Abstract
In eukaryotes the majority of mRNAs have an m7G cap that is added cotranscriptionally and that plays an important role in many aspects of mRNA metabolism. The nuclear cap-binding complex (CBC; consisting of CBP20 and CBP80) mediates the stimulatory functions of the cap in pre-mRNA splicing, 3′ end formation, and U snRNA export. As little is known about how nuclear CBC mediates the effects of the cap in higher eukaryotes, we have characterized proteins that interact with CBC in HeLa cell nuclear extracts as potential mediators of its function. Using cross-linking and coimmunoprecipitation, we show that eukaryotic translation initiation factor 4G (eIF4G), in addition to its function in the cytoplasm, is a nuclear CBC-interacting protein. We demonstrate that eIF4G interacts with CBC in vitro and that, in addition to its cytoplasmic localization, there is a significant nuclear pool of eIF4G in mammalian cells in vivo. Immunoprecipitation experiments suggest that, in contrast to the cytoplasmic pool, much of the nuclear eIF4G is not associated with eIF4E (translation cap binding protein of eIF4F) but is associated with CBC. While eIF4G stably associates with spliceosomes in vitro and shows close association with spliceosomal snRNPs and splicing factors in vivo, depletion studies show that it does not participate directly in the splicing reaction. Taken together the data indicate that nuclear eIF4G may be recruited to pre-mRNAs via its interaction with CBC and accompanies the mRNA to the cytoplasm, facilitating the switching of CBC for eIF4F. This may provide a mechanism to couple nuclear and cytoplasmic functions of the mRNA cap structure.
RNAs transcribed by RNA polymerase II (pol II) are characterized by an inverted m7G(5′)ppp(5′)N cap. The capping of the pre-mRNA occurs cotranscriptionally (50, 52) and is achieved by recruitment of the capping enzyme to the phosphorylated C-terminal domain of the largest subunit of pol II (5, 22, 36). The cap contributes to many aspects of pol II transcript metabolism, including protection against 5′–3′ exonucleases, facilitating efficient pre-mRNA splicing, 3′ end formation, U snRNA and mRNA nuclear export, and translation of mRNAs (32, 34).
Two distinct families of cap binding proteins (CBPs) mediate the stimulatory effects of the cap structure. In the nucleus, the cap structure interacts with the nuclear cap-binding complex (CBC), a heterodimer consisting of two highly conserved polypeptides, CBP80 and CBP20 (20, 23, 54). CBC plays a direct role in pre-mRNA splicing, 3′ end formation, and U snRNA export (reviewed in reference 32). In pre-mRNA splicing CBC promotes the association of U1 snRNP with the cap-proximal 5′ splice site (31, 33). In Saccharomyces cerevisiae, CBC interacts directly with yeast-specific components of the U1 snRNP (13, 15). This contrasts with the mammalian system, where there is no evidence of a direct interaction between CBC and U1 snRNP (33). Although CBC is required for efficient 3′ end cleavage of pre-mRNA, it is not required for polyadenylation per se (11). The cap also contributes to mRNA export, presumably through its interaction with CBC (25, 54) and with CBC-dependent export of U snRNAs from the nucleus to the cytoplasm mediated by nuclear export receptor CRM1 and by PHAX (12, 43).
The effect of the cap structure on mRNA translation is mediated by a trimeric complex termed eukaryotic translation initiation factor 4F (eIF4F; comprising eIF4E, the helicase eIF4A, and eIF4G), which recruits mRNA to the ribosome (19, 38). eIF4E interacts specifically with the mRNA cap structure and is essential for cap-dependent translation (19). eIF4G, which exists in two isoforms, plays an essential role in the mechanism of translation by acting as a molecular bridge between other components of the ribosomal initiation complex (reviewed in references 19, 21, 38, 39, and 48). Within the sequence of eukaryotic eIF4G there are domains that interact with eIF4E, eIF4A, eIF3, the poly(A) binding protein (PABP), and the eIF4E kinase, Mnk1 (reviewed in references 19, 21, 39, and 48). It has been suggested that interaction between PABP and eIF4G facilitates the functional association of the 3′ end of an mRNA with the 5′ end to promote mRNA translation (21), while the association between eIF4G and eIF4E markedly enhances the binding of the latter to the mRNA cap (19). During the course of our studies reported here, Fortes et al. (14) used a combination of biochemical and genetic approaches to demonstrate that a defined region of S. cerevisiae eIF4G also has the ability to interact with CBP80. Furthermore, this interaction was antagonized by eIF4E, suggesting that the exchange of nuclear for cytoplasmic CBPs may be mediated by eIF4G (14).
To gain more insight into the role of nuclear proteins that mediate the effects of CBC in pre-mRNA splicing and 3′ end formation, we have investigated nuclear proteins that specifically interact with capped RNA in a CBC-dependent manner. Using immunofluorescence and biochemical analysis, we demonstrate that, as with eIF4E (9), there is a nuclear pool of eIF4G. Nuclear eIF4G exhibits partial colocalization with spliceosomal snRNPs and stably associates with CBC, pre-mRNA, and the spliceosome. These data, together with genetic studies with S. cerevisiae CBP80 (14), further strengthen the possibility that eIF4G has a role in coupling RNA-processing events in the nucleus with mRNA translation in the cytoplasm.
MATERIALS AND METHODS
Chemicals and biochemicals.
Materials for tissue culture were from Gibco Life Technologies (Paisley, United Kingdom), T7 RNA polymerase was from New England Biolabs (Hitchin, United Kingdom), [α-32P]GTP and protein A-Sepharose were from Amersham Pharmacia Biotech (Little Chalfont, United Kingdom), 4-thio-UTP was from United States Biochemicals, cap analogue was from Kedar (Warsaw, Poland), and cytofectine was from Bio-Rad (Hemel Hempstead, United Kingdom). Unless otherwise stated, all other chemicals were from Sigma (Gillingham, United Kingdom).
4-Thio-U-substituted capped RNA and UV cross-linking.
4-Thio-U capped RNA was synthesized basically as described by Milligan et al. (37). The oligonucleotides used were T7P (TAATACGACTCACTATA) and UVXL (ATTATGCTGAGTGATATCCCAGACACATCTCCCCGCGCTTCC). For UV cross-linking, reaction mixtures (20 μl) comprised 5 μl of HeLa nuclear extract, 10 μg of Escherichia coli tRNA, 150 mM NaCl, and 4 × 104 cpm of α-32P-labeled capped RNA and cap analogue as indicated in the figure legend. Reaction mixtures were irradiated on ice-water for 10 min, approximately 4 cm from a 360-nm light source (BLAK-RAY long-wave UV lamp, model B100AP). One hundred nanograms of RNase A was added for a further 10 min; samples were then denatured, and the cross-linked products were resolved by Tris-Tricine sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (51), dried, and exposed to X-ray film.
Extracts, immunoprecipitation, immunodepletion, and in vitro splicing.
HeLa cell nuclear extracts, prepared according to the method of Dignam (7), were purchased from 4C (Mons, Belgium). Immunodepletion of nuclear extracts for CBC using anti-CBP80 antibodies and splicing reactions were performed as described previously (24); depletion of eIF4G was performed in parallel using rabbit antiserum against the C-terminal domain of eIF4G (3, 16). For immunoprecipitations, samples were diluted fivefold in IP150 buffer and incubated with a 30-μl packed volume of antibody beads for 1 h at 4°C, with rotation. The beads were then washed three times with 1 ml of ice-cold IP150 buffer, and the recovered proteins were eluted with SDS-PAGE sample buffer. The proteins were resolved by SDS-PAGE; gels were dried and subjected to autoradiography, or proteins were transferred to a membrane (either nitrocellulose or polyvinylidene difluoride) and probed with an antibody specific for CBP80 (24) or affinity-purified anti-eIF4GI antibody (3).
