Abstract
Simian virus 40 large T antigen has been shown to inhibit p53-mediated transcription once tethered to p53-responsive promoters through interaction with p53. In this study we report that p53 stimulates transcription by enhancing the recruitment of the basal transcription factors, TFIIA and TFIID, on the promoter (the DA complex) and by inducing a conformational change in the DA complex. Significantly, we have demonstrated that T antigen inhibits p53-mediated transcription by blocking this ability of p53. We investigated the mechanism for this inhibition and found that DA complex formation was resistant to T-antigen repression when the TFIID-DNA complex was formed prior to addition of p53-T antigen complex, indicating that the T antigen, once tethered to the promoter by p53, targets TFIID. Further, we have shown that the p53-T antigen complex prevents the TATA binding protein from binding to the TATA box. Thus, these data suggest a detailed mechanism by which p53 activates transcription and by which T antigen inhibits p53-mediated transcription.
The tumor suppressor p53 protein responds to DNA damage, resulting in cell cycle arrest or apoptosis (15, 27). The biochemical activity of p53 that is required for this relies on its ability to function as a sequence-specific DNA binding transcription factor (26). A number of oncoproteins have been discovered to transform cells by binding to p53 and inactivating its transcriptional activity. The inactivation can be achieved by a mechanism in which the transactivation domain of p53 is inhibited while DNA binding is unaffected (21, 28, 29, 31). In the case of the adenovirus early 1B (E1B) 55K protein, once it is brought to the promoter through interaction with p53 the E1B 55K protein can function as an active repressor to inhibit p53-responsive genes (31) and inhibit acetylation of p53 by PCAF (20). In the case of the cellular oncoprotein MDM2, once it is tethered to the promoter MDM2 can conceal the activation domain of p53 from the transcription machinery (22) and function as an active repressor (29).
Simian virus 40 (SV40) T antigen has been shown to bind to p53 and inhibit p53-dependent transcription via two mechanisms. In the first, T antigen prevents human p53 binding to its cognate DNA binding sequence (1). Recently we and others proposed a second mechanism by which T antigen forms a DNA binding complex with p53 that is transcriptionally inactive (28). Because this mechanism occurs in mouse cells but not in human cells, we proposed a model for T-antigen transformation (28). In human cells, latent p53 and T antigen form a complex that is unable to bind to DNA. Upon activation of p53, a possible conformational change in p53 allows p53 to dissociate from the p53-T complex and bind to DNA. Therefore, p53-activated transcription that is required for growth arrest and apoptosis would not be lost. In mouse cells, again latent p53 and T antigen form a complex that cannot bind DNA. Upon activation, however, the p53-T complex binds DNA as a transcriptionally inactive complex. p53-responsive promoters therefore would be completely blocked. Because SV40 T antigen is known to cause tumors in rodents but has not been shown to be a complete carcinogen in humans (3, 4), we speculate that our model may provide an explanation at the molecular level. Therefore, it will be of interest to elucidate the mechanism by which T antigen negatively affects p53-mediated transcription once tethered to the promoter.
Transcription initiation by RNA polymerase II involves the assembly of TFIIA, TFIIB, TFIID, TFIIE, TFIIF, and TFIIH (23). One of the first steps in preinitiation complex assembly is the association of TFIID with the TATA element. This is followed by the binding of the general transcription factor TFIIA to form the TFIIA-TFIID-promoter (DA) complex. The formation of the DA complex can be a major rate-limiting step in gene expression (5, 13, 17, 30). The DA complex assembly activity can be assayed in vitro by an electrophoretic mobility shift assay (EMSA) using Mg-agarose gels (Mg-EMSA). Using this assay system, several activators, such as the herpes simplex virus VP16 and the Epstein-Barr virus Zta, have been found to stimulate polymerase II transcription at least in part by promoting DA complex formation (6, 14).
In the present study we show that, like Zta and VP16, p53 also stimulates DA complex formation. Analysis of a transcriptionally inactive mutant form of p53 (a deletion at amino acids 1 to 92 [Δ92]) reveals that the binding of the DA complex to the TATA box region is necessary but not sufficient for transcription activation. It appears that a conformational change in the DA complex is also required for transactivation. Importantly, we have demonstrated that the transcriptionally inactive p53-T complex blocks the formation of the DA complex by targeting TFIID. Further, we have found that the p53-T complex is capable of preventing the TATA binding protein (TBP) from binding to the TATA box. Taken together, these results suggest that p53 stimulates transcription at least in part by enhancing the recruitment of the basal transcription factors, TFIIA and TFIID, and by inducing a conformational change in the DA complex. The transcriptionally inactive p53-T complex inhibits p53-mediated DA complex formation by preventing TBP from binding to the TATA element.
MATERIALS AND METHODS
Protein purification.
The TFIID complex was affinity purified from the LTRα3 cell line, a HeLa cell line that expresses hemagglutinin (HA)-tagged TBP, as described previously (18). Recombinant HA-tagged TBP was prepared essentially as described previously (19). The TFIIAαβ and TFIIAγ subunits were expressed in Escherichia coli BL21 cells from the plasmids pET-mycTFIIAαβ (13) and pQIIA-γ (24), respectively, and recombinant TFIIA containing the fused αβ subunit and the γ subunit was purified and renatured essentially as described previously (24).
p53-T complex was immunopurified from scid cell nuclear extracts as described previously (28). Briefly, 1 ml of scid nuclear extract (7 mg of protein/ml) was incubated with 100 μl of packed protein A-Sepharose beads to which Pab 421, a monoclonal antibody specific for p53, was covalently linked. Beads were then washed and p53 was eluted with 100 μl of Pab 421 peptide (KKGQSTSRHKK) at a 1-mg/ml concentration in 0.1 M KCl D buffer (20 mM HEPES [pH 7.9], 20% glycerol, 0.2 mM EDTA, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride). To purify mouse p53 in the absence of T antigen (p53-T–T), Pab 421 anti-p53 beads were incubated with nuclear extract. T antigen was eluted from the bound p53 by being washed with 2 M urea in 0.1 M KCl D buffer. The beads were then washed overnight with 0.1 M KCl D buffer, after which p53 was eluted as described above. Recombinant T antigen was prepared from SF21 insect cells infected with baculovirus expressing T antigen (a gift from C. Prives, Columbia University) as described by Bargonetti et al. (1). To purify p53 and ΔN92, HeLa cells were infected with recombinant vaccinia viruses expressing either an HA-tagged p53 (30) or an HA-tagged ΔN92 (ΔN92 [18]). The p53 proteins were affinity purified from the nuclear extract of infected cells by being bound to a matrix of anti-HA antibody, 12CA5, and eluted with HA epitope oligopeptide as described previously (18).
