Keywords: Donnai-Barrow syndrome, endocytosis, renal proximal tubule cell, sodium-glucose cotransporter isoform 2, transcriptional profiling
Abstract
The multiligand receptors megalin (Lrp2) and cubilin (Cubn) and their endocytic adaptor protein Dab2 (Dab2) play essential roles in maintaining the integrity of the apical endocytic pathway of proximal tubule (PT) cells and have complex and poorly understood roles in the development of chronic kidney disease. Here, we used RNA-sequencing and CRISPR/Cas9 knockout (KO) technology in a well-differentiated cell culture model to identify PT-specific transcriptional changes that are directly consequent to the loss of megalin, cubilin, or Dab2 expression. KO of Lrp2 had the greatest transcriptional effect, and nearly all genes whose expression was affected in Cubn KO and Dab2 KO cells were also changed in Lrp2 KO cells. Pathway analysis and more granular inspection of the altered gene profiles suggested changes in pathways with immunomodulatory functions that might trigger the pathological changes observed in KO mice and patients with Donnai-Barrow syndrome. In addition, differences in transcription patterns between Lrp2 and Dab2 KO cells suggested the possibility that altered spatial signaling by aberrantly localized receptors contributes to transcriptional changes upon the disruption of PT endocytic function. A reduction in transcripts encoding sodium-glucose cotransporter isoform 2 was confirmed in Lrp2 KO mouse kidney lysates by quantitative PCR analysis. Our results highlight the role of megalin as a master regulator and coordinator of ion transport, metabolism, and endocytosis in the PT. Compared with the studies in animal models, this approach provides a means to identify PT-specific transcriptional changes that are directly consequent to the loss of these target genes.
NEW & NOTEWORTHY Megalin and cubilin receptors together with their adaptor protein Dab2 represent major components of the endocytic machinery responsible for efficient uptake of filtered proteins by the proximal tubule (PT). Dab2 and megalin expression have been implicated as both positive and negative modulators of kidney disease. We used RNA sequencing to knock out CRISPR/Cas9 cubilin, megalin, and Dab2 in highly differentiated PT cells to identify PT‐specific changes that are directly consequent to knockout of each component.
INTRODUCTION
The multiligand receptor megalin is a member of the low-density lipoprotein receptor family that plays an essential and complex role in kidney function. In the proximal tubule (PT), megalin and the cubilin/amnionless (AMN) receptor mediate the uptake of filtered proteins that escape the glomerular filtration barrier. Binding of the clathrin adaptor protein Dab2 to peptide motifs within the cytoplasmic tails of megalin and amnionless is essential for their endocytic uptake in clathrin-coated vesicles that form at the base of microvilli and the delivery of internalized ligands to lysosomes (1, 2). Knockout (KO) of the genes that encode megalin (Lrp2), cubilin (Cubn), or Dab2 (Dab2) results in tubular proteinuria caused by impaired recovery of filtered ligands by the PT and their consequent excretion in the urine (3).
Mutations in the LRP2 gene, which encodes megalin, result in autosomal recessive Donnai-Barrow/facio-oculo-acoustico-renal (DB/FOAR) syndrome (4). In addition to tubular proteinuria, this syndrome is characterized by facial abnormalities, hearing loss, and developmental delays. Strikingly, in contrast to normal PT cells, the subapical region of PT cells in these patients is largely devoid of the densely packed array of apical endocytic compartments needed to maintain the efficient recovery of filtered proteins (5). Similarly, KO of megalin in mice and zebrafish also impairs the formation of the apical endocytic pathway (6, 7). How megalin expression controls the elaboration of the apical endocytic pathway is unknown. The cytoplasmic tail of megalin has been demonstrated to undergo regulated intramembrane proteolysis and to regulate Lrp2 gene expression in PT cells (8, 9). However, expression of the soluble intracellular domain in place of one endogenous copy of full-length megalin had no discernable effect on the PT function or the renal transcriptome (10).
In addition to the PT, megalin is also expressed in podocytes. Megalin function in these cells may contribute to the progressive chronic kidney disease, recently documented in Lrp2 KO mice and patients with DB/FOAR (11). Although megalin KO during embryogenesis did not apparently affect nephrogenesis, KO mice rapidly began losing glomeruli, with a 19% reduction relative to control quantified as early as 8–10 wk of age (11). In addition, both human patients and Lrp2 KO mice had increased urinary excretion of kidney injury molecule-1, consistent with tubular injury and inflammation (11). Significant variability in kidney pathology and immune infiltration of Lrp2 KO mice has been noted, however, making it challenging to correlate loss of megalin expression with downstream sequelae (12).
In contrast to LRP2, mutations in CUBN or amnionless (AMN) genes (collectively resulting in Imerslund-Gräsbeck syndrome) have only modest consequences on the human health (13). In addition to its first-described function in mediating the uptake of intrinsic factor vitamin B12, cubilin also binds to several other carrier proteins, including vitamin D-binding protein, albumin, myoglobin, and transferrin. More recently, polymorphisms in the CUBN gene were found to correlate with chronic proteinuria, but these do not grossly impair renal function (14). Moreover, absence of functional cubilin expression does not apparently perturb PT apical endocytic pathway integrity (15, 16).
Although global KO of Dab2 in mice is embryonically lethal, conditional KO in kidney causes a striking reduction in PT apical endocytic compartments, similar to that observed upon megalin KO and a concomitant redistribution of megalin to the plasma membrane (17). Although Dab2 binds to NPXY and NPXF endocytic motifs within the cytoplasmic tails of megalin and amnionless, respectively, loss of megalin expression alone is sufficient to displace Dab2 from the apical surface (17). Whether the effects of Dab2 KO on the PT apical endocytic pathway are mediated through megalin is not known.
In addition to its role in endocytic uptake of megalin and cubilin/amnionless, Dab2 has been demonstrated to modulate numerous cellular signaling cascades, including the transforming growth factor-β and Wnt pathways (18–21). Strikingly, Dab2 has been implicated in both the development of chronic kidney disease as well as in successful recovery. In one study, increased expression of Dab2 was identified as a causal factor of chronic kidney disease, and reduction of Dab2 expression protected mice from chronic kidney disease (22). On the other hand, expression of Dab2 was specifically identified in a subset of cells associated with successful repair after kidney injury (23).
The rapid development of pathology in Lrp2 KO mice, which likely involves glomerular, immunological, and tubular changes, makes it challenging to identify the transcriptional changes that contribute directly to megalin-dependent functions in the PT. Consistent with this, a previous microarray study comparing mRNA expression in Lrp2 KO versus wild-type (WT) mice identified differential expression of only 19 genes, several of which are known transforming growth factor-β1 target genes (24).
Here, we used an RNA-sequencing (RNA-Seq) approach in a highly differentiated PT cell culture model to examine shared and independent changes in gene expression resulting from CRISPR/Cas9 KO of Cubn, Lrp2, or Dab2. We previously demonstrated that culture of opossum kidney (OK) cells under continuous orbital shear stress results in expansion of the apical endocytic pathway, increased ion transporter expression, and a shift to oxidative metabolism (25–27). Moreover, quantitation of mRNA abundance using droplet digital PCR revealed comparable levels of Lrp2 and Cubn transcripts in these cells compared with the mouse kidney cortex (28). As predicted, CRISPR/Cas9 KO of each of these components dramatically reduced the endocytic uptake of albumin by these cells. Moreover, KO of Lrp2 or Dab2 also resulted in nearly complete loss of apical endocytic compartments, whereas a smaller reduction in endocytic compartment abundance was observed in Cubn KO cells. Compared with the studies in animal models, this approach provides a means to identify PT transcriptional changes that result directly from KO of the target gene.
MATERIALS AND METHODS
Generation and Characterization of CRISPR/Cas9 KO Clones
All guide and PCR sequences used in this study are provided in Supplemental Table S1 (all Supplemental material is available at https://doi.org/10.6084/m9.figshare.14933340.v1). CRISPR/Cas9 KO clones in OK cells were generated using the protocol in Invitrogen’s “TrueGuide Synthetic Guide RNA User Guide” (Pub. No. MAN0017058) with minor modifications. Guide sequences were designed using our previously obtained OK cell transcriptomic data (where possible) or the Monodelphis domestica genomic database and were subcloned into the pHRS surrogate reporter vector as previously described to enable subsequent selection of gene-edited clones (29, 30). CRISPR RNA (CrRNA):transactivating crRNA (tracrRNA) duplexes were generated by annealing 2 nmol of target-specific CRISPR RNA (crRNA; TrueGuide, Thermo Fisher Scientific) and 5 nmol transactivating crRNA (TracrRNA; TrueGuide, Thermo Fisher Scientific) and then mixed in a 1:2 molar ratio with recombinant Cas9 (TrueCut Cas9 Protein v2, A36499, Thermo Fisher Scientific) to make the ribonucleoprotein (RNP) complex (75 pmol Cas9:150 pmol sgRNA). OK cells (1 × 106 cells per electroporation, RRID:CVCL_0472, OK-P subclone originally obtained from Moshe Levi, Georgetown University) in 100 µL of electroporation solution (MIR 50112, Ingenio Mirus) were combined with the RNP complex and 2.5 µg of pHRS and electroporated in an Amaxa electroporator (Lonza Nucleofector II) using program T-020. Control clones were generated from OK cells electroporated with recombinant Cas9 and empty pHRS vector. Cells were treated for 48 h with hygromycin B starting 36–48 h postelectroporation, and clones were isolated by single-cell dilution. DNA was isolated from individual CRISPR clones using QuickExtract DNA extraction solution (Lucigen.com) following the manufacturer’s protocol and amplified using gene-specific primers. PCR products were purified using the Wizard SV gel and PCR clean-up system (A9281, Promega), ligated into the pGEM-T Easy Vector System (A1360, Promega), and then transformed. DNA was isolated from single colonies and submitted to the University of Pittsburgh Genomics Research Core for Sanger sequencing. The sequences were analyzed in CLC Genomics Workbench. The sequences from at least eight bacterial colonies were used to confirm allele sequences.
