Abstract
Foodborne pathogens significantly impact public health globally. Excessive antimicrobial use plays a significant role in the development of the public health crisis of antibiotic resistance. Here, we determined the prevalence and antimicrobial resistance profiles of E. coli O157, Salmonella, L. monocytogenes, and Campylobacter isolated between 2016 and 2020 from small scale agricultural settings that were amended with dairy cattle or poultry manure in Northeastern Ohio. The total prevalence of the foodborne pathogens was 19.3%: Campylobacter 8%, Listeria monocytogenes 7.9%, Escherichia coli O157 1.8%, and Salmonella 1.5%. The prevalence was significantly higher in dairy cattle (87.7%) compared to poultry (12.2%) manure amended farms. Furthermore, the prevalence was higher in manure samples (84%) compared to soil samples (15.9%; p < 0.05). Multiple drug resistance was observed in 73%, 77%, 100%, and 57.3% of E. coli O157, Salmonella, L. monocytogenes, and Campylobacter isolates recovered, respectively. The most frequently observed resistance genes were mphA, aadA, and aphA1 in E. coli O157; blaTEM, tet(B), and strA in Salmonella; penA, ampC, lde, ermB, tet(O), and aadB in L. monocytogenes and blaOXA-61, tet(O), and aadE in Campylobacter. Our results highlight the critical need to address the dissemination of foodborne pathogens and antibiotic resistance in agricultural settings.
Keywords: Campylobacter, Salmonella, L. monocytogenes, E. coli O157, foodborne pathogens, antimicrobial resistance, diversity, phenotypic and genotypic, correlation
1. Introduction
Foodborne illnesses have a major public health impact in the USA and around the world. They affect approximately one in six Americans annually, leading to approximately 128,000 hospitalizations and 3000 deaths [1]. The most common foodborne pathogens include Campylobacter, Salmonella, Escherichia coli O157, Listeria monocytogenes, and Clostridium perfringens [2,3]. Campylobacter, Salmonella, E. coli O157, and L. monocytogenes result in about 37,000 hospitalizations and 750 deaths annually [1]. In the USA, the estimated annual cost of foodborne illness is about $90 billion per year [3,4]. For example, Salmonella spp. alone results in about one million infections and 400 deaths annually with a total of $4.4 billion in medical costs and lost productivity [3,5,6]. However, L. monocytogens infection causes the highest mortality rate compared to other foodborne pathogens with a case fatality rate of 20–30% [6] with an annual cost of about $2.6 billion [6,7]. Furthermore, E. coli O157:H7 causes approximately 265,000 illnesses with an estimated cost of $405 million [8,9]. Campylobacter infections cause 1.3 million illnesses, 13,240 hospitalizations with an estimated cost of $1.56 billion per year [5]. Foodborne pathogens cause a self-limiting gastroenteritis and do not require the use of antimicrobials, except in severe cases such as persistent enteritis, bacteremia, and in immunocompromised individuals [1,10]. However, in severe cases, antimicrobial treatments have become limited due to the rise of antimicrobial resistance among these foodborne pathogens, which complicate their treatment and have become a significant public health concern [2,11]. In the USA between 2009–2015, 5760 foodborne outbreaks were reported and the implicated bacteria were resistant to at least one antibiotic [12,13]. Out of these outbreaks, 896 were caused by Salmonella, 191 caused by Shiga toxin producing E. coli (STEC), 155 caused by Campylobacter, and 35 caused by L. monocytogenes [13]. Most of these outbreaks were associated with dairy and poultry products and vegetables [12,13]. Therefore, the USA Centers for Disease Control and Prevention (CDC) has classified drug-resistant Campylobacter and drug-resistant non-typhoidal Salmonella as serious threats to human health [2,14].
The number of antibiotic resistant bacteria isolated from humans, animals, and the environment has increased globally over the last two decades due to the overuse and misuse of antibiotics [15,16]. Currently, antibiotic resistance infections result in about 700,000 deaths worldwide. However, by 2050, if no action is taken to reduce the spread of antimicrobial resistance, the estimated number of deaths will increase by up to 10 million with more than USA $100 trillion in economic losses [17]. In the USA alone, more than 2 million infections occur due to antibiotic resistant bacteria with $20 billion in economic losses each year [17,18]. Antibiotics are often used in food animals as therapeutics or prophylactics, especially in poultry [19,20,21,22,23,24]. Additionally, several environmental components such as soil and water can act as reservoirs of antimicrobial resistance genes (ARGs) [25,26,27]. The agricultural soil may naturally contain pathogenic bacteria or receive them during soil amendment using animal manure [28]. Soil amendment with animal manure increases crop yield, but it can potentially increase the spread of foodborne pathogens and ARGs in the environment [29,30,31]. Therefore, the USA Food and Drug Administration has recently limited the use of antimicrobials on farms in the USA due to the growing impact of antibiotic resistance in clinical practice and to reduce the selection pressure on the emergence of resistance bacteria [32,33]. Here, we investigated the prevalence and phenotypic and genotypic antimicrobial resistance profiles of E. coli O157, Salmonella, L. monocytogenes, and Campylobacter isolated between 2016 and 2020 from small scale agricultural settings that were amended with dairy or poultry manure in Northeastern Ohio, USA.
2. Material and Methods
2.1. Study Area and Sample Collection
A total of 844 longitudinal manure and soil samples were collected monthly between October 2016 and October 2020 from eleven farms; five farms were amended with poultry manure, three farms were amended with dairy manure, and three farms were amended with both dairy and poultry manure. Samples were collected from a small-scale agricultural setting located in Northeastern Ohio (USA). Farms were selected based on their availability for longitudinal sampling throughout the study period and the types of manure applied (poultry or dairy). The dairy manure was obtained from open dairy heifers housed on a bedded pack during the winter and raised on pasture in the summer. The poultry manure came from boiler houses from various stages of composting. A total of 379 manure samples and 465 soil samples were collected. Six to eight fresh manure pats were collected from each farm and pooled to make one manure sample, whereas three manure samples were collected from poultry storage piles from each farm and pooled to make one manure sample. Similarly, soil samples were collected from three random sites per each field and pooled to make one soil sample. Samples were collected aseptically into Nasco Whirl-Pak™ (Fisher Scientific, Waltham, MA, USA) and stored in a cool box before transportation to the laboratory for further analyses. The numbers of the collected samples each year, their source, and the amendment type are listed in Table 1.
Table 1.
Total number of samples collected between October 2016 and October 2020.
| Year | No. of Farms | Total No of Samples | Farm Amendment Type | Sample Type | ||
|---|---|---|---|---|---|---|
| Dairy Manure | Poultry Manure | Manure | Soil | |||
| 2016/2017 | 11 | 252 | 131 | 121 | 112 | 140 |
| 2018 | 11 | 262 | 142 | 120 | 120 | 142 |
| 2019 | 11 | 221 | 113 | 108 | 95 | 126 |
| 2020 | 10 | 109 | 55 | 54 | 52 | 57 |
| Total | 844 | 441 | 403 | 379 | 465 | |
2.2. Bacterial Isolation, Enrichment, and Growth Conditions
Twenty-five grams of the samples (soil or manure) were suspended in 225 mL phosphate buffered saline (PBS; Fisher Scientific, Waltham, MA, USA) and mixed by shaking to form slurry. One mL of the slurry was then ten-fold serially diluted using PBS, and 100 µL of each serial dilution was plated onto modified charcoal cefoperazone deoxycholate agar (mCCDA; Fisher Scientific, Waltham, MA, USA), RAPID’ L. mono (Bio Rad, Hercules, CA, USA), sorbitol MacConkey agar plates containing cefixine and tellurite (SMACct; Becton, Dickinson; BD, San Jose, CA, USA), and xylose lysine tergitol-4 agar (XLT-4; Bio Rad, Hercules, CA, USA) plates for the direct isolation of Campylobacter, L. monocytogenes, E. coli O157, and Salmonella, respectively. Samples that did not show any bacterial growth due to direct plating were subjected to enrichment using pathogen specific media [34,35,36,37,38].
The enrichment and isolation of E. coli O157 was performed as previously described [34]. Briefly, 10 mL of the slurry was enriched in 90 mL of buffered peptone water (BPW; Thermo Scientific, Waltham, MA, USA) and incubated at 42 °C for 18–24 h. Automated immunomagnetic separation (AIMS) was performed on BPW enriched samples using anti-O157 specific immunomagnetic beads (Invitrogen, Waltham, MA, USA). The recovered beads were plated on SMACct and incubated at 37 °C for 18–24 h. An E. coli O157 Latex test (Oxoid Ltd., Cambridge, UK) was used to confirm the presence of E. coli O157. The isolated colonies were preserved at −80 °C in brain heart infusion broth (BHI; BD Difco, Franklin Lakes, NJ, USA) containing 30% glycerol (v/v) for further analysis.
To isolate L. monocytogenes, 10 mL of the slurry was added to 90 mL universal pre-enrichment broth (UPB; Oxoid Ltd., Cambridge, UK) and incubated at 35 °C for 24 h. One mL of the enriched culture was transferred to nine mL of Fraser broth (Oxoid Ltd., Cambridge, UK) and incubated at 35 °C for 24 h. A loopful (10 µL) of darkened Fraser broth was streaked on PALCAM plates (Neogen, Lansing, MI, USA) and incubated at 35 °C for 48 h. The grown colonies were subcultured on Rapid’ L. mono plate (Neogen, Lansing, MI, USA) and incubated at 37 °C for 24 h. Blue colonies without a yellow halo were collected and preserved at −80 °C in BHI broth containing 30% glycerol (v/v) for further analysis [35,36].
For Salmonella isolation, 10 mL of the slurry was enriched in 90 mL tetrathionate broth (Oxoid Ltd., Cambridge, UK) and incubated at 37 °C for 24 h. One mL of the grown culture was then transferred to ten mL of Rappaport-Vassiliadis broth (RV; Oxoid Ltd., Cambridge, UK) and incubated at 37 °C for 24 h. A loopful of bacteria grown in RV were plated on XLT-4 plates and incubated at 37 °C for 24 h [37]. Black suspect colonies were preserved at −80 °C in BHI broth containing 30% glycerol (v/v) for further analysis.
To isolate Campylobacter spp., 1 mL of manure or soil slurry was suspended in 9 mL of Preston enrichment broth containing Campylobacter growth supplements (CM067, SR048, SR117, and SR232; Oxoid Ltd., Cambridge, UK) [38]. The suspensions were incubated under microaerobic conditions (5% O2, 10% CO2, and 85% N2) at 42 °C for 48 h. After enrichment, 100 µL was plated on mCCDA containing a selective supplement (SR0155; Oxoid Ltd., Cambridge, UK) and incubated under microaerobic conditions at 42 °C for 48 h. The isolated colonies from mCCDA plate were then subcultured onto Mueller–Hinton (MH) agar containing a selective supplement (SR0117; Oxoid Ltd., Cambridge, UK) and incubated at 42 °C for 48 h under microaerobic conditions [39,40,41]. The grown Campylobacter cultures were frozen at −80 °C in MH broth supplemented with 30% glycerol (v/v) until further use.
2.3. DNA Extraction and PCR Analysis for Bacterial Species Identification
Bacterial DNA was extracted using the boiling method [39]. Briefly, a half loop from bacterial cultures were suspended in 100 μL of sterile DNase-free water, heated at 95 °C for 10 min, cooled, and centrifuged at 4000× g for 10 min. The supernatants containing the nucleic acids were collected in new tubes and stored at −20 °C. In cases where no PCR products were detected, template DNA was prepared using a MasterPure™ Complete DNA and RNA Purification Kit (Epicenter, Madison, WI, USA) according to the manufacturer’s instructions. Confirmation of bacterial spp. for E. coli O157, Salmonella L. monocytogenes, and Campylobacter was performed using a multiplex-PCR assay (mPCR) [39,42,43,44]. PCR products were visualized using gel-electrophoresis on a 2% agarose gel containing 0.5 µg/mL ethidium bromide. The mPCR conditions, target gene, primers sequence, and amplicon size are listed in Table S1.
2.4. Antimicrobial Susceptibility Testing
Antimicrobial susceptibility testing was carried out using the broth microdilution method [45]. E. coli O157 and Salmonella isolates were tested for their susceptibility to aminoglycosides: kanamycin (Kan), streptomycin (Str), and gentamicin (Gen); β–lactam combination agents: amoxicillin-clavulanic acid (Amo); cephems: cefoxitin (Cefo) and ceftriaxone (Ceft); folate pathway antagonists: sulfisoxazole (Sul) and trimethoprim- sulfamethoxazole (Tri); macrolides: azithromycin (Azi); penems: meropenem (Mer); penicillin: ampicillin (Amp); phenicols: chloramphenicol (Chl); quinolones: ciprofloxacin (Cip), nalidixic acid (Nal); tetracyclines: tetracycline (Tet); and polymyxins: colistin (Col) (Sigma-Aldrich, St. Louis, MO, USA). L. monocytogenes isolates were screened for susceptibility to aminoglycosides: kanamycin (Kan), streptomycin (Str), and gentamicin (Gen); cephems: cefoxitin (Cefo) and ceftriaxone (Ceft); folate pathway antagonists: trimethoprim- sulfamethoxazole (Tri); macrolides: azithromycin (Azi), erythromycin (Ery); penems: meropenem (Mer); penicillin: ampicillin (Amp) and penicillin G (Pen); rifamycins: rifampicin (Rif); phenicols: chloramphenicol (Chl); quinolones: ciprofloxacin (Cip); nalidixic acid (Nal); tetracyclines: tetracycline (Tet); nitrofuran: nitrofurantoin (Nit); oxazolidinone: linezolid (Lin); fluoroquinolone: levofloxacin (Lev); lincomycin: clindamycin (Clin); and glycopeptide: vancomycin (Van). Campylobacter isolates were screened for susceptibility to aminoglycosides: kanamycin (Kan), streptomycin (Str), and gentamicin (Gen); macrolides: azithromycin (Azi) and erythromycin (Ery); penicillin: ampicillin (Amp) and penicillin G (Pen); phenicols: chloramphenicol (Chl); quinolones: ciprofloxacin (Cip), nalidixic acid (Nal); tetracyclines: tetracycline (Tet); lincomycin: clindamycin (Cli); ketolides: telithromycin (Tel); and amphenicol: florfenicol (Flo). These antimicrobials are representatives of the drugs used for humans and in the animal industry and were chosen according to the National Antimicrobial Resistance Monitoring System (NARMS) records [46].