All purified proteins were analyzed by electrophoresis on sodium dodecyl sulfate (SDS)-polyacrylamide gels followed by Western blotting or silver staining.
DA complex assembly assays.
The radiolabeled 250-bp DNA fragment was prepared from plasmid p5RGCE4CAT (18), which contains five consecutive p53 binding sites and the adenovirus E4 TATA box. EMSA was performed in a total volume of 12.5 μl in 12.5 mM HEPES (pH 7.9), 60 mM KCl, 12.5% glycerol, 5 mM MgCl2, 1.6 mg of bovine serum albumin per ml, and 6 fmol of the radiolabeled probe essentially as described previously (14). The amounts of protein used in different reactions were as follows: 1 μl of TFIID that contained 2 to 4 ng of TBP, 20 ng of TFIIA, 10 ng of TBP, 50 ng of p53, 50 ng of ΔN92, or 3 μl of p53-T complex that contained 100 ng of p53. The reaction mixtures were incubated at 30°C for 30 min and analyzed on a 1.4% low EEO-agarose gel (Fisher) containing 5 mM MgAc as described previously (17). The DNA-protein complexes were visualized with a PhosphorImager using Adobe Photoshop software. In the supershift experiments, 100 ng of anti-TBP antibody (N-12; Santa Cruz, Santa Cruz, Calif.) was added at completion of DNA binding (30 min at 30°C). Samples were incubated for another 30 min at 30°C, and the reactions were stopped by loading the samples on the gel.
The DNase I footprinting assays were performed in a final volume of 25 μl, with reaction mixtures containing 6 fmol of the radiolabeled fragments one-end labeled. Binding reactions were performed as described above. Digestion reactions were performed with 1 or 2 ng of DNase I (BRL) for 60 s. The reactions were stopped by the addition of 50 μl of stop solution (200 mM NaCl, 30 mM EDTA, 1% SDS, 100 mg of yeast tRNA per ml), phenol-chloroform extracted twice, and ethanol precipitated in the presence of carrier RNA. The pellets were washed with 70% ethanol, dried, and resuspended in 3 μl of formamide loading buffer. The samples were then resolved on a 6% denaturing polyacrylamide gel.
E1B TATA-oligo probe (top; 5′-TCGACTTAAAGGGTATATAATGCGCCGTG-3′; bottom, 5′-TCGACACGGCGCATTATATACCCTTTAAG-3′) was labeled in the presence of [α-32P]dGTP using Klenow enzyme. Binding reactions were performed as described above. The reaction mixtures were analyzed on a 3% polyacrylamide gel. Mutant E1B TATA-oligo probe (5'-TCGACTTAAAGGGGAGAGAATGCGCCGTG-3') was included in some EMSA reactions as indicated.
In vitro protein-protein interaction assays.
Approximately 20 μl of glutathione-agarose beads were incubated with 2 μg of glutathione S-transferase (GST) or GST-TFIIA and then washed with 0.1 M KCl D buffer. p53 and ΔN92 were in vitro transcribed and translated in the presence of [35S]methionine. p53-T complex was radiolabeled in vivo by incubating scid cells in 1 ml of low-methionine medium containing 0.1 mCi of [35S]methionine for 3 h. After being labeled, the cells were washed with phosphate-buffered saline and lysed with radio immunoprecipitation assay buffer (10 mM NaPO4 [pH 7.2], 150 mM NaCl, 1% NP-40, 0.05% SDS, 0.5 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride). p53-T complex was purified using Pab 421-conjugated beads as described above. The radiolabeled p53 proteins were incubated with the GST and GST-TFIIA beads for 1 h at room temperature in a total volume of 0.2 ml in 0.1 M KCl D buffer. The beads were then washed with buffer D containing 500 mM KCl and 0.2% NP-40 and then with buffer D containing 100 mM KCl. The bound proteins were eluted by boiling the beads in SDS-polyacrylamide gel electrophoresis (PAGE) loading buffer, resolved on a SDS–10% PAGE gel, and visualized by autoradiography. GST-TBP protein interactions were performed using the same procedure described above.
TFIID in 250 μl of nuclear extract was immobilized on 20 μl of packed protein A-Sepharose beads using 2 μl of TAFII250 antibody (6B3; Santa Cruz). The protein pull-down assays were performed as described above.
RESULTS
p53 stimulates the TFIID-TFIIA-promoter complex formation.
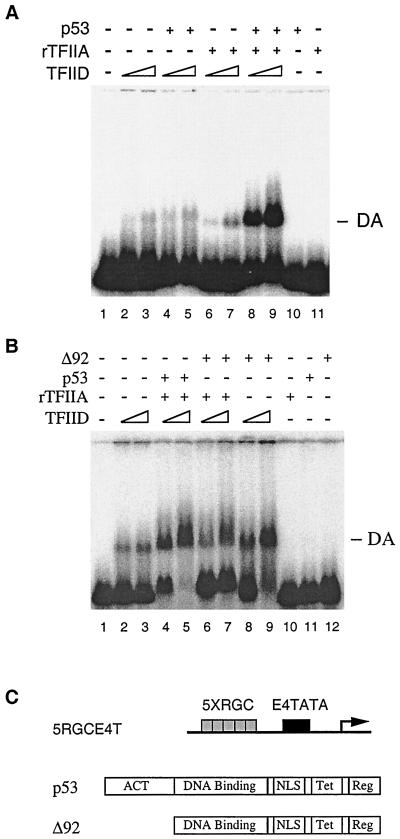
To better understand how T antigen inhibits p53-mediated transcription once brought to the promoter, we studied the transcription function of p53 at the molecular level. As several sequence-specific DNA binding transcription factors have been shown to function by stimulating TFIID-TFIIA-promoter complex (the DA complex) formation, we initially used the in vitro Mg-agarose gel shift assay (Mg-EMSA) to determine whether this is also true for p53. When a DNA fragment bearing five RGC p53 binding sites upstream of a high-affinity TATA box (Fig. 1C) was incubated with increased amounts of affinity-purified TFIID complex, a retardation of only a small fraction of the probe in a complex with TFIID was observed (Fig. 1A, lanes 2 and 3). Addition of purified p53 and TFIIA, however, resulted in a marked stimulation in the shifted TFIID-DNA complex (lanes 8 and 9). Under these conditions, neither p53 (lanes 4 and 5) nor TFIIA alone (lanes 6 and 7) significantly stimulated the TFIID-DNA complex assembly. Using identical conditions to those employed with wild-type p53, we have shown that a transcription-inactive mutant of p53, ΔN92 (18 and Fig. 1C), resulted in reduced DA complex formation (Fig. 1B, lanes 6 and 7). These results suggest that p53 activates transcription at least in part by stimulating DA complex formation. Of note, the addition of ΔN92 alone appeared to stimulate TFIID-DNA complex assembly in the absence of TFIIA in Mg-EMSA (Fig. 1B, lanes 8 and 9). The significance of this observation is presently unknown.
