Abstract
As an important bio-based chemical, methyl levulinate (ML) can be produced via enzymatic esterification of levulinic acid with methanol. A kinetic model is developed in this work based on the law of mass action and reaction reversibility, to investigate the effect of enzyme loading, alcohol/acid ratio and temperature on ML yield. Data analysis shows that newly developed binary regression is apparently more persuasive than the commonly used unitary regression. Kinetic study reveals: (1) rate constants of esterification/hydrolysis increase with increasing enzyme loading, while their ratio (equilibrium constant) remains invariant. (2) Methanol has no toxicity towards lipase, and hence, neither the rate constants of esterification/hydrolysis nor the equilibrium constant are affected by alcohol/acid ratio. (3) Both rate constants of esterification/hydrolysis and the equilibrium constant increase with temperature elevation, and their relationships agree with Arrhenius equation and Van't Hoff equation, respectively. (4) The esterification is endothermic and spontaneous. In total, the application of binary regression analysis for the developed model to study the enzymatic esterification kinetics is quite successful.
Based on reaction reversibility and the law of mass action, a mathematical model was developed. By the developed model, the effect of enzyme loading, molar alcohol/acid ratio, and temperature on methyl levulinate yield was kinetically analyzed.
1. Introduction
As an important bio-based chemical, methyl levulinate (ML) can be used as a gasoline/diesel additive, a green solvent, and a plasticizing and antifreeze agent.1–3 ML is mostly produced from enzymatic or chemical esterification of levulinic acid (LA).1,4 Compared to chemical catalysis, enzymatic catalysis has many benefits such as mild operation conditions, high product specificity and low pollution.5–7 Esterification is reversible, and H2O is also a product, which can promote reverse esterification (hydrolysis).8 Hence, in order to obtain high ML yield, the esterification should be carried out in non-aqueous systems (organic phase).7,9 However, organic solvents and alcohols are always toxic toward lipase, hence, direct use of free lipase cannot obtain efficient esterification. Lipase must be immobilized to improve its tolerance.10–12 Moreover, immobilization can provide the recyclability of the biocatalyst for possible-reuse, which can sharply reduce the lipase cost. Therefore, lipase immobilization gains more and more attention, and the enzymatic esterification becomes a heterogeneous reaction owing to the insolubility of the immobilized lipase.
To quantitively describe the effect of lipase loading, alcohol/acid ratio and temperature on ML yield, it is desirable to study the enzymatic esterification kinetics.13–15 Besides, the development of kinetic models always helps to predict the reaction results and gain insight into the mechanism.14,16 However, complex heterogeneous systems make it difficult to develop a kinetic model for reversible esterification. The typical Michaelis–Menten theory based on homogeneous systems cannot been applied to the heterogeneous process.
Based on reaction reversibility and law of mass action, a second-order model has been developed and it has gained much popularity.8,17–21 However, during the application of the model, either the rate constants vary with substrate concentration or the equilibrium constant varies with substrate concentration and enzyme loading.17–19,21 Further, irregular variation in the rate and equilibrium constants always occurs when the temperature increases.18,20,21 Obviously, it is unreasonable. Toward this, some constraint conditions are added in this study to revise the data analysis, in which the relationship between rate/equilibrium constants and various factors exhibits rationale rules.
2. Experimental
2.1. Materials
LA, ML and methanol were purchased from Aladdin (Shanghai, China). An organic solvent namely 1-butyl-3-methylimidazolium hexafluorophosphate ([bmim][PF6]) was purchased from Zhejiang Xinming Chemical Co., Ltd. (Ningbo, China). Immobilized lipase CAL-B (Candida Antarctica Lipase B) was purchased from Novozymes Co., Ltd. (Tianjin, China). 3Å molecular sieves were purchased from Tianjin Kemiou Chemical Reagent Co., Ltd. (Tianjin, China).
2.2. Dehydration of organic solvent
3Å molecular sieves were activated in a muffle furnace at 550 °C for 5 h, and cooled to room temperature in a desiccator. In order to dehydrate the organic solvent, the solvent and the activated molecular sieves were mixed at 150 rpm and room temperature. After 24 h, the molecular sieves were separated, and the obtained organic solvent was used as the dehydrated solvent for esterification reaction.