For experiments, 4 × 105 cells were seeded onto 12-mm Transwell permeable supports (No. 3401, Costar) in 12-well dishes. After overnight incubation, the filters were transferred to an orbital platform shaker in the incubator and rotated at 146 rpm for 72 h with daily medium changes. Albumin uptake and Western blot analysis were performed as previously described using anti-megalin antibody generously provided by Dr. Daniel Biemesderfer and Dr. Peter Aronson (Yale University, MC-220, 1:20,000; 9), polyclonal anti-cubilin antibody (No. 27445, validated in Ref. 28, 1:5,000), and anti-Dab-2 antibody (No. 12906, Cell Signaling, 1:2,000).
RNA Sequencing
RNA-Seq was performed essentially as previously described by Ren et al. (27). Briefly, 4 × 105 control, Cubn KO, Lrp2 KO, and Dab2 KO cells were plated at superconfluence on 12-mm permeable supports in 0.5-mL apical and 1.5-mL basolateral DMEM-F12 medium (D6421, Sigma) with 10% FBS (Atlanta Biologicals) and 5 mM GlutaMAX (Gibco). After 18 h (time = 0 h), the filters were transferred to an orbital platform shaker rotating at 146 rpm, and the incubation continued for 72 h with daily medium changes. Cells were collected using Accutase (BD Biosciences), and RNA was extracted using the Ambion PureLink RNA mini kit (Thermo Fisher Scientific). RNA from three independent experiments was sequenced for each clone. Library preparation and RNA-Seq were performed by the Health Sciences Sequencing Core at UPMC Children’s Hospital of Pittsburgh. Sequencing was carried out on a NextSeq 500 (Illumina) to generate 75-bp paired-end reads. An average of 40 million reads was analyzed per sample. Raw sequence reads were trimmed of adapter sequences using Cutadapt (31) and mapped to the Monodelphis domestica reference genome (MonDom5) using TopHat2 (32), allowing for a base-pair mismatch value of six to enable cross-species alignment. Reads aligned to mitochondrial genes and rRNA were removed before raw count values were calculated with the Subread package feature Counts (33). Libraries were normalized using the total of transcript-aligned sequence reads. Sequencing files were deposited in the Gene Expression Omnibus (GSE176090). An interactive spreadsheet enabling a comparison of normalized gene quantification for all samples is provided in Supplemental Table S2.
Identification of Differentially Expressed Genes and Pathway Analysis
The data provided in Supplemental Table S2 were filtered to identify differentially expressed genes as described in each table. The low-expression filter was set at 350 to exclude genes expressed at 0.1% of average maximal levels for all analyses. The resulting gene lists were analyzed using the Kyoto Encyclopedia of Genes and Genomes (KEGG) and the Gene Ontology cellular component (CC) functional annotation tools available on the Database for Annotation, Visualization and Integrated Discovery (DAVID) version 6.8 Bioinformatics Resource Tool (david.ncifcrf.gov).
Generation of Stably Transfected Cell Lines
Parental and Lrp2 KO OK cells were transfected with expression plasmids encoding bovine RAB guanine nucleotide exchange factor 1-green fluorescent protein (Rabgef1-GFP, also known as Rabex5-GFP; provided by Juan Bonifacino, National Institutes of Health) and hemagglutinin (HA)-tagged murine stromal membrane-associated protein 1 (Smap1; provided by Shunsuke Kon, Tokyo University of Science). After selection in 400 µg/mL geneticin, individual colonies were isolated using cloning cylinders and verified using immunofluorescence. Cells were incubated with 40 µg/mL Alexa Fluor-647 albumin for 15 min, and cell-associated albumin was assessed by spectrofluorimetry or fixed and processed for confocal imaging to detect albumin uptake and the stably expressed protein.
Lrp2 EMX-Cre Mice
All animal protocols conform to the National Institutes of Health “Guide for the Care and Use of Laboratory Animals” and were approved by the University of Pittsburgh Institutional Animal Care and Use Committee. Lrp2 lox/lox mice (34; originally provided by Thomas Willnow, Max Delbrück Center for Molecular Medicine) crossed with EMX-Cre mice were provided by Cecilia Lo (University of Pittsburgh). Mice with Lrp2 conditional kidney deletion were generated as follows: mice with loxP recombination sites introduced into the megalin gene locus were bred with mice expressing Cre recombinase under control of the EMX promoter. Experimental mice and littermates were generated in a C57Bl/6 background. Kidneys from 16-wk-old female mice were formalin-fixed for microscopy. Sections were cut by the Pitt Biospecimen Core at the University of Pittsburgh and stained with hematoxylin and eosin and Masson’s trichrome.
Quantitative PCR of Lrp2 EMX-Cre Mice
Kidneys were harvested from six control and six Lrp KO cohorts of male mice between 12 and 37 wk of age and flash frozen. RNA was isolated with TRIzol reagent (Ambion), and cDNA was generated with the RevertAid reverse transcriptase kit (Thermo Fisher Scientific). Dye-based quantitative PCR was performed with iTAQ Universal SYBR Green Supermix (Bio-Rad Laboratories), and the primer sets are provided in Supplemental Table S1 (35, 36). Results were normalized to β-actin (Actb) and expression was determined with the comparative 2−ΔΔCt method (where Ct is threshold cycle; 37). Similar data were obtained using Gapdh as a housekeeping gene.
Immunofluorescence of Mouse Kidney Cortex Sections
Mouse kidneys (Lrp2WT/WT; EMX-CreWT/WT and Lrp2flox/flox; EMX-Cremut/WT) were fixed in 4% paraformaldehyde overnight at 4°C. The tissue was quenched (0.2 M ammonium chloride), dehydrated in 30% sucrose, and embedded in resin at −20°C. Cryostat sections (8 µm) were placed on slides, rehydrated in PBS for 30 min, permeabilized with 0.1% Triton X-100 for 10 min, and blocked with 1% BSA for 1 h at ambient temperature (room temperature). Sections were incubated overnight at 4°C with rabbit anti-megalin antibody (1:1,000, MC-220), washed three times for 5 min each in wash buffer (1× PBS and 0.5% BSA), and incubated for 1 h at room temperature with Alexa Fluor 488 goat anti-rabbit IgG (1:500, A11034, Invitrogen) and Acti-Stain 555 phalloidin (1:140, PHDH1, Cytoskeleton). Sections were rinsed in wash buffer as before and incubated for 10 min with TO-PRO-3 Iodide (642/661, T3605, Invitrogen). After being washed three times in wash buffer and once in PBS, sections were mounted using Prolong Gold antifade (P10144, Invitrogen). Digital images were acquired using a Leica TCA SP5 confocal microscope with a ×63 oil immersion objective.
RESULTS AND DISCUSSION
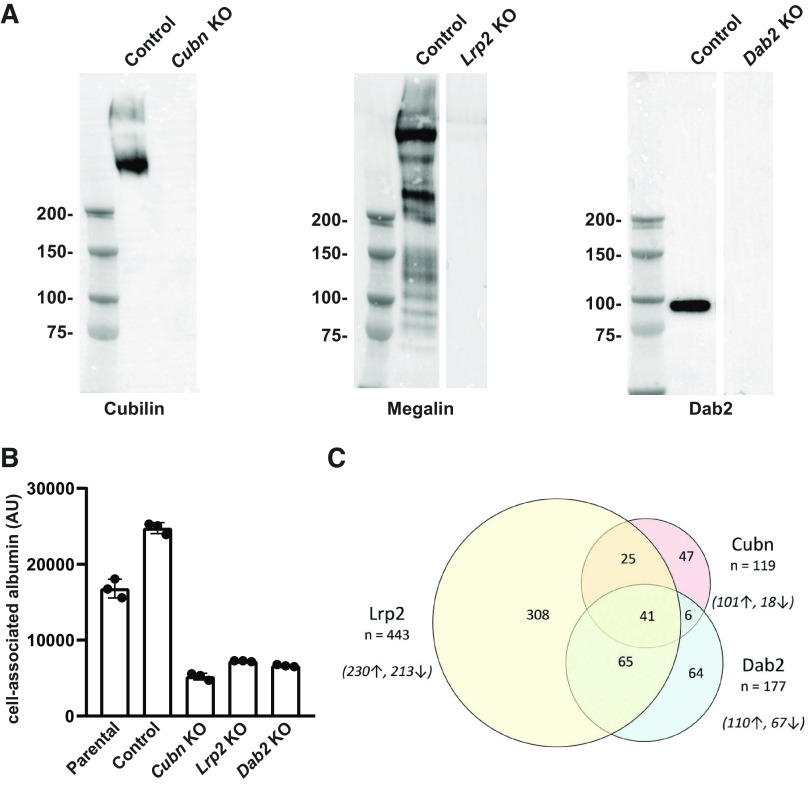
We used CRISPR/Cas9 technology to generate KO clones of Cubn, Lrp2, and Dab2 in OK cells. A control clone was isolated from cells transfected without guide sequences. Western blot analysis confirmed that each KO clone lacked detectable expression of the targeted protein (Fig. 1A). Similar to our previous observations using siRNA knockdown (28), albumin uptake in all clones was significantly reduced compared with control (Fig. 1B).
Figure 1.
