Abstract
In many normal and transformed cell types, the intracellular second messenger cyclic AMP (cAMP) blocks the effects of growth factors and serum on mitogenesis, proliferation, and cell cycle progression. cAMP exerts these growth-inhibitory effects via inhibition of the mitogen-activated protein (MAP) kinase cascade. Here, using Hek293 and NIH 3T3 cells, we show that cAMP's inhibition of the MAP kinase cascade is mediated by the small G protein Rap1. Activation of Rap1 by cAMP induces the association of Rap1 with Raf-1 and limits Ras-dependent activation of ERK. In NIH 3T3 cells, Rap1 is required not only for cAMP's inhibition of ERK activation but for inhibition of cell proliferation and mitogenesis as well.
Hormones that elevate intracellular cyclic adenosine monophosphate (cAMP) have a wide range of cell-type-specific effects on cell growth and differentiation (25, 75). In many cell types, cAMP inhibits the physiological actions of growth factors (45) and blocks the transformation phenotype in selected malignant cells (13). For example, in fibroblasts and smooth muscle cells, cAMP inhibits mitogen-activated protein (MAP) kinase activation by growth factors (17, 36) and during anchorage-independent cell growth (42). These growth-inhibitory actions are thought to be mediated by G protein pathways that regulate cAMP (13, 53) and the cAMP-dependent protein kinase PKA (45).
One of the ways that PKA can oppose the actions of growth factors is to block growth factor activation of the MAP kinase cascade. The mechanism of cAMP action has been proposed in a number of model systems including adipocytes (88), smooth muscle cells (36), and fibroblasts (17). MAP kinases (or ERKs [extracellular signal-regulated kinases]) are required for a broad array of biological processes, including two that are tightly linked to cellular transformation: mitogenesis (57, 97) and anchorage-independent cell growth (42). ERKs are required for the proliferative actions of growth factors in many cell types (8, 19, 60, 64) through multiple mechanisms (37), including the regulation of the levels of critical cell cycle proteins (108) including cyclin D1 (50, 58) and p27kip1 (110). ERKs are activated by growth factors through consecutive cascades of tyrosine and serine/threonine phosphorylations (20). The activation of the small G protein Ras serves as a link between this phosphorylation cascade and the receptors to which growth factors bind (20, 59). This cascade is initiated by Ras's association with (103, 117) and activation of Raf-1, a ubiquitously expressed protein kinase (2, 68). Raf-1, in turn, activates MAP kinase kinase (mitogen and extracellular signal-regulated kinase [MEK]), which then activates ERK (20).
It has previously been established that cAMP's inhibition of ERKs maps to a site downstream of Ras and upstream of Raf-1 (17). cAMP-dependent inhibition of the MAP kinase cascade requires PKA (36, 88). MEK and ERK are not targets of PKA, nor is Ras activation by growth factors itself a target of PKA inhibition (10, 17, 36, 111). In contrast, Raf-1 is both phosphorylated and inhibited by PKA in vivo. Thus, direct phosphorylation of Raf-1 by PKA has been proposed to be the site of action of PKA's antimitogenic effects (111). Two potential sites for PKA phosphorylation on Raf-1 have received significant attention: serine 43 (70, 111) and serine 621 (38, 67). Phosphorylation of serine 43 has been shown to interfere with Raf-1's interactions with Ras in vitro (111). Although the serine 43 site may participate in the inhibitory actions of other kinases, for example, PKG (92), recent studies have shown that phosphorylation of this site is not required for PKA's inhibition of Raf-1 signaling (89). Serine 621 phosphorylation has also been proposed to be a site of PKA-dependent inhibition of Raf-1 activity (38, 67). However, the ability of PKA to phosphorylate serine 621 in vivo (67) has recently been challenged (89). Therefore, it is likely that additional mechanisms of PKA's inhibition of Raf-1 may play a role in regulating the action of this kinase.
Another candidate protein that may function to antagonize Ras-dependent activation of Raf-1 is the small G protein Rap1. Rap1 was first cloned based on its ability to revert Ras-dependent transformation of fibroblasts and was initially named Krev-1 (Kirsten Ras revertant 1) (54). It shares 50% amino acid sequence homology with Ras, which is greatest in the effector and the GTPase domains. Two human proteins, Rap1a and Rap1b, share 97% homology within their amino acid sequence (79), and both antagonize Ras-induced activation of mitogenesis and MAP kinase in multiple cell types (12, 18). For example, the constitutively active mutant of Rap1b, RapV12, has been shown to block Ras-dependent activation of ERK2 in Rat-1 fibroblasts (18) and to potentiate the transformation-reverting effect of Rap1 (54). Although Rap1 can be activated by cAMP (23, 49) and is phosphorylated by PKA (3, 4), the role of Rap1 in cAMP-dependent growth inhibition has not been examined. The ability of Rap1 to block Ras-dependent signals to ERK has generally been examined in studies using transfected Rap1 proteins (18, 44, 54, 61). Indeed, it has been suggested that endogenous Rap1 does not antagonize Ras signaling (9, 118, 119). In this study, we examined whether endogenous Rap1 is required for cAMP's inhibitory actions on the MAP kinase cascade and cellular proliferation.
MATERIALS AND METHODS
Materials.
Antibodies to Rap1, Raf-1, ERK2 (c-14), and c-Myc (9E10) and agarose-conjugated antibodies to hemagglutinin (HA) (F-7) and Myc-ERK were purchased from Santa Cruz Biotechnology Inc (Santa Cruz, Calif.). Antibodies to HA (12CA5) were purchased from Boehringer Mannheim (Indianapolis, Ind.). Anti-Ras antibody was purchased from Upstate Biotechnology (Lake Placid, N.Y.). Phosphorylation-specific AKT antibodies that recognize phosphorylated AKT (pAKT)/PKB at threonine 308 was purchased from New England Biolabs (Beverly, Mass.). Phosphorylation-specific ERK antibodies that recognize phosphorylated ERK1 and ERK2 (pERK1/2) at residues threonine 183 and tyrosine 185 were purchased from New England Biolabs. Isoproterenol, Flag (M2) antibody, and 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrozolium bromide (MTT) were purchased from Sigma (St. Louis, Mo.). Forskolin, PD98059, epidermal growth factor (EGF), and N-[2-(p-bromocinnamylamino) ethyl]-5-isoquinolinesulfonamide (H89) were purchased from Calbiochem (Riverside, Calif.). Nickel-nitrilotriacetic acid-agarose (Ni-NTA-agarose) was purchased from Qiagen Inc. (Chatsworth, Calif.). [3H]thymidine was purchased from NEN Life Science Products (Boston, Mass.).
Cell culturing conditions and treatments.
Hek293 and NIH 3T3 cells were cultured in Dulbecco modified Eagle medium (DMEM) plus 10% fetal calf serum at 37°C in 5% CO2. Cells were maintained in serum-free DMEM for 16 h at 37°C in 5% CO2 prior to treatment with various reagents for coimmunoprecipitation assays, Western blotting, and MTT and [3H]thymidine labeling. In all experiments, cells were treated with isoproterenol (10 μM), EGF (100 ng/ml), or forskolin (10 μM) for 5 min unless otherwise indicated. Where indicated, cells were pretreated with isoproterenol or forskolin for 5 min and then stimulated with EGF for 5 min. H89 (10 μM) and PD98059 (10 μM) were added to cells 20 min prior to treatment.
Western blotting.
Cell lysates were prepared as described (87, 105). Cell lysate protein concentrations were quantified by the Bradford protein assay. For detection of Raf-1, Myc–Raf-1, ERK2, Myc-ERK2, HA, Flag, Ras, Rap1, pAKT, and pERK1/2, equal protein amounts of cell lysate per treatment condition were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), blotted onto polyrinylidene difluoride (PVDF) (Millipore Corporation, Bedford, Mass.) membranes, and probed with the corresponding antibodies according to the manufacturer's guidelines.
Plasmids and transfections.
Seventy to eighty percent confluent Hek293 or NIH 3T3 cells were cotransfected with the indicated cDNAs by using a Lipofectamine kit (Gibco BRL) according to the manufacturer's instructions. The control vector, pcDNA3 (Invitrogen Corp.), was included in each set of transfections to ensure that each plate received the same amount of DNA. The control vector pMACS 14.1 (Miltenyi Biotec, Auburn, Calif.) was used for MACS (magnetic bead cell sorting) selection. Following transfection, cells were allowed to recover in serum containing medium for 24 h. Cells were then starved overnight in serum-free DMEM before treatment and lysis.
Nickel affinity chromatography.
NIH 3T3 and Hek293 cells were transfected using Lipofectamine reagent with polyhistidine-tagged Rap1 (His-Rap1 and His-RapV12) and Ras (His-Ras) as previously described (87). Briefly, cells were lysed in ice-cold buffer containing 1% NP-40, 10 mM Tris (pH 8.0), 20 mM NaCl, 30 mM MgCl2, 1 mM phenylmethylsulfonyl fluoride, and aprotinin (0.5 mg/ml), and supernatants were prepared by low-speed centrifugation. Transfected His-tagged proteins were precipitated from supernatants containing equal amounts of protein using Ni-NTA-agarose and washed with 20 mM imidazole in lysis buffer and eluted with 500 mM imidazole and 5 mM EDTA in phosphate-buffered saline. The eluates containing His-tagged proteins were separated by SDS-PAGE, and Raf-1 proteins were detected by Western blotting (87, 105). Western blots for Raf-1 were scanned and analyzed using NIH Image (version 1.57) to quantitate the amount of Raf-1 protein. Equal amounts of His-Rap1 and His-Ras were confirmed by Western blotting. Results are presented as means ± standard deviations (SD) of multiple experiments.
Affinity assay for Rap1 activation in NIH 3T3 cells.
A glutathione S-transferase (GST) fusion protein of the Rap1 binding domain of RalGDS was expressed in Escherichia coli following induction by isopropyl-1thio-β-d-galactopyranoside (GST-RalGDS was a gift from J. L. Bos, Utrecht University, Utrecht, The Netherlands to P. J. S. Stork). NIH 3T3 cells grown as described above were stimulated at 37°C for the indicated times and immediately lysed in ice-cold lysis buffer (50 mM Tris-HCl [pH 8.0], 10% glycerol, 1% NP-40, 200 mM NaCl, 2.5 mM MgCl2, 1 mM phenylmethylsulfonyl fluoride, 1 μM leupeptin, 10 μg of soybean trypsin inhibitor/ml, 10 mM NaF, 0.1 μM aprotinin, 1 mM NaVO4). Active Rap1 was isolated as described by Franke et al. (31). Briefly, cell lysates were cleared by centrifugation, and equivalent amounts of supernatants (500 μg) were incubated with GST-RalGDS Rap1 binding domain precoupled to glutathione beads. Following a 1-h incubation at 4°C, beads were pelleted and rinsed threes times with ice-cold lysis buffer, and protein was eluted from the beads in 2× Laemmli buffer and applied to an SDS–12% polyacrylamide gel. Proteins were transferred to a PVDF membrane, blocked in 5% milk for 1 h, and probed with either Rap1/Krev-1 or Flag antibody overnight at 4°C, followed by a horseradish peroxidase-conjugated anti-rabbit secondary antibody (or anti-mouse secondary for anti-Flag blots). Proteins were detected by enhanced chemiluminescence.
Affinity assay for Ras activation in NIH 3T3 cells.
NIH 3T3 cells grown as described above were stimulated at 37°C for the indicated times and immediately lysed in ice-cold lysis buffer. Activated Ras was isolated according to the manufacturer's recommended protocol from stimulated lysates, using agarose-coupled GST-Raf1 Ras-binding domain (RBD) provided in an Upstate Biotechnology Ras activation assay kit. Proteins were eluted from the beads in 2× Laemmli buffer and applied to an SDS–12% polyacrylamide gel. Proteins were transferred to a PVDF membrane, blocked in 5% milk for 1 h, and probed with either Ras or Flag antibody as indicated overnight at 4°C, followed by a horseradish peroxidase-conjugated anti-mouse secondary antibody. Proteins were detected by enhanced chemiluminescence.
