Abstract
Simple Summary
Triple negative breast cancer lacks targeted therapies and has poor prognosis; chemotherapy is currently the main treatment modality. Growing evidence has shown that breast cancer stem cells are associated with tumor initiation and metastasis and may play a critical role in chemoresistance. Multiple targets against breast cancer stem cells are now under investigation. Recent advances in the role of breast cancer stem cells in triple negative breast cancer and the identification of cancer stem cell biomarkers have paved the way for the development of new targeted therapies. The discovery of potential molecular signaling pathways targeting breast cancer stem cells to overcome chemoresistance and prevent metastasis will improve the overall survival of patients with triple negative breast cancer.
Abstract
Triple negative breast cancer (TNBC) remains an aggressive disease due to the lack of targeted therapies and low rate of response to chemotherapy that is currently the main treatment modality for TNBC. Breast cancer stem cells (BCSCs) are a small subpopulation of breast tumors and recognized as drivers of tumorigenesis. TNBC tumors are characterized as being enriched for BCSCs. Studies have demonstrated the role of BCSCs as the source of metastatic disease and chemoresistance in TNBC. Multiple targets against BCSCs are now under investigation, with the considerations of either selectively targeting BCSCs or co-targeting BCSCs and non-BCSCs (majority of tumor cells). This review article provides a comprehensive overview of recent advances in the role of BCSCs in TNBC and the identification of cancer stem cell biomarkers, paving the way for the development of new targeted therapies. The review also highlights the resultant discovery of cancer stem cell targets in TNBC and the ongoing clinical trials treating chemoresistant breast cancer. We aim to provide insights into better understanding the mutational landscape of BCSCs and exploring potential molecular signaling pathways targeting BCSCs to overcome chemoresistance and prevent metastasis in TNBC, ultimately to improve the overall survival of patients with this devastating disease.
Keywords: cancer stem cells, breast cancer stem cells, tumor biomarker, triple negative breast cancer, chemoresistance, metastasis
1. Introduction
Stem cells are characterized by their self-renewal capacity and potential to differentiate into many more specialized mature cell types. A subpopulation of stem-like cells within tumors, known as cancer stem cells (CSCs), have been identified to exhibit characteristics of both stem cells and cancer cells. CSCs in human breast tumors were initially identified in 2003 by Al-Hajj et al. [1]. Since then, a number of cell surface markers such as the clusters of differentiation (CD) markers CD44 and CD24 and aldehyde dehydrogenase 1 (ALDH1) have been utilized to isolate/enrich CSCs in order to further characterize their behavior [2]. Marker panels have been studied in various tissue subtypes to isolate organ-specific tumor stem cells. The CD44+/CD24−/ALDH+ phenotype in breast cancer cells has been recognized with an increased potential of tumorigenicity [3]. Other CSC markers, including ATP Binding Cassette Subfamily G Member 2 (ABCG2) and CD133, have been investigated [4,5,6]. Breast CSCs (BCSCs) play a major role in breast cancer invasion and metastasis [7], with the tumor microenvironment being a critical factor for growth signaling and proliferation [8,9]. In addition to breast cancer, CSCs have been associated with the prognosis of other tumors such as medulloblastoma, lung cancer and prostate cancer [10].
Breast cancer consists of four major molecular subtypes: luminal A (Hormone receptor (HR)+/HER2), luminal B (HR+/HER2+), HER2-enriched (HR−/HER2+) and triple-negative or basal-like (HR−/HER2−) [11,12,13]. Triple negative breast cancer (TNBC) accounts for about 15–20% of all breast cancers. TNBC often has basal-like morphology, has an epidemiological predilection for younger female and African American or Hispanic ethnicity [12]. Patients with TNBC usually have poor prognosis compared to ER+ and/or HER2+ breast cancers. However, like other breast cancer subtypes, chemotherapy is still the primary treatment option for TNBC. Studies have shown that patients undergoing neoadjuvant chemotherapy have significantly worse survival rates in TNBC subtypes as compared to non-TNBC subtypes, particularly in the first 3 years after initial diagnosis [14]. TNBC also has been found to be associated with an increased risk for early metastasis, compared to other types [15]. There is a pressing need to study the tumor biology of TNBC, the mechanisms of metastases, and therapy resistance. More so, there is a need for effective targeted therapies against chemotherapy-resistant TNBC.
It has been suggested that CSCs play an important role in the tumorigenesis and tumor biology of TNBC. Like other organs and tissues, CSCs originate from normal stem cells in breast tissue with increasing capacity for self-renewal and proliferation. CSCs, if present in TNBC, can enhance a tumor’s capacity for metastasis and risk of invasion [8]. Studies have shown that CD44+/CD24− and ALDH1+ breast cancer stem cells are enriched in TNBC and may contribute to the propensity of TNBC for chemoresistance and tumor metastasis [16,17]. This tumor evasion mechanism from chemotherapy is likely to play a more important role in tumorigenesis and outcome in TNBC compared with non-TNBCs such as luminal A type breast cancer. CSCs can potentially serve as key regulators, driving the aggressiveness of TNBC [18].
Although uncertainties about BCSCs exist, it has been suggested that CSCs should be considered as possible therapeutic targets. Emerging evidence demonstrates that CSCs usually escape current anti-cancer therapies, which may partly contribute to metastasis dissemination, tumor recurrence and poor prognosis [18,19]. Eliminating breast cancer stem cells may potentially improve the prognosis of TNBC. Recent research has focused on the immunohistochemical and genomic signatures for potential biomarkers and targets specific to BCSCs. These potential markers could be the crucial information needed to guide treatment choices and predict sensitivity and response to therapy. Several clinical trials have aimed to target breast cancer stem cells with different proposed approaches. Specific drugs to Notch, Sonic hedgehog (Shh) and Wnt signaling pathways of BCSCs are under investigation [20]. Immunotherapies, such as monoclonal antibody against CD44, and gene-edit technology such as Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) are other potential treatment options that specifically target BCSCs [21].
2. Cancer Stem Cells in Breast Cancer
BCSCs are a small cell subpopulation among all tumor cells, with unique stem cell characteristics such as self-renewal, high proliferation and differentiation potential to multiple lineages within the breast. BCSCs interact with their tumor microenvironment (TME) as well as on the inducing factors and elements. Markers that have been identified in BCSCs include ALDH1+, CD24− and CD44+. Cells which simultaneously express these markers have the highest tumor initiating and proliferative capacity. Several theories have been proposed about the origin of BCSCs. One of them suggests mutations in dormant normal stem cells results in their transformation to cancer stem cells, a process similar to that of non-stem cells [22,23].
Several markers have been used for the isolation and identification of BCSCs. In 2003, Al-Hajj M et al. first described and isolated the BCSC phenotypes CD24− and CD44+ [1]. CD24 is a cell surface glycoprotein, which has been shown to play a role in tumor progression and metastasis. As a ligand for P-selectin, CD24 is proposed to interact with endothelial cells and platelets during metastasis and ultimately has been linked to poor prognosis and decreased survival [24,25]. As another cell surface glycoprotein and specific receptor to hyaluronan, CD44 plays a key role in breast cancer adhesion, motion, migration and invasion, all of which have a significant impact on early tumor metastasis. [26] ALDH1 is a recently described BCSC marker which belongs to a family of cytosolic enzymes involved in oxidation of intracellular aldehydes that convert retinaldehydes to retinoic acid [27].
Six molecular subtypes of TNBC have been proposed: immunomodulatory, mesenchymal, mesenchymal stem-like, luminal androgen receptor and two basal-like subtypes [28]. The CD44+/CD24− lineage cells generally possess a mesenchymal or myoepithelial-like phenotype and are found more peripherally at the tumor edge. ALDH1-expressing BCSCs have a more epithelial or luminal phenotype and are located more centrally in tumors. These characteristics enable effective epithelial-mesenchymal transition and vice versa [29]. This explains the increased rate of metastasis in BCSC enriched tumors.
Cancer cells with the CD44+/CD24−/ALDH1+ phenotype in breast cancer can therefore be distinguished from other cancer cells by their stem-like features, ability to maintain survival and the role in cancer invasion and metastasis, particularly in TNBC.
3. Breast Cancer Stem Cell Regulation Pathways in Triple Negative Breast Cancer
TNBC tumors are known as being enriched for BCSCs. Transcription factors, signaling pathways and tumor suppressor genes have played a pivotal role to maintain the state of stemness of BCSCs. The Wnt, Notch, and Shh pathways have been identified as playing an important role in BCSC and TNBC biology [20,30,31]. Like in many other epithelia, Wnt pathway activity is critical for stem cell self-renewal and multipotency in the breast [32]. Although canonical Wnt signaling is the most well-known path to β-catenin stabilization, numerous other signaling pathways also regulate β-catenin stability and transcriptional activity [33]. For instance, the β-catenin mediated Wnt pathway has several downstream targets including MMP7 and PTEN, as characterized by Dey and colleagues [34]. In TNBC, differential activation of the Wnt pathway correlates with increased MMP7 and decreased expression of PTEN. Patients with increases in MMP7 have a lower rate of pathologic complete response and a high residual disease burden [35]. Inhibition of the Wnt pathway leads to decreased proliferation and induction of apoptosis, bringing to light possible therapeutic targets [36]. Additionally, wnt10B/β-catenin modulates HMGA2, and expression of HMGA2 has been shown to predict the metastasis rate and relapse-free survival [37]. Sulaiman and colleagues found that increased Wnt and histone deacetylase (HDAC) activities are associated with loss of ER and PR expression, poor survival, and increased relapse in patients with invasive breast cancer. Furthermore, in a subset of TNBC cell lines, Wnt signaling and the repression of ER and PR was found to be inversely correlated [38].