2.3. Esterification reaction
LA and methanol were mixed in a molar ratio of 1 : 3 in a brown Erlenmeyer flask, and then [bmim][PF6] was added with methanol volume of 3 times. To start the reaction, 10 g L−1 CAL-B was added. The reaction temperature and rotation speed were 30 °C and 150 rpm, respectively. At 3 h, 5 h, 7 h, 9 h, 18 h and 24 h, 0.4 g samples were taken out and diluted with methanol in a 5 mL volumetric flask. The diluted samples were then filtered with 0.22 μm filters and used for ML analysis by gas chromatography as per our previous report.6
2.4. Model development
Based on the law of mass action and reaction reversibility, eqn (1) was firstly proposed by Han et al.:8
 |
1 |
Solving eqn (1) with the boundary condition (Y = 0, at t = 0), Y can be expressed as shown in eqn (2) and (3):
 |
2 |
 |
3 |
3. Results and discussion
3.1. Kinetic study on the effect of enzyme loading
The amount of enzyme loading CE directly decides the reaction rate and the time for the reaction to reach equilibrium. CAL-B predominantly exhibits esterification activity over hydrolytic activity.6 Therefore, the more CE is, the higher Y is at the same reaction time (Fig. 1 (symbol points)). With increase in CE, the binding of enzyme and substrate becomes more and more saturated, and hence the trend of increase in Y become progressively lower. Over 50% ML was produced within the first 5 h except for CE = 2 g L−1. The slowdown of reaction rate could be caused by enzyme deactivation,22,23 product inhibition24 and attainment of chemical equilibrium13 as others have demonstrated. Increase in CE can cause high cost even if Y is improved. Therefore, CE = 8 g L−1 may be the optimum choice.
Fig. 1. Experimental data versus fitted lines of ML yield for lipase-catalysed esterification of LA and methanol at different enzyme loadings. Temperature: 303.15 K; molar alcohol/acid ratio: 3.
Firstly, unitary regression was used to analyse the experimental Y, as shown in Fig. 1 (symbol points), where only t was taken as the independent variable and experimental Y at each CE were separately fitted by eqn (2). As shown in Fig. 1 (dotted lines), the fitting accuracy is very high, as verified by R2 (Table 1 (unitary regression)). As shown in Table 1, k2 decreases with the increase in CE although k1 increases with its increase, which has also been reported by others.19 Normally, both k1 and k2 should present a positive correlation with CE. Since the present results show the opposite, therefore, a significant problem exists for unitary regression. Besides, equilibrium constant Ke, defined by eqn (4), varies with CE (Table 1 (unitary regression)). Clearly, it is also unreasonable because Ke is related only to temperature. Similar issues exists in other reports, but further elaboration and analysis is still lacking.17–19,21 Both k1 and k2 increase with the increase in CE as per Tomke and Rathod's report, however, Ke at each CE is still different.21
 |
4 |
Fitted values of k1 and k2 from unitary and binary regression analyses of experimental data at each CE.
| Fitting method | Parameter | Unit | C E (g L−1) | ||||
|---|---|---|---|---|---|---|---|
| 2 | 4 | 6 | 8 | 10 | |||
| Unitary regression | k 1 | L h−1 mol−1 | 0.007354 | 0.03006 | 0.06844 | 0.09960 | 0.1142 |
| k 2 | L h−1 mol−1 | 0.1925 | 0.08964 | 0.05690 | 0.03965 | 0.02193 | |
| K e | 0.034 | 0.34 | 1.204 | 2.514 | 5.21 | ||
| R 2 | 0.9937 | 0.9909 | 0.9935 | 0.9974 | 0.9997 | ||
| Binary regression | k 1 | L h−1 mol−1 | 0.006006 | 0.02109 | 0.04399 | 0.07409 | 0.1110 |
| k 2 | L h−1 mol−1 | 0.001337 | 0.004695 | 0.009790 | 0.01649 | 0.02471 | |
| K e | 4.49 | ||||||
| R 2 | 0.9411 | 0.9112 | 0.9289 | 0.9770 | 0.9996 | ||
To solve the above problems, the fitting has been revised in the present work by adding some constraint conditions as shown in eqn (5):
| ki = ki,E × CEm, i = 1 or 2 | 5 |
The enzymatic promotion factor m must be identical for esterification and hydrolysis reactions. Otherwise, Ke will become different at different CE, as reported in our previous study.13 Binary regression is proposed by combining eqn (2) and (5), where both CE and t are taken as independent variables.
