −70°C. C. pneumoniae titrations were performed on frozen stocks in duplicate, and titers were expressed as inclusion-forming units (IFU) per ml.
Infection of U-937 cells.
U-937 cells were infected with C. pneumoniae in 25-cm2 flasks at a multiplicity of infection of 1. Briefly, 5 × 106 cells were suspended in 3 ml of RPMI 1640 medium and seeded into 25-cm2 flasks. Cells were infected by incubating them with C. pneumoniae for 40 to 48 h at 37°C in a 5% CO2 atmosphere. Following incubation, infected U-937 cells were harvested by centrifugation, washed twice in Hanks balanced salt solution (Gibco BRL), and counted in a Neubauer chamber.
Preparation of the mock-infected blood sample.
U-937 cells from C. pneumoniae-infected cultures (7 × 106 cells in a 0.5-ml volume) were added to 8 ml of whole blood collected into a Vacutainer CPT Cell Preparation Tube (Becton Dickinson, Franklin Lakes, N.J.). CPT tubes were centrifuged according to the manufacturer's instructions, and mononuclear cells were collected from the monocyte cell layer for DNA purification.
Staining C. pneumoniae-infected U-937 cells.
Infected U-937 cells were serially diluted in phosphate-buffered saline, and duplicate 35-μl volumes of each dilution were spotted onto flat-bottomed wells of glass microscope slides. Samples were dried at 37°C for 30 min and then fixed for 10 min in absolute ethanol. Cells were stained with a fluorescein isothiocyanate-labeled Chlamydia genus-specific monoclonal antibody (Pathfinder culture confirmation tube; Kallestad, Chaska, Minn.), and the number of infected U-937 cells from each dilution was determined by using an Olympus BH2-RFCA microscope equipped with an epifluorescence attachment.
Specimens.
Whole-blood specimens (8 ml each) from patients undergoing coronary angiography or angioplasty procedures between July and October 1999 were collected into monocyte preparation (CPT) tubes containing sodium citrate (Becton Dickinson, Franklin Lakes, N.J.). Within 2 h of collection, the blood samples were centrifuged at a relative centrifugal force of 1,500 to 1,800 for 30 min and transported to the laboratory. Upon arrival at the laboratory, specimens were remixed by gently inverting the tube 8 to 10 times immediately prior to recentrifugation. With a Pasteur pipette, the resulting mononuclear cell layer was transferred to a viral for extraction of DNA.
DNA extraction.
C. pneumoniae DNA was extracted from purified elementary bodies by proteinase K digestion followed by chloroform-phenol extraction as described previously (24). Mononuclear cells (200 μl) from CPT tubes were pelleted by centrifugation at 500 × g, and their DNA was extracted by using a Qiagen DNA Mini-kit according to the manufacturer's instructions. DNA was eluted in a final volume of 100 μl, aliquoted, and stored at −20°C.
PCR.
Five separate assays—three targeting the 16S rRNA gene, one targeting the ompA gene, and one targeting the PstI cloned gene fragment—were used for the detection of C. pneumoniae. In all cases, the amplification reactions were performed in a volume of 25 μl containing 2.5 μl of extracted DNA, 10 mM Tris-HCl (pH 8.3), 50 mM KCl, and a 200 μM concentration of four deoxynucleoside triphosphates. For all five assays, the ratio of volume of template DNA to volume of reaction mix was as indicated in the original publication (7, 13, 14, 21, 36). Concentrations of Taq polymerase (AmpliTaq Gold; Perkin-Elmer, Branchburg, N.J.), primers, and MgCl2 as well as the cycling conditions on an MJ Research PTC-200 thermocycler differed with each PCR. The PCR primer sets that were tested included HL1-HR1 (7), CpnA-CpnB (13), CpnA-CpnB with nested TW50-TW51 (14), CP1-CP2 with nested CPC-CPD (36), and CPN90-CPN91 (21). The sequences of these primers are shown in Table 1. All PCRs were performed according to published protocols. Briefly, the CpnA-CpnB PCR used a 0.5 μM concentration of each primer, 2.5 mM MgCl2, and 0.75 U of Taq polymerase and involved 40 cycles of 15 s at 94°C, 15 s at 55°C, and 1.1 min at 72°C. For the PCR using CP1-CP2 with nested primer pair TW50-TW51, both amplification rounds employed 2.5 mM MgCl2, a 0.5 μM