For cell fractionation studies, HeLa cells were grown in suspension to a density of 5 × 105 cells/ml and separated into nuclear and cytoplasmic fractions, as described previously (35). For Western blotting, equal cell equivalents of the nucleoplasmic and cytoplasmic fractions were resolved by SDS-PAGE and proteins were visualized with serum specific for eIF4G(Ct), CBP80, γ-tubulin or BiP (Santa Cruz Biotechnology), and ECL. For quantification of the distribution of the proteins between the nuclear and cytoplasmic compartments, a secondary Cy5-labeled antibody was used with a Storm 860 PhosphorImager (Molecular Dynamics) in red fluorescence mode; signals were analyzed using ImageQuant software.
Mammalian cell culture, confocal microscopy, and subcellular fractionation.
HeLa cells were cultured as monolayers in RPMI medium supplemented with 10% fetal calf serum (FCS), and diploid human fibroblasts (WI-38; American Type Culture Collection) were cultured in minimum essential Eagle's medium supplemented with nonessential amino acids and 10% FCS. For immunofluorescence analysis cells were grown on glass coverslips for a minimum of 24 h and collected when 70 to 90% confluent. Cells were briefly rinsed in phosphate-buffered saline (PBS) and immediately fixed in 2% formaldehyde in PBS for 10 min at room temperature; all formaldehyde solutions were freshly prepared from a 40% stock solution of paraformaldehyde in water (Electron Microscopy Sciences, Fort Washington, Wash.). After fixation cells were rinsed in PBS and subsequently permeabilized in 0.1% saponin (Sigma)–0.1% NP-40 (Fluka) in PBS at room temperature for an additional 6 to 7 min. In some experiments cells were simultaneously fixed and extracted in a solution containing 3.7% formaldehyde in HPEM buffer {30 mM HEPES, 65 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid), pH 6.9], 10 mM EGTA, 2 mM MgCl2} and 0.5% Triton X-100 for 10 min at room temperature as previously described (10); alternatively, cells were fixed and permeabilized in −20°C acetone for 2 min (41).
eIF4GI was labeled with affinity-purified antipeptide antibody eIF4GI(Nt) or eIF4GI(Ct) (3). Splicing snRNPs were labeled with anti-Sm monoclonal antibody Y12 (45), splicing factor SC-35 was detected with an anti-SC-35 monoclonal antibody (18), and other proteins of the SR family were labeled with monoclonal antibodies 104 and 16H3 (41, 42). Anti-mouse immunoglobulin G (IgG), anti-mouse IgM, and antirabbit secondary antibodies coupled to either fluorescein isothiocyanate, Texas red, or Alexa 488 were purchased from Jackson Immunoresearch Laboratories and were used at final dilutions ranging from 1/75 to 1/150. Leptomycin B (LMB) was used at a final concentration of 10 μg/ml for the times indicated in the figure legends. NIH 3T3 cells were transfected with vectors encoding FLAG-eIF4GI or myc-MNK using cytofectine according to the manufacturer's instructions. Recombinant proteins were detected with anti-FLAG-biotin conjugate and anti-biotin-Cy3 (Sigma) and anti-myc-cy3 (Sigma). The samples were examined using a Leica confocal microscope with three lasers giving excitation lines at 380, 488, and 543 nm. The data from the channels were collected separately using narrow-band-pass filter settings; in multiple staining experiments, the laser intensities and data collection settings were adjusted to avoid overlap (bleedthrough) between channels. Data were collected with four- to eightfold averaging at a resolution of 512 by 512 pixels using pinhole settings between 1.05 and 1.10 airy units. The coupled microscope was a Leica DMIBRE equipped with a 63× water immersion objective lens (numerical aperture, 1.4). Data sets were processed using the Leica TCS NT, version 1.4.338, software package and were subsequently exported for preparation for printing using Adobe Photoshop, version 5.5, and Deneba Canvas, version 7.02.
RESULTS
Characterization of CBC-dependent cross-links.
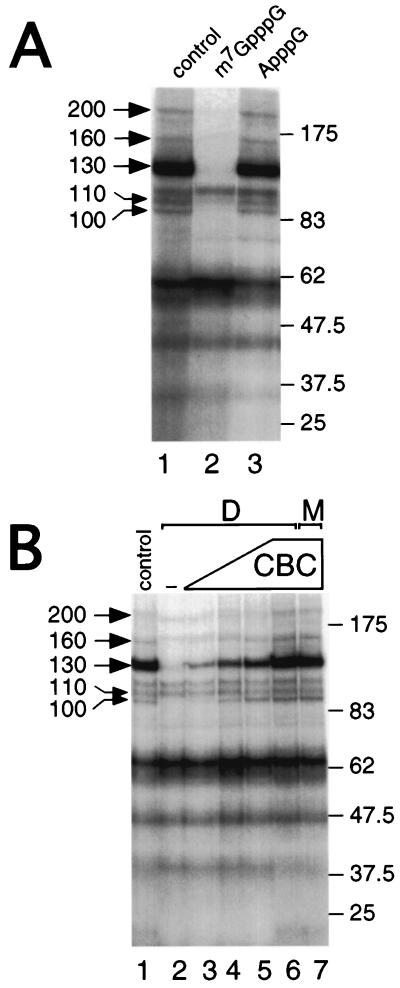
To identify novel CBC-interacting polypeptides that may mediate its function, we used a sensitive UV cross-linking strategy employing photoactivatable nucleotide 4-thio-U (53), incorporated close to the m7GpppG cap of a 32P-labeled synthetic RNA (m7GsU). m7GsU was incubated with HeLa nuclear extracts and UV-irradiated and cross-linked polypeptides were resolved by SDS-PAGE. Figure 1A (lane 1) shows that several cross-linked proteins, ranging from >20 to approximately 200 kDa, can be resolved. To determine which of these proteins interact with m7GsU in a cap-dependent manner, m7GpppG (lane 2) or ApppG (lane 3) was added prior to UV irradiation. While the cap analogue abolishes the cross-linking of several proteins (especially the 100-, 110-, 130-, 160-, and 200-kDa proteins), ApppG had little effect, demonstrating that these interactions were cap specific. No cross-links corresponding to CBP80 or CBP20 were observed using this technique. The most likely explanation is that, in m7GsU mRNA, the body of the mRNA is labeled (the first 4-thio-U being four nucleotides from the cap) and that CBP20 and CBP80 are not amenable to efficient cross-linking. Previous studies using cap (radioactively)-labeled mRNA gave a strong cross-linking of CBP20 (24, 49). To determine whether any of the observed cap-dependent interactions were also CBC dependent, the cross-linking profiles of CBC-depleted and mock-depleted extracts were compared. Compared to the control extract, depletion of CBC caused a strong reduction in cross-linking efficiency of the 100-, 130-, 160-, and 200-kDa proteins (Fig. 1B, lane 2 versus lane 1); in contrast, the 110-kDa cross-link was not affected by depletion of CBC. Interaction of these proteins with the mRNA cap could be restored by addition of increasing amounts of recombinant, purified CBC to the depleted extract (lanes 3 to 6). Recombinant CBC alone did not give rise to any cross-linked proteins with molecular weights comparable to those observed (data not shown). At the largest amount of added CBC, the recovery of CBC-interacting proteins (CIPs) was virtually indistinguishable from that observed in the nondepleted extract (lane 6 versus lane 1). Furthermore, addition of the largest amount of CBC to a mock-depleted extract did not increase the efficiency of cross-linking of CIPs (lane 7 versus lane 1). These data strongly suggest that these polypeptides bind capped RNA in a CBC-dependent manner, and we refer to these proteins as CIP followed by the relative molecular mass in kilodaltons (e.g., CIP200).
FIG. 1.