The proposed binary regression can ensure the increase in the rate constant and invariance of Ke with CE increase. Table 2 lists the parametric values fitted through binary regression, where k1,E is larger than k2,E. Hence, k1 is always larger than k2 at any CE according to eqn (5), and their ratio is the constant Ke. The value of m quantitatively describes the effect of CE on k1 and k2. The larger m is, the faster k1 and k2 increase with the increase in CE. As shown in Fig. 1 (solid lines), the experimental Y is also very close to the fitted lines, as verified by R2 (Table 1 (binary regression)), although the fitting accuracy is no better than that obtained through unitary regression. What's more, the proposed binary regression ensures the regular variance of k1 and k2, and invariance of Ke, compared to unitary regression.
Fitted parametric values of intrinsic rate constant and enzymatic promotion factor from binary regression analysis of experimental data at various enzyme loadings.
| Parameter | k 1,E | k 2,E | m | R 2 |
| Unit | g L−1 mol−1 | g L−1 mol−1 | ||
| 0.001710 | 0.0003806 | 1.8124 | 0.9679 |
3.2. Effect of alcohol/acid ratio
Alcohol, as a substrate of esterification, can favourably shift the reaction toward ML production (esterification). Hence, increasing Rm accelerates the esterification process and enhances Y as shown in Fig. 2 (symbol points). However, the increase in the trend becomes progressively smaller with increasing Rm. The increase in Y is very significant when Rm increases from 0.5 to 1.0 and from 1.0 to 1.5, while only a slight increase is observed when Rm increases from 1.5 to 2, from 2 to 2.5, and from 2.5 to 3.
Fig. 2. Experimental data versus fitted lines of Y for lipase-catalyzed esterification of LA and methanol at different Rm. Enzyme loading: 10 g L−1; temperature: 303.15 K.
Besides, Rm increase results in the decrease in the conversion rate of alcohol, although the conversion rate of LA increases. Therefore, Rm = 1.5 may be the best choice. Similarly, unitary regression was initially used to fit the experimental Y in Fig. 2 (symbol points), where only t was taken as the independent variable and experimental Y at each Rm were separately substituted to eqn (2).
Fig. 2 (dotted lines) shows a very high fitting accuracy, as verified by R2 > 0.98 in Table 3 (unitary regression). The table also shows that both k1 and k2 do not present a corresponding relationship with Rm. The variance in both k1 and k2 with the increase in Rm seems to be irregular, and even k2 = 0 when Rm = 1.5. Similar problems have been presented in other reports, but no major discussion or resolution has been proposed.18,20,21 Although both k1 and k2 decease as Rm increases as reported by Alves et al. (heptane as the solvent), the value of Ke is not identical at each Rm.18 Therefore, the fitting should be revised to overcome the problem.
Fitted values of k1 and k2 from unitary and binary regression analyses of experimental data for each Rm.