Western blot analysis and albumin uptake in CRISPR/Cas9 knockout (KO) clones. A: equal amounts of control, cubilin (Cubn) KO, megalin (Lrp2) KO, and Dab2 (Dab2) KO lysates were blotted for the indicated proteins. The migration of molecular mass standards (in kDa) is shown on the left of the blots. B: parental opposum kidney cells, CRISPR/Cas9 control, and KO clones were incubated with 40 µg/mL Alexa Fluor-647 albumin for 30 min, washed, and then solubilized. Cell-associated albumin was quantified by spectrofluorimetry. Means ± SD of triplicate samples are plotted. C: Venn diagram showing the overlap of differentially upregulated (↑) and downregulated (↓) genes in Cubn KO, Lrp2 KO, and Dab2 KO cells. AU, arbitrary units.
Differential Gene Expression in CRISPR/Cas9 KO Clones
We performed RNA-Seq analysis on control and KO cell clones to identify selective and coordinate changes in gene expression. An interactive spreadsheet containing the normalized gene expression, as well as ratiometric changes relative to the control clone, is provided in Supplemental Table S2. Lrp2 was the most abundantly represented gene in the control clone, whereas Cubn transcripts ranked 12th. Dab2 transcripts were also highly expressed, ranking 209th in control cells (Supplemental Table S2). Knockout of Cubn, Lrp2, or Dab2 resulted in a significant and selective reduction in transcripts for the targeted gene with no concomitant change in the expression of the other two mRNAs (Supplemental Table S2).
Using a relatively stringent cutoff (±70% of control values), we then identified genes whose expression was differentially upregulated or downregulated in each clone. As shown in Fig. 1C, Lrp2 KO cells had the greatest number of differentially expressed genes, with 230 upregulated genes and 213 downregulated genes compared with control. In contrast, Cubn KO and Dab2 KO cells had only 101 and 110 upregulated genes and 18 and 67 downregulated genes, respectively, compared with control cells.
Next, we performed pairwise comparisons to identify genes, whose expression was selectively upregulated or downregulated relative to the other KO clones. To this end, we first selected those genes in the “comparison” KO clone, whose expression was unchanged (±10%) relative to control cells and then queried this list to identify upregulated and downregulated genes (±70%) in the other KO cell lines (Table 1). This analysis revealed that most of the genes, whose expression was differentially regulated by Cubn KO or Dab2 KO, were also represented within the Lrp2 KO data set. There were only four upregulated genes and two downregulated genes (not including the gene targeted by CRISPR/Cas9) in Cubn KO and Dab2 KO cells that were expressed at control levels in Lrp2 KO cells. Similarly, there were no downregulated genes in Cubn KO cells relative to Dab2 KO other than Cubn itself. Thus, KO of Lrp2 has a considerably greater effect on gene expression compared with Cubn or Dab2 KO.
Table 1.
Pairwise comparison of upregulated and downregulated transcripts in CRISPR/Cas9 KO clones
| Comparison Set | Up/Downregulated Transcripts Relative to the Comparison Set |
||
|---|---|---|---|
| Cubn KO | Lrp2 KO | Dab2 KO | |
| Control (1.0) | 101/18 | 230/212 | 110/66 |
| Cubn KO (0.9–1.1) | 38/80 | 3/16 | |
| Lrp2 KO (0.9–1.1) | 4/2 | 4/2 | |
| Dab2 KO (0.9–1.1) | 19/0 | 44/63 | |
Transcripts with an average count of <350 (∼0.1% of the highest expressed gene) were excluded from the analysis. To compare knockout (KO) clones against one another, we first generated comparison sets by collecting “unaffected” transcripts that were expressed at 0.9- to 1.1-fold of control values in each KO clone. Next, we identified transcripts in KO clones that were expressed at ≥1.7-fold greater/less than these values. The number of upregulated/downregulated transcripts is shown for each pairwise comparison. The gene that was knocked out in each CRISPR/Cas9 clone was not included in the counts of downregulated transcripts [e.g., the only transcript downregulated in cubilin (Cubn) KO cells relative to Dab2 (Dab2) KO cells was cubilin itself]. Lrp2, megalin.
Identification of Coordinately Regulated Genes and Pathways in Lrp2 KO and Dab2 KO Clones
KO of either Lrp2 or Dab2 results in dramatic reductions in the density of subapical compartments. We reasoned that transcripts that were coordinately upregulated or downregulated in Lrp2 KO and Dab2 KO cells would include genes that regulate apical endocytic pathway integrity as well as genes involved in cellular responses to the impairment in endocytosis. The effect of progressively increasing the cutoff value from ±30% to ±70% of control on the number of upregulated and downregulated transcripts is provided in Supplemental Fig. S1. In all cases, there were more transcripts that were coordinately upregulated in the two KO clones than downregulated. Setting the cutoff to ±30% of control yielded the 174 downregulated genes and 298 upregulated genes provided in Supplemental Tables S3 and S4. KEGG pathway analysis of the combined list of upregulated and downregulated proteins highlighted pathways consistent with the known functions of megalin in PT cell endocytosis, signaling, and metabolism, including lysosomal, phosphatidylinositol 3-kinase (PI3K)-Akt, metabolic, and estrogen signaling pathways (Table 2; 38–40).
Table 2.
KEGG analysis of coordinately expressed transcripts in Lrp2 knockout and Dab2 knockout cells
| Term | Count | P Value | Genes |
|---|---|---|---|
| Sphingolipid signaling pathway | 11 | 0.00087 | PPP2CA, NRAS, CERS6, SPTLC2, PPP2R3C, SMPD1, GNAI3, AKT1, PPP2R5C, S1PR2, SGPP1 |
| Sphingolipid metabolism | 7 | 0.00102 | ARSA, CERS6, SPTLC2, SMPD1, B4GALT6, GLA, SGPP1 |
| Lysosome | 10 | 0.00291 | MFSD8, ARSA, NPC1, ATP6AP1, SORT1, SMPD1, NAGA, ACP5, AP4B1, GLA |
| Estrogen signaling pathway | 8 | 0.00993 | NRAS, HSP90AA1, CREB3L3, MMP2, GNAI3, AKT1, SOS2, HBEGF |
| Axon guidance | 9 | 0.01354 | EFNA1, SEMA5A, ITGB1, NRP1, NRAS, EPHA6, SEMA3C, CFL2, GNAI3 |
| Mineral absorption | 5 | 0.01459 | SLC6A19, HMOX1, MT2, ATP1A1, SLC5A1 |
| TNF signaling pathway | 8 | 0.01712 | NFKBIA, CXCL10, EDN1, CASP8, CREB3L3, CCL20, AKT1, TNFAIP3 |
| Prostate cancer | 7 | 0.02040 | NFKBIA, NRAS, HSP90AA1, CREB3L3, AKT1, IGF1, SOS2 |
| Ascorbate and aldarate metabolism | 4 | 0.02732 | ALDH3A2, MIOX, UGDH, RGN |
| Arginine and proline metabolism | 5 | 0.03116 | ALDH3A2, GAMT, ARG2, P4HA2, L3HYPDH |
| Metabolic pathways | 42 | 0.03291 | CDA, DDC, HAAO, MSMO1, PYGL, PIGV, TM7SF2, EXTL2, MTHFD1L, SPTLC2, MAN1A2, SMPD1, DSE, PHGDH, HMGCS2, CEPT1, MGAT2, PTGDS, NDUFV1, GAMT, GALNT7, FDPS, ARG2, PLA2G12B, CERS6, HSD3B2, HMGCS1, ATP6AP1, AGXT2, URAD, ACSL5, AMPD2, COQ6, ALDH3A2, UGDH, SQLE, UCK2, P4HA2, DPYD, RGN, B4GALT6, HPD |
| Biosynthesis of antibiotics | 11 | 0.03678 | ALDH3A2, FDPS, SQLE, ARG2, HMGCS1, RGN, PHGDH, AMPD2, MSMO1, HMGCS2, TM7SF2 |
| Phosphatidylinositol 3-kinase-Akt signaling pathway | 15 | 0.04826 | ITGB1, HSP90AA1, COL27A1, ITGB4, LAMA3, IGF1, PPP2R5C, EFNA1, PPP2CA, NRAS, CREB3L3, PPP2R3C, AKT1, SGK1, SOS2 |
Transcripts that were coordinately upregulated or downregulated by 30% (0.75–1.3 times control) were collected from Supplemental Table S2, and the merged list was subjected to Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis. Pathways with a P value of <0.05 are shown along with the number and names of the genes represented. Dab2, Dab2; Lrp2, megalin.
Several of the KEGG pathways identified using the merged list of transcripts were also represented in separate KEGG analyses of the upregulated and downregulated gene cohorts (Supplemental Tables S5 and S6). Comparing the genes that were upregulated versus downregulated in these pathways provided additional insights into the consequences of Dab2 or Lrp2 KO. For example, all of the genes represented in the PI3K-Akt pathway were upregulated except those encoding integrin [integrin-β1 (Itgb1) and integrin-β4 (Itgb4)] and laminin (Lama3) subunits and insulin-like growth factor-1 (Igf1). IGF-1 binding to integrins, including the Itgb4 product, is essential for IGF-1-mediated signaling (41). Thus, this module for PI3K-Akt signaling appears to be coordinately dampened in Lrp2 KO and Dab2 KO cells.
Within the lysosome pathway, expression of genes that encode enzymes involved in carbohydrate and lipid hydrolysis was uniformly downregulated [Arylsulfatase A (Arsa), acid sphingomyelinase (Smpd1), α-d-galactosidase A (Gla), and α-N-acetylgalactosaminidase (Naga)], along with proteins required for lysosomal acidification (Atp6ap1, which encodes an accessory subunit of the proton-transporting vacuolar-ATPase protein pump), cholesterol efflux [intracellular cholesterol transporter 1 (Npc1)], and transport of other molecules (Mfsd8, which encodes the solute transporter CLN7). In contrast, sortilin (Sort1) and AP-4 complex subunit-β1 (Ap4b1), which encode components of the machinery that deliver cargo to lysosomes, were upregulated.