MACS selection of transfected NIH 3T3 cells.
NIH 3T3 cells were transfected with either Flag-Rap1, Rap1GAP1 (see Results), or the pMACS 14.1 control vector along with the pMACS Kk.II positive selection plasmid as specified by the manufacturers (Miltenyi Biotec) (46, 96). Cells were also transfected with green fluorescent protein to monitor both selection and protein expression. Briefly, cells were transfected using a Lipofectamine kit (Gibco BRL) for 5 h and then allowed to recover for 24 h. Cells were prepared according to the manufacturer's guidelines and incubated with MACSelect Kk microbeads for 20 min with gentle rocking. Cells were then passed over sterile-prepared MACS separation columns (type BS) while using a Vario MACS separation magnet. Columns were washed four times with phosphate-buffered saline supplemented with 0.5% bovine serum albumin and 5 mM EDTA. Columns were then removed from the Vario MACS separation magnet, and cells were eluted in DMEM plus 10% fetal calf serum and recovered on 10-cm-diameter plates. Selected cells were then used for Rap1 assay, the MTT assay, and [3H]thymidine incorporation (described below) at the indicated times.
MTT assay for cell proliferation in NIH 3T3 cells.
NIH 3T3 cells were grown as described above, serum starved overnight and plated onto 96-well plates. Cells were then treated and incubated as indicated; 2.5 h prior to lysis, 20 μl of sterile MTT (2.5 μg/ml) was added, and the cells were allowed to incubate at 37°C. At the appropriate time, cells were lysed and proteins were solubilized in 50% (vol/vol) H2O and N,N,-dimethylformamide containing 20% SDS, 0.5% 80% acetic acid, and 0.4% 1 M HCl. Results were read using a microplate reader and are presented as the difference between optical densities at 570 and 650 nm.
Thymidine uptake assay for DNA synthesis and proliferation in NIH 3T3 cells.
Serum-starved NIH 3T3 cells were plated at 2 × 104 cells/well on 96-well plates. Cells were then treated and incubated with serum or EGF in the presence or absence of forskolin pretreatment. The cells were labeled with [3H]thymidine (1 μCi/well; NEN) 12 h prior to lysis. Cells were then lysed in 50% (vol/vol) H2O and N,N,-dimethylformamide containing 20% SDS, 0.5% 80% acetic acid, and 0.4% 1 M HCl. Unincorporated counts were removed with an automated harvester, and incorporated counts were harvested onto filters. The amount of [3H]thymidine incorporated was determined with an automated TopCount NXT version 2.13 scintillation and luminescence counter and software (Packard Instrument Co., Meriden, Conn.).
RESULTS
Rap1 inhibition of the association of Ras and Raf-1 in Hek293 cells.
EGF stimulates cell growth in a variety of cell types, via activation of a Ras-dependent signaling cascade to ERKs. Following stimulation by growth factors, Ras is activated and subsequently recruits Raf-1 to initiate the MAP kinase cascade. Association with Ras can be used as an assay for Raf-1 activation. The association of endogenous Raf-1 with polyhistidine-tagged Ras (His-Ras) was measured within lysates harvested from transfected cells by nickel affinity chromatography, elution, and Western blotting for Raf-1. The Raf-1 antibody is specific for Raf-1 and does not cross-react with B-Raf isoforms (87). In Hek293 cells, EGF, but not isoproterenol, stimulated the recruitment of endogenous Raf-1 to Ras. Importantly, EGF's recruitment of Raf-1 to Ras was inhibited by isoproterenol (Fig. 1A), demonstrating that isoproterenol's inhibition of Raf-1 activation occurs at the level of Ras recruitment of Raf-1, consistent with previous reports in other cell types (17, 36, 111). In Hek293 cells, isoproterenol can signal through endogenous β2-adrenergic receptors to activate PKA via Gs-coupled signaling pathways (21, 87). In these cells, isoproterenol can activate Ras, but this action does not require PKA (87). In contrast, isoproterenol's inhibition of Raf-1–Ras association was completely blocked by pretreatment with H89, an inhibitor of the cAMP-dependent protein kinase PKA (14) (Fig. 1A), suggesting the involvement of PKA via the intracellular second messenger cAMP.
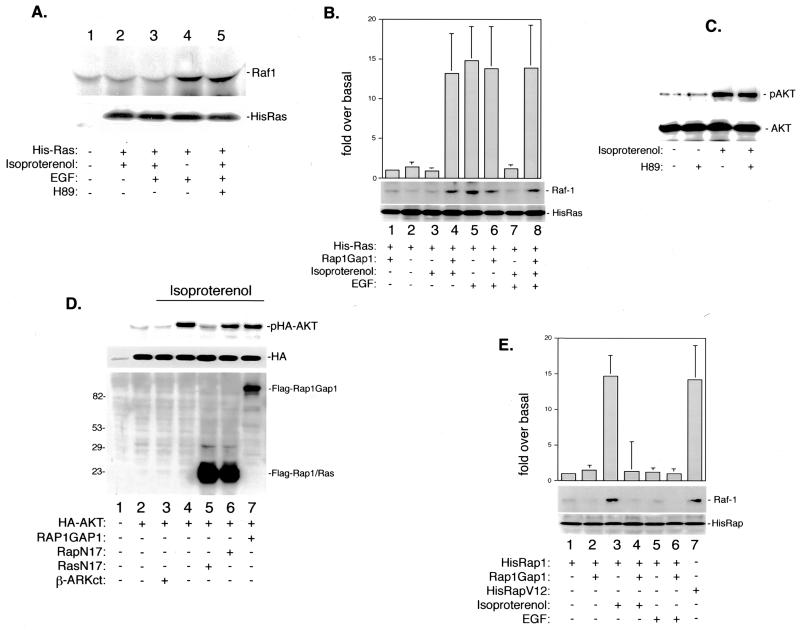
FIG. 1.
cAMP's inhibition of Ras–Raf-1 association by Rap1 in Hek293 cells. (A) Isoproterenol-mediated inhibition of Ras–Raf-1 association requires PKA. Cells were serum starved overnight and treated with isoproterenol and/or EGF or pretreated with isoproterenol and/or H89 and then treated with EGF. Top, Western blot of Raf-1 within purified eluates, prepared from equal amounts of His-Ras-transfected Hek293 cell lysates. Bottom, Western blot of Ras within purified eluates, demonstrating equal amounts of His-Ras proteins within Hek293 cell lysates. (B) Rap1 mediates the ability of isoproterenol to block Ras's association with endogenous Raf-1. Hek293 cells were transfected with cDNAs encoding His-Ras and Rap1GAP1 as indicated, serum starved, and stimulated with isoproterenol or EGF or pretreated with isoproterenol and then treated with EGF. Lysates were then purified using an Ni-NTA-agarose column, and eluates were examined for the presence of either Raf-1 or His-Ras by Western blotting. The bar graph represents data (±SD) from three pulldown experiments. Bars indicate fold Raf-1 binding over basal. (C) Isoproterenol induced phosphorylation of AKT does not require PKA. Hek293 cells were stimulated with isoproterenol in the presence or absence of H89 pretreatment, as indicated. Lysates containing equivalent amounts of protein were probed by Western blotting for pAKT (top) or AKT (bottom). (D) Rap1 does not regulate AKT activation by Ras and isoproterenol. Hek293 cells were cotransfected with the indicated cDNAs, serum starved, and stimulated with isoproterenol. Lysates containing equivalent amounts of protein were immunoprecipitated using HA-conjugated antibody, and purified proteins were probed by Western blotting for either pAKT (top) or HA as a loading control for AKT (middle). Lysates were also probed by Western blotting for Flag-tagged proteins, Ras, Rap1, and Rap1GAP1 (bottom). Sizes are indicated in kilodaltons. (E) Active Rap1 binds to endogenous Raf-1. Hek293 cells were transfected with cDNAs encoding His-Rap1, HisRapV12, and Rap1GAP1, serum starved, and stimulated with isoproterenol or EGF. Lysates were then purified using a nickel column, and eluates were examined for the presence of either Raf-1- or His-Rap-containing proteins (Rap1 or RapV12) by Western blotting. The bar graph represents data (±SD) from three pulldown experiments. Bars indicate fold Raf-1 binding over basal.
Previously, it was shown that isoproterenol and cAMP could activate the small G protein Rap1 (87, 106). Endogenous Rap1 activity can be selectively inhibited by the expression of Rap1GAP1 (80), a Rap1-specific GTPase-activating protein that can block Rap1 pathways when expressed ectopically within cells (6, 85, 113). To test the hypothesis that Rap1 was involved in the inhibition of Raf-1, we examined Raf-1 recruitment to Ras in cells transfected with Rap1GAP1. EGF stimulated Raf-1 recruitment to Ras, which was unaffected by Rap1 inhibition (Fig. 1B, lanes 5 and 6), suggesting that endogenous Rap1 does not participate in EGF signaling to Ras and Raf-1 in these cells. However, isoproterenol's ability to inhibit EGF's actions was completely blocked following Rap1 inhibition (Fig. 1B, lanes 7 and 8), demonstrating that endogenous Rap1 mediates isoproterenol's inhibitory effects in these cells. Since Rap1 is activated by isoproterenol through the actions of cAMP and PKA (87), these results show that inhibition of Raf-1 recruitment to Ras by cAMP and PKA required Rap1. Interestingly, although isoproterenol by itself was incapable of stimulating the recruitment of Raf-1 to Ras, blocking Rap1 action with Rap1GAP1 revealed isoproterenol's ability to recruit Raf-1 to Ras (Fig. 1B, lanes 3 and 4). We have previously shown that despite isoproterenol's ability to activate Ras in these cells, isoproterenol does not permit signaling from activated Ras to Raf-1 and ERK (87). The data presented here demonstrate that isoproterenol's activation of Rap1 uncouples isoproterenol's activation of Ras signaling from Raf-1 and provide an explanation for isoproterenol's inability to activate Raf-1 while activating Ras.
The mechanism of Rap1 antagonism of Ras-dependent signaling pathways is not well understood. To determine whether or not Rap1 antagonism of Ras function could be generalizable to other Ras effectors, we examined another Ras effector in these cells, phosphoinositide 3-kinase (PI3-K) (26). PI3-K activation can be monitored by the phosphorylation of the kinase AKT (PKB) at threonine 308, through the action of PDK1 (33, 77). Isoproterenol activated AKT in a PKA-independent manner (Fig. 1C). This suggests that the activation of at least one Ras effector (Raf-1) is blocked by PKA while that of another (PI3-K) is not. We have shown that isoproterenol's activation of Ras in these cells is PKA independent (87), and others have shown that this requires the action of the βγ subunits of the heterotrimeric G proteins that couple to the β-adrenergic receptor (55). To determine whether the βγ subunits of G proteins are necessary for AKT phosphorylation in response to isoproterenol, a peptide derived from the carboxyl terminus of the β-adrenergic receptor kinase 1 (β-ARKct) that sequesters βγ subunits (56) was transfected into cells. β-ARKct has previously been shown to block βγ-dependent signaling downstream of G protein-coupled receptors (62). AKT phosphorylation was blocked by β-ARKct expression (Fig. 1D), which suggests that like isoproterenol's activation of Ras, isoproterenol's activation of AKT is not mediated by Gsα signaling to PKA but utilizes the βγ subunits instead. The Ras dependence of isoproterenol's activation of AKT was confirmed by the ability of RasN17, an interfering mutant of Ras (90), to completely block the activation of AKT. In contrast, Rap1GAP1 (Fig. 1D) did not block this activation. The inability of Rap1 to interfere with other Ras-dependent signaling pathways suggests that Rap1 antagonism is selective for Raf-1.
