Abstract
The poor stability of antibacterial peptide to protease limits its clinical application. Among these limitations, trypsin mainly exists in digestive tract, which is an insurmountable obstacle to orally delivered peptides. OM19R is a random curly polyproline cationic antimicrobial peptide, which has high antibacterial activity against some gram-negative bacteria, but its stability against pancreatin is poor. According to the structure-activity relationship of OM19R, all cationic amino acid residues (l-arginine and l-lysine) at the trypsin cleavage sites were replaced with corresponding d-amino acid residues to obtain the designed peptide OM19D, which not only maintained its antibacterial activity but also enhanced the stability of trypsin. Proceeding high concentrations of trypsin and long-time (such as 10 mg/mL, 8 h) treatment, it still had high antibacterial activity (MIC = 16–32 µg/mL). In addition, OM19D also showed high stability to serum, plasma and other environmental factors. It is similar to its parent peptide in secondary structure and mechanism of action. Therefore, this strategy is beneficial to improve the protease stability of antibacterial peptides.
Keywords: antimicrobial peptides, trypsin, d-amino acids, stability, mechanism
1. Introduction
In recent decades, bacterial resistance has become more and more serious due to the abuse of antibiotics [1,2]. The emergence of new antibiotic-resistant bacteria means that clinical treatment is facing a great crisis. Therefore, the development of new antibacterial agents has become an urgent task [3]. Antimicrobial peptides (AMPs) are considered ideal candidates to replace antibiotics because of their strong potential against multidrug-resistant bacteria and their unique mechanism of action; that is these peptides are currently the main focus of research [3,4,5]. At present, most of the studies on antimicrobial peptides have focused on finding new peptides with good activity, but few studies have focused on their behavior in the clinical environment [6,7]. However, there are a variety of proteases in organisms, such as trypsin, which can lead to the degradation and inactivation of antimicrobial peptides. These proteases are one of the barriers preventing the clinical application of antimicrobial peptides [6,8]. Although some newly designed and discovered antimicrobial peptides have also been studied on protease resistance, most of them were only studied for the stability and hydrolysis rate of antimicrobial peptides at low concentrations of protease (e.g., 0.1 mg/mL) [9,10]. Generally, few studies have been conducted on the stability and activity changes of antimicrobial peptides at high concentrations (e.g., 10 mg/mL). In fact, the protease concentration in vivo may be far more than 0.1 mg/mL. In recent years, Wang et al. designed an antimicrobial peptide resistant to high concentrations of protease and carried out related research [11].
Trypsin is an important protease in mammals. Like most proteases, trypsin poses a great threat to the application of antimicrobial peptides [12,13]. Trypsin preferentially lyses Lys and Arg cationic residues located at P1. However, these two residues are critical to the activity of AMPs [11,14]. This is because they can produce electrostatic interaction with some charged organelles of bacteria, which affects the normal life activities of bacteria. Therefore, Arginine and Lysine are not only the cleavage sites of an enzyme, but also affect antibacterial activity [10,15,16,17]. Therefore, a question arises: how can we retain the activities of AMPs and improve their stability to trypsin? In order to improve the stability of antimicrobial peptides, many strategies have been proposed, such as cyclization [18], peptidomimetic [19], N-methylation [20,21], side-chain modification [17], multimerization [22], drug delivery methods [23] and the incorporation of unnatural amino acid substitutions [24]. In comparison to the above methods, d-amino acid substitution is a simple and inexpensive strategy to improve antimicrobial peptide–proteinase resistance [9]. d-amino acids are enantiomers of l-amino acids and belong to a group of non-natural amino acids [25]. Their molecular weights, formulas and side chains are the same, except for their spatial conformations. d-type amino acids can retain the physicochemical properties of corresponding l-type amino acids to the greatest extent. The incorporation of d-type amino acids makes it difficult for protease to recognize sensitive amino acids, which can improve protease resistance [26,27]. For example, replacing the cleavage sites of Pseudomonas aeruginosa elastase and Staphylococcus aureus metalloproteinase in the LL-37 turned sequence with d-amino acid can effectively resist protease degradation [28].
In this study, we tried to use this strategy to increase the resistance of trypsin to antimicrobial peptides. OM19R is a polyproline cationic antimicrobial peptide designed by our team. It has high antibacterial activity against some gram-negative bacteria and low toxicity in vitro. It has high stability against some environmental factors such as physiological salt and serum, but its resistance to trypsin needs to be improved [29]. We replaced all l-Arginine and l-Lysine in OM19R with d-Arginine and d-Lysine to obtain a new peptide OM19D. It not only maintained all its advantages, including high antibacterial activity but also improved its resistance to trypsin, especially at high concentrations of trypsin. In addition, the antibacterial mechanism of OM19D was investigated, and we found that its target was not the cell membrane but intracellular, similar to its parent peptide OM19R.
2. Results
2.1. The Structure-Activity Relationship (SAR) of OM19R
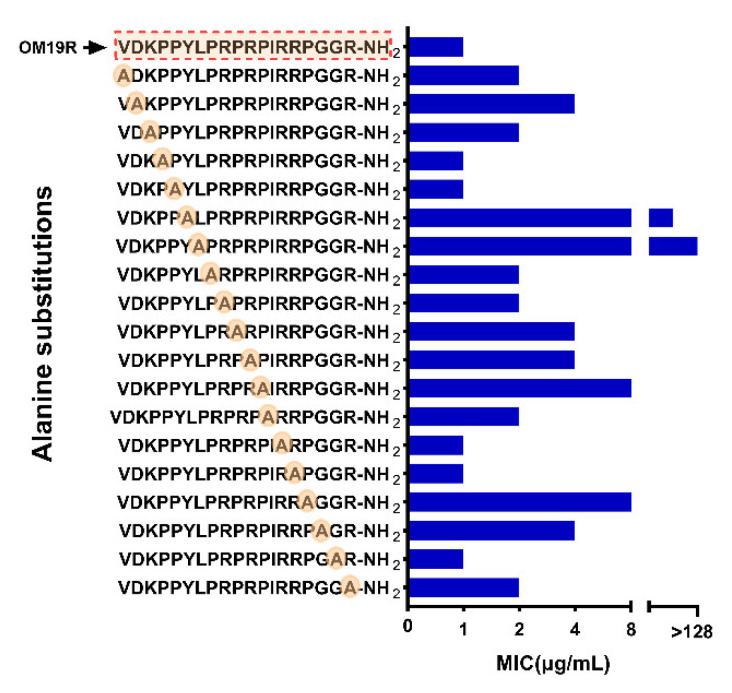
The antimicrobial peptide OM19R, VDKPPYLPRPRPIRRPGGR-NH2, consisted of 19 l-amino acids, including 1 l-Lysine (Lys, K) and 5 l-Arginine (Arg, R). The effect of each amino acid residue on the antibacterial activity of OM19R was studied by the l-alanine (Ala, A) scanning method. As shown in Figure 1, the antibacterial activity of OM19R disappeared completely when the tyrosine at position six and the leucine at position seven were replaced, while the antibacterial activity of OM19R decreased only 0–8 folds when other sites were replaced. This suggests that amino acids at positions six and seven were important and irreplaceable in the whole structure.
Figure 1.
Antibacterial activity of l-Alanine scanner derivatives of OM19R against Escherichia coli ATCC25922. The red dotted box is the sequence of the parent peptide OM19R.
2.2. Peptide Design and Antibacterial Activity
All l-Arginine and l-Lysine in OM19R were replaced by d-Arginine and d-Lysine to obtain a new peptide named OM19D. The sequence and key physicochemical parameters of the peptides are shown in Table 1. According to the results, the molecular weight and charge number of OM19R and OM19D were the same. The measured molecular weights of all peptides were consistent with the theoretical values, which prove that they are synthesized. The ESI-MS mass spectrometry results are presented in Supplementary Materials Figure S1. Their purity is over 98%, and the HPLC chromatographic profiles of the peptides are shown in the supplementary diagram of Supplementary Materials Figure S2.
Table 1.
Peptides and their key physicochemical parameters.
| Peptides | Sequence 1 | Theoretical MW |
Measured MW 2 |
Net Charge |
|---|---|---|---|---|
| OM19D | VDkPPYLPrPrPIrrPGGr-NH2 | 2227.65 | 2226.63 | 6 |
| OM19R | VDKPPYLPRPRPIRRPGGR-NH2 | 2227.65 | 2226.63 | 6 |
1 Lowercase letters indicate the d-enantiomer. 2 Molecular weight (MW) was determined by mass spectrometry.
2.3. Circular Dichroism (CD) Spectroscopy
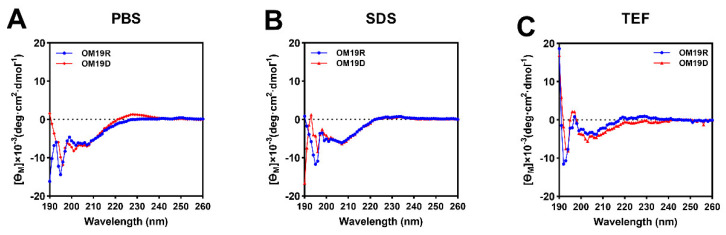
The secondary structures of OM19D and OM19R were detected by CD spectroscopy in phosphate buffer (10 mM, PBS), trifluoroethanol (50% v/v, TFE) and sodium dodecyl sulfate (30 mM, SDS). A 50% TFE solution simulates the hydrophobic environment of the bacterial cell membrane, while SDS solution simulates the negatively charged environment of the prokaryotic cell membrane [30]. Both of them can increase the helicity of peptides. As shown in Figure 2, the secondary structure patterns of the two peptides in the three environments were consistent with the characteristics of random coiling (strong negative peak below 200 nm and a small and wide positive peak around 220 nm). In comparison with the parent peptide OM19R, the secondary structure of the designed peptide OM19D did not change significantly.