When we compared differentially expressed genes in metabolic pathways, we noted increased expression of several proteins required for the transport of cholesterol and bile salts [ATP-binding cassette subfamily B member 11 (Abcb11), fatty acid-binding protein 1 (Fabp1), apolipoprotein A-I (Apoa1), apolipoprotein A-II (Apoa2), and apolipoprotein M (Apom)] as well as genes required for cholesterol and steroid biosynthesis [transmembrane 7 superfamily member 2 (Tm7sf2), 3-hydroxy-3-methylglutaryl-CoA synthase 1 (Hmgcs1), 3-hydroxy-3-methylglutaryl CoA synthase 2 (Hmgcs2), farnesyl diphosphate synthase (Fdps), hydroxy-Δ-5-steroid dehydrogenase, 3β- and steroid Δ-isomerase (Hsd3b2), and squalene epoxidase (Sqle)] in Lrp2 KO and Dab2 KO cells. Together with the reduction in transcripts for lysosomal and other lipid hydrolases [lipoprotein lipase (Lpl), aldehyde dehydrogenase 3 family member A2 (Aldh3a2), and sphingosine-1-phosphate phosphatase 1 (Sgpp1)], the data suggest that compared with control or Cubn KO cells, Lrp2 KO and Dab2 KO cells may rely more on endogenous synthesis to maintain lipid and cholesterol pools rather than recovery of these nutrients from the medium. A caveat to this conclusion is that OK cells cultured in serum may have access to megalin-binding ligands not normally present at appreciable levels in the ultrafiltrate. In this regard, some aspects of the metabolic or signaling transcriptional profile of control OK cells might be more “abnormal” than those of our CRISPR/Cas9 KO clones.
Inflammatory Response Gene Expression in Lrp2 and Dab2 KO Cells
In aggregate, the genes affected by KO of Lrp2 and/or Dab2 are consistent with changes in NF-κB, peroxisome proliferator-activated receptor-ɣ, and other signaling pathways that exert both pro- and anti-inflammatory responses in cells. Among the most highly upregulated genes in Lrp2 KO cells that were also (slightly) upregulated in Dab2 KO cells were those encoding prostaglandin D2 synthase (Ptgds; upregulated 27.64- and 2.37-fold in Lrp2 and Dab2 KO cells, respectively) and lactoferrin (Ltf; upregulated 21.98- and 1.37-fold in Lrp2 and Dab2 KO cells, respectively). 15-Deoxy-Δ12,14-prostaglandin J (PGJ2) is a nonenzymatically generated metabolite of PGD2 that activates peroxisome proliferator-activated receptor-ɣ and independently blocks NF-κB activation (42, 43). Similarly, lactoferrin expression in the kidney may play a role in immune defense, and lactoferrin-mediated inhibition of the Akt pathway has been shown to play a renoprotective role in acute kidney injury (44, 45). Transcripts encoding serum glucocorticoid kinase 1 (Sgk1), which also regulates MAPK and NF-κB signaling, were also upregulated (1.33- and 1.36-fold in Lrp2 and Dab2 KO cells, respectively; 46). We also observed a significant reduction in transcripts encoding dual-specificity phosphatase 5 (Dusp5; 0.65- and 0.58-fold of control in Lrp2 and Dab2 KO cells, respectively), a phosphatase that targets ERK and NF-κB signaling pathways to mediate anti-inflammatory responses (47). In addition, among the most downregulated genes was TNF-α-induced protein 3 (Tnfaip3, also known as A20; 0.20- and 0.24-fold of control in Lrp2 and Dab2 KO cells, respectively), which encodes a zinc finger protein and ubiquitin-editing enzyme that inhibits NF-κB activation and dampens inflammatory responses in injured kidneys and PT cells (48–50). On the other hand, we also observed a reduction in transcripts for a proinflammatory chemokine, chemokine (C-C motif) ligand 20 (Ccl20; 0.25- and 0.28-fold of control in Lrp2 and Dab2 KO cells, respectively). Many of these transcripts were also coordinately altered in Cubn KO cells, where there is a partial blockade of endocytic pathway integrity. We speculate that changes in these signaling pathways may contribute to the progressive chronic kidney disease observed in patients with Donnai-Barrow syndrome and Lrp2 KO mice (11).
Identification of Megalin-Dependent Genes Not Affected by Dab2 KO
We used a similar approach as that described earlier to identify genes whose expression was selectively altered in Lrp2 KO cells (±50%) but not in Dab2 KO (±25%) or Cubn KO (±10%) cells. This analysis yielded the annotated list of 65 upregulated genes and 116 downregulated genes provided in Supplemental Table S7. KEGG pathway analysis of this list highlighted PI3K-Akt signaling, endocytosis, and insulin and Rap1 signaling pathways (Supplemental Table S8). Cellular component terms retrieved by Gene Ontology analysis were primarily related to endocytic traffic and transcription (Supplemental Table S9).
Interestingly, the most highly downregulated gene that was selectively affected in Lrp2 KO cells was Slc5a2, which encodes Na+-glucose cotransporter isoform 2 (Sglt2). Slc5a2 transcripts were expressed at only 15% of control levels in Lrp2 KO cells but were present at 97% and 83% of control levels in Cubn KO and Dab2 KO cells, respectively (Supplemental Table S2). Inhibition of SGLT2 improves blood glucose levels in patients with diabetes and is also increasingly recognized as an effective treatment to reduce the risk and severity of acute kidney injury via independent mechanisms (51). We also observed a reduction in MLX interacting protein-like transcript (Mlxipl; 58% of control), which encodes the glucose-sensing transcription factor carbohydrate response element-binding protein (ChREBP). ChREBP is modulated by O-GlcNAcylation, which, in turn, is responsive to cellular glucose levels (52, 53). Levi and colleagues have shown that Sglt2 inhibitors mitigate renal lipid accumulation in a diabetic mouse model and that this is mediated in part via ChREBP (55, 56). Although data are conflicting, some reports have suggested that KO of Lrp2 may be protective against kidney injury (39, 57, 58). Quantitative PCR confirmed a reduction in Slc5a2 and Mlxipl transcript levels in our Lrp2 KO cells and an additional Lrp2 KO clone with distinct allelic mutations (Fig. 2). Our data suggest the possibility that a reduction in Sglt2 and/or ChREBP could contribute to these effects in Lrp2 KO models.
Figure 2.
Quantitative PCR confirmation of reduced Slc5a2 and MLX interacting protein-like (Mlxipl) transcripts in megalin (Lrp2) knockout (KO) cells. Slc5a2 and Mlxipl transcripts were quantified by quantitative PCR in control, cubilin (Cubn) KO, Lrp2 KO, and Dab2 (Dab2) KO cells. A second Lrp2 KO clone (Lrp2 KO cl3) with distinct allelic mutations was included to ensure that changes were not clone specific. Data were normalized to control cell values.
Selective Changes in Membrane Trafficking Genes in Lrp2 KO Cells
In contrast to transcripts that were coordinately altered in Dab2 KO and Lrp2 KO cells, the list of genes that were selectively affected by Lrp2 KO also included many that encode regulatory components important for membrane traffic (Table 3). The majority of these transcripts were downregulated in Lrp2 KO cells and encoded proteins involved in clathrin-dependent endocytosis [e.g., synaptosome associated protein 91 (Snap91; which encodes the clathrin adapter AP180/CALM), Huntingtin interacting protein 1 (Hip1), stonin-2 (Ston2), and small Arfgap1 (Smap1; an Arf6 GTPase activating protein)], early endosome function [e.g., Rabgef1 (the GTP exchange factor for Rab5, also known as Rabex5), syntaxin 7 (Stx7), and the rab effector microtubule-associated monooxygenase, calponin and LIM domain containing 1 (Mical1)], endosomal sorting [e.g., sortin nexin 3 (Snx3), sortin nexin 14 (Snx14), sortin nexin 10 (Snx10), vesicle trafficking 1 (Vta1), Rab23, and Rab32], and lysosomal delivery and autophagosome formation [e.g., vesicle-associated membrane protein 8 (Vamp8) and autophagy-related 5 (Atg5)]. We also observed reductions in the expression of cargo receptors that function in trans-Golgi network to endosome sorting, including insulin-like growth factor-2 receptor (Igf2r, which encodes the cation-independent mannose 6-phosphate receptor) and sortilin-related receptor (Sorl1). These constituents may represent essential components of the PT apical endocytic pathway required to efficiently recover filtered proteins.
Table 3.
