Abstract
Human immunodeficiency virus type 1 (HIV-1) RNA levels in female genital tract and peripheral blood samples were compared using two commercial amplification technologies: the Roche AMPLICOR HIV-1 MONITOR test and either the Organon Teknika nucleic acid sequence-based amplification (NASBA-QT) assay or the NucliSens assay. Estimates of HIV-1 RNA copy number were derived from internal kit standards and analyzed unadjusted and adjusted to a common set of external standards. We found a discordance rate of approximately 18% between the two technologies for the detection of HIV-1 in either the genital tract or peripheral blood samples. Detection discordance was not consistent among specimens or among women. There were no significant differences in adjusted or unadjusted estimates of HIV-1 RNA copy number in the genital tract samples using the AMPLICOR HIV-1 MONITOR test and either the NASBA-QT assay or the NucliSens assay. In addition, the estimated HIV-1 RNA copy number in peripheral blood samples did not differ when tested with the NucliSens assay and the AMPLICOR HIV-1 MONITOR test using kit standards. However, there was a significant difference in estimated RNA copy number between the NASBA-QT assay and the AMPLICOR HIV-1 MONITOR test for internal kit standards, which, as we have previously shown, was eliminated after adjustment with the external standards. Our results suggest that the Roche and Organon Teknika assays are equivalent for quantifying HIV-1 RNA in female genital tract specimens, although variation in detection does exist.
Monitoring human immunodeficiency virus type 1 (HIV-1) RNA in genital tract specimens has become of primary importance with our growing understanding of the issues surrounding compartmentalization (4, 11). Measuring HIV-1 RNA has been more complicated in genital tract than in peripheral blood samples. In semen, nonspecific inhibitors have been associated with loss of signal (4, 5, 7). However, in reconstruction experiments with seronegative subjects, no equivalent nonspecific inhibitors were observed for the female genital tract (8). That study did not rule out the presence of inhibitors in the genital tracts of HIV-seropositive women that may be lacking in HIV-seronegative women.
Genital tract variation has been shown to be greater in women (P. S. Reichelderfer, R. W. Coombs, D. Wright, D. Burns, and A. Kovacs for the WHS 001 Study Group, Abstr. 38th Intersci. Conf. Antimicrob. Agents Chemother., abstr. I-251, 1998) than in men (4). Total variation is a composite of biologic, assay, and sampling variation. Sampling methods used for the female genital tract contribute strongly to variation (Reichelderfer et al., 38th ICAAC). Interlaboratory and interassay variation in patient or spiked samples of peripheral blood could be significantly reduced by using a common set of standards (3, 14). However, variation may be increased in different matrices, and the findings in peripheral blood samples may therefore differ from those in genital tract samples. There has been no direct comparison of differences in HIV-1 RNA levels in the female genital tract among test kits that use the same sampling method. The objective of this study was to assess the differences in estimated HIV-1 RNA levels obtained with two commonly used gene amplification technologies in genital tract samples collected using the same sampling method.
MATERIALS AND METHODS
Study population.
This cross-sectional study involved 338 HIV-1-seropositive women enrolled in a longitudinal epidemiologic cohort study, the Women's Interagency HIV Study (1). Genital tract specimens and peripheral blood were obtained during one regularly scheduled 6-month visit between January 1997 and July 1998. The study was limited to women who were not pregnant, who either had not received therapy or had been on stable antiretroviral therapy for at least 2 months, and who were not experiencing an active opportunistic infection. Approximately 40% of the 338 women were on no therapy, while 60% were receiving combination therapy with or without a protease inhibitor.
Specimen collection.
Peripheral blood plasma specimens were collected in acid dextrose citrate tubes, frozen at −70°C, and shipped to a central repository. Female genital tract specimens were obtained by cervical vaginal lavage (CVL) with 10 ml of normal saline. Aliquots (1 ml) of lavage material, containing both cells and supernatant, were frozen at −70°C and distributed as described for the peripheral blood samples. The frozen samples were subsequently shipped to two independent laboratories.
Quantitative assays.