The canonical Notch signaling pathway is initiated by activating Notch receptors upon binding to Serrate- and Delta-like ligands present on the cell membranes of adjacent cells. Notch receptors have intracellular domains that are releases into the nucleus after proteolytic cleavages. These Notch intracellular domains can recruit MAML1 and histone acetyltransferase p300 to form active transcriptional complexes, which is the final regulator of the Notch target genes [39]. It was found that the Notch 4, one of the four Notch receptors, is involved in the constitutive ligand-independent activation of Notch 4, and could facilitate the development of mammary adenocarcinoma [40]. BCSCs ectopically expressing Notch 4 in TNBC have shown increased proliferation and invasiveness, whereas inhibition/knockdown of Notch4 decreases cell proliferation, invasion, tumor volume, and tumorigenicity [39].
The canonical Shh pathway is activated by releasing its ligands to bind and inhibit transmembrane receptor Patched-1 (PTCH1). PTCH1 can subsequent initiate an intracellular signal cascade to activate of the downstream transcription factors of glioma-associated oncogene (GLI) [41]. Common Shh target genes include Cyclin D1/2 [42], PDGFR, MYC [43], BCL2 [44], VEGF [45], MMP9 [46] and SOX2 [47]. The non-canonical Shh pathway, on the other hand, does not reply on the PTCH1 to activate GLI. Instead, it can cross talk with a variety of other signaling cascades [41]. Of note, Shh pathway activation is involved in the TME of the CD44+/CD24−/ALDH1+-expressing BCSCs. Shh pathway activation has been found to mediate the self-renewal of BCSCs after radiation or chemotherapy, which, not surprisingly, results in therapy resistance.
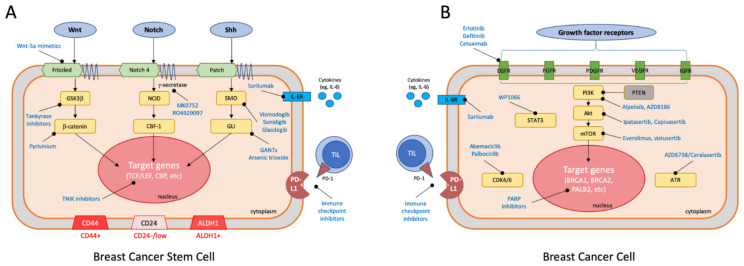
The self-renewal mechanism of CSCs, involving the signal transduction pathways noted above and including Wnt, Notch, and Shh [48], is illustrated in Figure 1A. Further investigation of these signaling pathway functions and their interactions with other pathways can aid the development of CSC-targeted therapy for the treatment of TNBC.
Figure 1.
Summary of CSC specific mechanisms and pathways targeting breast cancer stem cells (A) and non-CSC specific mechanisms and pathways targeting breast cancer cells (B).
4. Tumor Metastasis in Triple Negative Breast Cancer
Using CSCs as a prognostic marker in patients with breast cancer has profound clinical significance and has been discussed in one of our prior reviews [17]. The prognostic value of these CSC markers in breast cancer, especially the association with metastasis occurrence and survivals, has been summarized in Table 1. Based on the accumulating evidence, it appears as if combining the three stem cell markers CD44, CD24, and ALDH1 has tremendous value in predicting prognosis, including risk of metastasis and overall survival.
Table 1.
Summary of studies on metastasis occurrence and survivals in stem like breast cancer.
| Study | TNBC | All Cases | Metastasis Occurrence | Survival |
|---|---|---|---|---|
| (Abraham et al., 2005) [66] | NA | 112 | 12 (80%) cases with CD44+/CD24−/low phenotype had distant metastasis (p = 0.04). All 5 cases with more than 20% CD44+/CD24−/low tumor cells had osseous metastasis (p = 0.02). | The percentage of CD44+/CD24−/low tumor cells had no influence on DFS or OS |
| (Liu et al., 2007) [10] | N/A | 581 | The IGS * in CD44+/CD24−/low cancer cells was significantly associated with the risk of metastasis regardless of tumor size or lymph-node status (p < 0.05). | Of patients treated with chemotherapy, IGS in CD44+/CD24−/low cancer cells was associated with lower 10-year metastasis-free survival (p < 0.001). |
| (Lin et al., 2012) [67] | 62 | 147 | The proportion of CD44+/CD24− tumor cells was correlated with lymph node involvement (p = 0.026). | The proportion of CD44+/CD24− tumor cells was significantly associated with DFS (p = 0.002) and OS (p = 0.001). |
| (Adamczyk et al., 2014) [68] | 35 | 156 | NA | In patients treated with anthracyclines and taxanes, significantly longer survival was associated with CD44+ phenotype (DFS p = 0.019, OS p = 0.062) and CD44+/CD24− phenotype (DFS p=0.006, OS p = 0.019). |
| (Chen et al., 2015) [69] | 21 | 140 | The proportion of CD44+/CD24− tumor cells was significantly associated with lymph node involvement (p = 0.016), distant metastasis (p = 0.001), and recurrence (p = 0.013) | High CD44+/CD24− phenotype had worse response to chemotherapy (p = 0.001), and worse DFS (p = 0.0012) and OS (p = 0.017) |
| (Collina et al., 2015) [70] | 160 | 160 | Only CD44, not CD24, CD133, ALDH1 and ABCG2, was significantly associated with metastases (p = 0.011). | Among CD44, CD24, CD133, ALDH1 and ABCG2, only CD44 was significantly associated with DFS (p = 0.051). |
| (Wang et al., 2017) [71] | 67 | 67 | CD44+/CD24− subtype possessed slightly increased risk of metastasis or recurrence compared with CD44−/CD24− subtype. | CD44+/CD24− tumor cells were associated with worse OS (p = 0.005). |
| (Ma et al., 2017) [72] | 158 | 158 | ALDH1 expression was significantly correlated with tumor stage (p = 0.04). | ALDH1 expression was associated with shorter RFS (p = 0.01) and OS (p = 0.001). |
| (Lee & Kim, 2018) [73] | 1 | 2 | Both cutaneous metastatic cases had high expression of CD44+/CD24− and ALDH1+. | NA |
| (Rabinovich et al., 2018) [74] | 31 | 144 | NA | CD44+/CD24− phenotype was associated with a greater risk of relapse (p = 0.011) and a worse outcome (p = 0.019). TNBC was associated with ALDH+ (p = 0.039). |
| (Althobiti et al., 2020) [75] | 178 | 930 | NA | The high expression of ALDH1 was significantly associated with poor survival (p < 0.001), and particularly in the luminal B (p = 0.042) and TNBC (p = 0.003) subtypes. |
* IGS: invasiveness gene signature; DFS: disease-free survival; OS: overall survival; RFS: relapse-free survival; NA: not available.
The epithelial–mesenchymal transition (EMT) has been recently recognized to have the ability to convert epithelial cells to mesenchymal cells and subsequently acquire motile and migratory propensity [49]. This propensity, as expected, plays a key role for CSCs to metastasize to distant tissues or organs regardless of chemotherapy [50]. Several signaling pathways in the TME, including, but not limited to, Wnt, Notch, and Shh, as mentioned earlier, produce transcription factors to regulate tumor growth, invasion, and metastasis. If these tumor cells are present in patient vessels and/or lymphatics, they are called circulating tumor cells (CTCs) and have been hypothesized to be involved in the metastatic process [51,52]. Increasing evidence has demonstrated that CSC markers are conserved without alteration during tumor metastasis, with CTCs serving as intermediate cells from primary tumor cells and the distant metastatic tumor cells [53]. BCSCs have been identified in a CTC population among patient peripheral blood samples [54]. Accordingly, this finding makes it possible to detect CTCs by using stem cell markers mentioned above [55,56]. These markers, while indicating the stemness of the CTCs, may be used for the early diagnosis of tumor metastasis, prognosis prediction and therapeutic effect monitoring.
It has been shown that TNBC exhibits more traits possessed by CSC than other breast cancer subtypes and is more likely to develop metastases [57,58]. Driven by the aggregation of CD44+ CSC, more CTC clusters and polyclonal metastasis of TNBC were found to be associated with an unfavorable prognosis [59]. The epidermal growth factor receptor (EGFR) family has been found to be involved in the regulation of cancer metastasis, including TNBC [57,60]. Interactions between the receptor tyrosine kinases EGFR and metastasis with extracellular matrix (ECM)-binding integrins enhance metastatic colonization in model systems [60,61,62]. The current evidence supports dynamic crosstalk between CSCs and metastatic site TME, with contributions by surrounding non-stem cells, including stromal cells, immune cells, and ECM, which are crucial for tumor growth after CTCs seed in the metastatic site [63,64]. CSCs in TNBC have been shown to have characteristic their high invasiveness and metastatic behavior, with increased expression of pro-invasive genes, including those for interleukin (IL)-1, IL-6, IL-8, and urokinase plasminogen activator [64,65].