Briefly, E. coli O157 and Salmonella cultures were suspended in LB (Luria-Bertani) broth while L. monocytogenes cultures were suspended in UPB broth and Campylobacter cultures were suspended in MH broth to achieve an optical density (OD600) of 0.05. One hundred microliters of a suspension were added to each well of the 96-well plate containing two-fold serial dilutions of the antimicrobial agents. Positive and negative control wells contained bacterial suspensions without antimicrobials, and sterile broth containing each of the antimicrobials, respectively. The E. coli O157 and Salmonella plates were then incubated under aerobic conditions at 37 °C for 24 h [47], while the plates for L. monocytogenes were incubated for 48 h under aerobic condition at 37 °C and the Campylobacter plates were incubated at 42 °C under microaerobic condition [47,48,49]. The plates were assessed visually and using a spectrophotometer (Tecan Group Ltd., San Jose, CA, USA) to determine growth inhibition. Minimum inhibitory concentration (MIC) values were defined as the lowest concentration of an antimicrobial agent that produced no visible growth. The isolates that possessed resistance to three or more classes of antimicrobials were considered multi-drug resistant (MDR) [50]. The test was performed in accordance with the recommendations of the Clinical Laboratory Standards Institute (CLSI) criteria by using the available CLSI breakpoint interpretive criteria (Table S2).
2.5. Detection of Antibiotic Resistance Genes (ARGs)
The most prevalent ARGs for each antibiotic were selected and screened for their presence in the foodborne pathogens. E. coli O157 isolates were screened for six ARGs: tetracycline resistant gene tet(A), aminoglycoside acetyltransferase gene aac (3)-IV, streptomycin adenyl transferase gene aadA, aminoglycoside 3′-phosphotransferase gene aphA1, sulfonamide resistant gene sulII, and macrolide resistance gene mphA. Salmonella isolates were screened for six ARGs: beta-lactamase gene bla TEM, streptomycin resistant gene strA, tetracycline resistant gene tet (B), aminoglycoside acetyltransferase gene aac (3)-Iva, sulfonamide resistant gene sulII, and macrolide resistant gene mphA. L. monocytogenes isolates were screened for nine ARGs: ampicillin resistant gene ampC, ciprofloxacin resistant gene lde, erythromycin resistant gene ermB, tetracycline resistant gene tet(O), gentamicin resistant gene aadB, penicillin G resistant gene penA, cefoxitin resistance gene cfxA, macrolide resistant gene mefA, and sulfonamide resistant gene sulI. Campylobacter isolates were screened for ampicillin resistant gene blaOXA-61, streptomycin resistant gene aadE, tetracycline resistant gene tet(O), and gentamycin resistant gene aph-3-1. Amplification was performed in a 25 µL reaction volume containing 12.5 µL of GoTaq Green Master Mix (2X; 2.5 units) (Promega, Madison, WI, USA), 3 µL of template DNA, and 0.2 µM of each forward and reverse primer. The PCR was performed with 3 min of initial denaturation at 95 °C, followed by 35 cycles of denaturation at 95 °C for 1 min, annealing at optimal annealing temperature for 45 s, extension at 72 °C for 1 min, and final extension at 72 °C for 5 min. Primer sequence, expected amplicon size, and annealing temperatures are described in Table S3. Nuclease-free water was used as a negative control. PCR products were visualized on a 2% agarose gel containing 0.5 µg/mL ethidium bromide under UV light.
2.6. Statistical Analysis
Statistical analysis was performed in IBM SPSS 26.0 using one-way analysis of variance (ANOVA) followed by Tukey’s posttest. Student’s t-test and the chi-square test were used for pairwise comparisons of differences in the resistance rates for each antimicrobial agent between poultry and dairy manure amended farms. A p-value of <0.05 was considered statistically significant difference. Linear regression analysis was used for the trend analysis of the prevalence and antimicrobial resistance during the study period. JMP Pro 15 was used to plot heatmap representation with dendrogram of each foodborne pathogen. Principal component analysis (PCA) was used to visualize the distribution of phenotypic and genotypic resistance. The statistical analysis of the correlation between phenotypic and genotypic resistance was performed in the vegan package on R studio (SAS institute Inc., Cary, NC, USA).
3. Results
3.1. Prevalence and Distribution of Campylobacter, E. coli O157, Salmonella, and L. monocytogenes
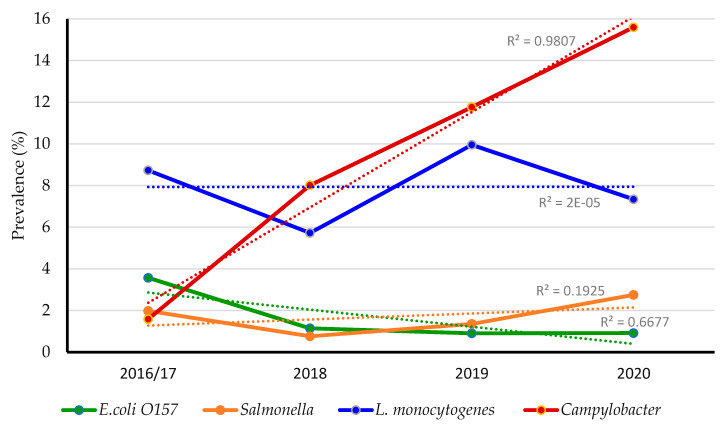
A total of 19.3% (163 of 844) of the collected samples were positive for at least one of the foodborne pathogens. Out of these, 82.2% (134 of 163) of samples had a single pathogen, while 17.8% (29 of 163) of samples had more than one pathogen. Notably, the total prevalence of Campylobacter (8%; 68 of 844) and L. monocytogenes (7.9%; 67 of 844) was higher than E. coli O157 (1.8%; 15 of 844) and Salmonella (1.5%; 13 of 844). However, 93.1% (27 of 29 isolates) had contamination with different pathogens; L. monocytogenes + Campylobacter (44.8%; 13 of 29 isolates), Campylobacter + Salmonella (13.7%; 4 of 29 isolates), Campylobacter + E. coli O157 (10.3%; 3 of 29 isolates), L. monocytogenes + Salmonella (6.8%; 2 of 29 isolates), L. monocytogenes + E. coli O157 (13.7%; 4 of 29 isolates), E. coli O157 + Salmonella (3.4%; 1 of 29 isolates), and Salmonella + L. monocytogenes + Campylobacter (6.8%; 2 of 29 isolates). None of the samples were positive for all four tested foodborne pathogens. Notably, the prevalence of the foodborne pathogens had not increased from 2016 to 2020 except for Campylobacter, which showed significant increase throughout the study years (R of 0.9; p < 0.05; Figure 1). This increase might be attributed to climate changes, rainfall, and soil properties [51,52,53]. Moreover, out of 67 L. monocytogenes isolates recovered, 5.9% (4 of 67) were 1/2a serotype, 26.8% (18 of 67) were 1/2b serotype, 19.4% (13 of 67) were 4b serotype, and the remaining 47.7% (32 of 67) were untypable.
Figure 1.
Prevalence of foodborne pathogens in dairy cattle and poultry manure amended farms between 2016 and 2020.
Notably, the prevalence of the foodborne pathogens was significantly higher in dairy cattle (87.7%) compared to poultry manure amended farms (12.3%; p < 0.05; Table 2). In dairy cattle manure amended farms, Campylobacter (14.9%) was the most prevalent compared to L. monocytogenes (13.1%), E. coli O157 (2.9%), and Salmonella (1.4%), whereas in poultry manure amended farms, L. monocytogenes (2.2%) was the most prevalent compared to Salmonella (1.7%) and Campylobacter and E. coli O157 (0.5%). The prevalence of all pathogens was higher in manure samples (84%) compared to soil samples (15.9%; p < 0.05; Table 2). In manure samples, Campylobacter (17.7%) was the most prevalent than L. monocytogenes (12.4%), Salmonella (3.2%), and E. coli O157 (2.9%), whereas in soil samples, L. monocytogenes (4.3%) was the most observed foodborne pathogen followed by E. coli O157 (0.9%), Salmonella, and Campylobacter (0.2%; Table 2).
Table 2.
Prevalence of foodborne pathogen from different sources.
| Pathogen | No. of Isolates |
Farm Amendment Type | Sample Type | ||
|---|---|---|---|---|---|
| Dairy Manure (n = 441) No. (%) |
Poultry Manure (n = 403) No. (%) |
Manure (n = 379) No. (%) | Soil (n = 465) No. (%) |
||
| E. coli O157 | 15 | 13 (2.9) | 2 (0.5) | 11 (2.9) | 4 (0.9) |
| Salmonella | 13 | 6 (1.4) | 7 (1.7) | 12 (3.2) | 1 (0.2) |
| L. monocytogenes | 67 | 58 (13.1) | 9 (2.2) | 47(12.4) | 20 (4.3) |
| Campylobacter | 68 | 66 (14.9) | 2 (0.5) | 67 (17.7) | 1 (0.2) |
| Total positive | 163 | 143 (87.7) * | 20 (12.2) | 137 (84) * | 26 (15.9) |
* Significantly higher prevalence of foodborne pathogens (p < 0.05).
3.2. Antimicrobial Resistance Phenotypical Profile of E. coli O157, Salmonella, L. monocytogenes and Campylobacter
All isolates that were confirmed positive for E. coli O157, Salmonella, L. monocytogenes, and Campylobacter were used for broth microdilution assay. Collectively, all the tested isolates possessed resistance to at least one of the tested antimicrobials. E. coli O157, Salmonella, and L. monocytogenes isolates possessed 100% resistance to Tri. Additionally, E. coli O157 and Salmonella isolates showed 100% resistance to Sul, whereas Campylobacter isolates possessed 100% resistance to Pen. Interestingly, most of the E. coli O157 isolates were susceptible to the tested antibiotics, however, most of the L. monocytogenes isolates were resistant to the tested antibiotics. For example, E. coli O157 isolates possessed 100% susceptibility to Chl, Cip, Nal, Ceft, and Mer, while L. monocytogenes isolates possessed 100% resistant to Ceft, Cefo, Cli, Rif, Tri, Mer, and Azi. The antimicrobial resistance profile of different foodborne pathogens is shown in Table 3.
Table 3.
Antimicrobial resistance profile of the foodborne pathogens.
