Abstract
Atypical rough cell filaments of Listeria monocytogenes (designated FR variants), isolated from clinical and food samples, form long filaments up to 96 μm in length and demonstrated wild-type levels of adherence, invasion, and cytotoxicity to human epithelial HEp-2, Caco-2, and HeLa cells. Unlike previously described avirulent rough mutants of L. monocytogenes that secrete diminished levels of the major extracellular protein p60 and that form long chains that consist of multiple cells of similar size (designated MCR variants), FR variants secreted wild-type or greater levels of p60. This study shows that virulent filamentous forms of L. monocytogenes occur in clinical and food environments and have atypical morphological characteristics compared to those of the wild-type form.
Listeria monocytogenes is a facultative intracellular bacterial pathogen that can cause severe food-borne infections such as meningitis, encephalitis, and septicemia in humans (16). The major risk groups are immunocompromised individuals and pregnant women (16). Virulent strains of L. monocytogenes are able to survive and multiply within host macrophages and can invade, replicate, and multiply in nonprofessional phagocytes such as mouse 3T6 fibroblasts, hepatocytes, and human colon carcinoma Caco-2 epithelial cells (9, 10, 11). Epidemiological observations and electron microscopic studies of tissues of infected guinea pigs (20) provided evidence that the gastrointestinal tract is an important route of infection and that the epithelial cells of the intestine may be the primary site of entry for these bacteria.
L. monocytogenes requires a 60-kDa major extracellular housekeeping protein (termed p60) for normal cell division (6). This p60 protein is transcribed independently of the central virulence regulator PrfA, and it is also involved in the attachment of L. monocytogenes to certain eukaryotic cells and the internalization of L. monocytogenes into certain eukaryotic cells (6, 14). Spontaneously occurring, rough mutants of L. monocytogenes that secrete greatly reduced levels of p60 and that form long chains of cells (designated MCR variants in this study) were described previously (13, 14, 25). Although septum formation still occurs, separation of the divided cells does not take place (25). MCR variants were shown to have reduced virulence in the mouse model of infection and did not efficiently invade mouse 3T6 fibroblasts (14). Treatment of MCR variants with partially purified cell-free p60 led to disaggregation of cell chains to normal-sized single bacterial cells whose invasiveness was restored (6, 14, 25). Thus, for these MCR variants, cell-free p60 not only causes decay of cell chains but participates actively in the invasion process.
Here, we report on the identification and characterization of atypical rough filaments of L. monocytogenes (designated FR variants) from clinical specimens (isolated from patients with sepsis and pyrexia) and food samples that show wild-type levels of adherence, invasion, and cytotoxicity to human epithelial HEp-2, Caco-2, and HeLa cells. In addition, we show that these virulent FR isolates differ from previously reported avirulent MCR strains in that they secrete wild-type or greater levels of cell-free p60.
MATERIALS AND METHODS
Bacterial strains and growth media.
The Listeria strains used in the study were, if not otherwise indicated, derived or obtained from the Special Listeria Culture Collection (SLCC) of H. P. R. Seeliger, Würzburg, Germany, or from the National Collection of Type Cultures (NCTC), Public Health Laboratory Service (PHLS), Central Public Health Laboratory, Colindale, United Kingdom (Table 1). Two autoagglutinable blood culture and food isolates of L. monocytogenes that exhibited a rough phenotype were obtained from the Food Safety Microbiology Laboratory, PHLS, Colindale, United Kingdom. Clinical strains PHLRIII and PHLRIV were blood culture isolates from a 76-year-old female and a 72-year-old male, respectively, both of whom had sepsis and pyrexia. Strain RII (SLCC 5779) was originally obtained from J. Potel (Institute for Medical Microbiology, Medical Academy, Hannover, Germany). The rough variants SURI, SURII, and SURIII were derived from parent strains S1, S2, and S3, respectively, via heating studies (23). Isolation of spontaneous rough variant RI and L. monocytogenes rough variants RII and RIV, derived from L. monocytogenes Mackaness and EGD, respectively, was reported elsewhere (14). L. monocytogenes RIII, another rough mutant derived from a smooth strain of serovar 1/2a, was obtained from J. Potel (Institute for Medical Microbiology).
TABLE 1.