Identification of cap-dependent cross-links and CIPs in HeLa nuclear extracts. (A) HeLa nuclear extracts were incubated in the absence (lane 1) or presence of 50 μM m7GpppG (lane 2) or ApppG (lane 3) prior to UV cross-linking. Following UV cross-linking, labeled proteins were resolved by SDS-PAGE. Left, cap-dependent cross-links; right, molecular weight markers. (B) HeLa nuclear extracts were either depleted of CBC (D; lanes 2 to 6) or mock-depleted (M; lanes 1 and 7), as described in Materials and Methods. Increasing amounts of purified, recombinant CBC were added to the depleted extract (60, 125, 250, and 500 ng; lanes 3 to 6, respectively) before the addition of m7GsU and UV cross-linking. CBC (500 ng) was also added to a mock-depleted extract as a control (lane 7). Labeled proteins were resolved by SDS-PAGE and visualized by autoradiography. Left, cap-dependent cross-links; right, molecular weight markers.
Identification of CIP200 as eIF4G.
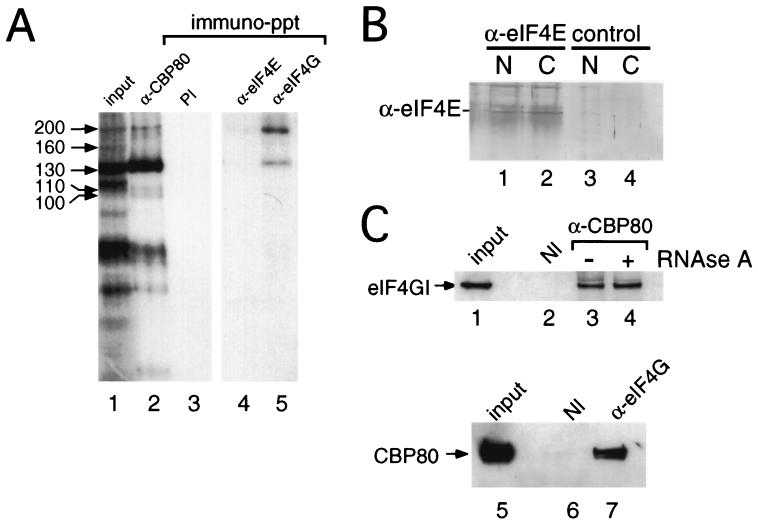
eIF4G plays an essential role in the mechanism of translation in the cytoplasm by acting as a molecular bridge between mRNA, eIF4E, and components of the ribosomal initiation complex (reviewed in references 19, 21, 38, 39, and 48). The size of CIP200 was consistent with that of eIF4G; indeed, eIF4G could be specifically detected in our cell fractions by Western blotting (see Fig. 3B). In order to investigate whether CIP200 was in fact eIF4G, cross-linking reaction mixtures (Fig. 1) were subjected to immunoprecipitation using antibodies specific to eIF4G. Figure 2A shows that anti-eIF4GI serum specifically precipitated proteins that comigrated with CIP200 and to a lesser extent CIP130 (lane 5). In contrast, anti-CBP80 serum efficiently precipitated all CIPs with the exception of CIP160 (lane 2 versus lane 1), and none of these proteins were recovered with preimmune serum (lane 3). Little or no coprecipitation of eIF4G from the nuclear fraction was observed using anti-eIF4E antiserum (lane 4), although this serum was able to recover eIF4G from cytoplasmic extracts (16, 40) (data not shown). Furthermore, eIF4E was precipitated from both nuclear and cytoplasmic fractions with anti-eIF4E serum (Fig. 2B, lanes 1 and 2); the background smear is due to the light chains from the IgGs that comigrate. These data suggest that, although present in the nuclear fraction (9), eIF4E is either not associated with eIF4G or does not interact with the cap structure to an appreciable level in our nuclear extracts. Furthermore, depletion of eIF4E or digestion of the cross-linked products with RNase A prior to immunoprecipitation did not significantly affect recovery of CIP200, but the latter treatment reduced the amount of CIP130 precipitated (data not shown).
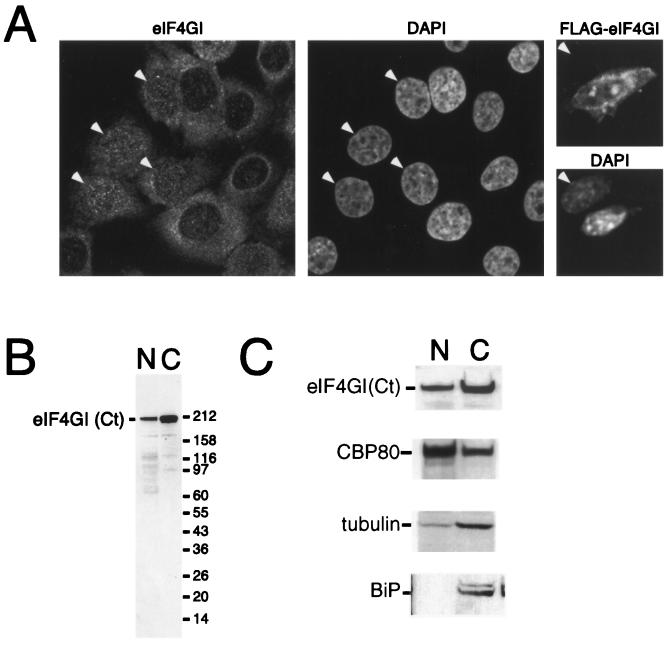
FIG. 3.
eIF4GI is distributed between the nucleus and cytoplasm. (A) HeLa cells were cultured on coverslips and fixed, endogenous eIF4G was localized by using affinity-purified anti-eIF4GI(Ct) antiserum (left), and DNA was stained with DAPI (4′,6′-diamidino-2-phenylindole) (middle). Note that eIF4GI localizes to both the nucleus and the cytoplasm and that some cells display strong nuclear staining (arrowheads). NIH 3T3 cells were grown on coverslips (right) and transfected with a vector encoding FLAG-eIF4G. Expressed eIF4G was localized using an anti-FLAG antibody (upper), and DNA was visualized by DAPI staining (lower). A transfected and a nontransfected (arrowhead) cell are shown side by side; epitope-tagged eIF4GI can be detected in both the nucleus and the cytoplasm of the transfected cell. (B) HeLa cells were separated into nuclear (N) and cytoplasmic (C) fractions, as described in Materials and Methods. The recovery of eIF4G in the nuclear and cytoplasmic fractions was visualized by SDS-PAGE and immunoblotting using an affinity-purified antibody. (C) HeLa cells were separated into nuclear and cytoplasmic fractions as described above. Cell equivalents of the nuclear and cytoplasmic fractions were loaded, separated by SDS-PAGE, and blotted. The recovery of eIF4G [using antisera specific to eIF4G(Ct)], CBP80, BiP, and γ-tubulin in the nuclear and cytoplasmic fractions was monitored by SDS-PAGE and Western blotting using ECL.
FIG. 2.
eIF4G (CIP200) interacts with nuclear CBC. (A) HeLa nuclear extracts were incubated with m7GsU and subjected to UV cross-linking. Cross-linked proteins were immunoprecipitated (immuno-ppt) using immobilized antibodies to CBP80 (lane 2), eIF4E (lane 4), or eIF4G (lane 5) or those derived from a preimmune (PI) bleed (lane 3). Lane 1, input to the immunoprecipitation. (B) HeLa nuclear (N) or cytoplasmic extracts (C) were subjected to immunoprecipitation in the absence (lanes 3 and 4) or presence (lanes 1 and 2) of antiserum specific for eIF4E. The proteins recovered in immunocomplexes were resolved by SDS-PAGE, and recovery of eIF4E was visualized by Western blotting using an alkaline phosphatase-conjugated secondary antibody. (C) HeLa nuclear extract was subjected to immunoprecipitation using either preimmune antiserum (lane 2) or an anti-CBP80 antibody (lanes 3 and 4), as described in Materials and Methods. Lane 1, input to the immunoprecipitation. Prior to being washed, the recovered resin was incubated for 10 min in the absence (lane 3) or presence (lane 4) of 50 μg of RNase A/ml and then processed as described in Materials and Methods. The proteins were recovered from immunocomplexes using nondenaturing conditions and then denatured and resolved by SDS-PAGE, and eIF4GI was visualized using anti-eIF4GI(Ct) and Western blotting using ECL. In the reciprocal experiment immunoprecipitations were performed using either preimmune antiserum (lane 6) or an anti-eIF4GI antibody (lane 7), as described above. Lane 5, input to the immunoprecipitation. The proteins were recovered as described in Materials and Methods and probed using anti-CBP80 antiserum.