All the above mechanisms have been shown to be possibly involved in the high propensity for invasiveness and metastasis of BCSCs in TNBC. Targeting these mechanisms can potentially prevent metastasis and therefore improve survival in TNBC.
5. Chemoresistance in Triple Negative Breast Cancer
TNBC, the most aggressive and deadliest type of breast cancer, lacks a targeted therapy and has therapy-resistance to the majority of chemotherapy regimens. Since none of the current chemotherapies specifically targets CSCs, CSCs serve as a tumor reservoir for self-renewal and proliferation. This causes inevitable tumor invasion and metastasis, which is more common in TNBC than non-TNBC, ultimately resulting in poor prognosis. Studies have shown CSCs are associated with chemoresistance, particularly in TNBC due to the higher propensity of developing stemness compared to those found in non-TNBC [20,76].
Doxorubicin (Dox), for instance, is widely used in the treatment of TNBC. However, resistance by tumor cells limits its effectiveness. CSCs have been found to be associated with Dox resistance. Signal transducer and activator of transcription 3 (Stat3) and its downstream pathway can convert non-CSCs to CSCs [77]. More recently, a novel signaling pathway that involves Stat3, Oct-4 and c-Myc has been demonstrated to regulate stemness-mediated Dox resistance in TNBC [16,78]. To overcome the mechanism of Dox resistance, WP1066, a Stat3 inhibitor that was more widely studied to treat CNS tumors, is being explored to reduce proliferation of BCSCs in Dox-resistant TNBC [79,80].
Since the CD44+/CD24−/ALDH1+ phenotypes have been recognized as BCSC markers in many studies, a hypothetical approach to targeting CSCs against cell surface membrane antigens such as CD44 would be justified [81,82]. Target options include monoclonal, competitive protein/peptide, HD-CD44 crosstalk, and others [81]. In addition, ALDH1 can be a possible therapeutic target [83], although only limited in vitro data is available through ALDH1 knockdown [84].
Immune evasion has also been suggested to result in chemoresistance in TNBC, as well as tumorigenesis in other tumor types. Tumor-infiltrating lymphocytes (TILs) have been shown to be present in some TNBC and non-TNBC breast cancers and have been associated with favorable prognosis [85,86]. Programmed cell death-1 (PD-1) receptor expression on tumor cells and programmed death-ligand 1 (PD-L1) expression on TILs are therefore indicators of the enrichment of the adaptive immune response against tumor cells. Lack of TIL or PD-1/PD-L1 expression in the tumor microenvironment has been suggested to be associated with less favorable prognosis, especially in early-stage breast cancer [87]. The blockade of PD-1/PD-L1 with checkpoint inhibitors has become a promising immunotherapy to enhance anti-tumor immunity in TNBC after success in treating other types of cancer and is being widely investigated in many ongoing studies.
The involvement of any of the above mechanisms may result in chemoresistance in TNBC. It is essential to take these mechanisms into account when developing novel targeted therapies to overcome chemoresistance.
6. Potential Mechanisms/Pathways of Eradicating Breast Cancer Stem Cells and Development of Targeted Therapies
Recent studies have demonstrated the role of BCSCs as the source of metastatic disease and drug resistance in TNBC. Understanding the mechanisms and pathways that stimulate cancer cell proliferation can lead to the development of effective targeted treatment strategies. More importantly, in TNBC, as treatment failure is likely to be associated with BCSCs that possess unique survival mechanisms and pathways that bypass or evade the conventional chemotherapy, developing cancer stem cell-targeted therapies should focus on the specific signaling pathways unique to BCSCs. The three signaling pathways reviewed above, namely the Wnt, Notch, and Shh pathways are potential treatment targets [83]. More and more drugs have been investigated and developed that directly target the Wnt, Notch, and Shh pathways [88]. Treatment with a small molecule β-catenin/TCF inhibitor inhibits β-catenin-mediated transcription, resulting in a reduction in stem cell proliferation and tumor bulking in TNBC cell lines and animal models [89,90]. A selective inhibitor of Wnt, HDAC, and ESR1 has been also shown to be able to suppress the stemness of BSCSs and therefore to convert BCSCs to non-BCSCs in TNBC cells while normal cells in the breast are not affected [38]. Blocking antibodies against the Notch pathway can be categorized into two groups, i.e., those directed to the negative regulatory region (NRR) that enable γ-secretase mediated-cleavage and those that block receptor-ligand interactions by hindering EGF repeats [91,92]. Shh-targeted therapies mostly consist of inhibitors directed against one of the pathway targets, SMO. Although the clinical benefit from SMO inhibitors such as Vismodegib has been established in basal cell carcinoma and medulloblastoma, its clinical effectiveness or benefits towards other solid-type tumors have been limited [93]. GLI transcription factors are the terminal effectors of the Shh-SMO signaling pathway, as discussed above, and are more likely to target CSCs by GANTS (a GLI inhibitor), which is under investigation and has shown some promising in vitro results [88,94]. A summary of the potential pathways specifically targeting BCSCs (CSC-specific mechanisms) is illustrated in Figure 1A.
Non-CSC specific molecular signaling pathways have been proposed to be able to potentially eradicate CSCs and other cancer cells in TNBC. Due to the proven association between the EGFR pathway and TNBC metastasis [95], EGFR inhibitors, such as cetuximab, have been under investigation as a potential adjunct treatment option for metastatic TNBC [96,97]. Notably, although EGFR is relatively non-specific for CSCs, one of its downstream regulators, Stat3, as mentioned earlier, is involved in regulating the self-renewal and stemness of cells, and therefore can be a promising candidate of BCSC-targeted therapy [18,98]. PI3K/Akt/mTOR is another classical pathway that has been found to be associated with many types of cancer. The mechanisms for gene or pathway activation include loss of tumor suppressor gene PTEN function, amplification or mutation of PI3K and/or Akt, and the activation of other inducers such as growth factor receptors and carcinogens [99]. Therapy targeting the PI3K/Akt/mTOR pathway has been suggested to overcome the drug resistance in TNBC by regulating apoptosis in BCSCs in addition to cancer cells [100]. Metformin, a conventional medication for diabetic treatment, has been shown to be able to block the mTOR pathway by activating AMPK adenosinemonophosphate-activated protein kinase (AMPK) [101,102]. Interestingly, it has also been shown to particularly target the CSC population in breast cancer cell lines [103,104,105].
There are a few other mechanisms that have been proposed to tackle therapy-resistant TNBC. These mainly include the blockade or reversal of the EMT of cancer cells and the disruption of TME or the metabolism [106,107,108,109]. Tumors enriched in the carcinoma-associated fibroblasts involved in TME can secrete hepatocyte growth factor (HGF, ligand for c-Met receptor), and C-X-C motif chemokine 12 (CXCL12, THE ligand for chemokine receptor CXCR4) [110]. These factors may contribute to chemoresistance [111]. Dual HDAC and 3-hydroxy-3-methylglutaryl coenzyme A reductase inhibitor, JMF3086, and several other compound drugs have been investigated to alter the EMT of cancer cells and subsequently reduce their plasticity [110]. While therapy leveraging these tumorigenesis mechanisms such as Entinostat may show effectiveness against TNBC [112], it is noted that they are not CSC-specific.
When being treated with conventional chemotherapy, therapy resistant BCSCs, non-BCSC tumor cells, stromal cells and immune cells can result in the minimal residual disease [113,114]. Unlike non-BCSC tumor cells, the stemness of BCSCs allows them to differentiate into multilineages and therefore present with different heterogeneous components within the tumor. The relapsed component, however, can evolve to be more aggressive, evading human immune defense mechanism and the conventional chemotherapy mechanism of action. This acquired characteristics result in early metastasis, particularly in TNBC. To develop targeted therapies for TNBC, a focus-target shifting from non-BCSCs towards BCSCs (the majority of tumor cells) may help overcome multi-drug resistance [111]. In principle, the tumor biological characteristics, BCSC signaling pathways, drug-resistance mechanisms, and the TMEs surrounding the BCSCs are the key factors that can be modified to become a potential treatment options. Several new targets against BCSCs have been identified as promising. Oncotargets such as the cyclin family (D1 and D3) are critical for BCSCs to maintain their clonogenicity. BCSCs have been also shown to be dependent on survival induction factors such as survivin and Myeloid Cell Leukemia 1(MCL1) to extend their survival advantages. Both of these two factors play a role in chemoresistance [73,115]. All these targets have an mRNA sequence that can be modified by mRNA helicase eIF4A [116]. The same authors have shown that eIF4A could downregulate CSC stemness and the levels of ATP-binding cassette transporters that export xenobiotics were significantly reduced [115]. Therefore, eFT226, which targets eIF4A, has become a promising new treatment target option in BCSCs. It may overcome drug resistance by downregulating those drug transporters [111].
Although BCSCs can evade the immune system, resulting in insufficient adaptive and inmate immune responses and subsequent chemoresistance, they are strongly antigenic so that naïve CD8+ effector T-cells, once activated, can still eliminate BCSCs. There is more and more evidence suggesting that empowering the CD8+ effector T-cells through the engagement of their programmed death receptor-1 (PD-1) with the PD1 ligand (PD-L1) would synergize with BCSC-directed therapy and in a combination with other target therapies [117,118]. A summary of the non-CSC specific mechanisms and pathways targeting breast cancer cell is illustrated in Figure 1B.