concentration of each primer, and 0.75 U of Taq polymerase. For the first amplification, reactions were run for 20 cycles of 15 s at 94°C, 15 s at 65°C minus 1°C per cycle, and 1.1 min at 72°C plus an additional 20 cycles of 15 s at 94°C, 15 s at 45°C, and 1.1 min at 72°C. The inner amplification consisted of 30 cycles of 15 s at 94°C, 15 s at 55°C, and 1 min at 72°C. The HL1-HR1 PCR employed 2.5 mM MgCl2, a 0.5 μM concentration of each primer, and 0.75 U of Taq polymerase and involved 40 cycles of 30 s at 94°C, 30 s at 57°C, and 1 min at 72°C. For the PCR using CP1-CP2 with nested primer pair CPC-CPD, the conditions were as follows: the first round of amplification employed 1.5 mM MgCl2, 0.4 μM primers, and 0.625 U of Taq polymerase and involved 20 cycles of 1 min at 94°C, 1 min at 65°C minus 0.5°C per cycle, and 1 min at 72°C plus an additional 20 cycles of 1 min at 94°C, 1 min at 55°C, and 1 min at 72°C. The PCR products amplified by the outer primers (CP1-CP2) were diluted 1:10, and a volume of 2.5 μl was added to a new 25-μl PCR mixture for a second amplification with nested primer pair CPC-CPD. The second round of amplification employed 3 mM MgCl2, 1 μM primers, and 0.625 U of Taq polymerase and involved 30 cycles of 1 min at 94°C, 1 min at 50°C, and 1 min at 72°C. For the TETR PCR, the CPN90-CPN91 primer pair was used with 2.5 mM MgCl2, 0.25 μM primers, and 0.5 U of Taq polymerase. This touchdown PCR applied an enzyme time release protocol (21). Cycling consisted of 75 s at 95°C followed by 60 cycles of 45 s at 94°C, 45 s beginning at 62°C and ending at 52°C, and 1 min at 72°C. The annealing temperature was lowered 1°C every four cycles until reaching 52°C, at which point it was kept constant until the end of the cycling process. All amplification products were analyzed by agarose gel electrophoresis followed by ethidium bromide staining.
TABLE 1.
Comparison of five PCR assays for detection of C. pneumoniae DNA
| Assay | Target | Assay format | Primers | Amplicon size (bp) | No. of amplification cycles | Reference |
|---|---|---|---|---|---|---|
| 1 | Cloned PstI | Standard | HL1 5′ GTT GTT CAT GAA GGC CTA CT 3′ | 437 | 40 | 7 |
| HR1 5′ TGC ATA ACC TAC GGT GTG TT 3′ | ||||||
| 2 | 16S rRNA | Standard | CpnA 5′ TGA CAA CTG TAG AAA TAC AGC 3′ | 463 | 40 | 13 |
| CpnB 5′CGC CTC TCT CCT ATA AAT 3′ | ||||||
| 3 | 16S rRNA | Nested | CpnA 5′ TGA CAA CTG TAG AAA TAC AGC 3′ | 463 | 40 | 14 |
| CpnB 5′ CGC CTC TCT CCT ATA AAT 3′ | ||||||
| TW50 5′ AGT CCC GCA ACG AGC GCA 3′ | 270 | 30 | ||||
| TW51 5′ GCT GAC ACG CCA TTA CTA 3′ | ||||||
| 4 | ompA | Nested | CP1 5′ TTA CAA GCC TTG CCT GTA GG 3′ | 333 | 40 | 36 |
| CP2 5′ GCG ATC CCA AAT GTT TAA GGC 3′ | ||||||
| CPC 5′ TTA TTA ATT GAT GGT ACA ATA 3′ | 207 | 30 | ||||
| CPD 5′ ATC TAC GGC AGT AGT ATA GTT 3′ | ||||||
| 5 | 16S rRNA | TETR | CPN90 5′ GGT CTC AAC CCC ATC CGT GTC GG 3′ | 197 | 60 | 21 |
| CPN91 5′ TGC GGA AAG CTG TAT TTC TAC AGT T 3′ |
Exhaustive measures were applied for all PCR assays to ensure that carryover contamination of specimens by preamplified product did not occur (24). These included the use of (i) separate biosafety containment hoods for preparing specimens, setting up PCRs, and analyzing products; (ii) plugged pipette tips or positive-displacement pipettors; (iii) several negative controls interspersed with clinical specimens; and (iv) periodic swabbing of work areas to detect amplified DNA.
Data analysis.
SPSS version 10.0 for Windows (SPSS Inc., Chicago, Ill.) was used for statistical testing. All PCR results were dichotomized as positive or negative. For comparing diagnostic assays, the Cochran Q test statistic for binary related variables was used. For comparisons of results of pairs of assays, the two highest-ranking diagnostic assays were compared with all others by using McNemar's test of two related variables (for a total of seven comparisons). For the Cochran Q test, which was used to compare the results of all five diagnostic assays simultaneously, an alpha level of 0.05 was set as the level of significance. For pairwise comparisons, an alpha level of 0.01 was set to account for multiple-comparison testing.