L. monocytogenes strains used and their characteristic morphological and physiological properties identified in this study
| Strain | Serotype | Reference strain no. | Origin | Cell morphology
|
Colony area (μm2)a
|
Titer by ELISA (A492) with anti-p60 MAbb | |||
|---|---|---|---|---|---|---|---|---|---|
| Form | Designation | Length (μm)c | TSYEA | LSA | |||||
| S1 | 4b | NCTC 11994 | Adult meningitis | Single, paired cells | Wild-type smooth | 3.0 ± 1.0 A | (6.1 ± 0.6) × 105 A | (9.4 ± 0.6) × 104 A | 0.63 ± 0.06 B |
| S2 | 4b | NCTC 9863 | Infantile meningitis | Single, paired cells | Wild-type smooth | 3.7 ± 1.2 A | (6.4 ± 0.4) × 105 A | (2.1 ± 1.1) × 105 B | 0.53 ± 0.09 B |
| S3 | 1/2a | NCTC 10357 | Type strain (rabbit) | Single, paired cells | Wild-type smooth | 3.3 ± 0.9 A | (5.9 ± 0.3) × 105 A | (1.7 ± 0.7) × 105 B | 0.55 ± 0.04 B |
| RId | 1/2a | SLCC 5764 | Kathariou et al. (13) | Long cell chains | MCR variant | 71 ± 18.9 B | (1.1 ± 0.8) × 106 B | (1.2 ± 0.7) × 106 C | 0.26 ± 0.08 D |
| RIIe | 1/2a | Kathariou et al. (13) | Long cell chains | MCR variant | 80 ± 28.1 B | (1.6 ± 1.3) × 106 B | (1.8 ± 0.8) × 106 C | 0.28 ± 0.02 D | |
| RIII | 1/2a | SLCC 5779 | J. Potel (see text) | Long cell chains | MCR variant | 81.5 ± 19.1 B | (1.8 ± 0.9) × 106 B | (2.1 ± 0.8) × 106 C | 0.23 ± 0.03 D |
| PHLRII | L7071 | Dried custard powder | Long cell chains | MCR variant | 68.8 ± 22.4 B | (9.1 ± 1.1) × 105 B | (9.4 ± 0.7) × 105 C | 0.31 ± 0.05 D | |
| SURIf | 4b | Rowan and Anderson (23) | Long filaments | FR variant | 56.1 ± 31 B | (9.5 ± 0.5) × 105 B | (9.8 ± 1.3) × 105 C | 0.60 ± 0.06 B | |
| SURIIg | 4b | Rowan and Anderson (23) | Long filaments | FR variant | 59.2 ± 37.9 B | (9.7 ± 1.1) × 105 B | (1.0 ± 0.8) × 106 C | 0.49 ± 0.08 C | |
| SURIIIh | 1/2a | This study | Long filaments | FR variant | 61.7 ± 42.1 B | (9.5 ± 0.5) × 105 B | (9.7 ± 0.6) × 105 C | 0.58 ± 0.05 B | |
| PHLRI | L6705 | Dried custard powder | Long filaments | FR variant | 49.9 ± 34.1 B | (9.2 ± 0.6) × 105 B | (9.3 ± 0.6) × 106 C | 0.75 ± 0.04 A | |
| PHLRIII | L7346 | Blood culture | Long filaments | FR variant | 58.2 ± 25.3 B | (8.9 ± 0.7) × 105 B | (9.1 ± 1.0) × 105 C | 0.71 ± 0.06 B | |
| PHLRIV | L1342 | Blood culture | Long filaments | FR variant | 73.5 ± 18.3 B | (9.9 ± 0.5) × 105 B | (1.4 ± 1.1) × 106 C | 0.65 ± 0.04 B | |
| RIVd | 1/2a | Kathariou et al. (13) | Cells and long filaments | IR variant | 62 ± 28.3 B | (9.6 ± 1.0) × 105 B | (8.5 ± 1.6) × 105 C | 0.72 ± 0.06 A | |
Mean of area measurements for 20 colonies.
ODs at 492 nm of greater than 0.1 were considered a positive result. BHI broth controls gave a value of 0.03 ± 0.01.
Average length from 20 measurements. Single and paired cells were measured and averaged for the smooth-cell forms. Values in the same column followed by the same letter did not differ at the level of P < 0.05, while values with different letters differed at the level of P < 0.05.
Derived from L. monocytogenes Mackaness (SLCC 5764).
Derived from L. monocytogenes EGD.
Derived from L. monocytogenes NCTC 11994.
Derived from L. monocytogenes NCTC 9863.
Derived from L. monocytogenes NCTC 10357.
Listeria cells were grown in brain heart infusion (BHI) broth (Oxoid Ltd., Basingstoke, England) at 37°C with agitation. For adhesion and invasion assays, the bacteria were harvested in the logarithmic growth phase and were washed three times by centrifugation (MSE Centaur 1) at 3,000 rpm for 10 min in Dulbecco's minimum essential medium (DMEM; Gibco BRL, Life Technologies Ltd., Paisley, Scotland). The stored bacteria were kept at −70°C in phosphate-buffered saline (PBS) with 20% (vol/vol) glycerol until they were used.
Biochemical and physiological methods.
Catalase production was determined by applying a drop of 30% H2O2 to the colonies and observing the occurrence of O2 bubbles, as described elsewhere (4). The CAMP test was performed by standard procedures by streaking out bacteria perpendicular to Staphylococcus aureus on 5% sheep blood agar plates and observing zones of augmented hemolysis, as described elsewhere (4). The characteristic blue-green sheen from the colonies as detected with obliquely transmitted light and the tumbling motility of Listeria cells were determined as described elsewhere (23). The test strains were examined for arsenic and cadmium sensitivities as described elsewhere (17). The commercial biochemical API Listeria test (bioMerieux, Marcy l'Etoile, France) was used according to the manufacturer's instructions.
Adhesion and invasion assays.