FIG. 1.
p53 stimulates TFIID-TFIIA-promoter complex assembly. (A) Mg-EMSA of p53-induced DA complex formation. Purified TFIID complex, recombinant TFIIA (rTFIIA), and p53 (as indicated above each lane) were incubated with the 5RGCE4T probe for 30 min at 30°C. The amount of each factor added to the Mg-EMSA reactions corresponds to 0.5 or 1 μl of TFIID, 20 ng of rTFIIA, and 100 ng of p53. The specific position of the DA complex is indicated on the right. (B) ΔN92 (Δ92) fails to induce DA complex formation. The amount of each factor added to the Mg-EMSA reactions corresponds to 0.5 or 1 μl of TFIID, 20 ng of rTFIIA, 100 ng of p53, or 100 ng of ΔN92. The position of the DA complex is indicated on the right. (C) Schematic diagram of the 5RGCE4T template (transcription initiation site designated by the right-angle arrow), p53, and ΔN92 used Act, activation domain; NLS, nuclear localization signal; Tet, tetramerization; Reg, regulatory domain.
The conformational change of the DA complex is necessary for p53 transactivation.
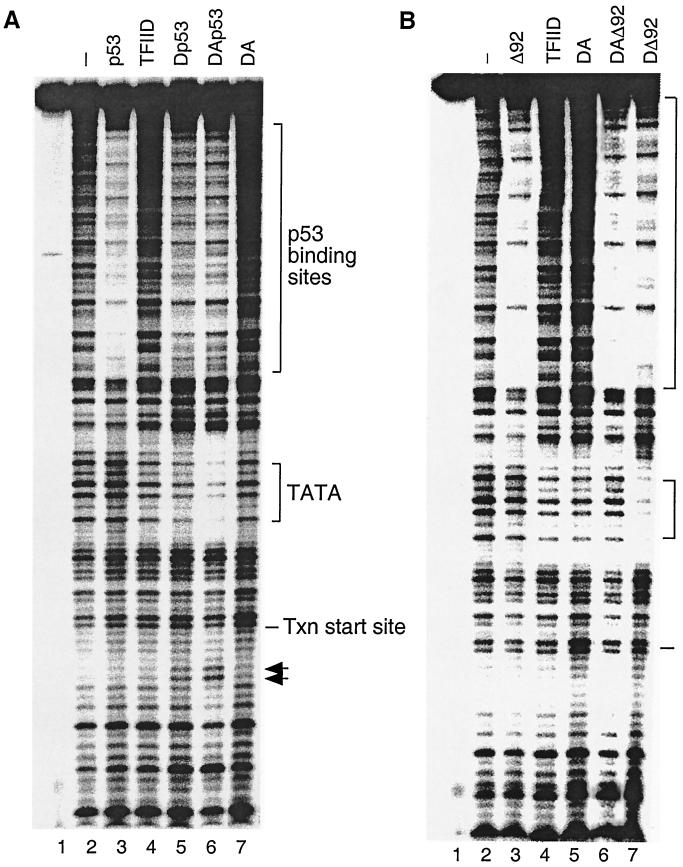
To further study the stimulation of DA complex formation by p53, in vitro DNase I footprinting was carried out using the same promoter (Fig. 2A). In the absence of p53, the purified TFIID complex resulted in a weak protection of the TATA box region as expected (Fig. 2A, lane 4). Addition of purified p53 alone slightly enhanced protection of the TATA box (lane 5). A less complete protection of the p53 binding sites, however, was observed in lane 5. This may be due to a cooperative binding between p53 and TFIID. When all three proteins were present simultaneously, a significant protection of the TATA box was observed (lane 6). In addition to the alteration of the protection of the TATA box, at least two hypersensitive sites downstream of the transcription initiation site were generated (lane 6). The appearance of the hypersensitive cleavage sites indicates that p53 induced a conformational change over the promoter region, which may lead to enhanced assembly of the rest of the transcription machinery. Taken together, these results suggest that p53 may result in a more stable and active TFIIA-TFIID-DNA complex.
FIG. 2.
The conformational change of the DA complex is necessary for p53 transactivation. (A) p53, TFIIA, and TFIID cooperate to form a stable TFIID footprint on the promoter. In vitro DNase I footprint reactions were carried out as described in Materials and Methods. The amount of each factor added to the DNase I footprint reactions (as indicated above each lane) corresponds to 5 μl of TFIID, 100 ng of recombinant TFIIA, and 500 ng of p53. The positions of the p53 binding sites, the TATA box, and the transcription (Txn) start site are indicated on the right. Arrows denote hypersensitive sites. (B) ΔN92 (Δ92) fails to induce the conformational change of the DA complex on the promoter. The amount of each factor added to the DNase I footprint reactions (as indicated above each lane) corresponds to 5 μl of TFIID, 100 ng of recombinant TFIIA, 500 ng of p53, or 500 ng of ΔN92. The positions of the p53 binding sites, the TATA box, and the transcription start site are indicated on the right.
Compared to p53, ΔN92 was impaired in its ability to stimulate DA complex formation as expected (Fig. 2B, lane 6). However, in agreement with Mg-EMSA results (Fig. 1B, lanes 8 and 9), the addition of ΔN92 in the absence of TFIIA stimulated TFIID-DNA complex assembly (Fig. 2B, lane 7). Despite the enhanced TFIID-DNA complex assembly, no hypersensitive cleavage sites were observed in the downstream region in DNase I footprinting. Thus, a combination of recruitment and conformational change of the DA complex appears to be necessary for p53 transactivation.
p53-T complex is incapable of stimulating DA complex assembly.
