Abstract
Clostridium difficile-associated diarrhea (CAD) is a very common nosocomial infection that contributes significantly to patient morbidity and mortality as well as to the cost of hospitalization. Previously, strains of toxin A-negative, toxin B-positive C. difficile were not thought to be associated with clinically significant disease. This study reports the characterization of a toxin A-negative, toxin B-positive strain of C. difficile that was responsible for a recently described nosocomial outbreak of CAD. Analysis of the seven patient isolates from the outbreak by pulsed-field gel electrophoresis indicated that this outbreak was due to transmission of a single strain of C. difficile. Our characterization of this strain (HSC98) has demonstrated that the toxin A gene lacks 1.8 kb from the carboxy repetitive oligopeptide (CROP) region but apparently has no other major deletions from other regions of the toxin A or toxin B gene. The remaining 1.3-kb fragment of the toxin A CROP region from strain HSC98 showed 98% sequence homology with strain 1470, previously reported by M. Weidmann in 1997 (GenBank accession number Y12616), suggesting that HSC98 is toxinotype VIII. The HSC98 strain infecting patients involved in this outbreak produced the full spectrum of clinical illness usually associated with C. difficile-associated disease. This pathogenic spectrum was manifest despite the inability of this strain to alter tight junctions as determined by using in vitro tissue culture testing, which suggested that no functional toxin A was produced by this strain.
Clostridium difficile-associated diarrhea (CAD) is the most common infectious cause of nosocomial diarrhea (6, 8, 15, 18, 23, 30). The incidence of this disease ranges from 20 to 60 cases per 100,000 patient days (2, 30), and the incidence appears to be increasing (14, 30). C. difficile isolates associated with human disease are thought usually to produce both toxin A and toxin B (4, 6, 7, 9, 13, 15, 22, 24), and genetic evaluations suggest coregulation of the two genes (10, 35, 40). Toxin B acts as a potent cytotoxin, but it does not cause plasma membranes of T84 cells to become leaky, nor does it disrupt tight junctions of these cells, whereas toxin A is an enterotoxin that has potent ability to disrupt the tight junctions of cultured human intestinal epithelial monolayers (11, 12, 16, 17, 28). Evidence suggests that toxin A and toxin B may act synergistically (21, 24), but how this may occur is yet to be completely elucidated.
Strains of C. difficile that have defects in the toxin A gene have been described (3, 5, 9, 19, 26, 31, 32, 36). The elegant molecular typing and toxinotyping scheme reported by Rupnik et al. (32) provides detailed analyses of the known isolates of C. difficile that have defects in toxin A and/or toxin B production. The reported data indicate that strains that have deletions in the toxin A gene of greater than 800 bp have no detectable toxin A (32), and such strains were not associated with clinically significant human disease (19, 32). The lack of toxin A in serogroup F strains of C. difficile has been suggested to account for the high carriage of toxigenic strains in neonates, who do not often develop C. difficile-associated illness (5, 9, 20). Alternatively, it has been suggested that neonates who carry toxigenic C. difficile have predominantly the spore form, and therefore, any toxin production would not be expected (29). However, the spore theory does not explain the high rate of detectable cytotoxicity in stools from asymptomatic neonates.
Recently, however, a nosocomial outbreak of CAD caused by a toxin A-negative, toxin B-positive strain of C. difficile has been described (1). This is the first reported outbreak of clinically significant disease attributable to a strain of C. difficile that was negative by enzyme immunoassays (EIAs) that utilized a monoclonal antibody to the carboxy repetitive oligopeptide (CROP) region of toxin A. Understanding more about this strain may enhance our understanding of the pathogenesis of toxigenic C. difficile. The purpose of this investigation was to characterize the toxin A-negative, toxin B-positive strain of C. difficile that was responsible for the outbreak.
MATERIALS AND METHODS
EIAs for detection of toxin A or toxin A and B.
The two EIAs used for detection of toxin A in stool samples were the Prima Toxin A test (Bartels Inc., Issaquah, Wash.) and the Tox-A test (Techlab Inc., Blacksburg, Va.). The EIA used for detection of both toxin A and toxin B was the Toxin A/B test (Techlab Inc.). All assays were performed using fresh stool samples according to the manufacturer's instructions.
Cytotoxin assay for toxigenic C. difficile.
Human foreskin fibroblast (HFF) cells were grown as monolayers in 96-well tissue culture trays. Stool samples were used at a final dilution of 1:24, and the filter-sterilized filtrate was tested with and without neutralization treatment using polyclonal anti-C. difficile antitoxin (Techlab Inc.). Cytopathic effect (CPE), characterized by rounding of at least 50% of the HFF cells within 48 h, that was neutralized by polyclonal antiserum was considered a positive test result for the presence of C. difficile cytotoxin. C. difficile toxin B and anti-C. difficile toxin (Techlab Inc.) were used as positive (toxin alone) and negative (toxin plus antitoxin) controls that were included in each test panel.
To determine the cytotoxicity titers of the seven C. difficile clinical isolates, dialysis culture filtrate was assayed for cytotoxic activity against Chinese hamster ovary K-1 (CHO) cells that were grown in 96-well tissue culture trays. For the assay, 20-μl aliquots were serially diluted using a new pipette tip for each transfer. The cytotoxic titer was expressed as the reciprocal of the highest dilution that caused rounding of >90% of the CHO cells at 24 h.
Culture of C. difficile strains.
Culture for C. difficile from stool samples was performed by inoculating thioglycolate broth supplemented with cycloserine (500 μg/ml) and cefoxitin (16 μg/ml), incubating at 35°C for 24 h, and then subculturing the enrichment broth to blood agar containing vitamin K and hemin (BAK) plates which were then incubated anaerobically. Suspect nonhemolytic colonies from BAK were Gram stained and tested for fluorescence and by Microscreen latex agglutination (Microgen Bioproducts, Camberley, United Kingdom) for C. difficile antigen. Isolated C. difficile strains were stored at −70°C in skim milk. Culture of C. difficile for optimal toxin production was done by growing the strain overnight (18 h) in brain heart infusion broth. This culture was then used to inoculate brain heart infusion dialysis flasks as described by Sullivan et al. (33). The flasks were incubated at 37°C for 72 h, and the supernatant fluid was collected by centrifugation (10,000 × g for 15 minutes at 4°C). Supernatant fluids were filtered through 0.2-μm-pore-size membranes, and the filter-sterilized supernatant fluid was stored at 4°C. Culture dialysis supernatants were prepared for all seven clinical isolates as well as a toxin A-negative, toxin B-positive strain designated 8864 and a toxin A-positive, toxin B-positive strain designated 10463 (strains 8864 and 10463 were part of the culture collection of D. Lyerly).