| Antimicrobial Class | Antimicrobials | E. coli O157 (n = 15) | Salmonella (n = 13) | L. monocytogenes (n = 67) | Campylobacter (n = 68) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| R No. (%) |
I No. (%) |
S No. (%) |
R No. (%) |
I No. (%) |
S No. (%) |
R No. (%) |
I No. (%) |
S No. (%) |
R No. (%) |
I No. (%) |
S No. (%) |
||
| Penicillin | Amp | 6 (40) | 0 | 9 (60) | 6 (46) | 2 (15.3) | 5 (38.4) | 60 (89.5) | 0 | 7 (10.5) | 63 (97) | 0 | 5 (3) |
| Pen | NA | NA | NA | NA | NA | NA | 32 (47.7) | 0 | 35 (52.3) | 68 (100) | 0 | 0 | |
| Phenicol | Chl | 0 | 0 | 15 (100) | 1 (7.8) | 0 | 12 (92.2) | 42 (61.7) | 8 (11.7) | 17 (25) | 6 (5.9) | 0 | 62 (94.1) |
| Flo | NA | NA | NA | NA | NA | NA | NA | NA | NA | 2 (2.9) | 0 | 66 (97) | |
| Quinolones | Cip | 0 | 5 (33.3) | 10 (66.6) | 1 (7.6) | 7(46) | 5 (38.4) | 53 (79) | 1 (1.4) | 13 (19.6) | 4 (5.8) | 0 | 64 (94.2) |
| Nal | 0 | 0 | 15 (100) | 7 (53.8) | 1(7.8) | 5 (38.4) | 64 (95.5) | 0 | 3 (4.5) | 23 (33.8) | 0 | 45 (66.2) | |
| Aminoglycosides | Kan | 9 (60) | 1 (6.7) | 5 (33.3) | 6 (46) | 2 (15.3) | 5 (38.4) | 59 (88) | 0 | 8 (12) | 33 (48.5) | 4 (5.8) | 31 (45.5) |
| Gen | 13 (86.6) | 1 (6.7) | 1 (6.7) | 10 (76.9) | 1 (7.8) | 2 (15.3) | 52 (77.6) | 0 | 15 (22.3) | 17 (25) | 0 | 51 (75) | |
| Str | 8 (54) | 0 | 7 (46) | 11 (84.6) | 0 | 2 (15.3) | 66 (98.5) | 1 (1.5) | 0 | 15 (22) | 12 (17.6) | 41 (60.2) | |
| Tetracyclines | Tet | 2 (23) | 2 (13) | 11 (73.3) | 3 (23) | 1 (7.8) | 9 (69.2) | 23 (34.3) | 0 | 44 (65.6) | 43 (63.2) | 0 | 25 (36.8) |
| Macrolides | Ery | NA | NA | NA | NA | NA | NA | 25 (37.3) | 0 | 42 (62.7) | 67 (98.5) | 0 | 1 (1.5) |
| Azi | 11(73.3) | 0 | 4 (26.6) | 10 (77) | 0 | 3 (23) | 67 (100) | 0 | 0 | 6 (5.9) | 0 | 62 (94.1) | |
| Cephems | Ceft | 0 | 0 | 15 (100) | 1 (7.8) | 1 (7.8) | 11 (84.6) | 67 (100) | 0 | 0 | NA | NA | NA |
| Cefo | 1 (6.7) | 0 | 14 (93.1) | 1 (7.8) | 1 (7.8) | 11 (84.6) | 67 (100) | 0 | 0 | NA | NA | NA | |
| β–lactam | Amo | 1 (6.7) | 0 | 14 (93.1) | 1 (7.8) | 0 | 12 (92.2) | NA | NA | NA | NA | NA | NA |
| Folate pathway Antagonists |
Tri | 15 (100) | 0 | 0 | 13 (100) | 0 | 0 | 67 (100) | 0 | 0 | NA | NA | NA |
| Sul | 15 (100) | 0 | 0 | 13 (100) | 0 | 0 | NA | NA | NA | NA | NA | NA | |
| Polymyxins | Col | 1 (6.7) | 0 | 14 (93.1) | 2 (15.3) | 0 | 11 (84.6) | NA | NA | NA | NA | NA | NA |
| Penem | Mer | 0 | 0 | 15 (100) | 0 | 0 | 13 (100) | 67 (100) | 0 | 0 | NA | NA | NA |
| Glycopeptide | Van | NA | NA | NA | NA | NA | NA | 45 (67) | 22 (33) | 0 | NA | NA | NA |
| Oxazolidinone | Lin | NA | NA | NA | NA | NA | NA | 39 (58) | 28 (42) | 0 | NA | NA | NA |
| Nitrofuran | Nit | NA | NA | NA | NA | NA | NA | 6 (8.9) | 59 (86) | 2 (2.9) | NA | NA | NA |
| Lincomycin | Cli | NA | NA | NA | NA | NA | NA | 67 (100) | 0 | 0 | 8 (11.7) | 0 | 60 (80.8) |
| Rifamycins | Rif | NA | NA | NA | NA | NA | NA | 67 (100) | 0 | 0 | NA | NA | NA |
| Fluoroquinolone | Lev | NA | NA | NA | NA | NA | NA | 61 (91) | 6 (9) | 0 | NA | NA | NA |
| Ketolides | Tel | NA | NA | NA | NA | NA | NA | NA | NA | NA | 4 (5.8) | 0 | 64 (94.2) |
S, susceptible; I, intermediate; R, resistance; NA, not applicable. The antimicrobial resistance was determined using the broth microdilution method [45]. Results are shown as number of isolates with the percentage given in parentheses. Amp: ampicillin, Chl: chloramphenicol, Cip: ciprofloxacin, Kan: kanamycin, Nal: nalidixic acid, Str: streptomycin, Tet: tetracycline, Gen: gentamycin, Sul: sulfisoxazole, Ceft: ceftriaxone, Amo: amoxicillin, Cefo: cefoxitin, Azi: azithromycin, Tri: trimethoprim sulfamethoxazole, Mer: meropenem, Col: colistin, Pen: penicillin G, Ery: erythromycin, Van: vancomycin, Lin: linezolid, Nit: nitrofurantoin, Cli: clindamycin, Rif: rifampicin, Lev: levofloxacin, Tel: telithromycin, Flo: florfenicol.
Our results also showed that E. coli O157 isolates from poultry manure amended farms were 100% resistant to Amp, Str, Gen, Sul, and Tri, whereas in dairy cattle manure amended farms, 100% of E. coli O157 isolates were resistant to Sul and Tri (Table S4). Interestingly, 73% (11 of 15) of the E. coli O157 isolates revealed MDR to aminoglycosides (100%), macrolides (73%; 11 of 15), and folate antagonists (100%). Further, Salmonella isolates recovered from poultry manure amended farms possessed 100% resistant to Gen, Sul, Tri, and Azi, while isolates recovered from dairy cattle manure amended farms were 100% resistant to Sul and Tri (Table S5). A total of 77% (10 of 13) of the Salmonella isolates possessed MDR to aminoglycosides (84%; 11 of 13), macrolides (77%; 10 of 13), and folate antagonists (100%). Notably, all L. monocytogenes isolates recovered from poultry manure amended farms were resistant to Kan, Nal, and Lev. However, more than 50% of the L. monocytogenes isolates recovered from dairy cattle manure amended farms were resistant to Kan, Nal, and Lev (Table S6). We also found that all L. monocytogenes isolates showed MDR to cephems, lincomycin, antimycobacterial, folate antagonist, penem, and macrolides with 100% resistance for each antimicrobial class. Additionally, Campylobacter isolates recovered from poultry manure amended farms were 100% resistant to Amp, Nal, and Pen, while isolates recovered from dairy cattle manure amended farms were 100% resistant to Ery and Pen (Table S7). A total of 57.3% (39 of 68) of the Campylobacter isolates showed MDR to aminoglycosides (54%; 37 of 68), tetracyclines (63%; 43 of 68), and penicillin (100%). The resistance of the foodborne pathogens to the tested antimicrobials did not increase throughout the study period from 2016 to 2020 (Figure S1). On the other hand, manure samples contained more resistant isolates compared to soil samples. Details about the antimicrobial profile of the E. coli O157, Salmonella, L. monocytogenes, and Campylobacter isolates from manure and soil samples collected from dairy cattle and poultry manure amended farms are shown in Tables S4–S7.
3.3. Genotypic Profile of Antimicrobial Resistance in E. coli O157, Salmonella L. monocytogenes and Campylobacter Isolates
Phenotypically antimicrobial resistant isolates of different foodborne pathogens were tested for the presence of the corresponding ARGs. Regardless of the type of the isolated pathogen, more ARGs were detected in dairy cattle than poultry manure amended farms. Our results showed that the E. coli O157 isolates contained 100% of aphA1 and mph A genes. Further, aadA, tetA, aac (3)-IV, and Sul II genes were detected in 75% (6/8), 50% (1/2), 84% (11/13), and 63% (7/15) of the E. coli O157 isolates, respectively. Notably, none of the Salmonella isolates contained the Azi resistant ermB gene. However, 66% (4/6), 66% (2/3), 63% (7/11), 40% (4/10), and 15% (2/13) of Salmonella isolates contained blaTEM, tetB, strA, aac (3)-Iva, and sul II genes, respectively (Figure S2).
None of the L. monocytogenes isolates contained the cefoxitin resistance gene cfxA, macrolide resistance gene mefA, and sulfonamide resistance gene sulI. Regardless of the source of the isolates, the most frequently detected ARGs within L. monocytogenes isolates were lde 86.7% (46/53) followed by ampC 66% (40/60), aadB 51.9% (27/52), penA 50% (16/32), ermB 28% (7/25), and tet(O) 8% (2/23). However, the penA gene was detected only in dairy cattle manure amended farms. Notably, none of the Campylobacter isolates contained the gentamycin resistance aph-3-1 gene. However, Campylobacter isolates contained 90% (57/63), 79% (34/43), and 73% (11/15) of blaOXA-61, tet(O), and aadE genes, respectively. The prevalence of ARGs was higher in the foodborne pathogens isolated from dairy cattle compared to poultry manure amended farms. Regardless of the type of pathogen, 88% (260 of 304) of the resistance genes were detected in dairy manure amended farms, while 11% (36 of 304) were detected in poultry manure amended farms (p < 0.05). The prevalence of ARGs in E. coli O157, Salmonella, L. monocytogenes, and Campylobacter isolates recovered from different sources is shown in Figure S2.
3.4. Correlation between Phenotypic and Genotypic Resistance of the Foodborne Pathogens
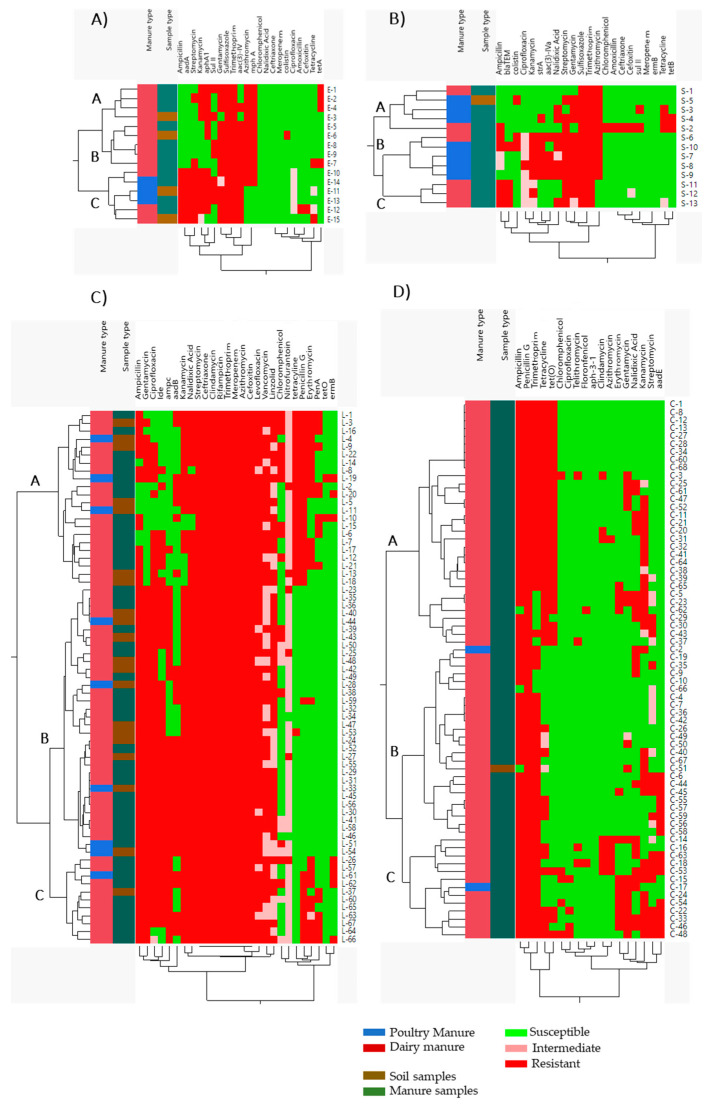
Hierarchical clustering, correlation matrix analysis, and PCA were used to determine the associations between the phenotypic and genotypic characteristics and the source of the isolates. The hierarchical clustering showed that four E. coli O157 isolates that possessed resistance to Kan, Gen, Sul, Tri, and Azi, susceptibility to Chl, Nal, Cef, Mer, Col, Cip, Amo, Ceft, and Tet, and contained aphA1 and mphA genes were clustered together (Cluster A). Further, another five isolates were clustered together and they showed resistance to Gen, Sul, Tri, and Azi, susceptibility to Amp, Chl, Nal, Ceft, Mer, Cip, Amo, and Ceft, and contained aac(3)-IV and mphA genes (Cluster B). Cluster A and B isolates originated from dairy manure amended farms. On the other hand, six of the E. coli O157 isolates were clustered together and showed resistance to Amp, Str, Sul, and Tri, susceptibility to Chl, Nal, Ceft, Mer, and Col, and contained aadA and aac(3)-IV genes (Cluster C), however, they originated from both poultry and dairy cattle manure amended farms (Figure 2A). Similarly, five isolates of Salmonella were clustered together and showed resistance to Azi and susceptibility to Cip, Kan, and Mer (Cluster A; Figure 2B). In cluster B, five isolates possessed resistance to Kan, Str, Gen, Sul, Tri, and Azi, susceptibility to Chl, Amo, Ceft, Cefo, Mer, and Tet, and contained the strA gene, while in cluster C, three isolates that showed resistance to Amp, Sul, and Tri, susceptibility to Chl, Amo, Ceft, and Mer, and contained the blaTEM gene were clustered together. Cluster A and B isolates originated from both poultry and dairy manure amended farms, whereas in cluster C isolates originated only from dairy manure amended farms (Figure 2B).
Figure 2.
Hierarchical clustering of (A) E. coli O157, (B) Salmonella, (C) L. monocytogenes, and (D) Campylobacter. Sample clustered according to their manure amendment type and phenotypic and genotypic antibiotic resistance profile. Red color, phenotypic resistance and presence of ARGs; green color, susceptible and absence of ARGs; blue color, poultry manure amendment; dark red color, dairy manure amendment; dark green color, manure samples; and brown color, soil samples.
Notably, 22 of L. monocytogenes isolates were clustered together and possessed resistance to Ceft, Cli, Rif, Tri, Mer, Azi, Cefo, Tet, and Pen (Cluster A; Figure 2C). However, in cluster B, 34 isolates were resistant to Amp, Gen, Ceft, Cli, Rif, Tri, Mer, Azi, Cefo, and Cip, and in cluster C, eleven isolates that were resistant to Amp and Gen, susceptible to Tet, and contained the ampC gene were clustered together. Isolates in cluster A and B contained variable ARGs. Cluster A, B, and C isolates originated from both poultry and dairy manure amended farms (Figure 2C). Similarly, 32 Campylobacter isolates that were resistant to Pen, Tet, and possessed the tet(O) gene (only one isolate did not have tet(O)) were clustered together (Cluster A; Figure 2D). Within this cluster, eight isolates were phenotypically and genotypically similar. All isolates in this cluster originated from dairy cattle manure amended farms. However, in cluster B, 23 isolates were resistant to Pen and susceptible to Chl, Cip, Tel, and Flo, whereas in cluster C, 13 isolates resistant to Amp and Pen were clustered together. Cluster B and C isolates originated from both poultry and dairy manure amended farms (Figure 2D).
