Abstract
Simple Summary
The incidence of young people <50 years old who are diagnosed with colorectal cancer (CRC), termed as early onset colorectal cancer (EOCRC), accounted for nearly 30% of the total CRC patients in Indonesia, which is about three times higher than what is being reported in Europe, the UK and USA. Lynch syndrome (LS) is a hereditary type of CRC that is associated with a younger age of onset. Detecting LS has been long reported to be a cost-effective strategy to provide aid in the diagnosis or management of the individual or at-risk family members. The aim of this retrospective study was to screen for Lynch Syndrome in Indonesian CRC patients using simple and robust polymerase chain reaction (PCR)-based molecular testing, known as N_LyST (Nottingham Lynch Syndrome Test). To our knowledge, we are the first to study and observe a potentially higher frequency of LS (13.85%) among CRC patients in Indonesia (n = 231). This may partially contribute to the reported much higher rate of EOCRC found in the country.
Abstract
There is about three times higher incidence of young patients <50 years old with colorectal cancer, termed EOCRC, in Indonesia as compared to Europe, the UK and USA. The aim of this study was to investigate the frequency of Lynch Syndrome (LS) in Indonesian CRC patients. The previously described Nottingham Lynch Syndrome Test (N_LyST) was used in this project. N_LyST is a robust high-resolution melting (HRM)-based test that has shown 100% concordance with standard reference methods, including capillary electrophoresis and Sanger sequencing. The test consisted of five mononucleotide microsatellite markers (BAT25, BAT26, BCAT25, MYB, EWSR1), BRAF V600E mutation and MLH1 region C promoter for methylation (using bisulphite-modified DNA). A total of 231 archival (2016–2019) formalin-fixed, paraffin-embedded (FFPE) tumour tissues from CRC patients collected from Dr. Sardjito General Hospital Yogyakarta, Indonesia, were successfully tested and analysed. Among those, 44/231 (19.05%) were MSI, 25/231 (10.82%) were harbouring BRAF V600E mutation and 6/231 (2.60%) had MLH1 promoter methylation. Almost all—186/197 (99.45%)—MSS cases were MLH1 promoter unmethylated, while there were only 5/44 (11.36%) MSI cases with MLH1 promoter methylation. Similarly, only 9/44 (20.45%) of MSI cases were BRAF mutant. There were 50/231 (21.65%) EOCRC cases, with 15/50 (30%) regarded as MSI, as opposed to 29/181 (16.02%) within the older group. In total, 32/231 patients (13.85%) were classified as “Probable Lynch” (MSI, BRAF wildtype and MLH1 promoter unmethylated), which were enriched in EOCRC as compared to older patients (24% vs. 11.05%, p = 0.035). Nonetheless, 30/50 (76.00%) cases among the EOCRC cases were non-LS (sporadic) and were significantly associated with a left-sided tumour. The overall survival of both “Probable Lynch” and non-LS (sporadic) groups (n = 227) was comparable (p = 0.59), with follow up period of 0–1845 days/61.5 months. Stage, node status, histological grading and ECOG score were significantly associated with patient overall survival (p < 0.005), yet only ECOG was an independent factor for OS (HR: 4.38; 95% CI: 1.72–11.2; p = 0.002). In summary, this study is the first to reveal a potentially higher frequency of LS among CRC patients in Indonesia, which may partially contribute to the reported much higher number of EOCRC as compared to the incidence in the West.
Keywords: microsatellite instability (MSI), BRAF mutation, MLH1 promoter methylation, high Resolution melting (HRM), early onset colorectal cancer (EOCRC)
1. Introduction
Colorectal cancer (CRC) is one of the leading causes of cancer-related mortality. The estimated incidence of CRC worldwide in 2018 was about 1.8 million cases, comprising 11% of all cancer diagnoses, with approximately 881,000 deaths, making it the second deadliest cancer worldwide [1]. Colorectal cancer is also a major health burden in Indonesia, with an incidence of 35,000 cases per year [2]. It is the fourth most prevalent cancer, with an age-standardized annual incidence rate of 12.4/100,000 individuals and a mortality rate of 6.7/100,000 individuals [3].
In the past decade, there has been an increase of the proportion of young people (<50 years old) being diagnosed with CRC, termed as Early Onset Colorectal Cancer (EOCRC), despite the decreased incidence in the population over 50 years old [4,5,6]. This may occur in the context of inherited diseases such as Lynch Syndrome (LS), although only around 15% of EOCRC is associated with known hereditary forms of the disease [7,8,9]. Limited studies have reported that the frequency of EOCRC in Indonesia is about 30% of total CRC cases [10,11]. Our current study (under review) also confirms this interesting finding, showing approximately three times higher frequency of EOCRC in Indonesia as compared to ~10% found in western countries [12,13,14]. Nonetheless, to our knowledge, no study has been conducted to determine the frequency of Lynch Syndrome, the most common inherited type of CRC, which has been linked to a higher risk of EOCRC.
The inactivating germline mutation of the DNA MMR (mismatch repair) genes, including MLH1, MLH2, MSH2, MSH3, MSH6 or PMS2, is reported to be responsible for LS [15]. Loss of MMR results in changes in the length of microsatellites (normal segments of DNA consisting of multiple repeats of sequences 1–6 nucleotides in length, known as microsatellite instability (MSI) [16,17,18]. While germline mutations of one of the MMR genes are responsible for LS, hypermethylation silencing of MLH1 is the most common mechanism for MMR inactivation, and it is responsible for sporadic CRC tumours [19,20,21]. Thus, sporadic tumours with deficient MMR/MSI can be distinguished from tumours arising in LS by demonstrating methylation of the MLH1 promoter. Similarly, somatic mutation of BRAF is common in sporadic tumours with MSI but very rarely occurs in tumours arising in LS. BRAF V600E mutation, through inhibition of apoptosis, is an early event in pre-malignant lesions and leads to the induction of methylation in the promoter region of the MLH1 gene, resulting in the MSI phenotype and increased invasiveness of serrated pathways of sporadic CRC carcinogenesis [22,23,24].
Guidance from the National Institute of Clinical and Healthcare Excellence (NICE-UK) and National Comprehensive Cancer Network (NCCN-USA) recommends that all CRC should be screened for the possibility of LS [25,26]. The NICE pathway involves two steps: first, identify cases with deficient MMR or MSI, and then filter out sporadic cases by testing for BRAF mutation and MLH1 promoter methylation. This approach has been reported to be a cost-effective strategy, with important clinical benefits for CRC patients, particularly under 70 years old, and their relatives by implementation of appropriate surveillance pathways for early diagnosis of associated cancers. In addition, prevention measures can also be applied, such as taking a low dose of aspirin, bowel removal surgery, and lifestyle modification (smoking, weight and diet control) [27,28].
We previously introduced Nottingham Lynch Syndrome Test (N_LyST), a simple and robust screening test for LS. This single closed-tube screening panel comprises tests for microsatellite instability (MSI) and BRAF mutation MLH1 methylation promoter using polymerase chain reaction (PCR) followed by high-resolution melting (HRM) analysis [29]. N_LyST incorporates the three components of LS screening, which are testing for MSI, BRAF V600E mutation and MLH1 promoter methylation into a single PCR run. A panel of five microsatellite markers were utilised, which includes two established markers (BAT25, BAT26) and three novel markers (BCAT25, MYB and EWSR1). The MSI testing of N_LyST showed 100% concordance with immunohistochemistry (IHC) for mismatch repair protein (MMR) and capillary electrophoresis (comparable limit of detection of ~ 6.25%). The BRAF V600E mutation and MLH1 promoter methylation tests were also in agreement with Sanger sequencing [29].
In this paper, utilising the N_LyST test, we sought to screen for Lynch Syndrome among Indonesian CRC patients.
2. Method
2.1. CRC Clinical Samples
Data on 1276 consecutive cases of CRC who visited Dr. Sardjito General Hospital Yogyakarta, Indonesia, from January 2016 to December 2019 were identified from the hospital-based cancer registry. Only a total of 271 formalin-fixed, paraffin-embedded (FFPE) CRC tumour samples were available from the archives of the Anatomical Pathology Department. This was due to the hospital being the national tertiary referral hospital, where many of the patients received treatment following resection procedures that had been conducted in secondary/regional hospitals elsewhere. Patient data that were retrieved were sociodemographic (age and sex), tumour pathology (location/side, tour morphology, histological grade, TNM stage, lymphovascular status, tumour infiltrating lymphocytes/TILs) and clinical parameters (haemoglobin/Hb, albumin, body mass index/BMI and Eastern Cooperative Oncology Group/ECOG scale).
2.2. DNA Extraction
Genomic DNA was extracted using the QIAamp DNA FFPE tissue kit (Qiagen, USA) following the manufacturer’s protocol. DNA samples were quantified using a NanoDrop™ spectrophotometer (Thermo Scientific, Waltham, MA, USA). Samples with adequate concentration and quality were adjusted to a concentration of 20 ng/µL for PCR application.
2.3. Bisulphite Conversion
In order to test for methylation of the MLH1 promoter, it was necessary to modify the DNA. Bisulphite conversion of 400 ng of genomic DNA from each sample was carried out using the EZ-DNA Methylation-Lightning Kit (Zymo Research, Irvine, CA, USA), according to the manufacturer’s protocol.