PCR analysis for C. difficile pathogenicity gene locus fragments.
The PCRs for the CROP region of toxin A were performed by two separate laboratories (those of M. J. Alfa and D. Lyerly) to ensure reproducibility of findings. The PCR mix (50-μl total reaction volume) for the A3 fragment described by Rupnik et al. (31) consisted of a 0.5 μM concentration of the A3 primers (31), a 0.2 mM concentration of each dinucleoside triphosphate (including A, C, G, and T) (Life Technologies, Gaithersburg, Md.), 2.5 U of Taq polymerase (Life Technologies), 3 mM MgCl2 (Life Technologies), and 5 μl of C. difficile DNA extract. The C. difficile DNA extract was prepared by a Triton X-100 extraction protocol (A. Petrich, B. Page, K. Luinstra, S. Callery, D. Stevens, A. Gafni, D. Groves, M. Chernesky, and J. Mahony. 1998. Presented at the 66th meeting of the Canadian Association for Clinical Microbiology and Infectious Diseases). Briefly, this method consisted of lysing about half a loopful of bacterial colony by suspending the organism in 10 mM Trizma base–1 mM EDTA, 1% (wt/vol) Triton X-100 (pH 8.0) and heating for 20 min at 95°C, followed by vortex mixing for 10 min. The suspension was then chilled on ice for 1 min, and debris was removed by centrifugation at 14,000 rpm for 1 min in a Microfuge. The upper portion of the suspension was diluted 1:40, and 10 μl of this dilution was used per 50 μl of PCR mixture. The PCR was performed using a PTC-2000 thermal cycler (MJ Research, Watertown, Mass.), and the cycles were as described by Rupnik et al. (31). The expected PCR product from this reaction is a 3.1-kb fragment (31). The PCR product size was determined using 1% agarose gel electrophoresis and a 100-bp DNA ladder (GenSura Laboratories Inc., Del Mar, Calif.) as well as a 500-bp DNA ladder (Life Technologies). The amplified product from this PCR protocol was purified using the Wizard PCR Preps DNA purification system (Promega Corp., Madison, Wis.), and this purified preparation was used for DNA sequencing.
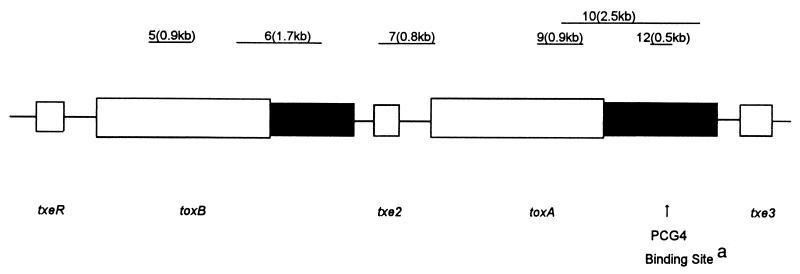
The second PCR assessment utilized 6 sets of primers that bound the regions shown in Table 2. The expected PCR product size for each primer set tested is indicated. The DNA template was prepared by taking a single colony from a plate culture of the C. difficile strain and suspending this in 100 μl of distilled deionized water using a wood applicator stick. After one cycle of freezing and thawing, the suspension was heated at 95°C for 5 min and then stored at −20°C prior to use. The PCR mix consisted of 10 μl of this template DNA sample, 15 μl of a solution of the two primers, and a Ready-To-Go PCR Bead (Pharmacia Inc.). After heating at 94°C for 1 min, 45 cycles of reaction (each cycle consisted of 30 s at 94°C, 1 min at a suitable annealing temperature, and 1 to 2.5 min at 72°C) were carried out. The PCR product size was determined using 1% agarose gel electrophoresis and a 100-bp DNA ladder (GenSura Laboratories Inc.) as well as a 500-bp DNA ladder (Life Technologies).
TABLE 2.
Pathogenicity gene locus analysis of C. difficile HSC98 isolates from different patients
| Isolateb | Amplification result with PCR region:
|
|||||
|---|---|---|---|---|---|---|
| 5 | 6 | 7 | 9 | 10 | 12 | |
| VPI 11186 | − | − | − | − | − | − |
| VPI 10463 | + | + | + | + | 2.5 kb | + |
| Patient 1 | + | + | + | + | 0.8 kb | − |
| Patient 5 | + | + | + | + | 0.8 kb | − |
| Patient 6 | + | + | + | + | 0.8 kb | − |
| Patient 7 | + | + | + | + | 0.8 kb | − |
| Patient 9 | + | + | + | + | 0.8 kb | − |
| Patient 10 | + | + | + | + | 0.8 kb | − |
| Patient 11 | + | + | + | + | 0.8 kb | − |
PCG4 is the monoclonal antibody used in the EIAs that detected toxin A only.
Strain VPI 11186 is a nontoxigenic strain of C. difficile (i.e., it does not carry the gene for either toxin A or toxin B), and strain VPI 10463 is a C. difficile strain that produces both toxin A and toxin B.
DNA sequencing.
Purified amplicon was sequenced using the Prism 310 Genetic Analyzer (Applied Biosystems Inc., Foster City, Calif.). Sequencing was performed using the Big Dye Terminator Cycle Sequencing kit (Applied Biosystems Inc.) according to the manufacturer's recommended procedure.
Pulsed-field gel electrophoresis (PFGE).
C. difficile was grown in 10 ml of prereduced brain heart Infusion broth for 5 h anaerobically at 35°C and centrifuged at 3,500 × g, and the bacterial pellet was suspended in PIV buffer (10 mM Tris-HCl, 1 M NaCl, pH 8.0). The bacterial suspension was mixed 1:1 with molten low-melting-point agarose (final concentration, 0.8% [wt/vol] agarose) and used to prepare plugs. Lysis was achieved using lysis solution containing 0.5% (wt/vol) N-lauroyl-sarcosine (Sigma Chemical Co., St. Louis, Mo.), 0.5% (wt/vol) Brij 58 (Sigma), 2.5 μg of mutanolysin (Sigma) per ml, 20 μg of RNase (Sigma) per ml, 2 mg of lysozyme (Sigma) per ml, 1 M NaCl (Sigma), 100 mM EDTA (pH 9.0) (Sigma), 0.2% (wt/vol) sodium deoxycholate (Sigma), and 6 mM Tris-HCl (pH 8.0) (Sigma) at 35°C overnight. The plugs were washed for 30 min in TE buffer (10 mM Tris [Sigma], 0.1 mM EDTA [Sigma], pH 9.0). ESP buffer, containing 50 μg of proteinase K per ml (Sigma) and 1% (wt/vol) N-lauroyl-sarcosine (Sigma), was added and then incubated at 50°C overnight. The plugs were then washed with TE buffer three times for 1 h each at 35°C. Digestion was done using 30 U of SmaI (Life Technologies Inc.) per plug at 30°C for 4 h. The DNA fragments were separated using a Mapper unit (Bio-Rad Laboratories, Hercules, Calif.). The run conditions consisted of a 22-h run time, 1- to 40-s switch times, and linear ramping at 6 V/cm in a 1% agarose gel using 0.5× TBE buffer, consisting of 40 mM Tris, 25 mM EDTA (pH 8.0), and 40 mM boric acid (Sigma).