Lastly, gene therapy using advanced gene editing technology such as CRISPR targeting CSCs in TNBC could be a potential treatment option. The CRISPR/Cas9 system has been emerged as a leading technology to more quickly target specific genetic loci and more accurately modify genes of interest [119]. Castro et al. developed a novel TNBC model in mice and discovered Cripto-1 as a target for TNBC tumor cells that can be genetically modified by the CRISPR/Cas9 system [21]. Hypothetically, the system can target any genes that involve cancer cell stemness and therefore serves as a versatile treatment strategy for TNBC.
Table 2 summarizes the current clinical trials in chemoresistant breast cancer excluding PD-1/PD-L1 trials since they have been extensively reviewed in the literature [120,121,122,123]. Also note that poly (ADP-ribose) polymerase (PARP) and cyclin-dependent kinase (CDK) 4/6 play a key role in cell cycle, and are neither specific to breast cancer cells or CSCs. Although several trials found on clinicaltrials.gov use PARP inhibitors and CDK4/6 inhibitors, their mechanism is less relevant to the theme of this review and is therefore not discussed.
Table 2.
Clinical trials treating chemoresistant breast cancer registered on clinicaltrials.gov.
| NCT Number | Conditions | Drug of Interest | Drug Category | Phases | Enrollment | Study Designs | Country |
|---|---|---|---|---|---|---|---|
| NCT02158507 | Metastatic TNBC | Veliparib | PARP inhibitor | 23 | Single group | USA | |
| NCT04134884 | Metastatic Breast Cancer | Talazoparib | PARP inhibitor | 1 | 38 | Sequential | USA |
| NCT01477060 | Metastatic Breast Cancer | Metformin | AMPK agonist | 2 | 32 | RCT # | Italy |
| NCT02299635 | TNBC | PF-03084014 | γ-secretase inhibitor | 2 | 19 | Multiple | |
| NCT03361800 | TNBC | Entinostat | HDAC inhibitor | 1 | 5 | Single group | USA |
| NCT04333706 | TNBC | Sarilumab | IL-6R | 1/2 | 65 | Non-Randomized; Parallel | USA |
| NCT04360941 | Advanced Breast Cancer | Palbociclib | CDK4/6 inhibitor | 1 | 45 | Single group | UK |
| NCT03218826 | Advanced Breast Cancer | AZD8186 | PI3K inhibitor | 1 | 58 | Single group | USA |
| NCT03853707 | Advanced Breast Cancer | Ipatasertib | Akt inhibitor | 1/2 | 40 | RCT | USA |
| NCT03979508 | Breast Cancer | Abemaciclib | CDK4/6 inhibitor | 2 | 100 | Non-Randomized; Parallel | USA |
| NCT03740893 | Breast Cancer | AZD6738/Olaparib | ATR * kinase inhibitor/PARP inhibitor | 2 | 81 | RCT | UK |
| NCT01617668 | Breast Cancer | LCL161 | SMAC ^ mimetic | 2 | 209 | RCT | Multiple |
| NCT01266486 | Breast Cancer | Metformin | AMPK agonist | 2 | 41 | Single group | UK |
| NCT04092673 | Solid Tumor (w/Breast Cancer) | eFT226 | eIF4A Inhibitor | 1/2 | 45 | Sequential | USA |
* ATR: ataxia telangiectasia and Rad3 related; ^ second mitochondria-derived activator of caspases; # RCT: randomized controlled trial.
7. Conclusions
TNBC remains an aggressive disease due to the lack of targeted treatment and low rate of response to chemotherapy. BCSCs are a small subpopulation of breast tumors that play a vital role in metastatic disease and drug resistance. TNBC tumors are characterized as being enriched for BCSCs.
Recent advances in BCSC biology in TNBC and the identification of cancer stem cell biomarkers have paved the way for the development of cancer stem cell-targeted therapies.
Multiple targets against BCSCs are currently under investigation with the goals of either selectively targeting BCSCs or co-targeting BCSCs and non-BCSCs (majority of tumor cells).
We hope this review provides insights into mutational landscape of BCSCs and the discovery of potential molecular signaling pathways targeting BCSCs to overcome chemoresistance and prevent metastasis, ultimately to improve the prognosis of TNBC.
8. Future Direction
With the growing evidence of the role of BCSCs in TNBC, it is important to better define their biological characteristics, molecular pathways that sustain stemness, and mechanisms of drug resistance to discover effective cancer stem cell targeted therapies.
The identification and quantification of existence of BCSCs using stem cell markers and deployment of CSC-targeted therapies may revolutionize the TNBC treatment paradigm. BCSCs may become potential prognostic and predictive biomarkers for metastasis and chemoresistance, and eradicating BCSCs in the TNBC may improve the overall survival of these patients. Although there have been more promising results recently, the shortcomings of the current research on BCSCs include a limited number of biomarkers and therapeutic targets. More clinical trials are also needed to demonstrate effectiveness and safety profiles of the new investigational drugs.
Author Contributions
Conceptualization, Y.P.; methodology, Y.P.; investigation, L.H., N.W., S.K.G., Y.P.; resources, Y.P.; data curation, L.H., N.W., Y.P.; writing—original draft preparation, L.H., N.W., Y.P.; writing—review and editing, Y.P.; visualization, L.H.; supervision, Y.P.; project administration, Y.P. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Department of Pathology at UT Southwestern medical center, Dallas, TX, USA.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Al-Hajj M., Wicha M.S., Benito-Hernandez A., Morrison S.J., Clarke M.F. Prospective Identification of Tumorigenic Breast Cancer Cells. Proc. Natl. Acad. Sci. USA. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yu Z., Pestell T.G., Lisanti M.P., Pestell R.G. Cancer Stem Cells. Int. J. Biochem. Cell Biol. 2012;44:2144–2151. doi: 10.1016/j.biocel.2012.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu M., Liu Y., Deng L., Wang D., He X., Zhou L., Wicha M.S., Bai F., Liu S. Transcriptional Profiles of Different States of Cancer Stem Cells in Triple-Negative Breast Cancer. Mol. Cancer. 2018;17:65. doi: 10.1186/s12943-018-0809-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Britton K.M., Eyre R., Harvey I.J., Stemke-Hale K., Browell D., Lennard T.W.J., Meeson A.P. Breast Cancer, Side Population Cells and ABCG2 Expression. Cancer Lett. 2012;323:97–105. doi: 10.1016/j.canlet.2012.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nadal R., Ortega F.G., Salido M., Lorente J.A., Rodríguez-Rivera M., Delgado-Rodríguez M., Macià M., Fernández A., Corominas J.M., García-Puche J.L., et al. CD133 Expression in Circulating Tumor Cells from Breast Cancer Patients: Potential Role in Resistance to Chemotherapy. Int. J. Cancer. 2013;133:2398–2407. doi: 10.1002/ijc.28263. [DOI] [PubMed] [Google Scholar]
- 6.Zhao P., Lu Y., Jiang X., Li X. Clinicopathological Significance and Prognostic Value of CD133 Expression in Triple-Negative Breast Carcinoma. Cancer Sci. 2011;102:1107–1111. doi: 10.1111/j.1349-7006.2011.01894.x. [DOI] [PubMed] [Google Scholar]
- 7.Nie G., Cao X., Mao Y., Lv Z., Lv M., Wang Y., Wang H., Liu C. Tumor-Associated Macrophages-Mediated CXCL8 Infiltration Enhances Breast Cancer Metastasis: Suppression by Danirixin. Int. Immunopharmacol. 2021;95:107153. doi: 10.1016/j.intimp.2020.107153. [DOI] [PubMed] [Google Scholar]
- 8.Bozorgi A., Khazaei M., Khazaei M.R. New Findings on Breast Cancer Stem Cells: A Review. J. Breast Cancer. 2015;18:303–312. doi: 10.4048/jbc.2015.18.4.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Velasco-Velázquez M.A., Popov V.M., Lisanti M.P., Pestell R.G. The Role of Breast Cancer Stem Cells in Metastasis and Therapeutic Implications. Am. J. Pathol. 2011;179:2–11. doi: 10.1016/j.ajpath.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu R., Wang X., Chen G.Y., Dalerba P., Gurney A., Hoey T., Sherlock G., Lewicki J., Shedden K., Clarke M.F. The Prognostic Role of a Gene Signature from Tumorigenic Breast-Cancer Cells. N. Engl. J. Med. 2007;356:217–226. doi: 10.1056/NEJMoa063994. [DOI] [PubMed] [Google Scholar]