Figure 2.
The circular dichroism (CD) spectra of OM19D and OM19R were dissolved in (A) 10 mM phosphate-buffered saline (PBS), (B) 30 mM sodium dodecyl sulfate (SDS), and (C) 50% trifluoroethanol (TFE). The average value after three scans of every sample is shown. The CD spectrum of the buffer was subtracted.
2.4. Antibacterial Activity
As shown in Table 2, OM19D is similar to its parent peptide OM19R in its antimicrobial spectrum and antimicrobial activity. For standard strains, the MIC values of OM19D were 1–4 folds lower than OM19R. For the clinically isolated drug-resistant strains, some strains were slightly sensitive to OM19D, and some were slightly sensitive to OM19R. These drug-resistant strains were resistant to more than three antibiotics with different mechanisms, and their MICs for antibiotics were shown in Supplementary Materials Table S1.
Table 2.
Minimum inhibitory concentrations (MICs) (µg/mL).
| Bacterial Species | OM19D | OM19R |
|---|---|---|
| Sensitive strains | ||
| Escherichia coli ATCC25922 | 2 | 1 |
| Escherichia coli k88 | 4 | 2 |
| Salmonella enterica ATCC 14028 | 4 | 2 |
| Salmonella typhimurium CMCC50115 | 2 | 1 |
| Shigella flexneri ATCC12022 | 2 | 1 |
| Shigella dysenteriae CMCC51252 | 4 | 2 |
| Klebsiella pneumoniae ATCC25955 | >128 | >128 |
| Staphylococcus aureus ATCC25923 | >128 | >128 |
| Clinical isolated strains | ||
| Escherichia coli SN5 | 2 | 8 |
| Escherichia coli S1N1 | 2 | 8 |
| Escherichia coli w136 | 4 | 1 |
| Escherichia coli w122 | 8 | 2 |
| Escherichia coli w123 | 4 | 1 |
| Escherichia coli w124 | 4 | 1 |
| Escherichia coli QY | 4 | 4 |
| Shigella flexneri QY1 | 8 | 8 |
| Methicillin-resistant Staphylococcus aureus HP | >128 | >128 |
2.5. Stability toward Tryptic Degradation
The results of the antibacterial activity of peptides treated with trypsin at different concentrations and different times are shown in Table 3. In short, OM19D showed high antibacterial activity (MIC = 16 µg/mL), even at high concentration of trypsin (10 mg/mL), while the parent peptide OM19R and control melittin had lost their antibacterial activity at 5 mg/mL trypsin (MIC > 128 µg/mL). When compared with untreated, the MIC reduced by 8–16 folds. Moreover, OM19D still had high antibacterial activity (16–32 µg/mL) after being treated with 10 mg/mL trypsin for 8 h, while OM19R and melittin had already lost their antibacterial activity. Therefore, the designed peptide OM19D was resistant to long-term treatment with a high concentration of trypsin.
Table 3.
MIC Values (µg/mL) of peptides against Escherichia coli ATCC 25922 in the presence of trypsin.
| Peptides | Control 1 | Trypsin (mg/mL) | Trypsin (10mg/mL) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 10 | 5 | 2.5 | 1.25 | 0.625 | 1 h | 2 h | 4 h | 8 h | ||
| OM19D | 2 | 16 | 16 | 8 | 8 | 4 | 16 | 16 | 16 | 32 |
| OM19R | 1 | >128 | >128 | 64 | 64 | 64 | >128 | >128 | >128 | >128 |
| Melittin | 2 | >128 | >128 | 64 | 32 | 16 | >128 | >128 | >128 | >128 |
1 The control MIC values were determined in the absence of proteases.
2.6. Condition Sensitivity Assays
As shown in Table 4, Table 5 and Table 6, OM19D still has high antibacterial activity in different physiological salt environments, high-temperature environments and acid-base environments, which is not much different from before treatment. In addition, OM19D still maintained antibacterial activity in serum and plasma. Parent peptide OM19R and melittin were the control groups.
Table 4.
The MICs (µg/mL) of the designed peptides against Escherichia coli ATCC25922 in the presence of physiological salts.
| Peptides | Control 1 | Physiological Salts 2 | |||||
|---|---|---|---|---|---|---|---|
| NaCl | KCl | NH4Cl | MgCl2 | ZnCl2 | FeCl3 | ||
| OM19D | 2 | 4 | 2 | 2 | 4 | 2 | 4 |
| OM19R | 1 | 4 | 2 | 2 | 4 | 2 | 2 |
| Melittin | 2 | 4 | 2 | 2 | 2 | 2 | 2 |
1 The control MICs were determined in Mueller–Hinton broth MHB medium without physiological salts. 2 The final concentrations of NaCl, KCl, NH4Cl, MgCl2, CaCl2, ZnCl2, and FeCl3 were 150 mM, 4.5 mM, 6 μM, 1 mM, 2 mM, 8 μM, and 4 μM, respectively.
Table 5.
The MICs (µg/mL) of the designed peptides against Escherichia coli ATCC25922 after different temperature and pH treatment.
| Peptides | Control 1 | Temperature | pH | ||||||
|---|---|---|---|---|---|---|---|---|---|
| 0 °C | 42 °C | 65 °C | 100 °C | pH4 | pH6 | pH8 | pH10 | ||
| OM19D | 2 | 2 | 2 | 2 | 2 | 2 | 4 | 2 | 4 |
| OM19R | 1 | 1 | 1 | 1 | 1 | 0.5 | 0.5 | 1 | 1 |
| Melittin | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
1 The control MICs were determined at 37 °C and pH 7.4.
Table 6.
MIC Values (µg/mL) of the Peptides against Escherichia coli ATCC 25922 in the presence of Serum and Plasma.
| Peptides | Control 1 | Serum Concentrations (%) | Plasma Concentrations (%) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| 100 | 50 | 25 | 12.5 | 100 | 50 | 25 | 12.5 | ||
| OM19D | 2 | 32 | 32 | 16 | 8 | 64 | 64 | 32 | 16 |
| OM19R | 1 | 32 | 32 | 32 | 16 | 64 | 64 | 64 | 32 |
| Melittin | 2 | >128 | 64 | 64 | 32 | >128 | >128 | >128 | >128 |
1 The control MIC values were determined in the absence of serum and plasma.
2.7. In Vitro Safety Evaluation
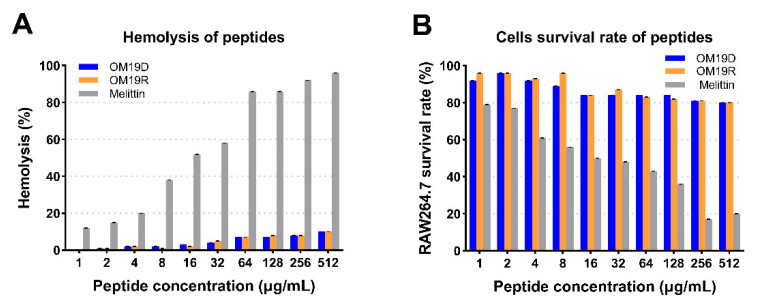
The safety was evaluated by hemolysis rate and cytotoxicity in vitro. The hemolytic activity and cytotoxicity of peptides were measured in the range of 1–512 µg/mL. As shown in Figure 3, compared with the control group melittin, even when the concentration is higher than the minimum inhibitory concentration, the hemolysis rate of rabbit red blood cells caused by OM19D is lower than 10%, and the survival rate of mouse macrophages (RAW 264.7) can reach more than 80%.
Figure 3.
(A) Cytotoxicity of peptides against RAW 264.7 cells. (B) Hemolytic activity of peptides against rabbit red blood cells. The peptide concentration range is 1–512 µg/mL, melittin and OM19R performed as controls.
2.8. Antimicrobial Mechanism Study
2.8.1. Growth Curves of Bacteria and Time-Kill Curve
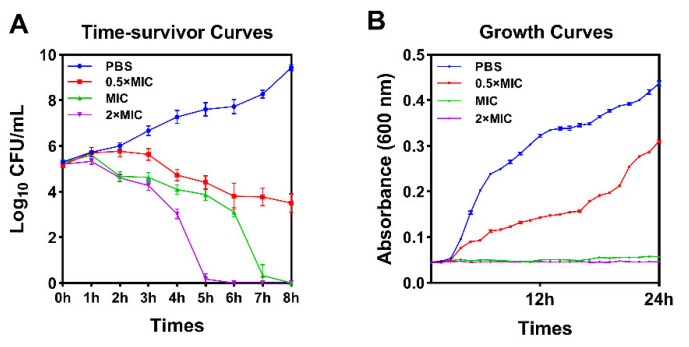
As shown in Figure 4, according to the growth curve, OM19D can completely inhibit the growth of bacteria when the concentration is greater than or equal to MIC. According to the bactericidal kinetics curve, almost all bacteria could be killed by OM19D within 8 h, and the bactericidal rate was proportional to the concentration.
Figure 4.
Time-killing curve (A) and Growth curves (B) for OM19D against Escherichia coli ATCC25922. Cells suspensions were incubated with a final concentration of peptides of 0.5 × MIC, MIC, 2 × MIC and controls were performed without the peptide.