Tight-junction analysis using CaCo-2 polarized cell culture.
CaCo-2 cells (cell line HTB-37; American Type Culture Collection, Manassas, Va.) were seeded into Transwell inserts (Corning Glassworks, Corning, N.Y.) at 5 × 105 cells/insert and allowed to incubate until confluent as indicated by a high stable electrical resistance (>400 Ω · cm2) as described by Eveillard et al. (11). Filter-sterilized preparations of the dialysis culture fluid (described in “Culture of C. difficile strains” above) were inoculated onto the apical side of the confluent polarized CaCo-2 monolayer in the Transwell insert, and electrical resistance was tested hourly using a MILLICELL-ERS probe (Millipore, Bedford, Mass.). Each test assessment was performed using triplicate inserts, and for each insert resistance measurements were performed in triplicate (i.e., a total of nine readings per test). The resistance probe was rinsed with sterile distilled water, rinsed with 70% alcohol, and then equilibrated in sterile tissue culture medium between each reading on a different insert.
RESULTS
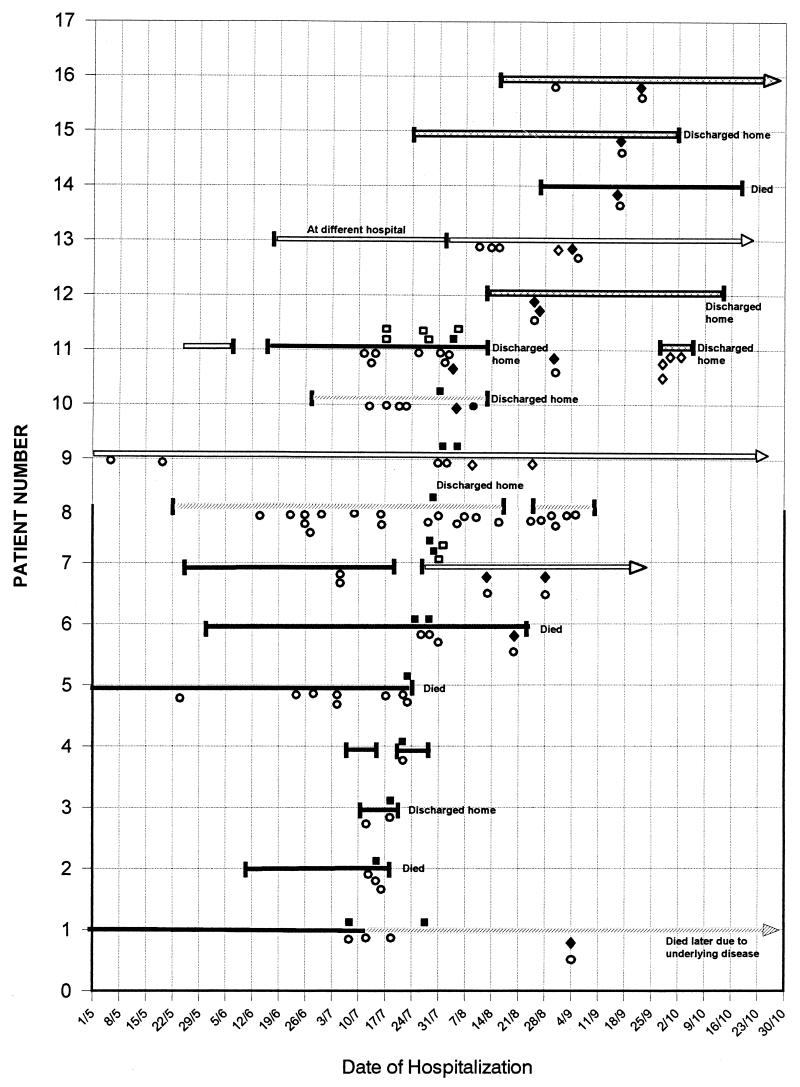
The outbreak of CAD due to the HSC98 strain was reported by Al-Barrak et al. (1). This outbreak initially involved 16 patients on four medical wards of an 800-bed acute-care tertiary teaching hospital. The range of presenting symptoms for the 16 patients is shown in Table 1. The two patients who were not treated for CAD had been screened because they were roommates of patients with CAD. One of these two untreated patients was asymptomatic, and the other had mild diarrhea that resolved without specific treatment. This center used an EIA that detected toxin A only as their routine diagnostic test. As shown in Fig. 1, all 16 patients' stool specimens were consistently negative for C. difficile toxin using the toxin A EIA, indicating that this finding was not due to test sensitivity but rather was due to a lack of detectable toxin A target. Testing by cytotoxin assay and by an EIA that detected both toxins A and B revealed that these 16 patients had cytotoxin in their stool specimens, despite the negative result with the toxin A EIA.
TABLE 1.
C. difficile-associated-illness: range of presenting symptoms in patients involved in the outbreak
| Case | Age (yr) | Sexa | Diseaseb | Treatment | Recurrence |
|---|---|---|---|---|---|
| 1 | 38 | M | PMC | Yes | Yes (died) |
| 2 | 71 | F | AAD and THICK | Yes | No (died)c |
| 3 | 50 | F | Diarrhea (resolved) | No | No |
| 4 | 78 | M | No symptoms | No | No |
| 5 | 62 | M | PMC | Yes | No (died)c |
| 6 | 63 | F | AAD | Yes | Yes (died) |
| 7 | 77 | F | AAD | Yes | Yes |
| 8 | 3 | F | AAD | Yes | Yes |
| 9 | 74 | M | AAD | Yes | No |
| 10 | 16 | M | AAD | Yes | No (died) |
| 11 | 57 | F | AAD | Yes | No |
| 12 | 75 | F | AAD | Yes | No |
| 13 | 78 | F | PMC | Yes | Yes |
| 14 | 67 | M | AAD | Yes | No (died) |
| 15 | 35 | M | AAD | Yes | No |
| 16 | 77 | M | AAD | Yes | No |
M, male; F, female.
