Abstract
An association between (unculturable) gastrospirillum-like organisms (GLO) and ulcerative lesions in the pars oesophagea in stomachs of swine has been claimed. In dogs GLO detected by microscopy may represent several Helicobacter species or subspecies. Therefore we investigated which Helicobacter spp. are present in stomachs of swine and their possible association with ulcerative lesions of the pars oesophagea. The presence of Helicobacter spp. in the antrum and pars oesophagea in 122 stomachs of slaughter swine was determined by microscopy (n = 122), by culture on selective and nonselective media (n = 112), and by a genus-specific 16S ribosomal DNA (rDNA) PCR (n = 80). GLO could not be cultured. Phylogenetic analysis of 43 16S rDNA fragments (out of 54 PCR-positive biopsy specimens) revealed the presence of Helicobacter heilmannii type 1 in 42 of them. This correlated with the presence of bacteria with GLO morphology. Helicobacter bilis 16S rDNA was amplified directly from one sample harboring bacteria with H. bilis morphology. The association between Helicobacter spp. and gastric lesions was investigated with a second group of 41 pigs with (n = 21 cases) or without (n = 20 controls) gastric lesions. Fifteen of the 21 cases were positive by PCR or microscopy, compared to 7 of 20 of the controls (P = 0.03). 16S rDNA sequence analysis of 7 of 14 PCR-positive cases revealed the presence of H. heilmannii type 1. Microscopy showed bacteria with GLO morphology. One sample (cases) was culture negative but PCR positive for Helicobacter pullorum-related 16S rDNA. In conclusion, our findings indicate that H. heilmannii type 1 is the predominant Helicobacter spp. in the stomachs of pigs and that its presence is associated with ulcerative lesions in the pars oesophagea.
Gastric ulceration in the pars oesophagea is a well-known problem in swine which can lead to growth retardation, bleeding, and death (6, 20). In The Netherlands ulcerative lesions are observed in up to 25% of slaughterhouse swine (6, 9). Many factors have been considered as possible causes, e.g., nutrition, stress, and infection. However, the etiology of gastric ulcer formation in swine is still unknown.
A spirally shaped bacterium, Helicobacter pylori, has been identified as the major cause of peptic ulcer disease in humans (4). It has also been known for some time that spirally shaped gastrospirillum-like organisms (GLO) are present in the stomachs of pigs (15). In view of this knowledge several investigators have studied and confirmed the association between GLO, as detected by microscopy, and ulcerative lesions in the pars oesophagea of stomachs of swine (1, 2, 17). Despite several attempts these GLO could not be grown in vitro, and detection of them was based only on microscopy. In dogs it has been demonstrated that bacteria with GLO morphology can represent several species (5, 10), which may differ in their disease-inducing properties. Determination of whether this also holds true for GLO observed in swine requires a technique that discriminates between these potentially different GLO strains. Cloned 16S ribosomal DNA (rDNA) from GLO derived from one pig appeared to be 99.5% similar to human Helicobacter heilmannii type 1 (13, 18). This enables the detection of Helicobacter spp. (including GLO) in pigs based on a 16S rDNA Helicobacter genus-specific PCR. Sequence analysis of the amplified fragments can than be used for species or subspecies analysis.
We analyzed gastric biopsy specimens from swine for the presence of different Helicobacter species or subspecies by direct 16S rDNA PCR and sequence analysis. Helicobacter-specific primers were selected based on 16S rDNA of different Helicobacter spp. and some other bacterial genera. In addition, we attempted to culture GLO directly from stomach samples. Finally, the association between Helicobacter spp. and ulcerative lesions in the pars oesophagea was investigated.
MATERIALS AND METHODS
Animals.
In the first series 122 swine stomachs collected at three different slaughterhouses in The Netherlands were investigated for the presence of different Helicobacter spp. All stomachs were processed within 3 h after collection, mostly at the slaughterhouse in order to avoid loss of viability of bacteria due to delay by transportation. The stomachs were opened along the greater curvature and washed gently with tap water. Mucus from the antrum and/or esophagus was collected for microscopic examination (n = 122), culture (n = 112), and PCR (n = 80). For microscopy, mucus was smeared directly on glass slides and air dried. Mucus was inoculated on different agar media for culture immediately after collection. For PCR analysis, samples were stored in 100 μl of buffer (10 mM Tris-HCl [pH 8], 1 mM EDTA, 0.5% Tween 20) at −20°C.