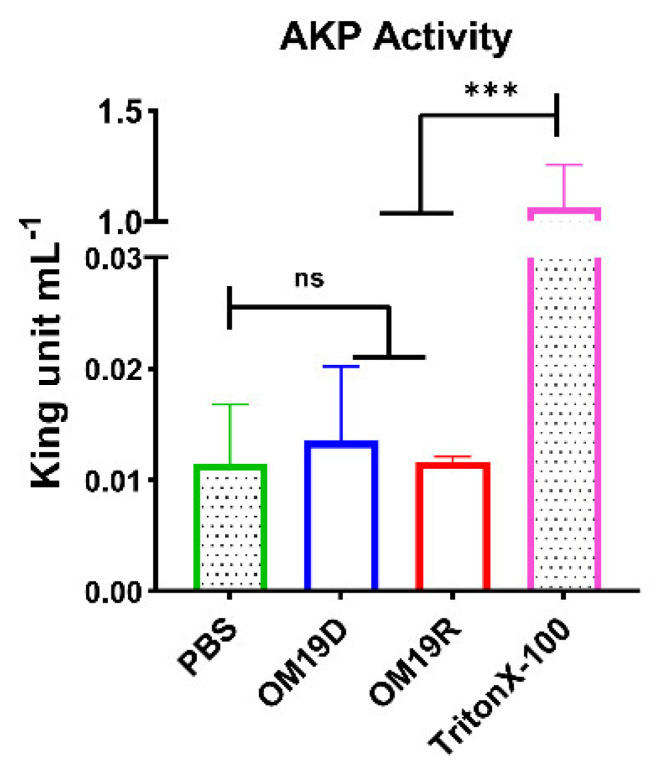
2.8.2. AKP Activity Determination
AKP (alkaline phosphatase) is found between bacterial cell walls and cell membranes [31]. When the cell wall was intact, its activity was almost undetectable in the bacterial fluid. Once the cell wall is damaged, AKP leaks into the bacterial fluid, where it can be detected. Therefore, the integrity of the cell wall can be reflected by the change of AKP content in bacterial fluid. As shown in Figure 5, there was no significant outflow of AKP after 4 h of the peptide treatment compared with negative control group PBS. This shows that the cell integrity has not been destroyed.
Figure 5.
The AKP activity of Escherichia coli ATCC25922 was treated with MIC of OM19D and OM19R, the control group was bacteria treated without any peptide. All data are presented as mean ± SD and the significances were determined by nonparametric one-way ANOVA (*** p < 0.001, ns represents insignificant).
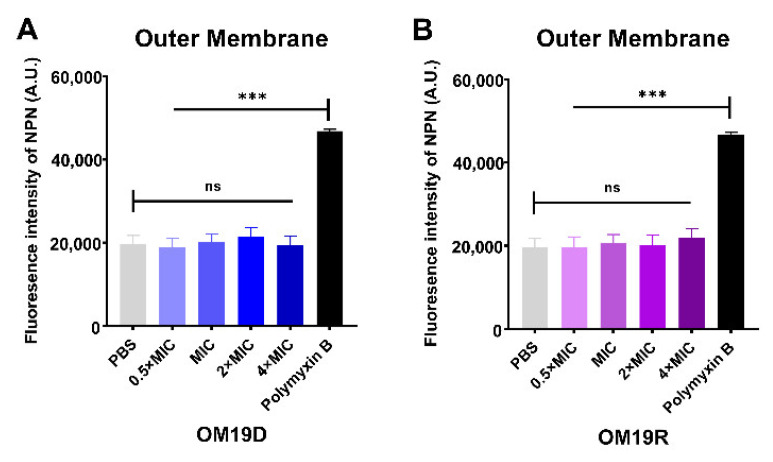
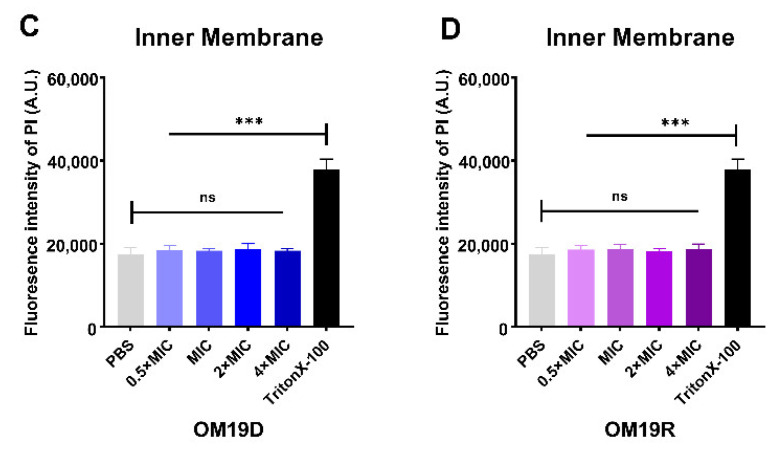
2.8.3. Outer Membrane and Inner Membrane Permeability Assay
As shown in Figure 6, the effects of OM19D and OM19R on the permeability of the bacterial outer membrane and inner membrane were compared by fluorescence intensity analysis. Compared with PBS in the negative control group, there was no obvious change in fluorescence intensity in each group. However, when compared with the positive control group (outer membrane: polymyxin; inner membrane: Triton X-100), the fluorescence intensity of each group had significant changes. Therefore, the designed peptide OM19D was similar to the parent peptide OM19R and had no significant effect on the permeability of the bacterial outer membrane and inner membrane.
Figure 6.
Outer membrane permeability of OM19D (A) and OM19R (B) at the concentrations from 0.5 × MIC to 2 × MIC. Inner membrane permeability of OM19D (C) and OM19R (D) at the concentrations from 0.5 × MIC to 2 × MIC. All data are presented as mean ± SD and the significances were determined by nonparametric one-way ANOVA *** p < 0.001, ns represents insignificant).
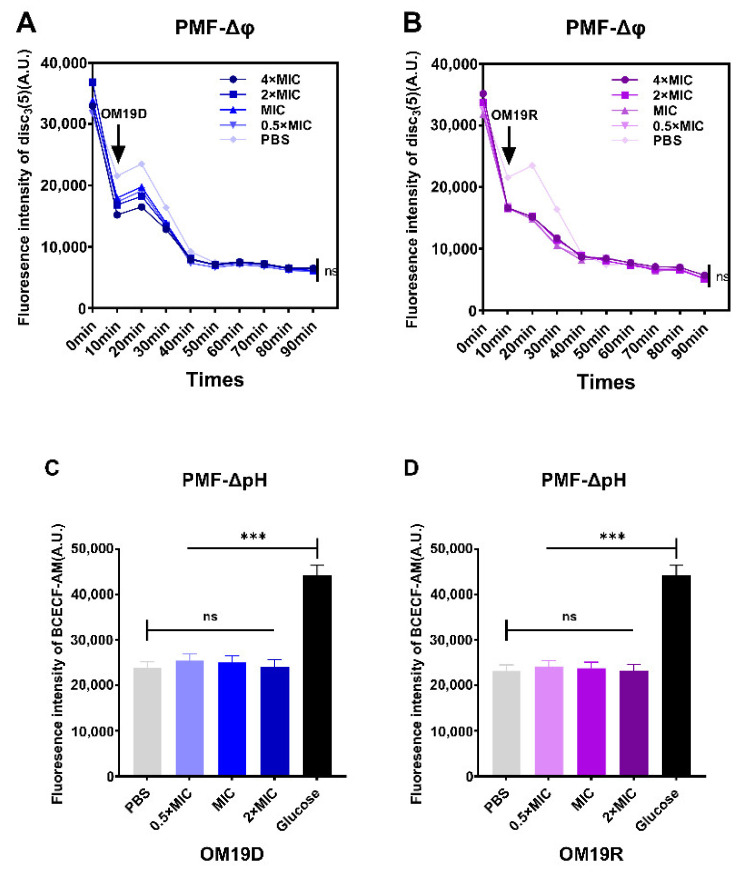
2.8.4. Proton Motive Force
Proton motive force (PMF) is composed of the pH gradient (ΔpH) and the potential gradient (Δφ), which together constitute the electrochemical gradient(ΔP) [32]. Fluorescent probe disc3(5) was used to detect membrane potential, and fluorescent probe BCECF-AM was used to detect intracellular pH changes. As shown in Figure 7, when compared with the negative control group PBS, the membrane potential and pH of OM19D and OM19R did not change significantly. This suggests that the designed peptide OM19D was similar to the parent peptide OM19R, as they did not disrupt the PMF of the bacteria.
Figure 7.
Effects of OM19D (A) and OM19R (B) on bacterial membrane potential (Δφ) in the concentration range of 0.5 × MIC to 4 × MIC, controls were performed without the peptide. Effect of OM19D (C) and OM19R (D) on bacterial intracellular pH (ΔpH) in the concentration range of 0.5 × MIC to 4 × MIC, PBS and glucose were the control groups. All data are presented as mean ± SD and the significances were determined by nonparametric one-way ANOVA (*** p < 0.001, ns represents insignificant).