- 11.Patel A., Unni N., Peng Y. The Changing Paradigm for the Treatment of HER2-Positive Breast Cancer. Cancers. 2020;12:2081. doi: 10.3390/cancers12082081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perou C.M., Sørlie T., Eisen M.B., van de Rijn M., Jeffrey S.S., Rees C.A., Pollack J.R., Ross D.T., Johnsen H., Akslen L.A., et al. Molecular Portraits of Human Breast Tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 13.Sorlie T., Tibshirani R., Parker J., Hastie T., Marron J.S., Nobel A., Deng S., Johnsen H., Pesich R., Geisler S., et al. Repeated Observation of Breast Tumor Subtypes in Independent Gene Expression Data Sets. Proc. Natl. Acad. Sci. USA. 2003;100:8418–8423. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liedtke C., Mazouni C., Hess K.R., André F., Tordai A., Mejia J.A., Symmans W.F., Gonzalez-Angulo A.M., Hennessy B., Green M., et al. Response to Neoadjuvant Therapy and Long-Term Survival in Patients with Triple-Negative Breast Cancer. J. Clin. Oncol. 2008;26:1275–1281. doi: 10.1200/JCO.2007.14.4147. [DOI] [PubMed] [Google Scholar]
- 15.Foulkes W.D., Smith I.E., Reis-Filho J.S. Triple-Negative Breast Cancer. N. Engl. J. Med. 2010;363:1938–1948. doi: 10.1056/NEJMra1001389. [DOI] [PubMed] [Google Scholar]
- 16.Moreira M.P., da Conceição Braga L., Cassali G.D., Silva L.M. STAT3 as a Promising Chemoresistance Biomarker Associated with the CD44+/High/CD24−/Low/ALDH+ BCSCs-like Subset of the Triple-Negative Breast Cancer (TNBC) Cell Line. Exp. Cell Res. 2018;363:283–290. doi: 10.1016/j.yexcr.2018.01.018. [DOI] [PubMed] [Google Scholar]
- 17.O’Conor C.J., Chen T., González I., Cao D., Peng Y. Cancer Stem Cells in Triple-Negative Breast Cancer: A Potential Target and Prognostic Marker. Biomark. Med. 2018;12:813–820. doi: 10.2217/bmm-2017-0398. [DOI] [PubMed] [Google Scholar]
- 18.Park S.-Y., Choi J.-H., Nam J.-S. Targeting Cancer Stem Cells in Triple-Negative Breast Cancer. Cancers. 2019;11:965. doi: 10.3390/cancers11070965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vikram R., Chou W.C., Hung S.-C., Shen C.-Y. Tumorigenic and Metastatic Role of CD44-/Low/CD24−/Low Cells in Luminal Breast Cancer. Cancers. 2020;12:1239. doi: 10.3390/cancers12051239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Palomeras S., Ruiz-Martínez S., Puig T. Targeting Breast Cancer Stem Cells to Overcome Treatment Resistance. Molecules. 2018;23:2193. doi: 10.3390/molecules23092193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Castro N.P., Fedorova-Abrams N.D., Merchant A.S., Rangel M.C., Nagaoka T., Karasawa H., Klauzinska M., Hewitt S.M., Biswas K., Sharan S.K., et al. Cripto-1 as a Novel Therapeutic Target for Triple Negative Breast Cancer. Oncotarget. 2015;6:11910–11929. doi: 10.18632/oncotarget.4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang R.-A., Li Z.-S., Zhang H.-Z., Zheng P.-J., Li Q.-L., Shi J.-G., Yan Q.-G., Ye J., Wang J.-B., Guo Y., et al. Invasive Cancers Are Not Necessarily from Preformed in Situ Tumours—An Alternative Way of Carcinogenesis from Misplaced Stem Cells. J. Cell Mol. Med. 2013;17:921–926. doi: 10.1111/jcmm.12078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hartwig F.P., Nedel F., Collares T., Tarquinio S.B.C., Nör J.E., Demarco F.F. Oncogenic Somatic Events in Tissue-Specific Stem Cells: A Role in Cancer Recurrence? Ageing Res. Rev. 2014;13:100–106. doi: 10.1016/j.arr.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 24.Kristiansen G., Winzer K.-J., Mayordomo E., Bellach J., Schlüns K., Denkert C., Dahl E., Pilarsky C., Altevogt P., Guski H., et al. CD24 Expression Is a New Prognostic Marker in Breast Cancer. Clin. Cancer Res. 2003;9:4906–4913. [PubMed] [Google Scholar]
- 25.Schabath H., Runz S., Joumaa S., Altevogt P. CD24 Affects CXCR4 Function in Pre-B Lymphocytes and Breast Carcinoma Cells. J. Cell Sci. 2006;119:314–325. doi: 10.1242/jcs.02741. [DOI] [PubMed] [Google Scholar]
- 26.Herrera-Gayol A., Jothy S. Adhesion Proteins in the Biology of Breast Cancer: Contribution of CD44. Exp. Mol. Pathol. 1999;66:149–156. doi: 10.1006/exmp.1999.2251. [DOI] [PubMed] [Google Scholar]
- 27.Balicki D. Moving Forward in Human Mammary Stem Cell Biology and Breast Cancer Prognostication Using ALDH1. Cell Stem Cell. 2007;1:485–487. doi: 10.1016/j.stem.2007.10.015. [DOI] [PubMed] [Google Scholar]
- 28.Lehmann B.D., Bauer J.A., Chen X., Sanders M.E., Chakravarthy A.B., Shyr Y., Pietenpol J.A. Identification of Human Triple-Negative Breast Cancer Subtypes and Preclinical Models for Selection of Targeted Therapies. J. Clin. Investig. 2011;121:2750–2767. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu S., Cong Y., Wang D., Sun Y., Deng L., Liu Y., Martin-Trevino R., Shang L., McDermott S.P., Landis M.D., et al. Breast Cancer Stem Cells Transition between Epithelial and Mesenchymal States Reflective of Their Normal Counterparts. Stem Cell Rep. 2014;2:78–91. doi: 10.1016/j.stemcr.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lindvall C., Bu W., Williams B.O., Li Y. Wnt Signaling, Stem Cells, and the Cellular Origin of Breast Cancer. Stem Cell Rev. 2007;3:157–168. doi: 10.1007/s12015-007-0025-3. [DOI] [PubMed] [Google Scholar]
- 31.Rangel M.C., Bertolette D., Castro N.P., Klauzinska M., Cuttitta F., Salomon D.S. Developmental Signaling Pathways Regulating Mammary Stem Cells and Contributing to the Etiology of Triple-Negative Breast Cancer. Breast Cancer Res. Treat. 2016;156:211–226. doi: 10.1007/s10549-016-3746-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zeng Y.A., Nusse R. Wnt Proteins Are Self-Renewal Factors for Mammary Stem Cells and Promote Their Long-Term Expansion in Culture. Cell Stem Cell. 2010;6:568–577. doi: 10.1016/j.stem.2010.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Incassati A., Chandramouli A., Eelkema R., Cowin P. Key Signaling Nodes in Mammary Gland Development and Cancer: β-Catenin. Breast Cancer Res. 2010;12:213. doi: 10.1186/bcr2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dey N., Young B., Abramovitz M., Bouzyk M., Barwick B., De P., Leyland-Jones B. Differential Activation of Wnt-β-Catenin Pathway in Triple Negative Breast Cancer Increases MMP7 in a PTEN Dependent Manner. PLoS ONE. 2013;8:e77425. doi: 10.1371/journal.pone.0077425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dey N., Barwick B.G., Moreno C.S., Ordanic-Kodani M., Chen Z., Oprea-Ilies G., Tang W., Catzavelos C., Kerstann K.F., Sledge G.W., et al. Wnt Signaling in Triple Negative Breast Cancer Is Associated with Metastasis. BMC Cancer. 2013;13:537. doi: 10.1186/1471-2407-13-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bilir B., Kucuk O., Moreno C.S. Wnt Signaling Blockage Inhibits Cell Proliferation and Migration, and Induces Apoptosis in Triple-Negative Breast Cancer Cells. J. Transl. Med. 2013;11:280. doi: 10.1186/1479-5876-11-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wend P., Runke S., Wend K., Anchondo B., Yesayan M., Jardon M., Hardie N., Loddenkemper C., Ulasov I., Lesniak M.S., et al. WNT10B/β-Catenin Signalling Induces HMGA2 and Proliferation in Metastatic Triple-Negative Breast Cancer. EMBO Mol. Med. 2013;5:264–279. doi: 10.1002/emmm.201201320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sulaiman A., Sulaiman B., Khouri L., McGarry S., Nessim C., Arnaout A., Li X., Addison C., Dimitroulakos J., Wang L. Both Bulk and Cancer Stem Cell Subpopulations in Triple-Negative Breast Cancer Are Susceptible to Wnt, HDAC, and ERα Coinhibition. FEBS Lett. 2016;590:4606–4616. doi: 10.1002/1873-3468.12496. [DOI] [PubMed] [Google Scholar]