AAD, antibiotic-associated diarrhea; PMC, pseudomembranous colitis; THICK, thickened bowel lining on computerized tomography scan of abdomen.
Death was attributable to CAD.
FIG. 1.
Diagnostic tests performed on samples from patients involved in the outbreak. Each solid horizontal line represents the time line for the length of hospitalization for one of the 16 patients involved in the initial outbreak. Gaps in the line indicate when the patient was discharged and then was readmitted. Arrows indicate that the patient was still in the hospital beyond this date. There were four wards involved in this outbreak; ward 1 ( ), ward 2 (
), ward 2 ( ), and ward 4 (
), and ward 4 ( ) were general medical wards, and ward 3 (
) were general medical wards, and ward 3 ( ) was an oncology ward. The EIA tests used detected toxin A alone (a solid circle represents a positive test and an open circle represents a negative test) or toxin A and toxin B (a solid diamond represents a positive test and an open diamond represents a negative test). The cytotoxin test using HFF-cell culture was also performed (a solid square represents a positive test and an open square represents a negative test).
) was an oncology ward. The EIA tests used detected toxin A alone (a solid circle represents a positive test and an open circle represents a negative test) or toxin A and toxin B (a solid diamond represents a positive test and an open diamond represents a negative test). The cytotoxin test using HFF-cell culture was also performed (a solid square represents a positive test and an open square represents a negative test).
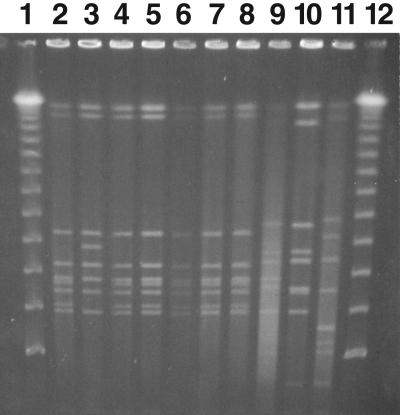
To determine if this was a point source outbreak and if there were genetic defects in the infecting organism's ability to produce toxin A, stool samples from the 16 patients were cultured for C. difficile. Some of the patients had had or were receiving metronidazole or vancomycin therapy when culture was attempted. C. difficile therefore could be isolated from only 7 of the 16 patients despite repeated attempts by three separate laboratories (those of M. J. Alfa, A. Kabani, and D. Lyerly). The PFGE analysis shown in Fig. 2 demonstrates that all seven patients who had toxigenic C. difficile grown from their stool had strains with identical PFGE banding patterns. Patient 11 had two C. difficile strains that had different colony morphologies. One strain had a PFGE banding pattern that was identical to that of the other six patient strains, and the second C. difficile strain had three band differences compared to the outbreak pattern (Fig. 2, lanes 3 and 4).
FIG. 2.
PFGE of C. difficile outbreak isolates. Isolates of C. difficile were detected in 7 of the 16 patients involved in the initial outbreak. PFGE was done using SmaI as the restriction enzyme by the method described in Materials and Methods. Lanes 1 and 12, lambda ladder size markers; lanes 2 to 8, isolates from patients 1, 11, 11, 2, 7, 6, and 9, respectively. The two isolates from patient 11 had different colony morphologies. Lanes 9 to 11, C. difficile isolates that were toxin A positive and toxin B positive that were not part of the outbreak.
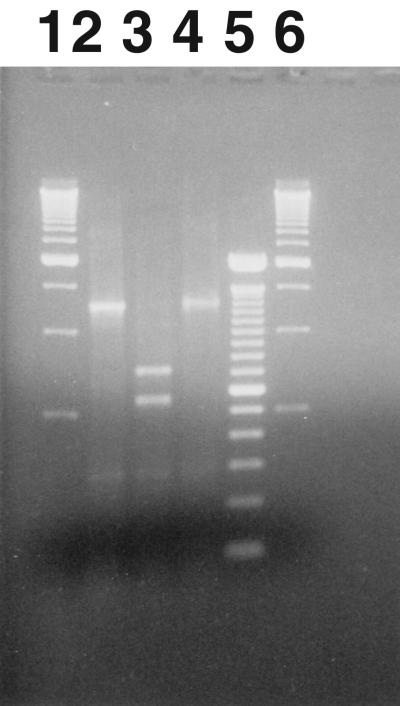
The organisms were assessed using PCR to amplify various regions of the pathogenicity locus to determine if there were any genetic defects in the C. difficile isolates causing the outbreak. The initial PCR set utilized the primers described by Rupnik et al. (31) to assess the CROP region of toxin A. The expected amplicon size for this set of primers is 3.1 kb (31). Figure 3 shows the 1,250-bp fragment (designated A3′) that was amplified when the A3C and A4N primers, bounding the CROP region of toxin A, were used on the HSC98 isolate. From this assessment there appeared to be a 1.8-kb deletion from the CROP region of toxin A. To determine the characteristics of the HSC98 A3′ amplicon, the 1,250-bp amplicon was assessed for EcoRI and SpeI restriction sites. Rupnik et al. (32) have reported that those strains of C. difficile that have deletions in the portion of the toxin A gene that codes for the CROP region often lose one or both of these restriction sites from the A3 region. As shown in Fig. 3, the A3′ amplicon retains its SpeI restriction site but has lost the EcoRI site. The A3′ amplicon was also sequenced. A BLAST search of GenBank indicated that this A3′ sequence was a 98% match to the sequence for the tcdA gene CROP region (1,230 bp) from C. difficile strain 1470, submitted by M. Weidmann in 1997 (accession number Y12616).
FIG. 3.
Restriction endonuclease digestion patterns for the A3′ amplicon of the CROP region of the toxin A gene for HSC98. The PCR amplicon product A3′ (lane 2) obtained from HSC98 using the A3 primer set (product was 1,250 bp) was exposed to SpeI (lane 3) and EcoI (lane 4) restriction endonucleases to determine restriction fragment length patterns. The A3′ amplicon (lane 2) had no restriction site for EcoRI (lane 4) and had one restriction site for SpeI (lane 3). A 500-bp DNA ladder is shown in lanes 1 and 6, and a 100-bp ladder is shown in lane 5.