RESULTS
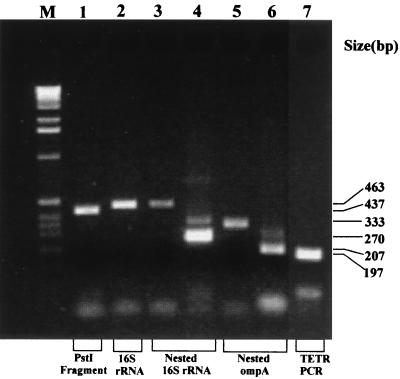
We compared the performances of five different PCR assays, including two nonnested assays, two nested assays, and a novel TETR assay, targeting three different genes, in detecting C. pneumoniae DNA. The nonnested assays targeted either the cloned PstI fragment or the 16S rRNA gene, the nested assays targeted either the 16S rRNA gene or the ompA gene, and the TETR assay targeted the 16S rRNA gene (Table 1). All five assays were performed according to their original published protocols, with the only change being a reduced reaction volume of 25 μl containing 2.5 μl of sample DNA. Both nonnested PCRs used 40 cycles of amplification, while both nested PCRs employed 40 cycles for the first and 30 cycles for the second round of amplification; the TETR PCR involved a total of 60 cycles of amplification (Table 1). Typical results for these five PCR assays are shown in Fig. 1. All assays gave amplification products of the expected size (Table 1) with virtually no mispriming when done under these conditions.
FIG. 1.
Agarose gel electrophoresis analysis of PCR products of five PCR assays. Gels were stained with ethidium bromide and photographed under UV light at 305 nm. PCR assay products are as follows: lane 1, nonnested PstI target, amplicon size 437 bp; lane 2, nonnested 16S rRNA gene target, amplicon size 463 bp; lanes 3 and 4, nested 16S rRNA gene target, amplicon sizes 463 bp (for the first round of amplification) (lane 3) and 270 bp (for the second round) (lane 4); lanes 5 and 6, nested ompA target, amplicon sizes 333 bp (for first round of amplification) (lane 5) and 207 bp (for the second round) (lane 6); and lane 7, TETR PCR, 16S rRNA gene target, amplicon size 197 bp. M, molecular size standards.
The analytical sensitivity of each assay was determined by testing serial dilutions of a C. pneumoniae stock, containing from 0.1 to 0.0005 IFU. The five PCRs had similar sensitivities, with all assays being able to detect 0.005 IFU. The ompA nested PCR and the TETR PCR were slightly more sensitive and could detect 0.001 IFU (Table 2). The reproducibility of each assay was examined by testing a dilution series of C. pneumoniae elementary bodies in replicates of 10 for each dilution. The dilution series was prepared from a freshly harvested culture (48 h), and replicate aliquots were titrated, without freezing, on HEp-2 cell cultures to determine their infectious titers. The PCR results for the replicate dilutions containing from 0.01 to 0.0001 IFU of C. pneumoniae are shown in Table 3. All five assays had reproducibility scores of 10 for 10 for 0.01 IFU, but scores decreased for smaller numbers of IFU. The total numbers of positive results for all dilutions were combined for each assay, and the five PCR protocols were compared. There was a significant difference between tests (Cochran Q = 32.0, P < 0.001). Pairwise comparisons with test 5, which identified 37 of 60 samples, were as follows: test 1, P < 0.001; test 2, P = 0.002; test 3, P = 0.146; and test 4, P = 1.0. Against test 4, which identified 36 of 60 samples, the results were as follows: test 1, P < 0.001; test 2, P = 0.004; and test 3, P = 0.125. Other comparisons were not significant at the P = 0.01 level. These results showed that reproducibility decreased with decreasing numbers of bacteria and that no single assay demonstrated a reproducibility that was statistically higher than that of any of the other four assays.
TABLE 2.
Sensitivities of five PCR assays for detecting C. pneumoniae DNAa
| No. of IFU detected | PCR results for assay:
|
||||
|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | |
| 1 | ++ | ++ | ++ | ++ | ++ |
| 0.1 | ++ | ++ | ++ | ++ | ++ |
| 0.01 | +− | −+ | ++ | ++ | ++ |
| 0.005 | −+ | ++ | −+ | ++ | ++ |
| 0.001 | −− | −− | −− | −+ | +− |
| 0.0005 | −− | −− | −− | −− | −− |
A 1-ml aliquot of C. pneumoniae seed strain VR1310, containing 106 IFU, was extracted with a Qiagen DNA Mini-kit, and 2.5-μl aliquots of eluted DNA were tested in duplicate by five PCR assays as shown in Table 1 and described in Materials and Methods.
TABLE 3.