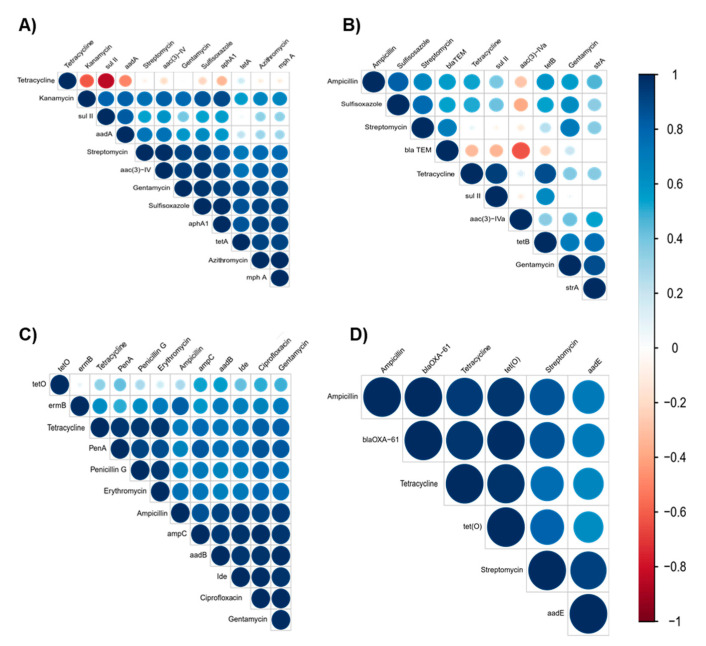
The correlation analysis revealed that the tested ARGs were significantly positively correlated to their corresponding antimicrobials (p < 0.05). There was a strong positive correlation between Azi and mph A (r = 0.8), Kan and aphA (r = 0.8), Str and aadA (r = 0.7), and Gen and aac 3 IV (r = 0.9) in E. coli O157 isolates (Figure 3A), whereas in Salmonella isolates, Tet and tet B were positively corelated (r = 0.9; p < 0.05; Figure 3B). Similarly, in L. monocytogenes isolates, there were strong positive correlations between Cip and Ide (r = 0.9), Pen and pen A (r = 0.9), Ery and ermB (r = 0.7), Amp and ampC (r = 0.8), and Gen and aadB (r = 0.9) (p < 0.05; Figure 3C). In Campylobacter isolates, Tet and tetO (r = 0.9) and Amp and blaOX-61 (r = 0.9) showed significant positive correlation (Figure 3D). This indicates that the detected resistance genes were the determinants for the observed phenotypic resistance in these pathogens [54]. We also observed a strong positive correlation between ARGs and their non-corresponding antimicrobials (p < 0.05). For example, there were positive correlations between Kan and aac 3 IV (r = 0.8) and Str and aac 3 IV (r = 0.9) in E. coli O157 isolates and Gen and strA (r = 0.8) in Salmonella isolates. The observed correlation might be due to the action of the ARGs to antimicrobials that belong to the same class [55]. Additionally, there was a positive correlation between ARGs and antimicrobials from different classes. For example, Gen and mphA (r = 0.8) in E. coli O157, Gen and TetB (r = 0.7) in Salmonella isolates, Ery and penA (r = 0.8), Gen and penA (r = 0.8), Amp and ermB (r = 0.8), and Ery and Ide (r = 0.7) in L. monocytogenes isolates, and Amp and aadE (r = 0.7) and Gen and aadE (r = 0.5) in Campylobacter isolates were positively correlated (p < 0.05; Figure 3). We also found a strong correlation between different antibiotics. For instance, a strong positive correlation was observed between Sul and Azi (r = 0.9), Str and Gen (r = 0.8), and Str and Sul (r = 0.9) in E. coli O157 isolates, Gen and Sul (r = 0.7) and Gen and Str (0.7) in Salmonella isolates, Cip and Gen (r = 0.9) and Pen and Tet (r = 0.9) in L. monocytogenes isolates, and Amp and Str (r = 0.8) and Tet and Amp (r = 0.9) in Campylobacter isolates (Figure 3). This observed strong correlation could be associated with the occurrence of MDR in these isolates [56]. These results suggest that manure amended farms that contain resistance pathogens could be important hotspots for the spread of antimicrobial resistance and ARGs between isolates and can cause a significant public health risk [57,58,59].
Figure 3.
Correlation matrix of phenotypic and genotypic antibiotic resistance of (A) E. coli O157, (B) Salmonella, (C) L. monocytogenes, and (D) Campylobacter. Blue is positive correlation and red is negative correlation (1 = positive correlation, 0 = no correlation, and −1 = negative correlation). Size and strength of the color represent numerical value of correlation coefficient.
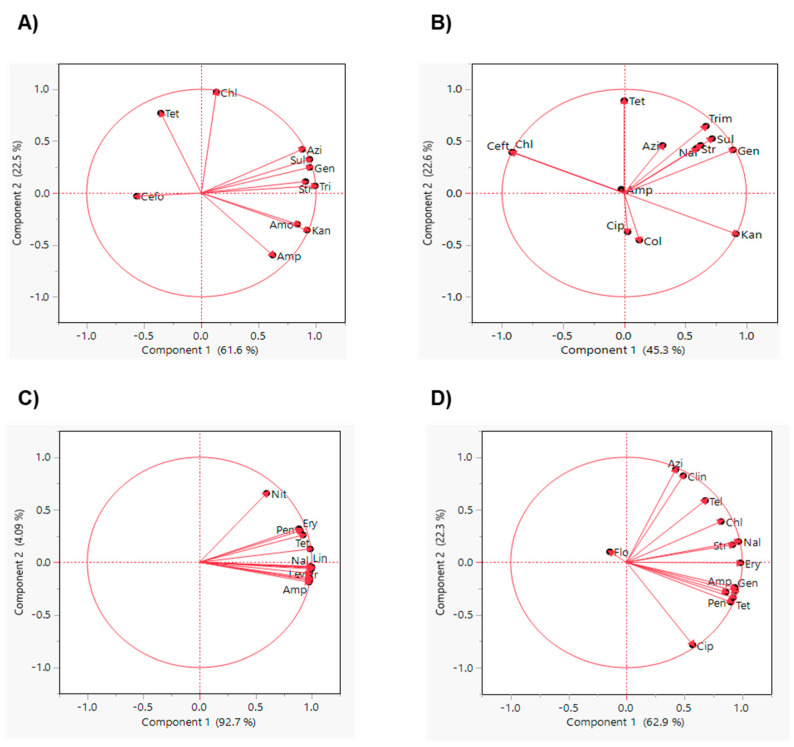
The PCA analysis showed that Sul, Tri, Gen, Str, and Azi resistant isolates and Amo and Kan resistant isolates of E. coli O157 were grouped together, showing their similar trend of occurrence within these isolates. However, Tet, Cefo, and Chl stand separately as they do not have a similar trend of occurrence to other antibiotics (Figure 4A). In Salmonella, Sul, Tri, Gen, Nal, and Str resistant isolates were grouped together, emphasizing their similar trends of occurrence within these isolates (Figure 4B). Similarly, Chl and Ceft and Cip and Col were close to each other, but the Tet resistant isolates stood separately from these antimicrobials (Figure 4B). These PCA results were supported by the correlation analysis where Sul, Gen, and Str strongly corelated in both E. coli O157 and Salmonella isolates (Figure 3A,B). On the other hand, except for Nit, the other antibiotics were grouped together in L. monocytogenes isolates (Figure 4C), suggesting that Nit does not have a similar trend of occurrence to the other antibiotics. A similar result was also shown in the hierarchical clustering (Figure 2C). Additionally, Ery, Amp, Gen, Tet, and Pen resistant Campylobacter isolates were close to each other (Figure 4D), explaining their similar trend of occurrence and strong correlation (Figure 3D). Furthermore, Nal and Str and Azi and Cli resistant isolates were also grouped together, while Cip resistant isolates were far from the other antibiotics and possessed a different occurrence trend compared to the other antibiotics (Figure 3D).
Figure 4.
Principal component analysis of relative abundance of phenotypic antimicrobial resistance of (A) E. coli O157, (B) Salmonella, (C) L. monocytogenes, and (D) Campylobacter. Amp: ampicillin, Chl: chloramphenicol, Cip: ciprofloxacin, Kan: kanamycin, Nal: nalidixic acid, Str: streptomycin, Tet: tetracycline, Gen: gentamycin, Sul: sulfisoxazole, Ceft: ceftriaxone, Amo: amoxicillin, Cefo: cefoxitin, Azi: azithromycin, Tri: trimethoprim sulfamethoxazole, Mer: meropenem, Col: colistin, Pen: penicillin G, Ery: erythromycin, Van: vancomycin, Lin: linezolid, Nit: nitrofurantoin, Cli: clindamycin, Lev: levofloxacin, Tel: telithromycin, Flo: florfenicol.
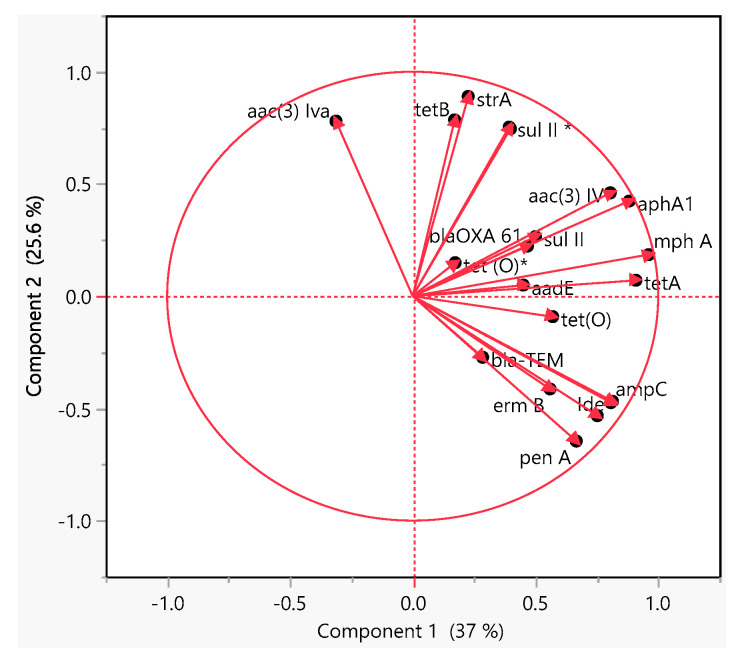
The PCA analysis of the ARGs showed that mphA and tetA and aac (3)-IV and aphA1 in E. coli O157 isolates were grouped together (Figure 5), indicating their similar trend of occurrence as shown in the hierarchical clustering (Figure 2A). However, tetB and strA genes in Salmonella isolates were grouped together (Figure 5) and have strong correlation (Figure 4B). Additionally, penA, Ide and ampC in L. monocytogenes isolates and blaOXA-61 and tet(O) in Campylobacter isolates (Figure 5) showed similar occurrence and they clustered together. Notably, L. monocytogenes isolates possessed strong positive correlation (Figure 3C; p < 0.05), while Campylobacter isolates clustered together (Figure 2D).
Figure 5.
Principal component analysis of the abundance of ARGs in all foodborne pathogens: for E. coli O157, tet(A), aac(3)-IV, aphA1, sulII, aadA, and mphA genes; for Salmonella, strA, tet (B), sulII*, blaTEM, and aac(3)-Iva; for L. monocytogenes, ampC, lde, ermB, tet(O)*, aadB, and penA; and for Campylobacter, blaOXA-61, aadE, and tet(O).
4. Discussion
The widespread use and misuse of antibiotics in food animal production systems has resulted in the emergence of antibiotic resistant zoonotic bacteria that can be transmitted to humans through the food chain [60]. Infection with antibiotic resistant bacteria negatively impacts the public health due to an increased incidence of treatment failure and severity of disease [61].
Interestingly, the total prevalence of foodborne pathogens in this study (19.3%) is lower than in previous reports [62,63,64]. The prevalence of E. coli O157 (1.8%) and Salmonella (1.5%) (Table 2) is lower than in previous reports conducted in California [65,66,67]. The total prevalence of L. monocytogenes (7.9%; Table 2) is lower than in previous reports in Ohio, New York and other states of the US [68,69,70]. Furthermore, the prevalence of Campylobacter (8%; Table 2) is lower than the detected prevalence in Michigan [71]. In our study, E. coli O157 was detected only in 2.9% of dairy cattle manure amended farms. The previously detected prevalence of E. coli O157 in cattle in Ohio increased from 2.1% in 2002 [72] to 24% in 2009 [73]. However, at the global level, the estimated prevalence of E. coli O157 in cattle ranged between 0.13% and 61.8% [74]. The prevalence of Salmonella (1.4%) in dairy cattle manure is lower than in previous reports from Texas [75]. Additionally, the detected prevalence of Salmonella in poultry manure amended farms (1.7%) is lower than the detected prevalence in Georgia (6%) and North Carolina (26%) [76,77]. L. monocytogenes prevalence in poultry (2.2%) and dairy cattle (13.1%) manure amended farms (Table 2) is lower than the detected prevalence in 2010 in Ohio, New York, and southeastern US [78,79,80]. The prevalence of Campylobacter among dairy cattle manure amended farms in our study (14.9%; Table 2) is higher than the findings reported in dairy cattle feces in other localities (northeastern, north, east, midwest, and south) in the US [68,71,81]. Interestingly, the prevalence of Campylobacter in poultry manure amended farms (0.5%) in northeastern Ohio (Table 2) is lower than the detected prevalence (69%) in northwestern Ohio [82] and Maryland [83]. Our result revealed that 1/2a serotype of L. monocytogenes isolates is less prevalent (5.9%) than similar studies previously conducted in Ohio and other states of the US [67,69,84]. Regardless of the detected bacteria, the variation in the prevalence estimates might be due to differences in the husbandry and management systems, type of feed, geographical locations, and the concentration of the farms in each location [85,86], which can have an impact on the dissemination and transmission of pathogens. Additionally, in poultry this variation might also be due to seasonal effects and differences in the hatchery sources, feed composition, vaccination programs, and flock-disease status [77]. Contamination of agricultural farms with different foodborne pathogens might be attributed to using either dairy or poultry manure amendments [87]. Other factors might contribute to the contamination of agricultural farms by pathogens including weather parameters, rainfall, and soil properties (hydraulic properties, texture, soil cover slope), however, these factors were not studied in our study [51,52,53].