The ability of Listeria test strains to adhere to and invade HeLa, HEp-2, and Caco-2 cells was determined by previously described procedures (24), with minor modifications. HeLa, HEp-2, and Caco-2 cell monolayers were grown overnight in a 5% CO2 atmosphere at 37°C in DMEM supplemented with 10% fetal calf serum (FCS; Gibco BRL) in 24-well tissue culture plates seeded with approximately 3 × 105 cells per well. Prior to adhesion and invasion assays, the monolayers were washed three times in DMEM followed by the addition of 1 ml of DMEM containing 10% FCS to each well. Bacterial cultures were resuspended in 1 ml of DMEM to optical densities (ODs; model UV-120-02 spectrophotometer; Shimadzu Corp., Kyoto, Japan) at 580 nm of 2.0 for wild-type cells, 2.2 for FR-type cells, and 2.3 for MCR-type cells (corresponding to cell populations of ∼3.5 × 109 CFU/ml). For adherence assays, triplicate monolayers were infected with 0.1 ml of bacterial culture, followed by a 2-h incubation at 37°C in a 5% CO2 atmosphere. After incubation, nonadherent bacteria were removed by three washes with 3 ml of DMEM. The tissue culture cells were lysed with 1 ml of 1% (vol/vol) Triton X-100 (in distilled water) for 5 min at 37°C, followed by serial dilution in 0.9 ml of PBS, with subsequent enumeration by plating 0.1 ml of appropriate 10-fold dilutions on BHI agar. For invasion and intracellular growth assays, 1 ml of fresh DMEM containing 10% FCS and 100 μg of gentamicin per ml was added to the infected tissue culture monolayers, followed by a 2-h incubation at 37°C. The tissue culture cells were washed three times in 3 ml of DMEM and were then lysed with 1 ml of 1% (vol/vol) Triton X-100 (in distilled water) for 5 min at 37°C. The surviving bacteria were quantified by the same method outlined above.
Cytotoxicity assay.
Assessment of the cytotoxic effect was made by measuring total cellular metabolic activity with the tetrazolium salt 3-(4,5-dimethylthiazole-2-yl)-2,5-phenyl tetrazolium bromide (MTT; Sigma). The cytotoxicity assay of Coote and Arain (8) was used, with minor modifications. HEp-2 and HeLa cell monolayers were grown overnight at 37°C in a 5% CO2 atmosphere in DMEM supplemented with 10% FCS in 96-well microplates seeded with ∼5 × 104 cells per well. Bacterial culture supernatants, prepared as described above, were added in triplicate to the test plates immediately or after heating of the supernatants at 95°C for 10 min. Positive and negative assay controls were 1% Triton X-100 (Sigma) and PBS, respectively. Microplates that contained the infected tissue culture monolayers were incubated overnight at 37°C in a 5% CO2 atmosphere, followed by the addition of PBS containing 0.5% MTT (Sigma) to each well for 4 h at 37°C. The suspension in the wells was then removed and the formazan product was solubilized by the addition of 100 μl of 0.04 N HCl in dimethyl sulfoxide (Sigma). The contents of the wells were measured spectrophotometrically at 620 nm in a microplate reader (Bio-Rad). The toxic effect of the cell-free bacterial culture supernatant on the HEp-2 and HeLa cell lines was calculated from the following equation: (1 − OD of test sample/OD of negative control) × 100.
ELISA for detection of p60 protein.
Detection of p60 protein by an indirect enzyme-linked immunosorbent assay (ELISA) involved the addition of 100 μl of cell-free supernatant (supernatant from an overnight culture harvested by centrifugation) to each well of the microtiter plates and incubation for 2 h at 37°C. Coating proteins were washed three times with wash buffer (PBS containing 0.1% [vol/vol] Tween 20), and the L. monocytogenes-specific anti-p60 monoclonal antibody (MAb) K3A7 (7) was added. This MAb was generated against the L. monocytogenes-specific epitope, peptide D, of the p60 protein, which has been described previously (6, 12). The anti-p60 MAb was prepared as a tissue culture supernatant that was diluted 1/200 (vol/vol) in wash buffer and incubated for 1 h at room temperature. The microtiter wells were washed three times with wash buffer, sheep anti-mouse horseradish peroxidase conjugate (Sigma) was added at 100 μl well−1 and a dilution of 1/1,000 in wash buffer, and the plates were incubated for 1 h at room temperature. Excess conjugate was washed five times with wash buffer, as the substrate SIGMA FAST OPD tablets (Sigma) were added at 100 μl/well, and the plates were incubated for 0.5 h at room temperature. The A492 was measured after the addition of 50 μl of 3 M H2SO4 to each well.
Biological assay for p60.