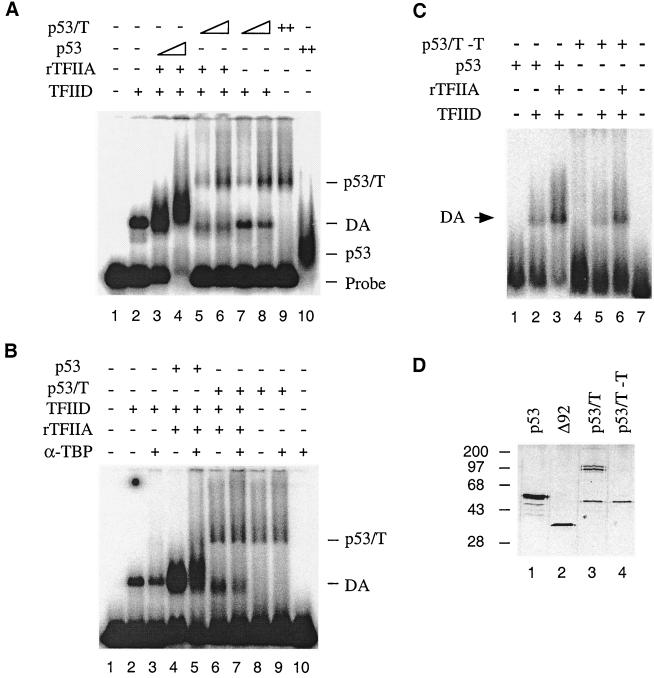
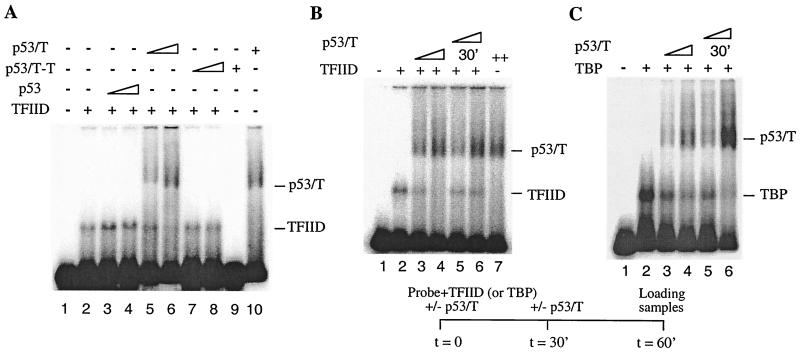
Having established that p53 induces DA complex formation, we next tested if T antigen affects this function of p53. Because p53-T complex purified from scid cells has been demonstrated previously to bind to DNA as a transcriptionally inactive complex (28), the effect of T antigen on the ability of p53 to stimulate DA complex formation was tested using this complex. When the p53-T complex was incubated with the same DNA template as that used with the wild-type p53 assays, it produced a slowly migrating p53-T–DNA complex (Fig. 3A, lane 9). This result suggested that the p53-T complex could bind to the 5RGCE4T probe in the Mg-EMSA assay. However, by comparison to p53 (lanes 3 and 4) shows that p53-T complex failed to stimulate DA complex assembly (lanes 5 and 6) at the expected shift position. The amounts of p53-T complex have been tested over a range of concentrations corresponding from one half to eight times the amount of p53 used in lane 3 of Fig. 3A. At each concentration tested, no stimulation of DA complex assembly was observed (data not shown). These results indicated that T antigen affected the ability of p53 to stimulate DA complex formation.
FIG. 3.
p53-T complex (p53/T) is incapable of stimulating DA complex assembly. (A) p53-T complex fails to induce DA complex formation. Mg-EMSA reactions were carried out as described in Materials and Methods. The amount of each factor added to the Mg-EMSA reactions corresponds to 1 μl of TFIID, 20 ng of recombinant TFIIA (rTFIIA), 50 or 100 ng of p53, and 3 or 6 μl of p53-T complex (containing 100 or 200 ng of p53). The positions of the DA complex, p53-T–DNA, and p53-DNA are indicated on the right. (B) Experiments were performed as for panel A but in the presence of 100 ng of anti-TBP antibody (α-TBP, N-12). (C) p53 purified from mouse cells is capable of stimulating DA complex assembly. Mg-EMSA reactions were carried out as described. The amount of each factor added to the Mg-EMSA reactions (as indicated above each lane) corresponds to 1 μl of TFIID, 20 ng of rTFIIA, 100 ng of p53, and 100 ng of p53-T without T antigen (p53/T −T). The position of the DA complex is indicated on the left. (D) A silver-stained SDS-PAGE gel is shown. Lane 1, 200 ng of HA-tagged human p53; lane 2, 100 ng of HA-tagged ΔN92; lane 3, 1 μl of purified p53-T antigen complex corresponding to 30 ng of p53; lane 4, 1 μl of purified p53-T complex without T antigen corresponding to 30 ng of p53. The sizes (in kilodaltons) of molecular mass standards are indicated on the left.
We considered the possibility that the shifted DA complex might exist but that it migrated at a different position. This notion, however, cannot be tested using DNase I footprinting assays because of technical difficulties in obtaining concentrated p53-T and TFIID complexes. To exclude the possibility that the shifted DA complex might migrate at the same positions as the p53-T–DNA complex, we carried out Mg-EMSA in the presence of an anti-TBP antibody, N-12. While incubation of anti-TBP antibody resulted in a partial inhibition of TFIID binding to DNA either alone (Fig. 3B, lane 3) or in the presence of p53 and TFIIA (Fig. 3B, lane 5), there was no alteration of the gel shift pattern at the p53-T–DNA complex position. These results support the view that T antigen, when in a complex with p53 and DNA, affects the ability of p53 to stimulate DA complex formation.
To determine if these results were specific for the mouse p53 protein, a further Mg-EMSA was performed with mouse p53 purified from scid cells without T antigen (p53-T–T [see Materials and Methods for purification]) (Fig. 3D). Results are shown in Fig. 3C. The addition of p53-T–T resulted in a generation of a shifted complex (lane 4) at a position similar to that of the human p53-DNA complex. Importantly, the addition of p53-T–T also resulted in a stimulation of DA complex formation (lane 6), similar to that of human p53 (lane 3). Notably, we consistently observed that mouse p53 was less efficient than human p53 in promoting DA complex formation under these conditions (cf. lanes 3 and 6). This may be due to a number of reasons. For instance, human p53 may interact with human transcription factors better. Alternatively, mouse p53 may partially lose its activity during purification. The fact that the purified mouse p53 protein without T antigen is less active in an in vitro transcription assay (28) supports this hypothesis. Nevertheless, the mouse p53 protein is capable of stimulating the DA complex, and therefore it can be concluded that it is the associated T antigen that inhibits DA complex assembly.
T antigen targets TFIID to inhibit DA complex assembly.
On the basis of our results, it seemed probable that TFIIA and/or TFIID were likely targets with which p53 interacted to promote DA complex assembly. Therefore, to characterize the components required for T-antigen inhibition, we performed protein-protein interaction experiments to test if T antigen can affect the interaction of p53 with TFIID or TFIIA.