One model for Rap1's action is that it binds to Raf-1, sequestering it from Ras (72). Indeed, as isoproterenol blocked Raf-1's recruitment to Ras, it increased Raf-1 binding to Rap1 (Fig. 1E, lane 3). Activation of Rap1 was necessary and sufficient for this effect, since Rap1GAP1 blocked this effect (Fig. 1E, lane 4) and RapV12 (a constitutively active mutant of Rap1 [105]) but not wild-type Rap1 mimicked this effect (Fig. 1E, lanes 2 and 7). EGF did not trigger Raf-1 recruitment to Rap1 (Fig. 1E), nor did it stimulate GTP loading (data not shown), confirming that EGF did not activate Rap1 in these cells.
Rap1 inhibition of the association of Ras and Raf-1 in NIH 3T3 cells.
The best-studied consequence of Raf-1's recruitment to Ras is the subsequent activation of the MAP kinase kinase MEK and ERK (1, 7, 15) to stimulate cell proliferation (64, 74). However, the ability of Rap1 to antagonize the activation of MEK and ERK and cell growth is cell type specific (5, 29, 105, 112, 115) and depends on the specific Raf isoform(s) expressed in a given cell (72). For example, both cAMP (105) and Rap1 can activate B-Raf (71), a potent activator of MEK, and both cAMP and Rap1 are activators of ERK signaling in B-Raf-expressing cells (105). Hek293 cells express the 95-kDa isoform of B-Raf, which confers regulation by PKA (63, 83). In these cells, both isoproterenol and cAMP-PKA can activate ERK through Rap1 (87). To examine cAMP's inhibition of ERK in a B-Raf-negative cell type, we chose NIH 3T3 cells. These cells do not express B-Raf (105), and they have been a model system for the study of cAMP on ERKs and cell growth (13, 111).
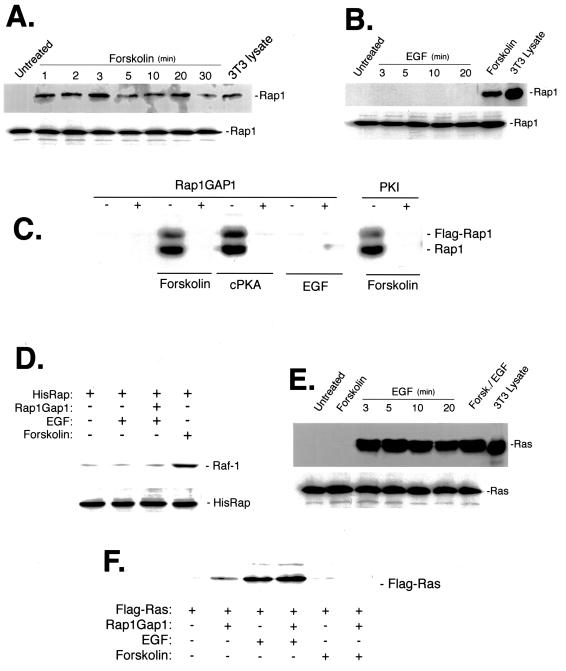
Forskolin is a potent activator of adenylyl cyclase and rapidly increases cAMP levels and PKA activity in all cell types. In NIH 3T3 cells, forskolin rapidly activated Rap1 (Fig. 2A and B), as measured by a GST-RalGDS assay (31). In contrast, EGF did not activate Rap1 (Fig. 2B and C), nor did it stimulate the association between Rap1 and Raf-1 (Fig. 2D). As an independent test for the requirement of PKA in forskolin's actions, we transfected the cDNA encoding PKI, a specific inhibitor of PKA (22). Briefly, cells were transfected with Flag-Rap1, Rap1GAP1, PKI, cPKA (catalytic subunit of PKA), or vector DNA along with cDNA encoding the truncated mouse major histocompatibility class I molecule H-2Kk, and transfected cells were separated using anti-H-2Kk coupled to MACSelect Kk microbeads (Miltenyi Biotec) as described in Materials and Methods. Importantly, either PKI or Rap1GAP1 blocked the actions of both transfected and endogenous Rap1 (Fig. 2C). These results demonstrate that cAMP's activation of endogenous Rap1 in these cells required PKA and was blocked by Rap1GAP1. Binding of GTP-Ras to the Raf-1 RBD in vitro can be used to monitor EGF's stimulation of Ras GTP loading (Fig. 2E). While EGF potently activated Ras, forskolin did not, nor did forskolin alter EGF's activation of Ras (Fig. 2E). This suggests that forskolin inhibits Ras-dependent activation of Raf-1, without affecting Ras activation (GTP loading) itself. Moreover, Rap1GAP1 did not block Ras activation (Fig. 2F), suggesting that its actions on Rap1 (Fig. 2C and D) were selective (86).
FIG. 2.
Rap1 activation by cAMP in NIH 3T3 cells. (A) cAMP stimulation activates Rap1. NIH 3T3 cells were serum starved and stimulated with forskolin for the indicated times. Lysates containing equivalent amounts of protein (500 μg) were incubated with GST-RalGDS and probed by Western blotting for active Rap1. NIH 3T3 lysate (3T3 lysate; 10μg) was used as a control to indicate the position of Rap1 (top). Total lysates were probed for Rap1 as a control for protein loading (bottom). (B) EGF does not activate Rap1 in NIH 3T3 cells. Serum-starved NIH 3T3 cells were treated with EGF for the indicated times or with forskolin (5 min). Lysates containing equivalent amounts of protein were incubated with GST-RalGDS and probed by Western blotting for active Rap1. NIH 3T3 lysate (3T3 lysate; 50 μg) was used as a control to indicate the position of Rap1 (top). Total lysates were probed for Rap1 to control for protein loading (bottom). (C) Rap1GAP1 and PKI block forskolin's activation of both transfected and endogenous Rap1. NIH 3T3 cells were transfected with Rap1GAP1, PKI, cPKA, or vector along with pMACS Kk.II, and transfected cells were positively selected as described in Materials and Methods. Transfected cells were treated with forskolin or EGF or left untreated, and Rap1 assays were performed with GST-RalGDS. Western blotting with anti-Rap1 antibodies identified activation of endogenous Rap1 (bottom) and transfected Flag-Rap1 (top). (D) EGF does not induce the association of Rap1 with endogenous Raf-1. NIH 3T3 cells were transfected with His-Rap1 and Rap1GAP1, serum starved, and stimulated with EGF or forskolin as a positive control. Lysates containing equivalent amounts of protein were then purified using a nickel column, and eluates were examined for the presence of Raf-1 (upper panel) and His-Rap1 (lower panel) by Western blotting using Raf-1 and Rap1 antisera, respectively. (E) EGF, but not cAMP, activates Ras in NIH 3T3 cells. Cells were serum starved and stimulated with forskolin or EGF for the indicated times or were pretreated with forskolin and then stimulated with EGF. Lysates containing equivalent amounts of protein were incubated with GST-Raf1 RBD and probed by Western blotting for active Ras (top). Total lysates were probed for Ras to control for protein loading (bottom). NIH 3T3 lysate (3T3 lysate; 50 μg) was used as a control to indicate the position of Ras. (F) Rap1GAP1 does not interfere with Ras activation. NIH 3T3 cells were cotransfected with Flag-Ras and Rap1GAP1 or Flag-Ras alone. Transfected cells were left untreated or treated with forskolin or EGF as indicated, and Ras assays were performed with GST-Raf1 RBD. Western blotting with Flag antibody was performed to identify activated Flag-Ras.
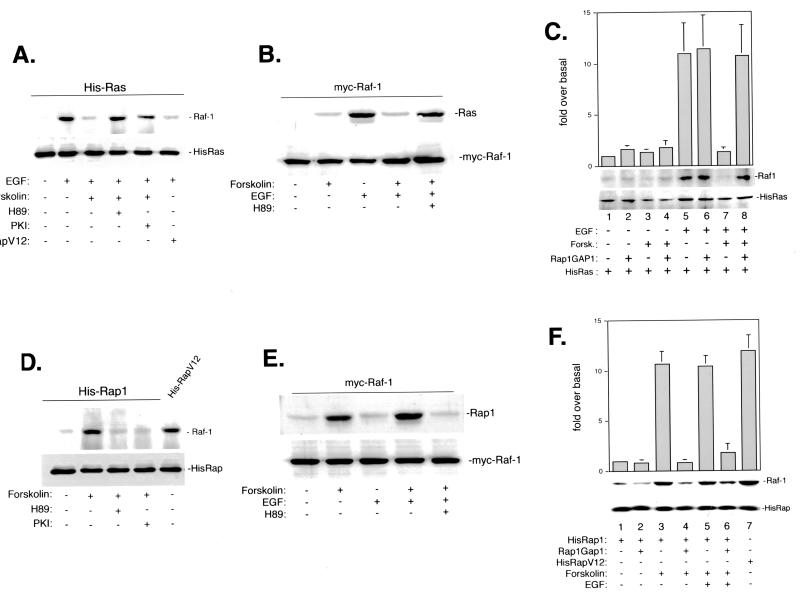
Forskolin inhibited Ras recruitment of endogenous Raf-1 in NIH 3T3 cells, as shown for Hek293 cells (Fig. 3A and C). The action of forskolin was blocked by pretreatment with H89 or transfection of PKI and was mimicked by RapV12 (Fig. 3A). Examining endogenous Ras association with Myc-tagged Raf-1 showed the same results; forskolin blocked EGF's action, and this was blocked by H89 (Fig. 3B). The requirement of Rap1 activation for forskolin's inhibition of Ras–Raf-1 association is shown in Fig. 3C. Inactivating endogenous Rap1 by expressing Rap1GAP1 in these cells prevented forskolin's inhibitory actions (Fig. 3C, lanes 7 and 8). The activation of Rap1 by forskolin not only uncoupled Raf-1 from Ras activation but also triggered the association of Rap1 with endogenous Raf-1 (Fig. 3D and F). This association required PKA, since it was blocked by either H89 or PKI (Fig. 3D). In addition, forskolin stimulated the association of endogenous Rap1 with Myc-tagged Raf-1 in the presence or absence of EGF, and this was blocked by H89 (Fig. 3E). Moreover, it required activated Rap1, as it was blocked by the ectopic expression of Rap1GAP1 (Fig. 3F) and could be mimicked by RapV12 (Fig. 3D and F). Thus, in both Hek293 and NIH 3T3 cells, cAMP's inhibition of Raf-1 binding to Ras required Rap1 and PKA.
FIG. 3.