2.4. MSI, BRAF and MLH1 Analysis Using N_LyST Panel
N_Lyst was described extensively in our previous study [29]. The method consists of detection of a panel of five mononucleotide microsatellite repeats, BRAF V600E mutations, and MLH1 region C promoter methylation status. Single-plex (single reaction for each marker) reaction was mixed with the following for a final volume of 10 µL: 1.75 µL nuclease free water (Qiagen, Germantown, MD, USA), 5 µL PCR Hot Shot Diamond PCR master mix (client Life Science, Stourbridge, UK), 1 µL Evagreen dye (Biotium, Fremont, CA, USA), 0.25 µL primers, and 2 µL DNA template. Analysis was carried out in single-step using a CFX-Connect real-time PCR instrument (Bio-Rad, Hercules CA, USA). PCR was performed under the following temperature protocol: 1 cycle of 95 °C/5 min; 45 cycles of 95 °C/10 s, 55 °C/30 s and 72 °C/30 s; and 1 cycle of 72 °C/2 min. Prior to melting analysis, PCR products underwent heating to 95 °C for 15 s, rapid cooling to 60 °C and maintenance at 60 °C for 1 min. High-resolution melting analysis was carried out by increasing temperature with an increment of 0.3 °C from 60 °C to 95 °C. The melting data were analysed following normalisation using Precision Melt Analysis (Bio-Rad, Hercules, CA, USA). For MSI analysis, samples were regarded as MSI if ≥2 markers (40%) showed instability; otherwise, they were regarded as MSS tumours. Samples showing MSI, BRAF mutation and MLH1 promoter methylation were classified as “Probable Lynch”.
2.5. Statistical Analysis
Correlation between variables was calculated using the Fisher’s exact test with a significance level of α = 0.05. The Kaplan-Meier method was used to calculate the overall survival (OS), and comparisons between groups of interest were carried out using a log-rank test. A multivariate analysis of the factors that influence the OS was performed using the Cox proportional hazards regression model. All analysis was performed using R version 4.0.3.
3. Results
3.1. Patient Clinicopathology Characteristics
As shown in Table 1, there were mostly adenocarcinoma (97.84%) among the 231 CRC samples with complete molecular data, with an equal number of female and male (51.52% vs. 48.48%), enrichment of EOCRC (21.65%), higher frequency of left-sided tumours that included the rectum (77.92%), advanced stages of III (24.24%) and IV (39.83%) as well as T3 (64.94%) and T4 (22.94%). Yet, the majority of the samples had a lower histology grade of 1 and 2 (44.59% and 39.39%, respectively), with almost equal distribution between with and without nodal involvement and metastasis. Most cases showed an Hb level of ≥10 g/dL (84.85%) and ECOG scale of 0–1 (63.64%).
Table 1.
Patient clinicopathology characteristics.
| Characteristic | n = 231 |
|---|---|
| Age | |
| <50 | 50 (21.65%) |
| ≥50 | 181 (78.35%) |
| Sex | |
| Female | 119 (51.52%) |
| Male | 112 (48.48%) |
| Tumor Site | |
| Left | 180 (77.92%) |
| Right | 50 (21.65%) |
| Unknown | 1 (0.43%) |
| Stage | |
| I | 11 (4.76%) |
| II | 66 (28.57%) |
| III | 56 (24.24%) |
| IV | 92 (39.83%) |
| Unknown | 6 (2.60%) |
| T Status | |
| 1 | 2 (0.87%) |
| 2 | 25 (10.82%) |
| 3 | 150 (64.94%) |
| 4 | 53 (22.94%) |
| x | 1 (0.43%) |
| N Status | |
| 0 | 115 (49.78%) |
| 1 | 80 (34.63%) |
| 2 | 30 (12.99%) |
| x | 6 (2.60%) |
| Metastatic Status | |
| 0 | 134 (58.01%) |
| 1 | 91 (39.39%) |
| x | 6 (2.60%) |
| Histological Grading | |
| 1 | 103 (44.59%) |
| 2 | 91 (39.39%) |
| 3 | 32 (13.85%) |
| 4 | 2 (0.87%) |
| Unknown | 3 (1.30%) |
| Lymphovascular Status | |
| 0 | 51 (22.08%) |
| 1 | 58 (25.11%) |
| Unknown | 122 (52.81%) |
| Pathological Morphology | |
| Adenocarcinoma | 226 (97.84%) |
| Mucinous Carcinoma | 5 (2.16%) |
| TILs | |
| Low | 42 (18.18%) |
| Medium | 74 (32.03%) |
| High | 76 (32.90%) |
| Unknown | 39 (16.88%) |
| Hemoglobin level (g/dL) | |
| <10 | 27 (11.69%) |
| ≥10 | 196 (84.85%) |
| Unknown | 8 (3.46%) |
| Serum albumin (g/dL) | |
| <3.5 | 98 (42.42%) |
| >3.5 | 64 (27.71%) |
| Unknown | 69 (29.87%) |
| ECOG | |
| ECOG 0–1 | 147 (63.64%) |
| ECOG 2 | 36 (15.58%) |
| ECOG 3–4 | 19 (8.23%) |
| Unknown | 29 (12.55%) |
| BMI (kg/m2) | |
| <18.5 | 71 (30.74%) |
| 18.5–22.9 | 90 (38.96%) |
| 23–24.9 | 31 (13.42%) |
| ≥25 | 30 (12.99%) |
| Unknown | 9 (3.90%) |
3.2. Lynch Syndrome Screening Using N_Lyst
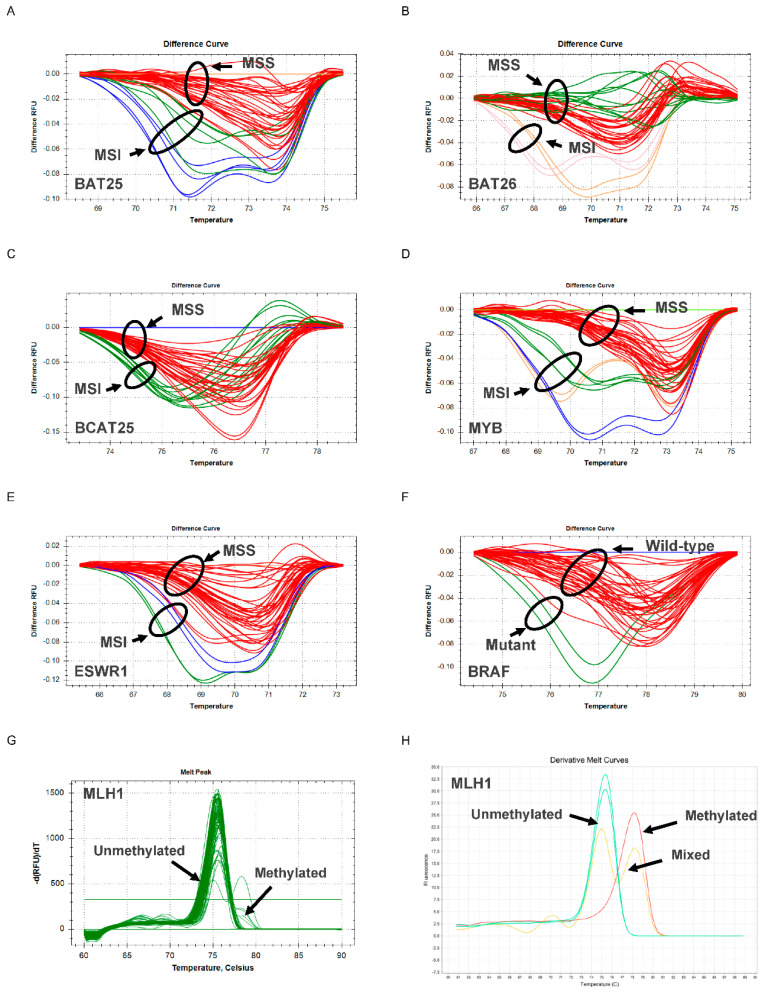
MSI, BRAF and MLH1 promoter methylation analysis was successfully done on 231 samples. The remaining 40 samples were not included in the PCR testing due to low quality of DNA following FFPE tissue extraction and bisulphite conversion. The typical HRM plots for the N_LyST panel are shown in Figure 1.
Figure 1.
Use of the N_LyST panel to screen for Lynch syndrome. Differential plots are shown for 5 microsatellite markers BAT25 (A), BAT26 (B), BCAT25 (C), MYB (D), and EWSR1 (E). The differential melt curves of the tumours with microsatellite instability (MSI) and those that are microsatellite stable (MSS) are indicated by the distinct black circles. The differential melt curves of the tumours with wild-type BRAF are different from those with mutant BRAF, as indicated by respective black circles (F). Derivative plots are shown for the CpG Island region C of MLH1 for tumour samples (G) and human DNA control (H). For tumour samples, two discrete melting forms are shown: ‘methylated’, comprising two melting peaks that represent methylated DNA (from tumour epithelium) and non-methylated DNA (from tumour stroma) or ‘non-methylated’, comprising one melting peak that characterises a completely non-methylated tumour and stroma cell population.
MSI status was determined based on a threshold of two unstable MSI markers. Among the 231 samples, the majority of the MSI samples, 40/44 (90.91%), showed instability on four to five markers. Likewise, most of the MSS samples, 166/181 (88.77%), showed no instability in any of the MSI markers (Table 2).
Table 2.
Microsatellite marker instability distribution (N = 231).
| MSI Status | Number of Unstable MSI Markers | |||||
|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | 5 | |
| MSI | (0%) | (0%) | 3 (6.82%) | 1 (2.27%) | 7 (15.91%) | 33 (75%) |
| MSS | 166 (88.77%) | 21 (11.23%) | (0%) | (0%) | (0%) | (0%) |
MSI, Microsatellite Instability; MSS, Microsatellite Stable.
Though there was a significant correlation between MSI and BRAF mutation (p = 0.031), only 9/44 (20.45%) of MSI cases were BRAF mutant. The mutation frequency was even less in MSS, accounting for 16/187 (8.56%). Almost all, 186/197 (99.45%), MSS cases were MLH1 promoter unmethylated, while 5/44 (11.36%) of MSI cases had MLH1 promoter methylation (p = 0.001) (Table 3).