Antimicrobial resistance in foodborne pathogens isolated from different sources appears to be increasing in many countries. Previous studies have indicated that both poultry and dairy cattle manure could be sources of antibiotic resistance bacteria [63,88,89]. In our study, 73%, 77%, 100%, and 57.3% of E. coli O157, Salmonella, L. monocytogenes, and Campylobacter isolates were resistant to at least one of the tested antimicrobial classes (Table 3), respectively. These results were higher than previous studies conducted on the same pathogens in Ohio and other states of the US [69,90,91,92]. The relatively high percentages of resistance to the tested antimicrobials might be due to differences in antimicrobial agents use [93]. The antimicrobial resistance in E. coli O157 isolates to aminoglycoside (Str) is higher among dairy cattle farms than poultry farms. A previous study showed that E. coli O157:H7 isolates from food animals have a higher rate of resistance to Amp, Str, Kan, and Tet [94]. The usage of these classes of antimicrobials has increased more than 20% between 2009 and 2016 [95]. The resistance of E. coli O157 isolates to Gen (84.6%) in dairy cattle feces (Table S4) is significantly higher than the previously reported prevalence in Ohio and the west, midwest, northeast, and southeast states of the US [96,97]. Additionally, the prevalence of Gen and Tet resistance in Salmonella isolates is higher than previously detected [98,99,100]. In the US, the resistance of Salmonella to Nal, Gen, and Cip has increased from 2.2%, 1.3%, and 0.3% in 2004 to 2.8%, 2.0%, and 0.5% in 2013, respectively [101]. Furthermore, the resistance of Salmonella isolates to Amp, Kan, Str, and Sul is higher in our study than previous reports from dairy feces in New York and other northeastern states of the US [92].
E. coli O157 isolates showed 73% MDR with higher resistance to aminoglycosides, macrolides, and folate antagonists (Table 3). These results were higher than previously detected from Colorado and southeastern US [102,103]. Similarly, the most detected MDR in Salmonella isolates (77%) was to aminoglycosides, macrolide, and folate antagonists (Table 3), which is higher than previously detected from Georgia and Washington [104,105]. The MDR resistance to Amp (penicillin)/Str (aminoglycosides)/Sul (folate antagonists) continued to increase from less than 1.5% in 1996 to 17% in 2010, 18.3% in 2011, 26.5% in 2012, and 45.5% in 2013 [106]. The rise in antimicrobial-resistant Salmonella might be attributed in part to the clonal spread of multidrug-resistance strains, differences in farming practices, or to the variations in antimicrobial use guidelines [107].
Interestingly, 100% of L. monocytogenes isolates recovered from poultry manure amended farms possessed resistance to Kan, Nal, and Lev, while 86%, 94.8%, and 89.6% of isolates recovered from dairy cattle manure amended farms possessed resistance to Kan, Nal, and Lev, respectively (Table S6). The resistance in L. monocytogenes isolates was significantly higher among tested antimicrobials than previously reported resistance in dairy farms in Tennessee [35]. Notably, 95.5% of L. monocytogenes isolates demonstrated resistance to nalidixic acid; however, L. monocytogenes has been previously reported to have intrinsic resistance to nalidixic acid [108].
Campylobacter isolates showed MDR (57.3%) to aminoglycosides, tetracycline, and penicillin (Table 3). This is higher than previous reports in North Carolina and southeastern US [103,109]. The resistance of Campylobacter isolates to Tet (63%), Cli (11.7%), Gen (25%), Kan (48.5%), and Str (22%) in this study is higher than the detected prevalence in the midwestern and northeastern US [110,111]. Campylobacter isolates recovered from poultry manure amended farms were not resistant to Tet and Cip (Table S7), however, resistance to these two antibiotics was higher in Ohio [82]. The resistance of Campylobacter isolates to Cip was lower than in the previous studies in the US, which increased from 12.8% in 2004 to 16.1% in 2012 [112]. Generally, the resistance trend of foodborne pathogens did not increase from 2016 to 2020 (Figure S1) compared to other studies [107,113,114,115,116]. Our study showed that antimicrobial resistance increased in dairy cattle more than in poultry manure amended farms regardless of the type of pathogen. However, other studies showed that there is increased antimicrobial resistance in both poultry and dairy cattle manure [24]. The observed trend might be due to improved animal management, change in the use of antimicrobials, the physicochemical property of the soil, or other environmental factors such as pH, temperature, oxygen, and the abundance of heavy metals [87,117,118].
In this study, mphA, aadA, aphA1, and tet(A) were the most frequently detected genes within E. coli O157 isolates and blaTEM, tet(B), and strA were the most frequently detected genes within Salmonella isolates. Furthermore, penA, ampC, lde, ermB, tet(O), and aadB were the most frequently detected genes from L. monocytogenes isolates, and blaOXA-61, tet(O), and aadE were the most frequently detected genes in Campylobacter isolates (Figure S2). These results were different from the results obtained in previous studies [35,119,120,121]. The prevalence of aadA (75%) and tet(A) (50%) genes in E. coli O157 isolates [122] and blaTEM (66.6%) and tet(B 66.6%) genes in Salmonella isolates (Figure S2) were lower than the detected prevalence by McMillan et al. [123], whereas the detected prevalence of penA (50%), ampC (66.6%), and ermB (28%) genes in L. monocytogenes isolates is higher in our study than the detected prevalence of the same genes in fecal and environmental samples collected from Tennessee [35,124]. Further, the prevalence of blaOXA-61 (90%) and tet(O) (79%) in Campylobacter isolates is higher than their prevalence in fecal samples collected from northwestern Ohio and Michigan [71,82]. Interestingly, aadA, aph(6)-I, bla, tet, tet(A), and sulII were the most prevalent ARGs found in different foodborne pathogens in Pennsylvania, Maryland, New York, New Mexico, Minnesota, and California [125]. However, in our study, aadA and blaTEM were more prevalent genes. Our results demonstrated that animal manure are important reservoirs of ARGs; thus, it is recommended to develop specific management practices such as aerobic and hyperthermophilic composting for farm amendment of different types of animal manure [126,127,128].
Resistance of Salmonella isolates to Azi cannot be explained by ermB; similarly, resistance of L. monocytogen isolates to Cefo, Azi, and Tri cannot be explained by the resistance genes cfxA, mefA, and sulI. Further, resistance of Campylobacter isolates to Gen cannot be explained by aph-3-1. The observed antimicrobial resistance could be due to different resistance mechanisms such as antibiotic modification and a multidrug efflux pump, which confers a broad spectrum of resistance [129]. Notably, specific alleles of aminoglycoside resistance have been detected in several studies of NARMS US food animal isolates [35,119,120,121,130]. Taken together, our data suggest that the E. coli O157, Salmonella, L. monocytogenes and Campylobacter occurring in dairy cattle and poultry farms have the genetic potential that is necessary for exhibiting resistance to antimicrobials. We found that there was a significant positive association between ARGs and the corresponding antimicrobials (Figure 3). However, the existence of significant correlations between ARGs and other unrelated antimicrobials belonging to different classes was also observed. This correlation could be due to the presence of multiple resistance genes on the same mobile genetic element, as many genes can play a role in the display of similar resistance phenotypes [131,132,133].
5. Conclusions
Livestock and poultry are the most important reservoirs for foodborne pathogens. They can transmit pathogenic bacteria to agricultural farms through manure amendments. Our results revealed that the prevalence and antimicrobial resistance of E. coli O157, Salmonella, L. monocytogenes, and Campylobacter varied among different farm types amended with animal manure in Northeastern Ohio. Dairy cattle manure amended farms were more frequently contaminated with the aforementioned pathogens and contained more resistant foodborne pathogens to the tested antimicrobials compared to poultry manure amended farms. Most of the bacterial isolates were resistant to multiple antimicrobials with genotypic diversity in ARGs. Therefore, there is a need to (1) track antibiotic use in different types of food animals, (2) restrict the use of antimicrobials in veterinary practices to reduce the antimicrobial resistant pathogens that might enter the agricultural farms through manure amendment and impact human health, and (3) control foodborne pathogens in agricultural farms to limit the transmission of these pathogens to humans. Further studies are needed to understand the impact of animal manure amendment in the food chain and in the environment.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/antibiotics10121450/s1, Figure S1: Antimicrobial resistance trend in different foodborne pathogens between 2016 and 2020; Figure S2: Antimicrobial resistance genes of; (A) E. coli O157 (B) Salmonella (C) L. monocytogenes (D) Campylobacter recovered from different manure type; Table S1: PCR primer used for the identification of different foodborne pathogens; Table S2: Interpretive Criteria for Minimum Inhibitory Concentrations; Table S3: Antimicrobial resistance gene primers used for the detection of antimicrobial resistance genes in E. coli O157, Salmonella L. monocytogenes and Campylobacter; Table S4: Antimicrobial resistance profile of the E. coli O157 isolates according to manure amendment type and the source of the isolates; Table S5: Antimicrobial resistance profile of the Salmonella isolates according to manure amendment type and the source of the isolates; Table S6: Antimicrobial resistance profile of the L. monocytogenes isolates according to manure amendment type and the source of the isolates; Table S7: Antimicrobial resistance profile of the Campylobacter isolates according to manure amendment type and the source of the isolates.
Author Contributions
Conceptualization, Y.A.H. and G.R.; methodology, W.H., Y.A.H., G.C.-K., M.K. and G.R.; software, W.H., Y.A.H. and G.C.-K.; validation, Y.A.H. and G.R.; formal analysis, W.H., Y.A.H. and G.C.-K.; investigation, W.H., Y.A.H., G.C.-K., M.K., D.F. and G.R.; data curation, W.H., Y.A.H., M.K., G.C.-K. and G.R.; writing—original draft preparation, W.H. and Y.A.H.; writing—review and editing, W.H., Y.A.H., G.C.-K., M.K., D.F. and G.R.; supervision, Y.A.H., D.F. and G.R.; funding acquisition, G.R. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by federal and state funds appropriated to the Ohio Agricultural Research and Development Center, The Ohio State University. Woinshet Hailu was supported by Sustainable One Health Research Training Capacity (OHEART): Molecular epidemiology of zoonotic foodborne and waterborne pathogens in Eastern Africa, funded by the NIH Fogarty International Center (D43TW008650), through the Global One Health initiative (GOHi).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.CDC . Centers for Disease Control and Prevention, United States Department of Health and Human Services; [(accessed on 23 November 2021)]. Burden of Foodborne Illness: Findings. Available online: https://www.cdc.gov/foodborneburden/2011-foodborne-estimates.html. [Google Scholar]
- 2.CDC . Centers for Disease Control and Prevention, United States Department of Health and Human Services; [(accessed on 23 November 2021)]. Antibiotic Resistance Threats in the United States, 2013. Available online: http://www.cdc.gov/drugresistance/threat-report-2013/ [Google Scholar]
- 3.Scharff R.L. Economic burden from health losses due to foodborne illness in the United States. J. Food Prot. 2012;75:123–131. doi: 10.4315/0362-028X.JFP-11-058. [DOI] [PubMed] [Google Scholar]
- 4.Scharff R.L. Food Attribution and Economic Cost Estimates for Meat- and Poultry-Related Illnesses. J. Food Prot. 2020;83:959–967. doi: 10.4315/JFP-19-548. [DOI] [PubMed] [Google Scholar]
- 5.Scallan E., Hoekstra R.M., Angulo F.J., Tauxe R.V., Widdowson M.A., Roy S.L., Jones J.L., Griffin P.M. Foodborne illness acquired in the United States—Major pathogens. Emerg. Infect. Dis. 2011;17:7–15. doi: 10.3201/eid1701.P11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoffman S., Maculloch B., Batz M. Economic Burden of Major Foodborne Illnesses Acquired in the United States. United States Department of Agriculture; Washington, DC, USA: 2015. [Google Scholar]
- 7.Lomonaco S., Nucera D., Filipello V. The evolution and epidemiology of Listeria monocytogenes in Europe and the United States. Infect. Genet. Evol. 2015;35:172–183. doi: 10.1016/j.meegid.2015.08.008. [DOI] [PubMed] [Google Scholar]
- 8.Henao O.L., Jones T.F., Vugia D.J., Griffin P.M. Foodborne Diseases Active Surveillance Network-2 Decades of Achievements, 1996–2015. Emerg. Infect. Dis. 2015;21:1529–1536. doi: 10.3201/eid2109.150581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iwu C.D., Okoh A.I. Preharvest Transmission Routes of Fresh Produce Associated Bacterial Pathogens with Outbreak Potentials: A Review. Int. J. Environ. Res. Public Health. 2019;16:4407. doi: 10.3390/ijerph16224407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kassem I.I., Kehinde O.O., Helmy Y.A., Kumar A., Chandrashekhar K., Pina-Mimbela R., Rajashekara G. Campylobacter in poultry: The conundrums of highly adaptable and ubiquitous foodborne pathogens. In: Soon J.M., Manning L., Wallace C.A., editors. Foodborne Diseases: Case Studies of Outbreaks in the Agri-Food Industries. CRC Press; Boca Raton, FL, USA: 2016. [Google Scholar]
- 11.Kassem I., Helmy Y.A., Kashoma I.P., Rajashekara G. The emergence of antibiotic resistance on poultry farms. In: Ricke S., editor. Achieving Sustainable Production of Poultry Meat Volume 1: Safety, Quality and Sustainability. Burleigh Dodds Science Publishing; Cambridge, UK: 2016. [Google Scholar]
- 12.CSPI Antibiotic Resistance in Foodborne Pathogens. [(accessed on 23 November 2021)]. Available online: https://cspinet.org/sites/default/files/attachment/outbreaks_antibiotic_resistance_in_foodborne_pathogens_2013.pdf.