Requirement of Rap1 for cAMP's ability to block Ras and Raf-1 association in NIH 3T3 cells. (A) cAMP blocked the association of Ras with endogenous Raf-1 via PKA. NIH 3T3 cells were transfected with cDNAs encoding His-Ras, Flag-RapV12, or PKI, serum starved, and stimulated with EGF in the presence or absence of forskolin after pretreatment with H89. Lysates were then purified using a nickel column, and eluates were examined for the presence of Raf-1 (top) or His-Ras (bottom) by Western blotting. (B) cAMP blocked the association of Raf-1 with endogenous Ras via PKA. NIH 3T3 cells were transfected with cDNA encoding Myc–Raf-1, serum starved, and stimulated with EGF in the presence or absence of forskolin after pretreatment with H89 as indicated. Lysates were then immunoprecipitated with Myc antibody and probed for Ras (top) or Myc–Raf-1 (bottom) by Western blotting. (C) cAMP blocked Ras–Raf-1 association via Rap1. NIH 3T3 cells were transfected with cDNA encoding His-Ras or Rap1GAP1, serum starved, and stimulated with forskolin or EGF or pretreated with forskolin and then treated with EGF. Lysates were then purified using a nickel column, and eluates were examined for the presence of either Raf-1 or His-Ras by Western blotting. The bar graph represents data (±SD) from three pulldown experiments. Bars indicate fold Raf-1 binding over basal (fold over basal). (D) cAMP blocked the association of Rap1 with endogenous Raf-1 via PKA. cAMP triggered the association between Rap1 and Raf-1 via PKA. NIH 3T3 cells were transfected with cDNAs encoding His-Rap1, HisRapV12, or PKI, serum starved, and stimulated with forskolin in the presence or absence of H89. Lysates were then purified using a nickel column, and eluates were examined for the presence of Raf-1 (top) or His-Rap-containing proteins (bottom) by Western blotting. (E) cAMP blocked the association of Raf-1 with endogenous Rap1 via PKA. NIH 3T3 cells were transfected with cDNA encoding Myc–Raf-1, serum starved, and stimulated with EGF in the presence or absence of forskolin after pretreated with H89 as indicated. Lysates were then immunoprecipitated with Myc antibody and probed for Rap1 (top) or Myc–Raf-1 (bottom) by Western blotting. (F) Active Rap1 binds to Raf-1. NIH 3T3 cells were transfected with cDNAs encoding His-Rap1, HisRapV12, and Rap1GAP1 as indicated, serum starved, and stimulated with forskolin or pretreated with forskolin prior to stimulation by EGF. Lysates were then purified using a nickel column, and eluates were examined for the presence of either Raf-1 or His-Rap-containing proteins (Rap1 or RapV12) by Western blotting. The bar graph represents data (±SD) from three pulldown experiments. Bars indicate fold Raf-1 binding over basal (fold over basal).
Rap1 and cAMP's inhibition of ERKs and cell proliferation in NIH 3T3 cells.
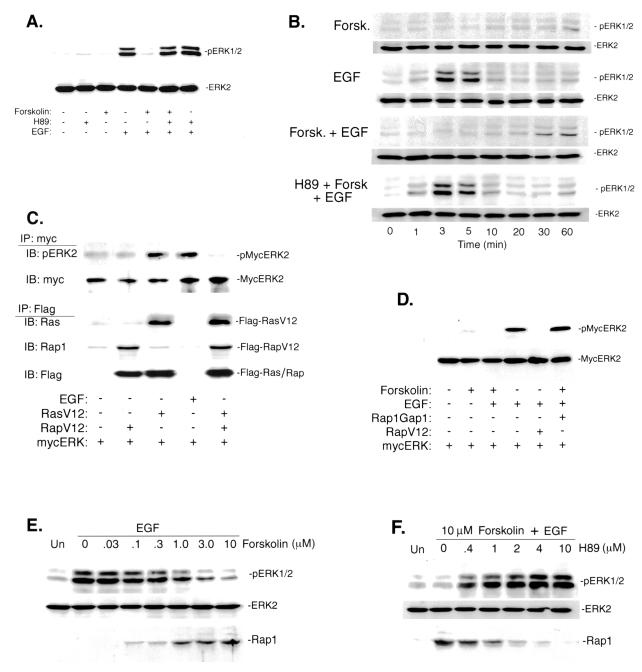
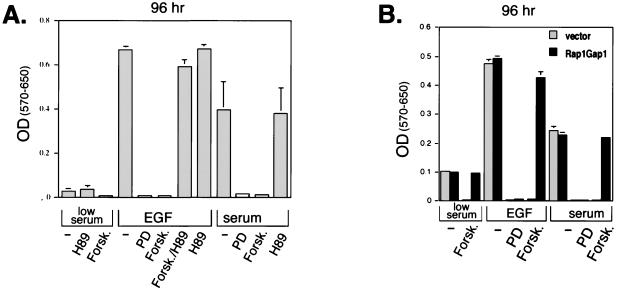
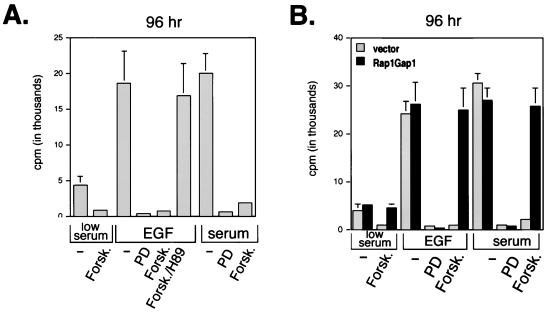
ERK activation can be measured by Western blotting using phosphospecific antibodies that recognize the activating phosphates within the ERK activation loop (116). Using this assay, forskolin inhibited activation of endogenous ERK by EGF in NIH 3T3 cells (Fig. 4A), confirming previous results (111), and this action of forskolin, but not EGF, was blocked by H89, demonstrating the specificity of H89 and confirming a role for PKA in forskolin's inhibition of ERKs. An extended time course of Forskolin's action is shown in Fig. 4B. Activated Rap1 (RapV12) did not stimulate ERKs in NIH 3T3 cells; rather, it blocked the activation of ERKs by activated Ras (RasV12) (Fig. 4C), suggesting that overexpression of Rap1 was sufficient to antagonize Ras-dependent signals in NIH 3T3 cells, as in other cell types (18). The proteins appeared to be expressed to equivalent levels, as judged by Flag Western blotting (lower panels). To determine whether endogenous Rap1 was necessary for Forskolin's inhibition of ERK activation, we examined ERK activation in the presence and absence of Rap1GAP1. The expression of Rap1GAP1 blocked the action of forskolin to inhibit EGF-dependent activation of ERK (Fig. 4D). These data demonstrate that cAMP's activation of Rap1 mediated cAMP's block of Ras-dependent signaling to Raf-1, MEK, and ERK in NIH 3T3 cells. To compare the relative effectiveness of forskolin to activate Rap1 and inhibit ERK, we examined the dose dependency of these actions of forskolin. For both actions, forskolin showed similar 50% effective concentrations (roughly 1.0 μM) (Fig. 4E). H89 blocked both actions of forskolin with a 50% inhibitory concentration of roughly 0.4 μM (Fig. 4F), a dose that is selective for PKA (14). At the highest dose used, H89 had no effect on the PKA-independent activation of Rap1 by thrombin (data not shown) (32, 87). These results and those for assays using PKI (Fig. 3) demonstrate that in NIH 3T3 cells, PKA is required for Rap1's activation by cAMP. To determine whether Rap1 mediates cAMP's inhibition of cell growth in NIH 3T3 cells, we first confirmed that forskolin blocked EGF's proliferative actions in these cells. Both EGF and serum (10%) stimulated cell proliferation, as measured 96 h later. Similar results were seen at 24 and 48 h (data not shown). All of these proliferative responses were blocked by inhibiting MEK with PD98059 and by pretreatment with forskolin (Fig. 5A). Forskolin's actions were blocked by H89, demonstrating that PKA was required. Forskolin did not induce apoptotic changes in these cells during the course of the experiment (data not shown). Taken together, the data confirm that EGF stimulates proliferation via ERK and that cAMP-PKA inhibition of ERK activation blocks EGF's proliferative effects (Fig. 5A). To test whether Rap1 activation was required for forskolin's antiproliferative actions, we examined proliferation in cells transfected with Rap1GAP1 and positively selected as described above. In all conditions where forskolin inhibited the proliferative effects of EGF or serum, these effects were completely blocked by the expression of Rap1GAP1 (Fig. 5B). Similar results were seen examining mitogenesis, as measured by [3H]thymidine uptake (Fig. 6). The ability of EGF or serum to stimulate [3H]thymidine uptake was blocked by PD98059, demonstrating the requirement of MEK for these effects. Forskolin also blocked the mitogenic actions of both EGF and serum. This inhibition required PKA, as forskolin's antimitogenic actions were blocked by H89 (Fig. 6A). H89 had no effect on the proliferation of untreated cells. As for cell proliferation, in all conditions where forskolin inhibited the mitogenic effects of EGF or serum, these effects were completely blocked by the expression of Rap1GAP1 (Fig. 6B). These data demonstrate that the activation of endogenous Rap1 is required to mediate cAMP's inhibition of cell growth and proliferation.
FIG. 4.
cAMP blockade of EGF's activation of ERKs via Rap1. (A) cAMP's inhibition of ERK phosphorylation by EGF requires PKA. NIH 3T3 cells were serum starved and treated with forskolin, H89, or EGF or pretreated with forskolin and/or H89 and then treated with EGF, as indicated. Lysates containing equivalent amounts of protein were probed for phosphorylation of endogenous ERK (pERK1/2) as well as total ERK (ERK2) to control for loading. (B) Extended time course of forskolin's inhibition of EGF's activation of ERK. Cells were treated with EGF, forskolin (Fork.), or H89 for the times indicated and analyzed as for panel A. (C) Rap1 antagonizes Ras's activation of ERKs. NIH 3T3 cells were transfected with cDNAs encoding Myc-ERK2, Flag-RasV12, and Flag-RapV12, serum starved, and treated with EGF as indicated. Lysates containing equivalent amounts of protein were immunoprecipitated (IP) with Myc-conjugated antibody and immunoblotted (IB) by Western blotting for pERK (p-MycERK2) or Myc (MycERK2) to control for protein loading. The levels of both transfected proteins Flag-RasV12 and Flag-RapV12 are also shown, following Flag immunoprecipitation (IP) and Ras, Rap1, or Flag immunoblotting (IB), as indicated (lower panels). (D) Rap1 mediates cAMP's ability to inhibit EGF stimulation of ERK phosphorylation. NIH 3T3 cells were transfected with cDNAs encoding Myc-ERK2, RapV12, and Rap1GAP1, serum starved, and treated with forskolin or EGF or pretreated with forskolin and then treated with EGF, as indicated. Lysates containing equivalent amounts of protein were immunoprecipitated with Myc-conjugated antibody and probed by Western blotting for pERK (p-MycERK2) or Myc (MycERK2) to control for protein loading. (E) Forskolin activates Rap1 and inhibits ERKs in a dose-dependent manner. NIH 3T3 cells were serum starved, pretreated with the indicated concentrations of forskolin and then treated with EGF as indicated, or untreated (Un). Lysates containing equivalent amounts of protein were either probed for phosphorylation of endogenous ERK (pERK1/2) (top) or total ERK (ERK2) to control for protein loading (middle) or used for Rap1 activation assay (bottom). (F) H89 blocks forskolin's activation of Rap1 and its inhibition of ERKs in a dose-dependent fashion. NIH 3T3 cells were serum starved, pretreated with the indicated concentration of H89, treated with forskolin, and then stimulated with EGF as indicated. Lysates containing equivalent amounts of protein were either probed for phosphorylation of endogenous ERK (pERK1/2) (top) or total ERK (ERK2) to control for protein loading (middle) or used for Rap1 activation assay (bottom).
FIG. 5.
Requirement of Rap1 for cAMP's ability to block EGF-induced cell proliferation. (A) Forskolin blocked cell proliferation induced by either serum or EGF. NIH 3T3 cells were serum starved, plated into 96-well plates, and placed into either 0.5% serum (low serum), EGF, or 10% serum (serum) plus forskolin (Forsk.), PD98059 (PD), or H89, as indicated. Forskolin was applied 5 min prior to either EGF or serum addition, as indicated. MTT assay buffer was added to the cells 2.5 h prior to the indicated time of lysis, the results were read using a microplate reader and are presented as the difference between optical densities (OD) at 570 and 650 nm [OD(570–650)]. The bar graph represents data (±SD) from five independent experiments. (B) Rap1GAP1 reverses cAMP's inhibitory effects on NIH 3T3 cell proliferation. NIH 3T3 cells were transfected with cDNAs encoding pMACS 14.1 or Rap1GAP1 and pMACS Kk.II and then positively selected as described in Materials and Methods. Selected cells were serum starved, plated into 96-well plates, and placed into either 0.5% serum (low serum), EGF (100 ng/ml), or 10% serum (serum) plus forskolin, PD98059, or H89, as indicated. MTT assay buffer was added to the cells 2.5 h prior to the indicated time of lysis; the results were read using a microplate reader and are presented as for panel A.