Table 3.
Association between MSI, BRAF mutation and MLH1 promoter methylation (n = 231).
| Characteristic | Microsatellite Instability | ||
|---|---|---|---|
| MSI, N = 44 | MSS, N = 187 | p-Value 1 | |
| BRAF Exon 15 | 0.031 * | ||
| Mutant | 9 (20.45%) | 16 (8.56%) | |
| Wild-type | 35 (79.55%) | 171 (91.44%) | |
| MLH1 Methylation | 0.001 ** | ||
| Methylated | 5 (11.36%) | 1 (0.53%) | |
| Unmethylated | 39 (88.64%) | 186 (99.47%) | |
| BRAF Exon 15 | |||
| Mutant, N = 25 | Wild-type, N = 206 | p-Value | |
| MLH1 Methylation | 0.13 | ||
| Methylated | 2 (8.00%) | 4 (1.94%) | |
| Unmethylated | 23 (92.00%) | 202 (98.06%) | |
1 Fisher’s exact test; * p < 0.05; ** p < 0.01.
The results summarised in Table 4 show that among 231 samples with complete molecular information, 44/231 (19.05%) were MSI, 25/231 (10.82%) were harbouring BRAF V600E mutation and 6/231 (2.60%) had MLH1 promoter methylation. Based on the workflow recommended by NICE, samples that have MSI, wild-type BRAF and unmethylated MLH1 promoter based on the N_LyST test were classified “Probable Lynch”. Only 3/35 (8.6%) of MSI cases with no mutation of BRAF V600E showed MLH1 promoter methylation. Thus, there were 32 samples (13.85%) identified as “Probable Lynch”. The frequency of MSI and “Probable Lynch” status among those of EOCRC (under 50 years old) were significantly higher compared to the older patients (30% vs. 16.02%, p = 0.040 and 24% vs. 11.05%, p = 0.035, respectively). Nonetheless, there were 38/50 (76.00%) cases among the EOCRC that were considered as non-LS (sporadic) CRC.
Table 4.
Molecular features by N_LyST panel.
| Characteristic | Total | Age Group | ||
|---|---|---|---|---|
| N = 231 | <50, N = 50 1 | ≥50, N = 181 | p-Value 1 | |
| Microsatellite Instability Status | 0.040 * | |||
| MSI | 44 (19.05%) | 15 (30.00%) | 29 (16.02%) | |
| MSS | 187 (80.95%) | 35 (70.00%) | 152 (83.98%) | |
| BRAF Exon 15 | 0.6 | |||
| Mutant | 25 (10.82%) | 4 (8.00%) | 21 (11.60%) | |
| Wild-type | 206 (89.18%) | 46 (92.00%) | 160 (88.40%) | |
| MLH1 Methylation | >0.9 | |||
| Methylated | 6 (2.60%) | 1 (2.00%) | 5 (2.76%) | |
| Unmethylated | 225 (97.40%) | 49 (98.00%) | 176 (97.24%) | |
| Probable Lynch | 0.035 * | |||
| No | 199 (86.15%) | 38 (76.00%) | 161 (88.95%) | |
| Yes | 32 (13.85%) | 12 (24.00%) | 20 (11.05%) | |
1 Fisher’s exact test; * p < 0.05; MSI, Microsatellite Instability; MSS, Microsatellite Stable.
3.3. Clinicopathology-Molecular Characteristic Association and Survival
In general, as shown in Table 5, there was no significant association between many patients’ clinicopathology characteristics and the molecular features (n = 231). Nevertheless, this study found a significant association of right-sided tumour with MSI (p < 0.001), methylated MLH1 promoter (p = 0.047), and less frequent “Probable Lynch” status (p = 0.003). Higher histology grading (3 and 4) was associated with MSI (p = 0.003) and MLH1 promoter methylation (p = 0.004). Although observed in only small number of samples, mucinous tumours were positively associated with MSI (p = 0.049) and lower frequency of “Probable Lynch” status (p = 0.020). Similarly, a low level of Hb (<10 g/dL) was also associated with MSI (p = 0.044) and lower frequency of “Probable Lynch” status (p = 0.010). On the contrary, higher ECOG was associated with MSI and higher frequency of “Probable Lynch” status (p = 0.010).
Table 5.
Association between clinicopathology characteristics and molecular features (n = 231).
| Characteristic | Microsatellite Instability Status | BRAF Exon 15 | MLH1 Promoter | Probable Lynch | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MSI N = 44 |
MSS N = 187 |
p-Value 1 | Mutant N = 25 |
Wild-Type N = 206 |
p-Value 1 | Methylated N = 6 |
Unmethylated N = 225 |
p-Value 1 | No N = 199 |
Yes N = 32 |
p-Value 1 | |
| Sex | 0.4 | 0.7 | 0.7 | 0.13 | ||||||||
| Female | 20 (45.45%) | 99 (52.94%) | 14 (56.00%) | 105 (50.97%) | 4 (66.67%) | 115 (51.11%) | 107 (53.77%) | 12 (37.50%) | ||||
| Male | 24 (54.55%) | 88 (47.06%) | 11 (44.00%) | 101 (49.03%) | 2 (33.33%) | 110 (48.89%) | 92 (46.23%) | 20 (62.50%) | ||||
| Tumor Site | <0.001 *** | 0.14 | 0.047 * | 0.003 ** | ||||||||
| Left | 23 (52.27%) | 157 (83.96%) | 19 (76.00%) | 161 (78.16%) | 2 (33.33%) | 178 (79.11%) | 162 (81.41%) | 18 (56.25%) | ||||
| Right | 20 (45.45%) | 30 (16.04%) | 5 (20.00%) | 45 (21.84%) | 4 (66.67%) | 46 (20.44%) | 36 (18.09%) | 14 (43.75%) | ||||
| Unknown | 1 (2.27%) | 0 (0.00%) | 1 (4.00%) | 0 (0.00%) | 0 (0.00%) | 1 (0.44%) | 1 (0.50%) | 0 (0.00%) | ||||
| Stage | 0.4 | 0.7 | 0.7 | 0.6 | ||||||||
| I | 3 (6.82%) | 8 (4.28%) | 0 (0.00%) | 11 (5.34%) | 0 (0.00%) | 11 (4.89%) | 8 (4.02%) | 3 (9.38%) | ||||
| II | 10 (22.73%) | 56 (29.95%) | 6 (24.00%) | 60 (29.13%) | 1 (16.67%) | 65 (28.89%) | 58 (29.15%) | 8 (25.00%) | ||||
| III | 14 (31.82%) | 42 (22.46%) | 6 (24.00%) | 50 (24.27%) | 3 (50.00%) | 53 (23.56%) | 48 (24.12%) | 8 (25.00%) | ||||
| IV | 15 (34.09%) | 77 (41.18%) | 12 (48.00%) | 80 (38.83%) | 2 (33.33%) | 90 (40.00%) | 80 (40.20%) | 12 (37.50%) | ||||
| Unknown | 2 (4.55%) | 4 (2.14%) | 1 (4.00%) | 5 (2.43%) | 0 (0.00%) | 6 (2.67%) | 5 (2.51%) | 1 (3.12%) | ||||
| T Status | 0.7 | >0.9 | 0.4 | 0.6 | ||||||||
| 1 | 1 (2.27%) | 1 (0.53%) | 0 (0.00%) | 2 (0.97%) | 0 (0.00%) | 2 (0.89%) | 1 (0.50%) | 1 (3.12%) | ||||
| 2 | 4 (9.09%) | 21 (11.23%) | 3 (12.00%) | 22 (10.68%) | 0 (0.00%) | 25 (11.11%) | 22 (11.06%) | 3 (9.38%) | ||||
| 3 | 30 (68.18%) | 120 (64.17%) | 16 (64.00%) | 134 (65.05%) | 6 (100.00%) | 144 (64.00%) | 130 (65.33%) | 20 (62.50%) | ||||
| 4 | 9 (20.45%) | 44 (23.53%) | 6 (24.00%) | 47 (22.82%) | 0 (0.00%) | 53 (23.56%) | 45 (22.61%) | 8 (25.00%) | ||||