To determine whether we could observe an interaction between nuclear CBC and eIF4G in the absence of cross-linking, immobilized antibodies to CBP80 or preimmune serum were incubated with HeLa nuclear extracts. Following extensive washing of the isolated resin, proteins that coprecipitated with CBC were resolved by SDS-PAGE and analyzed by Western blotting. Figure 2C shows that eIF4G was recovered with anti-CBP80 serum (lane 3) and not with preimmune serum (lane 2). Treatment of the isolated resin with RNase A prior to washing did not reduce the recovery of eIF4G (lane 4 versus lane 3), suggesting a close interaction between CBP80 and eIF4G in the nuclear extracts. The reciprocal experiment demonstrated that CBP80 could be coprecipitated with anti-eIF4G antibody (Fig. 2C, lane 7) and not with preimmune serum (lane 6) from nuclear extracts. Again, this interaction was unaffected by RNase treatment of the resin prior to washing (data not shown).
eIF4GI is distributed between the cytoplasm and nucleus.
Although the role of eIF4G in the initiation of protein synthesis has been extensively investigated, little is known about its subcellular localization. To further investigate the localization of eIF4GI, we used indirect immunofluorescence followed by analysis using confocal microscopy. HeLa cells, human diploid fibroblasts, and mouse NIH 3T3 cells were fixed and permeabilized according to three distinct protocols and labeled with two independent affinity-purified antibodies directed against eIF4GI. As expected, both antibodies displayed widespread intense cytoplasmic staining in the majority of the cells during interphase (Fig. 3A, left, and data not shown). In addition to this cytoplasmic labeling, an easily identifiable nuclear staining was observed in the HeLa cells, nontransformed human fibroblasts, and NIH 3T3 cells. This comprised numerous bright foci of higher concentrations of eIF4GI superimposed on a more-diffuse nucleoplasmic labeling; nucleolar staining could not be detected to any significant extent. We note that a similar staining pattern was observed irrespective of the fixation and permeabilization protocol used (data not shown). In addition, we have consistently observed that in a minor, but still significant, proportion of cells in a population the nucleoplasmic labeling was more prominent than its cytoplasmic counterpart (Fig. 3A, eIF4GI and DAPI panels). The specificity of the antibody used for the immunostainings shown was assayed on a Western blot of nuclear and cytoplasmic extracts and recognized predominantly a single band corresponding to eIF4GI in both extracts (Fig. 3B); the lower species probably correspond to degradation products. In addition, we transfected NIH 3T3 cells with a FLAG-tagged eIF4GI construct and determined the localization of expressed eIF4GI using an anti-FLAG antibody. The staining pattern of expressed eIF4G was similar to that observed for the endogenous protein, with little staining using the anti-FLAG antiserum in the absence of eIF4G expression (Fig. 3A, right, and data not shown).
To examine more directly the subcellular distribution of eIF4GI, exponentially growing HeLa cells were separated into nuclear and cytoplasmic fractions. Cell equivalents of both fractions were resolved by SDS-PAGE, and the distribution of eIF4GI, BiP, γ-tubulin, and CBP80 was determined by immunoblotting and enhanced chemiluminescence (ECL) detection (Fig. 3C) or, alternatively, the distribution was quantified using Cy5-labeled secondary antibodies and fluorescence. In agreement with the immunolocalization data presented in Fig. 3A, eIF4GI was predominantly cytoplasmic (Fig. 3C). Quantification of these data showed that approximately 22% of the eIF4GI was nuclear at steady state. Consistent with the ability of CBC to shuttle between the nucleus and cytoplasm, CBP80 was present in both the cytoplasmic and nuclear fractions, with approximately 74% being in the nuclear fraction. The finding that γ-tubulin and BiP show only minor contamination in the nuclear fraction (approximately 11 and 7%, respectively) indicates that there is a bona fide nuclear pool of eIF4GI in mammalian cells, a fraction of which is associated with CBC.
eIF4GI specifically associates with the spliceosome in vitro.
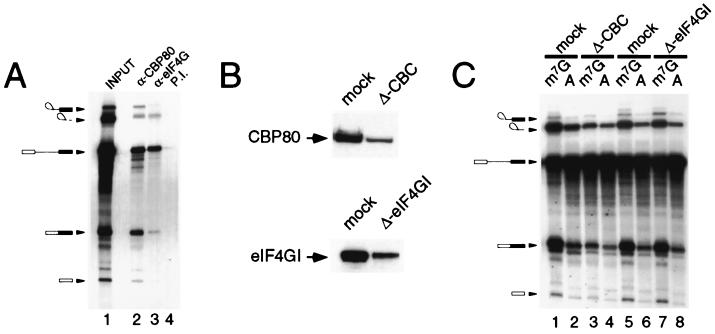
To determine whether eIF4GI plays a role in pre-mRNA splicing, we first asked whether eIF4GI associates with splicing complexes. In vitro splicing reaction mixtures were immunoprecipitated using antiserum specific for CBP80, eIF4G, or preimmune serum, and the recovered radiolabeled RNA was visualized by gel electrophoresis and autoradiography. Figure 4A shows that anti-CBP80 (lane 2) coprecipitated the pre-mRNA, the mature RNA products, and the exon lariat and 5′ exon (33). Anti-eIF4G precipitated a similar set of RNA products (lane 3) with decreased, but still significant, recovery of intermediates of the splicing reaction, indicating the association of eIF4G with the splicing complexes. This interaction is specific, as eIF4G preimmune serum showed very low recovery of RNA (lane 4 versus lane 3). These data suggest that eIF4GI stably associates with the spliceosome and might participate directly in the splicing reaction. To test this more directly, splicing extracts were immunodepleted of CBC using anti-CBP80 (33) or of eIF4G using immobilized antisera. As shown in Fig. 4B, depletion of both proteins was extensive but incomplete; repeated attempts to further deplete these extracts were not successful, indicating a population of these proteins which were refractory to this process (data not shown). When assayed in a splicing reaction with Ad1 pre-mRNA, depletion of CBC resulted in a severe reduction in capped-mRNA splicing efficiency (Fig. 4C, lane 3 versus lane 1), with a lesser effect on cap-independent splicing (lane 4 versus lane 2), in agreement with published data (33). However, immunodepletion of eIF4GI to levels similar to those observed for CBP80 from nuclear extracts had no effect on the efficiency of capped (lane 7 versus 5) or cap-independent pre-mRNA (lanes 8 versus lane 6) splicing in vitro. Similar results were observed for mock-depleted and eIF4GI-depleted extracts when assayed with a number of pre-mRNA templates (data not shown). Although this makes it unlikely that eIF4GI mediates the effects of CBC in facilitating the association of U1 snRNP with the cap-proximal 5′ splice site, we cannot exclude the possibility that it may be required for splicing specific transcripts (see below).