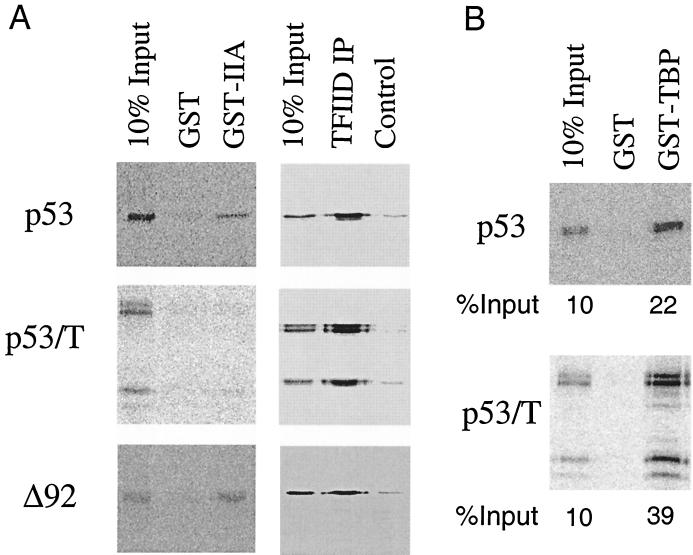
As several activators, such as VP16 and Zta, have been shown to stimulate DA complex formation through direct interaction with TFIIA (6, 14), we first determined if this was so for p53. GST-TFIIA binding assays were carried out with 35S-labeled p53, and an interaction of p53 with TFIIA was observed (Fig. 4, upper left panel). Next, we asked if the p53-T complex and ΔN92 that severely reduced DA complex assembly would alter their ability to interact with TFIIA. Our results show that under identical conditions, although p53-T complex greatly reduced its interaction with TFIIA, ΔN92 binds efficiently to TFIIA (bottom left panel). Therefore, interaction with TFIIA may be required, but is not sufficient, for the inhibition of DA complex assembly by p53-T antigen complex. Of note, p53-T antigen complex in the absence of T antigen (as shown in Fig. 3D) restores the ability of p53 to bind to TFIIA (data not shown).
FIG. 4.
Interaction with TFIIA is not sufficient for the inhibition of DA complex assembly. (A) p53 and ΔN92 (Δ92) were translated in vitro with [35S]methionine, and p53-T complex (p53/T) was radiolabeled in vivo in the presence of [35S]methionine and purified with Pab 421 antibody as described in Materials and Methods. The labeled proteins were either incubated with GST or GST-TFIIA immobilized on glutathione Sepharose beads (left panel) or with TFIID which was immobilized on protein A-Sepharose beads using TAFII250 antibody (6B3, right panel, TFIID IP). After being washed, bound proteins were analyzed by SDS-PAGE followed by autoradiography of the proteins retained on the beads. (B) Experiments were performed as for panel A but using GST-TBP. The relative amount of bound protein was quantitated using a PhosphorImager and ImageQuant software and is presented as a percentage of the total radiolabeled protein.
In earlier work, a direct interaction between p53 and the TFIID complex was found (19). We next addressed the possibility that p53 was able to stimulate DA complex formation through direct interaction with TFIID. We therefore compared the interaction of p53, ΔN92, and p53-T complex with TFIID complex by immunoprecipitation assays with an anti-TAFII250 antibody, 6B3. Results of representative immunoprecipitation assays are shown in Fig. 4A, right panels. As expected, p53 interacted with TFIID. Mutant ΔN92 and p53-T complex, however, appeared to interact with TFIID indistinguishably from p53. These results suggest that interaction with TFIID may not be sufficient to promote DA complex assembly. Previously, we also found a direct interaction between p53 and TBP (19). To test the possibility that p53-T complex may affect the interaction with TBP, we performed GST-TBP pull-down experiments and showed that p53-T complex interacts with TBP to an extent at least similar to that of p53 (Fig. 4B).
Although similar binding between TFIID and p53, ΔN92, or p53-T complex was observed, we considered the possibility that the interaction between p53-T complex and TFIID may affect the ability of TFIID to bind DNA. To test this, we performed the Mg-EMSA (Fig. 5A), which showed that when increased amounts of p53-T complex were included in the reactions, the TFIID DNA binding was reduced (lanes 5 and 6). However, under identical conditions the addition of p53 (lanes 3 and 4) and p53-T–T (lanes 7 and 8) did not affect the ability of TFIID to bind DNA. To further confirm these results, we carried out order-of-addition experiments in which p53-T complex was added to the Mg-EMSA either during or after the assembly of the TFIID-DNA complex (Fig. 5B). Our results showed that TFIID binding became partially resistant to inhibition by p53-T complex if the DNA template was first incubated with TFIID followed by addition of p53-T complex (lanes 5 and 6). In contrast, when the p53-T complex was introduced during the assembly of the TFIID-DNA complex, inhibition of binding was observed (lanes 3 and 4). Collectively, these data demonstrate that preincubation of TFIID and the DNA template prevents p53-T complex repression, thereby suggesting that T antigen, once brought to the promoter through interaction with p53, inhibits DA complex assembly via TFIID.
FIG. 5.
T antigen targets TFIID to inhibit DA complex assembly. (A) p53-T complex (p53/T) prevents TFIID from binding to the promoter. Mg-EMSA reactions were carried out with TFIID complex, p53, and p53-T complex (as indicated above each lane) using the 5RGCE4T probe. (B) A preassembled complex containing DNA and TFIID is partially resistant to p53-T complex inhibition. TFIID was incubated with the probe for 30 min at 30°C in either the presence or absence of increasing amounts of p53-T complex. p53-T complex was then added to the reaction mixtures in lanes 5 and 6 and was incubated for an additional 30 min prior to the Mg-EMSA. (C) A preassembled complex containing DNA and TBP is not resistant to p53-T complex inhibition. Experiments were performed as for panel A but with 10 ng of TBP instead of TFIID. All preincubation Mg-EMSA reactions were carried out for 60 min.
T antigen prevents TBP from binding to the TATA box.