Reproducibility of five PCR assays for detecting C. pneumoniaea
| No. of IFU | No. of aliquots testing positive for PCR assay:
|
||||
|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | |
| 0.01 | 10 | 10 | 10 | 10 | 10 |
| 0.005 | 5 | 6 | 6 | 8 | 10 |
| 0.002 | 3 | 4 | 5 | 6 | 4 |
| 0.001 | 4 | 4 | 4 | 7 | 9 |
| 0.0004 | 1 | 3 | 6 | 5 | 4 |
| 0.0001 | 0 | 0 | 0 | 0 | 0 |
| Total | 23 | 27 | 31 | 36 | 37 |
To further address the question of sensitivity, a mock-infected blood specimen was prepared by spiking a C. pneumoniae DNA-negative whole-blood sample in a Vacutainer CPT tube with 7 × 106 C. pneumoniae-infected U-937 cells. Mononuclear cells were prepared according to the manufacturer's instructions, their DNA was extracted, and duplicate aliquots were tested in each PCR assay. Infected mononuclear cells were counted following direct fluorescent antibody (DFA) staining with a monoclonal antibody to C. pneumoniae. All PCRs detected 15 C. pneumoniae-infected cells; three of the five assays—the nonnested 16S rRNA PCR, the nested 16S rRNA PCR, and the TETR PCR—detected the next dilution, containing 3 infected cells, and all assays were negative for 1.5 infected cells (Table 4).
TABLE 4.
Sensitivities of five PCR assays for detecting C. pneumoniae-infected U-937 mononuclear cellsa
| No. of infected U-937 cells | PCR result for assayb:
|
||||
|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | |
| 30 | ++ | ++ | ++ | ++ | ++ |
| 15 | ++ | ++ | ++ | ++ | ++ |
| 3 | −− | −+ | +− | −− | ++ |
| 1.5 | −− | −− | −− | −− | −− |
A 25-cm2 flask containing 7 × 106 U-937 cells was inoculated with C. pneumoniae, and the infected cells were spiked into 8 ml of whole blood in a Vacutainer CPT tube. Mononuclear cells were collected, DNA was extracted by using a Qiagen DNA Mini-kit and eluted in 0.1 ml of elution buffer, and aliquots (2.5 μl) of DNA were tested in duplicate by five PCR assays. The number of infected cells was calculated from the total number of cells that stained by DFA with a fluorescein isothiocyanate-labeled anti-C. pneumoniae monoclonal antibody, assuming a CPT tube cell recovery rate of 71% as indicated by the manufacturer.
PCR assays as described in Table 1.
To compare the clinical performances of the five PCR assays, 148 CPT specimens collected from patients attending a coronary angiography clinic were tested. All specimens were tested by the five PCR assays, and all positive specimens were first reextracted (equal aliquot) and then reamplified in triplicate. Only specimens that were positive after repeat extraction and amplification were considered positive for purposes of comparison. There were striking differences in the performances of the five assays in terms of their abilities to detect C. pneumoniae DNA (Table 5). The ompA nested PCR detected C. pneumoniae DNA in 11 of 148 (7.4%) specimens, while the 16S rRNA nested assay detected C. pneumoniae DNA in only 2 of 148 (1.4%) specimens (P < 0.001). The other three assays all failed to detect even a single positive (0 of 148; P < 0.001). All 11 positives detected by the nested ompA PCR were confirmed positive by repeat PCR testing of a reextracted sample and by hybridization with an internal oligonucleotide probe. There was a significant difference found when all five tests were compared with one another simultaneously (Cochran Q = 32.0, P < 0.001). Pairwise comparisons with test 4 (nested ompA) revealed statistically significant differences: for test 1, P = 0.000; for test 2, P = 0.004; for test 5, P = 1.0; and for test 3, P = 0.125. In addition, the magnitude of the increased sensitivity of test 4 is likely clinically important (11 positives, versus 2 or 0 for the other tests). Test 3 (nested 16S rRNA gene) detected two, but this was not a statistically significant increase from detecting zero positive specimens (P = 0.500).
TABLE 5.
Detection of C. pneumoniae in PBMCs from patients undergoing coronary angioplasty, using five PCR assaysa
| Assayb | PCR results
|
|
|---|---|---|
| No. positive/no. tested | % Positive | |
| 1 | 0/148 | 0 |
| 2 | 0/148 | 0 |
| 3 | 2/148 | 1.4 |
| 4 | 11/148 | 7.4 |
| 5 | 0/148 | 0 |
An 8-ml CPT tube specimen was collected from individual patients undergoing a coronary angioplasty procedure. Mononuclear cells were prepared according to the manufacturer's instructions, and DNA was extracted by using a Qiagen DNA Mini-kit. Aliquots of eluted DNA (2.5 μl) were tested by five PCR assays, and positives were confirmed by repeat PCR and hybridization with an internal oligonucleotide probe as described in Materials and Methods.