For this analysis, 338 samples were tested with the Roche AMPLICOR HIV-1 MONITOR test (10) (Roche Diagnostic Corporation, Indianapolis, Ind.), 177 samples were tested with the Organon Teknika Corporation (OTC) nucleic acid sequence-based amplification (NASBA-QT) assay (13) (Organon Teknika Corporation, Durham, N.C.), and 162 samples were tested with the OTC NucliSens assay (6). Specimens were tested at the Retrovirology Laboratory of Rush-Presbyterian St. Luke's Medical Center (Roche AMPLICOR HIV-1 MONITOR assay) and the University of Southern California Medical Center (OTC NASBA-QT and NucliSens assays). The laboratories used both the kit standards and standards prepared by the National Institute of Allergy and Infectious Diseases Virology Quality Assurance Program (VQA; Chicago, Ill.) (9). VQA copy standards at 0, 1,500, 15,000, and 150,000 copies/ml were included in the runs to permit the calculations. The assay methods (6, 10, 13) and the VQA standards (9, 14) have been previously described.
For the Roche AMPLICOR HIV-1 MONITOR assay, 0.2 ml of both plasma and CVL samples was processed using standard Roche plasma processing procedures. Extracted samples (50 μl) were amplified in a GeneAmp PCR System 9600 and detected according to the manufacturer's recommendations. Results were calculated according to the manufacturer's recommendations. The limit of assay sensitivity provided by the manufacturer was 400 copies/ml for all specimens.
For the OTC NASBA-QT and NucliSens assays, 0.2 ml of peripheral blood plasma and 0.8 to 1.0 ml of CVL specimens were processed in accordance with the manufacturer's instructions using the silica gel extraction methodology (2). For the NASBA-QT assay, 10-fold-diluted calibrators were used to increase sensitivity (12). The NucliSens assay was run according to the manufacturer's instructions. In accordance with the manufacturer's recommendations, the limits of detection for the NASBA-QT assay were 500 copies/ml in blood samples and 125 copies/ml in genital tract samples; for the NucliSens assay, the limits of detection were 400 copies/ml in blood samples and 80 copies/ml in genital tract samples.
Statistical analysis.
Analyses included independent comparisons of the Roche AMPLICOR HIV-1 MONITOR test with the OTC NASBA-QT and NucliSens assays, as well as a composite comparison of the AMPLICOR MONITOR and NASBA-QT and NucliSens assays. Comparisons between assays were based on samples for which estimates were above the limit of detection.
The analyses reported here assessed the differences among the assays and the extent to which a common set of standards reduced the differences among the assays. Therefore, the results obtained with the assays were compared both before and after adjustment to the VQA standards. All assays used internal standards; therefore, adjustment was made using regressions of estimated RNA concentration on nominal log10 concentration for the VQA standard estimates.
Comparisons included nominal copy numbers that were above the limits of detection for the assays under study. The number of specimens falling outside the dynamic range of the assay was noted for each assay system. The mean, median, and number of values above the assay cutoff were calculated for the population as a whole for each assay.
The analysis involved pairwise comparisons among assays rather than three-way comparisons. Paired t tests were used to test the null hypothesis that the average difference in estimated RNA concentration between assays is 0. Linear regression analysis was used to illustrate the effect of sample concentration of HIV-1 RNA on the correlations among assays and to assess the overall effect of the external standard versus the kit standard on HIV-1 RNA copy number estimates.
RESULTS
Qualitative differences among assays.