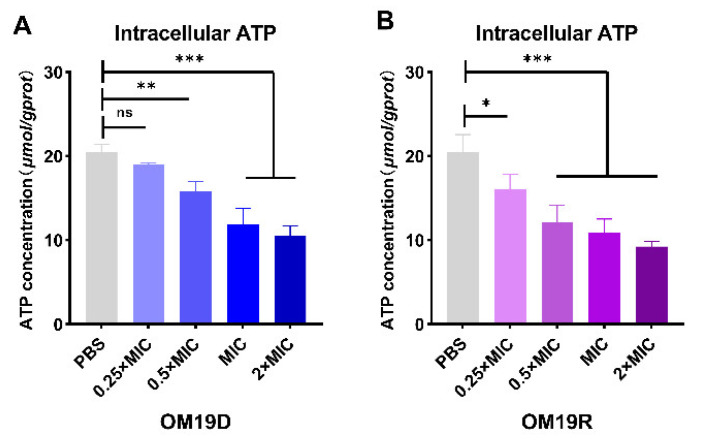
2.8.5. Intracellular ATP
As shown in Figure 8, the intracellular ATP of bacteria treated with OM19D decreased significantly and showed a concentration-dependent decrease. Likewise, OM19R showed a similar trend.
Figure 8.
Effects of different concentrations of OM19D (A) and OM19R (B) on intracellular ATP in Escherichia coli ATC25922. All data are presented as mean ± SD and the significances were determined by nonparametric one-way ANOVA (* p < 0.05, ** p < 0.01, *** p < 0.001, ns represents insignificant).
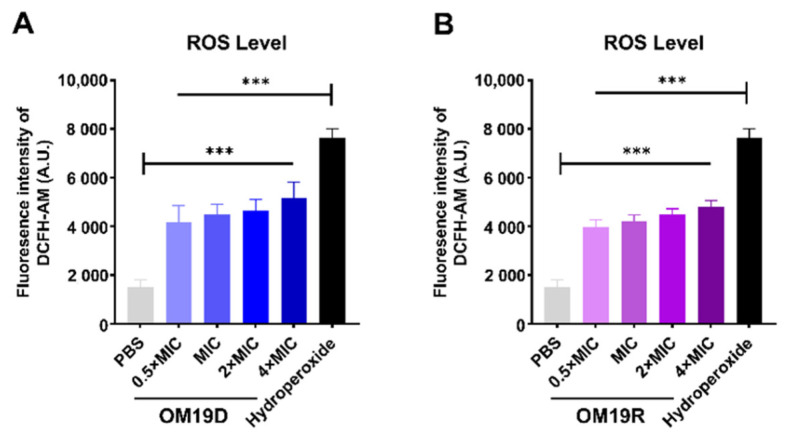
2.8.6. Total ROS Level
The intracellular ROS of bacteria treated with the peptides was detected by fluorescence probe DCFH-DA. As shown in Figure 9, the levels of ROS in OM19D treated bacteria were significantly higher than those in untreated bacteria, which was similar to the parental peptide OM19R.
Figure 9.
Effects of intracellular reactive oxygen species in Escherichia coli ATCC 25922 treated with OM19D (A) and OM19R (B) at concentrations ranging from 0.5 × MIC to 4 × MIC. The negative control was PBS, and the positive control was hydroperoxide. All data are presented as mean ± SD and the significances were determined by nonparametric one-way ANOVA *** p < 0.001.
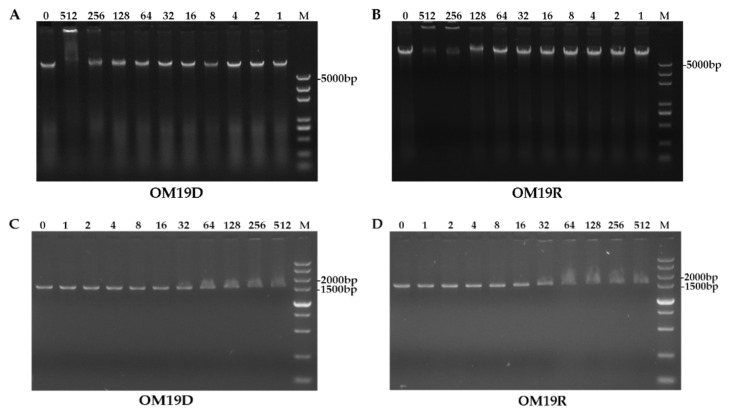
2.8.7. DNA Binding Assay
As shown in Figure 10A,B, the designed peptide OM19D showed the ability to bind to genomic DNA at a concentration of 512 μg/mL, and the parent peptide OM19R showed the ability to bind to DNA at a concentration greater than or equal to 256 μg/mL Their concentration of binding to DNA was much higher than the MIC. As shown in Figure 10C,D, OM19D and OM19R showed the ability to bind to small DNA fragments at the concentration of 16 μg/mL. Therefore, OM19D was similar to OM19R, and interference with bacterial genetic material was not its main mechanism of action.
Figure 10.
Gel electrophoresis of OM19D (A) and OM19R (B) from 0–512 μg/mL binding to genomic DNA of Escherichia coli ATCC25922, Gel electrophoresis of OM19D (C) and OM19R (D) from 0–512 μg/mL binding to small DNA fragments of Escherichia coli ATCC25922, the letter M stands for individual gene markers, and the number stands for concentration values.
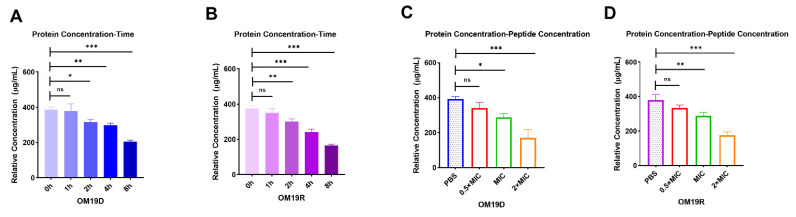
2.8.8. Quantification of Intracellular Protein Concentration
As shown in Figure 11, OM19D treatment caused bacterial protein concentration reduction in a time-dependent and a dose-dependent manner, respectively. The parental peptide OM19R also showed a similar trend. Therefore, OM19D showed the ability to affect the synthesis of bacterial proteins, as did the parent peptide OM19R.
Figure 11.
The trend of intracellular protein concentration of bacteria treated with 0.5 × MIC of OM19D (A) and OM19R (B) followed by time. The trend of intracellular protein concentration of bacteria followed the concentration of OM19D (C) and OM19R (D). The concentration of peptides ranged from 0.5 × MIC to 2 × MIC. All data are presented as mean ± SD and the significances were determined by nonparametric one-way ANOVA (* p < 0.05, ** p < 0.01, *** p < 0.001, ns represents insignificant).
2.8.9. Non-Specific Efflux Pumps
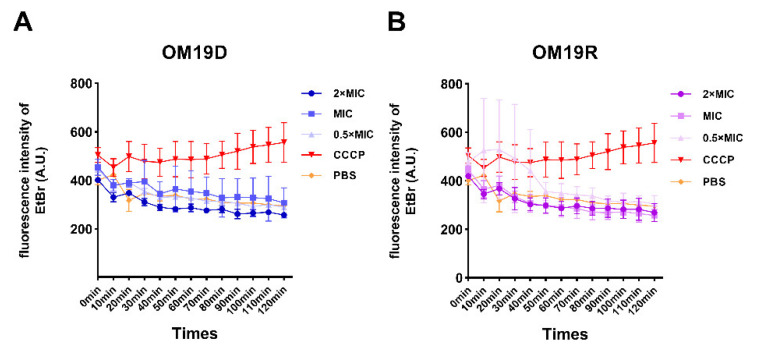
The effects of the peptides on the non-specific bacterial pumping system were detected by fluorescence probe Ethidium bromide (EtBr), and the EtBr efflux from cells was monitored for 120 min. As shown in Figure 12, compared with the control group (decoupling agent CCCP), EtBr efflux from bacteria treated with OM19D showed a decreasing trend in a time-dependent manner, and so did OM19R. The results showed that OM19D and OM19R had no inhibitory effect on the bacterial efflux pump.
Figure 12.
Effects of different concentrations of OM19D (A)and OM19R (B) on the efflux pump of Escherichia coli ATCC 25922.
3. Discussion
Up to now, more than 4500 AMPs have been stored in the antimicrobial peptide database (APD), and the number is still increasing [33,34,35]. However, there are only a few available for clinical use, such as bacitracin and polymyxin [36]. Antimicrobial peptides are easily degraded by protease and lose their antibacterial activity, which leads to the limitations of clinical application [3]. Among them, trypsin is a very important digestive enzyme ubiquitous in mammals, which mainly attacks the arginine and lysine of proteins or polypeptides [37]. In this study, we focused on trypsin and focused on overcoming its degradation of antibacterial peptides. According to our previous reports, OM19R has high activity against some intestinal pathogenic bacteria such as Escherichia coli and Salmonella [29]. Therefore, it is likely to be used as an intestinal microbicide in the future. Therefore, trypsin is one of the obstacles that must be overcome.