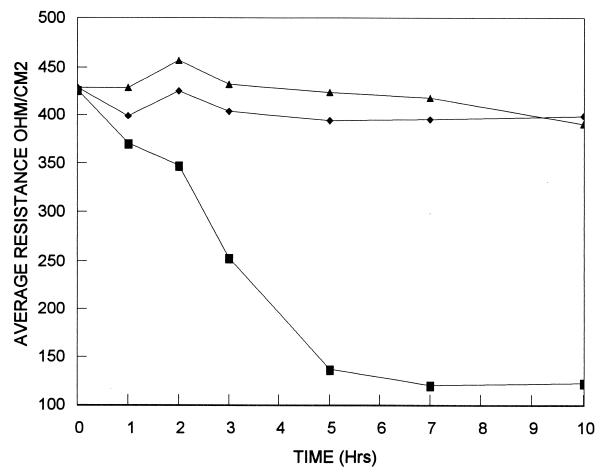
To assess the level of toxin production and to determine if there was any detectable truncated toxin A produced by the HSC98 strain, culture supernates were tested by EIA for the presence of toxin A alone and toxins A and B together. The supernatant fluids from the seven isolates from this outbreak all had absorbances of <0.08 and >0.5 in the toxin A and toxin A-B EIAs, respectively (data not shown). The same supernatant fluids from these seven outbreak strains produced 104 to 105 CPE units/ml when tested on CHO cells. Although the EIA for toxin A was negative, it is possible that a truncated form of toxin A could be produced by the HSC98 strain. Such a truncated form of toxin A would not be detected in the available toxin A EIAs, as it lacks the CROP region that is the recognition site of the monoclonal antibody used in the commercial assays that is specific for toxin A. To further assess whether a functional truncated form of toxin A was produced, the culture supernatant fluid from HSC98 was tested for any ability to cause tight-junction alterations when tested against polarized CaCo-2 cells. Hecht et al. (16, 17) have shown that purified toxin A is able to perturb tight junctions of cultured human intestinal epithelial monolayers within a few hours, while purified toxin B takes 24 to 48 h to elicit resistance drops of a similar magnitude. The HSC98 strain was compared to the toxin A-positive, toxin B-positive strain 10463 and the toxin A-negative, toxin B-positive strain 8864. The data presented in Fig. 4 indicate that the HSC98 strain lacks biologically functional toxin A as evidenced by its lack of ability to perturb the tight junctions of polarized CaCo-2 monolayers within 10 h. Similarly to the HSC98 strain, the 8864 strain (also toxin A negative and toxin B positive) did not cause decreases in electrical resistance within 10 h of exposure.
FIG. 4.
Effect of HSC98 culture supernatant fluid on CaCo-2 tight junctions. Filter-sterilized culture medium from dialysis membrane culture (as described in Materials and Methods) was used to inoculate the apical side of the CaCo-2 monolayer. C. difficile strains 10463 (toxin A positive and toxin B positive) (■), 8864 (toxin A negative and toxin B positive) (⧫), and HSC98 (toxin A negative and toxin B positive) (▴) were tested. Each point plotted represents the average of nine electrical resistance readings consisting of triplicate readings from each of three separate inserts.
To determine if there were other regions within the C. difficile pathogenicity locus that had gene deletions, a series of PCR amplification tests were performed to test the regions indicated in Table 2. Table 2 demonstrates that within the regions tested there were no deletions large enough to be detected by such techniques other than the one in the CROP region of toxin A. Lack of binding of the monoclonal antibody PCG4, which is used in the diagnostic EIA that detects only toxin A (Fig. 1), correlates with the genetic deletion in the HSC98 toxin A CROP region.
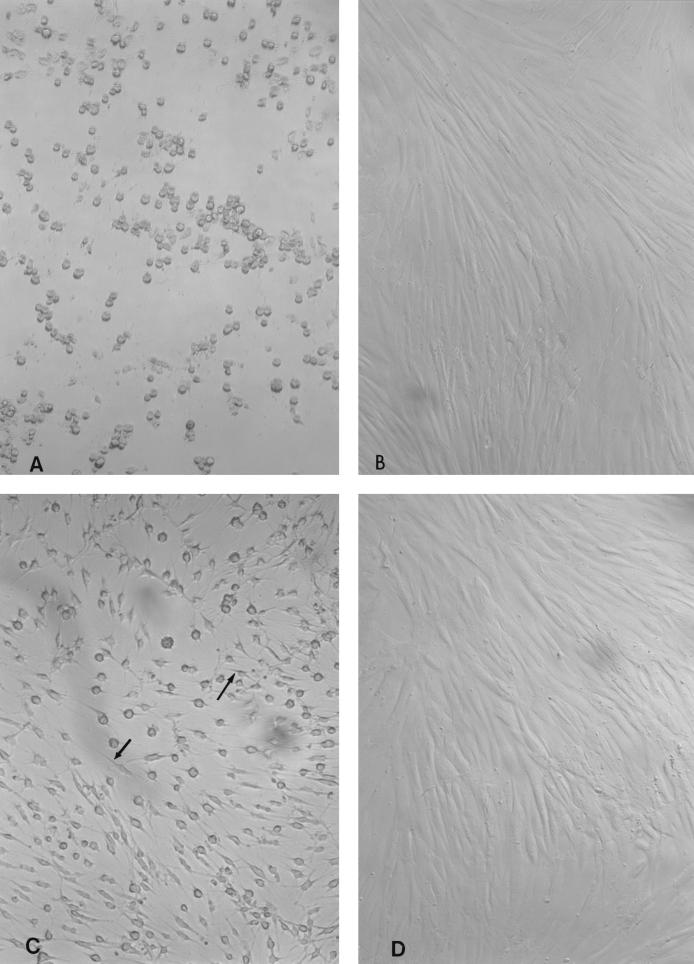
The CPE produced by HSC98 culture supernatant on HFF monolayers was distinctive in that it caused HFF cell rounding that did not have spindle formation (Fig. 5). Culture supernatant from C. difficile strain 10463, which produced both toxins A and B, elicited rounding CPE with spindle formation (Fig. 5C). Of interest, purified toxin B from a toxin B-positive, toxin A-positive strain of C. difficile produces CPE identical to that shown in Fig. 5C (results not shown).
FIG. 5.
Comparison of CPE on HFF cells elicited by C. difficile HSC98 and C. difficile 10463. Culture supernatant from the dialysis culture method described in Materials and Methods was filter sterilized for C. difficile strains HSC98 and 10463. HSC98 supernatant was inoculated in the absence (A) or presence (B) of C. sordellii antitoxin, and similarly, strain 10463 was inoculated in the absence (C) or presence (D) of C. difficile antitoxin. CPE was produced by both HSC98 and 10463, which was neutralized by C. difficile antitoxin (B and D); however, the CPE produced by HSC98 showed rounding with no spindle formation, whereas strain 10463 produced rounded cells that had spindles (arrows).