The cell chain disruption activity of p60 was tested with rough L. monocytogenes strains (Table 1) as described elsewhere (25), with minor modifications. Strains were grown in BHI broth to the early stationary phase; cell chains were washed with PBS, resuspended to ODs of 2.0, 2.2, and 2.3 at 580 nm (for reasons described earlier) in PBS containing 20% (vol/vol) glycerol, and stored at −20°C until use. The culture supernatant from L. monocytogenes S1, which contained extracellular proteins including p60, was concentrated 100-fold with a 10-kDa-molecular-mass-cutoff Omega membrane (Microsep Microconcentrators; Filtron Technology Corp., Northborough, United Kingdom), after which the culture supernatant was filter sterilized (pore size, 0.2 μm; Sartorius). To test for chain disruption activity, 30 μl of concentrated supernatant was diluted serially twofold in PBS containing 2 mM dithiothreitol (Sigma) and was separately incubated with 30 μl of the Listeria test strains for 1 to 3 h (samples were assessed at 30-min intervals). At the indicated time points, 10 μl of the mixture was examined by phase-contrast microscopy to evaluate the lengths of the bacteria.
Cell chain length and colony appearance determination.
Overnight cultures of all L. monocytogenes strains described in Table 1 were separately incubated in BHI broth at 37°C with aeration. At various time intervals, the lengths of the cells were determined by image analysis (with a Nikon Optiphot-2 microscope that was connected to a Solitaire 512 Image Analyzer [Seescan Plc.]). Twenty cells were measured per sample. Overnight cultures were also grown at 30°C on Listeria selective agar (LSA; Oxford formulation; Oxoid) and on Tryptone soya yeast extract agar (TSYEA; Oxoid) to investigate differences in colony appearance. The areas (in square micrometers) of 20 colonies per sample were measured with the image analysis system mentioned above.
Electron microscopy.
The cells were grown to the mid-log phase, washed twice with PBS, and resuspended in sterile-distilled water before application to Formvar-coated grids. After the grid was dried, 1 drop of a solution containing 3% (vol/vol) tungstophosphoric acid and 0.3% (vol/vol) sucrose (pH 6.8 to 7.4) was added. The solution was removed after 30 to 60 s, and the grid was dried and examined on a Zeiss 902 transmission electron microscope.
Statistical methods.
All experiments in this study were performed in triplicate, and the results are reported as averages. Differences in secreted levels of cell-free p60, cell chain or filament length, colony appearance, and putative virulence properties such as levels of cytotoxicity, adherence, and penetration into human epithelial HeLa, HEp-2, and Caco-2 cells were calculated at the 95 or 99.9% confidence intervals by analysis of variance (one-way or balanced model) with Minitab software, release 11 (Minitab Inc., State College, Pa.).
RESULTS
Identification of rough variants of L. monocytogenes from clinical and food samples.
All of the bacterial strains described in Table 1 were identified as L. monocytogenes by establishing the characteristic morphological, physiological, and biochemical properties associated with this bacterial pathogen. All cultures produced catalase, were CAMP test positive with S. aureus, and were identified as L. monocytogenes by the API Listeria tests. Confirmation of the species identification occurred by analysis of culture supernatant for p60 protein by indirect ELISA with an L. monocytogenes-specific anti-p60 MAb (Table 1).
The cell and colony appearances of all rough variants were shown to differ from those of the wild-type L. monocytogenes strains, which had the typical smooth morphology. Unlike wild-type smooth strains, whose cells have the characteristic coccobacillus appearance (approximately 0.5 μm in diameter by 2 μm in length), cell types associated with the rough variants were shown to be atypically long. Some rough variants consisted of unseptated or paired filaments that measured up to 96 μm in length (designated FR variants), whereas others formed long chains that were up to 110 μm in length and that consisted of multiple cells of similar size (designated MCR variants) (Table 1). Rough variants isolated from clinical specimens and food samples or derived under conditions of heat stress predominately showed an FR filamentous phenotype (Table 1). MCR and FR variants were found to be incapable of characteristic tumbling motility and formed irregular or rough colonies that no longer produced a blue-green sheen upon oblique illumination. Image analysis data also showed that irregular rough-form colonies consistently had different areas (in square micrometers) and appearances (P < 0.05) compared to those of the smaller, wild-type smooth-form colonies after 48 h of growth on both TSYEA and LSA plates (Table 1). The MCR and FR variants of L. monocytogenes were not shown to differ in colony size or appearance on either plating medium (Table 1).
It is unlikely that the FR variants PHLRI, PHLRIII, and PHLRIV were epidemiologically linked, as they differed in their sensitivities to arsenic and cadmium on biotyping. The sensitivities to arsenic and cadmium were as follows: PHLRIII and PHLRIV, arsenic sensitive and cadmium resistant; PHLRI, arsenic and cadmium resistant. Serotyping of these particular strains was not possible as they autoagglutinate.
FR variants of L. monocytogenes secrete wild-type levels of p60 and vary in the degree of cell septation.