FIG. 4.
eIF4G associates with splicing complexes in vitro. (A) 32P-labeled m7G-capped Ad1 pre-mRNA was spliced in HeLa nuclear extract using standard conditions for 90 min. Immunoprecipitations were performed using an anti-CBP80 antibody (lane 2), an eIF4G antibody (lane 3), or preimmune (P.I.) serum (lane 4). RNA was recovered from the immune complex and resolved on a denaturing urea-polyacrylamide gel; lane 1, input to the immunoprecipitation. (B) HeLa nuclear splicing extracts were depleted with an immobilized antibody specific to either CBP80 (upper) or eIF4G (lower). The extent of depletion was monitored by Western blotting using specific antibodies against CBP80 (upper) or eIF4G (lower). (C) CBP80-depleted and eIF4G-depleted extracts, with their respective mock-depleted controls, were assayed for their ability to splice either m7G-capped (odd lanes) or A-capped Ad1 pre-mRNA (even lanes). The RNA was recovered from the splicing reaction and resolved on a denaturing urea-polyacrylamide gel, as described for panel A.
eIF4G colocalizes with spliceosomal snRNPs in cultured cells.
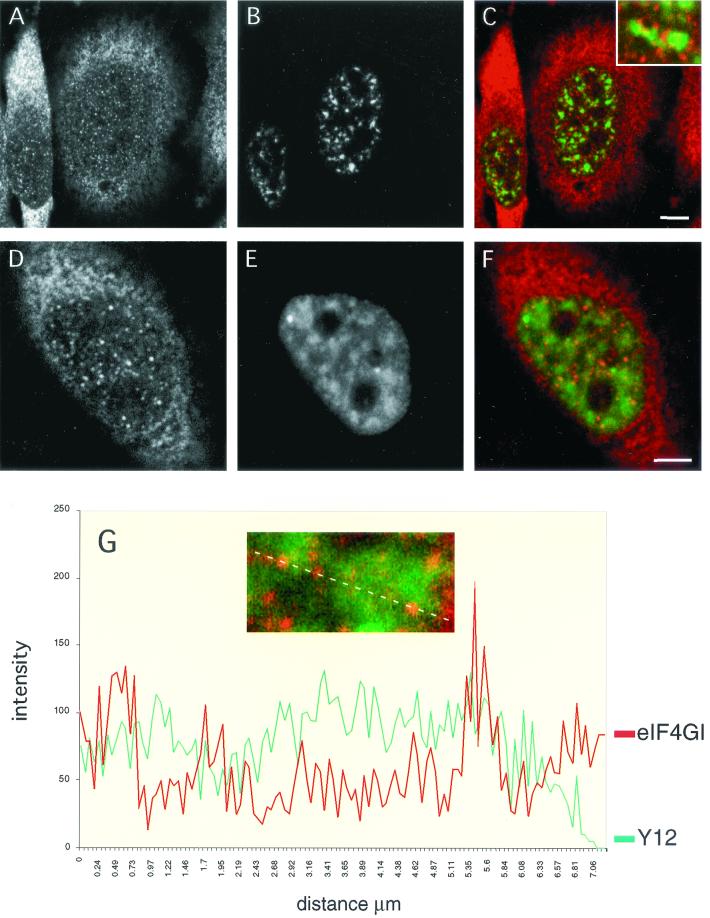
Given the association of eIF4GI with CBC and spliceosomes, we were particularly interested in understanding the relationship between the intranuclear distribution of eIF4GI and that of components of the splicing machinery. To achieve this, we performed double-labeling experiments where eIF4GI-specific affinity-purified antiserum was used in combination with monoclonal antibodies directed against components of the splicing apparatus (Fig. 5A and D). As previously described, the anti-SC-35 monoclonal antibody revealed a prominent speckled nucleoplasmic pattern (B) (18). When the confocal images of the SC-35 and eIF4GI staining were superimposed, it was apparent that, although the two patterns were dissimilar, there was a close spatial relationship between sites of more intense eIF4GI staining and nuclear speckles (Fig. 5C). Indeed, a large proportion of the eIF4GI foci localized to, and partially overlapped with, the periphery of SC-35 speckles (C, inset). In agreement with earlier studies (4, 45), the Y12 monoclonal antibody, which recognizes an epitope common to all splicing snRNPs, also elicits a speckled pattern on a diffuse nucleoplasmic staining, with a small number of very bright foci being discerned (E). In merged images of the Y12 and eIF4GI staining patterns a significant proportion of the eIF4GI foci localized to snRNP-enriched regions, which again mostly corresponded to the periphery of nuclear speckles (F). However, although concentration of eIF4GI within a speckle was a rare event, partial overlap between eIF4GI and snRNP labeling at the borders of these structures was frequently observed. The nuclear eIF4GI staining is orange compared to the cytoplasmic red staining, indicating colocalization with the diffuse Y12 staining. This was further demonstrated using a quantitative line scan through an optical section of the nucleus and representing the data graphically (Fig. 5G). The line that was quantified is shown above the graph. Several peaks of eIF4G and Y12 fluorescence colocalize; however, in contrast, many of the foci are spatially very close, and overlap at the periphery of peaks is observed. This colocalization of eIF4G staining with a subset of snRNP foci is consistent with the possibility that eIF4G may be required only for the processing of a subset of pre-mRNAs.
FIG. 5.
eIF4G partially colocalizes with spliceosomal snRNPs in vivo. HeLa cells were grown on coverslips and double labeled with an affinity-purified anti-eIF4G(Ct) antibody (A and D) and a monoclonal antibody specific for the SC-35 splicing factor (B). In the merge of the two channels (C and inset) note the close spatial relationship between eIF4G-rich sites (red signal) and the nuclear speckles (green signal). Double labelings with anti-eIF4GI (D) and the Y12 monoclonal antibody (E) show that nuclear staining of eIF4G partially colocalizes with the snRNP distribution; this is evidenced in the merged image (F; eIF4G staining, red signal; snRNP staining, green signal), where the nuclear staining of eIF4GI is mostly orange due to a partial overlap with the snRNP staining pattern. This is further shown in panel G (inset); the graph depicts the intensities of the signals from eIF4GI (red line) and snRNP (green line) staining across the dotted line (inset). Note that there is a substantial codistribution of the eIF4G and snRNP labeling, with specific foci observed at the edges of nuclear speckles.
Cellular distribution of eIF4GI is insensitive to LMB.
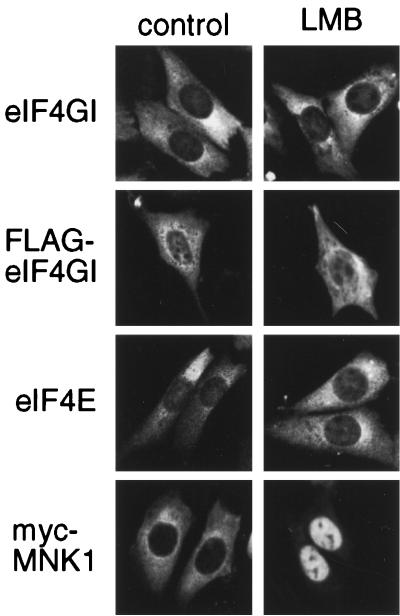
Within the sequence of eIF4GI there are a number of putative nuclear localization signals and also putative leucine-rich nuclear export signals, indicative of a protein that shuttles via the CRM1/exportin 1 pathway. To examine whether eIF4G, eIF4E, and the eIF4E kinase, Mnk1, actively shuttle between the nucleus and the cytoplasm using this pathway, we treated cells with LMB, an inhibitor of CRM1-/exportin 1-mediated nuclear export (28, 55). NIH 3T3 cells were transfected with vectors containing either FLAG-tagged eIF4GI (46) or a myc-tagged version of Mnk1 (56). After 48 h, cells were treated with LMB for 3 h and the endogenous eIF4E and eIF4GI or transfected FLAG-eIF4GI or myc-Mnk1 was localized using the appropriate antibody (Fig. 6). While both endogenous eIF4GI protein and transfected FLAG-eIF4GI were predominantly cytoplasmic, they also exhibited detectable levels of nuclear staining. However, the distribution of eIF4GI was not significantly altered by treatment of cells with LMB (Fig. 6). A similar result was obtained with endogenous eIF4E. In contrast, LMB treatment of cells caused a clear accumulation of myc-Mnk1 in the nucleus, consistent with its reported genetic interaction with importin-α (55). These data suggest that, while Mnk1 is exported from the nucleus using the CRM1-/exportin 1-dependent export pathway, eIF4GI and eIF4E must shuttle using a different export mechanism.