Membrane trafficking genes whose expression is altered in Lrp2 KO cells
| Lrp2 KO/Control | Gene | Protein |
|---|---|---|
| 0.27 | CLIP2 | CAP-Gly domain containing linker protein 2 |
| 0.49 | HIP1 | Huntingtin interacting protein 1 |
| 0.50 | COPS9 | COP9 signalosome subunit 9 |
| 0.54 | SNAP91 | Clathrin coat-associated protein AP180 |
| 0.55 | STXBP5 | Syntaxin-binding protein 5 |
| 0.57 | RAB32 | RAB32, member of the RAS oncogene family |
| 0.57 | RABGEF1 | RAB guanine nucleotide exchange factor 1 |
| 0.58 | SORL1 | Sortilin-related receptor 1 |
| 0.59 | STX7 | Syntaxin 7 |
| 0.59 | IGF2R | Insulin-like growth factor 2 receptor |
| 0.60 | IPCEF1 | Interaction protein for cytohesin exchange factors 1 |
| 0.62 | VTA1 | Vesicle trafficking 1 |
| 0.62 | SMAP1 | Small ArfGAP 1 |
| 0.62 | HACE1 | HECT domain and ankyrin repeat containing E3 ubiquitin protein ligase 1 |
| 0.62 | RAB23 | RAB23, member of the RAS oncogene family |
| 0.64 | MYO7A | Myosin VIIA |
| 0.65 | SNX3 | Sorting nexin 3 |
| 0.66 | MICAL1 | Microtubule-associated monooxygenase, calponin and LIM |
| 0.66 | ATG5 | Autophagy related 5 |
| 0.67 | SNX14 | Sorting nexin 14 |
| 1.52 | SNX10 | Sorting nexin 10 |
| 1.60 | CLINT1 | Clathrin interactor 1 |
| 1.62 | ARAP3 | ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 3 |
| 1.63 | SH3KBP1 | SH3 domain containing kinase binding protein 1 |
| 1.97 | PIK3R1 | Phosphoinositide 3-kinase regulatory subunit 1 |
| 2.30 | VAMP8 | Vesicle-associated membrane protein 8 |
| 5.63 | SYNPR | Synaptopodin 2 |
Genes that encode proteins with known functions in membrane traffic identified in the list of transcripts that are selectively downregulated or upregulated in megalin (Lrp2) knockout (KO) cells are listed, along with the ratio of expression relative to control cells.
The absence of changes in Rab proteins with known roles in apical endocytic traffic is consistent with our recent observations that protein levels of Rab11a do not correlate with endocytic capacity (26). We reasoned that small changes in the expression of regulatory proteins that we identified, such as guanine nucleotide exchange factors and GTPase-activating proteins, which regulate the activity of Rabs and other GTPases, may have a greater effect on endocytic traffic than changes in Rab proteins themselves. To this end, we selected two candidates of interest and examined whether heterologous expression of these could restore endocytic traffic in Lrp2 KO cells. Rabgef1 (aka Rabex-5) activates Rab5 as well as other Rab proteins (59–61) and has also been suggested to function as a negative regulator of Ras activation (62). In complex with rabaptin, Rabgef1 promotes maturation of early endosomes and inhibits the generation of Rab4-positive recycling vesicles (63). We generated stable transfectants of parental and Lrp2 KO cells stably expressing bovine Rabgef1-GFP and evaluated the effect on albumin endocytosis by immunofluorescence and spectrofluorimetry. As shown in Fig. 3, A and C, heterologous expression of Rabgef1 did not appreciably affect uptake in either cell line.
Figure 3.
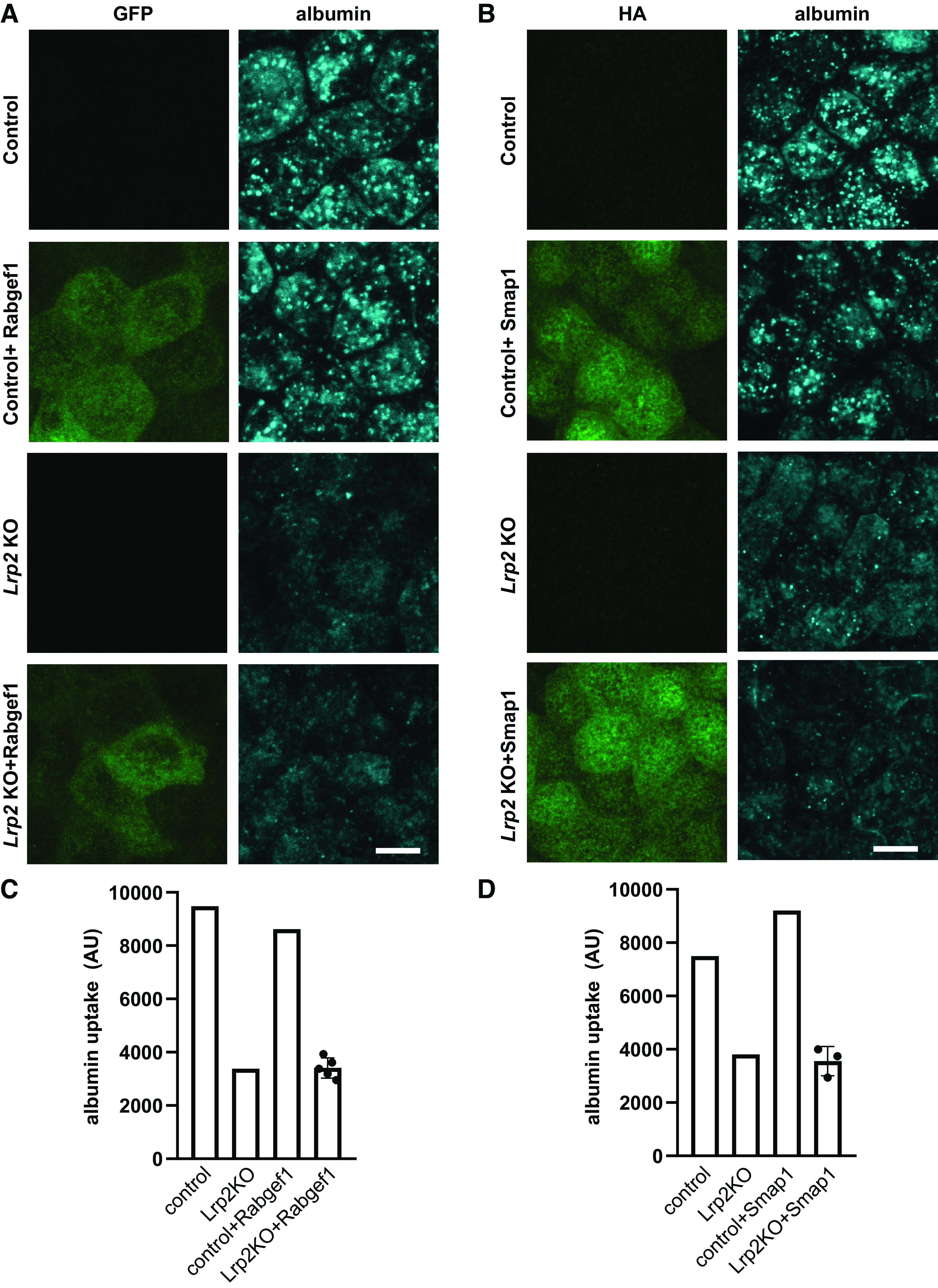
Heterologous expression of RAB guanine nucleotide exchange factor 1 (Rabgef1) or stromal membrane-associated protein 1 (Smap1) does not restore endocytic traffic in megalin (Lrp2) knockout (KO) cells. Control and Lrp2 KO cells were stably transfected with constructs encoding green fluorescent protein (GFP)-Rabgef1 (A) and hemagglutinin (HA)-Smap1 (B). Cells were incubated with Alexa Fluor-647 albumin for 15 min, washed, and then fixed. Cells in B were labeled using anti-HA antibody. Images were captured by confocal microscopy and maximum projections of representative fields are shown. Scale bars = 10 µm. Albumin uptake was assessed by spectrofluorimetry in control and Lrp2 KO cells transfected with GFP-Rabgef-1 (C) or HA-Smap1 (D). The individual points plotted for transfected Lrp2 KO cells represent uptake in four independent clones for Rabgef1 and three independent clones for Smap1. Similar results were obtained in two experiments. AU, arbitrary units.
We also examined the effect of expressing Smap1 on endocytic uptake in Lrp2 KO and control cells. Smap1 is thought to have a selective role in Arf6 activation at the plasma membrane, whereas other Arf GTPase-activating proteins function at distinct intracellular sites (64). Arf6 is expressed at the apical membrane of PT cells and is known to regulate apical endocytosis in these cells (65, 66). Smap1 binds to clathrin heavy chain (67) and activates phosphatidylinositol 5-kinase to enable adaptor protein complex-2 (AP-2) recruitment. Both overexpression and knockdown of SMAP1 profoundly inhibited endocytosis of transferrin receptor in HeLa cells (67). Albumin uptake in control cells stably expressing HA-tagged Smap1 appeared slightly higher than in untransfected cells, but uptake in Lrp2 KO cells was unaffected in multiple clones (Fig. 3, B and D). These data confirm a role for Arf6 in apical endocytic uptake in PT cells and also suggest that reduced expression of Smap1 in Lrp2 cells is unlikely to be the critical factor in the loss of endocytic pathway integrity. Given the complexity in generating and maintaining the membrane flux necessary to maintain high capacity and efficient endocytosis in PT cells, it is perhaps not surprising that heterologous expression of individual proteins in Lrp2 KO cells did not restore endocytic function.
Gene Expression in Lrp2 KO Mice
We used quantitative PCR to quantify transcript changes in control and Lrp2 EMX-Cre mice. Immunofluorescence staining of kidney sections confirmed high-efficiency (>95%) KO of Lrp2 in PTs (Fig. 4A). Hematoxylin and eosin staining revealed intraluminal casts, and Masson’s trichrome stain confirmed the presence of immune infiltration and renal fibrosis only in KO mice (Fig. 4B). By quantitative PCR, we reproducibly observed decreased Lrp2 (Fig. 4C) and Slc5a2 (P = 0.0158; Fig. 4D) transcripts in Lrp2 KO mice, consistent with Lrp2 KO OK cells. However, we did not detect significant differences in Rabgef1 (Fig. 4E), Smap1 (Fig. 4F), Mlxipl (Fig. 4G), Ptgds (Fig. 4H), or Ltf (Fig. 4I) transcripts in Lrp2 KO compared with control mouse kidney cortexes. The incomplete KO of megalin in mice compared with our CRISPR/Cas9 KO clones may contribute to the differences we observed. Moreover, expression of these proteins in other cell types in the kidney cortex or their selective expression in subsegments of the PT could obscure detection of physiologically relevant changes. For example, deep sequencing of microdissected mouse nephron segments found that Rabgef1 transcripts are expressed at much higher levels in the distal tubule than the PT (68). In addition, variability in expression across the PT may obscure physiologically relevant changes in PT subsegments. In this regard, it may be relevant that only 8% of mouse PT Mlxipl transcripts are expressed in the S1 segment (68). Together, these data highlight the complexity of RNA-Seq analysis to identify cell-specific alterations in kidney function and also confirm the disease pathology previously described in Lrp2 KO mice (11, 12).