- 13.Dewey-Mattia D., Manikonda K., Hall A.J., Wise M.E., Crowe S.J. Surveillance for Foodborne Disease Outbreaks—United States, 2009–2015. [(accessed on 23 November 2021)];MMWR Surveill. Summ. 2018 67:1–11. doi: 10.15585/mmwr.ss6710a1. doi: 10.15585/mmwr.ss6710a1. Available online: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.CDC . Centers for Disease. Control and Prevention, United States Department of Health and Human Services; [(accessed on 23 November 2021)]. Antibiotic Resistance Threats in the United States, 2019. Available online: https://www.cdc.gov/drugresistance/pdf/threats-report/2019-ar-threats-report-508.pdf. [Google Scholar]
- 15.Levy S.B. Active efflux, a common mechanism for biocide and antibiotic resistance. J. Appl. Microbiol. 2002;92:65S–71S. doi: 10.1046/j.1365-2672.92.5s1.4.x. [DOI] [PubMed] [Google Scholar]
- 16.Kraemer S.A., Ramachandran A., Perron G.G. Antibiotic Pollution in the Environment: From Microbial Ecology to Public Policy. Microorganisms. 2019;7:180. doi: 10.3390/microorganisms7060180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O’Neill J. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations. [(accessed on 23 November 2021)]. Available online: https://amr-review.org/sites/default/files/160518_Final%20paper_with%20cover.pdf.
- 18.Li B., Webster T.J. Bacteria antibiotic resistance: New challenges and opportunities for implant-associated orthopedic infections. J. Orthop. Res. 2018;36:22–32. doi: 10.1002/jor.23656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hamer D.H., Gill C.J. From the farm to the kitchen table: The negative impact of antimicrobial use in animals on humans. Nutr. Rev. 2002;60:261–264. doi: 10.1301/002966402320289395. [DOI] [PubMed] [Google Scholar]
- 20.Hao H., Cheng G., Iqbal Z., Ai X., Hussain H.I., Huang L., Dai M., Wang Y., Liu Z., Yuan Z. Benefits and risks of antimicrobial use in food-producing animals. Front. Microbiol. 2014;5:288. doi: 10.3389/fmicb.2014.00288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bacanlı M., Başaran N. Importance of antibiotic residues in animal food. Food Chem. Toxicol. 2019;125:462–466. doi: 10.1016/j.fct.2019.01.033. [DOI] [PubMed] [Google Scholar]
- 22.White D.G., Zhao S., Simjee S., Wagner D.D., McDermott P.F. Antimicrobial resistance of foodborne pathogens. Microbes Infect. 2002;4:405–412. doi: 10.1016/S1286-4579(02)01554-X. [DOI] [PubMed] [Google Scholar]
- 23.Founou L.L., Founou R.C., Essack S.Y. Antibiotic Resistance in the Food Chain: A Developing Country-Perspective. Front. Microbiol. 2016;7:1881. doi: 10.3389/fmicb.2016.01881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Y.-J., Hu H.-W., Chen Q.-L., Singh B.K., Yan H., Chen D., He J.-Z. Transfer of antibiotic resistance from manure-amended soils to vegetable microbiomes. Environ. Int. 2019;130:104912. doi: 10.1016/j.envint.2019.104912. [DOI] [PubMed] [Google Scholar]
- 25.Economou V., Gousia P. Agriculture and food animals as a source of antimicrobial-resistant bacteria. Infect. Drug Resist. 2015;8:49–61. doi: 10.2147/IDR.S55778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nnadozie C.F., Odume O.N. Freshwater environments as reservoirs of antibiotic resistant bacteria and their role in the dissemination of antibiotic resistance genes. Environ. Pollut. 2019;254:113067. doi: 10.1016/j.envpol.2019.113067. [DOI] [PubMed] [Google Scholar]
- 27.Westphal-Settele K., Konradi S., Balzer F., Schönfeld J., Schmithausen R. Die Umwelt als Reservoir für Antibiotikaresistenzen. Bundesgesundheitsblatt-Gesundh.-Gesundh. 2018;61:533–542. doi: 10.1007/s00103-018-2729-8. [DOI] [PubMed] [Google Scholar]
- 28.Rajwar A., Srivastava P., Sahgal M. Microbiology of Fresh Produce: Route of Contamination, Detection Methods, and Remedy. Crit. Rev. Food Sci. Nutr. 2016;56:2383–2390. doi: 10.1080/10408398.2013.841119. [DOI] [PubMed] [Google Scholar]
- 29.Litt Pushpinder K., Kelly A., Omar A., Johnson G., Vinyard Bryan T., Kniel Kalmia E., Sharma M., Dudley Edward G. Temporal and Agricultural Factors Influence Escherichia coli Survival in Soil and Transfer to Cucumbers. Appl. Environ. Microbiol. 2021;87:e02418–e02420. doi: 10.1128/AEM.02418-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang F.-H., Qiao M., Chen Z., Su J.-Q., Zhu Y.-G. Antibiotic resistance genes in manure-amended soil and vegetables at harvest. J. Hazard. Mater. 2015;299:215–221. doi: 10.1016/j.jhazmat.2015.05.028. [DOI] [PubMed] [Google Scholar]
- 31.Neher D.A., Cutler A.J., Weicht T.R., Sharma M., Millner P.D. Composts of poultry litter or dairy manure differentially affect survival of enteric bacteria in fields with spinach. J. Appl. Microbiol. 2019;126:1910–1922. doi: 10.1111/jam.14268. [DOI] [PubMed] [Google Scholar]
- 32.FDA Guidance for Industry. [(accessed on 12 April 2016)]; Available online: www.fda.gov/downloads/AnimalVeterinary/GuidanceComplianceEnforcement/GuidanceforIndustry/UCM299624.pdf.
- 33.FDA Timeline of FDA Action on Antimicrobial Resistance. [(accessed on 23 November 2021)]; Available online: https://www.fda.gov/animal-veterinary/antimicrobial-resistance/timeline-fda-action-antimicrobial-resistance.
- 34.LeJeune J.T., Hancock D.D., Besser T.E. Sensitivity of Escherichia coli O157 detection in bovine feces assessed by broth enrichment followed by immunomagnetic separation and direct plating methodologies. J. Clin. Microbiol. 2006;44:872–875. doi: 10.1128/JCM.44.3.872-875.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Srinivasan V., Nam H.M., Nguyen L.T., Tamilselvam B., Murinda S.E., Oliver S.P. Prevalence of antimicrobial resistance genes in Listeria monocytogenes isolated from dairy farms. Foodborne Pathog. Dis. 2005;2:201–211. doi: 10.1089/fpd.2005.2.201. [DOI] [PubMed] [Google Scholar]
- 36.Kersting A.L., Medeiros L.C., LeJeune J.T. Differences in Listeria monocytogenes contamination of rural Ohio residences with and without livestock. Foodborne Pathog. Dis. 2010;7:57–62. doi: 10.1089/fpd.2009.0318. [DOI] [PubMed] [Google Scholar]
- 37.Rybolt M.L., Wills R.W., Bailey R.H. Use of secondary enrichment for isolation of Salmonella from naturally contaminated environmental samples. Poult. Sci. 2005;84:992–997. doi: 10.1093/ps/84.7.992. [DOI] [PubMed] [Google Scholar]
- 38.Kassem I.I., Helmy Y.A., Kathayat D., Candelero-Rueda R.A., Kumar A., Deblais L., Huang H.C., Sahin O., Zhang Q., Rajashekara G. Nonculturability Might Underestimate the Occurrence of Campylobacter in Broiler Litter. Foodborne Pathog. Dis. 2017;14:472–477. doi: 10.1089/fpd.2017.2279. [DOI] [PubMed] [Google Scholar]
- 39.Kashoma I.P., Kassem I.I., John J., Kessy B.M., Gebreyes W., Kazwala R.R., Rajashekara G. Prevalence and Antimicrobial Resistance of Campylobacter Isolated from Dressed Beef Carcasses and Raw Milk in Tanzania. Microb. Drug Resist. 2016;22:40–52. doi: 10.1089/mdr.2015.0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Helmy Y.A., Kassem I.I., Kumar A., Rajashekara G. In Vitro Evaluation of the Impact of the Probiotic E. coli Nissle 1917 on Campylobacter jejuni’s Invasion and Intracellular Survival in Human Colonic Cells. Front. Microbiol. 2017;8:1588. doi: 10.3389/fmicb.2017.01588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Deblais L., Helmy Y.A., Kumar A., Antwi J., Kathayat D., Acuna U.M., Huang H.C., de Blanco E.C., Fuchs J.R., Rajashekara G. Novel narrow spectrum benzyl thiophene sulfonamide derivatives to control Campylobacter. J. Antibiot. 2019;72:555–565. doi: 10.1038/s41429-019-0168-x. [DOI] [PubMed] [Google Scholar]
- 42.Zhai L., Yu Q., Bie X., Lu Z., Lv F., Zhang C., Kong X., Zhao H. Development of a PCR test system for specific detection of Salmonella Paratyphi B in foods. FEMS Microbiol. Lett. 2014;355:83–89. doi: 10.1111/1574-6968.12443. [DOI] [PubMed] [Google Scholar]
- 43.Li B., Liu H., Wang W. Multiplex real-time PCR assay for detection of Escherichia coli O157:H7 and screening for non-O157 Shiga toxin-producing E. coli. BMC Microbiol. 2017;17:215. doi: 10.1186/s12866-017-1123-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meghdadi H., Khosravi A.D., Sheikh A.F., Alami A., Nassirabady N. Isolation and characterization of Listeria monocytogenes from environmental and clinical sources by culture and PCR-RFLP methods. Iran. J. Microbiol. 2019;11:7–12. doi: 10.18502/ijm.v11i1.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ge B., Wang F., Sjolund-Karlsson M., McDermott P.F. Antimicrobial resistance in Campylobacter: Susceptibility testing methods and resistance trends. J. Microbiol. Methods. 2013;95:57–67. doi: 10.1016/j.mimet.2013.06.021. [DOI] [PubMed] [Google Scholar]
- 46.NARMS National Antimicrobial Resistance Monitoring System for Enteric Bacteria (NARMS) [(accessed on 23 November 2021)]; Available online: https://www.cdc.gov/narms/antibiotics-tested.html.