DISCUSSION
We have demonstrated that this outbreak was caused by a single strain of C. difficile, designated HSC98. The affected patients exhibited the full range of classical CAD symptoms, ranging from asymptomatic carriage to fulminant pseudomembranous colitis and death. The rate of recurrence of clinical symptoms posttreatment for CAD was 35.7% (5 of 14). Despite the apparent lack of functional toxin A (as assayed by immunological methods and by lack of tight-junction disruption), this HSC98 strain appears to be as virulent as other reported clinical strains of C. difficile which produce both toxin A and toxin B. Based on the toxinotype categorization scheme of Rupnik et al. (32), HSC98 appears to be a toxinotype VIII strain. It has the same size of gene deletion in the CROP region of toxin A as described for C. difficile strain 1470. Furthermore, the sequences of the A3′ amplicon from strain HSC98 and the A3′ amplicon from strain 1470 were 98% identical, and the EcoRI and SpeI restriction digest patterns of these amplicons were identical. This evidence suggests that the HSC98 strain most likely has the same genetic alterations in its pathogenicity locus (based on the analysis to date) as C. difficile strain 1470.
This was unexpected, as strain 1470 was originally isolated from an asymptomatic infant in Belgium in 1981 (32). Strain 1470 is a serotype F strain that does not produce detectable toxin A as assessed by immunological methods (32), although Von Eichel-Streiber et al. (36) reported that strain 1470 does produce a variant form of toxin A. Toxinotype VIII was the most commonly reported alteration (25 strains found) within the 47 isolates with genetic defects of the 219 isolates that were studied by Rupnik et al. (32). This is similar to the frequency of serogroup F strains (44 toxin A-negative, toxin B-positive isolates of 421 isolates tested) described by Kato et al. (19). It has been suggested that the toxin A-negative, toxin B-positive strains of C. difficile may represent up to 10% of clinical isolates (38). Recent studies by Von Eichel-Streiber et al. (36) have indicated that C. difficile strain 1470 was cytotoxic but did not induce the typical disease in the hamster model. According to this group, both toxin A and toxin B from strain 1470 could be isolated from culture supernatant. Furthermore, they demonstrated that the CPE caused by 1470 was more like that of the heat-labile toxin of Clostridium sordellii (i.e., rounding with no cytoplasmatic extensions) than that of toxin B of C. difficile (36). Based on the data they presented, they hypothesized that 1470 may have resulted from a recombinational exchange between toxB of C. difficile and the gene for heat-labile toxin of C. sordellii. Our data suggest that if HSC98 produced toxin A, it does not have the typical biological function of unmodified toxin A. Similar to the data presented by Von Eichel-Streiber et al. (36), the HSC98 strain produces CPE on HFF cells that was distinct from that produced by regular toxin A- and toxin B-producing strains (Fig. 5) in that the affected HFF cells round up and do not have any spindle formation. Our data indicated that this type of CPE (rounding with no spindle formation) was also apparent for strain 8864 (toxin A negative and toxin B positive) but was not apparent for highly purified toxin B from a regular toxigenic (toxin A-positive, toxin B-positive) strain of C. difficile. Why a C. difficile strain with a deletion in the toxin A gene should elicit a CPE different from that of a strain with an intact toxin A gene is not readily explainable. Toxin B is the most potent cytotoxin produced, and its activity would not be expected to be altered by the presence or absence of functional toxin A.
Torres (34) and Lyerly et al. (26) have characterized strain 8864, which is a toxin A-negative toxin B-positive strain of C. difficile. Similar to the findings with strain 1470 reported by Von Eichel-Streiber et al. (36), both Torres (34) and Lyerly et al. (26) found that the toxin B produced by 8864 has cytopathic characteristics that were very similar to those of C. sordellii lethal toxin. Although Von Eichel-Streiber et al. (36) have reported that strain 1470 does produce a form of toxin A, neither strain 8864 (26, 34) nor our strain HSC98 appears to produce a detectable form of functional toxin A. The tight-junction assay results cannot rule out the possibility that a truncated version of Toxin A is produced that is nonfunctional due to lack of the binding region. However, these data do suggest that whatever role that bound or internalized toxin A normally plays in pathogenesis will not be manifested in those strains having this CROP region defect. Despite the lack of functional toxin A, HSC98 produced the full spectrum of CAD in humans, and C. difficile strain 8864 was able to cause disease in antibiotic-treated hamsters and was lethal on injection into mice. The characterization of strain 8864 by Borriello et al. (5) indicated that crude culture filtrate from 8864 could cause enterocyotoxicity in rabbit intestinal loops, whereas purified Toxin B from 8864 did not. Those authors suggested that strain 8864 produced a non-toxin A enterotoxic factor. These data raise questions about the current thoughts regarding pathogenesis of this toxigenic organism. Previous data suggested that toxins A and B likely functioned in a synergistic fashion to produce disease in humans (25), indeed, animal studies have demonstrated that neutralization of both toxins was required to prevent disease and relapse of disease (21). It has now been demonstrated, however, that a clinical isolate that does not have the tight-junction-altering capacity normally associated with biologically active toxin A is capable of producing disease in humans. The similarity of the CPE produced by HSC98, 8864, 1470, and C. sordellii raises questions about whether there may be another factor produced by these strains that may act to modulate the CPE produced by toxin B, thereby resulting in no spindle formation, or whether the toxin B in these strains is actually C. sordellii lethal toxin (TcsL). This differentiation may be difficult, since the sizes of TcsL and C. difficile toxin B (TcdB) are the same (270 kDa) and the genes have a high degree of sequence homology (37).
A recent point prevalence survey (data to be published elsewhere) indicated that this strain is still prevalent in the hospital where the outbreak occurred. In the past it was thought that all clinically relevant strains of C. difficile which were toxigenic produced both toxin A and toxin B, and therefore, diagnostic tests detecting either toxin were acceptable. Prevalence testing for toxin A-negative, toxin B-positive strains suggested that routine diagnostic testing for such strains was not of concern in the Buffalo, N.Y., region, as none were found in the 300 patients screened (3). Indeed, some reports suggested that detection of toxin A was more important than detection of toxin B, as toxin A was felt to be most important in eliciting disease in humans (4). A recent survey in Canada indicated that EIAs were commonly used (38.4%) for diagnostic testing (2) and that over half of these sites were using EIAs that detected toxin A only. Unlike the data reported in 1991 by Altaie et al. (3), recent reports suggest that such toxin A-negative, toxin B-positive strains may represent 10% of all C. difficile toxigenic strains (38). The value of diagnostic tests that detect both toxins has also been reported by McGowan and Kader (27), who reported that testing for only toxin A or only toxin B in children would have resulted in detection of C. difficile in only 50 and 82% of the time, respectively. They recommended using a test that detected both toxin A and toxin B to provide the optimal diagnostic yield. Al-Barrak et al. (1) suggested that when patients experience symptoms compatible with CAD but toxin A is negative, consideration should be given to an assay capable of detecting toxin B. Given that the incidence of CAD is on the rise (30) and that there is an apparent link between increased risk of death and CAD (14, 39), there is a need to ensure rapid and accurate testing to facilitate the diagnosis of CAD.