FIG. 6.
Requirement of Rap1 for cAMP's ability to block EGF-induced DNA synthesis. (A) Forskolin blocked DNA synthesis induced by either serum or EGF. NIH 3T3 cells were serum starved, plated into 96-well plates, and placed into either 0.5% serum (low serum), EGF, or 10% serum (serum) plus forskolin (Forsk.), PD98059 (PD), or H89. Forskolin was applied 5 min prior to either EGF or serum addition. [3H]thymidine uptake assays were performed as described in Materials and Methods, and the data are presented as means ±SD (n = 5). (B) Rap1GAP1 reverses cAMP's inhibitory effects on NIH 3T3 cell mitogenesis. NIH 3T3 cells were transfected with cDNAs encoding pMACS 14.1 or Rap1GAP1 and pMACS Kk.II and then positively selected as described in Materials and Methods. Selected cells were serum starved, plated into 96-well plates, and placed into either 0.5% serum (low serum), EGF, or 10% serum (serum) plus forskolin, PD98059, or H89. Forskolin was applied 5 min prior to either EGF or serum addition, and [3H]thymidine uptake was measured as described in Materials and Methods. The data are presented as for panel A.
DISCUSSION
Most studies examining cAMP's inhibition of ERK have focused on cAMP's regulation of either Ras (10) or Raf-1 signaling (17, 38, 67, 89, 111). This is largely because of the critical role of Ras signaling in proliferation (30) and in oncogenesis and metastasis (107). Ras is thought to stimulate cell cycle progression by multiple mechanisms (34). One of the earliest consequences of Ras action on cell proliferation is the stimulation of G1/S transition (34, 94, 95). Although Raf-1/ERK is the best-studied stimulator of cellular proliferation in NIH 3T3 cells (19, 74) as well as other fibroblast cells (91), oncogenic Ras activation of non-Raf-1/ERK pathways is sufficient to cause malignant transformation (51). Specific Ras effectors distinct from Raf-1 have been implicated in Ras's actions on cell growth and proliferation in selected cell types, including Rho (51, 82), Rac-1 (16, 47, 69, 73), PI3-K (27, 93, 98), and RalGDS (24, 40, 66, 109). Although the stimulation of cell growth clearly involves multiple Ras effectors, Raf-1/ERK appears to mediate the initial events in this transition, by stimulating the expression of cyclin D1 (34) and possibly by decreasing the levels of the cell cycle inhibitor p27 (48, 50, 110). Therefore, cAMP's inhibition of Ras/ERK signaling may inhibit cell growth in a variety of cell types where cell cycle proteins are tightly regulated by ERK. In NIH 3T3 cells, growth factor stimulation of cell growth requires Ras and MEK (19, 64), suggesting that Raf-1 is a critical effector of Ras' proliferative actions in these cells.
The ability of cAMP to block Ras-dependent signals was examined in both Hek293 and NIH 3T3 cells. In Hek293 cells, both Raf-1 and PI3-K are downstream of Ras (Fig. 1). Although cAMP inhibited Ras recruitment of Raf-1, it did not block the Ras-dependent activation of PI3-K, ruling out the general inhibition of Ras function as a mechanism for cAMP's actions. Indeed, recent reports suggest that under certain circumstances, cAMP can actually stimulate Ras (11, 78, 100). The studies presented here using Hek293 cells demonstrate that cAMP can selectively block Raf-1-dependent processes without affecting Ras activation and function.
A number of potential models by which cAMP can inhibit Raf-1 signaling have been proposed, some of which postulate a direct action of PKA on Raf-1 itself (38, 67, 111). Most studies have examined the phosphorylation of Raf-1 at serine 43 within the amino terminus (111). In one study, recombinant Raf-1 phosphorylated by PKA at this site in vitro bound less well to GTP-loaded Ras than did unphosphorylated Raf-1, although this difference was not greater than 50%, and was absent when higher concentrations of unphosphorylated and phosphorylated Raf-1 proteins were examined (52). Indeed, contrary conclusions have been drawn from studies performed in vivo. Studies in both Hek293 and NIH 3T3 cells have demonstrated that phosphorylation at serine 43 in Raf-1 is dispensable for cAMP's inhibitory effects (89), and the role of serine 43 phosphorylation in Raf-1 regulation remains unknown.
Support for additional mechanisms of PKA's ability to regulate Raf-1 comes from examination of the Raf isoform B-Raf. It has been previously shown that the association of B-Raf with Ras, like that of Raf-1, is also blocked following cAMP stimulation (76, 87, 89, 102). B-Raf lacks a PKA consensus site at the residue analogous to serine 43. Therefore, the ability of cAMP to block B-Raf–Ras association suggests that the direct phosphorylation of Raf cannot fully account for PKA's inhibition of the Raf family of kinases (89). We propose that the same protein or proteins upstream of both Raf-1 and B-Raf may mediate these effects. Interestingly, as for Ras–Raf-1, this block of Ras–B-Raf association by cAMP is also inhibited by the expression of Rap1GAP1 (data not shown). However, this action does not block ERK activation, since ERKs are activated by Rap1 in B-Raf-expressing cells (87, 105, 112), via the activation of B-Raf by Rap1 itself (71).
Our data suggest that PKA's actions do not require direct phosphorylation of Raf-1. It is possible that cAMP's activation of Rap1 prevents Ras from associating with Raf-1 by direct binding as suggested by other studies (72, 117), although additional studies may be necessary to establish this model in vivo. For example, biochemical experiments have shown that Ras has a higher affinity for Raf-1 than does Rap1 (41), suggesting that sequestration of Raf-1 by Rap1 in vivo may occur only when the level of activated Rap1 protein exceeds that of Ras. Given that the transfection of Rap1 and Ras result in similar levels of expression (Fig. 4C), it is possible that the ability of Rap1 to block Ras signals to ERKs in vivo may proceed by another mechanism. We propose that Rap1 activation by cAMP may block the activation of Raf-1 by Ras, thereby limiting signals downstream of Raf-1, including MEK and ERK, resulting in the inhibition of cell proliferation and mitogenesis. These studies implicating cAMP-PKA activation of Rap1 are distinct from those studies in other cell types identifying a PKA-independent activation of Rap1 (28, 101, 104), presumably via cAMP-guanine nucleotide exchange factors (23, 49).
Studies examining PKA actions on Rap1 have focused on the direct phosphorylation of Rap1 by PKA. PKA phosphorylates both Rap1a and Rap1b at a site within the carboxyl terminus (serine 179 in Rap1b). Rap1 is a target of phosphoryation following cAMP elevation in vivo (84, 105) and in vitro (3). PKA phosphorylation of Rap1 at this site has been proposed to enhance Rap1 activation (39), inhibit Rap1 coupling (43), or have only modest effects (3).
Although the regulation of Rap1 by direct phosphorylation remains controversial, direct phosphorylation of Rap1 has been proposed to influence its association with specific effectors (43). Hu et al. showed that PKA decreases the association of Rap1 and Raf-1 (43). In contrast, using transfected Rap1b, we show that in both Hek293 cells and NIH 3T3 cells PKA increases the association of Rap1b with Raf-1 (Fig. 1E and 3D to F) via the activation of Rap1 itself. In that study, the decrease in affinity of Rap1 and Raf-1 was due to the PKA-dependent phosphorylation of serine 180 in Rap1a, which decreases Rap1's affinity for the cysteine-rich domain of Raf-1. Both Rap1a and Rap1b are targets of PKA phosphorylation in vivo. Therefore, it is unlikely that the differences in our finding with Hu et al. relate to differences between the two Rap1 isoforms. These differences may be due to distinct biochemical properties of truncated Raf-1 fragments used in those studies or may reflect cell-type-specific effects. In any case, the precise mechanism of Rap1 stimulation by PKA remains unknown, and the role of PKA's phosphorylation of Rap1 in this process is not well understood. Importantly, targets of PKA that lie upstream of Rap1 itself have not been ruled out. These targets will be subjects of future research.
Because Hek293 cells express B-Raf, examination of the inhibitory role of Rap1 in cAMP inhibition of ERKs is complicated by Rap1's simultaneous activation of B-Raf and ERKs in these cells (87, 112). To enable us to examine the role of Rap1 in cAMP-mediated inhibition of ERKs, we used NIH3T3 cells. These cells express little or no B-Raf (105) and have been previously used as a model for cAMP-dependent inhibition of cell growth. One study showed that expression of constitutive activation of Gsα suppresses Ras-dependent proliferation and cellular transformation (13). In another study, PKA activation during anchorage-independent growth of these cells blocked ERK activation by growth factors, in part, via PKA's inhibition of PAK (42). However, inhibition of PAK could not entirely account for PKA's inhibitory actions, and additional targets of PKA were proposed (42). Interestingly, in studies examining signaling pathways during anchorage-dependent cell growth and cell adhesion, Rap1 regulation has been reported (81, 85, 99).
As with other members of the MAP kinase family (35), the kinetics of ERK activation are critical for determining the consequence of ERK signaling (65, 110, 114), and modest reductions in ERK activation may be physiologically significant. For example, in CCL39 fibroblast cells, PKA inhibition of ERK is transient (65). We show in NIH 3T3 cells that sustained activation of cAMP blocks ERK activation completely and for an extended period of time, suggesting that the extent of cAMP's inhibition may be cell type specific.
In NIH 3T3 cells, EGF stimulated ERK phosphorylation, cell proliferation, and mitogenesis. EGF's stimulation of cell proliferation and mitogenesis was blocked by PD98059, consistent with the requirement for MEK and ERK for growth factor-induced cell growth in these cells (19, 64, 74). cAMP blocked all three aspects of EGF's actions—stimulation of ERKs, cell proliferation, and mitogenesis—suggesting that cAMP's inhibition of mitogenesis was dictated, in part, by its inhibition of ERKs. We show here that Rap1 is required for cAMP's inhibitory actions of Raf-1, ERK activation, cellular proliferation, and mitogenesis. The ability of cAMP and PKA to block ERK activation by growth factors inhibits two central aspects of malignant transformation, cellular proliferation and anchorage-independent cell growth (42). We show here that Rap1 is a critical component of PKA's inhibition of ERK-dependent mitogenesis.
ACKNOWLEDGMENTS
We thank Kendall Carey (Vollum Institute, Oregon Health Sciences University) for scientific discussions and experimental assistance and Johannes Bos (University of Utrecht) for providing valuable reagents. We thank Kendall Carey, Tara Dillon, and Hong Yao (Vollum Institute, Oregon Health Sciences University) for critical reading of the manuscript and sharing technical and scientific expertise.
This work was supported by the NCI (P.J.S.S.).