The number of samples testing positive on each assay is given in Table 1. In peripheral blood samples, there were 250 samples above the lower limit of detection for the AMPLICOR HIV-1 MONITOR test, 122 for the NASBA-QT assay, and 96 for the NucliSens assay, for a total of 218 samples above the lower limit of detection by one or the other OTC assay. For genital tract specimens, there were 77 samples above the lower limit of detection for the AMPLICOR MONITOR test, 48 for the NASBA-QT assay, and 32 for the NucliSens assay, for a total of 80 samples above the lower limit of detection by one or the other OTC assay. The levels of discordance between the Roche and OTC assays were 18.6% (63 women) for the genital tract samples and 17.7% (60 women) for peripheral blood samples. For the 60 discordant blood samples, the RNA values ranged from 402 to 95,000 copies/ml (mean = 4,017). For the 63 discordant CVL samples, the RNA values ranged from 98 to 100,000 copies/ml (mean = 5,395). Only six of the discordants could be explained by OTC values in CVL samples below the limit of detection of the Roche assay (400 copies/ml). There was no obvious bias to discordance based on antiretroviral therapy. Of the 60 and 63 samples discordant in blood and CVL, 23 and 44%, respectively, were from women not on therapy. Discordance between assays was not consistent between the two compartments. Of the 60 and 63 women who were discordant in peripheral blood and the genital tract, respectively, only 9 were discordant in both compartments. There was no pattern to the discordance between assays for these nine women. Four were positive in both compartments by OTC and negative in both compartments by Roche; two were positive in both compartments by Roche and negative by OTC; two were positive in blood by OTC and positive in CVL only by Roche; and one was positive in CVL by OTC and positive in blood by Roche. Of these nine discordant samples, only two could be explained by OTC values in CVL samples below the limit of detection of the Roche assay (400 copies/ml). These two women were on antiretroviral therapy.
TABLE 1.
Qualitative assay concordance
| Roche result | No. of samples with OTC result for compartment:
|
|||
|---|---|---|---|---|
| Genital tracta
|
Peripheral bloodb
|
|||
| + | − | + | − | |
| + | 47 | 30 | 204 | 46 |
| − | 33a | 228 | 14b | 74 |
NASBA-QT was positive for 23 samples, and NucliSens was positive for 10 samples.
NASBA-QT was positive for 11 samples, and NucliSens was positive for 3 samples.
Quantitative differences among assays.
Table 2 shows the descriptive statistics for both the kit-based and VQA-based estimates of HIV-1 RNA copy number. In general, HIV-1 RNA copy number estimates were lower for the AMPLICOR HIV-1 MONITOR test than for either the NASBA-QT or NucliSens assay; this difference was more pronounced in peripheral blood specimens. Adjustment with the VQA standards further lowered the estimates for both methods and both compartments; this was especially true for genital tract RNA estimates obtained with the AMPLICOR MONITOR test.
TABLE 2.
Descriptive statistics for all assays with kit-based and VQA-based HIV-1 log10 RNA copy number estimates
| Estimate type for test | Genital tract
|
Peripheral blood
|
||||
|---|---|---|---|---|---|---|
| No. positive | Median | Range | No. positive | Median | Range | |
| Kit based | ||||||
| Roche | 77 | 3.24 | 2.61–5.27 | 250 | 3.61 | 2.60–6.05 |
| OTC | 80 | 3.31 | 1.95–5.90 | 222 | 3.87 | 2.66–6.56 |
| NASBA-QT | 48 | 3.36 | 2.18–5.90 | 122 | 3.90 | 2.74–5.98 |
| NucliSens | 32 | 3.27 | 1.95–4.65 | 100 | 3.84 | 2.66–6.56 |
| VQA based | ||||||
| Roche | 77 | 2.97 | 1.66–5.36 | 250 | 3.61 | 2.21–6.20 |
| OTC | 80 | 3.26 | 0.60–5.13 | 222 | 3.88 | 1.18–6.59 |
| NASBA-QT | 48 | 3.24 | 0.60–5.13 | 122 | 3.93 | 1.18–6.29 |
| NucliSens | 32 | 3.38 | 1.79–4.85 | 100 | 3.81 | 1.92–6.59 |
Table 3 provides summary statistics for absolute RNA copy number for specimens above the lower limit of detection for all assays. A positive median indicates that values from the first assay in the comparison were, on average, higher than values for the second assay. The P values are the results of tests of the null hypothesis that the average difference between assays is 0. Kit-based estimates of HIV-1 RNA copy number in peripheral blood from the AMPLICOR MONITOR and NASBA-QT assays were significantly different (P = 0.005), but the estimates adjusted to the VQA standards were not (P = 0.96). There was no significant difference between the AMPLICOR MONITOR and NucliSens assays for either kit-based or VQA-based estimates (P = 0.42 and 0.28, respectively). In contrast to peripheral blood, estimates of HIV-1 RNA copy number in genital tract specimens were not significantly different for any assay method using any set of standards for calculation.