FIG. 6.
Nuclear localization of endogenous eIF4G and eIF4E is insensitive to LMB. NIH 3T3 cells grown on coverslips were either nontransfected or transfected with vectors encoding Myc-Mnk1 or FLAG-eIF4GI, as indicated. After 48 h cells were treated with either LMB (right) or control buffer (left) for 3 h before being fixed and stained, as indicated. In nontransfected cells, endogenous eIF4GI and eIF4E were detected using affinity-purified antibodies and the transfected proteins were visualized using commercial antibodies.
DISCUSSION
Using UV cross-linking assays and coprecipitation studies, we have shown that there are several CIPs in HeLa nuclear extracts (Fig. 1). Although the identity and function of many of these proteins remain to be determined (e.g., CIP160, CIP130, and CIP100), we have identified CIP200 as eIF4GI (Fig. 2). Cellular fractionation and indirect immunofluorescence have shown that significant amounts of eIF4GI are present in the nucleus in addition to the expected cytoplasmic localization (Fig. 3 and 5). In the cytoplasm, eIF4G plays an essential role in translation by acting as an adapter molecule during the initiation phase of protein synthesis. Within its sequence, it contains domains that interact with eIF4E, eIF4A, eIF3, PABP, and Mnk1 (reviewed in references 19, 21, 39, and 48). More recently, S. cerevisiae eIF4G has also been shown to interact with the large subunit of CBC, CBP80, via a domain between those interacting with eIF4E and eIF3 (14). Our data suggest that, in contrast to the cytoplasmic eIF4F complex (39), nuclear eIF4G in mammalian cells may not interact with eIF4E, but rather is closely associated with CBC and CIP130 (Fig. 2). At this time it is not known which region(s) of mammalian eIF4GI plays a role in its interaction with CBC, as sequence analysis failed to reveal any strong homology between the identified domain of S. cerevisiae eIF4G (14) and human eIF4G. However, these data suggest that the interaction between CBP80 and eIF4G may be evolutionarily conserved. Unfortunately, we have been unable to perform colocalization studies with CBP80 and eIF4G as the primary antibodies were raised in the same species. Although the association of CBC with eIF4G was insensitive to RNase treatment, suggesting that this interaction did not require RNA, these data do not discount the possibility that the eIF4G interaction with CBC is indirect and mediated by other nuclear proteins.
We show that, in addition to its interaction with the nuclear CBC, eIF4GI is stably associated with capped RNA throughout pre-mRNA splicing in vitro (Fig. 4). Consistent with this, we find a significant, but incomplete overlap between eIF4G and Y12 nuclear staining in vivo (Fig. 5). While the putative role of eIF4G in splicing is unclear, our data support a model whereby eIF4GI from the nuclear pool may be recruited to nascent pre-mRNA transcripts via its interaction with CBC and possibly also RNA (27), and this may be important for downstream events in gene expression. During the course of our work, it was reported that S. cerevisiae eIF4G can interact with CBP80 via a domain between those recruiting eIF4E and eIF3 (14). Furthermore, evidence from yeast two-hybrid studies have demonstrated that Prp11p, a component of yeast U2 snRNP, interacts with eIF4G (17), and we have shown that this interaction is direct (Y. Kafasla and J. D. Lewis, unpublished observations). Together, these data suggest that eIF4G may play a role in nuclear pre-mRNA processing prior to export and translation. Although immunodepletion of eIF4GI from nuclear extracts had little effect on the Ad1 pre-mRNA splicing efficiency in vitro (Fig. 4), we cannot rule out the possibility that the residual levels of eIF4GI supported efficient splicing or that not all pre-mRNAs require eIF4GI for this process. The latter would also be consistent with our data (Fig. 5), indicating that eIF4G partially colocalized with a proportion of snRNPs at discrete foci. An alternative explanation is that eIF4GII may substitute for eIF4GI in these assays. In support of this, preliminary studies have shown that eIF4GII has a cellular distribution similar to that of eIF4GI (data not shown). However, due to limiting amounts of suitable reagents, we have been unable to address biochemically whether eIF4GII interacts with CBP80 or functions in splicing.
At this time, it is not known how eIF4G is localized to the nuclear pool. Our results with LMB (Fig. 6) indicate that, in contrast to Mnk1, the CRM1/exportin 1-dependent pathway is not involved in the export of eIF4GI or eIF4E from the nucleus. As generic mRNA export is not sensitive to inhibition by LMB in mammalian cells (12, 44, 55), these data are consistent with a model whereby the eIF4G-CBC-mRNA complex is part of the export substrate. Dostie et al. (8) have recently characterized a nuclear import adapter for eIF4E and shown that LMB causes transfected eIF4E and the 4E transporter to accumulate in the nucleus. As the localization of endogenous eIF4E and eIF4G was insensitive to LMB treatment of cells (Fig. 6), it is not known how these proteins are recycled to the cytoplasm or whether they represent a novel, separate pool of initiation factors. Future efforts will be directed at determining the site of interaction of CBC with mammalian eIF4G, which cis-acting signals are responsible for the import of eIF4GI into the nucleus, and also which transporters are involved in its export.
Recently MIF4G, a domain that is conserved between eIF4G, NMD2/Upf2, and CBP80, has been described, implicating CBP80 and eIF4G in nonsense-mediated decay (47). One attractive model is that CBC bound to the cap of an mRNA interacts with eIF4G at the 5′ end, which interacts with PABP (a shuttling protein [1]) bound to the poly(A) tail at the 3′ end. This would circularize mRNA in the nucleus in a manner analogous to that described for mRNA in the cytoplasm (57). Studies on Balbiani ring mRNA in Chironomous tentans have shown that the nuclear CBC binds early to the nascent transcript and then accompanies the mRNP during nuclear export (54). This large mRNP has a highly structured crescent morphology that would juxtapose the 5′ and 3′ ends, indicating that the cap and poly(A) tail may well be in close physical contact in the nucleus. As such, the interaction of eIF4G with CBC would play a central role in allowing the cell to ensure that mRNAs are properly capped and polyadenylated prior to nuclear export, with defective RNAs being degraded in the nucleus; indeed recent work has implicated CBP80 in nuclear-RNA degradation (6). Work from a number of laboratories has shown that exon-exon junctions of spliced mRNAs are marked by nuclear proteins and that these may be implicated in coupling nuclear RNA splicing with export and recognition of premature stop codons (26, 29, 30, 58). Further work is needed to determine the exact role of CBP80 and eIF4G in nonsense-mediated mRNA decay and nuclear mRNA degradation
ACKNOWLEDGMENTS
Linda McKendrick and Elizabeth Thompson contributed equally to this work.
We thank D. Poncet for FLAG-eIF4GI, M. Yoshida for LMB, S. Kaufmann and T. Kottke for advice on cell fractionation and blots, G. Wilkie for initial help with microscopy, and members of our laboratories for helpful discussions.
Research in the laboratory of S.J.M. was supported by project and equipment grants from The Wellcome Trust (040800, 050703, 045619, and 056778), and S. J. Morley is a Senior Research Fellow of The Wellcome Trust. This research in the laboratory of J.D.L. was initially supported by the Wellcome Trust and is currently supported by the Medical Research Council. J. D. Lewis is an MRC Senior Fellow.