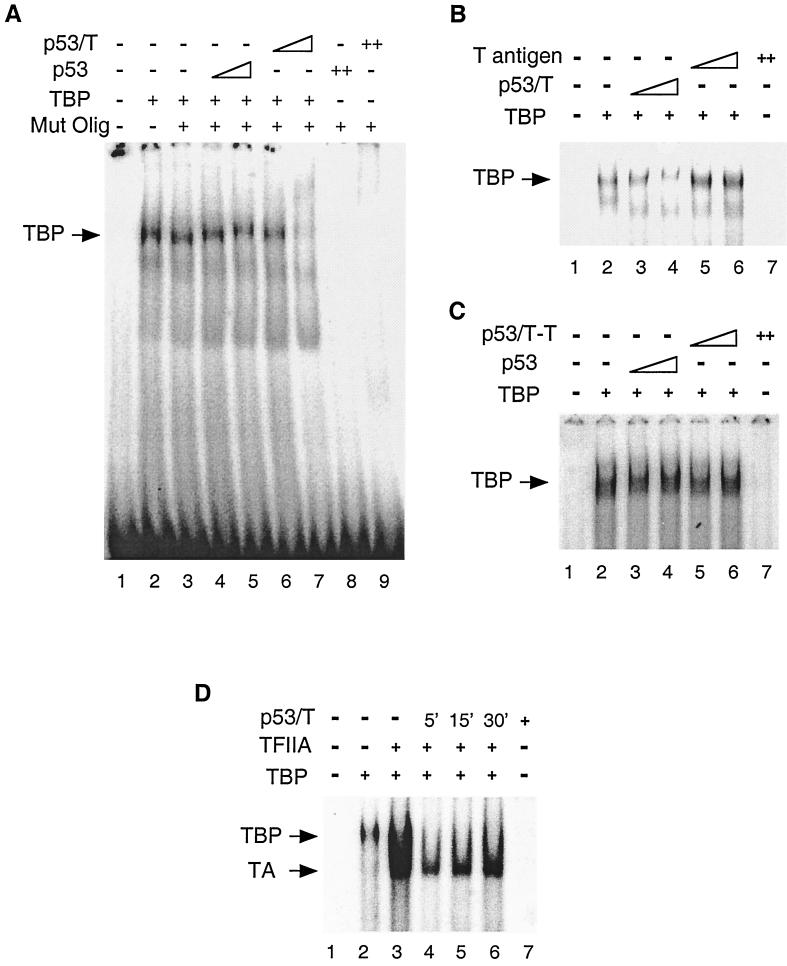
To extend the above studies regarding the effect of T antigen on the assembly of TFIID on a promoter, we next wanted to determine whether T antigen functions via TBP in the TFIID complex, as TBP makes direct contact with the DNA template. To address this, TFIID was replaced by bacterially expressed and purified TBP (Fig. 5C). TBP bound to the template and produced a retarded TBP-DNA complex at a position similar to that of TFIID-DNA in the Mg-EMSA (lane 2). When p53-T complex was included in the reactions, the ability of TBP to bind DNA was repressed (lanes 3 and 4). An equivalent amount of p53, however, had no effect on TBP binding (data not shown). This result argues strongly that p53-T complex targets TBP to inhibit DA complex formation, although it does not rule out the possibility that p53-T complex might make additional contacts with one or more TAFs. In contrast to TFIID, however, TBP DNA binding was also repressed by the later addition of p53-T complex (lanes 5 and 6), suggesting that preincubation of TBP and the DNA template is unable to prevent p53-T complex repression. Thus, our data provide evidence that TAFs function to stabilize TBP binding, and their presence reduces the inhibition of TBP binding by T antigen.
The data described above suggest a model for the inhibition of DA complex formation by p53-T complex in which p53-T complex contacts TBP and inhibits TBP binding to DNA. If this model is correct, addition of p53-T complex should also inhibit TBP in a probe lacking p53 binding sites. To test this possibility, we examined the effect of p53-T complex on TBP DNA binding with a probe containing the E1B TATA element. As expected, TBP bound to the E1B probe and produced a retarded TBP-DNA complex (Fig. 6A, lane 2). Addition of a 100-fold excess of cold wild-type, but not mutant (lane 3), E1B probe was sufficient to inhibit its formation (data not shown). This TBP-specific DNA binding was largely reduced when p53-T complex was included in the reaction mixture (lanes 6 and 7). Addition of an equivalent amount of p53 or p53-T–T (Fig. 6A, lanes 4 and 5, and 6C, lanes 3 to 6), however, had no effect on TBP DNA binding. These results suggest that p53-T complex inhibits the ability of TBP to bind to the TATA element.
FIG. 6.
T antigen prevents TBP from binding to the TATA box. (A) EMSA reactions were carried out with 10 ng of TBP, 10 or 20 ng of p53, and 0.3 or 0.6 μl of p53-T complex corresponding to 10 or 20 ng of p53 for 30 min at 30°C. The reaction mixtures were analyzed on a 3% polyacrylamide gel. The position of the TBP-DNA complex is indicated on the left. Mut Olig, E1B TATA-oligo probe. (B) T antigen alone has no effect on TBP binding. Experiments were performed as for panel A but with 10 or 20 ng of T antigen. (C) Mouse p53 alone has no effect on TBP binding. Experiments were performed as for panel A but with 10 or 20 ng of mouse p53 (p53/T−T). (D) A preassembled complex containing DNA, TFIIA, and TBP is partially resistant to p53-T complex inhibition. TBP, TFIIA, and the E1B probe were preincubated for 5, 15, or 30 min before the addition of p53-T complex. The time at which p53-T complex (p53/T) was added is indicated above. All preincubation EMSA reactions were carried out for 60 min. The arrows on the left indicate the positions of the TBP-DNA and TBP-IIA-DNA complexes.
The observation that p53-T complex inhibits TBP DNA binding, thereby affecting the ability of p53 to stimulate the DA complex, raised the question of whether this inhibition relies on p53-T configuration. T antigen has been previously demonstrated to stabilize TBP-TFIIA complex formation on the TATA element (10). We therefore determined if T antigen could affect TBP DNA binding under our assay conditions. Our results showed that in contrast to p53-T complex (Fig. 6B, lanes 3 and 4), T antigen alone was unable to prevent TBP from binding to the TATA element (lanes 5 and 6). Taken together, our data suggest that p53-T complex prevents TBP from binding to the TATA element and thus inhibits p53-mediated transcription.
DISCUSSION
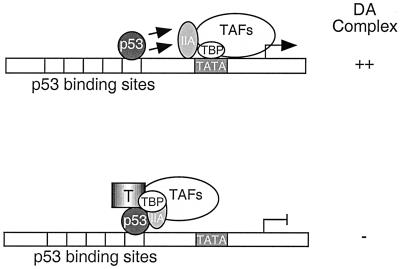
Although SV40 T antigen from mouse cells is known to form a transcription-inactive complex with p53 on p53-responsive promoters (28), the molecular mechanisms by which this occurs remained elusive. In this work, we present biochemical evidence that p53 stimulates transcription, at least in part, by enhancing assembly of the DA complex on the promoter and by inducing a conformational change. Importantly, we have shown that T antigen, once tethered to the promoter through interaction with p53, represses transcription by inhibiting DA complex assembly. Furthermore, the repression of DA complex formation by T antigen was largely reduced when the TFIID-DNA complex was formed prior to addition of T antigen. These results indicate that the T antigen targets TFIID to inhibit transcription. In support of this theory, we have found that the p53-T complex is capable of preventing TBP from binding to the TATA element. A model summarizing these results is shown in Fig. 7. On the basis of these results, we propose that p53-T complex inhibits DA complex formation by disrupting the activity of the general transcription factor TBP.
FIG. 7.
A model for how p53-T complex inhibits p53-mediated transcription. (Top) p53 stimulates the TFIID-TFIIA-promoter complex formation. (Bottom) T antigen represses transcription when tethered to the promoter by p53. This could occur through a conformational change of TBP within the TFIID-TFIIA-promoter complex or could be due to the exclusion of the ability of TFIIA to function as an antirepressor.