In conclusion, this report highlights two important findings: (i) either tight-junction modification due to toxin A is apparently not absolutely critical or there is another, as-yet-undescribed cofactor responsible for the pathogenesis of CAD caused by toxigenic C. difficile in humans and (ii) optimal diagnostic test methods should detect at least toxin B and preferably should detect both toxins A and B.
ACKNOWLEDGMENTS
The skilled technical assistance of Pat Degagne of St. Boniface General Hospital and Christine Turenne of the Health Sciences Centre is acknowledged. The support from the infection control practitioners from the Health Sciences Centre and the laboratory support from the microbiology laboratories at the St. Boniface General Hospital and the Health Sciences Centre are acknowledged.
REFERENCES
- 1.Al-Barrak A, Embil J, Dyck B, Olekson K, Nicoll D, Alfa M, Kabani A. An outbreak of Toxin A negative, Toxin B positive Clostridium difficile-associated diarrhea in a Canadian tertiary-care hospital. Canada Communicable Dis Rep. 1999;25-7:1–3. [PubMed] [Google Scholar]
- 2.Alfa M J, Du T, Beda G. Survey of incidence of Clostridium difficile infection in Canadian hospitals and diagnostic approaches. J Clin Microbiol. 1998;36:2076–2080. doi: 10.1128/jcm.36.7.2076-2080.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altaie S S, Penque P H, Mookherjee S, Evans D T. Should laboratories test for Toxin A-negative, toxin B-positive Clostridium difficile? Lab Med. 1996;27:468–471. [Google Scholar]
- 4.Bartlett J G. Clostridium difficile: clinical considerations. Rev Infect Dis. 1990;12(Suppl. 2):S243–S251. doi: 10.1093/clinids/12.supplement_2.s243. [DOI] [PubMed] [Google Scholar]
- 5.Borriello S P, Wren B W, Hyde S, Seddon S V, Sibbons P, Krishna M M, Tabaqhali S, Manek S, Price A B. Molecular, immunological, and biological characterization of a toxin A-negative, toxin B-positive strain of Clostridium difficile. Infect Immun. 1992;60:4192–4199. doi: 10.1128/iai.60.10.4192-4199.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borriello S P. Clostridial disease of the gut. Clin Infect Dis. 1995;20(Suppl. 2):S242–S250. doi: 10.1093/clinids/20.supplement_2.s242. [DOI] [PubMed] [Google Scholar]
- 7.Bradbury A W, Barrett S. Surgical aspects of Clostridium difficile colitis. Br J Surg. 1997;8:150–159. [PubMed] [Google Scholar]
- 8.Clabots C R, Johnson S, Olson M M, Peterson L R, Gerding D N. Acquisition of Clostridium difficile by hospitalized patients: evidence for colonized new admissions as a source of infection. J Infect Dis. 1992;166:561–567. doi: 10.1093/infdis/166.3.561. [DOI] [PubMed] [Google Scholar]
- 9.Depitre C, Delmee M, Avesani V, Haridon R L, Roels A, Popoff M, Corthier G. Serogroup F strains of Clostridium difficile produce toxin B but not toxin A. J Med Microbiol. 1993;38:434–441. doi: 10.1099/00222615-38-6-434. [DOI] [PubMed] [Google Scholar]
- 10.Dove C H, Wang S Z, Price S B, Phelbs C J, Lyerly D M, Wilkins T D, Johnson J L. Molecular characterization of the Clostridium difficile toxin A gene. Infect Immun. 1990;58:480–488. doi: 10.1128/iai.58.2.480-488.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eveillard M, Fourel V, Barc M C, Kerneis S, Coconier M H, Karjalainen T, Bourlioux P, Servin A L. Identification and characterization of adhesive factors of Clostridium difficile involved in adhesion to human colonic enterocyte-like Caco-2 and mucus-secreting HT20 cells in culture. Mol Microbiol. 1993;7:371–381. doi: 10.1111/j.1365-2958.1993.tb01129.x. [DOI] [PubMed] [Google Scholar]
- 12.Fiorentini C, Thelestan M. Clostridium difficile Toxin A and its effects on cells. Toxincon. 1991;29:543–567. doi: 10.1016/0041-0101(91)90050-2. [DOI] [PubMed] [Google Scholar]
- 13.Fluit A D C, Wolfhagen M J H M, Verdonk G P H T, Jansze M, Torensma R, Verhoef J. Nontoxigenic strains of Clostridium difficile lack the genes for both toxin A and toxin B. J Clin Microbiol. 1991;29:2666–2667. doi: 10.1128/jcm.29.11.2666-2667.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frost F, Craun G F, Calderon R L. Increasing hospitalization and death possible due to Clostridium difficile diarrheal disease. Emerging Infect Dis. 1998;4:619–625. doi: 10.3201/eid0404.980412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gerding D N, Johnson S, Peterson L R, Mulligan M E, Silva J., Jr Clostridium difficile-associated diarrhea and colitis: SHEA position paper. Infect Control Hosp Epidemiol. 1995;16:459–477. doi: 10.1086/648363. [DOI] [PubMed] [Google Scholar]
- 16.Hecht G, Pothoulakis C, Lamont J T, Madara J L. Clostridium difficile Toxin A perturbs cytoskeletal structure and tight junction permeability of culture human intestinal epithelial monolayers. J Clin Invest. 1988;82:1516–1524. doi: 10.1172/JCI113760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hecht G, Koutsouris A, Pothoulakis C, Lamont J T, Madara J L. Clostridium difficile Toxin B disrupts the barrier function of T84 monolayers. Gastroenterology. 1992;102:416–423. doi: 10.1016/0016-5085(92)90085-d. [DOI] [PubMed] [Google Scholar]
- 18.Johnson S, Gerding D N. Clostridium difficile-associated diarrhea. Clin Infect Dis. 1998;26:1027–1036. doi: 10.1086/520276. [DOI] [PubMed] [Google Scholar]