The association between Helicobacter spp. and ulcerative lesions was investigated with stomachs of a second series of pigs sent in for pathological examination to the Animal Health Service in Boxtel, The Netherlands. Stomachs were scored for the presence of macroscopically visible lesions according to a previously described classification (9). Stomachs were classified on a scale from 0 to 5, reflecting an increasing abnormality of the mucosa of the pars oesophagea. Score 0 means no abnormalities. Scores 1 and 2 imply increased thickening of the mucosa (hyperkeratosis). Scores 3 to 5 reflect an increasing damage of the mucosa from less than 5 small (length, <2.5-cm) to more than 10 large (length, >5-cm) erosions or frank ulcers. We defined all stomachs with score 3 or higher as stomachs with ulcerative lesions. Biopsy specimens of the pars oesophageae and the antra of 21 stomachs with ulcerative lesions (cases; 4 with score 3, 2 with score 4, and 15 with score 5) and of 20 stomachs without lesions (controls) were collected. Small specimens of the luminal sides of these biopsy specimens were cut out and stored in 200 μl of buffer (10 mM Tris-HCl [pH 8], 1 mM EDTA, 0.5% Tween 20) at −20°C for PCR analysis.
In addition, tissue specimens from the antrum, cardia, fundus, and pars oesophagea were collected for microscopic examination.
Detection of helicobacter bacteria by microscopy and culture.
Mucus samples from the stomachs of the first group of pigs (n = 122) were Gram stained and examined by light microscopy.
Culture of mucus was performed in an atmosphere of 85% N2–10% CO2–5% O2 on both selective and nonselective media. Selective media were Columbia agar (BBL)–7% lysed horse blood with Dent supplement (Oxoid), brain heart infusion agar (Difco)–10% lysed horse blood with Skirrow supplement (Oxoid), brain heart infusion agar–2% horse serum–0.0001% dextrin with Skirrow supplement, and brucella broth (Difco)–1% newborn calf serum with Dent supplement. As nonselective media Columbia agar–7% lysed horse blood, brain heart infusion agar–7% horse blood, and chocolate agar with 1% IsoVitaleX (BBL) were used. Cultures were checked daily for up to 2 weeks.
PCR analysis.
After the gastric specimens were thawed, protease (2 mg/ml) was added, and samples were heated for 60 min at 37°C and subsequently boiled for 10 min. The samples were centrifuged for 5 min at 12,000 × g, and PCR was performed on 2 μl of the supernatant in a final volume of 50 μl. PCR was performed in a Perkin-Elmer 9700 thermocycler, and the primers used for these PCRs (25 pmol per reaction) are listed in Table 1. Each PCR (final MgCl2 concentration, 1.5 mM) consisted of a denaturation step of 3 min at 94°C followed by 40 cycles of 1 min of denaturation at 94°C, 1 min of annealing at 65°C for the first cycle reduced by 1°C per cycle to 55°C, and 2 min of extension at 72°C. Finally, elongation was completed at 72°C for 10 min.
TABLE 1.
Primers
| Primer | Sequence (5′–3′) |
|---|---|
| 8FPL2 | TGC AGA GTT TGA TCC TGG CTC AG |
| 8F | AGA GTT TGA TCC TGG CTC AG |
| H16s6 | CAC CCT CTC AGG CCG GA |
| 274R | TCT CAG GCC GGA TAC CCG TCA TAG CCT |
| 782R | GTT TGC TCC CCA CGC TTT C |
| 1492R | GGT TAC CTT GTT ACG ACT T |
Two positive-control samples with H. pylori DNA equivalent to 100 or 1 CFU were always included with each run of PCRs. For each 10 samples a negative control consisting of 100 μl of distilled water that was processed in the same way as the biopsy specimens was included.