| x | 0 (0.00%) | 1 (0.53%) | 0 (0.00%) | 1 (0.49%) | 0 (0.00%) | 1 (0.44%) | 1 (0.50%) | 0 (0.00%) | ||||
| N Status | 0.8 | 0.7 | 0.3 | 0.8 | ||||||||
| 0 | 21 (47.73%) | 94 (50.27%) | 11 (44.00%) | 104 (50.49%) | 1 (16.67%) | 114 (50.67%) | 97 (48.74%) | 18 (56.25%) | ||||
| 1 | 15 (34.09%) | 65 (34.76%) | 10 (40.00%) | 70 (33.98%) | 4 (66.67%) | 76 (33.78%) | 70 (35.18%) | 10 (31.25%) | ||||
| 2 | 6 (13.64%) | 24 (12.83%) | 3 (12.00%) | 27 (13.11%) | 1 (16.67%) | 29 (12.89%) | 27 (13.57%) | 3 (9.38%) | ||||
| x | 2 (4.55%) | 4 (2.14%) | 1 (4.00%) | 5 (2.43%) | 0 (0.00%) | 6 (2.67%) | 5 (2.51%) | 1 (3.12%) | ||||
| Metastatic Status | 0.4 | 0.4 | >0.9 | >0.9 | ||||||||
| 0 | 27 (61.36%) | 107 (57.22%) | 12 (48.00%) | 122 (59.22%) | 4 (66.67%) | 130 (57.78%) | 115 (57.79%) | 19 (59.38%) | ||||
| 1 | 15 (34.09%) | 76 (40.64%) | 12 (48.00%) | 79 (38.35%) | 2 (33.33%) | 89 (39.56%) | 79 (39.70%) | 12 (37.50%) | ||||
| x | 2 (4.55%) | 4 (2.14%) | 1 (4.00%) | 5 (2.43%) | 0 (0.00%) | 6 (2.67%) | 5 (2.51%) | 1 (3.12%) | ||||
| Histological Grading | 0.003 ** | 0.8 | 0.004 ** | 0.14 | ||||||||
| 1 | 12 (27.27%) | 91 (48.66%) | 10 (40.00%) | 93 (45.15%) | 2 (33.33%) | 101 (44.89%) | 94 (47.24%) | 9 (28.12%) | ||||
| 2 | 18 (40.91%) | 73 (39.04%) | 10 (40.00%) | 81 (39.32%) | 0 (0.00%) | 91 (40.44%) | 76 (38.19%) | 15 (46.88%) | ||||
| 3 | 14 (31.82%) | 18 (9.63%) | 5 (20.00%) | 27 (13.11%) | 3 (50.00%) | 29 (12.89%) | 24 (12.06%) | 8 (25.00%) | ||||
| 4 | 0 (0.00%) | 2 (1.07%) | 0 (0.00%) | 2 (0.97%) | 1 (16.67%) | 1 (0.44%) | 2 (1.01%) | 0 (0.00%) | ||||
| Unknown | 0 (0.00%) | 3 (1.60%) | 0 (0.00%) | 3 (1.46%) | 0 (0.00%) | 3 (1.33%) | 3 (1.51%) | 0 (0.00%) | ||||
| Lymphovascular Status | 0.3 | 0.8 | 0.7 | 0.3 | ||||||||
| 0 | 11 (25.00%) | 40 (21.39%) | 4 (16.00%) | 47 (22.82%) | 2 (33.33%) | 49 (21.78%) | 42 (21.11%) | 9 (28.12%) | ||||
| 1 | 14 (31.82%) | 44 (23.53%) | 6 (24.00%) | 52 (25.24%) | 1 (16.67%) | 57 (25.33%) | 48 (24.12%) | 10 (31.25%) | ||||
| Unknown | 19 (43.18%) | 103 (55.08%) | 15 (60.00%) | 107 (51.94%) | 3 (50.00%) | 119 (52.89%) | 109 (54.77%) | 13 (40.62%) | ||||
| Pathological Morphology | 0.049 * | >0.9 | >0.9 | 0.020 * | ||||||||
| Adenocarcinoma | 41 (93.18%) | 185 (98.93%) | 25 (100.00%) | 201 (97.57%) | 6 (100.00%) | 220 (97.78%) | 197 (98.99%) | 29 (90.62%) | ||||
| Mucinous Carcinoma | 3 (6.82%) | 2 (1.07%) | 0 (0.00%) | 5 (2.43%) | 0 (0.00%) | 5 (2.22%) | 2 (1.01%) | 3 (9.38%) | ||||
| TILs | 0.4 | 0.7 | 0.14 | 0.12 | ||||||||
| Low | 10 (22.73%) | 32 (17.11%) | 3 (12.00%) | 39 (18.93%) | 2 (33.33%) | 40 (17.78%) | 34 (17.09%) | 8 (25.00%) | ||||
| Medium | 15 (34.09%) | 59 (31.55%) | 7 (28.00%) | 67 (32.52%) | 0 (0.00%) | 74 (32.89%) | 61 (30.65%) | 13 (40.62%) | ||||
| High | 10 (22.73%) | 66 (35.29%) | 11 (44.00%) | 65 (31.55%) | 2 (33.33%) | 74 (32.89%) | 71 (35.68%) | 5 (15.62%) | ||||
| Unknown | 9 (20.45%) | 30 (16.04%) | 4 (16.00%) | 35 (16.99%) | 2 (33.33%) | 37 (16.44%) | 33 (16.58%) | 6 (18.75%) | ||||
| Hemoglobin level (g/dL) | 0.044 * | 0.8 | 0.12 | 0.010 ** | ||||||||
| <10 | 10 (22.73%) | 17 (9.09%) | 2 (8.00%) | 25 (12.14%) | 1 (16.67%) | 26 (11.56%) | 18 (9.05%) | 9 (28.12%) | ||||
| ≥10 | 33 (75.00%) | 163 (87.17%) | 22 (88.00%) | 174 (84.47%) | 4 (66.67%) | 192 (85.33%) | 173 (86.93%) | 23 (71.88%) | ||||
| Unknown | 1 (2.27%) | 7 (3.74%) | 1 (4.00%) | 7 (3.40%) | 1 (16.67%) | 7 (3.11%) | 8 (4.02%) | 0 (0.00%) | ||||
| Serum albumin (g/dL) | 0.4 | 0.9 | 0.3 | 0.9 | ||||||||
| <3.5 | 16 (36.36%) | 82 (43.85%) | 10 (40.00%) | 88 (42.72%) | 3 (50.00%) | 95 (42.22%) | 85 (42.71%) | 13 (40.62%) | ||||
| >3.5 | 11 (25.00%) | 53 (28.34%) | 8 (32.00%) | 56 (27.18%) | 0 (0.00%) | 64 (28.44%) | 56 (28.14%) | 8 (25.00%) | ||||
| Unknown | 17 (38.64%) | 52 (27.81%) | 7 (28.00%) | 62 (30.10%) | 3 (50.00%) | 66 (29.33%) | 58 (29.15%) | 11 (34.38%) | ||||
| ECOG | 0.043 * | 0.2 | 0.5 | 0.010 ** | ||||||||
| ECOG 0–1 | 24 (54.55%) | 123 (65.78%) | 18 (72.00%) | 129 (62.62%) | 4 (66.67%) | 143 (63.56%) | 130 (65.33%) | 17 (53.12%) | ||||
| ECOG 2 | 12 (27.27%) | 24 (12.83%) | 1 (4.00%) | 35 (16.99%) | 0 (0.00%) | 36 (16.00%) | 25 (12.56%) | 11 (34.38%) | ||||
| ECOG 3–4 | 1 (2.27%) | 18 (9.63%) | 1 (4.00%) | 18 (8.74%) | 1 (16.67%) | 18 (8.00%) | 19 (9.55%) | 0 (0.00%) | ||||
| Unknown | 7 (15.91%) | 22 (11.76%) | 5 (20.00%) | 24 (11.65%) | 1 (16.67%) | 28 (12.44%) | 25 (12.56%) | 4 (12.50%) | ||||
| BMI (kg/m2) | 0.2 | 0.4 | 0.4 | 0.2 | ||||||||
| <18.5 | 19 (43.18%) | 52 (27.81%) | 6 (24.00%) | 65 (31.55%) | 4 (66.67%) | 67 (29.78%) | 57 (28.64%) | 14 (43.75%) | ||||
| 18.5–22.9 | 16 (36.36%) | 74 (39.57%) | 14 (56.00%) | 76 (36.89%) | 1 (16.67%) | 89 (39.56%) | 79 (39.70%) | 11 (34.38%) | ||||
| 23–24.9 | 6 (13.64%) | 25 (13.37%) | 3 (12.00%) | 28 (13.59%) | 0 (0.00%) | 31 (13.78%) | 26 (13.07%) | 5 (15.62%) | ||||
| ≥25 | 2 (4.55%) | 28 (14.97%) | 1 (4.00%) | 29 (14.08%) | 1 (16.67%) | 29 (12.89%) | 29 (14.57%) | 1 (3.12%) | ||||
| Unknown | 1 (2.27%) | 8 (4.28%) | 1 (4.00%) | 8 (3.88%) | 0 (0.00%) | 9 (4.00%) | 8 (4.02%) | 1 (3.12%) | ||||
1 Fisher’s exact test; * p < 0.05; ** p < 0.01; *** p < 0.001.
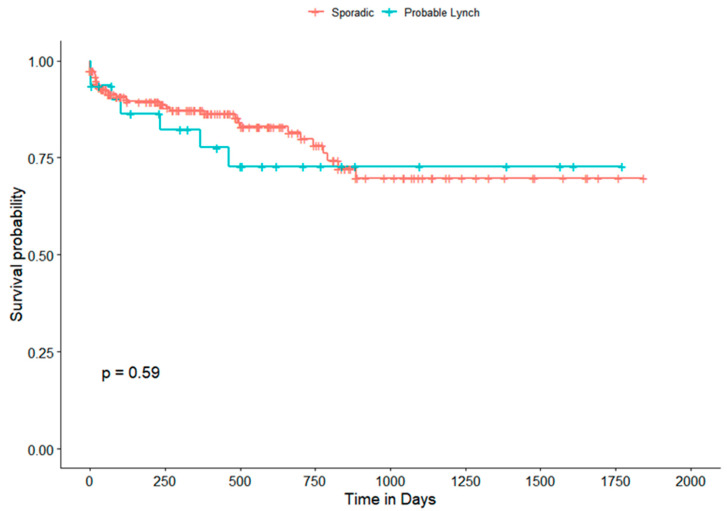
The overall survival (follow-up period of 0–1845 days/61.5 months) of both “Probable Lynch” and non-LS (sporadic) groups (n = 227) was comparable (p = 0.59) (Figure 2). In univariate analysis, there were significant associations between stage (p = 0.040), node status (p = 0.005), histological grading (p = 0.036) and ECOG (p ≤ 0.001) with patient overall survival. Multivariate analysis using the Cox proportional hazards model further showed ECOG (p = 0.002) as an independent survival predictor in the CRC population (Table 6).
Figure 2.