- 19.Kato H, Kato N, Watanabe K, Iwai N, Nakamura H, Yamamoto T, Suzuki K, Kim S-M, Chong Y, Wasito E. Identification of toxin A-negative, toxin B-positive Clostridium difficile by PCR. J Clin Microbiol. 1998;36:2178–2182. doi: 10.1128/jcm.36.8.2178-2182.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kato H, Kato N, Watanabe K, Ueno K, Ushijima H, Hashira S, Abe T. Application of typing by pulsed-field gel electrophoresis to the study of Clostridium difficile in a neonatal intensive care unit. J Clin Microbiol. 1994;32:2067–2070. doi: 10.1128/jcm.32.9.2067-2070.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kink J A, Williams J A. Antibodies to recombinant Clostridium difficile toxins A and B are an effective treatment and prevent relapse of C. difficile-associated disease in a hamster model of infection. Infect Immun. 1998;66:2018–2025. doi: 10.1128/iai.66.5.2018-2025.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knudsen J D, Tvede M. Demonstration of toxin A and B by polymerase chain reaction and McCoy cell assay in clinical isolates of Clostridium difficile from Denmark. APMIS. 1993;101:18–22. doi: 10.1111/j.1699-0463.1993.tb00074.x. [DOI] [PubMed] [Google Scholar]
- 23.Lai K K, Melvin Z S, Menard M J, Kotilainen H R, Baker S. Clostridium difficile-associated diarrhea: epidemiology, risk factors, and infection control. Infect Control Hosp Epidemiol. 1997;18:628–632. doi: 10.1086/647687. [DOI] [PubMed] [Google Scholar]
- 24.Lyerly D M, Krivan H C, Wilkins T D. Clostridium difficile: its disease and toxins. Clin Microbiol Rev. 1988;1:1–18. doi: 10.1128/cmr.1.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lyerly D M, Saum E K, MacDonald K D K, Wilkins T D. Effects of Clostridium difficile toxins given intragastrically to animals. Infect Immun. 1985;47:349–352. doi: 10.1128/iai.47.2.349-352.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lyerly D M, Barroso L A, Wilkins T D, Depitre C, Corthier G. Characterization of a toxin A-negative, toxin B-positive strain of Clostridium difficile. Infect Immun. 1992;60:4633–4639. doi: 10.1128/iai.60.11.4633-4639.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McGowan K L, Kader H A. Clostridium difficile infection in children. Clin Microbiol Newsl. 1999;21:49–53. [Google Scholar]
- 28.Mitchell T J, Ketley J M, Burdon D W, Candy D C A, Stephen J. Biological mode of action of Clostridium difficile toxin A: a novel enterotoxin. J Med Microbiol. 1987;23:211–219. doi: 10.1099/00222615-23-3-211. [DOI] [PubMed] [Google Scholar]
- 29.Miyazaki S, Matsunaga T, Kawasaki K, Kobayashi I, Tada H, Yamaguchi K, Goto S. Separate isolation of Clostridium difficile spores and vegetative cells from the feces of newborn infants. Microbiol Immunol. 1992;36:131–138. doi: 10.1111/j.1348-0421.1992.tb01650.x. [DOI] [PubMed] [Google Scholar]
- 30.Riley T V. Clostridium difficile: a pathogen of the nineties. Eur J Clin Microbiol Infect Dis. 1998;17:137–141. doi: 10.1007/BF01691108. [DOI] [PubMed] [Google Scholar]
- 31.Rupnik M, Braun V, Soehn F, Janc M, Hofstetter M, Laufenberg-Feldmann R, von Eichel-Streiber C. Characterization of polymorphisms in the toxin A and B genes of Clostridium difficile. FEMS Microbiol Lett. 1997;148:197–202. doi: 10.1111/j.1574-6968.1997.tb10288.x. [DOI] [PubMed] [Google Scholar]
- 32.Rupnik M, Avesani V, Janc M, von Eichel-Streiber C, Delmee M. A novel toxinotyping scheme and correlation of toxinotypes with serogroups of Clostridium difficile isolates. J Clin Microbiol. 1998;36:2240–2247. doi: 10.1128/jcm.36.8.2240-2247.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sullivan N M, Pellett S, Wilkins T D. Purification and characterization of toxins A and B of Clostridium difficile. Infect Immun. 1982;35:1032–1040. doi: 10.1128/iai.35.3.1032-1040.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Torres J F. Purification and characterization of toxin B from a strain of Clostridium difficile that does not produce toxin A. J Med Microbiol. 1991;34:40–44. doi: 10.1099/00222615-35-1-40. [DOI] [PubMed] [Google Scholar]
- 35.Von Eichel-Streiber C, Laufenberg-Feldmann R, Sartingen S, Schulze J, Sauerborn M. Comparative sequence analysis of the Clostridium difficile toxins A and B. Mol Gen Genet. 1992;233:260–268. doi: 10.1007/BF00587587. [DOI] [PubMed] [Google Scholar]
- 36.Von Eichel-Streiber C, Meyer zu Heringdorf D, Habermann E, Sartingen S. Closing in on the toxic domain through analysis of a variant Clostridium difficile cytotoxin B. Mol Microbiol. 1995;17:313–321. doi: 10.1111/j.1365-2958.1995.mmi_17020313.x. [DOI] [PubMed] [Google Scholar]
- 37.Von Eichel-Streiber C, Boquet P, Suerborn M, Thelestam M. Large clostridial cytotoxins—a family of glycosyltransferases modifying small GTP-binding proteins. Trends Microbiol. 1996;4:375–382. doi: 10.1016/0966-842X(96)10061-5. [DOI] [PubMed] [Google Scholar]
- 38.Wilcox M H. Summary Proceedings from the Satellite Symposia to the 38th Interscience Conference on Antimicrobial Agents and Chemotherapy, 22-23 September 1999. Oakville, Ontario. Canada: Pulsus Group Inc.; 1999. Clostridium difficile: an emerging threat; pp. 12–15. [Google Scholar]
- 39.Williamson J, editor. Clostridium difficile hospitalization and death may be a bigger problem than anticipated. Infect Control Prevention Rep. 1998;3:161–165. [Google Scholar]
- 40.Wren B W, Clayton C L, Castledine N B, Tabaqchali S. Identification of toxigenic Clostridium difficile strains by using a toxin A gene-specific probe. J Clin Microbiol. 1990;28:1808–1812. doi: 10.1128/jcm.28.8.1808-1812.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]