Primer combinations 8FPL2-274R and 8F-H16s6 were used to amplify fragments of approximately 280 bp of Helicobacter 16S rDNA directly from biopsy specimen lysates. As an inhibition control we spiked PCRs with a double-stranded DNA fragment of 339 bp flanked by sequences of primers 8F and H16s6; we used an amount that gave a clear signal on gel but that did not affect the sensitivity of the PCR.
Both the combinations 8F-782R and 715F-1492R were used to amplify the 16S rDNA of cultured Helicobacter bilis-like bacteria for sequence analysis. Amplified samples (10 μl) were analyzed by electrophoresis in a 1% agarose gel and visualized by ethidium bromide staining and long-wavelength UV light.
Cloning.
All DNA manipulations were performed according to standard techniques unless stated otherwise. PCR fragments were ligated into the pGEM-T vector according to the manufacturer's instructions (Promega) and introduced into competent DH5αF cells. Transformants were selected on Luria-Bertani plates supplemented with ampicillin at 100 μg/ml and isopropylthio-β-galactoside and 5-bromo-4-chloro-3-indolyl-β-d-galactoside. Several transformants were picked, and plasmids were isolated with a QIAprep plasmid kit. Restriction analysis with PstI and NcoI (New England BioLabs, Beverly, Mass.) was performed to confirm the presence and size of the insert.
Sequencing.
The DNA sequences of the pGEM-T plasmids containing the amplified 16S rDNA fragment were determined with the Thermo Sequenase premixed cycle sequencing kit and standard Texas red-labeled M13 forward and reverse primers on a Vistra system 725 DNA sequencer (Amersham Pharmacia Biotech, Essex, United Kingdom).
Sequences were aligned with ClustalX (21). The alignments were used to calculate a distance matrix from which the most likely phylogenetic tree was inferred with the programs of J. Felsenstein distributed as part of the PHILYP package (7).
Histopathological examination.
Gastric biopsy specimens of the second group of 41 animals were fixed in 10% buffered formalin, embedded in paraffin wax, cut at a thickness of 4 μm, and stained with hematoxylin and eosin and by the Warthin-Starry staining procedure. Gastritis was classified semiquantitatively as absent (only a few scattered mononuclear cells), mild, moderate, or severe, reflecting the presence of an increasing number of mononuclear cells.
Statistical analysis.
To compare proportions the Fisher exact test was used.
Nucleotide sequence accession numbers.
Sequences determined have been deposited in GenBank under the following accession numbers: AF252624 (H. pullorum), AF25625 (H. heilmannii type 1), AF252626 (H. bilis).
RESULTS
Primer selection.
In a pilot study primers 8F and 274R were used to detect Helicobacter spp. in stomach samples of pigs by PCR. However, our data (not shown) revealed that 247R was not as Helicobacter sp. specific as claimed (18). In order to select Helicobacter spp.-specific primers, alignments were performed with the following 16S rDNA sequences (GenBank accession numbers are in parentheses): Helicobacter pullorum (L36141), Helicobacter muridarum (M80205), Helicobacter cinaedi (M88150), Flexispira rappinii (M88137), H. pylori (U01330), Helicobacter acinonyx (M88148), Helicobacter felis (M57398), Gastrospirillum hominis type 1 (L10079), G. hominis type 2 (L10080), Wolinella succinogenes (M88159), Campylobacter coli (L04312), Campylobacter jejuni (L04315), Campylobacter lari (L04316), Escherichia coli (M24828), Pseudomonas aeruginosa (X06684), and Proteus vulgaris (X07652).
Criteria for the selection of primers were as follows: at least a two-base difference at the 3′ end between helicobacter 16S rDNA (including Gastrospirillum and W. succinogenes) and nonhelicobacter 16S rDNA, amplification of a relatively short 16S rDNA fragment of 200 to 300 bp to minimize the risk of missing positive samples due to DNA degradation, and amplification of a 16S rDNA region that was sufficiently variable to enable phylogenetic analysis of generated fragments.
H16s6 fulfills our specificity criteria. In combination with 8F (eubacterial primer) H16s6 amplifies a fragment of 279 bp in a highly variable region of the 16S rDNA.