Several activators have been found to stimulate the formation of the DA complex through a direct interaction with the general transcription factor TFIIA (13, 14). Our results suggested that p53, like other transcription factors, interacted with TFIIA. However, we have observed that a deletion mutant of p53 lacking the activation domain, ΔN92, which did not stimulate DA complex assembly, bound to TFIIA to an extent similar to that of p53 (Fig. 4). Furthermore, we failed to detect a correlation between the ability of p53 to interact with TFIID and its ability to promote DA complex assembly (Fig. 4). These results indicated that interaction with TFIIA or TFIID may be necessary but is not sufficient for the stimulation of DA complex formation. This view is supported by the demonstration that p53, but not ΔN92, induced a conformational change in the DA complex, as indicated by the formation of hypersensitive cleavage sites detected in DNase I footprinting assays. These data are consistent with the idea that the formation of the isomerized DA complex is necessary and sufficient for gene activation (5, 11). Thus, the conformational change and/or isomerization of the DA complex mediated by p53 is probably required for efficient assembly of a functional transcription complex.
We have shown that the p53-T complex inhibits the TBP-TATA interaction in the absence of p53 binding sites. Thus, the p53-T complex inhibits DA complex formation probably by disrupting the activity of the general transcription factor TBP. In this regard, T antigen, once tethered to the p53-responsive promoters, appears to function in a manner similar to that of the Drosophila melanogaster transcriptional repressor, Eve (16). Eve represses transcription by directly binding TBP in such a way that it blocks the interaction between TBP and DNA. However, Eve seems to repress transcription by competing with the TATA box for TBP binding. p53-T complex is distinct from Eve in that it does not specifically bind to the TATA element (Fig. 6). It may be similar in this regard to SAGA subunits, Spt3 and Spt8, which can downregulate TBP function through interaction with TBP (2). Alternatively, TBPs have been reported to exist as dimers, which must dissociate to bind to DNA (6–8). Perhaps p53-T complex prevents this required dissociation, thereby inhibiting stable association of TBP with the promoter. Indeed, we have observed that TBP binds to p53-T complex more stably than transcription activator p53 (Fig. 4B). Although the relevance of the interaction in regulating TBP dimers remains uncertain, the interaction of p53-T complex with TBP appears to play a role in the inhibition of TBP binding. It is possible that p53-T complex, like several other characterized repressors, may function by more than one mechanism (reviewed in reference 12). Nevertheless, the efficiency of TBP-mediated repression suggests that this is an important aspect of p53-T complex activity. Of note, despite the ability of p53-T complex to inhibit TBP on promoters without p53 binding sites, specific inhibition of transcription is likely to occur on the promoters containing p53 binding sites in cells.
SV40 T antigen has been previously shown to stabilize the TBP-TFIIA-promoter complex and to activate transcription (10). Thus, T antigen appears to play both negative and positive roles in transcription regulation. When bound to p53 and tethered to p53-responsive promoters, T antigen inhibits TBP-DNA interaction and represses transcription from p53-activated genes. When not bound to p53, however, T antigen may stabilize the TBP-TFIIA-promoter complex and activate transcription. Interestingly, T antigen has been reported to function like TAFII250 (9), which prevents TBP from binding to the TATA element (25). The question that arises is whether TFIIA can prevent p53-T complex inhibition of TBP-DNA binding, since TFIIA has been shown to counteract a number of TBP inhibitors (reviewed in reference 23), including TAFII250 (25). We therefore tested T-antigen inhibition in DNA binding reactions with or without TFIIA (Fig. 6D). Our results reveal that simultaneous addition of p53-T complex and TFIIA to TBP resulted in a reduction of DNA binding (Fig. 6D, lane 4). However, after 30 min of preincubation of TFIIA, TBP, and DNA, a stable complex formed that was partially resistant to p53-T inhibition (lane 6). Thus, TFIIA appears to compete with p53-T complex for interaction with TBP, resulting in a stable association of TBP with the promoter.
SV40 is known to cause tumors in rodents (3, 4) but has not been proven to do so in humans. In a recent study, we and others demonstrated that T antigen present in transformed mouse cells, but not in human cells, forms a DNA binding complex with p53 that is transcriptionally inactive (28). In the present study, we have defined a detailed mechanism of transcription repression of p53 by T antigen in mouse cells. These studies have extended our understanding of T-antigen inhibition of transcription from p53-activated genes. A number of cellular proteins have also been shown to bind to p53 and negatively affect its transcriptional activity. It will be interesting to determine whether the mechanism of T-antigen inhibition could resemble that of other cellular p53-interacting proteins.
ACKNOWLEDGMENTS
We are very grateful to A. Berk, N. Kobayashi (University of California, Los Angeles), P. Lieberman (Wistar), J. Ross, and B. Dynlacht (Harvard) for many helpful discussions. We thank A. Berk and N. Kobayashi for providing TFIIA expression vectors and anti-TFIIA antibody, L. Anton for LTRα3 nuclear extracts, and E. Martinez for valuable comments on the manuscript.
This work was supported by grants from the National Cancer Institute (CA75180) and the U.S. Army Breast Cancer Research Program (DAMD17-96-6076) to X.L. S.I.C is supported by a postdoctoral fellowship from the California Cancer Research Program.