- 39.Krishna B.M., Jana S., Singhal J., Horne D., Awasthi S., Salgia R., Singhal S.S. Notch Signaling in Breast Cancer: From Pathway Analysis to Therapy. Cancer Lett. 2019;461:123–131. doi: 10.1016/j.canlet.2019.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gallahan D., Callahan R. Mammary Tumorigenesis in Feral Mice: Identification of a New Int Locus in Mouse Mammary Tumor Virus (Czech II)-Induced Mammary Tumors. J. Virol. 1987;61:66–74. doi: 10.1128/jvi.61.1.66-74.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bhateja P., Cherian M., Majumder S., Ramaswamy B. The Hedgehog Signaling Pathway: A Viable Target in Breast Cancer? Cancers. 2019;11:1126. doi: 10.3390/cancers11081126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kasper M., Schnidar H., Neill G.W., Hanneder M., Klingler S., Blaas L., Schmid C., Hauser-Kronberger C., Regl G., Philpott M.P., et al. Selective Modulation of Hedgehog/GLI Target Gene Expression by Epidermal Growth Factor Signaling in Human Keratinocytes. Mol. Cell. Biol. 2006;26:6283–6298. doi: 10.1128/MCB.02317-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zuo M., Rashid A., Churi C., Vauthey J.-N., Chang P., Li Y., Hung M.-C., Li D., Javle M. Novel Therapeutic Strategy Targeting the Hedgehog Signalling and MTOR Pathways in Biliary Tract Cancer. Br. J. Cancer. 2015;112:1042–1051. doi: 10.1038/bjc.2014.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Regl G., Kasper M., Schnidar H., Eichberger T., Neill G.W., Philpott M.P., Esterbauer H., Hauser-Kronberger C., Frischauf A.-M., Aberger F. Activation of the BCL2 Promoter in Response to Hedgehog/GLI Signal Transduction Is Predominantly Mediated by GLI2. Cancer Res. 2004;64:7724–7731. doi: 10.1158/0008-5472.CAN-04-1085. [DOI] [PubMed] [Google Scholar]
- 45.Po A., Silvano M., Miele E., Capalbo C., Eramo A., Salvati V., Todaro M., Besharat Z.M., Catanzaro G., Cucchi D., et al. Noncanonical GLI1 Signaling Promotes Stemness FeatuRes. and in Vivo Growth in Lung Adenocarcinoma. Oncogene. 2017;36:4641–4652. doi: 10.1038/onc.2017.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Corte C.M.D., Bellevicine C., Vicidomini G., Vitagliano D., Malapelle U., Accardo M., Fabozzi A., Fiorelli A., Fasano M., Papaccio F., et al. SMO Gene Amplification and Activation of the Hedgehog Pathway as Novel Mechanisms of Resistance to Anti-Epidermal Growth Factor Receptor Drugs in Human Lung Cancer. Clin. Cancer Res. 2015;21:4686–4697. doi: 10.1158/1078-0432.CCR-14-3319. [DOI] [PubMed] [Google Scholar]
- 47.Merchant A.A., Matsui W. Targeting Hedgehog—A Cancer Stem Cell Pathway. Clin. Cancer Res. 2010;16:3130–3140. doi: 10.1158/1078-0432.CCR-09-2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Geng S.-Q., Alexandrou A.T., Li J.J. Breast Cancer Stem Cells: Multiple Capacities in Tumor Metastasis. Cancer Lett. 2014;349:1–7. doi: 10.1016/j.canlet.2014.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gupta P.B., Chaffer C.L., Weinberg R.A. Cancer Stem Cells: Mirage or Reality? Nat. Med. 2009;15:1010–1012. doi: 10.1038/nm0909-1010. [DOI] [PubMed] [Google Scholar]
- 50.Thiery J.P., Acloque H., Huang R.Y.J., Nieto M.A. Epithelial-Mesenchymal Transitions in Development and Disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 51.Baccelli I., Schneeweiss A., Riethdorf S., Stenzinger A., Schillert A., Vogel V., Klein C., Saini M., Bäuerle T., Wallwiener M., et al. Identification of a Population of Blood Circulating Tumor Cells from Breast Cancer Patients That Initiates Metastasis in a Xenograft Assay. Nat. Biotechnol. 2013;31:539–544. doi: 10.1038/nbt.2576. [DOI] [PubMed] [Google Scholar]
- 52.Magbanua M.J.M., Sosa E.V., Roy R., Eisenbud L.E., Scott J.H., Olshen A., Pinkel D., Rugo H.S., Park J.W. Genomic Profiling of Isolated Circulating Tumor Cells from Metastatic Breast Cancer Patients. Cancer Res. 2013;73:30–40. doi: 10.1158/0008-5472.CAN-11-3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cristofanilli M., Pierga J.-Y., Reuben J., Rademaker A., Davis A.A., Peeters D.J., Fehm T., Nolé F., Gisbert-Criado R., Mavroudis D., et al. The Clinical Use of Circulating Tumor Cells (CTCs) Enumeration for Staging of Metastatic Breast Cancer (MBC): International Expert Consensus Paper. Crit. Rev. Oncol. Hematol. 2019;134:39–45. doi: 10.1016/j.critrevonc.2018.12.004. [DOI] [PubMed] [Google Scholar]
- 54.Theodoropoulos P.A., Polioudaki H., Agelaki S., Kallergi G., Saridaki Z., Mavroudis D., Georgoulias V. Circulating Tumor Cells with a Putative Stem Cell Phenotype in Peripheral Blood of Patients with Breast Cancer. Cancer Lett. 2010;288:99–106. doi: 10.1016/j.canlet.2009.06.027. [DOI] [PubMed] [Google Scholar]
- 55.Savelieva O.E., Tashireva L.A., Kaigorodova E.V., Buzenkova A.V., Mukhamedzhanov R.K., Grigoryeva E.S., Zavyalova M.V., Tarabanovskaya N.A., Cherdyntseva N.V., Perelmuter V.M. Heterogeneity of Stemlike Circulating Tumor Cells in Invasive Breast Cancer. Int. J. Mol. Sci. 2020;21:2780. doi: 10.3390/ijms21082780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Strati A., Nikolaou M., Georgoulias V., Lianidou E.S. RNA-Based CTC Analysis Provides Prognostic Information in Metastatic Breast Cancer. Diagnostics. 2021;11:513. doi: 10.3390/diagnostics11030513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bryan S., Witzel I., Borgmann K., Oliveira-Ferrer L. Molecular Mechanisms Associated with Brain Metastases in HER2-Positive and Triple Negative Breast Cancers. Cancers. 2021;13:4137. doi: 10.3390/cancers13164137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ren D., Zhu X., Kong R., Zhao Z., Sheng J., Wang J., Xu X., Liu J., Cui K., Zhang X.H.-F., et al. Targeting Brain-Adaptive Cancer Stem Cells Prohibits Brain Metastatic Colonization of Triple-Negative Breast Cancer. Cancer Res. 2018;78:2052–2064. doi: 10.1158/0008-5472.CAN-17-2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu X., Taftaf R., Kawaguchi M., Chang Y.-F., Chen W., Entenberg D., Zhang Y., Gerratana L., Huang S., Patel D.B., et al. Homophilic CD44 Interactions Mediate Tumor Cell Aggregation and Polyclonal Metastasis in Patient-Derived Breast Cancer Models. Cancer Discov. 2019;9:96–113. doi: 10.1158/2159-8290.CD-18-0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Oskarsson T., Batlle E., Massagué J. Metastatic Stem Cells: Sources, Niches, and Vital Pathways. Cell Stem Cell. 2014;14:306–321. doi: 10.1016/j.stem.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Barkan D., Green J.E., Chambers A.F. Extracellular Matrix: A Gatekeeper in the Transition from Dormancy to Metastatic Growth. Eur. J. Cancer. 2010;46:1181–1188. doi: 10.1016/j.ejca.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Desgrosellier J.S., Cheresh D.A. Integrins in Cancer: Biological Implications and Therapeutic Opportunities. Nat. Rev. Cancer. 2010;10:9–22. doi: 10.1038/nrc2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gao X.-L., Zhang M., Tang Y.-L., Liang X.-H. Cancer Cell Dormancy: Mechanisms and Implications of Cancer Recurrence and Metastasis. OncoTargets Ther. 2017;10:5219–5228. doi: 10.2147/OTT.S140854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee K.-L., Chen G., Chen T.-Y., Kuo Y.-C., Su Y.-K. Effects of Cancer Stem Cells in Triple-Negative Breast Cancer and Brain Metastasis: Challenges and Solutions. Cancers. 2020;12:2122. doi: 10.3390/cancers12082122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sheridan C., Kishimoto H., Fuchs R.K., Mehrotra S., Bhat-Nakshatri P., Turner C.H., Goulet R., Badve S., Nakshatri H. CD44+/CD24− Breast Cancer Cells Exhibit Enhanced Invasive Properties: An Early Step Necessary for Metastasis. Breast Cancer Res. 2006;8:R59. doi: 10.1186/bcr1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Abraham B.K., Fritz P., McClellan M., Hauptvogel P., Athelogou M., Brauch H. Prevalence of CD44+/CD24−/Low Cells in Breast Cancer May Not Be Associated with Clinical Outcome but May Favor Distant Metastasis. Clin. Cancer Res. 2005;11:1154–1159. [PubMed] [Google Scholar]
- 67.Lin Y., Zhong Y., Guan H., Zhang X., Sun Q. CD44+/CD24− Phenotype Contributes to Malignant Relapse Following Surgical Resection and Chemotherapy in Patients with Invasive Ductal Carcinoma. J. Exp. Clin. Cancer Res. 2012;31:59. doi: 10.1186/1756-9966-31-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Adamczyk A., Niemiec J.A., Ambicka A., Mucha-Małecka A., Mituś J., Ryś J. CD44/CD24 as Potential Prognostic Markers in Node-Positive Invasive Ductal Breast Cancer Patients Treated with Adjuvant Chemotherapy. J. Mol. Histol. 2014;45:35–45. doi: 10.1007/s10735-013-9523-6. [DOI] [PubMed] [Google Scholar]