With serial dilution of H. pylori DNA and C. jejuni DNA the primer combination 8F and H16s6 proved to be highly sensitive and specific. H. pylori DNA was detected at an equivalent of 1 CFU, whereas a 106 excess of C. jejuni DNA was not amplified.
The usefulness of the 16S rDNA fragments generated by 8F and H16s6 for phylogenetic analysis was investigated by comparing phylogenetic trees generated by analysis of the 279-bp highly variable fragments and complete 16S rDNA of the different Helicobacter species and related genera. No major differences between the phylogenetic trees generated by both approaches were observed (data not shown).
Screening for the presence of Helicobacter species or subspecies.
Fifty-four of the 80 stomachs investigated (first series) were PCR positive. Sequence analysis of 43 fragments revealed the presence of H. heilmannii type 1 in 42 of them (Fig. 1). The corresponding positive smears revealed the presence of bacteria with GLO morphology. In one other (GLO-negative) biopsy sample bacteria with H. bilis morphology were observed but not cultured. 16S rDNA analysis of the amplified fragment revealed the presence of H. bilis-related bacteria (strain from pig 69). Isolation attempts were negative, except in one case where an H. bilis-related species (as determined by 16S rDNA analysis) was cultured (isolate from pig 63). In one case bacteria with GLO morphology were observed after 1 week of culture on brain heart infusion agar–7% horse blood with Skirrow supplement and amphotericin B. However, they were lost upon subsequent subculture.
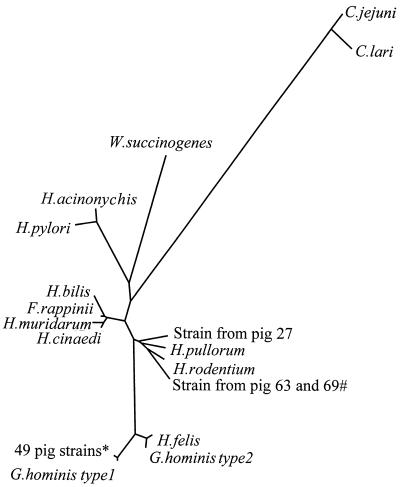
FIG. 1.
Inferred evolutionary distances of selected 16S rDNA gene sequences. The following 16S rDNA sequences were selected directly from GenBank (accession numbers are in parentheses): C. jejuni (Z29326), C. lari (L04316), W. succinogenes (M26636), H. acinonychis (M88148), H. pylori (U01330), H. cinaedi (M88150), H. muridarum (AF013464), H. pullorum (L36146), F. rappinii (AF034135), H. rodentium (U96300), H. bilis (AF04784), H. felis (U51870), G. hominis type 1, (L10079), G. hominis type 2 (L10080). An alignment was made from these sequences using the ClustalX programs (21). This alignment was then used to calculate a distance matrix from which an unrooted phylogenetic tree was inferred by the neighbor-joining method with the help of the programs of J. Felsenstein (University of Seattle) that are distributed as part of the PHYLIP package (7). ∗, 42 strains are from series 1, and 7 strains are from series 2; #, the sequence for the strain from pig 63 is from an isolate; all other pig strain sequences are from direct PCR on gastric biopsies containing unculturable GLO.
Association of Helicobacter spp. with ulcerative lesions.
Helicobacter spp. were found in the stomachs of 15 of the 21 cases, whereas 7 out of the 20 controls were positive (P = 0.03). PCR was inhibited in eight samples, but never in both gastric specimens of the same animal. Of the 22 positive animals, 14 were positive by PCR. 16S rDNA fragments of 8 of the 14 PCR-positive animals were sequenced. After phylogenetic analysis seven 16S rDNA fragments were identified as belonging to bacteria most related to H. heilmannii type 1 (Fig. 1). The corresponding microscopy samples showed bacteria with GLO morphology when positive. From one sample a fragment from a bacterium with 16S rDNA most closely related to H. pullorum 16S rDNA was generated (Fig. 1), but no bacteria were observed in the microscopic preparation from this animal. In the 22 H. heilmannii type 1-positive animals, these bacteria were found in various locations of the stomach and often at multiple sites (15, 9, 8, and 11 cases in the antrum, fundus, cardia, and pars oesophagea, respectively).