According to the structure–activity relationship of OM19R, the cleavage site of trypsin, and the structural characteristics of l-Arginine and l-Lysine, we chose the strategy of replacing l-amino acid at the cleavage site with d-amino acid, in other words, replacing all l-Arginine and l-Lysine in OM19R with d-Arginine and d-Lysine, and obtaining the designed peptide OM19D. According to the results, OM19D still maintained high activity (MIC = 16 μg/mL) after being treated with a high concentration of trypsin (10 mg/mL). When compared with the untreated, MIC decreased by only 8–16 folds. The reason for the slight loss of antibacterial activity of OM19D may be that a very small fraction of peptide is degraded during the operation of inactivating trypsin. In addition, there is a potential possibility that a fraction of peptide is co-precipitated with trypsin that resulted in an increased MIC. The trypsin concentration of 10 mg/mL was chosen because it was consistent with the simulated intestinal fluid (SIF) content specified in the United States Pharmacopoeia [38]. However, the parent peptide OM19R and the control group melittin have lost their antibacterial activity. In addition, OM19D still maintained its antibacterial activity after being treated with 10 mg/mL trypsin for 8 h. Therefore, it can be concluded that the strategy not only improves the resistance of the peptide to trypsin but also maintains its antibacterial activity. Importantly, it can resist high concentrations of trypsin for a long time.
d-type amino acid substitution strategy has the advantages of simplicity, low price and relatively low biological toxicity [3]. Polymyxin and daptomycin, which have been put on the market, all contain d-type amino acids in their structures [39,40]. In this study, l-Lysine and d-Lysine, and l-Arginine and d-Arginine, have the same molecular weight, molecular formula, primary amino (Lysine) and guanidine (Arginine) in the side chains, which play a critical role in antibacterial activity, but they are only different in spatial conformation. Therefore, the antibacterial activity of the peptide was almost unaffected by d-amino acid substitution; however, resistance to trypsin was significantly increased. According to the results of this experiment, the designed peptide OM19D is completely in line with the original expectations, which not only retained the antibacterial activity but also enhanced the stability of trypsin. This shows the feasibility of this strategy.
The stability of antimicrobial peptides in the blood is also very important, as no matter the method of administration, the drug will eventually be absorbed into the blood. We tested the stability of OM19D in bovine serum and rabbit plasma. After treatment, the peptide maintained antibacterial activity. In addition, we also tested that it can still have high antibacterial activity in the physiological salt environment, extreme temperatures and an acid-base environment. OM19D exhibits heat-cold and acid-base stability, which is important for many production and storage processes such as medicine, food or feed processing [35]. For in vitro safety, we evaluated its hemolytic activity and cytotoxicity. According to previous reports, and the results of this experiment, the cytotoxicity of both the designed and the parent peptide OM19R are low. In comparison to OM19R, the designed peptide OM19D has no increase in hemolytic activity and cytotoxicity even if it contains unnatural amino acids, which is within the reasonable range.
Finally, the action mechanism of OM19D was preliminarily discussed. The main targets of antimicrobial peptides include external targets that target the cell membrane (e.g., Melittin) or cell wall (e.g., Nisin), and intracellular targets such as nucleic acid synthesis (e.g., Buforin II), protein synthesis (e.g., Indolicidin), inhibition of efflux pump (e.g., PAβN) and ATP efflux (e.g., Histatins), etc. [41,42,43]. According to the target of antimicrobial peptides, the following studies were performed. Circular Dichroism showed that OM19D and OM19R had similar secondary structures, which correlated with the characteristics of a random coil. Therefore, the incorporation of d-amino acids did not cause significant changes in the secondary structure. According to previous reports, the mechanism of OM19R is not to destroy the bacterial cell membrane. We have also done related research and found that OM19D does not destroy the integrity of cells and does not affect the permeability of the cell membrane, which is similar to the parent peptide OM19R. This is similar to most polyproline antimicrobial peptides. The targets of the polyproline antimicrobial peptides are generally inside cells. Therefore, we did some intracellular research. We found that the bacteria treated with OM19D had no significant effect on the proton-powered and nonspecific effluent systems. It only binds to DNA when it is 8 folds higher than MIC. However, the intracellular reactive oxygen species increased significantly, which may be due to the entry of polypeptides into cells affecting their normal life activities, resulting in stress response or its normal redox response being affected. Intracellular ATP concentration decreased significantly, which may destroy the energy chain. The decrease of ATP may also affect protein synthesis. The results showed that OM19D had a significant effect on intracellular protein concentration, and the protein concentration decreased significantly with the increase of time and concentration. In all the experiments related to the antibacterial mechanism of OM19D, we used the parent peptide OM19R as the control group. We found that their antibacterial mechanisms are very similar. The main mechanism of OM19D action is to affect intracellular activity rather than to destroy cell integrity, as most antimicrobial peptides do. Therefore, the incorporation of d-amino acids did not affect the antibacterial mechanism.
4. Materials and Methods
4.1. Materials
4.1.1. Bacterial Species
Standard strains: Escherichia coli ATCC25922, Escherichia coli k88, Salmonella enterica ATCC 14028, Salmonella typhimurium CMCC50115, Shigella flexneri ATCC12022, Shigella dysenteriae CMCC51252, Klebsiella pneumoniae ATCC25955, Staphylococcus aureus ATCC25923. Clinically isolated strains: Escherichia coli SN5, Escherichia coli S1N1, Escherichia coli w136, Escherichia coli w122, Escherichia coli w123, Escherichia coli w124, Escherichia coli QY, Shigella flexneri QY1, Methicillin-resistant Staphylococcus aureus HP. RAW 264.7 cells. All strains were obtained from the College of Animal Science and Technology, Jilin Agricultural University (Changchun, China).
4.1.2. Reagent
Mueller–Hinton broth (MHB) and Mueller–Hinton agar (MHA) were purchased from GL Biochem (Shanghai, China). Triton X-100 and phosphate-buffered saline (PBS) solution were purchased from Macklin (Shanghai, China). BCECF-AM, EtBr, PI, diSC3(5), polymyxin B, 4-(2-hydroxyethyl) piperazine-1-ethanesulfonic acid (HEPES) and NPN were also purchased from Macklin (Shanghai, China). TransDetect Cell Counting Kit-8 (CCK-8) and Dulbecco’s modified Eagle’s medium with high glucose (DMEM) were purchased from TransGen Biotech (Beijing, China). Fetal bovine serum (FBS) was purchased from Gibco (Shanghai, China).
4.2. Synthesis and Sequence Analysis of Peptides
The alanine scan was performed by replacing each amino acid residue in the original peptide OM19R with alanine. OM19D was the replacement of l-arginine and l-lysine in the original peptide OM19R with d-arginine and d-lysine. The designed peptides were synthesized and purified by GL Biochem (Shanghai, China). The peptides were synthesized using standard Fmoc protocols. The Peptides were purified and determined by RP-HPLC with SHIMADZU Inertsil ODS-SP (4.6 mm × 250 mm × 5 μm, RP-C18) column, UV absorption wavelength 214 nm, volume 60 μL, eluted with water/acetonitrile gradient containing 0.1% trifluoroacetic acid, flow rate 1.0 mL/min. Finally, ESI-MS was used to determine the molecular weight.
4.3. MIC Measurements
Minimum inhibitory concentrations (MICs) of OM19R and its derivatives were determined by the broth micro-dilution method, which was in accordance with CLSI 2015 guidelines [44]. Briefly, bacteria were cultured in Mueller–Hilton broth (MHB) medium to logarithmic phase and diluted to 105 CFU/mL. Then, 50 μL of peptides of different concentrations and 50 μL bacterial solution were mixed in a sterile 96-well plate, and the plates were incubated for 16 h at 37 °C. MIC was examined by measuring the OD at 600 nm. The MIC was defined as the lowest peptide concentration that inhibited 95% of the bacterial growth.
4.4. CD Measurements
The test methods refer to previous reports [5]. The CD spectra of the peptides were detected on a J-810 spectropolarimeter (Jasco, Tokyo, Japan). The peptides were respectively dissolved in 10 mM PBS (pH 7.4), 50% TFE, and 30 mM SDS where the final concentration was 128 µM. The experimental parameters were as follows: 10 nm·min−1 scanning rate, 1 mm path length, 190–260 nm wavelength range, and 25 °C. Each sample was scanned at least three times.
4.5. Typsin Resistance Assays
The effect of different concentrations of trypsin on the antimicrobial activity of peptides was tested using previously reported methods [11]. Trypsin was prepared to the required concentration. In the sterile 96-well plates, the peptide was continuously twofold diluted to different concentrations in trypsin solution. The plates were incubated at 37 °C, and the incubation time was determined according to the experiment requirements. The peptide alone and protease alone were used as a control. Samples were taken at different times, heated at 100 °C for 15 min to inactivate the protease, and centrifuged at 13,000× g for 30 min to precipitate the protease. Finally, an equal volume of bacteria (105 CFU/mL) was added to the 96-well plates, the plates were incubated for 16 h at 37 °C, and then the MIC value of the peptide was observed. The indicator bacterium was Escherichia coli ATCC25922. This experiment was repeated three times.
4.6. Physiological Salt Sensitivity Assays
The peptide salt sensitivity was evaluated by measuring the MIC. The method is as previously described [11,45]. The peptides were diluted in deionized water containing different salts, and then an equal volume of Escherichia coli ATCC25922 solution (105 cfu/mL) was added. The mixture was incubated at 37 °C for 16 h. The final physiological concentrations of these salts were 150 mM NaCl, 4.5 mM KCl, 6 μM NH4Cl, 1 mM MgCl2, 8 μM ZnCl2 and 4 μM FeCl3, respectively. This experiment was repeated three times.
4.7. Temperature and pH Sensitivity Assays
The temperature and pH sensitivities of the peptides were determined by an MIC assay as described previously [30]. For temperature sensitivity, the diluted peptide solution was incubated at 0 °C, 37 °C, 42 °C, 65 °C and 100 °C, respectively, for 30 min, then stopped heating and returned to room temperature. An equal volume of Escherichia coli ATCC25922 (105 CFU/mL) was added, and the mixture was incubated at 37 °C for 16 h. MIC values were observed. This experiment was repeated three times.
For pH sensitivity, we adjusted the diluent to pH 4.0, 6.0, 8.0 and 10.0 with HCl or NaOH, and then diluted the peptide with this diluent. The peptide solutions of different pH were incubated for 1 h, and then the pH of the reaction solution was neutralized by HCl or NaOH to terminate the reaction. An equal volume of Escherichia coli ATCC25922 (105 CFU/mL) was added, and the mixture was incubated at 37 °C for 16 h. MIC values were observed. This experiment was repeated three times.