Assays are as indicated in Table 1.
DISCUSSION
To our knowledge, this is the first study to compare the abilities of different PCR assays to detect C. pneumoniae DNA in blood specimens of patients with CAD. We compared the performances of five different PCR assays for detecting C. pneumoniae DNA and found that analytical sensitivity does not predict an assay's ability to detect C. pneumoniae DNA in PBMCs. In comparing the assays, we used the same primer concentrations, cycling profiles, and magnesium chloride concentrations as were described in the original publication; the only changes in the assays were a reduction in the volume of the reaction mixture and the use of AmpliTaq Gold in all of the assays. In our hands, all five assays gave robust products of the expected size with little or no mispriming (Fig. 1).
The analytical sensitivities of the five assays were remarkably similar. In two separate experiments, all five PCRs detected between 0.005 and 0.0004 IFU (Tables 2 and 3). In our hands, no one assay was significantly more sensitive than the others. The minimal amount of C. pneumoniae that could be reproducibly detected by all five assays (10 of 10 testing positive) was 0.01 IFU. All assays detected 0.0004 IFU at frequencies between 1 and 6 of 10 when 10 replicates were tested (Table 3). These sensitivities compare favorably with sensitivities reported by others. Campbell et al. reported that the nonnested PstI PCR had a sensitivity of 0.5 IFU (7), and Gaydos et al. reported a sensitivity of 0.4 IFU for the nonnested 16S rRNA PCR (13). The sensitivity of the nested ompA PCR was originally reported as <1 elementary body by DFA staining by Tong and Sillis (36). Boman et al., who used this assay to detect C. pneumoniae DNA in PBMCs of angiography patients, did not present any data on the assay's sensitivity (4, 6). Gaydos et al. used the 16S rRNA nested PCR to detect C. pneumoniae in bronchoalveolar lavage specimens from immunocompromised patients but did not present data on the sensitivity of this assay (14). In the only report to date comparing the sensitivities of various PCRs for detecting C. pneumoniae, Gaydos et al. recently showed that a novel TETR PCR was more sensitive than two nested PCR assays (C. A. Gaydos, G. E. Madico, K. Crotchfelt, and T. C. Quinn, Abstr. 99th Gen. Meet. Am. Soc. Microbiol., abstr. C-491, 1999). The TETR PCR had a sensitivity of 0.004 IFU for C. pneumoniae strain A-03, compared with 0.06 IFU for the nested ompA PCR, 1 IFU for the nested 16S rRNA PCR, and 4 IFU for the nonnested PstI PCR.
In their study, Gaydos et al. determined sensitivities by testing a dilution series of C. pneumoniae in duplicate by four different PCRs, while we tested 10 replicates for each dilution. Gaydos et al. found that the smallest amount of C. pneumoniae that could be detected by all four assays was 4 IFU (Gaydos et al., Abstr. 99th Gen. Meet. Am. Soc. Microbiol., abstr. C-491, 1999). In our hands, the smallest amount of the bacterium that all five assays could reproducibly detect (10 of 10 times) was 0.01 IFU (Table 3). Differences in sensitivities between the two studies could be due to several factors, such as the use of different DNA extraction methods, differences in the ramping times of the different thermal cyclers used in the two studies, or differences in the number and/or homogeneity of replicates used in the two studies. Differences in DNA extraction efficiencies, clumping of elementary bodies, or differences in culture sensitivity would markedly affect sensitivity levels. Indeed, sensitivities of less than 1 IFU probably indicate the technical limitations of determining the sensitivity of PCR or other NAA tests based on IFU. Sensitivity differences among these assays should not be due to differences in target copy numbers, since only a single ompA gene, presumably a single copy of the PstI fragment, and only two rRNA operons are present in the C. pneumoniae genome (35).
The differences in the endpoint sensitivities of 0.01 IFU (Table 3) and 15 infected cells (Table 4), although not directly comparable, may reflect the presence of interfering factors in human blood which reduce the sensitivity of all PCR assays. In an unselected sample of specimens from patients attending an angiography clinic, we found 11 of 148 (7.4%) PBMC specimens to have C. pneumoniae DNA by one PCR assay while 2 were positive by a second PCR assay and 3 other assays detected no positives. Our criterion for positivity was a specimen that was repeat positive by PCR. The prevalence of C. pneumoniae DNA in other studies of PBMCs has ranged from a low of 8.8% to a high of 69% (6, 37). The wide range of positivity could be due to many factors, including differences in population studies, but might also be explained by differences in the clinical performances of PCR assays, as we have shown in the present study. The reason for the difference in sensitivities of the assays is not known; however, amplification inhibitors present in PBMC specimens that exert differential inhibition for various PCR primers may explain these differences. Differences in the sensitivities of various PCRs for detecting Chlamydia trachomatis in clinical specimens have been observed (23). Furthermore, we have recently reported that different NAA tests for C. trachomatis detection show different susceptibilities to endogenous inhibitors in clinical specimens (22). Therefore, further studies, using a variety of clinical specimens, will be required to explain the difference in assays for detecting C. pneumoniae.