In all cases gastritis was present as indicated by the presence of mononuclear cells. Strikingly, there was no significant association between gastritis and the presence of GLO (P > 0.05).
DISCUSSION
Spirally shaped bacteria with GLO morphology have been detected in the stomachs of pigs by light microscopy (15). However, bacteria with GLO morphology can represent different Helicobacter species (5, 10). This emphasizes the need for classification of these bacteria by criteria other than strictly morphological characteristics. Thus far culture of GLO from pigs has not been successful. In this study also GLO could not be cultured on artificial media which support the growth of various Helicobacter species. Therefore, comparison of GLO from pigs based on biochemical criteria is not possible. The phylogenetic relationship between bacteria is often established by sequence analysis of the 16S rDNA. 16S rDNA sequencing of DNA obtained from pig GLO has been performed once previously (13). In that study the authors claimed 99.5% homology with H. heilmannii type 1 (formerly G. hominis type 1) (13, 18). However, their sequence data are not publicly available, and the data were derived from GLO obtained from only one animal. We examined a large number of pigs from three distinct geographic locations for the presence of different Helicobacter species by direct PCR and sequence analysis of a highly variable part of the 16S rDNA. Our data demonstrate that H. heilmannii type 1 is the predominant Helicobacter species in stomachs of pigs. The detection of H. heilmannii type 1 by PCR and sequence analysis corresponded with the presence of bacteria with GLO morphology. Therefore, it is highly likely that the GLO associated with gastric ulcerative lesions found by others (1, 17) are H. heilmannii type 1. We also confirmed the association of H. heilmannii type 1 with the presence of ulcerative lesions in the pars oesophagea of the stomach. Not all animals with gastric ulceration were H. heilmannii type 1 positive, which correlates with the findings with respect to GLO of Barbosa et al. (1). This may be due to the detection technique used because a highly sensitive mouse inoculation test showed that all animals with ulcers were GLO positive (14, 17). Strikingly, H. heilmannii type 1 was mainly found in the glandular region of the stomach and to a lesser extent in the pars oesophagea, while ulcerative lesions were mostly observed in the nonglandular pars oesophagea. Should there be a role for H. heilmannii type 1 in the induction of ulcerative lesions, this finding suggests a direct or indirect influence on the glandular part of the stomach. This could be G-cell hyperfunction or a direct influence on parietal cells, both of which influence acid production. In addition, H. heilmannii type 1 might affect mucus production altering the protective capability of the mucosa.
Helicobacter species other than H. heilmannii type 1 were found in only three animals and seem not to be of major significance. In one animal we detected an H. pullorum-related strain. In two other pigs (pigs 63 and 69) H. bilis-related strains were found, one of which could be cultured (strain from pig 63). This H. bilis isolate was from an H. heilmannii type 1, GLO-positive biopsy specimen. This emphasizes the need to apply multiple techniques to study the diversity of Helicobacter species in pigs. In Fig. 1 both H. bilis-like strains (pigs 63 and 69) are clustered with H. pullorum and Helicobacter rodentium, whereas individual 16S rRNA sequences clearly classified both strains as being H. bilis because the amplified fragments of these strains contain a 170-bp insertion fragment that is present in H. bilis and absent in all other helicobacters. In the phylogenetic analysis, however, this 170-bp insertion/deletion is only scored as a single difference (and not as 170), because in the evolution of this bacterium it probably represented a single insertion/deletion event. Strikingly the sequence outside this 170-bp H. bilis-specific insertion/deletion is more similar to that of H. pullorum, and therefore in the evolutionary tree of Fig. 1 this isolate is clustered more closely to H. pullorum than to H. bilis.