| Fitting method | Parameter | Unit | R m | |||||
|---|---|---|---|---|---|---|---|---|
| 0.5 | 1.0 | 1.5 | 2.0 | 2.5 | 3.0 | |||
| Unitary regression | k 1 | L h−1 mol−1 | 0.1343 | 0.1035 | 0.08362 | 0.07394 | 0.1321 | 0.1142 |
| k 2 | L h−1 mol−1 | 0.02127 | 0.01203 | 0 | 0.006472 | 0.02711 | 0.02193 | |
| K e | 6.31 | 8.60 | +∞ | 11.42 | 4.87 | 5.21 | ||
| R 2 | 0.9966 | 0.9777 | 0.9837 | 0.9901 | 0.9915 | 0.9997 | ||
| Binary regression | k 1 | L h−1 mol−1 | 0.1110 | |||||
| k 2 | L h−1 mol−1 | 0.02471 | ||||||
| K e | 4.49 | |||||||
| R 2 | 0.9939 | 0.9778 | 0.9636 | 0.9615 | 0.9864 | 0.9996 | ||
Generally, neither k1 nor k2 is related to substrate concentration. However, alcohol is a unique substrate which always has some toxicity toward the enzyme. It has been reported that alcohols, especially short-chain alcohols can seriously inhibit the activity of some lipases.25,26 In this study, the inhibition of alcohol on lipase activity is described by rate constant as shown in eqn (6):
| ki = ki,Rm × CM0−n = ki,Rm × (Rm × CS0)−n, i = 1 or 2 | 6 |
From eqn (6), it can be concluded that both k1 and k2 decrease with the increase in Rm. Using eqn (6) as constraint condition, the binary regression is used to fit the experimental data in Fig. 2 (symbol points) by eqn (2), where both t and Rm are considered as independent variables.
Surprisingly, the fitted value of n is 0, which validates that alcohol doesn't have any toxicity towards CAL-B. Hence, k1 and k2 are identical at each Rm. This outcome may be due to CAL-B being an immobilized lipase, and the high tolerance against alcohol is attained via immobilization.10,12 Substituting the same values of k1 and k2 in eqn (2), the correlation coefficient (R2 > 0.96) shows a very high fitting accuracy. Of course, the fitting performance is not superior to that of unitary regression, but confirms the invariance of Ke at each Rm.
3.3. Effect of temperature
T is another important parameter for enzymatic reaction.27,28 Both esterification and hydrolysis rate constants can be improved by evaluating T. However, the improvement is limited within a certain range due to inevitable denaturation and deactivation of lipase at high temperatures.29 As shown in Fig. 3 (symbol points), Y increased with the increase in T.
Fig. 3. Experimental data versus fitted lines of Y for lipase-catalyzed esterification of LA and methanol at different T. Enzyme loading: 10 g L−1; molar alcohol/acid ratio: 3.
Similarly, unitary regression was initially used to fit the experimental Y in Fig. 3 (symbol points), where only t was taken as the independent variable. For the proposed unitary regression, experimental Y at each T was substituted into eqn (2). The fitted parametric values are listed in Table 4. Although k1 increases with T elevation, k2 decreases. Obviously, the fitting based on unitary regression is unreasonable. Similar issues were also reported by others, but further analysis and resolution is still lacking.17,19 Besides, completely irregular variance of k1 and k2 at different T, has also been reported.18,20,21 To this end, some constraint conditions must be added to revise the fitting.
Fitted values of k1 and k2 from unitary and binary regression analyses of experimental data at each molar alcohol/acid ratio.
| Fitting method | Parameter | Unit | T (K) | ||
|---|---|---|---|---|---|
| 293.15 | 298.15 | 303.15 | |||
| Unitary regression | k 1 | L h−1 mol−1 | 0.02935 | 0.03761 | 0.1142 |
| k 2 | L h−1 mol−1 | 0.09462 | 0.02751 | 0.02192 | |
| K e | 0.31 | 1.37 | 5.21 | ||
| R 2 | 0.9953 | 0.9916 | 0.9997 | ||
| Binary regression | k 1 | L h−1 mol−1 | 0.01720 | 0.04439 | 0.1110 |
| k 2 | L h−1 mol−1 | 0.004205 | 0.01035 | 0.02471 | |
| K e | 4.09 | 4.29 | 4.49 | ||
| R 2 | 0.8980 | 0.9902 | 0.9996 | ||
It is widely accepted that the relationship between the rate constant and temperature always agrees with Arrhenius equation, as shown in eqn (7):
 |
7 |
Using eqn (7) as the constraint condition, binary regression is proposed for eqn (2) to analyse the experimental Y at all T, where both t and Y are taken as independent variables. The fitted lines and parametric values are shown Fig. 3 (dotted lines) and Table 5, respectively.