Figure 4.

Transcriptional changes in megalin (Lrp2) knockout (KO) mouse kidneys. A: kidney sections from 16-wk-old female Lrp2WT/WT, EMX-CreWT/WT and Lrp2flox/flox, EMX-Cremut/WT mice were stained to detect megalin (green), actin (red), and nuclei (blue). Scale bars = 25 µm. B: hematoxylin and eosin staining (top) and Masson’s trichrome staining (bottom) of kidney sections from the same mice shown in A revealing tubular dilation and intraluminal casts (black arrows), immune infiltration (black arrowhead), and fibrosis (white arrowhead) consistent with injury in Lrp2 KO mice (arrows). Scale bars = 100 µm. Quantitative PCR of kidney lysates was used to compare transcript levels of the indicated genes in six wild-type (WT) and six Lrp2 KO male mice between 12 and 37 wk of age. A two-tailed unpaired t test was used to assess significance. C: Lrp2 (****P < 0.001). D: Slc5a2 (*P = 0.0158). E: RAB guanine nucleotide exchange factor 1 (Rabgef1; P = 0.0927). F: stromal membrane-associated protein 1 (Smap1; P = 0.4303). G: MLX interacting protein like (Mlxipl; P = 0.7831). H: prostaglandin D2 synthase (Ptgds; P = 0. 0355). I: lactoferrin (Ltf; P = 0.0416).
Why Does KO of Megalin Have a Greater Transcriptional Effect than Dab2 KO?
Compared with loss of Dab2 or Cubn expression, KO of Lrp2 had a much more pronounced transcriptional footprint, resulting in significant changes in mRNAs encoding transcription factors, ion transporters, and proteins that regulate membrane trafficking. Given the essential need for megalin and Dab2 expression in maintaining the apical endocytic pathway, it is perhaps surprising that we identified very few transcripts encoding membrane trafficking proteins, whose expression was altered in both Lrp2 KO and Dab2 KO cell lines. One possibility to explain this difference is that KO of megalin versus Dab2 may have discoordinate effects on the distribution of receptors and other cellular components to differentially affect spatial signaling. Compared with control or Cubn KO cells, we observed a dramatic accumulation of clathrin (and the loss of Dab2 staining) at the apical surface of Lrp2 KO cells. In contrast, no clathrin could be detected at the apical surface of Dab2 KO cells (Y. Rbaibi, K. R. Long et al., unpublished observations). We hypothesize that clustering of receptor tyrosine kinases, G protein-coupled receptors, or other signaling molecules in clathrin-coated pits may lead to aberrant signaling in Lrp2 KO cells (69).
Perspectives and Significance
The use of CRISPR/Cas9 technology in a well-differentiated cell culture model makes it easier to identify PT-specific responses to Lrp2 KO. Our data demonstrate that loss of megalin (as well as Dab2) expression alters transcription in PT cells. Transcriptional changes initiated within the PT may contribute to the renal inflammation observed in patients with Donnai-Barrow syndrome and Lrp2 KO mice (11). In addition, our study revealed unexpected and selective changes in Slc5a2 and Mlxipl transcripts in Lrp2 KO cells that might contribute to the protective effect of megalin KO observed in some kidney injury models. Finally, differential changes in gene expression in Dab2 KO versus Lrp2 KO cells may reflect differences in the distributions of signaling proteins trapped at the apical plasma membrane and suggest the possibility that differential targeting of these proteins could be leveraged for a therapeutic advantage.
SUPPLEMENTAL DATA
Supplemental Tables S1–S9 and Supplemental Fig. S1: https://doi.org/10.6084/m9.figshare.14933340.v1.
GRANTS
This work was supported by American Heart Association Postdoctoral Fellowship 20POST35200358 (to C.D.B.), an ASN Carl W. Gottschalk Research Scholar Award (to R.J.T.), and National Institutes of Health Grants T32CA082084 (to B.R.F.), R01AI153104 and R21AI135027 (to A.C.P.), K08DK118211 (to C.R.B-S.), and R01DK118726 (to O.A.W.).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
R.J.T. and O.A.W. conceived and designed research; K.R.L., Y.R., C.D.B., A.C.P., C.R.B-S., and R.J.T. performed experiments; K.R.L., C.D.B., B.R.F., A.C.P., C.R.B-S., R.J.T., J.D.L., and O.A.W. analyzed data; C.D.B., C.R.B-S., R.J.T., and O.A.W. interpreted results of experiments; C.D.B., J.D.L., and O.A.W. prepared figures; O.A.W. drafted manuscript; K.R.L., Y.R., C.D.B., A.C.P., C.R.B-S., R.J.T., J.D.L., and O.A.W. edited and revised manuscript; K.R.L., Y.R., C.D.B., B.R.F., A.C.P., C.R.B-S., J.D.L., and O.A.W. approved final version of manuscript.
ACKNOWLEDGMENTS
Our sincere thanks to Thomas Willnow, Cecilia Lo, Ivy Lin, and Angelo Arrigo for sharing the Lrp2-EMX Cre mouse strain and to Allison Marciszyn for the expert assistant in rodent handling. We thank the Pittsburgh Center for Kidney Research (P30DK079307) for access to core facilities, the Health Sciences Sequencing Core at UPMC Children’s Hospital of Pittsburgh for Next-Generation Sequencing Services, and the University of Pittsburgh Center for Research Computing for computing cluster access and support. Shunsuke Kon (Tokyo University of Science) and Juan Bonifacino (National Institutes of Health) kindly provided the HA-Smap1 and GFP-Rabgef1 plasmids, respectively, and we are indebted to Daniel Biemesderfer and Peter Aronson (Yale University) for the gift of anti-megalin antibody. Morphology support was provided by the Pittsburgh Center for Kidney Research.
REFERENCES
- 1.Oleinikov AV, Zhao J, Makker SP. Cytosolic adaptor protein Dab2 is an intracellular ligand of endocytic receptor gp600/megalin. Biochem J 347 : 613–621, 2000. doi: 10.1042/bj3470613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pedersen GA, Chakraborty S, Steinhauser AL, Traub LM, Madsen M. AMN directs endocytosis of the intrinsic factor-vitamin B12 receptor cubam by engaging ARH or Dab2. Traffic 11: 706–720, 2010. doi: 10.1111/j.1600-0854.2010.01042.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Christensen EI, Nielsen R. Role of megalin and cubilin in renal physiology and pathophysiology. Rev Physiol Biochem Pharmacol 158: 1–22, 2007. doi: 10.1007/112_0604. [DOI] [PubMed] [Google Scholar]
- 4.Kantarci S, Al-Gazali L, Hill RS, Donnai D, Black GCM, Bieth E, Chassaing N, Lacombe D, Devriendt K, Teebi A, Loscertales M, Robson C, Liu T, MacLaughlin DT, Noonan KM, Russell MK, Walsh CA, Donahoe PK, Pober BR. Mutations in LRP2, which encodes the multiligand receptor megalin, cause Donnai-Barrow and facio-oculo-acoustico-renal syndromes. Nat Genet 39: 957–959, 2007. doi: 10.1038/ng2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dachy A, Paquot F, Debray G, Bovy C, Christensen EI, Collard L, Jouret F. In-depth phenotyping of a Donnai-Barrow patient helps clarify proximal tubule dysfunction. Pediatr Nephrol 30: 1027–1031, 2015. doi: 10.1007/s00467-014-3037-7. [DOI] [PubMed] [Google Scholar]
- 6.Kur E, Christa A, Veth KN, Gajera CR, Andrade-Navarro MA, Zhang J, Willer JR, Gregg RG, Abdelilah-Seyfried S, Bachmann S, Link BA, Hammes A, Willnow TE. Loss of Lrp2 in zebrafish disrupts pronephric tubular clearance but not forebrain development. Dev Dyn 240: 1567–1577, 2011. doi: 10.1002/dvdy.22624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leheste JR, Rolinski B, Vorum H, Hilpert J, Nykjaer A, Jacobsen C, Aucouturier P, Moskaug JO, Otto A, Christensen EI, Willnow TE. Megalin knockout mice as an animal model of low molecular weight proteinuria. Am J Pathol 155: 1361–1370, 1999. doi: 10.1016/S0002-9440(10)65238-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Y, Cong R, Biemesderfer D. The COOH terminus of megalin regulates gene expression in opossum kidney proximal tubule cells. Am J Physiol Cell Physiol 295: C529–C537, 2008. doi: 10.1152/ajpcell.00037.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zou Z, Chung B, Nguyen T, Mentone S, Thomson B, Biemesderfer D. Linking receptor-mediated endocytosis and cell signaling: evidence for regulated intramembrane proteolysis of megalin in proximal tubule. J Biol Chem 279: 34302–34310, 2004. doi: 10.1074/jbc.M405608200. [DOI] [PubMed] [Google Scholar]
- 10.Christ A, Terryn S, Schmidt V, Christensen EI, Huska MR, Andrade-Navarro MA, Hübner N, Devuyst O, Hammes A, Willnow TE. The soluble intracellular domain of megalin does not affect renal proximal tubular function in vivo. Kidney Int 78: 473–477, 2010. doi: 10.1038/ki.2010.169. [DOI] [PubMed] [Google Scholar]