REFERENCES
- 1.Bargonetti J, Reynisdottir I, Friedman P, Prives C. Site-specific binding of wild-type p53 to cellular DNA is inhibited by SV40 T antigen and mutant p53. Genes Dev. 1992;6:1886–1898. doi: 10.1101/gad.6.10.1886. [DOI] [PubMed] [Google Scholar]
- 2.Belotserkovskaya R, Sterner D E, Deng M, Sayre M H, Lieberman P M, Berger S L. Inhibition of TATA-binding protein function by SAGA subunits Spt3 and Spt8 at Gcn4-activated promoters. Mol Cell Biol. 2000;20:634–647. doi: 10.1128/mcb.20.2.634-647.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carbone M, Rizzo P, Pass H I. Simian virus 40, poliovaccines and human tumors: a review of recent developments. Oncogene. 1997;15:1877–1888. doi: 10.1038/sj.onc.1201375. [DOI] [PubMed] [Google Scholar]
- 4.Carbone M, Rizzo P, Grimley P M, Procopio A, Mew D J Y, Shridhar V, de Bartolomeis A, Esposito V, Giuliano M T, Steinberg S M, Levine A, Giordano A, Pass H I. Simian virus-40 large T-antigen binds p53 in human mesotheliomas. Nat Med. 1997;3:908–912. doi: 10.1038/nm0897-908. [DOI] [PubMed] [Google Scholar]
- 5.Chi T, Lieberman P, Ellwood K, Carey M. A general mechanism for transcription synergy by eukaryotic activators. Nature. 1995;377:254–257. doi: 10.1038/377254a0. [DOI] [PubMed] [Google Scholar]
- 6.Chi T, Carey M. Assembly of the isomerized TFIIA-TFIID-TATA ternary complex is necessary and sufficient for gene activation. Genes Dev. 1996;10:2540–2550. doi: 10.1101/gad.10.20.2540. [DOI] [PubMed] [Google Scholar]
- 7.Coleman R A, Taggart A K, Benjamin S L R, Pugh B F. Dimerization of the TATA binding protein. J Biol Chem. 1995;270:13842–13849. doi: 10.1074/jbc.270.23.13842. [DOI] [PubMed] [Google Scholar]
- 8.Coleman R A, Taggart A K P, Burma S, Chicca II J J, Pugh B F. TFIIA regulates TBP and TFIID dimers. Mol Cell. 1999;4:451–457. doi: 10.1016/s1097-2765(00)80453-0. [DOI] [PubMed] [Google Scholar]
- 9.Damania B, Alwine J C. TAF-like function of SV40 large T antigen. Genes Dev. 1996;10:1369–1381. doi: 10.1101/gad.10.11.1369. [DOI] [PubMed] [Google Scholar]
- 10.Damania B, Lieberman P L, Alwine J C. Simian virus 40 large T antigen stabilizes the TATA-binding protein-TFIIA complex on the TATA element. Mol Cell Biol. 1998;18:3926–3935. doi: 10.1128/mcb.18.7.3926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Emami K H, Jain A, Smale S T. Mechanism of synergy between TATA and initiator: synergistic binding of TFIID following a putative TFIIA-induced isomerization. Genes Dev. 1997;11:3007–3019. doi: 10.1101/gad.11.22.3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hampsey M. Molecular genetics of the RNA polymerase II general transcriptional machinery. Microbiol Mol Biol Rev. 1998;62:465–503. doi: 10.1128/mmbr.62.2.465-503.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kobayashi N, Boyer T G, Berk A J. A class of activation domains interacts directly with TFIIA and stimulates TFIIA-TFIID-promoter complex assembly. Mol Cell Biol. 1995;15:6465–6473. doi: 10.1128/mcb.15.11.6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kobayashi N, Horn P J, Sullivan S M, Trizenberg S J, Boyer T G, Berk A J. DA-complex assembly activity required for VP16C transcriptional activation. Mol Cell Biol. 1998;18:4023–4031. doi: 10.1128/mcb.18.7.4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Levine A J. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 16.Li C, Manley J L. Even-skipped repressed transcription by binding TATA binding protein and blocking the TFIID-TATA box interaction. Mol Cell Biol. 1998;18:3771–3781. doi: 10.1128/mcb.18.7.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lieberman P M, Berk A J. A mechanism for TAFs in transcriptional activation: activation domain enhancement of TFIID-TFIIA-promoter DNA complex formation. Genes Dev. 1994;8:995–1006. doi: 10.1101/gad.8.9.995. [DOI] [PubMed] [Google Scholar]
- 18.Liu X, Berk A J. Reversal of in vitro p53 squelching by both TFIIB and TFIID. Mol Cell Biol. 1995;15:6474–6478. doi: 10.1128/mcb.15.11.6474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu X, Miller C W, Koeffler H P, Berk A J. The p53 activation domain binds the TATA box-binding polypeptide in holo-TFIID, and a neighboring p53 domain inhibits transcription. Mol Cell Biol. 1993;13:3291–3300. doi: 10.1128/mcb.13.6.3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Y, Colosimo A L, Yang X-J, Liao D. Adenovirus E1B 55-kilodalton oncoprotein inhibits p53 acetylation by PCAF. Mol Cell Biol. 2000;20:5540–5553. doi: 10.1128/mcb.20.15.5540-5553.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin M E D, Berk A J. Corepressor required for adenovirus E1B 55,000-molecular weight protein repression of basal transcription. Mol Cell Biol. 1999;19:3403–3414. doi: 10.1128/mcb.19.5.3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oliner J D, Pietenpol J A, Thiagalingam S, Gyuris J, Kinzler K W, Vogelstein B. Oncoprotein MDM2 conceals the activation domain of tumour suppressor p53. Nature. 1993;362:857–860. doi: 10.1038/362857a0. [DOI] [PubMed] [Google Scholar]
- 23.Orphanides G, Lagrange T, Reinberg D. The general transcription factors of RNA polymerase II. Genes Dev. 1996;10:2657–2683. doi: 10.1101/gad.10.21.2657. [DOI] [PubMed] [Google Scholar]
- 24.Ozer J, Moore P A, Bolde A H, Lee A, Rosen C A, Lieberman P M. Molecular cloning of the small (γ) subunit of human TFIIA reveals functions critical for activated transcription. Genes Dev. 1994;8:2324–2335. doi: 10.1101/gad.8.19.2324. [DOI] [PubMed] [Google Scholar]
- 25.Ozer J, Mitsouras K, Zerby D, Carey M, Lierberman P M. Transcription factor IIA derepresses TATA-binding protein (TBP)-associated factor inhibition of TBP-DNA binding. J Biol Chem. 1998;273:14293–14300. doi: 10.1074/jbc.273.23.14293. [DOI] [PubMed] [Google Scholar]
- 26.Pietenpol J A, Tokino T, Thiagalingam S, El-Deiry W S, Kinzler K W, Vogelstein B S. Sequence-specific transcriptional activation is essential for growth suppression by p53. Proc Natl Acad Sci USA. 1994;91:1998–2002. doi: 10.1073/pnas.91.6.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prives C. Signaling to p53: breaking the Mdm2–p53 circuit. Cell. 1998;95:5–8. doi: 10.1016/s0092-8674(00)81774-2. [DOI] [PubMed] [Google Scholar]
- 28.Sheppard H M, Corneillie S J, Espiritu C, Gatti A, Liu X. New insights into the mechanism of inhibition of p53 by simian virus 40 large T antigen. Mol Cell Biol. 1999;19:2746–2753. doi: 10.1128/mcb.19.4.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thut C J, Goodrich J A, Tijan R. Repression of p53-mediated transcription by MDM2: a dual mechanism. Genes Dev. 1997;11:1974–1986. doi: 10.1101/gad.11.15.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang W, Gralla J D, Carey M. The acidic activator GAL4-AH can stimulate polymerase II transcription by promoting assembly of a closed complex requiring TFIID and TFIIA. Genes Dev. 1992;6:1761–1772. doi: 10.1101/gad.6.9.1716. [DOI] [PubMed] [Google Scholar]
- 31.Yew P R, Liu X, Berk A J. Adenovirus E1B oncoprotein tethers a transcriptional repression domain to p53. Genes Dev. 1994;8:190–202. doi: 10.1101/gad.8.2.190. [DOI] [PubMed] [Google Scholar]