REFERENCES
- 1.Ahn N G. The MAP kinase cascade. Discovery of a new signal transduction pathway. Mol Cell Biochem. 1993;127–128:201–209. doi: 10.1007/BF01076771. [DOI] [PubMed] [Google Scholar]
- 2.Alest L V, Barr M, Marcus S, Polverino A, Wigler M. Complex formation between RAS and RAF and other protein kinases. Proc Natl Acad Sci USA. 1993;90:6213–6217. doi: 10.1073/pnas.90.13.6213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschuler D, Lapetina E G. Mutational analysis of the cAMP-dependent protein kinase-mediated phosphorylation site of Rap1b. J Biol Chem. 1993;268:7527–7531. [PubMed] [Google Scholar]
- 4.Altschuler D L, Peterson S N, Ostrowski M C, Lapetina E G. Cyclic AMP-dependent activation of Rap1b. J Biol Chem. 1995;270:10373–10376. doi: 10.1074/jbc.270.18.10373. [DOI] [PubMed] [Google Scholar]
- 5.Altschuler D L, Ribeiro-Neto F. Mitogenic and oncogenic properties of the small G protein Rap1b. Proc Natl Acad Sci USA. 1998;95:7475–7479. doi: 10.1073/pnas.95.13.7475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anneren C, Reedquist K A, Bos J L, Welsh M. GTK, a Src-related tyrosine kinase, induces nerve growth factor-independent neurite outgrowth in PC12 cells through activation of the Rap1 pathway. Relationship to Shb tyrosine phosphorylation and elevated levels of focal adhesion kinase. J Biol Chem. 2000;275:29153–29161. doi: 10.1074/jbc.M003926200. [DOI] [PubMed] [Google Scholar]
- 7.Avruch J, Zhang X-F, Kyriakis J M. Raf meets Ras: completing the framework of a signal transduction pathway. Trends Biochem Sci. 1994;19:279–283. doi: 10.1016/0968-0004(94)90005-1. [DOI] [PubMed] [Google Scholar]
- 8.Blenis J. Signal transduction via the MAP kinases: proceed at your own RSK. Proc Natl Acad Sci USA. 1993;90:5889–5892. doi: 10.1073/pnas.90.13.5889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bos J L. All in the family? New insights and questions regarding interconnectivity of Ras, Rap1 and Ral. EMBO J. 1998;17:6776–6782. doi: 10.1093/emboj/17.23.6776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burgering B M, Pronk G J, van Weeren P C, Chardin P, Bos J L. cAMP antagonizes p21ras-directed activation of extracellular signal-regulated kinase 2 and phosphorylation of mSos nucleotide exchange factor. EMBO J. 1993;12:4211–4220. doi: 10.1002/j.1460-2075.1993.tb06105.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Busca R, Abbe P, Mantoux F, Aberdam E, Peyssonnaux C, Eychene A, Ortonne J P, Ballotti R. Ras mediates the cAMP-dependent activation of extracellular signal-regulated kinases (ERKs) in melanocytes. EMBO J. 2000;19:2900–2910. doi: 10.1093/emboj/19.12.2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Campa M J, Chang K-J, Molina y Vedia L, Reep B R, Lapetina E G. Inhibition of ras-induced germinal vesicle breakdown in Xenopus oocytes by rap-1B. Biochem Biophys Res Commun. 1991;174:1–5. doi: 10.1016/0006-291x(91)90475-m. [DOI] [PubMed] [Google Scholar]
- 13.Chen J, Iyengar R. Suppression of Ras-induced transformation of NIH 3T3 cells by activated Gαs. Science. 1994;263:1278–1281. doi: 10.1126/science.8122111. [DOI] [PubMed] [Google Scholar]
- 14.Chijiwa T, Mishima A, Hagiwara M, Sano M, Hayashi K, Inoue T, Naito K, Toshioka T, Hidaka H. Inhibition of forskolin-induced neurite outgrowth and protein phosphorylation by a newly synthesized selective inhibitor of cyclic AMP-dependent protein kinase, N-[2-(p-bromocinnamylamino) ethyl]-5-isoquinolinesulfonamide (H-89), of PC12D pheochromocytoma cells. J Biol Chem. 1990;265:5267–5272. [PubMed] [Google Scholar]
- 15.Cobb M H, Goldsmith E J. How MAP kinases are regulated. J Biol Chem. 1995;270:14843–14846. doi: 10.1074/jbc.270.25.14843. [DOI] [PubMed] [Google Scholar]
- 16.Cobellis G, Missero C, Di Lauro R. Concomitant activation of MEK-1 and Rac-1 increases the proliferative potential of thyroid epithelial cells, without affecting their differentiation. Oncogene. 1998;17:2047–2057. doi: 10.1038/sj.onc.1202130. [DOI] [PubMed] [Google Scholar]
- 17.Cook S J, McCormick F. Inhibition by cAMP of Ras-dependent activation of Raf. Science. 1993;262:1069–1072. doi: 10.1126/science.7694367. [DOI] [PubMed] [Google Scholar]
- 18.Cook S J, Rubinfeld B, Albert I, McCormick F. RapV12 antagonizes Ras-dependent activation of ERK1 and ERK2 by LPA and EGF in Rat-1 fibroblasts. EMBO J. 1993;12:3475–3485. doi: 10.1002/j.1460-2075.1993.tb06022.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cowley S, Paterson H, Kemp P, Marshall C J. Activation of MAP kinase kinase is necessary and sufficient for PC12 differentiation and for transformation of NIH3T3 cells. Cell. 1994;77:841–852. doi: 10.1016/0092-8674(94)90133-3. [DOI] [PubMed] [Google Scholar]
- 20.Crews C M, Erikson R L. Extracellular signals and reversible protein phosphorylation: what to Mek of it all. Cell. 1993;74:215–217. doi: 10.1016/0092-8674(93)90411-i. [DOI] [PubMed] [Google Scholar]
- 21.Daaka Y, Luttrell L M, Lefkowitz R J. Switching of the coupling of the beta2-adrenergic receptor to different G proteins by protein kinase A. Nature. 1997;390:88–91. doi: 10.1038/36362. [DOI] [PubMed] [Google Scholar]
- 22.Day R N, Walder J A, Maurer R A. A protein kinase inhibitor gene reduces both basal and multihormone-stimulated prolactin gene transcription. J Biol Chem. 1989;264:431–436. [PubMed] [Google Scholar]
- 23.de Rooij J, Zwartkruis F J, Verheijen M H, Cool R H, Nijman S M, Wittinghofer A, Bos J L. Epac is a Rap1 guanine-nucleotide-exchange factor directly activated by cyclic AMP. Nature. 1998;396:474–477. doi: 10.1038/24884. [DOI] [PubMed] [Google Scholar]
- 24.de Ruiter N D, Wolthuis R M, van Dam H, Burgering B M, Bos J L. Ras-dependent regulation of c-Jun phosphorylation is mediated by the ral guanine nucleotide exchange factor-Ral pathway. Mol Cell Biol. 2000;20:8480–8488. doi: 10.1128/mcb.20.22.8480-8488.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dhanasekaran N, Heasley L E, Johnson G L. G protein-coupled receptor systems involved in cell growth and oncogenesis. Endocrine Rev. 1995;16:259–270. doi: 10.1210/edrv-16-3-259. [DOI] [PubMed] [Google Scholar]
- 26.Downward J. Ras signalling and apoptosis. Curr Opin Genet Dev. 1998;8:49–54. doi: 10.1016/s0959-437x(98)80061-0. [DOI] [PubMed] [Google Scholar]
- 27.Downward J. A target for PI(3) kinase. Nature. 1995;376:553–554. doi: 10.1038/376553a0. [DOI] [PubMed] [Google Scholar]
- 28.Dremier S, Vandeput F, Zwartkruis F J, Bos J L, Dumont J E, Maenhaut C. Activation of the small G protein Rap1 in dog thyroid cells by both cAMP-dependent and -independent pathways. Biochem Biophys Res Commun. 2000;267:7–11. doi: 10.1006/bbrc.1999.1919. [DOI] [PubMed] [Google Scholar]
- 29.Dugan L L, Kim J S, Zhang Y, Bart R D, Sun Y, Holtzman D M, Gutmann D H. Differential effects of cAMP in neurons and astrocytes. Role of B-raf. J Biol Chem. 1999;274:25842–25848. doi: 10.1074/jbc.274.36.25842. [DOI] [PubMed] [Google Scholar]
- 30.Feig L A, Cooper G M. Inhibition of NIH 3T3 cell proliferation by a mutant Ras protein with preferential affinity for GDP. Mol Cell Biol. 1988;8:3235–3243. doi: 10.1128/mcb.8.8.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Franke B, Akkerman J-W, Bos J L. Rapid Ca2+-mediated activation of rap1 in human platelets. EMBO J. 1997;16:252–259. doi: 10.1093/emboj/16.2.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Franke B, van Triest M, de Bruijn K M, van Willigen G, Nieuwenhuis H K, Negrier C, Akkerman J W, Bos J L. Sequential regulation of the small GTPase Rap1 in human platelets. Mol Cell Biol. 2000;20:779–785. doi: 10.1128/mcb.20.3.779-785.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Franke T F, Kaplan D R, Cantley L C. PI3K: downstream AKTion blocks apoptosis. Cell. 1997;88:435–437. doi: 10.1016/s0092-8674(00)81883-8. [DOI] [PubMed] [Google Scholar]
- 34.Gille H, Downward J. Multiple ras effector pathways contribute to G(1) cell cycle progression. J Biol Chem. 1999;274:22033–22040. doi: 10.1074/jbc.274.31.22033. [DOI] [PubMed] [Google Scholar]
- 35.Gong Q, Cheng A M, Akk A M, Alerola-Ila J, Gong G, Pawson T, Chan A C. Disruption of T cell signaling networks and development by Grb2 haploid insufficiency. Nat Immunol. 2000;2:29–36. doi: 10.1038/83134. [DOI] [PubMed] [Google Scholar]
- 36.Graves L M, Bornfeldt K E, Raines E W, Potts B C, Macdonald S G, Ross R, Krebs E G. Protein kinase A antagonizes platelet-derived growth factor-induced signaling by mitogen-activated protein kinase in human arterial smooth muscle cells. Proc Natl Acad Sci USA. 1993;90:10300–10304. doi: 10.1073/pnas.90.21.10300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Graves L M, Guy H I, Kozlowski P, Huang M, Lazarowski E, Pope R M, Collins M A, Dahlstrand E N, Earp III H S, Evans D R. Regulation of carbamoyl phosphate synthetase by MAP kinase. Nature. 2000;403:328–332. doi: 10.1038/35002111. [DOI] [PubMed] [Google Scholar]
- 38.Hafner S, Adler H S, Mischak H, Janosch P, Heidecker G, Wolfman A, Pippig S, Lohse M, Ueffing M, Kolch W. Mechanism of inhibition of Raf-1 by protein kinase A. Mol Cell Biol. 1994;14:6696–6703. doi: 10.1128/mcb.14.10.6696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hata Y, Kaibuchi K, Kawamura S, Hiroyoshi M, Shirataki H, Takai Y. Enhancement of the actions of smg p21 GDP/GTP exchange protein by the protein kinase A-catalyzed phosphorylation of smg p21. J Biol Chem. 1991;266:6571–6577. [PubMed] [Google Scholar]
- 40.Hernandez-Munoz I, Malumbres M, Leonardi P, Pellicer A. The Rgr oncogene (homologous to RalGDS) induces transformation and gene expression by activating Ras, Ral and Rho mediated pathways. Oncogene. 2000;19:2745–2757. doi: 10.1038/sj.onc.1203586. [DOI] [PubMed] [Google Scholar]
- 41.Herrmann C, Horn G, Spaargaren M, Wittinghofer A. Differential interaction of the ras family GTP-binding proteins H-Ras, Rap1A, and R-Ras with the putative effector molecules Raf kinase and Ral-guanine nucleotide exchange factor. J Biol Chem. 1996;271:6794–6800. doi: 10.1074/jbc.271.12.6794. [DOI] [PubMed] [Google Scholar]