TABLE 3.
Differences in log10 HIV-1 RNA copy number estimates for all assays for kit-based and VQA-based estimates
| Estimate type and tests compared | Genital tract
|
Peripheral blood
|
||||
|---|---|---|---|---|---|---|
| Median | SD | P value | Median | SD | P value | |
| Kit based | ||||||
| Roche vs OTC | 0.05 | 0.47 | 0.83 | −0.11 | 0.43 | 0.009 |
| Roche vs NASBA-QT | 0.03 | 0.45 | 0.83 | −0.13 | 0.43 | 0.009 |
| Roche vs NucliSens | 0.08 | 0.51 | 0.92 | −0.08 | 0.43 | 0.42 |
| VQA based | ||||||
| Roche vs OTC | −0.12 | 0.67 | 0.32 | −0.10 | 0.60 | 0.51 |
| Roche vs NASBA-QT | −0.12 | 0.64 | 0.57 | −0.10 | 0.62 | 0.96 |
| Roche vs NucliSens | −0.13 | 0.71 | 0.41 | −0.12 | 0.57 | 0.28 |
For the AMPLICOR MONITOR and NucliSens assays, the standard deviations (SD) were 0.16 and 0.31 log10 HIV-1 RNA copies per ml, respectively, based on the performance of the 1,500 VQA copy number standard. For the NASBA-QT assay, the SD was 0.30 log10 HIV-1 RNA copies per ml based on the 15,000 VQA copy number standard. Thus, the 95% confidence intervals were fairly broad (±0.8 log10). Virtually all samples, from both genital tract and peripheral blood, fell within ±2 SD of the assays (data not shown).
Effect of HIV-1 RNA level on assay correlation.
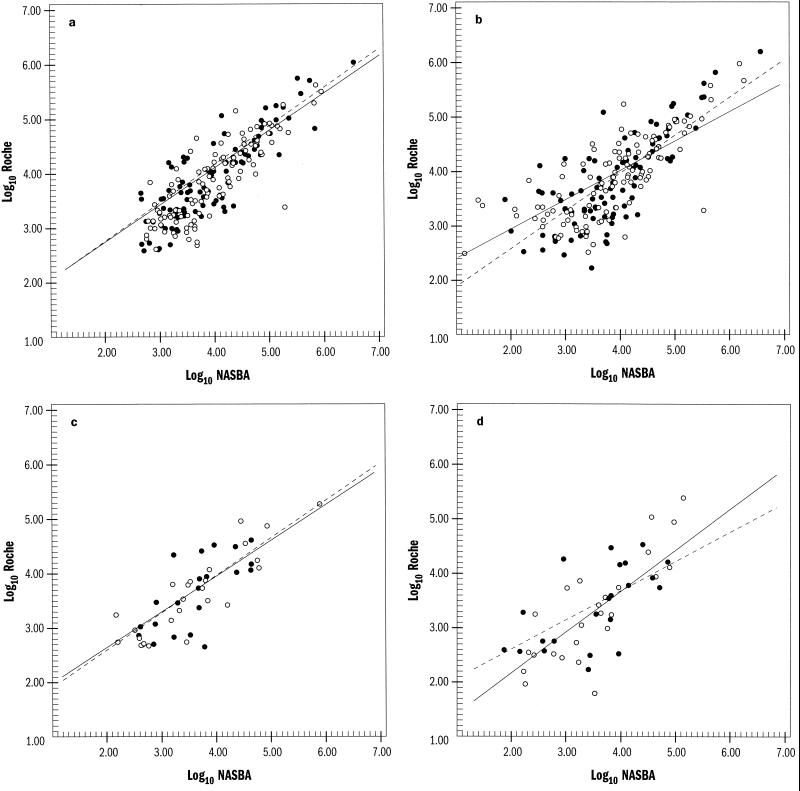
Figure 1 illustrates the linear regression analysis of the OTC and Roche assays for kit- and VQA-adjusted values in peripheral blood and genital tract samples. All estimates were highly correlated (P ≤ 0.001). For genital tract, the coefficients of correlation between the Roche AMPLICOR HIV-1 MONITOR test and the OTC NASBA-QT and NucliSens assays were 0.71 and 0.72, respectively, for kit-based determinations and 0.75 and 0.53, respectively, for VQA-based estimates. Similarly, for peripheral blood, the coefficients of correlation between the AMPLICOR MONITOR test and the NASBA-QT and NucliSens assays were 0.78 and 0.80, respectively, for kit-based determinations and 0.59 and 0.74, respectively, for VQA-based estimates. Differences in HIV-1 RNA copy numbers between the NASBA-QT and NucliSens assays relative to the AMPLICOR MONITOR test appear to be greater at the ends of the HIV-1 RNA concentration scales, and these differences were not reduced by adjustment with the external standards.