Overall survival curves for CRC patients in “Probable Lynch” and sporadic groups. Survival was evaluated using Kaplan–Meier curves and compared with the log-rank test.
Table 6.
Univariate and multivariate survival analysis (n = 231).
| Characteristic | Univariate | Multivariate | |||||||
|---|---|---|---|---|---|---|---|---|---|
| N | Event N | HR 1 | 95% CI 1 | p-Value | Event N | HR 1 | 95% CI 1 | p-Value | |
| Age | 227 | 40 | |||||||
| <50 | — | — | |||||||
| ≥50 | 0.85 | 0.42, 1.74 | 0.7 | ||||||
| Sex | 227 | 40 | |||||||
| Female | — | — | |||||||
| Male | 0.90 | 0.48, 1.69 | 0.7 | ||||||
| Tumor Site | 226 | 40 | |||||||
| Left | — | — | |||||||
| Right | 1.01 | 0.48, 2.12 | >0.9 | ||||||
| Stage | 223 | 40 | 30 | ||||||
| I–II | — | — | — | — | |||||
| III–IV | 2.18 | 1.03, 4.60 | 0.040 * | 1.82 | 0.58, 5.73 | 0.3 | |||
| T Status | 227 | 40 | |||||||
| 1–2 | — | — | |||||||
| 3–4 | 3.21 | 0.77, 13.3 | 0.11 | ||||||
| x | † | ||||||||
| Node Status | 227 | 40 | 30 | ||||||
| 0 | — | — | — | — | |||||
| 1 | 1.29 | 0.64, 2.61 | 0.5 | 0.79 | 0.27, 2.37 | 0.7 | |||
| 2 | 3.35 | 1.45, 7.72 | 0.005 ** | 1.97 | 0.62, 6.30 | 0.3 | |||
| x | † | ||||||||
| Metastatic Status | 227 | 40 | |||||||
| 0 | — | — | |||||||
| 1 | 1.86 | 1.00, 3.48 | 0.051 | ||||||
| x | † | ||||||||
| Histological Grading | 225 | 38 | 30 | ||||||
| 1–2 | — | — | — | — | |||||
| 3–4 | 2.23 | 1.05, 4.71 | 0.036 * | 1.27 | 0.51, 3.16 | 0.6 | |||
| Lymphovascular Status | 109 | 22 | |||||||
| 0 | — | — | |||||||
| 1 | 0.70 | 0.30, 1.65 | 0.4 | ||||||
| Pathological Morphology | 227 | 40 | |||||||
| Adenocarcinoma | — | — | |||||||
| Mucinous Carcinoma | 1.66 | 0.23, 12.1 | 0.6 | ||||||
| TILs | 189 | 33 | |||||||
| High | — | — | |||||||
| Medium | 0.85 | 0.38, 1.93 | 0.7 | ||||||
| Low | 2.29 | 0.95, 5.51 | 0.065 | ||||||
| Hemoglobin level (g/dL) | 220 | 40 | |||||||
| <10 | — | — | |||||||
| ≥10 | 0.62 | 0.22, 1.79 | 0.4 | ||||||
| Serum albumin (g/dL) | 160 | 35 | |||||||
| <3.5 | — | — | |||||||
| >3.5 | 0.62 | 0.31, 1.25 | 0.2 | ||||||
| ECOG | 198 | 32 | 30 | ||||||
| ECOG 0–1 | — | — | — | — | |||||
| ECOG 2 | 2.18 | 0.90, 5.27 | 0.083 | 1.70 | 0.64, 4.50 | 0.3 | |||
| ECOG 3–4 | 4.60 | 1.98, 10.7 | <0.001 *** | 4.38 | 1.72, 11.2 | 0.002 ** | |||
| BMI (kg/m2) | 219 | 38 | |||||||
| <18.5 | — | — | |||||||
| 18.5–22.9 | 0.87 | 0.41, 1.84 | 0.7 | ||||||
| 23–24.9 | 0.81 | 0.28, 2.29 | 0.7 | ||||||
| ≥25 | 0.73 | 0.26, 2.07 | 0.6 | ||||||
| Microsatellite Instability Status | 227 | 40 | |||||||
| MSI | — | — | |||||||
| MSS | 0.82 | 0.39, 1.73 | 0.6 | ||||||
| BRAF Exon 15 | 227 | 40 | |||||||
| Mutant | — | — | |||||||
| Wild-type | 0.89 | 0.35, 2.28 | 0.8 | ||||||
| MLH1 Methylation | 227 | 40 | |||||||
| Methylated | — | — | |||||||
| Unmethylated | 0.96 | 0.13, 6.96 | >0.9 | ||||||
| Probable Lynch | 227 | 40 | |||||||
| No | — | — | |||||||
| Yes | 1.25 | 0.55, 2.83 | 0.6 | ||||||
1 HR = Hazard Ratio, CI = Confidence Interval; † Too small a number to analyse; TILs, Tumour-infiltrating lymphocytes; ECOG, Eastern Cooperative Oncology Group Performance Status; BMI, Body Mass Index; MSI, Microsatellite Instability; MSS, Microsatellite Stable; * p < 0.05; ** p < 0.01; *** p < 0.001.
4. Discussion
The molecular cohort tested in this study was enriched for 21.65% of EOCRC (<50 years old). This was part of a larger study to assess the clinicopathology parameters of 1276 consecutive patients treated for CRC at Dr Sardjito Hospital, Yogyakarta, Indonesia, during 2016–2019. Despite only limited FFPE samples being available for molecular testing in this project, the patient clinicopathology characteristics of this sub-population for molecular testing were similar to the characteristics found in the overall CRC cohort (Supplementary Table S1). The frequency of EOCRC in the larger cohort was 27.6% [30], as similarly reflected in the molecular cohort. This high proportion of EOCRC was intriguing, as no existing data have been reported regarding the extent of LS, which is usually characterised by early onset, in the local context of Indonesia.
In this study, MSI and BRAF frequency (19.05% and 10.82%) among the CRC cases was within the range of previously reported data, at about ~20% and ~15%, respectively [31,32,33,34,35,36,37]. The well-established strong association (p = 0.031) between MSI and BRAF V600E mutation was also confirmed in this study, although the percentage (20.45%) was much lower than what usually has been reported: 50–70% [29,38,39]. Yet, the frequency of BRAF mutation in MSS cases (8.56%) was similar to that reported in the literature [24,40]. Lower BRAF mutation frequency in CRC in Asian countries such as Japan, Russia, Israel and China has been reported previously by many, ranging from 3–6% of all cases and about 15.4% of cases with loss of the expression of MLH1/PMS2 [33,41,42,43,44,45,46,47].
There was also a much lower frequency of MLH1 promoter methylation, at only 2.60%, compared to approximately 20% reported in the published literature [48,49]. Colorectal cancer showing somatic MLH1 methylation tends to occur at relatively advanced ages, is more common in women, is often located in the right colon (as also shown in this study), displays the somatic activating BRAF V600E mutation and occurs in sporadic tumours instead of in the inherited form of CRC, like LS [24,50,51]. However, it has been reported on very rare occasion that MLH1 promoter methylation could present as a constitutional defect, detectable in normal cells such as peripheral leukocytes, in patients presenting with phenotypic characteristics of LS [52].
This study reveals much higher “Probable Lynch” (13.85%) as compared to what has been reported in western countries, where LS accounts for about 2–3% of total CRC cases [53,54,55]. This was indicated by the presence of MSI with a much lower number of BRAF V600E mutations and MLH1 promoter methylations. It would be ideal to confirm the findings by analysing the germline MMR mutation. Nonetheless, we are confident of the robustness of the N_LyST tests as detailed in the previous publication, describing the high frequency of BRAF mutants and MLH1 methylation within the MSI of the UK CRC samples, with near perfect agreement with some standard methods [29]. Prior validation of the test using 50 Indonesian CRC samples with known MSI, BRAF and MLH1 promoter methylation status also showed 100% concordance (unpublished data). In this study, the fact that ~90% of MSI samples showed instability in more than four MSI markers and ~90% of MSS samples showed no instability in any of the markers implies that it was less likely that the instabilities observed were due to polymorphism. Further, BRAF V600E mutation, and MLH1 promoter ‘C region’ methylation specifically (as in the N_LyST panel), have been reported as strong predictors of negative MMR mutation status [56].
As expected, the “Probable Lynch” was more frequent in EOCRC as compared to the older group, accounting for 24% of all EOCRC, which is slightly higher than the 16.1% that was reported in a study conducted on Asian immigrants with CRC in the USA [57]. Yet, the majority of EOCRC was non-LS, referred as sporadic cases. Despite the global trend of increased incidence, studies comparing EOCRC and LOCRC show no difference in the profile or frequency of somatic mutation in the known cancer driver genes [9,58,59]. This study found that non-LS cases were associated with left-sided tumours, which has also been reported by others [60,61,62]. The sporadic cases also were associated with low levels of Hb, which has been suggested to be closely associated with a higher stage of disease at presentation and a higher risk of death [63].
Despite previously reports in the literature that LS patients have better long-term survival prognosis than sporadic CRC patients [64,65,66], comparable overall survival was observed in this study between the “Probable Lynch” and the non-LS (sporadic) groups. Consistent with the findings from the larger cohort of CRC (n = 1276) (Hutajulu et al., under review) and with previous reports on selected cases of CRC of local patients, ECOG index played an important role in predicting the OS [67].