Gastritis in pigs consisted predominantly of mononuclear cell infiltrates, as observed by others (1, 2, 17). This is in contrast with the chronic active gastritis found in association with H. heilmannii in humans (19). H. pylori induces active gastritis in humans but only a lymphocytic gastritis in gnotobiotic swine (11). Thus it seems that different hosts present different histopathological responses to the same Helicobacter spp. Conflicting data about the association between GLO and gastritis in pigs have been presented (1, 2, 11, 17). In our study GLO were not associated with the presence of gastritis. This corresponds with the findings of Barbosa et al. (1). However, the same group has demonstrated an association between GLO and antrum gastritis in other investigations (17), a finding also confirmed by others (2). In one study there was even a 100% association, in which antrum gastritis was present in all GLO-positive animals and absent from histologically normal stomachs (17). An explanation for these discrepancies may be differences in the sensitivities of detection of GLO and the use of different criteria for the classification of gastritis. Severe inflammation was observed only in the pars oesophagea in the presence of lesions, regardless of the presence of H. heilmannii type 1. In all other cases inflammation was mild or moderate. If H. heilmannii induces gastritis in pigs, then this gastritis is moderate at most. This corresponds to the relatively mild gastritis observed in humans in the presence of H. heilmannii.
H. heilmannii has been found in association with peptic ulcers in humans (3, 8, 19). In a large study seven out of eight ulcers found in 202 H. heilmannii-positive patients were from patients who also received NSAIDs (19). In contrast, Debongnie et al. demonstrated that characteristics of patients with ulcers who were H. heilmannii positive differed substantially from those of patients with H. pylori and those using NSAIDs (3). A recent study describes ulcer healing in patients with duodenal ulcers after eradication of H. heilmannii (8). These observations suggest a role for H. heilmannii in ulcerogenesis. Data about the role of GLO (which are probably H. heilmannii type 1) in ulcerogenesis in pigs are conflicting (12, 16). Whereas Landrace pigs developed ulcers after inoculation with GLO (16), gnotobiotic piglets did not (12). In the latter study ulcers developed only in animals infected with fermentative bacteria (Lactobacillus spp.) in combination with a high-carbohydrate diet. This suggests a primary role for locally induced bacterial acid production in ulcerogenesis.
In conclusion, H. heilmannii type 1 is the predominant Helicobacter species in swine stomachs and is associated with the presence of ulcerative lesions in the pars oesophagea.
ACKNOWLEDGMENT
Part of this work was supported by Intervet International B.V., Boxmeer, The Netherlands.
REFERENCES
- 1.Barbosa A J A, Silva J C P, Nogueira A M M F, Paulino E, Miranda C R. Higher incidence of Gastrospirillum sp. in swine with gastric ulcer of the pars oesophagea. Vet Pathol. 1995;32:134–139. doi: 10.1177/030098589503200206. [DOI] [PubMed] [Google Scholar]
- 2.Bedel A, Pichard F, Wuscher N, Labigne A, Huerre M. Prevalence of helicobacter infection and gastric-associated pathologies in swine originating from pork producers in the west of France. Gut. 1997;41(Suppl. 1):A124. [Google Scholar]
- 3.Debongnie J C, Donnay M, Mairesse J, Lamy V, Dekoninck X, Ramdani B. Gastric ulcers and Helicobacter heilmannii. Eur J Gastroenterol Hepatol. 1998;10:251–254. doi: 10.1097/00042737-199803000-00011. [DOI] [PubMed] [Google Scholar]