FIG. 1.
Linear regression analysis of the OTC and Roche assays for kit (a and c)- and VQA (b and d)-adjusted values for peripheral blood (a and b) and genital tract (c and d). Log10 RNA values from the Roche assay are plotted against those of the NASBA-QT (○) and NucliSens (●) assays. Linear regression correlation for the NASBA and NucliSens assays (——) is shown, compared to a slope of 1 (–––).
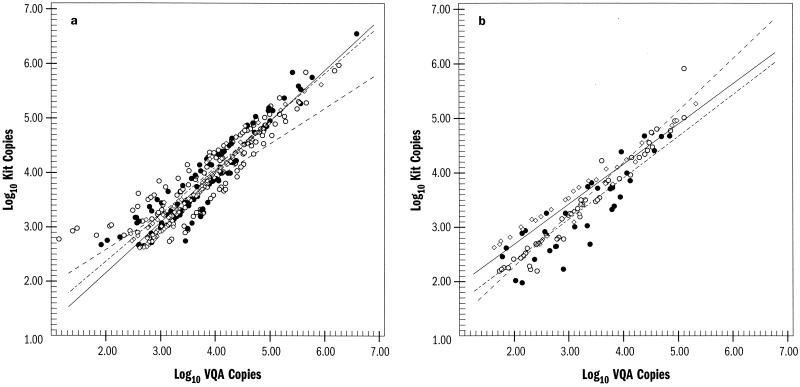
Figure 2 demonstrates the effect of VQA adjustment of any assay compared to the kit-determined HIV-1 RNA copy number. For all assays and both specimen types, the VQA-adjusted copy number estimates were higher at higher concentrations and lower at lower concentrations. This pattern was most noticeable for the NASBA-QT assay and peripheral blood specimens. In both peripheral blood and genital tract specimens, the effect of VQA adjustment on copy number estimates from the NucliSens assay more closely paralleled the effect observed for the AMPLICOR MONITOR assay than that observed for the NASBA-QT assay.
FIG. 2.
Effect of VQA adjustment compared to kit-determined HIV-1 copy number estimates for peripheral blood (a) and genital tract (b). Log10 RNA values for kit copies are plotted against VQA-adjusted copy numbers for the Roche (◊ and ——), NASBA-QT (○ and –––), and NucliSens (● and -–-–) assays.
DISCUSSION
The results of this study suggest that different gene amplification methods can be used to arrive at comparable determinations of HIV-1 RNA copy number in the female genital tract. In previous studies of blood plasma (3), we observed differences in estimated RNA copy number between the OTC NASBA-QT assay and the Roche AMPLICOR HIV-1 MONITOR assay; these differences could be eliminated by adjusting to a common set of external standards. In the current study, we confirmed our previous findings concerning the NASBA-QT and AMPLICOR MONITOR assays but found no significant differences between HIV-1 RNA copy number estimates obtained with the NucliSens and AMPLICOR MONITOR assays.
Importantly, there were no significant differences among the assays in terms of HIV-1 RNA quantification for the female genital tract. Thus, the use of a common set of external standards would not appear to be necessary when making comparisons between assays using genital tract specimens. The absence of a difference between assays using peripheral blood specimens and those using genital tract specimens could not be explained by a decrease in the dynamic range. The adjusted and unadjusted dynamic ranges for peripheral blood and genital tract HIV-1 RNA were equivalent. Similarly, although the assays use different processing technologies, each uses the same processing technology for peripheral blood and genital tract specimens.