Detecting LS has been long reported to provide aid in the diagnosis or management of the individual or at-risk family members, including the potential of non-invasive CRC prevention with aspirin [68]. It is also worth noting that there was a considerable number of “Probable Lynch” cases among the older patients (11.05%), which supports the cost-effectiveness of LS screening for all CRC patients up to 70 years old [27,28]. MSI testing is likely to increase, as it provides information that extends beyond LS testing; for example, MSI can be used to stratify patients into groups eligible for treatment with 5-fluorouracil-based therapy [69,70], which is the most frequent chemotherapy regimen for CRC in Indonesia, or the latest immunotherapy such as pembrolizumab [71].
We found only 3/35 (8.6%) cases of MSI with BRAF wildtype group showed MLH1 promoter methylation, which are less likely to be LS. This is in contrast to a recent study in China, where 37% of cases without BRAF V600E mutation had MLH1 promoter methylation [47]. This suggests, at least in Indonesia, the prospective of simplifying the diagnostic test workflow to MSI and BRAF testing only, without the need for performing the bisulphite conversion for MLH1 promoter methylation analysis, preferably with the inclusion of family history of CRC as additional criteria. This study showed that N_LyST has high utility to accommodate low-quality samples and portability, as it does not require expensive machines and has minimal infrastructure requirements, which fits the need for implementation in recourse limited settings. Obviously, further validation using larger cohorts and confirmation of the germline mutation of the relevant MMR genes are required.
5. Conclusions
In summary, despite some limitations, which include relatively a small number of samples from a single centre and the absence of germline mutation data on the MMR genes, to our knowledge, this study is the first to reveal a potentially higher incidence of Lynch Syndrome among CRC patients in Indonesia, which may partially contribute to the reported much higher number of EOCRC found in some studies. Further study is required to examine the extent of LS incidence in Indonesia, including in the population setting, to confirm the existing and novel germline pathological variants. Furthermore, a large proportion of EOCRC, as shown in this study, was considered sporadic (non-LS), for which the mechanism of the so-called “accelerated carcinogenesis” warrants further investigation.
Acknowledgments
The authors thank Aru W Sudoyo and Ahmad Utomo for critical input into study design and analysis of results, Wien Kusharyoto for support in setting up the test in Indonesia, and Yasjudan Rastrama Putra and Yana Suryani for technical assistance and coordination.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/cancers13246245/s1, Table S1: Clinicopathology characteristics of overall Jogjakarta CRC cohort and the sub-population used for molecular testing.
Author Contributions
S.S., D.S.H., S.H.H. and M.I.; Methodology: S.S., S.W., G.A., N.Y., D.S.H., A.M.R., S.H.H. and M.I.; Software: S.S., S.W., G.A. and A.M.R.; Validation: S.S., S.W., G.A., N.Y., D.S.H., A.M.R., S.H.H. and M.I.; Formal Analysis: S.S., S.W., G.A., N.Y., D.S.H., A.M.R., S.H.H. and M.I.; Investigation: S.S., S.W., G.A., N.Y., A.M.R. and H.H.; Resources: S.S., N.Y., D.S.H., A.Y.H., J.K., S.H.H. and M.I.; Data Curation: S.S., S.W., G.A., N.Y., A.M.R., H.H. and S.H.H.; Writing–Original Draft Preparation: S.S. and S.W.; Writing–Review and Editing: S.S., D.S.H., A.Y.H., J.K., S.H.H. and M.I.; Visualization: S.S., S.W. and G.A.; Supervision: S.S., N.Y., D.S.H., S.H.H. and M.I.; Project Administration: S.W., G.A. and A.M.R.; Funding Acquisition: S.S., S.H.H. and M.I.; Approval of the version to be published: all authors. All authors have read and agreed to the published version of the manuscript.
Funding
The authors received financial support from the Applied Research Grant 2020–2021, The Indonesian Ministry of Research, Technology, and Higher Education and the Institutional Links grant, ID 527558574, under the Newton Institutional Link-Indonesia KLN Fund partnership, funded by the UK Department for Business, Energy and Industrial Strategy and Indonesia Ministry of Research Technology & Higher Education and delivered by the British Council.
Institutional Review Board Statement
This study was conducted under Ethical approval (KE/FIV0549|ECD020; Medical and Health Research Ethics Committee (MHREC), FKKMK, UGM-Dr. Sardjito General Hospital).
Informed Consent Statement
The informed consents have been obtained for the use of tumour samples, clinical data and any other relevant data in the research for all subjects.
Conflicts of Interest
MI was appointed as specialist committee member to the Diagnostics Assessment Committee of the National Institute for Health and Care Excellence (NICE), which produced the guidance DG27 on Lynch Syndrome testing. MI and AMR are unpaid scientific advisors of PathGen Diagnostik Teknologi.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J., Ervik M., Lam F., Colombet M., Mery L., Piñeros M., Znaor A., Soerjomataram I., Bray F. Global Cancer Observatory: Cancer Today. International Agency for Research on Cancer; Lyon, France: 2020. [Google Scholar]
- 3.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 4.You Y.N., Xing Y., Feig B.W., Chang G.J., Cormier J.N. Young-Onset Colorectal Cancer: Is It Time to Pay Attention? Arch. Intern. Med. 2012;172:287–289. doi: 10.1001/archinternmed.2011.602. [DOI] [PubMed] [Google Scholar]
- 5.Murphy C.C., Lund J.L., Sandler R.S. Young-Onset Colorectal Cancer: Earlier Diagnoses or Increasing Disease Burden? Gastroenterology. 2017;152:1809–1812.e3. doi: 10.1053/j.gastro.2017.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Siegel R.L., Torre L.A., Soerjomataram I., Hayes R.B., Bray F., Weber T.K., Jemal A. Global patterns and trends in colorectal cancer incidence in young adults. Gut. 2019;68:2179–2185. doi: 10.1136/gutjnl-2019-319511. [DOI] [PubMed] [Google Scholar]
- 7.O’Connell J.B., Maggard M.A., Livingston E.H., Yo C.K. Colorectal cancer in the young. Am. J. Surg. 2004;187:343–348. doi: 10.1016/j.amjsurg.2003.12.020. [DOI] [PubMed] [Google Scholar]
- 8.Limburg P.J., Harmsen W.S., Chen H.H., Gallinger S., Haile R.W., Baron J.A., Casey G., Woods M.O., Thibodeau S.N., Lindor N.M. Prevalence of Alterations in DNA Mismatch Repair Genes in Patients With Young-Onset Colorectal Cancer. Clin. Gastroenterol. Hepatol. 2011;9:497–502. doi: 10.1016/j.cgh.2010.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stoffel E.M., Koeppe E., Everett J., Ulintz P., Kiel M., Osborne J., Williams L., Hanson K., Gruber S.B., Rozek L.S. Germline Genetic Features of Young Individuals with Colorectal Cancer. Gastroenterology. 2018;154:897–905.e1. doi: 10.1053/j.gastro.2017.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anthonysamy M.A., Maker L.P., Lin I., Gotra I.M., Saputra H. Prevalence of colorectal carcinoma based on microscopic type, sex, age and anatomical location in Sanglah General Hospital. Intisari Sains Medis. 2020;11:272–276. doi: 10.15562/ism.v10i2.171. [DOI] [Google Scholar]
- 11.Sudoyo A.W., Hernowo B., Krisnuhoni E., Reksodiputro A.H., Hardjodisastro D. Colorectal cancer among young native Indonesians: A clinicopathological and molecular assessment on microsatellite instability. Med. J. Indones. 2010;19:245–251. doi: 10.13181/mji.v19i4.411. [DOI] [Google Scholar]
- 12.Vuik F.E., Nieuwenburg S.A., Bardou M., Lansdorp-Vogelaar I., Dinis-Ribeiro M., Bento M.J., Zadnik V., Pellisé M., Esteban L., Kaminski M.F., et al. Increasing incidence of colorectal cancer in young adults in Europe over the last 25 years. Gut. 2019;68:1820–1826. doi: 10.1136/gutjnl-2018-317592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Siegel R.L., Jakubowski C.D., Fedewa S.A., Davis A., Azad N.S. Colorectal Cancer in the Young: Epidemiology, Prevention, Management. Am. Soc. Clin. Oncol. Educ. B. 2020;40:e75–e88. doi: 10.1200/EDBK_279901. [DOI] [PubMed] [Google Scholar]
- 14.Wei J., Ge X., Tang Y., Qian Y., Lu W., Jiang K., Fang Y., Hwang M., Fu D., Xiao Q., et al. An Autophagy-Related Long Noncoding RNA Signature Contributes to Poor Prognosis in Colorectal Cancer. J. Oncol. 2020;2020:4728947. doi: 10.1155/2020/4728947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lynch H.T., de la Chapelle A. Genetic susceptibility to non-polyposis colorectal cancer. J. Med. Genet. 1999;36:801–818. [PMC free article] [PubMed] [Google Scholar]
- 16.Marzouk O., Schofield J. Review of histopathological and molecular prognostic features in colorectal cancer. Cancers. 2011;3:2767–2810. doi: 10.3390/cancers3022767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boland C.R., Goel A. Microsatellite instability in colorectal cancer. Gastroenterology. 2010;138:2073–2087.e3. doi: 10.1053/j.gastro.2009.12.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peltomäki P. Role of DNA Mismatch Repair Defects in the Pathogenesis of Human Cancer. J. Clin. Oncol. 2003;21:1174–1179. doi: 10.1200/JCO.2003.04.060. [DOI] [PubMed] [Google Scholar]
- 19.Veigl M.L., Kasturi L., Olechnowicz J., Ma A., Lutterbaugh J.D., Periyasamy S., Li G.-M., Drummond J., Modrich P.L., Sedwick W.D., et al. Biallelic inactivation of hMLH1 by epigenetic gene silencing, a novel mechanism causing human MSI cancers. Proc. Natl. Acad. Sci. USA. 1998;95:8698–8702. doi: 10.1073/pnas.95.15.8698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herman J.G., Umar A., Polyak K., Graff J.R., Ahuja N., Issa J.P., Markowitz S., Willson J.K., Hamilton S.R., Kinzler K.W., et al. Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proc. Natl. Acad. Sci. USA. 1998;95:6870–6875. doi: 10.1073/pnas.95.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cunningham J.M., Christensen E.R., Tester D.J., Kim C.-Y., Roche P.C., Burgart L.J., Thibodeau S.N. Hypermethylation of the hMLH1 Promoter in Colon Cancer with Microsatellite Instability. Cancer Res. 1998;58:3455 LP–3460. [PubMed] [Google Scholar]
- 22.Deng G., Bell I., Crawley S., Gum J., Terdiman J.P., Allen B.A., Truta B., Sleisenger M.H., Kim Y.S. BRAF Mutation Is Frequently Present in Sporadic Colorectal Cancer with Methylated hMLH1, But Not in Hereditary Nonpolyposis Colorectal Cancer. Clin. Cancer Res. 2004;10:191 LP–195. doi: 10.1158/1078-0432.CCR-1118-3. [DOI] [PubMed] [Google Scholar]
- 23.Bouzourene H., Hutter P., Losi L., Martin P., Benhattar J. Selection of patients with germline MLH1 mutated Lynch syndrome by determination of MLH1 methylation and BRAF mutation. Fam. Cancer. 2010;9:167–172. doi: 10.1007/s10689-009-9302-4. [DOI] [PubMed] [Google Scholar]
- 24.Thiel A., Heinonen M., Kantonen J., Gylling A., Lahtinen L., Korhonen M., Kytölä S., Mecklin J.-P., Orpana A., Peltomäki P., et al. BRAF mutation in sporadic colorectal cancer and Lynch syndrome. Virchows Arch. 2013;463:613–621. doi: 10.1007/s00428-013-1470-9. [DOI] [PubMed] [Google Scholar]
- 25.National Comprehensive Cancer Network Genetic/Familial High-Risk Assessment: Colorectal (Version 3.2017) 2017. [(accessed on 20 June 2021)]. Available online: https://www.nccn.org/professionals/physician_gls/pdf/genetics_colon.pdf.