- 69.Chen Y., Song J., Jiang Y., Yu C., Ma Z. Predictive Value of CD44 and CD24 for Prognosis and Chemotherapy Response in Invasive Breast Ductal Carcinoma. Int. J. Clin. Exp. Pathol. 2015;8:11287–11295. [PMC free article] [PubMed] [Google Scholar]
- 70.Collina F., Di Bonito M., Li Bergolis V., De Laurentiis M., Vitagliano C., Cerrone M., Nuzzo F., Cantile M., Botti G. Prognostic Value of Cancer Stem Cells Markers in Triple-Negative Breast Cancer. BioMed Res. Int. 2015;2015:158682. doi: 10.1155/2015/158682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang H., Wang L., Song Y., Wang S., Huang X., Xuan Q., Kang X., Zhang Q. CD44+/CD24− Phenotype Predicts a Poor Prognosis in Triple-Negative Breast Cancer. Oncol. Lett. 2017;14:5890–5898. doi: 10.3892/ol.2017.6959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ma F., Li H., Li Y., Ding X., Wang H., Fan Y., Lin C., Qian H., Xu B. Aldehyde Dehydrogenase 1 (ALDH1) Expression Is an Independent Prognostic Factor in Triple Negative Breast Cancer (TNBC) Medicine. 2017;96:e6561. doi: 10.1097/MD.0000000000006561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lee J.S., Kim W.G. Cutaneous Metastases of Breast Cancer during Adjuvant Chemotherapy Correlates with Increasing CD44+/CD24− and ALDH-1 Expression: A Case Report and Literature Review. Stem Cell Investig. 2018;5:7. doi: 10.21037/sci.2018.03.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rabinovich I., Sebastião A.P.M., Lima R.S., Urban C.D.A., Junior E.S., Anselmi K.F., Elifio-Esposito S., De Noronha L., Moreno-Amaral A.N. Cancer Stem Cell Markers ALDH1 and CD44+/CD24− Phenotype and Their Prognosis Impact in Invasive Ductal Carcinoma. Eur. J. Histochem. 2018;62:2943. doi: 10.4081/ejh.2018.2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Althobiti M., El Ansari R., Aleskandarany M., Joseph C., Toss M.S., Green A.R., Rakha E.A. The Prognostic Significance of ALDH1A1 Expression in Early Invasive Breast Cancer. Histopathology. 2020;77:437–448. doi: 10.1111/his.14129. [DOI] [PubMed] [Google Scholar]
- 76.Nasr M., Farghaly M., Elsaba T., El-Mokhtar M., Radwan R., Elsabahy M., Abdelkareem A., Fakhry H., Mousa N. Resistance of Primary Breast Cancer Cells with Enhanced Pluripotency and Stem Cell Activity to Sex Hormonal Stimulation and Suppression. Int. J. Biochem. Cell Biol. 2018;105:84–93. doi: 10.1016/j.biocel.2018.10.005. [DOI] [PubMed] [Google Scholar]
- 77.Cheng C.-C., Shi L.-H., Wang X.-J., Wang S.-X., Wan X.-Q., Liu S.-R., Wang Y.-F., Lu Z., Wang L.-H., Ding Y. Stat3/Oct-4/c-Myc Signal Circuit for Regulating Stemness-Mediated Doxorubicin Resistance of Triple-Negative Breast Cancer Cells and Inhibitory Effects of WP1066. Int. J. Oncol. 2018;53:339–348. doi: 10.3892/ijo.2018.4399. [DOI] [PubMed] [Google Scholar]
- 78.Doherty M.R., Cheon H., Junk D.J., Vinayak S., Varadan V., Telli M.L., Ford J.M., Stark G.R., Jackson M.W. Interferon-Beta Represses Cancer Stem Cell Properties in Triple-Negative Breast Cancer. Proc. Natl. Acad. Sci. USA. 2017;114:13792–13797. doi: 10.1073/pnas.1713728114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hussain S.F., Kong L.-Y., Jordan J., Conrad C., Madden T., Fokt I., Priebe W., Heimberger A.B. A Novel Small Molecule Inhibitor of Signal Transducers and Activators of Transcription 3 Reverses Immune Tolerance in Malignant Glioma Patients. Cancer Res. 2007;67:9630–9636. doi: 10.1158/0008-5472.CAN-07-1243. [DOI] [PubMed] [Google Scholar]
- 80.Kong L.-Y., Wei J., Sharma A.K., Barr J., Abou-Ghazal M.K., Fokt I., Weinberg J., Rao G., Grimm E., Priebe W., et al. A Novel Phosphorylated STAT3 Inhibitor Enhances T Cell Cytotoxicity against Melanoma through Inhibition of Regulatory T Cells. Cancer Immunol. Immunother. 2009;58:1023–1032. doi: 10.1007/s00262-008-0618-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zöller M. CD44: Can a Cancer-Initiating Cell Profit from an Abundantly Expressed Molecule? Nat. Rev. Cancer. 2011;11:254–267. doi: 10.1038/nrc3023. [DOI] [PubMed] [Google Scholar]
- 82.Yang C., He Y., Zhang H., Liu Y., Wang W., Du Y., Gao F. Selective Killing of Breast Cancer Cells Expressing Activated CD44 Using CD44 Ligand-Coated Nanoparticles in Vitro and in Vivo. Oncotarget. 2015;6:15283–15296. doi: 10.18632/oncotarget.3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dittmer J. Breast Cancer Stem Cells: Features, Key Drivers and Treatment Options. Semin. Cancer Biol. 2018;53:59–74. doi: 10.1016/j.semcancer.2018.07.007. [DOI] [PubMed] [Google Scholar]
- 84.Pan M., Li M., Guo M., Zhou H., Xu H., Zhao F., Mei F., Xue R., Dou J. Knockdown of ALDH1A3 Reduces Breast Cancer Stem Cell Marker CD44 via the MiR-7-TGFBR2-Smad3-CD44 Regulatory Axis. Exp. Ther. Med. 2021;22:1093. doi: 10.3892/etm.2021.10527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Loi S., Sirtaine N., Piette F., Salgado R., Viale G., Van Eenoo F., Rouas G., Francis P., Crown J.P.A., Hitre E., et al. Prognostic and Predictive Value of Tumor-Infiltrating Lymphocytes in a Phase III Randomized Adjuvant Breast Cancer Trial in Node-Positive Breast Cancer Comparing the Addition of Docetaxel to Doxorubicin with Doxorubicin-Based Chemotherapy: BIG 02-98. J. Clin. Oncol. 2013;31:860–867. doi: 10.1200/JCO.2011.41.0902. [DOI] [PubMed] [Google Scholar]
- 86.Dieci M.V., Mathieu M.C., Guarneri V., Conte P., Delaloge S., Andre F., Goubar A. Prognostic and Predictive Value of Tumor-Infiltrating Lymphocytes in Two Phase III Randomized Adjuvant Breast Cancer Trials. Ann. Oncol. 2015;26:1698–1704. doi: 10.1093/annonc/mdv239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Loi S., Michiels S., Adams S., Loibl S., Budczies J., Denkert C., Salgado R. The Journey of Tumor-Infiltrating Lymphocytes as a Biomarker in Breast Cancer: Clinical Utility in an Era of Checkpoint Inhibition. Ann. Oncol. 2021;32:1236–1244. doi: 10.1016/j.annonc.2021.07.007. [DOI] [PubMed] [Google Scholar]
- 88.Krishnamurthy N., Kurzrock R. Targeting the Wnt/Beta-Catenin Pathway in Cancer: Update on Effectors and Inhibitors. Cancer Treat. Rev. 2018;62:50–60. doi: 10.1016/j.ctrv.2017.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jang G.-B., Hong I.-S., Kim R.-J., Lee S.-Y., Park S.-J., Lee E.-S., Park J.H., Yun C.-H., Chung J.-U., Lee K.-J., et al. Wnt/β-Catenin Small-Molecule Inhibitor CWP232228 Preferentially Inhibits the Growth of Breast Cancer Stem-like Cells. Cancer Res. 2015;75:1691–1702. doi: 10.1158/0008-5472.CAN-14-2041. [DOI] [PubMed] [Google Scholar]
- 90.Pohl S.-G., Brook N., Agostino M., Arfuso F., Kumar A.P., Dharmarajan A. Wnt Signaling in Triple-Negative Breast Cancer. Oncogenesis. 2017;6:e310. doi: 10.1038/oncsis.2017.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tamagnone L., Zacchigna S., Rehman M. Taming the Notch Transcriptional Regulator for Cancer Therapy. Molecules. 2018;23:431. doi: 10.3390/molecules23020431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Falk R., Falk A., Dyson M.R., Melidoni A.N., Parthiban K., Young J.L., Roake W., McCafferty J. Generation of Anti-Notch Antibodies and Their Application in Blocking Notch Signalling in Neural Stem Cells. Methods. 2012;58:69–78. doi: 10.1016/j.ymeth.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Habib J.G., O’Shaughnessy J.A. The Hedgehog Pathway in Triple-Negative Breast Cancer. Cancer Med. 2016;5:2989–3006. doi: 10.1002/cam4.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Infante P., Alfonsi R., Botta B., Mori M., Di Marcotullio L. Targeting GLI Factors to Inhibit the Hedgehog Pathway. Trends Pharmacol. Sci. 2015;36:547–558. doi: 10.1016/j.tips.2015.05.006. [DOI] [PubMed] [Google Scholar]