In contrast to previous findings (3, 4), however, this study found a higher SD for the OTC assays than for the Roche assay. Thus, the lack of differences between the estimated RNA copy numbers obtained with these assays may be confounded by the higher assay SD. Nonetheless, as previously reported (3), we observed similar differences when the NASBA-QT and AMPLICOR MONITOR assays were used to test peripheral blood, in spite of the higher SD.
The effect of VQA standard adjustment was greatest for the NASBA-QT assay and was more pronounced at higher and lower HIV-1 RNA concentrations. For both peripheral blood and genital tract specimens and for both amplification technologies, the copy number estimates obtained with the VQA standards were higher than the kit-based estimates at the high end and lower than the kit-based estimates at the low end.
An issue of concern is the high level of discordance in HIV-1 detection between the OTC and Roche gene amplification systems. This discordance was not confined to either compartment or to any group of women. Many of these differences were not at the level of the limit of detection for the assays. Sequence variation among the highly discordant isolates may have resulted in differences in primer pair efficiencies, which could explain the discordance.
In summary, the OTC NucliSens and Roche AMPLICOR HIV-1 MONITOR assays appear to be equivalent for determining HIV-1 RNA levels in either the female genital tract or peripheral blood, although the SD for the NucliSens assay was high. These equivalent determinations can be made without using an external set of common standards. However, the two amplification technologies do produce some discordant positive and negative results.
ACKNOWLEDGMENTS
We acknowledge the following individuals for their assistance with this study: Vinita Goveia, Shirley Lewis, Cheryl Jennings, John Nelson, Anna Soloviov, and the women enrolled in DATRI 009.
The data in this paper were collected by the Women's Interagency HIV Study Collaborative Study Group, with centers (principal investigators) at the New York City/Bronx Consortium (Kathryn Anastos); Brooklyn, N.Y. (Howard Minkoff); the Washington, D.C., Metropolitan Consortium (Mary Young); the Connie Wofsy Study Consortium of Northern California (Ruth Greenblatt and Herminia Palacio); the Los Angeles County/Southern California Consortium (Alexandra Levine); the Chicago Consortium (Mardge Cohen); and the Data Coordinating Center (Alvaro Muñoz and Stephen J. Gange).
This study was supported by the Division of AIDS Treatment Research Initiative, National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health, Bethesda, Md. (contract no. AI-15123); the Program Support Center, U.S. Department of Health and Human Services (contract 282-97-0015, task order 21); and the Women's Interagency HIV Study, funded by NIAID, with supplemental funding from the National Cancer Institute, the National Institute of Child Health and Human Development (NICHD), the National Institute on Drug Abuse, the National Institute of Dental Research, the Agency for Health Care Policy and Research, and the Centers for Disease Control and Prevention (UO1-AI-35004, UO1-AI-31834, UO1-AI-34994, AI-34989, UO1-HD-32632 [NICHD], UO1-AI-34993, and UO1-AI-42590).
REFERENCES
- 1.Barkin S, Melnick S, Preston-Martin S. The Women's Interagency HIV Study. Epidemiology. 1998;9:117–125. [PubMed] [Google Scholar]
- 2.Boom R, Sol C, Saliman M, Jansen C, Wertheim-van Dillen P, van der Noordaa J. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brambilla D, Leung S, Lew J, Todd J, Herman S, Cronin M, Shapiro D, Bremer J, Hanson C, Hillyer G, McSherry G, Sperling R, Coombs R, Reichelderfer P. Absolute copy number and relative change in determinations of human immunodeficiency virus type 1 RNA in plasma: effect of an external standard on kit comparisons. J Clin Microbiol. 1998;36:311–314. doi: 10.1128/jcm.36.1.311-314.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coombs R W, Speck C E, Hughes J P, Lee W, Sampoleo R, Ross S O, Dragavon J, Peterson G, Hooton T M, Collier A C, Corey L. Association between culturable human immunodeficiency virus type-1 (HIV-1) in semen and HIV-1 RNA levels in semen and blood: evidence for compartmentalization of HIV-1 between semen and blood. J Infect Dis. 1998;177:320–330. doi: 10.1086/514213. [DOI] [PubMed] [Google Scholar]