- 42.Howe A K, Juliano R L. Regulation of anchorage-dependent signal transduction by protein kinase A and p21-activated kinase. Nat Cell Biol. 2000;2:593–600. doi: 10.1038/35023536. [DOI] [PubMed] [Google Scholar]
- 43.Hu C D, Kariya K, Okada T, Qi X, Song C, Kataoka T. Effect of phosphorylation on activities of Rap1A to interact with Raf-1 and to suppress Ras-dependent Raf-1 activation. J Biol Chem. 1999;274:48–51. doi: 10.1074/jbc.274.1.48. [DOI] [PubMed] [Google Scholar]
- 44.Hu C D, Kariya K, Kotani G, Shirouzu M, Yokoyama S, Kataoka T. Coassociation of Rap1A and Ha-Ras with Raf-1 N-terminal region interferes with ras-dependent activation of Raf-1. J Biol Chem. 1997;272:11702–11705. doi: 10.1074/jbc.272.18.11702. [DOI] [PubMed] [Google Scholar]
- 45.Indolfi C, Avvedimento E V, Di Lorenzo E, Esposito G, Rapacciuolo A, Giuliano P, Grieco D, Cavuto L, Stingone A M, Ciullo I, Condorelli G, Chiariello M. Activation of cAMP-PKA signaling in vivo inhibits smooth muscle cell proliferation induced by vascular injury. Nat Med. 1997;3:775–779. doi: 10.1038/nm0797-775. [DOI] [PubMed] [Google Scholar]
- 46.Innocente S A, Abrahamson J L, Cogswell J P, Lee J M. p53 regulates a G2 checkpoint through cyclin B1. Proc Natl Acad Sci USA. 1999;96:2147–2152. doi: 10.1073/pnas.96.5.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Joyce D, Bouzahzah B, Fu M, Albanese C, D'Amico M, Steer J, Klein J U, Lee R J, Segall J E, Westwick J K, Der C J, Pestell R G. Integration of Rac-dependent regulation of cyclin D1 transcription through a nuclear factor-kappaB-dependent pathway. J Biol Chem. 1999;274:25245–25249. doi: 10.1074/jbc.274.36.25245. [DOI] [PubMed] [Google Scholar]
- 48.Kawada M, Yamagoe S, Murakami Y, Suzuki K, Mizuno S, Uehara Y. Induction of p27Kip1 degradation and anchorage independence by Ras through the MAP kinase signaling pathway. Oncogene. 1997;15:629–637. doi: 10.1038/sj.onc.1201228. [DOI] [PubMed] [Google Scholar]
- 49.Kawasaki H, Springett G M, Mochizuki N, Toki S, Nakaya M, Matsuda M, Housman D E, Graybiel A M. A family of cAMP-binding proteins that directly activate Rap1. Science. 1998;282:2275–2279. doi: 10.1126/science.282.5397.2275. [DOI] [PubMed] [Google Scholar]
- 50.Kerkhoff E, Rapp U R. Induction of cell proliferation in quiescent NIH 3T3 cells by oncogenic c-Raf-1. Mol Cell Biol. 1997;17:2576–2586. doi: 10.1128/mcb.17.5.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Khosravi-Far R, White M A, Westwick J K, Solski P A, Chrzanowska-Wodnicka M, Van Aelst L, Wigler M H, Der C J. Oncogenic Ras activation of Raf/mitogen-activated protein kinase-independent pathways is sufficient to cause tumorigenic transformation. Mol Cell Biol. 1996;16:3923–3933. doi: 10.1128/mcb.16.7.3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kikuchi A, Williams L T. Regulation of interaction of ras p21 with RalGDS and Raf-1 by cyclic AMP-dependent protein kinase. J Biol Chem. 1996;271:588–594. doi: 10.1074/jbc.271.1.588. [DOI] [PubMed] [Google Scholar]
- 53.Kim H A, DeClue J E, Ratner N. cAMP-dependent protein kinase A is required for Schwann cell growth: interactions between the cAMP and neuregulin/tyrosine kinase pathways. J Neurosci Res. 1997;49:236–247. [PubMed] [Google Scholar]
- 54.Kitayama H, Sugimoto Y, Matsuzaki T, Ikawa Y, Noda M. A ras-related gene with transformation suppressor activity. Cell. 1989;56:77–84. doi: 10.1016/0092-8674(89)90985-9. [DOI] [PubMed] [Google Scholar]
- 55.Koch W J, Hawes B E, Allen L F, Lefkowitz R J. Direct evidence that Gi-coupled receptor stimulation of mitogen-activated protein kinase is mediated by Gβγ activation of p21ras. Proc Natl Acad Sci USA. 1994;91:12706–12710. doi: 10.1073/pnas.91.26.12706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Koch W J, Inglese J, Stone W C, Lefkowitz R J. The binding site for the beta gamma subunits of heterotrimeric G proteins on the beta-adrenergic receptor kinase. J Biol Chem. 1993;268:8256–8260. [PubMed] [Google Scholar]
- 57.Kolch W, Heidecker G, Lloyd P, Rapp U R. Raf-1 protein kinase is required for growth of induced NIH/3T3 cells. Nature. 1991;349:426–428. doi: 10.1038/349426a0. [DOI] [PubMed] [Google Scholar]
- 58.Lavoie J N, L'Allemain G, Brunet A, Muller R, Pouyssegur J. Cyclin D1 expression is regulated positively by the p42/p44MAPK and negatively by the p38/HOGMAPK pathway. J Biol Chem. 1996;271:20608–20616. doi: 10.1074/jbc.271.34.20608. [DOI] [PubMed] [Google Scholar]
- 59.Leevers S J, Marshall C J. MAP kinase regulation—the oncogene connection. Trends Cell Biol. 1992;2:283–286. doi: 10.1016/0962-8924(92)90105-v. [DOI] [PubMed] [Google Scholar]
- 60.Lewis T S, Shapiro P S, Ahn N G. Signal transduction through MAP kinase cascades. Adv Cancer Res. 1998;74:49–139. doi: 10.1016/s0065-230x(08)60765-4. [DOI] [PubMed] [Google Scholar]
- 61.Lin Y, Mettling C, Chou C. Rap1-suppressed tumorigenesis is concomitant with the interference in ras effector signaling. FEBS Lett. 2000;467:184–188. doi: 10.1016/s0014-5793(00)01150-9. [DOI] [PubMed] [Google Scholar]
- 62.Lopez-Llasaca M, Crespo P, Pellicci P G, Gutkind J S, Wetzker R. Linkage of G protein-coupled receptors to the MAPK signaling pathway through PI 3-kinase γ. Science. 1997;275:394–397. doi: 10.1126/science.275.5298.394. [DOI] [PubMed] [Google Scholar]
- 63.MacNicol M C, MacNicol A M. Nerve growth factor-stimulated B-Raf catalytic activity is refractory to inhibition by cAMP-dependent protein kinase. J Biol Chem. 1999;274:13193–13197. doi: 10.1074/jbc.274.19.13193. [DOI] [PubMed] [Google Scholar]
- 64.Mansour S J, Matten W T, Hermann A S, Candia J M, Rong S, Fukasawa K, Woude G F V, Ahn N G. Transformation of mammalian cells by constitutively active MAP kinase kinase. Science. 1994;265:966–970. doi: 10.1126/science.8052857. [DOI] [PubMed] [Google Scholar]
- 65.McKenzie F R, Pouyssegur J. cAMP-mediated growth inhibition in fibroblasts is not mediated via mitogen-activated protein (MAP) kinase (ERK) inhibition. cAMP-dependent protein kinase induces a temporal shift in growth factor-stimulated MAP kinases. J Biol Chem. 1996;271:13476–13483. doi: 10.1074/jbc.271.23.13476. [DOI] [PubMed] [Google Scholar]
- 66.Miller M J, Prigent S, Kupperman E, Rioux L, Park S H, Feramisco J R, White M A, Rutkowski J L, Meinkoth J L. Ra1GDS functions in Ras- and cAMP-mediated growth stimulation. J Biol Chem. 1997;272:5600–5605. doi: 10.1074/jbc.272.9.5600. [DOI] [PubMed] [Google Scholar]
- 67.Mischak H, Seitz T, Janosch P, Eulitz M, Steen H, Schellerer M, Philipp A, Kolch W. Negative regulation of Raf-1 by phosphorylation of serine 621. Mol Cell Biol. 1996;16:5409–5418. doi: 10.1128/mcb.16.10.5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Moodie S A, Willumsen B M, Weber M J, Wolfman A. Complexes of ras.GTP with raf-1 and mitogen-activated protein kinase kinase. Science. 1993;260:1658–1661. doi: 10.1126/science.8503013. [DOI] [PubMed] [Google Scholar]
- 69.Moore K A, Sethi R, Doanes A M, Johnson T M, Pracyk J B, Kirby M, Irani K, Goldschmidt-Clermont P J, Finkel T. Rac1 is required for cell proliferation and G2/M progression. Biochem J. 1997;326:17–20. doi: 10.1042/bj3260017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Morrison D K, Heidecker G, Rapp U R, Copeland T D. Identification of the major phosphorylation sites of the Raf-1 kinase. J Biol Chem. 1993;268:17309–17316. [PubMed] [Google Scholar]
- 71.Ohtsuka T, Shimizu K, Yamamori B, Kuroda S, Takai Y. Activation of brain B-Raf protein kinase by Rap1B small GTP-binding protein. J Biol Chem. 1996;271:1258–1261. doi: 10.1074/jbc.271.3.1258. [DOI] [PubMed] [Google Scholar]
- 72.Okada T, Hu C D, Jin T G, Kariya K, Yamawaki-Kataoka Y, Kataoka T. The strength of interaction at the Raf cysteine-rich domain is a critical determinant of response of Raf to Ras family small GTPases. Mol Cell Biol. 1999;19:6057–6064. doi: 10.1128/mcb.19.9.6057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Olson M F, Ashworth A, Hall A. An essential role for Rho, Rac, and Cdc42 GTPases in cell cycle progression through G1. Science. 1995;269:1270–1272. doi: 10.1126/science.7652575. [DOI] [PubMed] [Google Scholar]
- 74.Pagés G, Lenormand P, L'Allemain G, Chambard J-C, Meloche S, Pouysségur J. Mitogen-activated protein kinases p42mapk and p44mapk are required for fibroblast proliferation. Proc Natl Acad Sci USA. 1993;90:8319–8323. doi: 10.1073/pnas.90.18.8319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pastan I H, Johnson G S, Anderson W B. Role of cyclic nucleotides in growth control. Annu Rev Biochem. 1975;44:491–522. doi: 10.1146/annurev.bi.44.070175.002423. [DOI] [PubMed] [Google Scholar]
- 76.Peraldi P, Frödin M, Barnier J V, Calleja V, Scimeca J-S, Filloux C, Calothy G, Van Obberghen E. Regulation of the MAP kinase cascade in PC12 cells: B-Raf activates MEK-1 (MAP kinase or ERK kinase) and is inhibited by cAMP. FEBS Lett. 1995;357:290–296. doi: 10.1016/0014-5793(94)01376-c. [DOI] [PubMed] [Google Scholar]
- 77.Peterson R T, Schreiber S L. Kinase phosphorylation: keeping it all in the family. Curr Biol. 1999;9:R521–R524. doi: 10.1016/s0960-9822(99)80326-1. [DOI] [PubMed] [Google Scholar]
- 78.Pham N, Cheglakov I, Koch C A, de Hoog C L, Moran M F, Rotin D. The guanine nucleotide exchange factor CNrasGEF activates ras in response to cAMP and cGMP. Curr Biol. 2000;10:555–558. doi: 10.1016/s0960-9822(00)00473-5. [DOI] [PubMed] [Google Scholar]
- 79.Pizon V, Chardin P, Tavitian A. Nucleotide sequence of a human cDNA encoding a ras-related protein (rap 1b) Nucleic Acids Res. 1990;16:7719. doi: 10.1093/nar/16.15.7719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Polakis P G, Rubinfeld B, Evans T, McCormick F. Purification of a plasma membrane-assoicated GTPase-activating protein specific for rap1/Krev-1 from HL60 cells. Proc Natl Acad Sci USA. 1991;88:239–243. doi: 10.1073/pnas.88.1.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Posern G, Weber C K, Rapp U R, Feller S M. Activity of Rap1 is regulated by bombesin, cell adhesion, and cell density in NIH3T3 fibroblasts. J Biol Chem. 1998;273:24297–24300. doi: 10.1074/jbc.273.38.24297. [DOI] [PubMed] [Google Scholar]