Spontaneously occurring MCR variants of L. monocytogenes with a rough colony appearance, namely, RI, RII, RIII, and RIV (14), and the food sample isolate PHLRII exhibited similar phenotypes; they tended to form cell chains in which septum formation between individual cells still occurred, but the cells did not separate. However, L. monocytogenes RIV differed from the other MCR variants by the random formation of septa along the length of the cell chains, resulting in cells of dissimilar lengths within the long chains. Indirect ELISA studies (Table 1) with an anti-p60 MAb showed that four of these MCR variants secrete a considerably reduced amount of the major extracellular protein p60, whereas the fifth one (variant RIV) synthesized a wild-type level or an even larger amount of this protein, as described previously (15). The addition of partially purified p60 to L. monocytogenes RI, RII, RIII, and PHLRII variants led to a decay of the cell chains (mean, 75.3 ± 22.1 μm) to the normal size (mean, 3.5 ± 1.1 μm) within 3 h of treatment. However, treatment of L. monocytogenes RIV with partially purified p60 of wild-type L. monocytogenes did not lead to a decay of the cell chains, as the cell chain lengths before (62 ± 28.3 μm) and after (66.4 ± 23.7 μm) 3 h of p60 treatment did not differ significantly (P < 0.05), which confirmed previous observations (14).
The two blood culture isolates PHLRIII and PHLRIV, the food sample isolate L. monocytogenes PHLRI, and the three L. monocytogenes variants SURI, SURII, and SURIII that we previously derived from wild-type smooth-form cultures under conditions of sublethal heat stress (Table 1) (23) also exhibited a rough colony appearance. However, these rough strains differed in cell morphology from the aforementioned MCR variants, as these FR variants formed long filaments that either were unseptated or contained a single septum (Table 1). These filaments were often of dissimilar size, as septum formation occurred at different locations. Unlike MCR variants that secrete diminished levels of cell-free p60, indirect ELISA studies showed that these FR variants produced wild-type and larger amounts of p60 (Table 1). The addition of partially purified p60 from wild-type L. monocytogenes to all FR variants did not decay the lengths of the filaments to the normal Listeria cell size.
FR variants are invasive and show wild-type L. monocytogenes levels of cytotoxicity to human epithelial HEp-2 and HeLa cells.
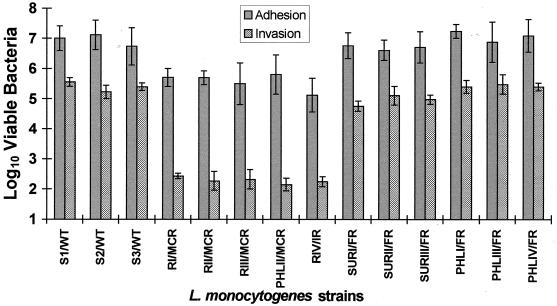
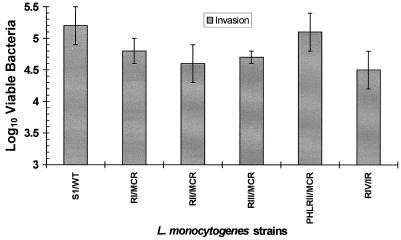
All MCR variants tested showed a considerably reduced ability to adhere to and invade HEp-2 and HeLa epithelial cells under conditions that were appropriate for the efficient attachment and uptake of the three wild-type smooth L. monocytogenes strains (Fig. 1). In marked contrast, FR variants showed levels of adherence to and invasiveness of HEp-2 and HeLa cells that were similar to those of wild-type smooth-form L. monocytogenes strains (Fig. 1). With the exception of L. monocytogenes RIV, neither epithelial cell line was more susceptible to adherence or penetration by the Listeria test strains (Fig. 1). Addition of partially purified p60 from wild-type L. monocytogenes S1 to the MCR variants L. monocytogenes RI, RII, RIII, and PHLRII not only caused a decay of cell chains but also resulted in a restoration of L. monocytogenes invasiveness to wild-type levels in HEp-2 and HeLa cells (Fig. 2). While treatment of the MCR variant L. monocytogenes RIV with p60 did not lead to a disruption of the cell chains, it did restore invasiveness to a significant extent in these human epithelial cell lines (Fig. 2). An interesting aspect of RIV is that it appears to be an intermediary morphological form (or IR variant), as it possesses both normal-sized bacterial cells and long filaments within its cell chain.
FIG. 1.
Adherence of different L. monocytogenes strains to HEp-2 cells and invasion of different L. monocytogenes strains into HEp-2 cells. Due to similarities in adhesion and invasion data for HEp-2 and HeLa cell lines (P < 0.05), only invasion results for HEp-2 cells are illustrated here.
FIG. 2.
Reconstitution of invasiveness of L. monocytogenes MCR variants by treatment of cells with p60 (concentrated supernatant of L. monocytogenes S1). Due to similarities in adhesion and invasion data for HEp-2 and HeLa cell lines (P < 0.05), only invasion results for HEp-2 cells are illustrated here.