The significant differences in performance of the five assays for detecting C. pneumoniae DNA in PBMCs from patients with CAD indicate the need for additional studies to assess the performance of these assays in different laboratories. One study, investigating the interlaboratory agreement of PCR results by using a blinded panel of specimens, reported that results from different laboratories were extremely variable and that there was little overall agreement in terms of which specimens tested positive (5; J. Boman et al., unpublished data). The differences in PCR performance between laboratories have not been adequately explained and require further study. In addition, there is a need for improved assays for the detection of C. pneumoniae nucleic acid in PBMCs by PCR or alternative amplification methods. With this in mind, we have recently developed a sensitive assay for the detection and quantitation of ompA RNA transcripts by nucleic acid sequence-based amplification that may be useful for studying C. pneumoniae persistence in vascular tissues (9). Interlaboratory standardization of NAA assays and/or development of commercially available assays will be absolutely required to precisely elucidate the etiologic role of C. pneumoniae in vascular disease.
ACKNOWLEDGMENTS
We thank Charlotte Gaydos and Guillermo Madico for providing details of their TETR PCR assay, M. Natarajen for recruitment of patients, Lynne Rainen (Becton Dickinson) for providing Vacutainer CPT tubes and Qiagen DNA Mini-kits, and Charles Goldsmith for statistical advice.
M. Smieja is a Research Fellow of the Heart and Stroke Foundation of Canada. B. K. Coombes is the recipient of a scholarship from the Father Sean O'Sullivan Research Centre, St. Joseph's Hospital, Hamilton, Ontario, Canada.
REFERENCES
- 1.Beatty W L, Morrison R P, Byrne G I. Persistent chlamydiae: from cell culture to a paradigm for chlamydial pathogenesis. Microbiol Rev. 1994;58:686–699. doi: 10.1128/mr.58.4.686-699.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beatty W L, Belanger T A, Desai A A, Morrison R P, Byrne G I. Tryptophan depletion as a mechanism of gamma interferon-mediated chlamydial persistence. Infect Immun. 1994;62:3705–3711. doi: 10.1128/iai.62.9.3705-3711.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blasi F, Boman J, Esposito G, Melissano G, Chiesa R, Cosentini R, Tarsia P, Tshomba Y, Betti M, Alessi M, Morelli N, Allegra L. Chlamydia pneumoniae DNA detection in peripheral blood mononuclear cells is predictive of vascular infection. J Infect Dis. 1999;180:2074–2076. doi: 10.1086/315126. [DOI] [PubMed] [Google Scholar]
- 4.Boman J, Allard A, Persson K, Lundborg M, Juto P, Wadell G. Rapid diagnosis of respiratory Chlamydia pneumoniae infection by nested touchdown polymerase chain reaction compared with culture and antigen detection by EIA. J Infect Dis. 1997;175:1523–1526. doi: 10.1086/516492. [DOI] [PubMed] [Google Scholar]
- 5.Boman J, Gaydos C A, Quinn T C. Molecular diagnosis of Chlamydia pneumoniae infection. J Clin Microbiol. 1999;37:3791–3799. doi: 10.1128/jcm.37.12.3791-3799.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boman J, Soderberg S, Forsberg J, Birgander L S, Allard A, Persson K, Jidell E, Kumlin U, Juto P, Waldenstrom A, Wadell G. High prevalence of Chlamydia pneumoniae DNA in peripheral blood mononuclear cells in patients with cardiovascular disease and in middle-aged blood donors. J Infect Dis. 1998;178:274–277. doi: 10.1086/517452. [DOI] [PubMed] [Google Scholar]
- 7.Campbell L A, Melgosa M P, Hamilton D J, Kuo C-C, Grayston J T. Detection of Chlamydia pneumoniae by polymerase chain reaction. J Clin Microbiol. 1992;30:434–439. doi: 10.1128/jcm.30.2.434-439.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campbell L A, Kuo C-C. Mouse models of Chlamydia pneumoniae infection and atherosclerosis. Am Heart J. 1999;138:S516–S518. doi: 10.1016/s0002-8703(99)70290-5. [DOI] [PubMed] [Google Scholar]