4.8. Serum and Plasma Sensitivity Assays
The serum sensitivities of the peptides were determined by an MIC assay as described previously [11,30]. Bovine serum and rabbit plasma solutions of different concentrations (12.5%, 25%, 50% and 100%) were used as the diluent of the peptide, and the peptides were twice diluted and incubated at 37 °C for 1 h. After that, the reaction was terminated after treatment at 100 °C for 15 min and cooled to room temperature. The same volume of Escherichia coli ATCC25922 solution (105 CFU/mL) was incubated for 16 h at 37 °C. The change of MIC value was observed. This experiment was repeated three times.
4.9. Measurement of Hemolysis Activity
We used fresh rabbit red blood cells and sheep blood red blood cells to test the hemolytic activity of the peptide, as previously reported [10]. The collected red blood cells were washed three times with sterile 0.9% NaCl solution (pH 7.4) and diluted to 2% (v/v). The peptides were two-fold diluted with 0.9% NaCl solution to different concentrations. The equal volume of red blood cell suspension and different concentrations of peptide solution was added to the 96-well plate and incubated at 37 °C for 1 h. Then, the mixture was centrifuged at 1000× g for 10 min at 4 °C, the supernatant was transferred to a new 96-well plate, and the absorbance was measured at OD 570 nm using a microplate reader (Microplate reader, TECAN GENios F129004, Tecan, Salzburg, Austria). A 0.9% NaCl erythrocyte suspension without peptide was used as negative control and 1% Triton X-100 as a positive control. The hemolysis rate was calculated according to the following equation:
| Hemolysis rate (%) = [(OD576 sample − OD576 blank)/(OD576 1%Triton X-100 − OD576 blank)] × 100% | (1) |
4.10. Cytotoxicity Assays
The CCK-8 assay was used to detect the cytotoxicity of the peptide, as previously reported [30,31]. RAW 264.7 cells were cultured in DMEM (containing 10% fetal bovine serum), and 10,000 cells were added to each well in 96-well plates and incubated at 37 °C in 5% CO2 for 6 h. The peptide was two-fold diluted in DMSO and added to 96-well plates in equal volume. The mixture was cultured for 16 h. Then, CCK-8 (10%, v/v) was added to the cell culture medium to each well and cultured for 4 h. The absorption was measured at OD 450 nm using a microplate reader. The blank medium was used as the negative control, and the medium containing only cells was used as the positive control. Cell survival rate was calculated according to the following equation:
| Cell survival rate (%) = [(OD450 sample − OD450 negative control)/(OD450 positive control − OD450 negative control)] × 100% | (2) |
4.11. Determination of AKP Outflow
The test methods refer to previous reports [46]. AKP of Escherichia coli ATCC 25922 were determined using an AKP Assay Kit (Jiancheng, Nanjing, China). Collected cells in the logarithmic growth phase were rinsed with 0.01M PBS (pH7.4) three times and diluted to OD600 = 0.5. Peptides of different concentrations were added and incubated for 4 h and then centrifuged at a low temperature. The PBS treatment group was a negative control group, and Triton X-100 treatment group was a positive control group. The supernatant was collected and placed in the working solution, following the instructions, and the absorbance was measured at OD520 nm. Then, the value was substituted into the formula to find the AKP concentration.
4.12. Outer Membrane Permeability Assay
Fluorescent dye NPN was used to examine outer membrane permeability according to previously reported methods [47]. Collected bacterial cells in the logarithmic growth phase were rinsed with 5mMHEPES buffer (pH7.4, contains 5 mM glucose) three times and diluted to OD600 = 0.5. Then, a 10 μM NPN was added to the bacterial suspension and incubated at room temperature for 30 min. An equal volume of different concentrations of peptides and the bacterial suspension was added to the black 96-well plate. The fluorescence intensity of the samples was measured by F4500 fluorescence spectrophotometer (emission λ = 420 nm, excitation λ = 350 nm). The negative control was the initial fluorescence value of Escherichia coli ATCC25922 bacterial suspension containing NPN, and the positive control was the fluorescence value produced by polymyxin B on Escherichia coli ATCC25922 cells.
4.13. Inner Membrane Permeability Assay
Fluorescent dye PI was used to examine outer membrane permeability according to previously reported methods [48]. The collected bacterial cells in the logarithmic growth phase were rinsed with 0.01 M PBS buffer (pH 7.4) three times and diluted to OD600 = 0.5. Then, a 10 μM PI was added to the bacterial suspension and incubated at room temperature for 10 min. The suspension was placed in each well of the 96-well plate and cultured at 37 °C with peptides ranging from 0.5 × MIC to 4 × MIC. The fluorescence intensity of the samples was measured by the F4500 fluorescence spectrophotometer (emission λ = 617 nm, excitation λ = 535 nm). The negative control was the initial fluorescence value of Escherichia coli ATCC25922 bacterial suspension containing PI, and the positive control was the fluorescence value produced by Triton X-100 on Escherichia coli ATCC25922 cells.
4.14. Proton Motive Force Assay
4.14.1. Membrane Potential Assay
The test methods refer to previous reports [48]. Escherichia coli ATCC25922 cells were washed and resuspended with 5 mM HEPES (pH 7.0, plus 5 mM Glucose). The potentiometric probe DiSC3(5) was added in the medium at a final concentration of 1 × 10−6 M. Peptides of different concentrations were added to the culture medium containing bacteria and DISC3(5), and incubated in the dark for 30 min. The excitation wavelength at 622 nm and emission wavelength at 670 nm, measured fluorescence intensity.
4.14.2. Intracellular pH Assay
The test methods refer to previous reports [48]. Escherichia coli ATCC25922 cells were washed and resuspended with 5 mM HEPES (pH 7.0). Fluorescent probe BCECF-AM was added to the medium at a final concentration of 20 μM. After the fluorescence stabilized, glucose (25 μM) or varying peptides were added. The excitation wavelength and emission wavelength were set at 488 nm and 522 nm, respectively.
4.15. Intracellular ATP Determination
Intracellular ATP levels of Escherichia coli ATCC25922 were determined using an ATP Assay Kit (Jiancheng, Nanjing, China). Collected cells in the logarithmic growth phase were rinsed with 0.01 M PBS (pH7.4) three times and diluted to OD600 = 0.05. Peptides of different concentrations were added and incubated for 1 h, then centrifuged at a low temperature. The supernatant was collected and placed in the ATP working solution, mixed, stood for 5 min, and the absorbance was measured at 636 nm. Then, the value was substituted into the formula to find the ATP concentration [48].
4.16. Total ROS Measurement
Total ROS levels of Escherichia coli ATCC 25922 were determined using a ROS Assay Kit (Jiancheng, Nanjing, China). Collected cells in the logarithmic growth phase were rinsed with 0.01 M PBS (pH7.4) three times and diluted to OD600 = 0.5. DCFH-DA dye (final concentration: 10 μM) was added into the bacterial suspension and incubated for 30 min at room temperature. Then, the cells were washed with PBS to remove fluorescent probes that had not entered the cells. After that, different concentrations of antimicrobial peptides were added and let stand for 1 h at 37 °C. Subsequently, the fluorescence intensity was immediately measured with the excitation wavelength at 488 nm and emission wavelength at 525 nm. Hydrogen peroxide as the hydrogen donor was used as a positive control of ROS production [49].
4.17. Efflux Pump Assay
Cells were co-incubated with 5μM EtBr and sub-MIC of peptides or known efflux pump inhibitor CCCP (100 μM) at 37 °C to OD600 = 0.5. After centrifuging at 8000× g at 4 °C for 10 min, the precipitate was collected and resuspended in MHB. Then, EtBr effluent was monitored within 120 min at an excitation wavelength of 525 nm and an emission wavelength of 605 nm [44,48].
4.18. Quantification of Intracellular Protein
4.18.1. Test of Protein Concentration Change with Peptides Concentration Change
A 5mL Escherichia coli ATCC25922 was cultured to the logarithmic growth stage. Bacteria were treated with the final concentration of 0.5 × MIC to 4 × MIC peptide and 0.01 M PBS (pH = 7.4) for 1 h. The bacteria were collected by centrifugation and washed with PBS. Then, 1mL PBS was added to resuspend the bacteria. The total number of bacteria at each concentration point was obtained by the flat colony counting method. Meanwhile, the bacterial resuspension was ultrasonically crushed in an ice bath for 5 min. The supernatant was collected and centrifuged at 4 °C for 30 min. Finally, the total protein concentration was determined using the BCA kit (Sangon, Shanghai, China) [49]. The bacterial survival rate of the PBS group was set as 100%, and the bacterial survival rate of the other concentration point was calculated. The actual total protein concentration, measured at each concentration point, was divided by the survival rate of each concentration point so as to get the relative total protein concentration when the survival rate of bacteria at each point was 100%.
4.18.2. Test of Protein Concentration Change with Action Time
A 5mL Escherichia coli ATCC25922 was cultured to the logarithmic growth stage. Bacteria were treated with peptides whose final concentration was subinhibitory concentrations for 0, 1, 2, 4 and 8 h, respectively. The total number of bacteria at each time point was obtained by the flat colony counting method. The remaining methods were as described above. The bacterial survival rate of 0-h treatment group was set as 100%, and the bacterial survival rate of the other time point was calculated. The actual total protein concentration measured at each time point was divided by the survival rate of each time point so as to get the relative total protein concentration when the survival rate of bacteria at each point was 100%.