It has also been reported that the ability of mutant RIII to adhere to Caco-2 cells is not affected; however, no data regarding the ability of this variant to invade this cell line were presented (5). Thus, the ability of a limited number of test L. monocytogenes strains that varied in their cell morphologies (namely, the wild-type strain S1, the MCR variants RIII and PHLRII, and the FR variant PHLRI) to adhere to and invade Caco-2 cells was investigated. These particular bacteria showed similar levels of adhesion (P < 0.05) to this epithelial cell line (approximately 1.4% ± 0.5% adherence for all bacterial cells challenged). However, the MCR variants L. monocytogenes RIII and PHLRII showed markedly reduced levels of invasion of Caco-2 cells (approximately 0.0009% ± 0.0003% and 0.0004% ± 0.0002% invasion of the cells that adhered, respectively) compared to the penetrating abilities of the wild-type L. monocytogenes S1 (2.3% ± 0.6% invasion) and the FR variant PHLRI (1.9% ± 0.4% invasion). This ability of the MCR variants of L. monocytogenes to show wild-type levels of adherence to Caco-2 cells is in marked contrast to the reduced adherence evident for these strains in other epithelial cell lines (i.e., 0.22% ± 0.04% and 0.18% ± 0.05% adherence to HEp-2 and HeLa cells, respectively), as described above (Fig. 1).
Due to earlier described differences in cell morphology and physiology between L. monocytogenes MCR and FR variants and smooth-form cells, the ability of these Listeria strains to elicit a cytotoxic response in HEp-2 and HeLa cells was investigated. All L. monocytogenes test strains showed similar levels of cytotoxicity for both HEp-2 and HeLa cells (P < 0.05), where the average toxicities for all 14 Listeria test strains were 68.3% ± 6.1% and 69.5% ± 4.7%, respectively. Both HEp-2 and HeLa cell lines were similarly affected by the levels of toxicity produced by the test L. monocytogenes strains (P < 0.05).
DISCUSSION
Due to the severity of listeriosis in predisposed individuals and the uncertainty as to the infectious dose of this pathogenic bacterium, the identification of atypical, virulent cell forms of L. monocytogenes in clinical or food samples is of paramount importance (16, 23). In this study we report on the characterization of atypical filaments of L. monocytogenes that were isolated from the blood of patients with pyrexia and sepsis and that showed wild-type levels of adherence, invasion, and cytotoxicity to human epithelial cells. Virulent filamentous cell forms of L. monocytogenes were also obtained from food samples and under conditions of sublethal environmental stress (23). These FR variants of L. monocytogenes not only had atypical cell and colony morphologies but were also unable to produce tumbling motility and failed to provide a characteristic blue-green sheen upon oblique illumination. While identification of irregular colonies as L. monocytogenes by conventional methods would appear to be problematic, once isolated, the identities of these atypical Listeria strains can be rapidly determined by conventional biochemical tests or by an indirect ELISA with an L. monocytogenes-specific MAb that recognizes a secreted p60 protein. The use of L. monocytogenes p60-specific antibodies for the unequivocal identification of this species, including other avirulent rough isolates, has been demonstrated in a series of publications (2, 4, 6). The test Listeria strains can also be successfully identified by indirect ELISA with the commercially available, polyclonal KPL antibody (Kirkegaard & Perry Laboratories, Gaithersburg, Md.) (data not shown).
Here, we have characterized rough variants of L. monocytogenes which differ from the wild-type smooth form of L. monocytogenes either by the formation of long chains that consist of multiple cells (MCR variants) or by the formation of single or paired filaments (FR variants). MCR variants are impaired in the synthesis of the major extracellular protein p60 and, consequently, tend to form cell chains in which septum formation between the individual cells still occurs but the cells do not separate. Other researchers have shown that p60 possesses a murein hydrolase activity which is required for a late step in cell division (3, 25). Cell-chain-disruption activity can be blocked by inhibition of the single cysteine residue that occurs in all p60 proteins at the same position in the C terminus (25).
All MCR variants that we tested showed a considerably reduced ability to invade the human epithelial HEp-2, Caco-2, and HeLa cell lines under conditions that were appropriate for efficient penetration by wild-type L. monocytogenes cells. This finding is in agreement with those of other researchers, who showed that spontaneously occurring MCR variants have a reduced ability to adhere to and invade embryo mouse fibroblast 3T6 cells (14). These data suggest that the reduced invasiveness of this MCR variant for HEp-2 and HeLa cells was due at least in part to the reduced adherence of this long-cell-chain variant. This reduction in adherence appears to be directly related to the reduced amount of cell-free p60 synthesized by these MCR mutants, since the addition of purified or partially purified p60 restored the invasiveness of this variant (14). It could be demonstrated that the addition of p60 to L. monocytogenes RIII restored adherence to and invasiveness into 3T6 fibroblasts to levels comparable to those demonstrated by wild-type L. monocytogenes (6, 14). Our study also corroborates the work of Kuhn and Goebel (14), who first described wild-type levels of p60 secretion by L. monocytogenes RIV and further showed that the addition of wild-type p60 did not disrupt these long cell chains but restored invasiveness into nonprofessional phagocytic 3T6 mouse fibroblast cells.