- 11.Charlton JR, Tan W, Daouk G, Teot L, Rosen S, Bennett KM, Cwiek A, Nam S, Emma F, Jouret F, Oliveira JP, Tranebjærg L, Frykholm C, Mane S, Hildebrandt F, Srivastava T, Storm T, Christensen EI, Nielsen R. Beyond the tubule: pathological variants of LRP2, encoding the megalin receptor, result in glomerular loss and early progressive chronic kidney disease. Am J Physiol Renal Physiol 319: F988–F999, 2020. doi: 10.1152/ajprenal.00295.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Janssens V, Gaide Chevronnay HP, Marie S, Vincent M-F, Van Der Smissen P, Nevo N, Vainio S, Nielsen R, Christensen EI, Jouret F, Antignac C, Pierreux CE, Courtoy PJ. Protection of cystinotic mice by kidney-specific megalin ablation supports an endocytosis-based mechanism for nephropathic cystinosis progression. J Am Soc Nephrol 30: 2177–2190, 2019. doi: 10.1681/ASN.2019040371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Christensen EI, Nielsen R, Birn H. From bowel to kidneys: the role of cubilin in physiology and disease. Nephrol Dial Transplant 28: 274–281, 2013. doi: 10.1093/ndt/gfs565. [DOI] [PubMed] [Google Scholar]
- 14.Bedin M, Boyer O, Servais A, Li Y, Villoing-Gaudé L, Tête M-J, et al. Human C-terminal CUBN variants associate with chronic proteinuria and normal renal function. J Clin Invest 130: 335–344, 2020. doi: 10.1172/JCI129937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Amsellem S, Gburek J, Hamard G, Nielsen R, Willnow TE, Devuyst O, Nexo E, Verroust PJ, Christensen EI, Kozyraki R. Cubilin is essential for albumin reabsorption in the renal proximal tubule. J Am Soc Nephrol 21: 1859–1867, 2010. doi: 10.1681/ASN.2010050492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kozyraki R, Fyfe J, Verroust PJ, Jacobsen C, Dautry-Varsat A, Gburek J, Willnow TE, Christensen EI, Moestrup SK. Megalin-dependent cubilin-mediated endocytosis is a major pathway for the apical uptake of transferrin in polarized epithelia. Proc Natl Acad Sci USA 98: 12491–12496, 2001. doi: 10.1073/pnas.211291398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nagai J, Christensen EI, Morris SM, Willnow TE, Cooper JA, Nielsen R. Mutually dependent localization of megalin and Dab2 in the renal proximal tubule. Am J Physiol Renal Physiol 289: F569–F576, 2005. doi: 10.1152/ajprenal.00292.2004. [DOI] [PubMed] [Google Scholar]
- 18.Finkielstein CV, Capelluto DGS. Disabled-2: a modular scaffold protein with multifaceted functions in signaling. Bioessays 38, Suppl 1: S45–S55, 2016. doi: 10.1002/bies.201670907. [DOI] [PubMed] [Google Scholar]
- 19.Hocevar BA, Smine A, Xu XX, Howe PH. The adaptor molecule disabled-2 links the transforming growth factor beta receptors to the Smad pathway. EMBO J 20: 2789–2801, 2001. doi: 10.1093/emboj/20.11.2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ogbu SC, Musich PR, Zhang J, Yao ZQ, Howe PH, Jiang Y. The role of disabled-2 (Dab2) in diseases. Gene 769: 145202, 2021. doi: 10.1016/j.gene.2020.145202. [DOI] [PubMed] [Google Scholar]
- 21.Tao W, Moore R, Smith ER, Xu X-X. Endocytosis and physiology: insights from disabled-2 deficient mice. Front Cell Dev Biol 4: 129, 2016. doi: 10.3389/fcell.2016.00129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qiu C, Huang S, Park J, Park Y, Ko Y-A, Seasock MJ, Bryer JS, Xu X-X, Song W-C, Palmer M, Hill J, Guarnieri P, Hawkins J, Boustany-Kari CM, Pullen SS, Brown CD, Susztak K. Renal compartment-specific genetic variation analyses identify new pathways in chronic kidney disease. Nat Med 24: 1721–1731, 2018. doi: 10.1038/s41591-018-0194-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kirita Y, Wu H, Uchimura K, Wilson PC, Humphreys BD. Cell profiling of mouse acute kidney injury reveals conserved cellular responses to injury. Proc Natl Acad Sci USA 117: 15874–15883, 2020. doi: 10.1073/pnas.2005477117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hilpert J, Wogensen L, Thykjaer T, Wellner M, Schlichting U, Orntoft TF, Bachmann S, Nykjaer A, Willnow TE. Expression profiling confirms the role of endocytic receptor megalin in renal vitamin D3 metabolism. Kidney Int 62: 1672–1681, 2002. doi: 10.1046/j.1523-1755.2002.00634.x. [DOI] [PubMed] [Google Scholar]
- 25.Long KR, Shipman KE, Rbaibi Y, Menshikova EV, Ritov VB, Eshbach ML, Jiang Y, Jackson EK, Baty CJ, Weisz OA. Proximal tubule apical endocytosis is modulated by fluid shear stress via an mTOR-dependent pathway. Mol Biol Cell 28: 2508–2517, 2017. doi: 10.1091/mbc.E17-04-0211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park HJ, Fan Z, Bai Y, Ren Q, Rbaibi Y, Long KR, Gliozzi ML, Rittenhouse N, Locker JD, Poholek AC, Weisz OA. Transcriptional programs driving shear stress-induced differentiation of kidney proximal tubule cells in culture. Front Physiol 11: 587358, 2020. doi: 10.3389/fphys.2020.587358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ren Q, Gliozzi ML, Rittenhouse NL, Edmunds LR, Rbaibi Y, Locker JD, Poholek AC, Jurczak MJ, Baty CJ, Weisz OA. Shear stress and oxygen availability drive differential changes in opossum kidney proximal tubule cell metabolism and endocytosis. Traffic 20: 448–459, 2019. doi: 10.1111/tra.12648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ren Q, Weyer K, Rbaibi Y, Long KR, Tan RJ, Nielsen R, Christensen EI, Baty CJ, Kashlan OB, Weisz OA. Distinct functions of megalin and cubilin receptors in recovery of normal and nephrotic levels of filtered albumin. Am J Physiol Renal Physiol 318: F1284–F1294, 2020. doi: 10.1152/ajprenal.00030.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eshbach ML, Sethi R, Avula R, Lamb J, Hollingshead DJ, Finegold DN, Locker JD, Chandran UR, Weisz OA. The transcriptome of the Didelphis virginiana opossum kidney OK proximal tubule cell line. Am J Physiol Renal Physiol 313: F585–F595, 2017. doi: 10.1152/ajprenal.00228.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pereira EM, Labilloy A, Eshbach ML, Roy A, Subramanya AR, Monte S, Labilloy G, Weisz OA. Characterization and phosphoproteomic analysis of a human immortalized podocyte model of Fabry disease generated using CRISPR/Cas9 technology. Am J Physiol Renal Physiol 311: F1015–F1024, 2016. doi: 10.1152/ajprenal.00283.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet j 17: 10, 2011. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 32.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36, 2013. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30: 923–930, 2014. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 34.Leheste JR, Melsen F, Wellner M, Jansen P, Schlichting U, Renner-Müller I, Andreassen TT, Wolf E, Bachmann S, Nykjaer A, Willnow TE. Hypocalcemia and osteopathy in mice with kidney-specific megalin gene defect. FASEB J 17: 247–249, 2003. doi: 10.1096/fj.02-0578fje. [DOI] [PubMed] [Google Scholar]
- 35.Ma C, de Baaij JHF, Millar PJ, Gault VA, de Galan BE, Bindels RJM, Hoenderop JGJ. Effect of dapagliflozin treatment on the expression of renal sodium transporters/channels on high-fat diet diabetic mice. Nephron 142: 51–60, 2019. doi: 10.1159/000496617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spandidos A, Wang X, Wang H, Seed B. PrimerBank: a resource of human and mouse PCR primer pairs for gene expression detection and quantification. Nucleic Acids Res 38: D792–D799, 2010. doi: 10.1093/nar/gkp1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nat Protoc 3: 1101–1108, 2008. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 38.Hammes A, Andreassen TK, Spoelgen R, Raila J, Hubner N, Schulz H, Metzger J, Schweigert FJ, Luppa PB, Nykjaer A, Willnow TE. Role of endocytosis in cellular uptake of sex steroids. Cell 122: 751–762, 2005. doi: 10.1016/j.cell.2005.06.032. [DOI] [PubMed] [Google Scholar]
- 39.Nielsen R, Christensen EI, Birn H. Megalin and cubilin in proximal tubule protein reabsorption: from experimental models to human disease. Kidney Int 89: 58–67, 2016. doi: 10.1016/j.kint.2015.11.007. [DOI] [PubMed] [Google Scholar]