Fitted parametric values of intrinsic rate constant and enzymatic promotion factor from binary regression analysis of experimental data at various temperatures.
| Parameter | E a,1 | E a,2 | A 1 | A 2 | R 2 |
| Unit | kJ mol−1 | kJ mol−1 | L h−1 mol−1 | L h−1 mol−1 | |
| 138 | 131 | 6.15 × 1022 | 8.79 × 1020 | 0.9611 |
E a,1 > Ea,2, indicates that the occurrence of esterification reaction requires a higher activation energy compared to the hydrolysis reaction. As a result of A1 being much larger than A2, the value of k1 is still higher than that of k2, although Ea,1 > Ea,2.
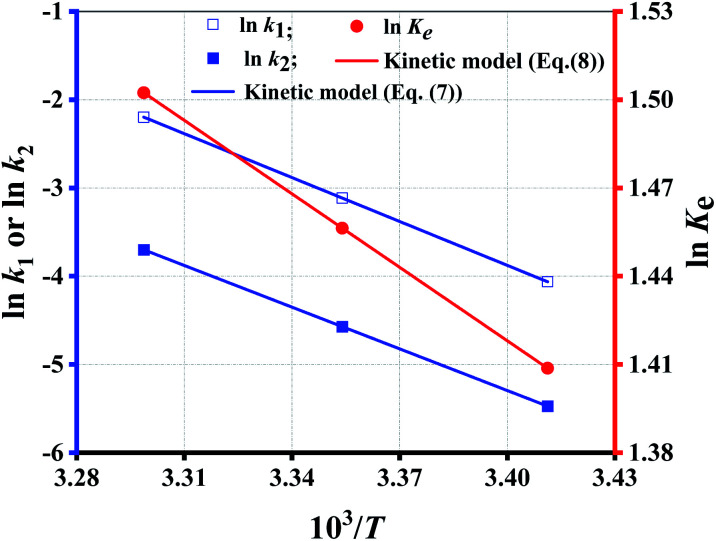
Since eqn (7) is a constraint condition, the relationship between ln k1 (or ln k2) and 1/T is fully linear (Fig. 4). Taking the data from Table 5, the values of k1 or k2 are calculated using eqn (7) (Table 4 (binary regression)). Thereafter, the kinetics lines are drawn using eqn (7) (Fig. 3 (dotted lines)). The figure shows that experimental data is very close to the kinetics lines, indicative of a very good fitting, as verified by the R2 values listed in Table 4. Overall, the fitting accuracy at high temperatures is apparently higher than at low temperatures.
Fig. 4. The fitting of rate/equilibrium constant with temperature.
Reaction equilibrium can be changed by T other than by CE and Rm. The effect of T on the reaction equilibrium is evaluated by Ke at each T, as defined by eqn (8):
 |
8 |
Taking the values of k1 and k2 listed in Table 4 (binary regression) into eqn (8), value of Ke is calculated at each T. According to Van't Hoff equation, the relationship between Ke and T can be described by eqn (9):
 |
9 |
Fig. 4 shows that the fitting is rather perfect, and the values of ΔH and ΔS are 6919 J mol−1, and 35.31 J mol−1 K−1, respectively. Ke increases with T elevation, and so ΔH > 0, which indicates that esterification is an endothermic reaction. Hence, the elevation of T can shift the equilibrium to esterification, which is beneficial to obtain higher Y. As result of Ke > 1, ΔG > 0 at any T, which demonstrates that the esterification occurs spontaneously.
4. Conclusions
Compared to unitary regression, binary regression has been demonstrated to be more suitable for the developed model to analyze the effect of CE, Rm and T on enzymatic esterification of LA with methanol. Both k1 and k2 increase with the increase in CE and T, while kept invariant at all Rm. Ke is not related to CE and Rm, but increases with T elevation. Besides, kinetic study also shows that the esterification is endothermic and spontaneous.