In contrast to MCR variants, we have described clinical and food isolates of L. monocytogenes that showed a long filamentous-like cell morphology and that secreted wild-type levels of cell-free p60. The appearance of FR variants was not altered by the addition of p60, and this atypical cell form demonstrated wild-type levels of invasiveness to HEp-2 and HeLa cells (and to Caco-2 cells for a limited number of FR variants tested). It has been observed that long filamentous forms of L. monocytogenes with a rough phenotype similar to that of lactobacilli or filamentous forms with a smooth phenotype can appear, in the latter case under the influence of suboptimal antibiotic concentrations (as cited in reference 4). However, to our knowledge, this is the only description of virulent filamentous forms of L. monocytogenes that have naturally occurred in clinical and food environments. It would appear that these autoagglutinable FR variants isolated from clinical (L. monocytogenes PHLRIII and PHLRIV) and food (L. monocytogenes PHLRI) samples are not epidemiologically linked, as the strains were isolated from different parts of the United Kingdom and have different biotypes, as a result of which they vary in their sensitivities to arsenic and cadmium. The FR phenotype is very stable on agar plates, and we assume that the FR phenotype was also presented by the clinical isolates during infection, since we also observed cell chain formation during multiplication inside host cells in our tissue culture studies (data not shown). Thus, it is not clear at which time point a mutation led to this phenotype. It must be appreciated, however, that while these clinical and food L. monocytogenes FR isolates exhibited wild-type L. monocytogenes levels of virulence in a variety of human epithelial cell lines, this does not necessarily allow one to infer that these cultures can cause disease in a complete animal.
However, in the laboratory it is possible to transform L. monocytogenes from the wild-type smooth form to the rough form. We previously showed that exposure of wild-type smooth forms of L. monocytogenes to environmental stress conditions, such as heat shock and growth at above-optimal temperatures, resulted in the generation of atypical cell forms of Listeria with an FR phenotype (23). Other researchers have also shown that exposure of L. monocytogenes to other environmental stresses results in a marked change in the lengths of treated cells (22). It is likely that the ability of L. monocytogenes to react and respond to changes in its surroundings is crucial to its survival (1, 18). It has recently been shown that exposure of bacterial enteropathogens such as Salmonella enterica serotype Typhimurium and verocytotoxigenic Escherichia coli to sublethal environmental stresses protects these problematic bacteria from lethal levels of the same and different stresses and may also increase their virulence (22). Acid-adapted L. monocytogenes cells demonstrated increased tolerance to thermal stress, osmotic stress, crystal violet, and ethanol; and emerging acid-tolerant L. monocytogenes cells were often shown to be more virulent when injected interperitoneally into mice (15). Recent evidence shows that there is expressional cross talk between the central virulence regulator PrfA and the stress response mediator ClpC in L. monocytogenes (21). The reasons for the difference in cell morphology and virulence of FR variants compared with those of the wild-type smooth form of L. monocytogenes and other types of R variants are still not known. However, other studies have shown that double mutations of the molecular chaperones ClpC and ClpE in Listeria cells also affect both cell division and virulence (19).
In summary, we have described the occurrence and unequivocal identification of three different rough forms of L. monocytogenes from clinical and food samples and from strains derived under conditions of sublethal heat stress. As the occurrence of virulent FR variants in clinical and food environments may be underestimated because of misidentification and underrecognition, we suggest that irregularly shaped bacterial cultures from clinical and food samples that produce a characteristic black precipitate on LSA plates should be further investigated.
ACKNOWLEDGMENTS
We thank the University of Strathclyde for funding this research.
We thank K. Deans for tissue culture assistance and V. Gill and D. McLaughlin for excellent technical support.
REFERENCES
- 1.Abee T, Wouters J A. Microbial stress response in minimal processing. Int J Food Microbiol. 1999;50:65–92. doi: 10.1016/s0168-1605(99)00078-1. [DOI] [PubMed] [Google Scholar]
- 2.Allerberger F, Dierich M, Petranyi G, Lalic M, Bubert A. Nonhemolytic strains of Listeria monocytogenes detected in milk products using VIDAS immunoassay kit. Zentbl Bakteriol Parasitenkd Infektkrankh Hyg Abt 1 Orig. 1997;200:189–195. [PubMed] [Google Scholar]
- 3.Bubert A, Kestler H, Götz M, Böckmann R, Goebel W. The Listeria monocytogenes iap gene as an indicator gene for the study of PrfA-dependent regulation. Mol Gen Genet. 1997;256:54–62. doi: 10.1007/s004380050545. [DOI] [PubMed] [Google Scholar]