- 95.Sirkisoon S.R., Carpenter R.L., Rimkus T., Miller L., Metheny-Barlow L., Lo H.-W. EGFR and HER2 Signaling in Breast Cancer Brain Metastasis. Front. Biosci. (Elite Ed.) 2016;8:245–263. doi: 10.2741/E765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Carey L.A., Rugo H.S., Marcom P.K., Mayer E.L., Esteva F.J., Ma C.X., Liu M.C., Storniolo A.M., Rimawi M.F., Forero-Torres A., et al. TBCRC 001: Randomized Phase II Study of Cetuximab in Combination with Carboplatin in Stage IV Triple-Negative Breast Cancer. J. Clin. Oncol. 2012;30:2615–2623. doi: 10.1200/JCO.2010.34.5579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bramati A., Girelli S., Torri V., Farina G., Galfrascoli E., Piva S., Moretti A., Dazzani M.C., Sburlati P., Verde N.M.L. Efficacy of Biological Agents in Metastatic Triple-Negative Breast Cancer. Cancer Treat. Rev. 2014;40:605–613. doi: 10.1016/j.ctrv.2014.01.003. [DOI] [PubMed] [Google Scholar]
- 98.Oh E., Kim Y.-J., An H., Sung D., Cho T.-M., Farrand L., Jang S., Seo J.H., Kim J.Y. Flubendazole Elicits Anti-Metastatic Effects in Triple-Negative Breast Cancer via STAT3 Inhibition. Int. J. Cancer. 2018;143:1978–1993. doi: 10.1002/ijc.31585. [DOI] [PubMed] [Google Scholar]
- 99.Mirzania M. Approach to the Triple Negative Breast Cancer in New Drugs Area. Int. J. Hematol. Oncol. Stem Cell Res. 2016;10:115–119. [PMC free article] [PubMed] [Google Scholar]
- 100.Jabbarzadeh Kaboli P., Salimian F., Aghapour S., Xiang S., Zhao Q., Li M., Wu X., Du F., Zhao Y., Shen J., et al. Akt-Targeted Therapy as a Promising Strategy to Overcome Drug Resistance in Breast Cance—A Comprehensive Review from Chemotherapy to Immunotherapy. Pharmacol. Res. 2020;156:104806. doi: 10.1016/j.phrs.2020.104806. [DOI] [PubMed] [Google Scholar]
- 101.Vallianou N.G., Evangelopoulos A., Kazazis C. Metformin and Cancer. Rev. Diabet. Stud. 2013;10:228–235. doi: 10.1900/RDS.2013.10.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Rattan R., Ali Fehmi R., Munkarah A. Metformin: An Emerging New Therapeutic Option for Targeting Cancer Stem Cells and Metastasis. J. Oncol. 2012;2012:928127. doi: 10.1155/2012/928127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cufí S., Corominas-Faja B., Vazquez-Martin A., Oliveras-Ferraros C., Dorca J., Bosch-Barrera J., Martin-Castillo B., Menendez J.A. Metformin-Induced Preferential Killing of Breast Cancer Initiating CD44+CD24−/Low Cells Is Sufficient to Overcome Primary Resistance to Trastuzumab in HER2+ Human Breast Cancer Xenografts. Oncotarget. 2012;3:395–398. doi: 10.18632/oncotarget.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Janzer A., German N.J., Gonzalez-Herrera K.N., Asara J.M., Haigis M.C., Struhl K. Metformin and Phenformin Deplete Tricarboxylic Acid Cycle and Glycolytic Intermediates during Cell Transformation and NTPs in Cancer Stem Cells. Proc. Natl. Acad. Sci. USA. 2014;111:10574–10579. doi: 10.1073/pnas.1409844111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Jung J.-W., Park S.-B., Lee S.-J., Seo M.-S., Trosko J.E., Kang K.-S. Metformin Represses Self-Renewal of the Human Breast Carcinoma Stem Cells via Inhibition of Estrogen Receptor-Mediated OCT4 Expression. PLoS ONE. 2011;6:e28068. doi: 10.1371/journal.pone.0028068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Soundararajan R., Fradette J.J., Konen J.M., Moulder S., Zhang X., Gibbons D.L., Varadarajan N., Wistuba I.I., Tripathy D., Bernatchez C., et al. Targeting the Interplay between Epithelial-to-Mesenchymal-Transition and the Immune System for Effective Immunotherapy. Cancers. 2019;11:714. doi: 10.3390/cancers11050714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Deepak K.G.K., Vempati R., Nagaraju G.P., Dasari V.R., Nagini S., Rao D.N., Malla R.R. Tumor Microenvironment: Challenges and Opportunities in Targeting Metastasis of Triple Negative Breast Cancer. Pharmacol. Res. 2020;153:104683. doi: 10.1016/j.phrs.2020.104683. [DOI] [PubMed] [Google Scholar]
- 108.Dattilo R., Mottini C., Camera E., Lamolinara A., Auslander N., Doglioni G., Muscolini M., Tang W., Planque M., Ercolani C., et al. Pyrvinium Pamoate Induces Death of Triple-Negative Breast Cancer Stem-Like Cells and Reduces Metastases through Effects on Lipid Anabolism. Cancer Res. 2020;80:4087–4102. doi: 10.1158/0008-5472.CAN-19-1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wang Z., Jiang Q., Dong C. Metabolic Reprogramming in Triple-Negative Breast Cancer. Cancer Biol. Med. 2020;17:44–59. doi: 10.20892/j.issn.2095-3941.2019.0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Boumahdi S., de Sauvage F.J. The Great Escape: Tumour Cell Plasticity in Resistance to Targeted Therapy. Nat. Rev. Drug Discov. 2020;19:39–56. doi: 10.1038/s41573-019-0044-1. [DOI] [PubMed] [Google Scholar]
- 111.Raman D., Tiwari A.K., Tiriveedhi V., Rhoades (Sterling) J.A. Editorial: The Role of Breast Cancer Stem Cells in Clinical Outcomes. Front. Oncol. 2020;10:299. doi: 10.3389/fonc.2020.00299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Maiti A., Qi Q., Peng X., Yan L., Takabe K., Hait N.C. Class I Histone Deacetylase Inhibitor Suppresses Vasculogenic Mimicry by Enhancing the Expression of Tumor Suppressor and Anti-Angiogenesis Genes in Aggressive Human TNBC Cells. Int. J. Oncol. 2019;55:116–130. doi: 10.3892/ijo.2019.4796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhao J. Cancer Stem Cells and Chemoresistance: The Smartest Survives the Raid. Pharmacol. Ther. 2016;160:145–158. doi: 10.1016/j.pharmthera.2016.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ross A.A. Minimal Residual Disease in Solid Tumor Malignancies: A Review. J. Hematother. 1998;7:9–18. doi: 10.1089/scd.1.1998.7.9. [DOI] [PubMed] [Google Scholar]
- 115.Sridharan S., Robeson M., Bastihalli-Tukaramrao D., Howard C.M., Subramaniyan B., Tilley A.M.C., Tiwari A.K., Raman D. Targeting of the Eukaryotic Translation Initiation Factor 4A Against Breast Cancer Stemness. Front. Oncol. 2019;9:1311. doi: 10.3389/fonc.2019.01311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Waldron J.A., Raza F., Le Quesne J. EIF4A Alleviates the Translational Repression Mediated by Classical Secondary Structures More than by G-Quadruplexes. Nucleic Acids Res. 2018;46:3075–3087. doi: 10.1093/nar/gky108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Khandekar D., Amara S., Tiriveedhi V. Immunogenicity of Tumor Initiating Stem Cells: Potential Applications in Novel Anticancer Therapy. Front. Oncol. 2019;9:315. doi: 10.3389/fonc.2019.00315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sridharan S., Howard C.M., Tilley A.M.C., Subramaniyan B., Tiwari A.K., Ruch R.J., Raman D. Novel and Alternative Targets Against Breast Cancer Stemness to Combat Chemoresistance. Front. Oncol. 2019;9:1003. doi: 10.3389/fonc.2019.01003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Chen Y., Zhang Y. Application of the CRISPR/Cas9 System to Drug Resistance in Breast Cancer. Adv. Sci. 2018;5:1700964. doi: 10.1002/advs.201700964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Berger E.R., Park T., Saridakis A., Golshan M., Greenup R.A., Ahuja N. Immunotherapy Treatment for Triple Negative Breast Cancer. Pharmaceuticals. 2021;14:763. doi: 10.3390/ph14080763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hall P.E., Schmid P. Emerging Drugs for the Treatment of Triple-Negative Breast Cancer: A Focus on Phase II Immunotherapy Trials. Expert Opin. Emerg. Drugs. 2021;26:131–147. doi: 10.1080/14728214.2021.1916468. [DOI] [PubMed] [Google Scholar]
- 122.Singh S., Numan A., Maddiboyina B., Arora S., Riadi Y., Md S., Alhakamy N.A., Kesharwani P. The Emerging Role of Immune Checkpoint Inhibitors in the Treatment of Triple-Negative Breast Cancer. Drug Discov. Today. 2021;26:1721–1727. doi: 10.1016/j.drudis.2021.03.011. [DOI] [PubMed] [Google Scholar]
- 123.Bianchini G., De Angelis C., Licata L., Gianni L. Treatment Landscape of Triple-Negative Breast Cancer—Expanded Options, Evolving Needs. Nat. Rev. Clin. Oncol. 2021 doi: 10.1038/s41571-021-00565-2. [DOI] [PubMed] [Google Scholar]