Nomenclature
- Y
ML yield (%)
- t
Reaction time (h)
- k 1
Rate constant of esterification (L h−1 mol−1)
- k 2
Rate constant of hydrolysis (L h−1 mol−1)
- C S0
Initial substrate LA concentration (mol L−1)
- C M0
Initial substrate methanol concentration (mol L−1)
- R m
Initial molar alcohol/acid ratio (CM0/CS0)
- K
Apparent rate constant (L h−1 mol−1)
- C E
Lipase loading (g L−1)
- K e
Equilibrium constant
- k 1,E
Intrinsic esterification rate constant of related to enzyme (L h−1 mol−1)
- k 2,E
Intrinsic hydrolysis rate constant of related to the enzyme (L h−1 mol−1)
- m
Enzymatic promotion factor
- k 1,Rm
Intrinsic esterification rate constant of related to alcohol(L h−1 mol−1)
- k 2,Rm
Intrinsic hydrolysis rate constant of related to alcohol (L h−1 mol−1)
- n
Alcohol inhibition factor
- A 1
Pre-exponential factor of esterification ((L h−1 mol−1))
- A 2
Pre-exponential factor of hydrolysis (L h−1 mol−1)
- E a,1
Activation energy of esterification (kJ mol−1)
- E a,2
Activation energy of hydrolysis (kJ mol−1)
- R
Molar gas constant (J mol−1 K−1)
- T
Temperature (K)
- ΔG
Gibbs free energy (J mol−1)
- ΔH
Enthalpy change (J mol−1)
- ΔS
Entropy change (J mol−1 K−1)
Author contributions
All authors contributed to the writing of the manuscript. All authors have approved the final version of the manuscript.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
This work is funded by the National Natural Science Foundation of China (21978288) and China Scholarship Council for her PhD fellowship (202008440326).
References
- Pileidis F. D. Titirici M. M. ChemSusChem. 2016;9:562–582. doi: 10.1002/cssc.201501405. [DOI] [PubMed] [Google Scholar]
- Raspolli Galletti A. M. Antonetti C. Fulignati S. Licursi D. Catalysts. 2020:10. [Google Scholar]
- Yan L. Yao Q. Fu Y. Green Chem. 2017;19:5527–5547. doi: 10.1039/C7GC02503C. [DOI] [Google Scholar]
- Badgujar K. C. Badgujar V. C. Bhanage B. M. Fuel Process. Technol. 2020;197:19. doi: 10.1016/j.fuproc.2019.106213. [DOI] [Google Scholar]
- Jia B. Y. Liu C. G. Qi X. H. Fuel Process. Technol. 2020:210. [Google Scholar]
- Di X. H. Zhang Y. Fu J. Y. Yu Q. Wang Z. M. Yuan Z. H. Process Biochem. 2019;81:33–38. doi: 10.1016/j.procbio.2019.03.024. [DOI] [Google Scholar]
- Stergiou P. Y. Foukis A. Filippou M. Koukouritaki M. Parapouli M. Theodorou L. G. Hatziloukas E. Afendra A. Pandey A. Papamichael E. M. Biotechnol. Adv. 2013;31:1846–1859. doi: 10.1016/j.biotechadv.2013.08.006. [DOI] [PubMed] [Google Scholar]
- Han D. Rhee J. S. Lee S. B. Biotechnol. Bioeng. 1987;30:381–388. doi: 10.1002/bit.260300309. [DOI] [PubMed] [Google Scholar]
- Dong B. Song H. Zhang W. He A. Yao S. Curr. Org. Chem. 2016;20:2894–2910. doi: 10.2174/1385272820666160902150343. [DOI] [Google Scholar]
- Ismail A. R. Baek K. H. Int. J. Biol. Macromol. 2020;163:1624–1639. doi: 10.1016/j.ijbiomac.2020.09.021. [DOI] [PubMed] [Google Scholar]