- 9.Coombes, B. K., and J. B. Mahony. Nucleic acid sequence based amplification (NASBA) of Chlamydia pneumoniae major outer membrane protein ompA mRNA with bioluminescent detection. Combinatorial Chem. High Throughput Screening, in press. [DOI] [PubMed]
- 10.Esposito G, Blasi F, Allegra L, Chiesa R, Melissano G, Cosentini R, Tarsia P, Dordoni L, Cantoni C, Arosio C, Fagetti L. Demonstration of viable Chlamydia pneumoniae in atherosclerotic plaques of carotid arteries by reverse transcriptase polymerase chain reaction. Ann Vasc Surg. 1999;13:421–425. doi: 10.1007/s100169900277. [DOI] [PubMed] [Google Scholar]
- 11.Fong I W, Chiu B, Viira E, Fong M W, Jang D, Mahony J. Rabbit model for Chlamydia pneumoniae infection. J Clin Microbiol. 1997;35:48–52. doi: 10.1128/jcm.35.1.48-52.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fong I W, Chiu B, Viira E, Jang D, Mahony J B. De novo induction of atherosclerosis by Chlamydia pneumoniae in a rabbit model. Infect Immun. 1999;67:6048–6055. doi: 10.1128/iai.67.11.6048-6055.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaydos C A, Quinn T C, Eiden J J. Identification of Chlamydia pneumoniae by DNA amplification of the 16S rRNA gene. J Clin Microbiol. 1992;30:796–800. doi: 10.1128/jcm.30.4.796-800.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaydos C A, Fowler C L, Gill V J, Eiden J J, Quinn T C. Detection of Chlamydia pneumoniae by polymerase chain reaction-enzyme immunoassay in an immunocompromised population. Clin Infect Dis. 1993;17:718–723. doi: 10.1093/clinids/17.4.718. [DOI] [PubMed] [Google Scholar]
- 15.Gaydos C A, Summersgill J T, Sahney N N, Ramirez J A, Quinn T C. Replication of Chlamydia pneumoniae in vitro in human macrophages, endothelial cells, and aortic artery smooth muscle cells. Infect Immun. 1996;64:1614–1620. doi: 10.1128/iai.64.5.1614-1620.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jackson L A, Campbell L A, Kuo C-C, Rodriquez D I, Lee A, Grayston J T. Isolation of Chlamydia pneumoniae from a carotid endarterectomy specimen. J Infect Dis. 1997;176:292–295. doi: 10.1086/517270. [DOI] [PubMed] [Google Scholar]
- 17.Juvonen J, Juvonen T, Laurila A, Alakarppa H, Lounatmaa K, Surcel H M, Leinonen M, Kairaluoma M I, Saikku P. Demonstration of Chlamydia pneumoniae in the walls of abdominal aortic aneurysms. J Vasc Surg. 1997;25:499–505. doi: 10.1016/s0741-5214(97)70260-x. [DOI] [PubMed] [Google Scholar]
- 18.Kuo C-C, Gown A M, Benditt E P, Grayston T J. Detection of Chlamydia pneumoniae in aortic lesions of atherosclerosis by immunocytochemical stain. Arterioscler Thromb. 1993;13:1500–1504. doi: 10.1161/01.atv.13.10.1501. [DOI] [PubMed] [Google Scholar]
- 19.Maass M, Dalhoff K. Transport and storage conditions for cultural recovery of Chlamydia pneumoniae. J Clin Microbiol. 1995;33:1793–1796. doi: 10.1128/jcm.33.7.1793-1796.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maass M, Bartels S C, Engel P M, Momat U, Sievers H H. Endovascular presence of viable Chlamydia pneumoniae is a common phenomenon in coronary artery disease. J Am Coll Cardiol. 1998;31:827–832. doi: 10.1016/s0735-1097(98)00016-3. [DOI] [PubMed] [Google Scholar]
- 21.Madico G, Quinn T C, Boman J, Gaydos C A. Touchdown enzyme time release-PCR for detection and identification of Chlamydia trachomatis, C. pneumoniae, and C. psittaci using the 16S and 16S-23S spacer rRNA genes. J Clin Microbiol. 2000;38:1085–1093. doi: 10.1128/jcm.38.3.1085-1093.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mahony J, Chong S, Jang D, Luinstra K, Faught M, Dalby D, Sellors J, Chernesky M. Urine specimens from pregnant and nonpregnant women inhibitory to amplification of Chlamydia trachomatis nucleic acid by PCR, ligase chain reaction, and transcription-mediated amplification: identification of urinary substances associated with inhibition and removal of inhibitory activity. J Clin Microbiol. 1998;36:3122–3126. doi: 10.1128/jcm.36.11.3122-3126.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mahony J B, Luinstra K E, Sellors J W, Chernesky M A. Comparison of plasmid- and chromosome-based polymerase chain reaction assays for detecting Chlamydia trachomatis nucleic acids. J Clin Microbiol. 