- 26.National Institute of Clinical and Healthcare Excellence Molecular Testing Strategies for Lynch Syndrome in People with Colorectal Cancer (NICE Diagnostics Guidance No. DG27) 2017. [(accessed on 20 June 2021)]. Available online: https://www.nice.org.uk/guidance/dg27.
- 27.Leenen C.H.M., Goverde A., de Bekker-Grob E.W., Wagner A., van Lier M.G.F., Spaander M.C.W., Bruno M.J., Tops C.M., van den Ouweland A.M.W., Dubbink H.J., et al. Cost-effectiveness of routine screening for Lynch syndrome in colorectal cancer patients up to 70 years of age. Genet. Med. 2016;18:966–973. doi: 10.1038/gim.2015.206. [DOI] [PubMed] [Google Scholar]
- 28.Kang Y.-J., Killen J., Caruana M., Simms K., Taylor N., Frayling I.M., Snowsill T., Huxley N., Coupe V.M.H., Hughes S., et al. The predicted impact and cost-effectiveness of systematic testing of people with incident colorectal cancer for Lynch syndrome. Med. J. Aust. 2020;212:72–81. doi: 10.5694/mja2.50356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Susanti S., Fadhil W., Ebili H.O., Asiri A., Nestarenkaite A., Hadjimichael E., Ham-Karim H.A., Field J., Stafford K., Matharoo-Ball B., et al. N-LyST: A simple and rapid screening test for Lynch syndrome. J. Clin. Pathol. 2018;71:713–720. doi: 10.1136/jclinpath-2018-205013. [DOI] [PubMed] [Google Scholar]
- 30.Hutajulu S.H., Putra Y.R., Susanti S., Heriyanto D.S., Yoshuantari N., Handaya A.Y., Utomo B.P., Dwidanarti S.R., Gusnanto A., Kurnianda J., et al. Early-onset versus late-onset colorectal cancer in Indonesia: A tertiary care center assessment. Manuscr. Submitt. Publ. 2021 in review. [Google Scholar]
- 31.Vilar E., Gruber S.B. Microsatellite instability in colorectal cancer-the stable evidence. Nat. Rev. Clin. Oncol. 2010;7:153–162. doi: 10.1038/nrclinonc.2009.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ashktorab H., Ahuja S., Kannan L., Llor X., Ellis N.A., Xicola R.M., Laiyemo A.O., Carethers J.M., Brim H., Nouraie M. A meta-analysis of MSI frequency and race in colorectal cancer. Oncotarget. 2016;7:34546–34557. doi: 10.18632/oncotarget.8945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seppälä T.T., Böhm J.P., Friman M., Lahtinen L., Väyrynen V.M.J., Liipo T.K.E., Ristimäki A.P., Kairaluoma M.V.J., Kellokumpu I.H., Kuopio T.H.I., et al. Combination of microsatellite instability and BRAF mutation status for subtyping colorectal cancer. Br. J. Cancer. 2015;112:1966–1975. doi: 10.1038/bjc.2015.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seligmann J.F., Fisher D., Smith C.G., Richman S.D., Elliott F., Brown S., Adams R., Maughan T., Quirke P., Cheadle J., et al. Investigating the poor outcomes of BRAF-mutant advanced colorectal cancer: Analysis from 2530 patients in randomised clinical trials. Ann. Oncol. 2017;28:562–568. doi: 10.1093/annonc/mdw645. [DOI] [PubMed] [Google Scholar]
- 35.Chiu V.K., Osman D., Belmonte J., Routh J.K., Soares H.P., Martin D.R., Hanson J.A., Gallupalli R.R., Kinney A., Vasef M., et al. Actionable genomic biomarkers in a low socioeconomic status (SES) population with gastrointestinal (GI) cancers. J. Clin. Oncol. 2018;36:e24310. doi: 10.1200/JCO.2018.36.15_suppl.e24310. [DOI] [Google Scholar]
- 36.Zhang X., Wu J., Wang L., Zhao H., Li H., Duan Y., Li Y., Xu P., Ran W., Xing X. HER2 and BRAF mutation in colorectal cancer patients: A retrospective study in Eastern China. PeerJ. 2020;8:e8602. doi: 10.7717/peerj.8602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang Y., Wang D., Jin L., Wu G., Bai Z., Wang J., Yao H., Zhang Z. Prognostic value of the combination of microsatellite instability and BRAF mutation in colorectal cancer. Cancer Manag. Res. 2018;10:3911–3929. doi: 10.2147/CMAR.S169649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roth A.D., Tejpar S., Delorenzi M., Yan P., Fiocca R., Klingbiel D., Dietrich D., Biesmans B., Bodoky G., Barone C., et al. Prognostic Role of KRAS and BRAF in Stage II and III Resected Colon Cancer: Results of the Translational Study on the PETACC-3, EORTC 40993, SAKK 60-00 Trial. J. Clin. Oncol. 2009;28:466–474. doi: 10.1200/JCO.2009.23.3452. [DOI] [PubMed] [Google Scholar]
- 39.Lochhead P., Kuchiba A., Imamura Y., Liao X., Yamauchi M., Nishihara R., Qian Z.R., Morikawa T., Shen J., Meyerhardt J.A., et al. Microsatellite instability and BRAF mutation testing in colorectal cancer prognostication. J. Natl. Cancer Inst. 2013;105:1151–1156. doi: 10.1093/jnci/djt173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fanelli G.N., Dal Pozzo C.A., Depetris I., Schirripa M., Brignola S., Biason P., Balistreri M., Dal Santo L., Lonardi S., Munari G., et al. The heterogeneous clinical and pathological landscapes of metastatic Braf-mutated colorectal cancer. Cancer Cell Int. 2020;20:30. doi: 10.1186/s12935-020-1117-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rozek L.S., Herron C.M., Greenson J.K., Moreno V., Capella G., Rennert G., Gruber S.B. Smoking, Gender, and Ethnicity Predict Somatic BRAF Mutations in Colorectal Cancer. Cancer Epidemiol. Biomarkers Prev. 2010;19:838–843. doi: 10.1158/1055-9965.EPI-09-1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hanna M.C., Go C., Roden C., Jones R.T., Pochanard P., Javed A.Y., Javed A., Mondal C., Palescandolo E., Van Hummelen P., et al. Colorectal Cancers from Distinct Ancestral Populations Show Variations in BRAF Mutation Frequency. PLoS ONE. 2013;8:e74950. doi: 10.1371/journal.pone.0074950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yanus G.A., Belyaeva A.V., Ivantsov A.O., Kuligina E.S., Suspitsin E.N., Mitiushkina N.V., Aleksakhina S.N., Iyevleva A.G., Zaitseva O.A., Yatsuk O.S., et al. Pattern of clinically relevant mutations in consecutive series of Russian colorectal cancer patients. Med. Oncol. 2013;30:686. doi: 10.1007/s12032-013-0686-5. [DOI] [PubMed] [Google Scholar]
- 44.Qiu T., Lu H., Guo L., Huang W., Ling Y., Shan L., Li W., Ying J., Lv N. Detection of BRAF mutation in Chinese tumor patients using a highly sensitive antibody immunohistochemistry assay. Sci. Rep. 2015;5:9211. doi: 10.1038/srep09211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ye J.-X. KRAS and BRAF gene mutations and DNA mismatch repair status in Chinese colorectal carcinoma patients. World J. Gastroenterol. 2015;21:1595. doi: 10.3748/wjg.v21.i5.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Natsume S., Yamaguchi T., Takao M., Iijima T., Wakaume R., Takahashi K., Matsumoto H., Nakano D., Horiguchi S.-I., Koizumi K., et al. Clinicopathological and molecular differences between right-sided and left-sided colorectal cancer in Japanese patients. Jpn. J. Clin. Oncol. 2018;48:609–618. doi: 10.1093/jjco/hyy069. [DOI] [PubMed] [Google Scholar]
- 47.Wang W., Ying J., Shi S., Ye Q., Zou S., Dong L., Lyu N. A modified screening strategy for Lynch syndrome among MLH1-deficient CRCs: Analysis from consecutive Chinese patients in a single center. Transl. Oncol. 2021;14:101049. doi: 10.1016/j.tranon.2021.101049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gay L.J., Arends M.J., Mitrou P.N., Bowman R., Ibrahim A.E., Happerfield L., Luben R., McTaggart A., Ball R.Y., Rodwell S.A. MLH1 Promoter Methylation, Diet, and Lifestyle Factors in Mismatch Repair Deficient Colorectal Cancer Patients From EPIC-Norfolk. Nutr. Cancer. 2011;63:1000–1010. doi: 10.1080/01635581.2011.596987. [DOI] [PubMed] [Google Scholar]