- 82.Qiu R-G, Chen J, McCormick F, Symons M. A role for Rho in Ras transformation. Proc Natl Acad Sci USA. 1995;92:11781–11785. doi: 10.1073/pnas.92.25.11781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Qiu W, Zhuang S, von Lintig F C, Boss G R, Pilz R B. Cell type-specific regulation of B-Raf kinase by cAMP and 14-3-3 proteins. J Biol Chem. 2000;275:31921–31929. doi: 10.1074/jbc.M003327200. [DOI] [PubMed] [Google Scholar]
- 84.Quilliam L A, Mueller H, Bohl B P, Prossnitz V, Sklar L A, Der C J, Bokoch G M. Rap1A is a substrate for cyclic AMP-dependent protein kinase in human neutrophils. J Immunol. 1991;147:1628–1635. [PubMed] [Google Scholar]
- 85.Reedquist K A, Ross E, Koop E A, Wolthuis R M, Zwartkruis F J, van Kooyk Y, Salmon M, Buckley C D, Bos J L. The Small GTPase, Rap1, mediates CD31-induced integrin adhesion. J Cell Biol. 2000;148:1151–1158. doi: 10.1083/jcb.148.6.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rubinfeld B, Munemitsu S, Clark R, Conroy L, Watt K, Crosier W J, McCormick F, Polakis P. Molecular cloning of a GTPase activating protein specific for the Krev-1 protein p21rap1. Cell. 1991;65:1033–1042. doi: 10.1016/0092-8674(91)90555-d. [DOI] [PubMed] [Google Scholar]
- 87.Schmitt J M, Stork P J S. β2-adrenergic receptor activates extracellular regulated kinases (ERKs) via the small G protein Rap1 and the serine/threonine kinase B-Raf. J Biol Chem. 2000;275:25342–25350. doi: 10.1074/jbc.M003213200. [DOI] [PubMed] [Google Scholar]
- 88.Sevetson B R, Kong X, Lawrence J C., Jr Increasing cAMP attenuates activation of mitogen-activated protein kinase. Proc Natl Acad Sci USA. 1993;90:10305–10309. doi: 10.1073/pnas.90.21.10305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sidovar M F, Kozlowski P, Lee J W, Collins M A, He Y, Graves L M. Phosphorylation of serine 43 is not required for inhibition of C-Raf kinase by the cAMP-dependent protein kinase. J Biol Chem. 2000;275:28688–28694. doi: 10.1074/jbc.M909351199. [DOI] [PubMed] [Google Scholar]
- 90.Stacey D W, Feig L A, Gibbs J B. Dominant inhibitory Ras mutants selectively inhibit the activity of either cellular or oncogenic Ras. Mol Cell Biol. 1991;11:4053–4064. doi: 10.1128/mcb.11.8.4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Stang S, Bottorff D, Stone J C. Interaction of activated Ras with Raf-1 alone may be sufficient for transformation of Rat2 cells. Mol Cell Biol. 1997;17:3047–3055. doi: 10.1128/mcb.17.6.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Suhasini M, Li H, Lohmann S M, Boss G R, Pilz R B. Cyclic-GMP-dependent protein kinase inhibits the Ras/mitogen-activated protein kinase pathway. Mol Cell Biol. 1998;18:6983–6994. doi: 10.1128/mcb.18.12.6983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Takuwa N, Fukui Y, Takuwa Y. Cyclin D1 expression mediated by phosphatidylinositol 3-kinase through mTOR-p70S6K-independent signaling in growth factor-stimulated NIH 3T3 fibroblasts. Mol Cell Biol. 1999;19:1346–1358. doi: 10.1128/mcb.19.2.1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Takuwa N, Takuwa Y. Ras activity late in G1 phase required for p27kip1 downregulation, passage through the restriction point, and entry into S phase in growth factor-stimulated NIH 3T3 fibroblasts. Mol Cell Biol. 1997;17:5348–5358. doi: 10.1128/mcb.17.9.5348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Taylor S J, Shalloway D. Cell cycle-dependent activation of Ras. Curr Biol. 1996;6:1621–1627. doi: 10.1016/s0960-9822(02)70785-9. [DOI] [PubMed] [Google Scholar]
- 96.Tetsu O, McCormick F. Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature. 1999;398:422–426. doi: 10.1038/18884. [DOI] [PubMed] [Google Scholar]
- 97.Thomas S M, DeMarco M, D'Arcangelo G, Halegoua S, Brugge J S. Ras is essential for nerve growth factor and phorbol ester-induced tyrosine phosphorylation of MAP kinases. Cell. 1992;68:1031–1040. doi: 10.1016/0092-8674(92)90075-n. [DOI] [PubMed] [Google Scholar]
- 98.Treinies I, Paterson H F, Hooper S, Wilson R, Marshall C J. Activated MEK stimulates expression of AP-1 components independently of phosphatidylinositol 3-kinase (PI3-kinase) but requires a PI3-kinase signal to stimulate DNA synthesis. Mol Cell Biol. 1999;19:321–329. doi: 10.1128/mcb.19.1.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tsukamoto N, Hattori M, Yang H, Bos J L, Minato N. Rap1 GTPase-activating protein SPA-1 negatively regulates cell adhesion. J Biol Chem. 1999;274:18463–18469. doi: 10.1074/jbc.274.26.18463. [DOI] [PubMed] [Google Scholar]
- 100.Tsygankova O M, Kupperman E, Wen W, Meinkoth J L. Cyclic AMP activates Ras. Oncogene. 2000;19:3609–3615. doi: 10.1038/sj.onc.1203680. [DOI] [PubMed] [Google Scholar]
- 101.Tsygankova O M, Saavedra A, Rebhun J F, Quilliam L A, Meinkoth J L. Coordinated regulation of Rap1 and thyroid differentiation by cyclic AMP and protein kinase A. Mol Cell Biol. 2001;21:1921–1929. doi: 10.1128/MCB.21.6.1921-1929.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Vaillancourt R R, Gardner A M, Johnson G L. B-Raf dependent regulation of the MEK-1/MAP kinase pathway in PC12 cells and regulation by cAMP. Mol Cell Biol. 1994;14:6522–6530. doi: 10.1128/mcb.14.10.6522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Vojitek A B, Hollenberg S M, Cooper J A. Mammalian ras interacts directly with the serine/threonine kinase raf. Cell. 1993;74:205–214. doi: 10.1016/0092-8674(93)90307-c. [DOI] [PubMed] [Google Scholar]
- 104.von Lintig F C, Pilz R B, Boss G R. Quantitative determination of Rap 1 activation in cyclic nucleotide-treated HL-60 leukemic cells: lack of Rap 1 activation in variant cells. Oncogene. 2000;19:4029–4034. doi: 10.1038/sj.onc.1203741. [DOI] [PubMed] [Google Scholar]
- 105.Vossler M, Yao H, York R, Rim C, Pan M-G, Stork P J S. cAMP activates MAP kinase and Elk-1 through a B-Raf- and Rap1-dependent pathway. Cell. 1997;89:73–82. doi: 10.1016/s0092-8674(00)80184-1. [DOI] [PubMed] [Google Scholar]
- 106.Wan Y, Huang X Y. Analysis of the Gs/mitogen-activated protein kinase pathway in mutant S49 cells. J Biol Chem. 1998;273:14533–14537. doi: 10.1074/jbc.273.23.14533. [DOI] [PubMed] [Google Scholar]
- 107.Webb C P, Van Aelst L, Wigler M H, Woude G F. Signaling pathways in Ras-mediated tumorigenicity and metastasis. Proc Natl Acad Sci USA. 1998;95:8773–8778. doi: 10.1073/pnas.95.15.8773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wilkinson M G, Millar J B. Control of the eukaryotic cell cycle by MAP kinase signaling pathways. FASEB J. 2000;14:2147–2157. doi: 10.1096/fj.00-0102rev. [DOI] [PubMed] [Google Scholar]
- 109.Wolthuis R M, Bauer B, van't Veer L J, de Vries-Smits A M, Cool R H, Spaargaren M, Wittinghofer A, Burgering B M, Bos J L. Ra1GDS-like factor (R1f) is a novel Ras and Rap 1A-associating protein. Oncogene. 1996;13:353–362. [PubMed] [Google Scholar]
- 110.Woods D, Parry D, Cherwinski H, Bosch E, Lees E, McMahon M. Raf-induced proliferation or cell cycle arrest is determined by the level of Raf activity with arrest mediated by p21Cip1. Mol Cell Biol. 1997;17:5598–5611. doi: 10.1128/mcb.17.9.5598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wu J, Dent P, Jelinek T, Wolfman A, Weber M J, Sturgill T W. Inhibition of the EGF-activated MAP kinase signaling pathway by adenosine 3′, 5′-monophosphate. Science. 1993;262:1065–1068. doi: 10.1126/science.7694366. [DOI] [PubMed] [Google Scholar]
- 112.Xing L, Ge C, Zeltser R, Maskevitch G, Mayer B J, Alexandropoulos K. c-Src signaling induced by the adapters Sin and Cas is mediated by Rap1 GTPase. Mol Cell Biol. 2000;20:7363–7377. doi: 10.1128/mcb.20.19.7363-7377.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.York R D, Molliver D C, Grewal S S, Stenberg P E, McCleskey E W, Stork P J. Role of phosphoinositide 3-kinase and endocytosis in nerve growth factor-induced extracellular signal-regulated kinase activation via Ras and Rap1. Mol Cell Biol. 2000;20:8069–8083. doi: 10.1128/mcb.20.21.8069-8083.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.York R D, Yao H, Dillon T, Ellig C L, Eckert S P, McCleskey E W, Stork P J S. Rap1 mediates sustained MAP kinase activation induced by nerve growth factor. Nature. 1998;392:622–625. doi: 10.1038/33451. [DOI] [PubMed] [Google Scholar]
- 115.Yoshida Y, Kawata M, Miura Y, Musha T, Sasaki T, Kikuchi A, Takai U. Microinjection of smg/rap1/Krev-1 p21 into Swiss 3T3 cells induces DNA synthesis and morphological changes. Mol Cell Biol. 1992;12:3407–3414. doi: 10.1128/mcb.12.8.3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yung Y, Dolginov Y, Yao Z, Rubinfeld H, Michael D, Hanoch T, Roubini E, Lando Z, Zharhary D, Seger R. Detection of ERK activation by a novel monoclonal antibody. FEBS Lett. 1997;408:292–296. doi: 10.1016/s0014-5793(97)00442-0. [DOI] [PubMed] [Google Scholar]
- 117.Zhang X-F, Settleman J, Kyriakis J M, Takeuchi-Suzuki E, Elledge S J, Marshall M S, Bruder J T, Rapp U R, Avruch J. Normal and oncogenic p21ras proteins bind to the amino-terminal regulatory domain of c-raf-1. Nature. 1993;364:308–313. doi: 10.1038/364308a0. [DOI] [PubMed] [Google Scholar]
- 118.Zwartkruis F J, Bos J L. Ras and Rap1: two highly related small GTPases with distinct function. Exp Cell Res. 1999;253:157–165. doi: 10.1006/excr.1999.4695. [DOI] [PubMed] [Google Scholar]
- 119.Zwartkruis F J, Wolthuis R M, Nabben N M, Franke B, Bos J L. Extracellular signal-regulated activation of Rap1 fails to interfere in Ras effector signalling. EMBO J. 1998;17:5905–5912. doi: 10.1093/emboj/17.20.5905. [DOI] [PMC free article] [PubMed] [Google Scholar]