1993;31:1753–1758. doi: 10.1128/jcm.31.7.1753-1758.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mahony J B, Luinstra K E, Sellors J W, Jang D, Chernesky M A. Confirmatory polymerase chain reaction testing for Chlamydia trachomatis in first-void urine from asymptomatic and symptomatic men. J Clin Microbiol. 1992;30:2241–2245. doi: 10.1128/jcm.30.9.2241-2245.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mehta S J, Miller R D, Ramirez J A, Summersgill J T. Inhibition of Chlamydia pneumoniae replication in HEp-2 cells by interferon-gamma: role of tryptophan catabolism. J Infect Dis. 1998;177:1326–1331. doi: 10.1086/515287. [DOI] [PubMed] [Google Scholar]
- 26.Moazed T C, Kuo C-C, Patton D, Grayston J T, Campbell L A. Experimental rabbit models of Chlamydia pneumoniae infection. Am J Pathol. 1996;148:667–676. [PMC free article] [PubMed] [Google Scholar]
- 27.Moazed T C, Kuo C-C, Grayston J T, Campbell L A. Evidence of systemic dissemination of Chlamydia pneumoniae via macrophages in the mouse. J Infect Dis. 1998;177:1322–1325. doi: 10.1086/515280. [DOI] [PubMed] [Google Scholar]
- 28.Moazed T C, Kuo C-C, Grayston J T, Campbell L A. Murine models of Chlamydia pneumoniae infection and atherosclerosis. J Infect Dis. 1997;175:883–890. doi: 10.1086/513986. [DOI] [PubMed] [Google Scholar]
- 29.Moulder J W. Interaction of chlamydiae and host cells in vitro. Microbiol Rev. 1991;55:143–190. doi: 10.1128/mr.55.1.143-190.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muhlestein J B, Anderson J L, Hammond E H, Zhao L, Trehan S, Schwabe E P, Carlquist J F. Infection with Chlamydia pneumoniae accelerates the development of atherosclerosis and the treatment with azithromycin prevents it in a rabbit model. Circulation. 1998;97:633–636. doi: 10.1161/01.cir.97.7.633. [DOI] [PubMed] [Google Scholar]
- 31.Petersen E, Boman J, Persson K, Arnerlov C, Wadell G, Juto P, Eriksson A, Dahlen G, Angquist K A. Chlamydia pneumoniae in human abdominal aortic aneurysms. Eur J Vasc Endovasc Surg. 1998;15:138–142. doi: 10.1016/s1078-5884(98)80134-2. [DOI] [PubMed] [Google Scholar]
- 32.Pruckler J M, Masse N, Stevens V A, Gang L, Yang Y, Zell E R, Dowell S F, Fields B S. Optimizing culture of Chlamydia pneumoniae by using multiple centrifugations. J Clin Microbiol. 1999;37:3399–3401. doi: 10.1128/jcm.37.10.3399-3401.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ramirez J A the Chlamydia pneumoniae-Atherosclerosis Study Group. Isolation of Chlamydia pneumoniae from the coronary artery of a patient with coronary atherosclerosis. Ann Intern Med. 1996;125:979–982. doi: 10.7326/0003-4819-125-12-199612150-00008. [DOI] [PubMed] [Google Scholar]
- 33a.Roblin P M, Dumornay W, Hammerschlag M R. Use of HEp-2 cells for improved isolation and passage of Chlamydia pneumoniae. J Clin Microbiol. 1992;30:1968–1971. doi: 10.1128/jcm.30.8.1968-1971.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saikku P, Matilla K, Nieminen M S, Makela P J, Huttunen J K, Valtonen V. Serological evidence of an association of a novel chlamydia, TWAR, with chronic coronary heart disease and acute myocardial infarction. Lancet. 1988;ii:983–986. doi: 10.1016/s0140-6736(88)90741-6. [DOI] [PubMed] [Google Scholar]
- 35.Stephens R S, Kalman S, Lammel C, Fan J, Marathe R, Aravind L, Mitchell W, Olinger L, Tatusov R L, Zhao Q, Koonin E V, Davis R W. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science. 1998;282:754–759. doi: 10.1126/science.282.5389.754. [DOI] [PubMed] [Google Scholar]
- 36.Tong C Y W, Sillis M. Detection of Chlamydia pneumoniae and Chlamydia psittaci in sputum samples by PCR. J Clin Pathol. 1993;46:313–317. doi: 10.1136/jcp.46.4.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wong Y-K, Dawkins K D, Ward M E. Circulating Chlamydia pneumoniae DNA as a predictor of coronary artery disease. J Am Coll Cardiol. 1999;34:1435–1439. doi: 10.1016/s0735-1097(99)00391-5. [DOI] [PubMed] [Google Scholar]