REFERENCES
- 1.Afonina E, Stauber R, Pavlakis G N. The human poly(A)-binding protein 1 shuttles between the nucleus and the cytoplasm. J Biol Chem. 1998;273:13015–13021. doi: 10.1074/jbc.273.21.13015. [DOI] [PubMed] [Google Scholar]
- 2.Boisvert F M, Hendzel M J, Bazett-Jones D P. Promyelocytic leukemia (PML) nuclear bodies are protein structures that do not accumulate RNA. J Cell Biol. 2000;148:283–292. doi: 10.1083/jcb.148.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bushell M, Wood W, Clemens M J, Morley S J. Changes in integrity and association of eukaryotic protein synthesis initiation factors during apoptosis. Eur J Biochem. 2000;267:1083–1091. doi: 10.1046/j.1432-1327.2000.01101.x. [DOI] [PubMed] [Google Scholar]
- 4.Carmo-Fonseca M, Tollervey D, Pepperkok R, Barabino S M, Merdes A, Brunner C, Zamore P D, Green M R, Hurt E, Lamond A I. Mammalian nuclei contain foci which are highly enriched in components of the pre-mRNA splicing machinery. EMBO J. 1991;10:195–206. doi: 10.1002/j.1460-2075.1991.tb07936.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cho E J, Takagi T, Moore C R, Buratowski S. mRNA capping enzyme is recruited to the transcription complex by phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev. 1997;11:3319–3326. doi: 10.1101/gad.11.24.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Das B, Guo Z, Russo P, Chartrand P, Sherman F. The role of nuclear cap binding protein Cbc1p of yeast in mRNA termination and degradation. Mol Cell Biol. 2000;20:2827–2838. doi: 10.1128/mcb.20.8.2827-2838.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dignam J D, Lebovitz R M, Roeder R G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dostie J, Ferraiuolo M, Pause A, Adam S A, Sonenberg N. A novel shuttling protein, 4E-T, mediates the nuclear import of the mRNA 5′ cap-binding protein, eIF4E. EMBO J. 2000;19:3142–3156. doi: 10.1093/emboj/19.12.3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dostie J, Lejbkowicz F, Sonenberg N. Nuclear eukaryotic initiation factor 4E (eIF4E) colocalizes with splicing factors in speckles. J Cell Biol. 2000;148:239–247. doi: 10.1083/jcb.148.2.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferreira J A, Carmo-Fonseca M, Lamond A I. Differential interaction of splicing snRNPs with coiled bodies and interchromatin granules during mitosis and assembly of daughter cell nuclei. J Cell Biol. 1994;126:11–23. doi: 10.1083/jcb.126.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flaherty S M, Fortes P, Izaurralde E, Mattaj I W, Gilmartin G M. Participation of the nuclear cap binding complex in pre-mRNA 3′ processing. Proc Natl Acad Sci USA. 1997;94:11893–11898. doi: 10.1073/pnas.94.22.11893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fornerod M, Ohno M, Yoshida M, Mattaj I W. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 13.Fortes P, Bilbao-Cortes D, Fornerod M, Rigaut G, Raymond W, Seraphin B, Mattaj I W. Luc7p, a novel yeast U1 snRNP protein with a role in 5′ splice site recognition. Genes Dev. 1999;13:2425–2438. doi: 10.1101/gad.13.18.2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fortes P, Inada T, Preiss T, Hentze M, Mattaj I W, Sachs A B. The yeast nuclear cap binding complex can interact with translation factor eIF4G and mediate translation initiation. Mol Cell. 2000;6:191–196. [PubMed] [Google Scholar]
- 15.Fortes P, Kufel J, Fornerod M, Polycarpou-Schwarz M, Lafontaine D, Tollervey D, Mattaj I W. Genetic and physical interactions involving the yeast nuclear cap-binding complex. Mol Cell Biol. 1999;19:6543–6553. doi: 10.1128/mcb.19.10.6543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fraser C S, Pain V M, Morley S J. The association of initiation factor 4F with poly(A)-binding protein is enhanced in serum-stimulated Xenopus kidney cells. J Biol Chem. 1999;274:196–204. doi: 10.1074/jbc.274.1.196. [DOI] [PubMed] [Google Scholar]
- 17.Fromont-Racine M, Rain J C, Legrain P. Toward a functional analysis of the yeast genome through exhaustive two-hybrid screens. Nat Genet. 1997;16:277–282. doi: 10.1038/ng0797-277. [DOI] [PubMed] [Google Scholar]
- 18.Fu X D, Maniatis T. Factor required for mammalian spliceosome assembly is localized to discrete regions in the nucleus. Nature. 1990;343:437–441. doi: 10.1038/343437a0. [DOI] [PubMed] [Google Scholar]
- 19.Gingras A C, Raught B, Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation Annu. Rev Biochem. 1999;68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- 20.Gorlich D, Kraft R, Kostka S, Vogel F, Hartmann E, Laskey R A, Mattaj I W, Izaurralde E. Importin provides a link between nuclear protein import and U snRNA export. Cell. 1996;87:21–32. doi: 10.1016/s0092-8674(00)81319-7. [DOI] [PubMed] [Google Scholar]
- 21.Hentze M. eIF4G: a multipurpose ribosome adapter. Science. 1998;275:500–501. doi: 10.1126/science.275.5299.500. [DOI] [PubMed] [Google Scholar]
- 22.Ho C K, Sriskanda V, McCracken S, Bentley D, Schwer B, Shuman S. The guanylyltransferase domain of mammalian mRNA capping enzyme binds to the phosphorylated carboxyl-terminal domain of RNA polymerase II. J Biol Chem. 1998;273:9577–9585. doi: 10.1074/jbc.273.16.9577. [DOI] [PubMed] [Google Scholar]
- 23.Izaurralde E, Lewis J, Gamberi C, Jarmolowski A, McGuigan C, Mattaj I W. A cap-binding protein complex mediating U snRNA export. Nature. 1995;376:709–712. doi: 10.1038/376709a0. [DOI] [PubMed] [Google Scholar]
- 24.Izaurralde E, Lewis J, McGuigan C, Jankowska M, Darzynkiewicz E, Mattaj I W. A nuclear cap binding protein complex involved in pre-mRNA splicing. Cell. 1994;78:657–668. doi: 10.1016/0092-8674(94)90530-4. [DOI] [PubMed] [Google Scholar]
- 25.Jarmolowski A, Boelens W C, Izaurralde E, Mattaj I W. Nuclear export of different classes of RNA is mediated by specific factors. J Cell Biol. 1994;124:627–635. doi: 10.1083/jcb.124.5.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kataoka N, Yong J, Kim V N, Velazquez F, Perkinson R A, Wang F, Dreyfuss G. Pre-mRNA splicing imprints mRNA in the nucleus with a novel RNA-binding protein that persists in the cytoplasm. Mol Cell. 2000;6:673–682. doi: 10.1016/s1097-2765(00)00065-4. [DOI] [PubMed] [Google Scholar]
- 27.Kim C Y, Takahashi K, Nguyen T B, Roberts J K, Webster C. Identification of a nucleic acid binding domain in eukaryotic initiation factor eIFiso4G from wheat. J Biol Chem. 1999;274:10603–10608. doi: 10.1074/jbc.274.15.10603. [DOI] [PubMed] [Google Scholar]
- 28.Kudo N, Wolff B, Sekimoto T, Schreiner E P, Yoneda Y, Yanagida M, Horinouchi S, Yoshida M. Leptomycin B inhibition of signal-mediated nuclear export by direct binding to CRM1. Exp Cell Res. 1998;242:540–547. doi: 10.1006/excr.1998.4136. [DOI] [PubMed] [Google Scholar]
- 29.Le Hir H, Izaurralde E, Maquat L E, Moore M J. The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon-exon junctions. EMBO J. 2000;19:6860–6869. doi: 10.1093/emboj/19.24.6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Le Hir H, Moore M J, Maquat L E. Pre-mRNA splicing alters mRNP composition: evidence for stable association of proteins at exon-exon junctions. Genes Dev. 2000;14:1098–1108. [PMC free article] [PubMed] [Google Scholar]