- 47.Obaidat M.M. Prevalence and antimicrobial resistance of Listeria monocytogenes, Salmonella enterica and Escherichia coli O157:H7 in imported beef cattle in Jordan. Comp. Immunol. Microbiol. Infect. Dis. 2020;70:101447. doi: 10.1016/j.cimid.2020.101447. [DOI] [PubMed] [Google Scholar]
- 48.Helmy Y.A., Kassem I.I., Rajashekara G. Immuno-modulatory effect of probiotic E. coli Nissle 1917 in polarized human colonic cells against Campylobacter jejuni infection. Gut Microbes. 2021;13:1–16. doi: 10.1080/19490976.2020.1857514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Terefe Y., Deblais L., Ghanem M., Helmy Y.A., Mummed B., Chen D., Singh N., Ahyong V., Kalantar K., Yimer G., et al. Co-occurrence of Campylobacter Species in Children from Eastern Ethiopia, and Their Association with Environmental Enteric Dysfunction, Diarrhea, and Host Microbiome. Front. Public Health. 2020;8:99. doi: 10.3389/fpubh.2020.00099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Magiorakos A.P., Srinivasan A., Carey R.B., Carmeli Y., Falagas M.E., Giske C.G., Harbarth S., Hindler J.F., Kahlmeter G., Olsson-Liljequist B., et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012;18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 51.Harris C.S., Tertuliano M., Rajeev S., Vellidis G., Levy K. Impact of storm runoff on Salmonella and Escherichia coli prevalence in irrigation ponds of fresh produce farms in southern Georgia. J. Appl. Microbiol. 2018;124:910–921. doi: 10.1111/jam.13689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gu G., Strawn L.K., Ottesen A.R., Ramachandran P., Reed E.A., Zheng J., Boyer R.R., Rideout S.L. Correlation of Salmonella enterica and Listeria monocytogenes in Irrigation Water to Environmental Factors, Fecal Indicators, and Bacterial Communities. Front. Microbiol. 2021;11:3426. doi: 10.3389/fmicb.2020.557289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alegbeleye O.O., Sant’Ana A.S. Manure-borne pathogens as an important source of water contamination: An update on the dynamics of pathogen survival/transport as well as practical risk mitigation strategies. Int. J. Hyg. Environ. Health. 2020;227:113524. doi: 10.1016/j.ijheh.2020.113524. [DOI] [PubMed] [Google Scholar]
- 54.Pornsukarom S., Thakur S., Schaffner Donald W. Horizontal Dissemination of Antimicrobial Resistance Determinants in Multiple Salmonella Serotypes following Isolation from the Commercial Swine Operation Environment after Manure Application. Appl. Environ. Microbiol. 2017;83:e01503–e01517. doi: 10.1128/AEM.01503-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ryu S.-H., Lee J.-H., Park S.-H., Song M.-O., Park S.-H., Jung H.-W., Park G.-Y., Choi S.-M., Kim M.-S., Chae Y.-Z., et al. Antimicrobial resistance profiles among Escherichia coli strains isolated from commercial and cooked foods. Int. J. Food Microbiol. 2012;159:263–266. doi: 10.1016/j.ijfoodmicro.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 56.Guruge K.S., Tamamura Y.A., Goswami P., Tanoue R., Jinadasa K.B.S.N., Nomiyama K., Ohura T., Kunisue T., Tanabe S., Akiba M. The association between antimicrobials and the antimicrobial-resistant phenotypes and resistance genes of Escherichia coli isolated from hospital wastewaters and adjacent surface waters in Sri Lanka. Chemosphere. 2021;279:130591. doi: 10.1016/j.chemosphere.2021.130591. [DOI] [PubMed] [Google Scholar]
- 57.Wang M., Liu P., Xiong W., Zhou Q., Wangxiao J., Zeng Z., Sun Y. Fate of potential indicator antimicrobial resistance genes (ARGs) and bacterial community diversity in simulated manure-soil microcosms. Ecotoxicol. Environ. Saf. 2018;147:817–823. doi: 10.1016/j.ecoenv.2017.09.055. [DOI] [PubMed] [Google Scholar]
- 58.Peng S., Feng Y., Wang Y., Guo X., Chu H., Lin X. Prevalence of antibiotic resistance genes in soils after continually applied with different manure for 30 years. J. Hazard. Mater. 2017;340:16–25. doi: 10.1016/j.jhazmat.2017.06.059. [DOI] [PubMed] [Google Scholar]
- 59.Martinez J.L., Coque T.M., Baquero F. Prioritizing risks of antibiotic resistance genes in all metagenomes. Nat. Rev. Microbiol. 2015;13:396. doi: 10.1038/nrmicro3399-c2. [DOI] [PubMed] [Google Scholar]
- 60.Rohr J.R., Barrett C.B., Civitello D.J., Craft M.E., Delius B., DeLeo G.A., Hudson P.J., Jouanard N., Nguyen K.H., Ostfeld R.S., et al. Emerging human infectious diseases and the links to global food production. Nat. Sustain. 2019;2:445–456. doi: 10.1038/s41893-019-0293-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Friedman N.D., Temkin E., Carmeli Y. The negative impact of antibiotic resistance. Clin. Microbiol. Infect. 2016;22:416–422. doi: 10.1016/j.cmi.2015.12.002. [DOI] [PubMed] [Google Scholar]
- 62.Chen S., Zhao S., White D.G., Schroeder C.M., Lu R., Yang H., McDermott P.F., Ayers S., Meng J. Characterization of multiple-antimicrobial-resistant salmonella serovars isolated from retail meats. Appl. Environ. Microbiol. 2004;70:1–7. doi: 10.1128/AEM.70.1.1-7.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Furtula V., Farrell E.G., Diarrassouba F., Rempel H., Pritchard J., Diarra M.S. Veterinary pharmaceuticals and antibiotic resistance of Escherichia coli isolates in poultry litter from commercial farms and controlled feeding trials. Poult. Sci. 2010;89:180–188. doi: 10.3382/ps.2009-00198. [DOI] [PubMed] [Google Scholar]
- 64.Williams A., Oyarzabal O.A. Prevalence of Campylobacter spp. in skinless, boneless retail broiler meat from 2005 through 2011 in Alabama, USA. BMC Microbiol. 2012;12:184. doi: 10.1186/1471-2180-12-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen Z., Biswas S., Aminabadi P., Stackhouse J.W., Jay-Russell M.T., Pandey P.K. Prevalence of Escherichia coli O157 and Salmonella spp. in solid bovine manure in California using real-time quantitative PCR. Lett. Appl. Microbiol. 2019;69:23–29. doi: 10.1111/lam.13156. [DOI] [PubMed] [Google Scholar]
- 66.Benjamin L., Atwill E.R., Jay-Russell M., Cooley M., Carychao D., Gorski L., Mandrell R.E. Occurrence of generic Escherichia coli, E. coli O157 and Salmonella spp. in water and sediment from leafy green produce farms and streams on the Central California coast. Int. J. Food Microbiol. 2013;165:65–76. doi: 10.1016/j.ijfoodmicro.2013.04.003. [DOI] [PubMed] [Google Scholar]
- 67.Cooley M.B., Quiñones B., Oryang D., Mandrell R.E., Gorski L. Prevalence of shiga toxin producing Escherichia coli, Salmonella enterica, and Listeria monocytogenes at public access watershed sites in a California Central Coast agricultural region. Front. Cell. Infect. Microbiol. 2014;4:30. doi: 10.3389/fcimb.2014.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pradhan A.K., Van Kessel J.S., Karns J.S., Wolfgang D.R., Hovingh E., Nelen K.A., Smith J.M., Whitlock R.H., Fyock T., Ladely S., et al. Dynamics of endemic infectious diseases of animal and human importance on three dairy herds in the northeastern United States. J. Dairy Sci. 2009;92:1811–1825. doi: 10.3168/jds.2008-1486. [DOI] [PubMed] [Google Scholar]
- 69.Sonnier J.L., Karns J.S., Lombard J.E., Kopral C.A., Haley B.J., Kim S.-W., Van Kessel J.A.S. Prevalence of Salmonella enterica, Listeria monocytogenes, and pathogenic Escherichia coli in bulk tank milk and milk filters from US dairy operations in the National Animal Health Monitoring System Dairy 2014 study. J. Dairy Sci. 2018;101:1943–1956. doi: 10.3168/jds.2017-13546. [DOI] [PubMed] [Google Scholar]
- 70.Strawn Laura K., Gröhn Yrjo T., Warchocki S., Worobo Randy W., Bihn Elizabeth A., Wiedmann M. Risk Factors Associated with Salmonella and Listeria monocytogenes Contamination of Produce Fields. Appl. Environ. Microbiol. 2013;79:7618–7627. doi: 10.1128/AEM.02831-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cha W., Mosci R.E., Wengert S.L., Venegas Vargas C., Rust S.R., Bartlett P.C., Grooms D.L., Manning S.D. Comparing the Genetic Diversity and Antimicrobial Resistance Profiles of Campylobacter jejuni Recovered from Cattle and Humans. Front. Microbiol. 2017;8:818. doi: 10.3389/fmicb.2017.00818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dodson K., LeJeune J. Escherichia coli O157:H7, Campylobacter jejuni, and Salmonella Prevalence in cull dairy cows marketed in northeastern Ohio. J. Food Prot. 2005;68:927–931. doi: 10.4315/0362-028X-68.5.927. [DOI] [PubMed] [Google Scholar]
- 73.Cernicchiaro N., Pearl D.L., McEwen S.A., Harpster L., Homan H.J., Linz G.M., Lejeune J.T. Association of wild bird density and farm management factors with the prevalence of E. coli O157 in dairy herds in Ohio (2007–2009) Zoonoses Public Health. 2012;59:320–329. doi: 10.1111/j.1863-2378.2012.01457.x. [DOI] [PubMed] [Google Scholar]
- 74.Islam M.Z., Musekiwa A., Islam K., Ahmed S., Chowdhury S., Ahad A., Biswas P.K. Regional variation in the prevalence of E. coli O157 in cattle: A meta-analysis and meta-regression. PLoS ONE. 2014;9:e93299. doi: 10.1371/journal.pone.0093299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rodriguez-Rivera L.D., Cummings K.J., Loneragan G.H., Rankin S.C., Hanson D.L., Leone W.M., Edrington T.S. Salmonella Prevalence and Antimicrobial Susceptibility Among Dairy Farm Environmental Samples Collected in Texas. Foodborne Pathog. Dis. 2016;13:205–211. doi: 10.1089/fpd.2015.2037. [DOI] [PubMed] [Google Scholar]
- 76.Liljebjelke K.A., Hofacre C.L., Liu T., White D.G., Ayers S., Young S., Maurer J.J. Vertical and horizontal transmission of salmonella within integrated broiler production system. Foodborne Pathog. Dis. 2005;2:90–102. doi: 10.1089/fpd.2005.2.90. [DOI] [PubMed] [Google Scholar]
- 77.Alali W.Q., Thakur S., Berghaus R.D., Martin M.P., Gebreyes W.A. Prevalence and distribution of Salmonella in organic and conventional broiler poultry farms. Foodborne Pathog. Dis. 2010;7:1363–1371. doi: 10.1089/fpd.2010.0566. [DOI] [PubMed] [Google Scholar]
- 78.Locatelli A., Lewis M.A., Rothrock M.J., Jr. The Distribution of Listeria in Pasture-Raised Broiler Farm Soils Is Potentially Related to University of Vermont Medium Enrichment Bias toward Listeria innocua over Listeria monocytogenes. Front. Vet. Sci. 2017;4:227. doi: 10.3389/fvets.2017.00227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ho A.J., Ivanek R., Grohn Y.T., Nightingale K.K., Wiedmann M. Listeria monocytogenes fecal shedding in dairy cattle shows high levels of day-to-day variation and includes outbreaks and sporadic cases of shedding of specific L. monocytogenes subtypes. Prev. Vet. Med. 2007;80:287–305. doi: 10.1016/j.prevetmed.2007.03.005. [DOI] [PubMed] [Google Scholar]
- 80.Nightingale K.K., Schukken Y.H., Nightingale C.R., Fortes E.D., Ho A.J., Her Z., Grohn Y.T., McDonough P.L., Wiedmann M. Ecology and transmission of Listeria monocytogenes infecting ruminants and in the farm environment. Appl. Environ. Microbiol. 2004;70:4458–4467. doi: 10.1128/AEM.70.8.4458-4467.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sanad Y.M., Kassem I.I., Abley M., Gebreyes W., LeJeune J.T., Rajashekara G. Genotypic and phenotypic properties of cattle-associated Campylobacter and their implications to public health in the USA. PLoS ONE. 2011;6:e25778. doi: 10.1371/journal.pone.0025778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kassem I.I., Kehinde O., Kumar A., Rajashekara G. Antimicrobial-Resistant Campylobacter in Organically and Conventionally Raised Layer Chickens. Foodborne Pathog. Dis. 2017;14:29–34. doi: 10.1089/fpd.2016.2161. [DOI] [PubMed] [Google Scholar]
- 83.Cui S., Ge B., Zheng J., Meng J. Prevalence and antimicrobial resistance of Campylobacter spp. and Salmonella serovars in organic chickens from Maryland retail stores. Appl. Environ. Microbiol. 2005;71:4108–4111. doi: 10.1128/AEM.71.7.4108-4111.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Borucki M.K., Call D.R. Listeria monocytogenes Serotype Identification by PCR. J. Clin. Microbiol. 2003;41:5537–5540. doi: 10.1128/JCM.41.12.5537-5540.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sahin O., Kassem I.I., Shen Z., Lin J., Rajashekara G., Zhang Q. Campylobacter in poultry: Ecology and potential interventions. Avian Dis. 2015;59:185–200. doi: 10.1637/11072-032315-Review. [DOI] [PubMed] [Google Scholar]
- 86.Humphrey T., O’Brien S., Madsen M. Campylobacters as zoonotic pathogens: A food production perspective. Int. J. Food Microbiol. 2007;117:237–257. doi: 10.1016/j.ijfoodmicro.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 87.Manyi-Loh C.E., Mamphweli S.N., Meyer E.L., Makaka G., Simon M., Okoh A.I. An Overview of the Control of Bacterial Pathogens in Cattle Manure. Int. J. Environ. Res. Public Health. 2016;13:843. doi: 10.3390/ijerph13090843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Edrington T.S., Fox W.E., Callaway T.R., Anderson R.C., Hoffman D.W., Nisbet D.J. Pathogen prevalence and influence of composted dairy manure application on antimicrobial resistance profiles of commensal soil bacteria. Foodborne Pathog. Dis. 2009;6:217–224. doi: 10.1089/fpd.2008.0184. [DOI] [PubMed] [Google Scholar]
- 89.Resende J.A., Silva V.L., de Oliveira T.L., de Oliveira Fortunato S., da Costa Carneiro J., Otenio M.H., Diniz C.G. Prevalence and persistence of potentially pathogenic and antibiotic resistant bacteria during anaerobic digestion treatment of cattle manure. Bioresour. Technol. 2014;153:284–291. doi: 10.1016/j.biortech.2013.12.007. [DOI] [PubMed] [Google Scholar]
- 90.Tadesse D.A., Bahnson P.B., Funk J.A., Thakur S., Morrow W.E.M., Wittum T., DeGraves F., Rajala-Schultz P., Gebreyes W.A. Prevalence and Antimicrobial Resistance Profile of Campylobacter Spp. Isolated from Conventional and Antimicrobial-Free Swine Production Systems from Different U.S. Regions. Foodborne Pathog. Dis. 2010;8:367–374. doi: 10.1089/fpd.2010.0665. [DOI] [PubMed] [Google Scholar]
- 91.McConnel C.S., Stenkamp-Strahm C.M., Rao S., Linke L.M., Magnuson R.J., Hyatt D.R. Antimicrobial Resistance Profiles in Escherichia coli O157 Isolates from Northern Colorado Dairies. J. Food Prot. 2016;79:484–487. doi: 10.4315/0362-028X.JFP-15-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cummings K.J., Perkins G.A., Khatibzadeh S.M., Warnick L.D., Altier C. Antimicrobial Resistance Trends Among Salmonella Isolates Obtained from Dairy Cattle in the Northeastern United States, 2004–2011. Foodborne Pathog. Dis. 2013;10:353–361. doi: 10.1089/fpd.2012.1285. [DOI] [PubMed] [Google Scholar]
- 93.Aarestrup F.M., Wegener H.C., Collignon P. Resistance in bacteria of the food chain: Epidemiology and control strategies. Expert Rev. Anti Infect. Ther. 2008;6:733–750. doi: 10.1586/14787210.6.5.733. [DOI] [PubMed] [Google Scholar]
- 94.Tadesse D.A., Zhao S., Tong E., Ayers S., Singh A., Bartholomew M.J., McDermott P.F. Antimicrobial drug resistance in Escherichia coli from humans and food animals, United States, 1950–2002. Emerg. Infect. Dis. 2012;18:741–749. doi: 10.3201/eid1805.111153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.FDA 2016 Summary Report on Antimicrobials Sold or Distributed for Use in Food-Producing Animals. [(accessed on 23 November 2021)]; Available online: https://www.fda.gov/downloads/forindustry/userfees/animaldruguserfeeactadufa/ucm588085.pdf.