- 49.Li X., Yao X., Wang Y., Hu F., Wang F., Jiang L., Liu Y., Wang D., Sun G., Zhao Y. MLH1 Promoter Methylation Frequency in Colorectal Cancer Patients and Related Clinicopathological and Molecular Features. PLoS ONE. 2013;8:e59064. doi: 10.1371/journal.pone.0059064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ogino S., Nosho K., Kirkner G.J., Kawasaki T., Meyerhardt J.A., Loda M., Giovannucci E.L., Fuchs C.S. CpG island methylator phenotype, microsatellite instability, BRAF mutation and clinical outcome in colon cancer. Gut. 2009;58:90–96. doi: 10.1136/gut.2008.155473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Haraldsdottir S., Hampel H., Wu C., Weng D.Y., Shields P.G., Frankel W.L., Pan X., de la Chapelle A., Goldberg R.M., Bekaii-Saab T. Patients with colorectal cancer associated with Lynch syndrome and MLH1 promoter hypermethylation have similar prognoses. Genet. Med. 2016;18:863–868. doi: 10.1038/gim.2015.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Crucianelli F., Tricarico R., Turchetti D., Gorelli G., Gensini F., Sestini R., Giunti L., Pedroni M., Ponz de Leon M., Civitelli S., et al. MLH1 constitutional and somatic methylation in patients with MLH1 negative tumors fulfilling the revised Bethesda criteria. Epigenetics. 2014;9:1431–1438. doi: 10.4161/15592294.2014.970080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.de la Chapelle A. The Incidence of Lynch Syndrome. Fam. Cancer. 2005;4:233–237. doi: 10.1007/s10689-004-5811-3. [DOI] [PubMed] [Google Scholar]
- 54.Hampel H., Frankel W.L., Martin E., Arnold M., Khanduja K., Kuebler P., Nakagawa H., Sotamaa K., Prior T.W., Westman J., et al. Screening for the Lynch Syndrome (Hereditary Nonpolyposis Colorectal Cancer) N. Engl. J. Med. 2005;352:1851–1860. doi: 10.1056/NEJMoa043146. [DOI] [PubMed] [Google Scholar]
- 55.Moreira L., Balaguer F., Lindor N., de la Chapelle A., Hampel H., Aaltonen L.A., Hopper J.L., Le Marchand L., Gallinger S., Newcomb P.A., et al. Identification of Lynch syndrome among patients with colorectal cancer. JAMA. 2012;308:1555–1565. doi: 10.1001/jama.2012.13088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Parsons M.T., Buchanan D.D., Thompson B., Young J.P., Spurdle A.B. Correlation of tumour BRAF mutations and MLH1 methylation with germline mismatch repair (MMR) gene mutation status: A literature review assessing utility of tumour features for MMR variant classification. J. Med. Genet. 2012;49:151–157. doi: 10.1136/jmedgenet-2011-100714. [DOI] [PubMed] [Google Scholar]
- 57.Lee J., Xiao Y.-Y., Sun Y.Y., Balderacchi J., Clark B., Desani J., Kumar V., Saverimuthu A., Win K.T., Huang Y., et al. Prevalence and characteristics of hereditary non-polyposis colorectal cancer (HNPCC) syndrome in immigrant Asian colorectal cancer patients. BMC Cancer. 2017;17:843. doi: 10.1186/s12885-017-3799-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chang D.T., Pai R.K., Rybicki L.A., Dimaio M.A., Limaye M., Jayachandran P., Koong A.C., Kunz P.A., Fisher G.A., Ford J.M., et al. Clinicopathologic and molecular features of sporadic early-onset colorectal adenocarcinoma: An adenocarcinoma with frequent signet ring cell differentiation, rectal and sigmoid involvement, and adverse morphologic features. Mod. Pathol. 2012;25:1128–1139. doi: 10.1038/modpathol.2012.61. [DOI] [PubMed] [Google Scholar]
- 59.Cercek A., Chatila W.K., Yaeger R., Walch H., Fernandes G.D.S., Krishnan A., Palmaira L., Maio A., Kemel Y., Srinivasan P., et al. A Comprehensive Comparison of Early-Onset and Average-Onset Colorectal Cancers. JNCI J. Natl. Cancer Inst. 2021;113:1683–1692. doi: 10.1093/jnci/djab124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mork M.E., You Y.N., Ying J., Bannon S.A., Lynch P.M., Rodriguez-Bigas M.A., Vilar E. High Prevalence of Hereditary Cancer Syndromes in Adolescents and Young Adults With Colorectal Cancer. J. Clin. Oncol. 2015;33:3544–3549. doi: 10.1200/JCO.2015.61.4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Abdelsattar Z.M., Wong S.L., Regenbogen S.E., Jomaa D.M., Hardiman K.M., Hendren S. Colorectal cancer outcomes and treatment patterns in patients too young for average-risk screening. Cancer. 2016;122:929–934. doi: 10.1002/cncr.29716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yeo H., Betel D., Abelson J.S., Zheng X.E., Yantiss R., Shah M.A. Early-onset Colorectal Cancer is Distinct From Traditional Colorectal Cancer. Clin. Colorectal Cancer. 2017;16:293–299.e6. doi: 10.1016/j.clcc.2017.06.002. [DOI] [PubMed] [Google Scholar]
- 63.Qiu Y., Cai G., Su M., Chen T., Liu Y., Xu Y., Ni Y., Zhao A., Cai S., Xu L.X., et al. Urinary Metabonomic Study on Colorectal Cancer. J. Proteome Res. 2010;9:1627–1634. doi: 10.1021/pr901081y. [DOI] [PubMed] [Google Scholar]
- 64.Stigliano V., Assisi D., Cosimelli M., Palmirotta R., Giannarelli D., Mottolese M., Mete L.S., Mancini R., Casale V. Survival of hereditary non-polyposis colorectal cancer patients compared with sporadic colorectal cancer patients. J. Exp. Clin. Cancer Res. 2008;27:39. doi: 10.1186/1756-9966-27-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.de Matos M.B., Barbosa L.E., Teixeira J.P. Narrative review comparing the epidemiology, characteristics, and survival in sporadic colorectal carcinoma/Lynch syndrome. J. Coloproctol. 2020;40:73–78. doi: 10.1016/j.jcol.2019.07.006. [DOI] [Google Scholar]
- 66.Xu Y., Feng Q., Chen Y., Mao Y., Zhang Z., Lv Y., Zheng P., Yu S., He G., Xu J. Significance of Surgery in the Treatment of Colorectal Cancer Ovarian Metastases: A Retrospective Case Matching Study. Cancer Manag. Res. 2021;13:1087–1097. doi: 10.2147/CMAR.S285451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wardhani Y., Hutajulu S.H., Ferianti V.W., Fitriani Z., Taroeno-Hariadi K.W., Kurnianda J. Effects of oxaliplatin-containing adjuvant chemotherapy on short-term survival of patients with colon cancer in Dr. Sardjito Hospital, Yogyakarta, Indonesia. J. Gastrointest. Oncol. 2019;10:226–234. doi: 10.21037/jgo.2018.12.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Burn J., Sheth H., Elliott F., Reed L., Macrae F., Mecklin J.-P., Möslein G., McRonald F.E., Bertario L., Evans D.G., et al. Cancer prevention with aspirin in hereditary colorectal cancer (Lynch syndrome), 10-year follow-up and registry-based 20-year data in the CAPP2 study: A double-blind, randomised, placebo-controlled trial. Lancet. 2020;395:1855–1863. doi: 10.1016/S0140-6736(20)30366-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Klingbiel D., Saridaki Z., Roth A.D., Bosman F.T., Delorenzi M., Tejpar S. Prognosis of stage II and III colon cancer treated with adjuvant 5-fluorouracil or FOLFIRI in relation to microsatellite status: Results of the PETACC-3 trial. Ann. Oncol. 2015;26:126–132. doi: 10.1093/annonc/mdu499. [DOI] [PubMed] [Google Scholar]
- 70.Jin Z., Sinicrope F.A. Prognostic and Predictive Values of Mismatch Repair Deficiency in Non-Metastatic Colorectal Cancer. Cancers. 2021;13:300. doi: 10.3390/cancers13020300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.André T., Shiu K.-K., Kim T.W., Jensen B.V., Jensen L.H., Punt C., Smith D., Garcia-Carbonero R., Benavides M., Gibbs P., et al. Pembrolizumab in Microsatellite-Instability–High Advanced Colorectal Cancer. N. Engl. J. Med. 2020;383:2207–2218. doi: 10.1056/NEJMoa2017699. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.