- 31.Lewis J D, Gorlich D, Mattaj I W. A yeast cap binding protein complex (yCBC) acts at an early step in pre-mRNA splicing. Nucleic Acids Res. 1996;24:3332–3336. doi: 10.1093/nar/24.17.3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lewis J D, Izaurralde E. The role of the cap structure in RNA processing and nuclear export. Eur J Biochem. 1997;247:461–469. doi: 10.1111/j.1432-1033.1997.00461.x. [DOI] [PubMed] [Google Scholar]
- 33.Lewis J D, Izaurralde E, Jarmolowski A, McGuigan C, Mattaj I W. A nuclear cap-binding complex facilitates association of U1 snRNP with the cap-proximal 5′ splice site. Genes Dev. 1996;10:1683–1698. doi: 10.1101/gad.10.13.1683. [DOI] [PubMed] [Google Scholar]
- 34.Lewis J D, Tollervey D. Like attracts like: getting RNA processing together in the nucleus. Science. 2000;288:1385–1389. doi: 10.1126/science.288.5470.1385. [DOI] [PubMed] [Google Scholar]
- 35.Martins L M, Mesner P W, Kottke T J, Basi G S, Sinha S, Tung J S, Svingen P A, Madden B J, Takahashi A, McCormick D J, Earnshaw W C, Kaufmann S H. Comparison of caspase activation and subcellular localization in HL-60 and K562 cells undergoing etoposide-induced apoptosis. Blood. 1997;90:4283–4296. [PubMed] [Google Scholar]
- 36.McCracken S, Fong N, Rosonina E, Yankulov K, Brothers G, Siderovski D, Hessel A, Foster S, Shuman S, Bently D Amgen EST Program. 5′-capping enzymes are targeted to pre-mRNA by binding to the phosphorylated carboxy-terminal domain of RNA polymerase II. Genes Dev. 1997;11:3306–3318. doi: 10.1101/gad.11.24.3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Milligan J F, Groebe D R, Witherell G W, Uhlenbeck O C. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987;15:8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morley S J, editor. Regulation of components of the translational machinery by protein phosphorylation. The Netherlands: Hardwood Academic Publishers, Amsterdam; 1996. [Google Scholar]
- 39.Morley S J, Curtis P S, Pain V M. eIF4G: translation's mystery factor begins to yield its secrets. RNA. 1997;3:1085–1104. [PMC free article] [PubMed] [Google Scholar]
- 40.Morley S J, McKendrick L. Involvement of stress-activated protein kinase and p38/RK mitogen-activated protein kinase signaling pathways in the enhanced phosphorylation of initiation factor 4E in NIH 3T3 cells. J Biol Chem. 1997;272:17887–17893. doi: 10.1074/jbc.272.28.17887. [DOI] [PubMed] [Google Scholar]
- 41.Neugebauer K M, Roth M B. Distribution of pre-mRNA splicing factors at sites of RNA polymerase II transcription. Genes Dev. 1997;11:1148–1159. doi: 10.1101/gad.11.9.1148. [DOI] [PubMed] [Google Scholar]
- 42.Neugebauer K M, Stolk J A, Roth M B. A conserved epitope on a subset of SR proteins defines a larger family of pre-mRNA splicing factors. J Cell Biol. 1995;129:899–908. doi: 10.1083/jcb.129.4.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ohno M, Segref A, Bachi A, Wilm M, Mattaj I W. PHAX, a mediator of U snRNA nuclear export whose activity is regulated by phosphorylation. Cell. 2000;101:187–198. doi: 10.1016/S0092-8674(00)80829-6. [DOI] [PubMed] [Google Scholar]
- 44.Otero G C, Harris M E, Donello J E, Hope T J. Leptomycin B inhibits equine infectious anemia virus Rev and feline immunodeficiency virus Rev function but not the function of the hepatitis B virus posttranscriptional regulatory element. J Virol. 1998;72:7593–7597. doi: 10.1128/jvi.72.9.7593-7597.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pettersson I, Hinterberger M, Mimori T, Gottlieb E, Steitz J A. The structure of mammalian small nuclear ribonucleoproteins. Identification of multiple protein components reactive with anti-(U1) ribonucleoprotein and anti-Sm autoantibodies. J Biol Chem. 1984;259:5907–5914. [PubMed] [Google Scholar]
- 46.Piron M, Vende P, Cohen J, Poncet D. Rotavirus RNA-binding protein NSP3 interacts with eIF4GI and evicts the poly(A) binding protein from eIF4F. EMBO J. 1998;17:5811–5821. doi: 10.1093/emboj/17.19.5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ponting C P. Novel eIF4G domain homologues linking mRNA translation with nonsense-mediated mRNA decay. Trends Biochem Sci. 2000;25:423–426. doi: 10.1016/s0968-0004(00)01628-5. [DOI] [PubMed] [Google Scholar]
- 48.Preiss T, Hentze M W. From factors to mechanisms: translation and translational control in eukaryotes. Curr Opin Genet Dev. 1999;9:515–521. doi: 10.1016/s0959-437x(99)00005-2. [DOI] [PubMed] [Google Scholar]
- 49.Rozen F, Sonenberg N. Identification of nuclear cap specific proteins in HeLa cells. Nucleic Acids Res. 1987;15:6489–6510. doi: 10.1093/nar/15.16.6489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Salditt-Georgieff M, Harpold M, Chen-Kiang S, Darnell J E., Jr The addition of 5′ cap structures occurs early in hnRNA synthesis and prematurely terminated molecules are capped. Cell. 1980;19:69–78. doi: 10.1016/0092-8674(80)90389-x. [DOI] [PubMed] [Google Scholar]
- 51.Schaegger H, von Jagow G. Tricine-sodium dodecyl sulphate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 52.Shatkin A J. Capping of eucaryotic mRNAs. Cell. 1976;9:645–653. doi: 10.1016/0092-8674(76)90128-8. [DOI] [PubMed] [Google Scholar]
- 53.Stade K, Rinke-Appel J, Brimacombe R. Site-directed cross-linking of RNA analogues to the Escherichia coli ribosome; identification of 30S ribosomal components that can be cross-linked to the mRNA at various points 5′ with respect to the decoding site. Nucleic Acids Res. 1989;17:9889–9990. doi: 10.1093/nar/17.23.9889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Visa N, Izaurralde E, Ferreira J, Daneholt B, Mattaj I W. A nuclear cap-binding complex binds Balbiani ring pre-mRNA cotranscriptionally and accompanies the ribonucleoprotein particle during nuclear export. J Cell Biol. 1996;133:5–14. doi: 10.1083/jcb.133.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Waskiewicz A J, Johnson J C, Penn B, Mahalingam M, Kimball S R, Cooper J A. Phosphorylation of the cap-binding protein eukaryotic translation initiation factor 4E by protein kinase Mnk1 in vivo. Mol Cell Biol. 1999;19:1871–1880. doi: 10.1128/mcb.19.3.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Watanabe M, Fukuda M, Yoshida M, Yanagida M, Nishida E. Involvement of CRM1, a nuclear export receptor, in mRNA export in mammalian cells and fission yeast. Genes Cells. 1999;4:291–297. doi: 10.1046/j.1365-2443.1999.00259.x. [DOI] [PubMed] [Google Scholar]
- 57.Wells S E, Hillner P E, Vale R D, Sachs A B. Circularization of mRNA by eukaryotic translation initiation factors. Mol Cell. 1998;2:135–140. doi: 10.1016/s1097-2765(00)80122-7. [DOI] [PubMed] [Google Scholar]
- 58.Zhou Z, Luo M J, Straesser K, Katahira J, Hurt E, Reed R. The protein Aly links pre-messenger-RNA splicing to nuclear export in metazoans. Nature. 2000;407:401–405. doi: 10.1038/35030160. [DOI] [PubMed] [Google Scholar]