- Matsumoto T. Yamada R. Ogino H. World J. Microbiol. Biotechnol. 2019;35:193. doi: 10.1007/s11274-019-2777-8. [DOI] [PubMed] [Google Scholar]
- Facin B. R. Melchiors M. S. Valerio A. Oliveira J. V. de Oliveira D. Ind. Eng. Chem. Res. 2019;58:5358–5378. doi: 10.1021/acs.iecr.9b00448. [DOI] [Google Scholar]
- Zhang Y. Di X. H. Wang W. Song M. J. Yu Q. Wang Z. M. Yuan Z. H. Chen X. Y. Xu H. J. Guo Y. Biochem. Eng. J. 2020;161:107587. doi: 10.1016/j.bej.2020.107587. [DOI] [Google Scholar]
- de Miranda A. S. Miranda L. S. M. de Souza R. O. M. A. Biotechnol. Adv. 2015;33:372–393. doi: 10.1016/j.biotechadv.2015.02.015. [DOI] [PubMed] [Google Scholar]
- Sikora A. Siodmiak T. Marszall M. P. Chirality. 2014;26:663–669. doi: 10.1002/chir.22362. [DOI] [PubMed] [Google Scholar]
- Seddigi Z. S. Malik M. S. Ahmed S. A. Babalghith A. O. Kamal A. Coord. Chem. Rev. 2017;348:54–70. doi: 10.1016/j.ccr.2017.08.008. [DOI] [Google Scholar]
- Rani K. N. P. Neeharika T. S. V. R. Kumar T. P. Satyavathi B. Sailu C. Prasad R. B. N. J. Taiwan Inst. Chem. Eng. 2015;55:12–16. doi: 10.1016/j.jtice.2015.04.011. [DOI] [Google Scholar]
- Alves M. D. Cren E. C. Mendes A. A. J. Mol. Catal. B: Enzym. 2016;133:S377–S387. doi: 10.1016/j.molcatb.2017.02.011. [DOI] [Google Scholar]
- Neeharika T. Bhimjiyani V. H. Dole B. R. Rani K. N. P. Karadbhajne V. Y. Prasadl R. B. N. Indian J. Chem. Technol. 2017;24:213–217. [Google Scholar]
- Lima L. C. D. Peres D. G. C. Mendes A. A. Bioprocess Biosyst. Eng. 2018;41:991–1002. doi: 10.1007/s00449-018-1929-9. [DOI] [PubMed] [Google Scholar]
- Tomke P. D. Rathod V. K. Bioprocess Biosyst. Eng. 2020;43:1659–1670. doi: 10.1007/s00449-020-02358-w. [DOI] [PubMed] [Google Scholar]
- Badgujar K. C. Bhanage B. M. Bioproc. Biosyst. Eng. 2017;40:741–757. doi: 10.1007/s00449-017-1740-z. [DOI] [PubMed] [Google Scholar]
- Park K. M. Kwon C. W. Choi S. J. Son Y. H. Lim S. Yoo Y. Chang P. S. J. Agric. Food Chem. 2013;61:9421–9427. doi: 10.1021/jf402539m. [DOI] [PubMed] [Google Scholar]
- Reis P. Holmberg K. Miller R. Leser M. E. Raab T. Watzke H. J. Cron. Chim. 2009;12:163–170. doi: 10.1016/j.crci.2008.04.018. [DOI] [Google Scholar]
- Yadav G. D. Kamble M. P. Int. J. Chem. React. Eng. 2018;16:20170179. [Google Scholar]
- Lopresto C. G. Calabrò V. Woodley J. M. Tufvesson P. J. Mol. Catal. B: Enzym. 2014;110:64–71. doi: 10.1016/j.molcatb.2014.09.011. [DOI] [Google Scholar]
- Elias M. Wieczorek G. Rosenne S. Tawfik D. S. Trends Biochem. Sci. 2014;39:1–7. doi: 10.1016/j.tibs.2013.11.001. [DOI] [PubMed] [Google Scholar]
- Senn H. M. Kastner J. Breidung J. Thiel W. Can. J. Chem. 2009;87:1322–1337. doi: 10.1139/V09-092. [DOI] [Google Scholar]
- Kumar R. Goomber S. Kaur J. Biochim. Biophys. Acta, Proteins Proteomics. 2019;1867:140261. doi: 10.1016/j.bbapap.2019.08.001. [DOI] [PubMed] [Google Scholar]