- 5.Dyer J R, Gilliam B, Eron J, Grosso L, Cohen M, Fiscus S A. Quantitation of human immunodeficiency virus type 1 RNA in cell free seminal plasma: comparison of NASBA and Amplicor reverse transcription-PCR amplification and correlation with quantitative culture. J Virol Methods. 1996;60:161–170. doi: 10.1016/0166-0934(96)02063-0. [DOI] [PubMed] [Google Scholar]
- 6.Dyer J R, Pilcher C, Shepard R, Schock J, Eron J, Fiscus S. Comparison of NucliSens and Roche Monitor assays for quantitation of levels of human immunodeficiency virus type 1 RNA in plasma. J Clin Microbiol. 1999;37:447–449. doi: 10.1128/jcm.37.2.447-449.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gupta P, Mellors J, Kingsley L, Singh M K, Riddler S, Schreiber S, Cronin M, Rinaldo C. High viral load in semen of human immunodeficiency virus type 1-infected men at all stages of disease and its reduction by therapy with protease and nonnucleoside reverse transcriptase inhibitors. J Virol. 1997;71:6271–6275. doi: 10.1128/jvi.71.8.6271-6275.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holodniy M, Anderson D, Wright D, Sharma O, Cohn J, Alexander N, Stratton P, Reichelderfer P DATRI 005 Study Team. HIV quantitation in spiked vaginocervical secretions: lack of non-specific inhibitory factors. J Virol Methods. 1998;72:185–195. doi: 10.1016/s0166-0934(98)00021-4. [DOI] [PubMed] [Google Scholar]
- 9.Lin H J, Myers L, Yen-Lieberman B, Hollinger F, Henrard D, Hooper C, Kokka R, Kwok S, Rasheed S, Vahey M, Winters M, McQuay L, Nara P, Reichelderfer P, Coombs R, Jackson J. Multicenter evaluation of methods for the quantitation of plasma HIV-1 RNA. J Infect Dis. 1994;170:553–562. doi: 10.1093/infdis/170.3.553. [DOI] [PubMed] [Google Scholar]
- 10.Mulder J, McKinney N, Christopherson C, Sninsky J, Greenfield L, Kwok S. Rapid and simple PCR assay for quantitation of human immunodeficiency virus type 1 RNA in plasma: application to acute retroviral infection. J Clin Microbiol. 1994;32:292–300. doi: 10.1128/jcm.32.2.292-300.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shepard R N, Schock J, Robertson K, Shugars D C, Dyer J, Vernazza P, Hall C, Cohen M S, Fiscus S A. Quantitation of human immunodeficiency virus type 1 RNA in different biological compartments. J Clin Microbiol. 2000;38:1414–1418. doi: 10.1128/jcm.38.4.1414-1418.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vandamme A-M, Schmit J-C, van Dooren S, van Laethem K, Gobbers E, Kok W, Goubau P, Witvrouw M, Peetermans W, de Clercq E, Desmyter J. Quantification of HIV-1 RNA in plasma: comparable results with NASBA HIV-1 RNA QT and the Amplicor HIV Monitor test. J Acquir Immune Defic Syndr. 1996;13:127–139. doi: 10.1097/00042560-199610010-00003. [DOI] [PubMed] [Google Scholar]
- 13.van Gemen B, Kievits T, Schukkink R, van Strijp D, Malek L, Sooknanan R, Huisman H, Lens P. Quantification of HIV-1 RNA in plasma using NASBA during HIV-1 primary infection. J Virol Methods. 1993;43:177–188. doi: 10.1016/0166-0934(93)90075-3. [DOI] [PubMed] [Google Scholar]
- 14.Yen-Lieberman B, Brambilla D, Jackson B, Bremer J, Coombs R, Cronin M, Herman S, Katzenstein D, Leung S, Lin H, Palumbo P, Rasheed S, Todd J, Vahey M, Reichelderfer P. Preliminary evaluation of a quality assurance program for quantitation of human immunodeficiency virus type 1 RNA in plasma by the AIDS Clinical Trials Group Virology Laboratories. J Clin Microbiol. 1996;34:2695–2701. doi: 10.1128/jcm.34.11.2695-2701.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]