4.19. DNA Binding Assay
DNA combined experiments were based on previous reports [30,50]. Collected Escherichia coli ATCC25922 cells in the logarithmic growth phase were rinsed with PBS (pH 7.4) three times and diluted to OD600 = 0.5. Then, the genomic DNA was extracted, or Small DNA fragments were obtained by PCR amplification of 16SrDNA. The primer sequences are shown in Supplementary Materials Table S2. A 400 ng genomic DNA or small DNA fragments was incubated with peptide solutions of different concentrations in the binding buffer at 37 °C for 2 h. After that, the samples were detected via 1% agarose gel electrophoresis.
4.20. Growth Curves of Bacteria and Time-Kill Curve
The bacterial growth curve and time-kill curve was measured as previously reported [51].
4.20.1. The Bacterial Growth Curve
The single colony of Escherichia coli ATCC25922 was selected and inoculated in 1 mL MH liquid medium and cultured at 37 °C to the logarithmic growth phase. The culture was standardized to meet the 0.5McFarland turbidity standard. Then, the bacterial liquid was diluted in an MHB medium 1:100. Finally, different concentrations of antimicrobial peptides were added to the 96-well plate, and then diluted bacterial solution of the same volume was added. The absorption was measured at 600 nm using a microplate reader every 1 h for 24 h.
4.20.2. Time-Kill Curve
Escherichia coli ATCC 25922 (105 CFU/mL) was treated with peptides of 0.5 × MIC and 2 × MIC concentrations. A 50 μL sample was taken out every 10 min and diluted 100-fold, then coated on MHA solid medium and cultured at 37 °C. Colonies were counted at each sampling time point, and the time-kill curve was drawn. The samples without antimicrobial peptides were the control group.
4.21. Statistical Analysis
Statistical analysis was performed using unpaired t-test between two groups and one-way ANOVA among multiple groups. SPSS22.0 was used for statistical analysis. Quantitative data were expressed as mean ± standard deviation, p-values was considered statistically significant (* p <0.05,** p < 0.01, *** p < 0.001, ns represents insignificant).
5. Conclusions
In this study, according to the structure–activity relationship, we successfully designed a polypeptide with high resistance to trypsin by replacing arginine and lysine at the trypsin restriction site with d-type amino acid and tested its biological activity. Compared with the parent peptide OM19R, the antimicrobial activity of the designed peptide OM19D was not significantly decreased but the stability of trypsin was significantly improved, especially in a high concentration of protease. OM19D still had an antibacterial activity after being treated with a high concentration of trypsin for 8 h and showed strong resistance to trypsin. In addition, OM19D is well tolerated in serum and plasma and is resistant to high temperatures, acid and alkali. It has no obvious hemolytic activity and cytotoxicity. According to CD spectra, the designed peptide OM19D and the parent peptide OM19R conform to the irregular curl characteristic, indicating that part of d-type amino acid substitution did not affect its secondary structure. Finally, the target of the designed peptide OM19D is not on the cell membrane, while the intracellular target may exist as a potential possibility, which is similar to the parent peptide OM19R. These results suggest that the substitution of d-type amino acids for amino acids at the cleavage site of protease is helpful to improve the resistance of antimicrobial peptides to protease, which is beneficial to future clinical studies.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/antibiotics10121465/s1, Figure S1: The ESI-MS of Peptides. Figure S2: The HPLC chromatographic profiles of Peptides. Table S1: Antimicrobial susceptibility test of clinical isolated strains used in this study (MIC, μg/mL). Table S2: PCR primer sequences.
Author Contributions
Conceptualization, X.Z.; funding acquisition, H.M.; supervision, H.M. and L.K.; project administration, H.M. and L.K.; investigation, M.Z., H.Z., I.M. and L.K.; validation, Q.C., Y.J. and Q.X. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (Grant number: 31872519) and the Open Research Fund of Engineering Research Center of Bioreactor and Pharmaceutical Development, Ministry of Education (Grant number: KF202003).
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare that no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Davies J. Where have All the Antibiotics Gone? Can. J. Infect. Dis. Med. Microbiol. 2006;17:287–290. doi: 10.1155/2006/707296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rosen T. Antibiotic Resistance: An Editorial Review With Recommendations. J. Drugs Dermatol. 2011;10:724–733. [PubMed] [Google Scholar]
- 3.Tan P., Fu H., Ma X. Design, optimization, and nanotechnology of antimicrobial peptides: From exploration to applications. Nano Today. 2021;39:101229. doi: 10.1016/j.nantod.2021.101229. [DOI] [Google Scholar]
- 4.Sato H., Feix J.B. Peptide–membrane interactions and mechanisms of membrane destruction by amphipathic α-helical antimicrobial peptides. Biochim. Biophys. Acta (BBA)-Biomembr. 2006;1758:1245–1256. doi: 10.1016/j.bbamem.2006.02.021. [DOI] [PubMed] [Google Scholar]
- 5.Wang J., Chou S., Yang Z., Yang Y., Wang Z., Song J., Dou X., Shan A. Combating Drug-Resistant Fungi with Novel Imperfectly Amphipathic Palindromic Peptides. J. Med. Chem. 2018;61:3889–3907. doi: 10.1021/acs.jmedchem.7b01729. [DOI] [PubMed] [Google Scholar]
- 6.Li Y., Liu T., Liu Y., Tan Z., Ju Y., Yang Y., Dong W. Antimicrobial activity, membrane interaction and stability of the D-amino acid substituted analogs of antimicrobial peptide W3R6. J. Photochem. Photobiol. B Biol. 2019;200:111645. doi: 10.1016/j.jphotobiol.2019.111645. [DOI] [PubMed] [Google Scholar]
- 7.Kim H., Jang J.H., Kim S.C., Cho J.H. De novo generation of short antimicrobial peptides with enhanced stability and cell specificity. J. Antimicrob. Chemother. 2013;69:121–132. doi: 10.1093/jac/dkt322. [DOI] [PubMed] [Google Scholar]
- 8.Rozek A., Powers J.-P.S., Friedrich C.L., Hancock R.E.W. Structure-Based Design of an Indolicidin Peptide Analogue with Increased Protease Stability. Biochemistry. 2003;42:14130–14138. doi: 10.1021/bi035643g. [DOI] [PubMed] [Google Scholar]
- 9.Jia F., Wang J., Peng J., Zhao P., Kong Z., Wang K., Yan W., Wang R. D-amino acid substitution enhances the stability of antimicrobial peptide polybia-CP. Acta Biochim. Biophys. Sin. 2017;49:916–925. doi: 10.1093/abbs/gmx091. [DOI] [PubMed] [Google Scholar]
- 10.Zhong C., Liu T., Gou S., He Y., Zhu N., Zhu Y., Wang L., Liu H., Zhang Y., Yao J., et al. Design and synthesis of new N-terminal fatty acid modified-antimicrobial peptide analogues with potent in vitro biological activity. Eur. J. Med. Chem. 2019;182:111636. doi: 10.1016/j.ejmech.2019.111636. [DOI] [PubMed] [Google Scholar]
- 11.Wang J., Song J., Yang Z., He S., Yang Y., Feng X., Dou X., Shan A. Antimicrobial Peptides with High Proteolytic Resistance for Combating Gram-Negative Bacteria. J. Med. Chem. 2019;62:2286–2304. doi: 10.1021/acs.jmedchem.8b01348. [DOI] [PubMed] [Google Scholar]
- 12.Vlieghe P., Lisowski V., Martinez J., Khrestchatisky M. Synthetic therapeutic peptides: Science and market. Drug Discov. Today. 2010;15:40–56. doi: 10.1016/j.drudis.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 13.Miura C., Ohta T., Ozaki Y., Tanaka H., Miura T. Trypsin is a multifunctional factor in spermatogenesis. Proc. Natl. Acad. Sci. USA. 2009;106:20972–20977. doi: 10.1073/pnas.0907631106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karstad R., Isaksen G., Brandsdal B.R.O., Svendsen J.S., Svenson J. Unnatural amino acid side chains as S1, S1′, and S2′ probes yield cationic antimicrobial peptides with stability toward chymotryptic degradation. J. Med. Chem. 2010;53:5558–5566. doi: 10.1021/jm1006337. [DOI] [PubMed] [Google Scholar]
- 15.Yeaman M.R., Yount N.Y. Mechanisms of Antimicrobial Peptide Action and Resistance. Pharmacol. Rev. 2003;55:27–55. doi: 10.1124/pr.55.1.2. [DOI] [PubMed] [Google Scholar]