- 40.Peruchetti DB, Cheng J, Caruso-Neves C, Guggino WB. Mis-regulation of mammalian target of rapamycin (mTOR) complexes induced by albuminuria in proximal tubules. J Biol Chem 289: 16790–16801, 2014. doi: 10.1074/jbc.M114.549717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cox OT, O'Shea S, Tresse E, Bustamante-Garrido M, Kiran-Deevi R, O'Connor R. IGF-1 receptor and adhesion signaling: an important axis in determining cancer cell phenotype and therapy resistance. Front Endocrinol (Lausanne) 6: 106, 2015. doi: 10.3389/fendo.2015.00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chana RS, Sidaway JE, Brunskill NJ. Statins but not thiazolidinediones attenuate albumin-mediated chemokine production by proximal tubular cells independently of endocytosis. Am J Nephrol 28: 823–830, 2008. doi: 10.1159/000137682. [DOI] [PubMed] [Google Scholar]
- 43.Lu Y, Zhou Q, Zhong F, Guo S, Hao X, Li C, Wang W, Chen N. 15-Deoxy-Δ12,14-prostaglandin J2 modulates lipopolysaccharide-induced chemokine expression by blocking nuclear factor-κB activation via peroxisome proliferator activated receptor-γ-independent mechanism in renal tubular epithelial cells. Nephron Exp Nephrol 123: 1–10, 2013. doi: 10.1159/000353232. [DOI] [PubMed] [Google Scholar]
- 44.Abrink M, Larsson E, Gobl A, Hellman L. Expression of lactoferrin in the kidney: implications for innate immunity and iron metabolism. Kidney Int 57: 2004–2010, 2000. doi: 10.1046/j.1523-1755.2000.00050.x. [DOI] [PubMed] [Google Scholar]
- 45.Hsu Y-H, Chiu I-J, Lin Y-F, Chen Y-J, Lee Y-H, Chiu H-W. Lactoferrin contributes a renoprotective effect in acute kidney injury and early renal fibrosis. Pharmaceutics 12: 434, 2020. doi: 10.3390/pharmaceutics12050434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Artunc F, Lang F. Mineralocorticoid and SGK1-sensitive inflammation and tissue fibrosis. Nephron Physiol 128: 35–39, 2014. doi: 10.1159/000368267. [DOI] [PubMed] [Google Scholar]
- 47.Seo H, Cho Y-C, Ju A, Lee S, Park BC, Park SG, Kim J-H, Kim K, Cho S. Dual-specificity phosphatase 5 acts as an anti-inflammatory regulator by inhibiting the ERK and NF-κB signaling pathways. Sci Rep 7: 17348, 2017. doi: 10.1038/s41598-017-17591-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kunter U, Daniel S, Arvelo MB, Choi J, Shukri T, Patel VI, Longo CR, Scali ST, Shrikhande G, Rocha E, Czismadia E, Mottley C, Grey ST, Floege J, Ferran C. Combined expression of A1 and A20 achieves optimal protection of renal proximal tubular epithelial cells. Kidney Int 68: 1520–1532, 2005. doi: 10.1111/j.1523-1755.2005.00564.x. [DOI] [PubMed] [Google Scholar]
- 49.Lutz J, Luong le A, Strobl M, Deng M, Huang H, Anton M, Zakkar M, Enesa K, Chaudhury H, Haskard DO, Baumann M, Boyle J, Harten S, Maxwell PH, Pusey C, Heemann U, Evans PC. The A20 gene protects kidneys from ischaemia/reperfusion injury by suppressing pro-inflammatory activation. J Mol Med (Berl) 86: 1329–1339, 2008. doi: 10.1007/s00109-008-0405-4. [DOI] [PubMed] [Google Scholar]
- 50.Shembade N, Harhaj EW. Regulation of NF-κB signaling by the A20 deubiquitinase. Cell Mol Immunol 9: 123–130, 2012. doi: 10.1038/cmi.2011.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vallon V, Verma S. Effects of SGLT2 inhibitors on kidney and cardiovascular function. Annu Rev Physiol 83: 503–528, 2021. doi: 10.1146/annurev-physiol-031620-095920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Krawczak J, Owczarek A, Winiarska K. Transcription factor ChREBP–the coordinator of carbohydrate and lipid metabolism. Postepy Biochem 66: 30–37, 2020. doi: 10.18388/pb.2019_282. [DOI] [PubMed] [Google Scholar]
- 53.Ortega-Prieto P, Postic C. Carbohydrate sensing through the transcription factor ChREBP. Front Genet 10: 472, 2019. doi: 10.3389/fgene.2019.00472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Proctor G, Jiang T, Iwahashi M, Wang Z, Li J, Levi M. Regulation of renal fatty acid and cholesterol metabolism, inflammation, and fibrosis in Akita and OVE26 mice with type 1 diabetes. Diabetes 55: 2502–2509, 2006. doi: 10.2337/db05-0603. [DOI] [PubMed] [Google Scholar]
- 56.Wang XX, Levi J, Luo Y, Myakala K, Herman-Edelstein M, Qiu L, Wang D, Peng Y, Grenz A, Lucia S, Dobrinskikh E, D’Agati VD, Koepsell H, Kopp JB, Rosenberg AZ, Levi M. SGLT2 protein expression is increased in human diabetic nephropathy: SGLT2 protein inhibition decreases renal lipid accumulation, inflammation, and the development of nephropathy in diabetic mice. J Biol Chem 292: 5335–5348, 2017. doi: 10.1074/jbc.M117.779520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kuwahara S, Hosojima M, Kaneko R, Aoki H, Nakano D, Sasagawa T, Kabasawa H, Kaseda R, Yasukawa R, Ishikawa T, Suzuki A, Sato H, Kageyama S, Tanaka T, Kitamura N, Narita I, Komatsu M, Nishiyama A, Saito A. Megalin-mediated tubuloglomerular alterations in high-fat diet-induced kidney disease. J Am Soc Nephrol 27: 1996–2008, 2016. doi: 10.1681/ASN.2015020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Motoyoshi Y, Matsusaka T, Saito A, Pastan I, Willnow TE, Mizutani S, Ichikawa I. Megalin contributes to the early injury of proximal tubule cells during nonselective proteinuria. Kidney Int 74: 1262–1269, 2008. doi: 10.1038/ki.2008.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Horiuchi H, Lippé R, McBride HM, Rubino M, Woodman P, Stenmark H, Rybin V, Wilm M, Ashman K, Mann M, Zerial M. A novel Rab5 GDP/GTP exchange factor complexed to Rabaptin-5 links nucleotide exchange to effector recruitment and function. Cell 90: 1149–1159, 1997. doi: 10.1016/s0092-8674(00)80380-3. [DOI] [PubMed] [Google Scholar]
- 60.Valsdottir R, Hashimoto H, Ashman K, Koda T, Storrie B, Nilsson T. Identification of rabaptin-5, rabex-5, and GM130 as putative effectors of rab33b, a regulator of retrograde traffic between the Golgi apparatus and ER. FEBS Lett 508: 201–209, 2001. doi: 10.1016/s0014-5793(01)02993-3. [DOI] [PubMed] [Google Scholar]
- 61.Zhu H, Liang Z, Li G. Rabex-5 is a Rab22 effector and mediates a Rab22-Rab5 signaling cascade in endocytosis. Mol Biol Cell 20: 4720–4729, 2009. doi: 10.1091/mbc.e09-06-0453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.El Abbas S, Radermecker C, Bai Q, Beguin C, Schyns J, Meunier M, Pirottin D, Desmet CJ, Meuwis M-A, Art T, Louis E, Tam S-Y, Tsai M, Bureau F, Galli SJ, Marichal T. Epithelial RABGEF1 deficiency promotes intestinal inflammation by dysregulating intrinsic MYD88-dependent innate signaling. Mucosal Immunol 13: 96–109, 2020. doi: 10.1038/s41385-019-0211-z. [DOI] [PubMed] [Google Scholar]
- 63.Pagano A, Spiess M. Reconstitution of rab4‐dependent vesicle formation in vitro. In: GTPases Regulating Membrane Targeting and Fusion, edited by Balch WE, Der CJ, Hall A.. San Diego, CA: Elsevier, 2005, p. 81–92. [DOI] [PubMed] [Google Scholar]
- 64.Tanabe K, Kon S, Ichijo N, Funaki T, Natsume W, Watanabe T, Satake M. A SMAP gene family encoding ARF GTPase-activating proteins and its implication in membrane trafficking. Meth Enzymol 438: 155–170, 2008. doi: 10.1016/S0076-6879(07)38011-7. [DOI] [PubMed] [Google Scholar]
- 65.El-Annan J, Brown D, Breton S, Bourgoin S, Ausiello DA, Marshansky V. Differential expression and targeting of endogenous Arf1 and Arf6 small GTPases in kidney epithelial cells in situ. Am J Physiol Cell Physiol 286: C768–C678, 2004. doi: 10.1152/ajpcell.00250.2003. [DOI] [PubMed] [Google Scholar]
- 66.Wolff NA, Lee W-K, Abouhamed M, Thévenod F. Role of ARF6 in internalization of metal-binding proteins, metallothionein and transferrin, and cadmium-metallothionein toxicity in kidney proximal tubule cells. Toxicol Appl Pharmacol 230: 78–85, 2008. doi: 10.1016/j.taap.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 67.Tanabe K, Torii T, Natsume W, Braesch-Andersen S, Watanabe T, Satake M. A novel GTPase-activating protein for ARF6 directly interacts with clathrin and regulates clathrin-dependent endocytosis. Mol Biol Cell 16: 1617–1628, 2005. doi: 10.1091/mbc.e04-08-0683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen L, Chou C-L, Knepper MA. A Comprehensive map of mRNAs and their isoforms across all 14 renal tubule segments of mouse. J. Am. Soc. Nephrol 32: 897–912, 2021. doi: 10.1681/ASN.2020101406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.von Zastrow M, Sorkin A. Mechanisms for regulating and organizing receptor signaling by endocytosis. Annu Rev Biochem 90: 709–737, 2021. doi: 10.1146/annurev-biochem-081820-092427. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Tables S1–S9 and Supplemental Fig. S1: https://doi.org/10.6084/m9.figshare.14933340.v1.