- 96.Lundin J.I., Dargatz D.A., Wagner B.A., Lombard J.E., Hill A.E., Ladely S.R., Fedorka-Cray P.J. Antimicrobial drug resistance of fecal Escherichia coli and Salmonella spp. isolates from United States dairy cows. Foodborne Pathog. Dis. 2008;5:7–19. doi: 10.1089/fpd.2007.0018. [DOI] [PubMed] [Google Scholar]
- 97.Oliver S.P., Murinda S.E., Jayarao B.M. Impact of antibiotic use in adult dairy cows on antimicrobial resistance of veterinary and human pathogens: A comprehensive review. Foodborne Pathog. Dis. 2011;8:337–355. doi: 10.1089/fpd.2010.0730. [DOI] [PubMed] [Google Scholar]
- 98.Blau D.M., McCluskey B.J., Ladely S.R., Dargatz D.A., Fedorka-Cray P.J., Ferris K.E., Headrick M.L. Salmonella in dairy operations in the United States: Prevalence and antimicrobial drug susceptibility. J. Food Prot. 2005;68:696–702. doi: 10.4315/0362-028X-68.4.696. [DOI] [PubMed] [Google Scholar]
- 99.Antunes P., Reu C., Sousa J.C., Peixe L., Pestana N. Incidence of Salmonella from poultry products and their susceptibility to antimicrobial agents. Int. J. Food Microbiol. 2003;82:97–103. doi: 10.1016/S0168-1605(02)00251-9. [DOI] [PubMed] [Google Scholar]
- 100.Liljebjelke K.A., Hofacre C.L., White D.G., Ayers S., Lee M.D., Maurer J.J. Diversity of Antimicrobial Resistance Phenotypes in Salmonella Isolated from Commercial Poultry Farms. Front. Vet. Sci. 2017;4:96. doi: 10.3389/fvets.2017.00096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Michael G.B., Schwarz S. Antimicrobial resistance in zoonotic nontyphoidal Salmonella: An alarming trend? Clin. Microbiol. Infect. 2016;22:968–974. doi: 10.1016/j.cmi.2016.07.033. [DOI] [PubMed] [Google Scholar]
- 102.Haberecht H.B., Nealon N.J., Gilliland J.R., Holder A.V., Runyan C., Oppel R.C., Ibrahim H.M., Mueller L., Schrupp F., Vilchez S., et al. Antimicrobial-Resistant Escherichia coli from Environmental Waters in Northern Colorado. J. Environ. Public Health. 2019;2019:3862949. doi: 10.1155/2019/3862949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rothrock M.J., Jr., Hiett K.L., Guard J.Y., Jackson C.R. Antibiotic Resistance Patterns of Major Zoonotic Pathogens from All-Natural, Antibiotic-Free, Pasture-Raised Broiler Flocks in the Southeastern United States. J. Environ. Qual. 2016;45:593–603. doi: 10.2134/jeq2015.07.0366. [DOI] [PubMed] [Google Scholar]
- 104.Xu Y., Tao S., Hinkle N., Harrison M., Chen J. Salmonella, including antibiotic-resistant Salmonella, from flies captured from cattle farms in Georgia, U.S.A. Sci. Total. Environ. 2018;616–617:90–96. doi: 10.1016/j.scitotenv.2017.10.324. [DOI] [PubMed] [Google Scholar]
- 105.Adhikari B., Besser T.E., Gay J.M., Fox L.K., Davis M.A., Cobbold R.N., Berge A.C.B., Hancock D.D. The role of animal movement, including off-farm rearing of heifers, in the interherd transmission of multidrug-resistant Salmonella. J. Dairy Sci. 2009;92:4229–4238. doi: 10.3168/jds.2008-1494. [DOI] [PubMed] [Google Scholar]
- 106.CDC National Antimicrobial Resistance Monitoring System for Enteric Bacteria (NARMS): Human Isolates Final Report, 2013 U.S. Department of Health and Human Services, Atlanta (2015) [(accessed on 23 November 2021)]; Available online: https://www.cdc.gov/narms/pdf/2013-annual-report-narms-508c.pdf.
- 107.Wang X., Biswas S., Paudyal N., Pan H., Li X., Fang W., Yue M. Antibiotic Resistance in Salmonella Typhimurium Isolates Recovered from the Food Chain Through National Antimicrobial Resistance Monitoring System between 1996 and 2016. Front. Microbiol. 2019;10:985. doi: 10.3389/fmicb.2019.00985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lampidis R., Kostrewa D., Hof H. Molecular characterization of the genes encoding DNA gyrase and topoisomerase IV of Listeria monocytogenes. J. Antimicrob. Chemother. 2002;49:917–924. doi: 10.1093/jac/dkf065. [DOI] [PubMed] [Google Scholar]
- 109.Hull D.M., Harrell E., van Vliet A.H.M., Correa M., Thakur S. Antimicrobial resistance and interspecies gene transfer in Campylobacter coli and Campylobacter jejuni isolated from food animals, poultry processing, and retail meat in North Carolina, 2018–2019. PLoS ONE. 2021;16:e0246571. doi: 10.1371/journal.pone.0246571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Oliver J.P., Gooch C.A., Lansing S., Schueler J., Hurst J.J., Sassoubre L., Crossette E.M., Aga D.S. Invited review: Fate of antibiotic residues, antibiotic-resistant bacteria, and antibiotic resistance genes in US dairy manure management systems. J. Dairy Sci. 2020;103:1051–1071. doi: 10.3168/jds.2019-16778. [DOI] [PubMed] [Google Scholar]
- 111.Obeng A.S., Rickard H., Sexton M., Pang Y., Peng H., Barton M. Antimicrobial susceptibilities and resistance genes in Campylobacter strains isolated from poultry and pigs in Australia. J. Appl. Microbiol. 2012;113:294–307. doi: 10.1111/j.1365-2672.2012.05354.x. [DOI] [PubMed] [Google Scholar]
- 112.Geissler A.L., Bustos Carrillo F., Swanson K., Patrick M.E., Fullerton K.E., Bennett C., Barrett K., Mahon B.E. Increasing Campylobacter Infections, Outbreaks, and Antimicrobial Resistance in the United States, 2004–2012. Clin. Infect. Dis. 2017;65:1624–1631. doi: 10.1093/cid/cix624. [DOI] [PubMed] [Google Scholar]
- 113.Tang Y., Fang L., Xu C., Zhang Q. Antibiotic resistance trends and mechanisms in the foodborne pathogen, Campylobacter. Anim. Health Res. Rev. 2017;18:87–98. doi: 10.1017/S1466252317000135. [DOI] [PubMed] [Google Scholar]
- 114.Ma L., Wang Y., Shen J., Zhang Q., Wu C. Tracking Campylobacter contamination along a broiler chicken production chain from the farm level to retail in China. Int. J. Food Microbiol. 2014;181:77–84. doi: 10.1016/j.ijfoodmicro.2014.04.023. [DOI] [PubMed] [Google Scholar]
- 115.Zhang P., Shen Z., Zhang C., Song L., Wang B., Shang J., Yue X., Qu Z., Li X., Wu L., et al. Surveillance of antimicrobial resistance among Escherichia coli from chicken and swine, China, 2008–2015. Vet. Microbiol. 2017;203:49–55. doi: 10.1016/j.vetmic.2017.02.008. [DOI] [PubMed] [Google Scholar]
- 116.Alonso-Hernando A., Prieto M., García-Fernández C., Alonso-Calleja C., Capita R. Increase over time in the prevalence of multiple antibiotic resistance among isolates of Listeria monocytogenes from poultry in Spain. Food Control. 2012;23:37–41. doi: 10.1016/j.foodcont.2011.06.006. [DOI] [Google Scholar]
- 117.Qian X., Gu J., Sun W., Wang X.-J., Su J.-Q., Stedfeld R. Diversity, abundance, and persistence of antibiotic resistance genes in various types of animal manure following industrial composting. J. Hazard. Mater. 2018;344:716–722. doi: 10.1016/j.jhazmat.2017.11.020. [DOI] [PubMed] [Google Scholar]
- 118.Alexander T.W., Yanke J.L., Reuter T., Topp E., Read R.R., Selinger B.L., McAllister T.A. Longitudinal characterization of antimicrobial resistance genes in feces shed from cattle fed different subtherapeutic antibiotics. BMC Microbiol. 2011;11:19. doi: 10.1186/1471-2180-11-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Glenn L.M., Lindsey R.L., Frank J.F., Meinersmann R.J., Englen M.D., Fedorka-Cray P.J., Frye J.G. Analysis of antimicrobial resistance genes detected in multidrug-resistant Salmonella enterica serovar Typhimurium isolated from food animals. Microb. Drug Resist. 2011;17:407–418. doi: 10.1089/mdr.2010.0189. [DOI] [PubMed] [Google Scholar]
- 120.Frye J.G., Fedorka-Cray P.J., Jackson C.R., Rose M. Analysis of Salmonella enterica with reduced susceptibility to the third-generation cephalosporin ceftriaxone isolated from U.S. cattle during 2000–2004. Microb. Drug Resist. 2008;14:251–258. doi: 10.1089/mdr.2008.0844. [DOI] [PubMed] [Google Scholar]
- 121.Glenn L.M., Englen M.D., Lindsey R.L., Frank J.F., Turpin J.E., Berrang M.E., Meinersmann R.J., Fedorka-Cray P.J., Frye J.G. Analysis of antimicrobial resistance genes detected in multiple-drug-resistant Escherichia coli isolates from broiler chicken carcasses. Microb. Drug Resist. 2012;18:453–463. doi: 10.1089/mdr.2011.0224. [DOI] [PubMed] [Google Scholar]
- 122.Evangelista A.G., Corrêa J.A.F., Pinto A.C.S.M., Luciano F.B. The impact of essential oils on antibiotic use in animal production regarding antimicrobial resistancea-review. Crit. Rev. Food Sci. Nutr. 2021;61:1–17. doi: 10.1080/10408398.2021.1883548. [DOI] [PubMed] [Google Scholar]
- 123.McMillan E.A., Gupta S.K., Williams L.E., Jové T., Hiott L.M., Woodley T.A., Barrett J.B., Jackson C.R., Wasilenko J.L., Simmons M., et al. Antimicrobial Resistance Genes, Cassettes, and Plasmids Present in Salmonella enterica Associated with United States Food Animals. Front. Microbiol. 2019;10:832. doi: 10.3389/fmicb.2019.00832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Escolar C., Gómez D., del Carmen Rota García M., Conchello P., Herrera A. Antimicrobial Resistance Profiles of Listeria monocytogenes and Listeria innocua Isolated from Ready-to-Eat Products of Animal Origin in Spain. Foodborne Pathog. Dis. 2017;14:357–363. doi: 10.1089/fpd.2016.2248. [DOI] [PubMed] [Google Scholar]
- 125.Zhang N., Liu E., Tang A., Ye M.C., Wang K., Jia Q., Huang Z. Data-Driven Analysis of Antimicrobial Resistance in Foodborne Pathogens from Six States within the US. Int. J. Environ. Res. Public Health. 2019;16:1811. doi: 10.3390/ijerph16101811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Huang C., Tang Z., Xi B., Tan W., Guo W., Wu W., Ma C. Environmental effects and risk control of antibiotic resistance genes in the organic solid waste aerobic composting system: A review. Front. Environ. Sci. Eng. 2021;15:1–12. [Google Scholar]
- 127.Liao H., Lu X., Rensing C., Friman V.P., Geisen S., Chen Z., Yu Z., Wei Z., Zhou S., Zhu Y. Hyperthermophilic composting accelerates the removal of antibiotic resistance genes and mobile genetic elements in sewage sludge. Environ. Sci. Technol. 2018;52:266–276. doi: 10.1021/acs.est.7b04483. [DOI] [PubMed] [Google Scholar]
- 128.Liu B., Yu K., Ahmed I., Gin K., Xi B., Wei Z., He Y., Zhang B. Key factors driving the fate of antibiotic resistance genes and controlling strategies during aerobic composting of animal manure: A review. Sci. Total. Environ. 2021;791:148372. doi: 10.1016/j.scitotenv.2021.148372. [DOI] [PubMed] [Google Scholar]
- 129.Iovine N.M. Resistance mechanisms in Campylobacter jejuni. Virulence. 2013;4:230–240. doi: 10.4161/viru.23753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lindsey R.L., Frye J.G., Fedorka-Cray P.J., Meinersmann R.J. Microarray-based analysis of IncA/C plasmid-associated genes from multidrug-resistant Salmonella enterica. Appl. Environ. Microbiol. 2011;77:6991–6999. doi: 10.1128/AEM.00567-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ramadan H., Jackson C.R., Frye J.G., Hiott L.M., Samir M., Awad A., Woodley T.A. Antimicrobial Resistance, Genetic Diversity and Multilocus Sequence Typing of Escherichia coli from Humans, Retail Chicken and Ground Beef in Egypt. Pathogens. 2020;9:357. doi: 10.3390/pathogens9050357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wales A.D., Davies R.H. Co-Selection of Resistance to Antibiotics, Biocides and Heavy Metals, and Its Relevance to Foodborne Pathogens. Antibiotics. 2015;4:567–604. doi: 10.3390/antibiotics4040567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Rosengren L.B., Waldner C.L., Reid-Smith R.J. Associations between antimicrobial resistance phenotypes, antimicrobial resistance genes, and virulence genes of fecal Escherichia coli isolates from healthy grow-finish pigs. Appl. Environ. Microbiol. 2009;75:1373–1380. doi: 10.1128/AEM.01253-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.