- 4.Bubert A, Riebe J, Schnitzler N, Schönberg A, Goebel W, Schubert P. Isolation of catalase-negative Listeria monocytogenes strains from listeriosis patients and their rapid identification by anti-p60 antibodies and/or PCR. J Clin Microbiol. 1997;35:179–183. doi: 10.1128/jcm.35.1.179-183.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bubert A, Kuhn M, Goebel W, Köhler S. Structural and functional properties of the p60 proteins from different Listeria species. J Bacteriol. 1992;174:8166–8171. doi: 10.1128/jb.174.24.8166-8171.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bubert A, Schubert P, Köhler S, Frank R, Goebel W. Synthetic peptides derived from the Listeria monocytogenes p60 protein as antigens for the generation of polyclonal antibodies specific for secreted cell-free L. monocytogenes p60 proteins. Appl Environ Microbiol. 1994;60:3120–3127. doi: 10.1128/aem.60.9.3120-3127.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chakraborty T, Leimeister-Wächter M, Domann E, Hartl M, Goebel W, Nichterlein T, Notermans S. Coordinate regulation of virulence genes in Listeria monocytogenes requires the product of the prfA gene. J Bacteriol. 1992;174:568–574. doi: 10.1128/jb.174.2.568-574.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coote J G, Arain T. A rapid, colourimetric assay for cytotoxin activity in Campylobacter jejuni. FEMS Immunol Med Microbiol. 1996;13:65–70. doi: 10.1111/j.1574-695X.1996.tb00217.x. [DOI] [PubMed] [Google Scholar]
- 9.Dramsi S, Biswas I, Maguin E, Braun L, Mastroeni P, Cossart P. Entry of Listeria monocytogenes into hepatocytes requires expression of inlB, a surface protein of the internalin multigene family. Mol Microbiol. 1995;16:251–261. doi: 10.1111/j.1365-2958.1995.tb02297.x. [DOI] [PubMed] [Google Scholar]
- 10.Drevets D A, Sawyer R T, Potter T A, Campbell P A. Listeria monocytogenes infects human endothelial cells by two distinct mechanisms. Infect Immun. 1995;63:4268–4276. doi: 10.1128/iai.63.11.4268-4276.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gaillard J L, Berche P, Mounier J, Richard S, Sansonetti P. In vitro model of penetration and intracellular growth of Listeria monocytogenes in the human enterocyte-like cell line Caco-2. Infect Immun. 1987;55:2822–2829. doi: 10.1128/iai.55.11.2822-2829.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 13.Kathariou S, Hacker J, Hof H, Then I, Wagner W, Kuhn M, Goebel W. Bacterial cytotoxins—extracellular proteins and virulence factors. In: Rott R, Goebel W, editors. Molecular basis of viral and microbial pathogenesis. Berlin, Germany: Springer-Verlag; 1987. pp. 141–150. [Google Scholar]
- 14.Kuhn M, Goebel W. Identification of an extracellular protein of Listeria monocytogenes possibly involved in intracellular uptake by mammalian cells. Infect Immun. 1989;57:55–61. doi: 10.1128/iai.57.1.55-61.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marron L, Emerson N, Gahan C G M, Hill C. A mutant of Listeria monocytogenes L028 unable to induce an acid tolerance response displays diminished virulence in a murine model. Appl Environ Microbiol. 1997;63:4945–4947. doi: 10.1128/aem.63.12.4945-4947.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McLauchlin J. The pathogenicity of Listeria monocytogenes: a public health perspective. Rev Med Microbiol. 1997;8:1–14. [Google Scholar]
- 17.McLauchlin J, Hampton M D, Shah S, Threlfall J, Wieneke A A, Curtis G D W. The subtyping of Listeria monocytogenes on the basis of plasmid profiles and arsenic and cadmium susceptibilities. J Appl Microbiol. 1997;83:381–388. doi: 10.1046/j.1365-2672.1997.00238.x. [DOI] [PubMed] [Google Scholar]
- 18.Mekalanos J J. Environmental signals controlling expression of virulence determinants in bacteria. J Bacteriol. 1992;174:1–7. doi: 10.1128/jb.174.1.1-7.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nair S, Frehel C, Nguyen L, Escuyer V, Berche P. ClpE, a novel member of the HSP100 family, is involved in cell division and virulence of Listeria monocytogenes. Mol Microbiol. 1999;31:185–196. doi: 10.1046/j.1365-2958.1999.01159.x. [DOI] [PubMed] [Google Scholar]
- 20.Racz P, Tenner K, Mero E. Experimental Listeria enteritis. 1. An electron microscopic study of the epithelial phase in experimental Listeria infection. Lab Investig. 1972;26:694–700. [PubMed] [Google Scholar]
- 21.Ripio M-T, Vázquez-Boland J-A, Vega Y, Nair S, Berche P. Evidence for expressional crosstalk between the central virulence regulator PrfA and the stress response mediator ClpC in Listeria monocytogenes. FEMS Microbiol Lett. 1998;158:45–50. doi: 10.1111/j.1574-6968.1998.tb12798.x. [DOI] [PubMed] [Google Scholar]
- 22.Rowan N J. Evidence that inimical food-preservation barriers alter microbial resistance, cell morphology and virulence. Trends Food Sci Technol. 1999;10:261–270. [Google Scholar]
- 23.Rowan N J, Anderson J G. Effects of above-optimum growth temperature and cell morphology on thermotolerance of Listeria monocytogenes cells suspended in bovine milk. Appl Environ Microbiol. 1998;64:2065–2071. doi: 10.1128/aem.64.6.2065-2071.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Strom M S. Phenotypic and genetic characterization of a non-hemolytic variant of Listeria monocytogenes from cold-smoked salmon. Food Microbiol. 1998;15:329–337. [Google Scholar]
- 25.Wuenscher M D, Köhler S, Bubert A, Gerike U, Goebel W. The iap gene of Listeria monocytogenes is essential for cell viability, and its gene product, p60, has bacteriolytic activity. J Bacteriol. 1993;175:3491–3501. doi: 10.1128/jb.175.11.3491-3501.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]