- 16.Bechinger B., Gorr S.-U. Antimicrobial Peptides: Mechanisms of Action and Resistance. J. Dent. Res. 2016;96:254–260. doi: 10.1177/0022034516679973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arias M., Piga K.B., Hyndman M.E., Vogel H.J. Improving the Activity of Trp-Rich Antimicrobial Peptides by Arg/Lys Substitutions and Changing the Length of Cationic Residues. Biomolecules. 2018;8:19. doi: 10.3390/biom8020019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bartoloni M., Jin X., Marcaida M.J., Banha J., Dibonaventura I., Bongoni S., Bartho K., Gräbner O., Sefkow M., Darbre T., et al. Bridged bicyclic peptides as potential drug scaffolds: Synthesis, structure, protein binding and stability. Chem. Sci. 2015;6:5473–5490. doi: 10.1039/C5SC01699A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheng R.P., Gellman S.H., DeGrado W.F. β-Peptides: From Structure to Function. Chem. Rev. 2001;101:3219–3232. doi: 10.1021/cr000045i. [DOI] [PubMed] [Google Scholar]
- 20.Chatterjee J., Gilon C., Hoffman A., Kessler H. N-Methylation of Peptides: A New Perspective in Medicinal Chemistry. Acc. Chem. Res. 2008;41:1331–1342. doi: 10.1021/ar8000603. [DOI] [PubMed] [Google Scholar]
- 21.Räder A.F., Reichart F., Weinmüller M., Kessler H. Improving oral bioavailability of cyclic peptides by N-methylation. Bioorg. Med. Chem. 2018;26:2766–2773. doi: 10.1016/j.bmc.2017.08.031. [DOI] [PubMed] [Google Scholar]
- 22.Chen Y.-F., Lai Y.-D., Chang C.-H., Tsai Y.-C., Tang C.-C., Jan J.-S. Star-shaped polypeptides exhibit potent antibacterial activities. Nanoscale. 2019;11:11696–11708. doi: 10.1039/C9NR02012H. [DOI] [PubMed] [Google Scholar]
- 23.Qu S., Liu Y., Hu Q., Han Y., Hao Z., Shen J., Zhu K. Programmable antibiotic delivery to combat methicillin-resistant Staphylococcus aureus through precision therapy. J. Control. Release. 2020;321:710–717. doi: 10.1016/j.jconrel.2020.02.048. [DOI] [PubMed] [Google Scholar]
- 24.Lu J., Xu H., Xia J., Ma J., Xu J., Li Y., Feng J. d- and Unnatural Amino Acid Substituted Antimicrobial Peptides with Improved Proteolytic Resistance and Their Proteolytic Degradation Characteristics. Front. Microbiol. 2020;11:563030. doi: 10.3389/fmicb.2020.563030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hamase K., Morikawa A., Etoh S., Tojo Y., Miyoshi Y., Zaitsu K. Analysis of Small Amounts of D-Amino Acids and the Study of Their Physiological Functions in Mammals. Anal. Sci. 2009;25:961–968. doi: 10.2116/analsci.25.961. [DOI] [PubMed] [Google Scholar]
- 26.Narancic T., Almahboub S.A., O’Connor K.E. Unnatural amino acids: Production and biotechnological potential. World J. Microbiol. Biotechnol. 2019;35:67. doi: 10.1007/s11274-019-2642-9. [DOI] [PubMed] [Google Scholar]
- 27.Pless S.A., Ahern C.A. Unnatural Amino Acids as Probes of Ligand-Receptor Interactions and Their Conformational Consequences. Annu. Rev. Pharmacol. Toxicol. 2013;53:211–229. doi: 10.1146/annurev-pharmtox-011112-140343. [DOI] [PubMed] [Google Scholar]
- 28.Stromstedt A.A., Pasupuleti M., Schmidtchen A., Malmsten M. Evaluation of Strategies for Improving Proteolytic Resistance of Antimicrobial Peptides by Using Variants of EFK17, an Internal Segment of LL-37. Antimicrob. Agents Chemother. 2009;53:593–602. doi: 10.1128/AAC.00477-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu L., Liu J., Cui Q., Jia B.-Y., Pei Z.-H., Odah K.A., Wang Y.-M., Dong W.-L., Kong L.-C., Ma H.-X. Design and Characterization of a Novel Hybrid Antimicrobial Peptide OM19R Based on Oncocin and MDAP-2. Int. J. Pept. Res. Ther. 2019;26:1839–1846. doi: 10.1007/s10989-019-09984-3. [DOI] [Google Scholar]
- 30.Jia B.-Y., Wang Y.-M., Zhang Y., Wang Z., Wang X., Muhammad I., Kong L.-C., Pei Z.-H., Ma H.-X., Jiang X.-Y., et al. High Cell Selectivity and Bactericidal Mechanism of Symmetric Peptides Centered on d-Pro–Gly Pairs. Int. J. Mol. Sci. 2020;21:1140. doi: 10.3390/ijms21031140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guo F., Liang Q., Zhang M., Chen W., Chen H., Yun Y., Zhong Q., Chen W. Antibacterial Activity and Mechanism of Linalool against Shewanella putrefaciens. Molecules. 2021;26:245. doi: 10.3390/molecules26010245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abad J.P. Proton Motive Force. Proton Motive Force; Heidelberg, Germany: 2015. [Google Scholar]
- 33.Wang G., Li X., Wang Z. APD2: The updated antimicrobial peptide database and its application in peptide design. Nucleic Acids Res. 2008;37:D933–D937. doi: 10.1093/nar/gkn823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kang X., Dong F., Shi C., Liu S., Sun J., Chen J., Li H., Xu H., Lao X., Zheng H. DRAMP 2.0, an updated data repository of antimicrobial peptides. Sci. Data. 2019;6:148. doi: 10.1038/s41597-019-0154-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Molchanova N., Hansen P.R., Franzyk H. Advances in Development of Antimicrobial Peptidomimetics as Potential Drugs. Molecules. 2017;22:1430. doi: 10.3390/molecules22091430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Svenson J., Stensen W., Brandsdal B.-O., Haug B.E., Monrad J., Svendsen J.S. Antimicrobial Peptides with Stability toward Tryptic Degradation. Biochemistry. 2008;47:3777–3788. doi: 10.1021/bi7019904. [DOI] [PubMed] [Google Scholar]
- 37.Moreton R.C. United States pharmacopeia-national formulary. J. Excip. Food Chem. 2016;6:925. [Google Scholar]
- 38.Rabanal F., Cajal Y. Recent advances and perspectives in the design and development of polymyxins. Nat. Prod. Rep. 2017;34:886–908. doi: 10.1039/C7NP00023E. [DOI] [PubMed] [Google Scholar]
- 39.Baltz R.H., Miao V., Wrigley S.K. Natural products to drugs: Daptomycin and related lipopeptide antibiotics. Nat. Prod. Rep. 2005;22:717–741. doi: 10.1039/b416648p. [DOI] [PubMed] [Google Scholar]
- 40.Lai Z., Tan P., Zhu Y., Shao C., Shan A., Li L. Highly Stabilized alpha-Helical Coiled Coils Kill Gram-Negative Bacteria by Multicomplementary Mechanisms under Acidic Condition. ACS Appl. Mater. Interfaces. 2019;11:22113–22128. doi: 10.1021/acsami.9b04654. [DOI] [PubMed] [Google Scholar]
- 41.Pasupuleti M., Schmidtchen A., Malmsten M. Antimicrobial peptides: Key components of the innate immune system. Crit. Rev. Biotechnol. 2011;32:143–171. doi: 10.3109/07388551.2011.594423. [DOI] [PubMed] [Google Scholar]
- 42.Askoura M., Mottawea W., Abujamel T., Taher I. Efflux pump inhibitors (EPIs) as new antimicrobial agents against Pseudomonas aeruginosa. Libyan J. Med. 2011;6 doi: 10.3402/ljm.v6i0.5870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Performance Standards for Antimicrobial Susceptibility Testing. Clinical and Laboratory Standards Institute; Wayne, PA, USA: 2015. [Google Scholar]
- 44.Wang J., Chou S., Xu L., Zhu X., Dong N., Shan A., Chen Z. High specific selectivity and Membrane-Active Mechanism of the synthetic centrosymmetric α-helical peptides with Gly-Gly pairs. Sci. Rep. 2015;5:15963. doi: 10.1038/srep15963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shao C., Li W., Tan P., Shan A., Dou X., Ma D., Liu C. Symmetrical Modification of Minimized Dermaseptins to Extend the Spectrum of Antimicrobials with Endotoxin Neutralization Potency. Int. J. Mol. Sci. 2019;20:1417. doi: 10.3390/ijms20061417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dong N., Zhu X., Chou S., Shan A., Li W., Jiang J. Antimicrobial potency and selectivity of simplified symmetric-end peptides. Biomaterials. 2014;35:8028–8039. doi: 10.1016/j.biomaterials.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 47.Liu Y., Jia Y., Yang K., Li R., Xiao X., Zhu K., Wang Z. Metformin Restores Tetracyclines Susceptibility against Multidrug Resistant Bacteria. Adv. Sci. 2020;7:1902227. doi: 10.1002/advs.201902227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ding X., Yang C., Moreira W., Yuan P., Periaswamy B., De Sessions P.F., Zhao H., Tan J., Lee A., Ong K.X., et al. A Macromolecule Reversing Antibiotic Resistance Phenotype and Repurposing Drugs as Potent Antibiotics. Adv. Sci. 2020;7:2001374. doi: 10.1002/advs.202001374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maisuria V.B., Hosseinidoust Z., Tufenkji N. Polyphenolic extract from maple syrup potentiates antibiotic susceptibility and reduces biofilm formation of pathogenic bacteria. Appl. Environ. Microbiol. 2015;81:3782–3792. doi: 10.1128/AEM.00239-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Greco I., Hansen J.E., Jana B., Molchanova N., Oddo A., Thulstrup P.W., Damborg P., Guardabassi L., Hansen P.R. Structure–Activity Study, Characterization, and Mechanism of Action of an Antimicrobial Peptoid D2 and Its d- and l-Peptide Analogues. Molecules. 2019;24:1121. doi: 10.3390/molecules24061121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Song M., Liu Y., Huang X., Ding S., Wang Y., Shen J., Zhu K. A broad-spectrum antibiotic adjuvant reverses multidrug-resistant Gram-negative pathogens. Nat. Microbiol. 2020;5:1040–1050. doi: 10.1038/s41564-020-0723-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.