- 4.Dunn B E, Cohen H, Blaser M J. Helicobacter pylori. Clin Microbiol Rev. 1997;10:720–741. doi: 10.1128/cmr.10.4.720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eaton K A, Dewhirst F E, Paster B J, Tzellas N, Coleman B E, Paola J, Sherding R. Prevalence and varieties of Helicobacter species in dogs from random sources and pet dogs: animal and public health implications. J Clin Microbiol. 1996;34:3165–3170. doi: 10.1128/jcm.34.12.3165-3170.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elbers A R W, Hessing M J C, Tielen M J M, Vos J H. Growth and oesophagogastric lesions in finishing pigs offered finely pelleted feed ad libitum. Vet Rec. 1995;136:588–590. doi: 10.1136/vr.136.23.588. [DOI] [PubMed] [Google Scholar]
- 7.Felsenstein J. PHYLIP (phylogeny inference package) version 3.5c. Evolution. 1989;39:783–791. [Google Scholar]
- 8.Goddard A F, Logan R P H, Atherton J C, Jenkins D, Spiller R C. Healing of duodenal ulcer after eradication of Helicobacter heilmannii. Lancet. 1997;349:1815–1816. doi: 10.1016/S0140-6736(05)61696-0. [DOI] [PubMed] [Google Scholar]
- 9.Hessing M J C, Geudeke M J, Scheepens C J M, Tielen M J M, Schouten W G P, Wiepkema P R. Slijmvliesveranderingen in de pars oesophagea bij varkens: prevalentie en de invloed van stress. Tijdschr Diergeneeskd. 1992;117:445–450. [PubMed] [Google Scholar]
- 10.Jalava K, Kaartinen M, Utriainen M, Happonen I, Hänninen M L. Helicobacter salomonis sp. nov., a canine gastric Helicobacter sp. related to Helicobacter felis and Helicobacter bizzozeroni. Int J Syst Bacteriol. 1997;47:975–982. doi: 10.1099/00207713-47-4-975. [DOI] [PubMed] [Google Scholar]
- 11.Krakowka S, Morgan D R, Kraft W G, Lunk R D. Establishment of gastric Campylobacter pylori infection in the neonatal gnotobiotic piglet. Infect Immun. 1987;55:2789–2796. doi: 10.1128/iai.55.11.2789-2796.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krakowka S, Eaton K A, Rings D M, Argenzio R A. Production of gastroesophageal erosions and ulcers (GEU) in gnotobiotic swine monoinfected with fermentative commensal bacteria and fed high-carbohydrate diet. Vet Pathol. 1998;35:274–282. doi: 10.1177/030098589803500406. [DOI] [PubMed] [Google Scholar]
- 13.Mendes E N, Queiroz D M M, Dewhirst F E, Paster B J, Rocha G A, Fox J G. Are pigs a reservoir for human helicobacter infection? Am J Gastroenterol. 1994;8:1296. [Google Scholar]
- 14.Mendes E N, Queiroz D M M, Rocha G A, Moura S B, Barbosa M T, Carvalhaes S M, Freitas M L P, Reis A C S. In vivo culture improves the diagnosis of “Gastrospirillum suis” infection. Ir J Med Sci. 1992;61:78. [Google Scholar]
- 15.Queiroz D M M, Rocha G A, Mendes E N, Lage A P, Carvalho A C T, Barbosa A J A. A spiral organism in the stomach of pigs. Vet Microbiol. 1990;24:199–204. doi: 10.1016/0378-1135(90)90067-6. [DOI] [PubMed] [Google Scholar]
- 16.Queiroz D M M, Rocha G A, Mendes E N, Moura S B, Oliveira A M R, Oliveira C A. Swine experimentally infected with helicobacter as an animal model of ulcerogenesis. Am J Gastroenterol. 1994;8:1320. [Google Scholar]
- 17.Queiroz D M M, Rocha G A, Mendes E N, Moura S B, Oliveira A M R, Miranda D. Association between Helicobacter and gastric ulcer disease of the pars oesophagea in swine. Gastroenterology. 1996;111:19–27. doi: 10.1053/gast.1996.v111.pm8698198. [DOI] [PubMed] [Google Scholar]
- 18.Solnick J V, O'Rourke J, Lee A, Paster B J, Dewhirst F E, Tompkins L S. An uncultured gastric spiral organism is a newly identified Helicobacter in humans. J Infect Dis. 1993;168:379–385. doi: 10.1093/infdis/168.2.379. [DOI] [PubMed] [Google Scholar]
- 19.Stolte M, Kroher G, Meining A, Morner A, Bayerdörffer E, Bethke B. A comparison of Helicobacter pylori and H. heilmannii gastritis. Scand J Gastroenterol. 1997;32:28–33. doi: 10.3109/00365529709025059. [DOI] [PubMed] [Google Scholar]
- 20.Taylor D J. Pig diseases. 4th ed. Cambridge, United Kingdom: Burlington Press; 1984. [Google Scholar]
- 21.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]